Abstract
The tumor suppressor protein p53 has been described “as the guardian of the genome” for its crucial role in regulating the transcription of numerous genes responsible for cells cycle arrest, senescence, or apoptosis in response to various stress signals. Although p53 promotes longevity by decreasing the risk of cancer through activation of apoptosis or cellular senescence, several findings suggest that an increase of its activity may have deleterious effects leading to selected aspects of the aging phenotype and neurodegenerative diseases. There is the link between p53 and oxidative stress, the latter a crucial factor that contributes to neurodegenerative processes like Alzheimer disease (AD). In the present study, using a proteomics approach, we analyzed the impact of lack of p53 on the expression of several brain mitochondrial proteins involved in different pathways, and how lack of p53 may present a target to restore neuronal impairments. Our investigation on isolated brain mitochondria from p53(−/−) mice also provides a better understanding of the p53-mitochondria relationship and its involvement in the development of many diseases.
Introduction
The p53 tumor suppressor protein plays a central role to preserve genomic integrity [1] with effect on cell fate [2]. p53 is involved in many cellular pathways, and when this protein becomes activated in response to stress signals [3] it can promote a transient cell cycle arrest, cell death (apoptosis) or permanent cell cycle arrest (senescence) [4]. p53 often is lost or mutated in cancers [5]. Both apoptosis and cellular senescence prevent the propagation of damaged DNA [6] with consequent reduction of the risk of cancer. However, both of these processes favor tissue atrophy and aging phenotype [7]. Therefore, p53 can exert both beneficial and deleterious effects depending on a delicate balance between tumor suppressor and longevity.
The interaction among p53 and oxidative stress is intriguing, since this latter is well known to be associated with several age-related diseases [8], [9]. Under normal conditions, p53 protein levels are low and regulated by IKK but prominently by Mdm2, an ubiquitin ligase responsible for p53 degradation. Cellular stress reduces the interaction between p53 and Mdm2 leading to accumulation of the former [10], and several reactive oxygen (ROS) and nitrogen species (RNS) also modify p53 and its activity [11]. Moreover, the activation of p53 leads to the generation of ROS as well [12], [13]. Thus, there is an intricate link between p53 and ROS, even though specific mechanisms of their interplay are still unclear. Several results show that cellular redox status is under control of p53, and p53 may exert opposite effects in ROS regulation depending on its levels [11]. Physiological levels of p53 maintain ROS at basal levels through transactivation of antioxidant genes such as SESN1 (mammalian sestrin homologue), SESN2, and glutathione peroxidase-1 (GPx1) [14]. In addition, constitutive levels of p53 link energy metabolism to ROS formation by regulating the expression of essential metabolic enzymes that are able to balance energy metabolism among mitochondrial respiration, glycolysis, and the pentose phosphate shunt [11], and mitochondrial respiration is a major source of ROS [15], [16].
High levels of p53 increase intracellular ROS by transactivation of genes encoding pro-oxidant proteins such as NQO1 (quinone oxidoreductase) [11] and proline oxidase (POX) [11], and for pro-apoptotic proteins, which include BAX and PUMA [11]. Further, the repression of antioxidant enzymes such as MnSOD by p53, is another means to increase intracellular ROS [11], [17].
Changes in mitochondrial ROS production may influence the p53 pathway [18], [19]. Also p53 can regulate ROS production in mitochondria [20]. This suggests that there is an interaction between mitochondria and p53 essential to allow normal cellular functions and its interruption may have severe consequences [21]. Consequently, understanding better the mechanisms underlying this interaction may be helpful to further comprehend the development and the progression of many diseases [21].
The aim of this study was to analyze the impact that the lack of p53 had on basal protein expression levels in mitochondria isolated from mice brain, to gain insight into the special link between p53 and oxidative stress, and its impact on neurodegenerative disorders, such as Alzheimer disease. A proteomics approach was used.
Materials and Methods
Chemicals
All chemicals used in this study were purchased from Bio-Rad (Hercules, CA).
Animals
Heterozygous mice p53(−/+) were maintained in our laboratory to generate p53(−/−) and wt littermates. p53(−/−) are in the C57BL/6 background and were initially produced in the laboratory of Dr. Tyler Jacks at the Center for Cancer Research and Department of Biology, Massachusetts Institute of Tecnology (Cambridge, MA). The targeted disrupted p53 genes do not yield p53 protein, because of 40% of their gene-coding region is eliminated by the induced mutation. Male mice with an age between 10 and 12 weeks old were used in our study. All animal experimental procedures were approved by the Institute Animal Care and Use Committe of the University of Kentucky and followed NIH Guidelines for the Care and Use of Laboratory Animals.
Sample preparation
Mice were humanely euthanized, and the brain was quickly removed. Mitochondria were promptly isolated from the brain by differential centrifugation methods using Percoll Gradientswith some modifications [22].
Isoelectric focusing (IEF)
Proteins from mitochondrial homogenates (200 μg) were precipitated by addition of ice-cold 100% trichloroacetic acid (TCA) (15% final concentration) and incubated on ice for 10 min. Samples were centrifuged at 14,000 rpm (23,700× g) for 5 min at 4°C. Pellets were washed three times with 0.5 mL of wash buffer [1∶1 (v/v) ethanol: ethyl acetate] to remove excess salts. After the final wash, pellets were dried at room temperature (RT) for ∼10 min and rehydrated for 2 h at RT in 200 μl of a rehydration buffer [8 M urea, 2 M thiourea, 50 mM DTT, 2.0% (w/v) CHAPS, 0.2% Biolytes, Bromophenol Blue], placed in agitation for 3 hours, and then sonicated for 10 s. Samples (200 μg) were applied to 11 cm pH 3–10 ReadyStrip™ IPG strips and after 2 h, 2 ml of mineral oil was added to prevent sample evaporation. Strips were actively rehydrated at 20°C for 18 h at 50 V, focused at a constant temperature of 20°C beginning at 300 V for 2 h, 500 V for 2 h, 1000 V for 2 h, 8000 V for 8 h, and finishing at 8000 V for 10 h rapidly. IPG strips were stored at −80°C until the second dimension of analysis was carried out.
Two-dimensional polyacrylamide gel electrophoresis (2D-PAGE)
2D-PAGE was performed to separate proteins on IEF strips based on molecular migration rate. IEF strips were thawed and equilibrated for 10 min in equilibration buffer A [50 mM Tris–HCl, pH 6.8, 6 M urea, 1% (w/v) SDS, 30% v/v glycerol, 0.5% DTT] and then re-equilibrated for 10 min in equilibration buffer B [50 mM Tris–HCl, pH 6.8, 6 M urea, 1% (w/v) SDS, 30% v/v glycerol, 4.5% IA]. Criterion precast linear gradient (8–16%) Tris–HCl polyacrylamide gels were uesd to perform second dimension electrophoresis. Precision Plus Protein™ All Blue Standards and samples were run at a constant voltage of 200 V for 65 min.
SYPRO Ruby® staining
After 2D-PAGE, gels were incubated in a fixing solution [7% (v/v) acetic acid, 10% (v/v) methanol] for 20 min at RT. Sypro Ruby® Protein Gel Stain (∼50 ml) was added to gels to stain them overnight at RT on a gently rocking platform. Gels then were placed in deionized water at RT until scanning. Gels were scanned into Adobe Photoshop 6.0 with a Molecular Dynamics STORM Phosphoimager (λex/λem: 470/618 nm) and stored in deionized water at 4 °C until further use.
Image Analysis
Differential expression
Spot intensities from SYPRO Ruby®-stained 2D-gel images of WT and p53(−/−) samples were quantified by densitometry according to the total spot density using PD Quest analysis software from Bio-Rad (Hercules, CA). Intensities were normalized to total gel densities and/or densities of all valid spots on the gels. Only spots with a 1.5-fold increase or decrease in normalized spot density in those samples and a statistically significant difference based on a Student's t-test at 95% confidence (i.e., p<0.05) were considered for MS/MS analysis.
In-gel trypsin digestion
In-gel trypsin digestion of selected gel spots was performed as previously described [23]. Briefly, protein spots identified as significantly altered were excised from 2D-gels with a clean, sterilized blade and transferred to Eppendorf microcentrifuge tubes. Gel plugs were then washed with 0.1 M ammonium bicarbonate NH4HCO3) at RT for 15 min, followed by incubation with 100% acetonitrile at RT for 15 min. After solvent removal, gel plugs were dried in their respective tubes under a flow hood at RT. Plugs were incubated for 45 min in 20 μl of 20 mM DTT in 0.1 M NH4HCO3 at 56°C. The DTT/NH4HCO3 solution was then removed and replaced with 20 μl of 55 mM iodoacetate (IA) solution in 0.1 M NH4HCO3 and incubated with gentle agitation at room temperature in the dark for 30 min. Excess IA solution was removed and plugs incubated for 15 min with 200 μl of 50 mM NH4HCO3 at RT. A volume of 200 μl of 100% acetonitrile was added to this solution and incubated for 15 min at room temperature. Solvent was removed and gel plugs were allowed to dry for 30 min at RT under a flow hood. Plugs were rehydrated with 20 ng/μl of modified trypsin (Promega, Madison, WI, USA) in 50 mM NH4HCO3 in a shaking incubator overnight at 37°C. Enough trypsin solution was added in order to completely submerge the gel plugs.
Mass spectrometry (MS)
Salts and contaminants were removed from tryptic peptide solutions using C18 ZipTips (Sigma-Aldrich, St. Louis, MO, USA), reconstituted to a volume of ∼15 μl in a 50∶50 water: acetonitrile solution containing 0.1% formic acid. Tryptic peptides were analyzed with an automated Nanomate electrospray ionization (ESI) [Advion Biosciences, Ithaca, NY, USA] Orbitrap XL MS (Thermo-Scientific, Waltham, MA, USA) platform. The Orbitrap MS was operated in a data-dependent mode whereby the eight most intense parent ions measured in the Fourier Transform (FT) at 60,000 resolution were selected for ion trap fragmentation with the following conditions: injection time 50 ms, 35% collision energy, MS/MS spectra were measured in the FT at 7500 resolution, and dynamic exclusion was set for 120 s. Each sample was acquired for a total of ∼2.5 min. MS/MS spectra were searched against the International Protein Index (IPI) database using SEQUEST with the following parameters: two trypsin miscleavages, fixed carbamidomethyl modification, variable methionine oxidation, parent tolerance 10 ppm, and fragment tolerance of 25 mmu or 0.01 Da. Results were filtered with the following criteria: Xcorr1.5, 2.0, 2.5, 3.0 for 1, 2, 3, and 4 charge states, respectively, Delta CN0.1, and P-value (protein and peptide) 0.01. IPI accession numbers were cross-correlated with Swiss Prot accession numbers for final protein identification.
Statistical analysis
All statistical analyses were performed using a Mann-Whitney U statistical test and a two-tailed Student's t-test. p<0,05 was considered significant for differential fold-change values. Only proteins with significant p-values from both tests were considered further for MS identification. Protein and peptide identifications obtained with the SEQUEST search algorithm with p<0.01 were considered statistically significant. To further validate SEQUEST identification, the location of protein spots (i.e., molecular weight [MW] and isoelectric point [pI]) on 2D-gels was manually checked based on expected MW and pI values from SwissProt database information.
Results
Proteomics
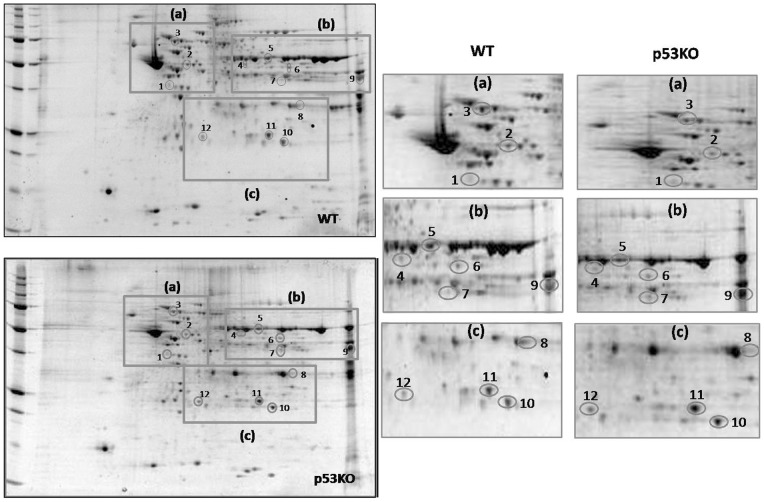
Proteomics analysis using 2-DE and Sypro Ruby staining was performed on proteins isolated from brain mitochondria of WT and p53(−/−) mice to determine proteins differently expressed. Fig. 1 shows 2D-gel images related to these analyses, with expanded images of protein spots significantly different (p<0.05) between WT and p53(−/−). Twelve proteins were identified as differently expressed between WT and p53(−/−) mice, and interestingly all twelve of these proteins were significantly over-expressed in p53(−/−) samples. Surprisingly, we did not find any mitochondrial proteins down-regulated in p53(−/−) mice relative to WT. The protein spots of interest were excised from the gels, and following digestion with the trypsin peptide were subjected to MS/MS analyses. Proteins identified are listed in Table 1 with the number of peptide sequences, the score, the coverage, MW, pI, fold-change levels, and p-value. All protein identifications were consistent with comparison of protein positions on the gel with MW and pI from databases.
Figure 1. Proteomic analysis of differential protein expression (WT vs. p53KO).
Proteomic profile of representative 2D-gels with proteins differently expressed between mitochondrial fraction isolated from the brain of WT mice and p53(−/−) (left); expanded images of protein spots that have significantly different levels (p<0.05) between WT and p53(−/−) (right).
Table 1. Proteins Expressed Differently in Mitochondrial Fraction Isolated from the Brain of WT and p53(−/−) mice.
| Spot | Protein Identified | Accession # | Coverage | Number of identified peptidesa | Score | MW (kDa) | pI | P valueb | Foldc |
| 1 | Guanine nucleotide-binding protein G (o) subunit alpha | P18872 | 12.15 | 3 | 24.11 | 40.1 | 5.53 | 0.0019 | 212 ↑ p53KO |
| 2 | ATP synthase subunit beta, mitochondrial | P56480 | 4.54 | 2 | 18.16 | 56.3 | 5.34 | 0.0035 | 125 ↑ p53KO |
| 3 | Heat shock cognate 71 kDa protein | P63017 | 37.31 | 20 | 196.60 | 70.8 | 5.52 | 0.002 | 212 ↑ p53KO |
| 4 | Aldehyde dehydrogenase family 5, subfamily A1 | B2RS41 | 14.72 | 6 | 36.70 | 55.9 | 8.25 | 0.0009 | 131 ↑ p53KO |
| 5 | Glutamate dehydrogenase 1, mitochondrial | P26443 | 26.34 | 13 | 78.69 | 61.3 | 8.00 | 0.0076 | 131 ↑ p53KO |
| 6 | Isoform mithocondrial of Fumarate hydratase | P97807-2 | 25.57 | 8 | 62.73 | 50.0 | 7.94 | 0.0019 | 325 ↑ p53KO |
| 7 | Acetyl-CoA acetyltransferase | Q8QZT1 | 26.89 | 8 | 50.64 | 44.8 | 8.51 | 0.00079 | 166 ↑ 53KO |
| 8 | Isoform Mt-VDAC1 of Voltage- dependent anion-selective channel protein 1 | Q60932-2 | 38.16 | 7 | 74.55 | 30.7 | 8.54 | 0.0027 | 201 ↑ p53KO |
| 9 | Aspartate aminotransferase | P05202 | 43.72 | 17 | 174.33 | 47.4 | 9.00 | 0.0037 | 210 ↑ p53KO |
| 10 | Mn Superoxide dismutase | P09671 | 13.96 | 4 | 43.39 | 24.6 | 8.62 | 0.0026 | 133 ↑ 53KO |
| 11 | Cytochrome b-c1 complex Rieske subunit | Q9CR68 | 26.28 | 7 | 70.31 | 29.3 | 8.70 | 0.0030 | 252 ↑ 53KO |
| 12 | Thioredoxin-dependent peroxide reductase | P20108 | 28.40 | 7 | 41.17 | 28.1 | 7.58 | 0.0015 | 253 ↑ 53KO |
The number of peptide sequences identified by nanospray ESI-MS/MS of tryptic peptides.
The fold-change in spot density from p53(−/−) mice compared to wt. The arrow indicates the direction of change.
The p-value associated with fold-change calculated using a Student's t-test.
The identified proteins were: guanine nucleotide-binding protein G (o) subunit alpha (212-fold ↑p53KO, *P<0.0019), ATP synthase subunit beta (125-fold ↑p53KO, *P<0.0035), heat shock cognate 71 (212-fold ↑p53KO, *P<0.002), aldehyde dehydrogenase family 5, subfamily A1 (131-fold ↑p53KO, *P<0.0009), glutamate dehydrogenase 1 (131-fold ↑p53KO, *P<0.0076), mitochondrial isoform of fumarate hydratase (325-fold ↑p53KO, *P<0.0019), acetyl-CoA acetyltransferase (166-fold ↑p53KO, *P<0.00079), isoform Mt-VDAC1 of voltage-dependent anion-selective channel protein 1 (201-fold ↑p53KO, *P<0.0027), aspartate aminotransferase (210-fold ↑p53KO, *P<0.0037), Mn superoxide dismutase (133-fold ↑p53KO, *P<0.0026), cytochrome b-c1 complex Rieske subunit (252-fold ↑p53KO, *P<0.0030), and thioredoxin-dependent peroxide reductase (253-fold ↑p53KO, *P<0.0015).
Discussion
Several studies have described p53, an important tumor suppressor protein, as the guardian of the genome [1], [2] for its critical role in regulating the transcription of numerous genes responsible for cells cycle arrest, senescence, or apoptosis in response to various stress signals [4]. Therefore, p53 is crucial in maintaining genetic stability [1]. What determines cell fate is unclear but different factors including the cell type, the particular insult, and the severity of damage are involved in this decision [24].
Undoubtedly p53 promotes longevity by decreasing the risk of cancer through activation of apoptosis or cellular senescence, but several reports suggest that an increase of its activity may have detrimental effects leading to selected aspects of the aging phenotype [7], [25] and neurodegenerative disease. Thus, there is a balance between cell death and survival that under normal conditions optimizes tumor suppression without accelerating aging.
Previous research from our laboratory found p53 over-expressed and oxidatively modified by oxidative and nitrosative stress in brain from subjects with mild cognitive impairment (MCI) and AD brain, compared to control samples [26], [27]. Conformational alterations of p53 in MCI and AD are known [19]. These observations are consistent with the role played by p53 in neuronal death detected in neurodegenerative conditions, and with an important link of p53 with oxidative stress. ROS and p53 appear to be interconnected at multiple levels in their signaling pathways. First, ROS are potent activators of p53, acting in different ways such as damaged DNA, and even by regulating the redox status of cysteines present in the DNA-binding domain of p53, affecting its DNA-binding activity [26], [28], [29]. Moreover, once activated p53 generates downstream ROS which mediate apoptosis [12], [30]. Therefore p53 appears to regulate cellular redox status [11].
Since oxidative stress has been considered a crucial factor that contributes to neurodegenerative processes like AD [31]–[33], p53 could be a therapeutic target to reduce the levels of ROS, and in this way prevent or attenuate neuronal death in neurodegenerative disorders such as MCI and AD.
In a previous study, we demonstrated for the first time that the lack of p53 significantly decreases basal levels of oxidative and nitrosative stress in mice brain, and that this loss of p53 could activate diverse protective pathways involved in maintaining cellular homeostasis in the brain of p53(−/−) mice [20]. In the present study using proteomics, we gained insight into the role of p53 in the CNS, and tested the hypothesis that knock out of p53 affected the expression of several brain mitochondrial proteins involved in different pathways; thus, loss of p53 may present a target to restore neuronal impairment. Since our investigation was performed on isolated brain mitochondria from p53(−/−) mice, our results conceivably could provide insights into progression of many mitochondrial-associated diseases. Hence, the identified proteins are involved in energy and mitochondrial alterations, signal transduction, antioxidant defense, and chaperone proteins, as shown in Table 2.
Table 2. Functionalities of Identified Proteins Differently Expressed.
| Functions | Proteins involved |
| Energy or mitochondrial alterations | ATP synthase subunit beta, mitochondrial Aldehyde dehydrogenase family 5, subfamily A1 Glutamate dehydrogenase 1, mitochondrial Isoform mitochondrial of Fumarate hydratase Acetyl-CoA acetyltransferase VDAC1 of Voltage-dependent anion-selective channel protein 1 Aspartate aminotransferase Mn Superoxide dismutase Cytochrome b-c1 complex Rieske subunit |
| Signal transduction | Guanine nucleotide-binding protein G (o) subunit alpha |
| Antioxidant defence/detoxification dysfunction | Mn Superoxide dismutase Thioredoxin-dependent peroxide reductase |
| Chaperone proteins | Heat shock cognate 71 kDa protein |
Antioxidant defense
Interestingly, MnSOD was significantly increased in mitochondria isolated from the brain of p53(−/−) mice compared to WT. This data was already shown in our prior study [20] and are consistent with the notion that MnSOD is transcriptionally repressed by p53 [34], [35] with consequent propagation of oxidative stress, since MnSOD provides critical antioxidant defense. Because the apoptotic programs require oxidative stress for their execution, an overexpression of MnSOD was shown to increase resistance to p53-dependent apoptosis [17], [34]. Drane et al. [34], and St. Clair and colleagues [18], further demonstrated that MnSOD has a mutual activity on p53 reducing its expression, and even negatively modulating its apoptotic function. Several studies indicate that overexpression of MnSOD protects neurons from oxidative damage thus exerting a defensive role during AD development [36]. St. Clair and co-workers [36], using APP-PS-1 neurons as a model of AD, found a reduction of MnSOD expression during neuronal maturation with high levels of oxidative stress. These researchers also indicated p53 as a possible factor for the suppression of MnSOD [36]. Therefore, an overexpression of MnSOD through the inhibition of p53 could be helpful to prevent or slow the progression of neurodegenerative processes such as AD.
Thioredoxin-dependent peroxide reductase, also called peroxiredoxin 3, is an antioxidant protein localized mainly in the matrix of mitochondria, and it regulates physiological levels of H2O2 [37]. The peroxiredoxin system requires a family of proteins called sestrins for its regeneration [38], and sestrin expression is regulated by p53 [39], [40]. Previous studies showed that p53 upregulates the expression of sestrins, including peroxiredoxin [14]. In contrast, in our study, we found an increase of Prdx3 levels in the mitochondrial of p53(−/−) mice, and a plausible explanation of this result could be, as proposed in our previous work [20], that the lack of p53 could disturb cellular homeostasis causing the activation of protective pathways by cells to combat cellular damage. Since H2O2 plays a central role in induction of apoptosis [41], the reduction of mitochondrial levels of H2O2by overexpression of Prdx3 seems to be antiapoptotic [42], and therefore beneficial for preserving cell survival. In addition Prdx3 was previously found down-regulated in AD brain [43].
Chaperone proteins
Heat shock cognate (HSC)-71, a member of the Hsp70 family of proteins [44], was found up-regulated in the mitochondrial fraction isolated from the brain of p53(−/−) mice compared to WT. Previously, Agoff [45] established that Hsp70 is repressed by p53, corroborating our result.
The Hsp family acts as chaperones assuring proper folding and assembly of proteins, and protects cells against apoptosis [46]. This latter function is prominently carried out by Hsp70. It is conceivable that the Hsp family exerts a crucial role in neuronal death linked with neurodegenerative disorders. HSC-71 is the constitutive isoform of the Hsp 70, activated by cells in adverse conditions. This chaperone protein is involved in the degradation of damaged proteins shuttling them for proteolysis [47]. In AD, the expression of Hsps seems to have a protective function to prevent the formation of amyloid fibrils [48], and previously, HSC-71 was found down regulated [49], and oxidatively modified in AD brain [50]. Therefore the increase of HSC-71 expression levels, induced by the lack of p53, conceivably could play a protective role in AD progression.
Energy dysfunction and mitochondrial alterations
Several findings suggest that p53 has a role in the regulation of pathways involved in glucose metabolism, supporting oxidative phosphorylation and the pentose phosphate shunt, and inhibiting glycolysis [11]. These activities of p53 prevent cancer development. In addition, mitochondria are a major site in which some constituents of these pathways play a role. Therefore, there is a connection between p53 and mitochondria [51], and a better understanding of this link conceivably could provide insight into the progression of mitochondria related disorders.
In our study VDAC was found up-regulated in mitochondria of p53(−/−) mice compared to mitochondria from WT mice. VDAC is a component of the mitochondria permeability transition pore (MPT), which allows the exchange of metabolities like ATP in and out of mitochondria, and it is also involved in synaptic communication and in the early phases of apoptosis [52]. Previous studies revealed the anti-apoptotic function of VDAC through its ability to bind BAK, a pro-apoptotic protein [53]. Likewise, VDAC may restrain p53, reducing its levels [54]. Therefore, these prior results suggest that VDAC and p53 are interconnected, and that lack of p53 could increase the expression of VDAC, in according with our results. The upregulation of VDAC conceivably could improve synaptic transmission and cell survival as well as modulate apoptotic events.
In addition, in our study we found several energy-related proteins: ATP synthase subunit beta, mitochondrial isoform of fumarate hydratase, and cytochrome bc1 complex Rieske subunit, over-expressed in brain mitochondrial of p53(−/−) mice. Since inhibition of p53 leads to dependence of cells on glycolysis and to considerable impairment of aerobic pathways [55], our data may reflect a stress response to compensate for this effect. Moreover the p53-dependent protein targets may be highly cellular type specific. Accordingly, our results also may reflect the high glycolytic metabolism in brain. The over-expression of these proteins, involved in energy metabolism, seems to confirm the hypothesis of this work, in which diminution of p53 may represent a target to restore mitochondrial dysfunction, since these proteins were found altered in models of aging and neurodegenerative diseases [56]–[58].
p53 plays an additional role in the regulation of glutamate metabolism activating the expression of glutaminase 2 which provides glutamate to promote the tricarboxylic acid (TCA) cycle and oxidative phosphorylation [59]. Glutamate may be oxidatively deaminated by glutamate dehydrogenase to form α-ketoglutarate, which can then enter the Krebs cycle and be oxidized to CO2 and H2O, or α-ketoglutarate can be transaminated by aspartate aminotransferase to form the neurotransmitter glutamate. Both of glutamate dehydrogenase and aspartate aminotransferase were shown up-regulated in mitochondrial brain of p53 knockout mice. These data are consistent with our previous results showing the enhancement of aerobic pathways in p53-deficient mice [20], and their contrast with the current literature [60] can be explained by the notion that p53-dependent effects cannot be reproduced in a particular cell system. Previously, glutamate dehydrogenase and aspartate aminotransferase have been shown to be oxidatively modified, and expressed differently in animal models of neurodegeneration [61]–[63]. Therefore, even these results strongly support the concept that inhibition of p53 may attenuate neurodegenerative disorders.
Another notable mitochondrial protein found to be basally up-regulated in brain mitochondria of p53(−/−) mice was aldehyde dehydrogenase family 5, subfamily A1, a member of the aldehyde dehydrogenase (ALDH) family known to participate in oxidizing a plethora of endogenous and exogenous aldehydes [64]. Previous studies showed a prominent role of ALDH family, including ALDH1, ALDH2, ALDH3A, and ALDH5A, in the oxidation of 4-hydroxy-trans-2-nonenal (HNE) to 4-hydroxy-trans-2-nonenoate (HNEAcid) [65], [66], a major pathway of HNE detoxification. In particular, in rat and in human brain, HNEAcid formation occurs in the mitochondria by ALDH5A [65]. The detoxification of these aldehydes is important for neurodegenerative disorders such as AD, since high levels of unsaturated lipid content increase brain vulnerability to oxidative damage [67]. Therefore up-regulation of ALDH5 could be protective against cell damage. Previous research identified ALDH4 as a p53-inducible gene [68]. The current study is the first to show that the lack of p53 increases ALDH5 expression levels, adding an additional p53-target gene.
An increasing body of evidence places p53 as a member of an intriguing network that includes tumor suppression and aging [69]. The tumor suppressor activity of p53 protects against malignant transformation but also enhances the aging process [69]. As the role of p53 in aging is still unclear, we designed this study gain insights into the role of p53 in the brain and its involvement in neuronal cell death.
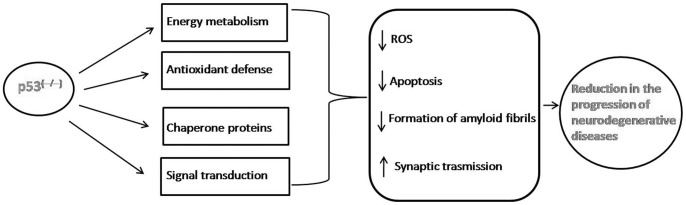
In conclusion, we identified brain mitochondrial proteins in p53 null mice that display crucial p53-dependent cellular functions in the central nervous system. Therefore, our results reinforce the concept that the lack of p53 could disturb cell homeostasis causing cells to stimulate defensive pathways. We also elaborated on the link between p53 and mitochondria as we used for the study mitochondria from p53 knock out mice. Since mitochondrial dysfunction is a key feature of neurodegenerative diseases such as AD, p53 conceivably could be a novel therapeutic target for the treatment of these disorders (Fig. 2).
Figure 2. Putative network of pathways regulated by p53KO.
A model of how the lack of p53 affects biological pathways that would attenuate progression of neurodegenerative disorders. Our result potentially makes p53 a novel therapeutic target for the delay, treatment, or prevention of these diseases.
Funding Statement
This work was supported in part by National Institutes of Health grants (www.nih.gov) to DAB [AG 05119] and DSC [CA 139843]. No additional external funding was received for this study. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Vousden KH, Lu X (2002) Live or let die: the cell's response to p53. Nat Rev Cancer 2: 594–604. [DOI] [PubMed] [Google Scholar]
- 2. Vousden KH (2006) Outcomes of p53 activation–spoilt for choice. J Cell Sci 119: 5015–5020. [DOI] [PubMed] [Google Scholar]
- 3. Horn HF, Vousden KH (2007) Coping with stress: multiple ways to activate p53. Oncogene 26: 1306–1316. [DOI] [PubMed] [Google Scholar]
- 4. Oren M (2003) Decision making by p53: life, death and cancer. Cell Death Differ 10: 431–442. [DOI] [PubMed] [Google Scholar]
- 5. Greenblatt MS, Bennett WP, Hollstein M, Harris CC (1994) Mutations in the p53 tumor suppressor gene: clues to cancer etiology and molecular pathogenesis. Cancer Res 54: 4855–4878. [PubMed] [Google Scholar]
- 6. Lim YP, Lim TT, Chan YL, Song ACM, Yeo BH, et al. (2006) The p53 knowledgebase: an integrated information resource for p53 research. Oncogene 26: 1517–1521. [DOI] [PubMed] [Google Scholar]
- 7. Tyner SD, Venkatachalam S, Choi J, Jones S, Ghebranious N, et al. (2002) p53 mutant mice that display early ageing-associated phenotypes. Nature 415: 45–53. [DOI] [PubMed] [Google Scholar]
- 8. Butterfield DA, Poon HF, St. Clair D, Keller JN, Pierce WM, et al. (2006) Redox proteomics identification of oxidatively modified hippocampal proteins in mild cognitive impairment: Insights into the development of Alzheimer's disease. Neurobiology of Disease 22: 223–232. [DOI] [PubMed] [Google Scholar]
- 9. Butterfield DA, Reed T, Newman SF, Sultana R (2007) Roles of amyloid β-peptide-associated oxidative stress and brain protein modifications in the pathogenesis of Alzheimer's disease and mild cognitive impairment. Free Radical Biology and Medicine 43: 658–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Momand J, Wu H-H, Dasgupta G (2000) MDM2 – master regulator of the p53 tumor suppressor protein. Gene 242: 15–29. [DOI] [PubMed] [Google Scholar]
- 11. Liu B, Chen Y, St. Clair DK (2008) ROS and p53: A versatile partnership. Free Radical Biology and Medicine 44: 1529–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Polyak K, Xia Y, Zweier JL, Kinzler KW, Vogelstein B (1997) A model for p53-induced apoptosis. Nature 389: 300–305. [DOI] [PubMed] [Google Scholar]
- 13. Johnson TM, Yu ZX, Ferrans VJ, Lowenstein RA, Finkel T (1996) Reactive oxygen species are downstream mediators of p53-dependent apoptosis. Proc Natl Acad Sci U S A 93: 11848–11852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sablina AA, Budanov AV, Ilyinskaya GV, Agapova LS, Kravchenko JE, et al. (2005) The antioxidant function of the p53 tumor suppressor. Nat Med 11: 1306–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Brookes PS, Yoon Y, Robotham JL, Anders MW, Sheu S-S (2004) Calcium, ATP, and ROS: a mitochondrial love-hate triangle. American Journal of Physiology – Cell Physiology 287: C817–C833. [DOI] [PubMed] [Google Scholar]
- 16. Raha S, Robinson BH (2000) Mitochondria, oxygen free radicals, disease and ageing. Trends in Biochemical Sciences 25: 502–508. [DOI] [PubMed] [Google Scholar]
- 17. Pani G, Bedogni B, Anzevino R, Colavitti R, Palazzotti B, et al. (2000) Deregulated manganese superoxide dismutase expression and resistance to oxidative injury in p53-deficient cells. Cancer Res 60: 4654–4660. [PubMed] [Google Scholar]
- 18. Holley AK, Dhar SK, St. Clair DK (2010) Manganese superoxide dismutase versus p53: the mitochondrial center. Annals of the New York Academy of Sciences 1201: 72–78. [DOI] [PubMed] [Google Scholar]
- 19. Buizza L, Cenini G, Lanni C, Ferrari-Toninelli G, Prandelli C, et al. (2012) Conformational altered p53 as an early marker of oxidative stress in Alzheimer's disease. PLoS One 7: e29789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Barone E, Cenini G, Sultana R, Di Domenico F, Fiorini A, et al. (2012) Lack of p53 decreases basal oxidative stress levels in the brain through upregulation of thioredoxin-1, biliverdin reductase-A, manganese superoxide dismutase, and nuclear factor kappa-B. Antioxid Redox Signal 16: 1407–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Holley AK, St. Clair DK (2009) Watching the watcher: regulation of p53 by mitochondria. Future Oncol 5: 117–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sims NR (1990) Rapid isolation of metabolically active mitochondria from rat brain and subregions using Percoll density gradient centrifugation. J Neurochem 55: 698–707. [DOI] [PubMed] [Google Scholar]
- 23.Sultana R, Perluigi M, Butterfield DA (2012) Lipid peroxidation triggers neurodegeneration: A redox proteomics view into the Alzheimer's disease brain. Free Radic Biol Med, in press. [DOI] [PMC free article] [PubMed]
- 24. Sionov RV, Haupt Y (1999) The cellular response to p53: the decision between life and death. Oncogene 18: 6145–6157. [DOI] [PubMed] [Google Scholar]
- 25. Maier B, Gluba W, Bernier B, Turner T, Mohammad K, et al. (2004) Modulation of mammalian life span by the short isoform of p53. Genes Dev 18: 306–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cenini G, Sultana R, Memo M, Butterfield DA (2008) Elevated levels of pro-apoptotic p53 and its oxidative modification by the lipid peroxidation product, HNE,in brain from subjects with amnestic mild cognitive impairment and Alzheimer's disease. Journal of Cellular and Molecular Medicine 12: 987–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cenini G, Sultana R, Memo M, Butterfield DA (2008) Effects of oxidative and nitrosative stress in brain on p53 proapoptotic protein in amnestic mild cognitive impairment and Alzheimer disease. Free Radical Biology and Medicine 45: 81–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Di Domenico F, Cenini G, Sultana R, Perluigi M, Uberti D, et al. (2009) Glutathionylation of the pro-apoptotic protein p53 in Alzheimer's disease brain: implications for AD pathogenesis. Neurochem Res 34: 727–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Martindale JL, Holbrook NJ (2002) Cellular response to oxidative stress: Signaling for suicide and survival*. Journal of Cellular Physiology 192: 1–15. [DOI] [PubMed] [Google Scholar]
- 30. Minamino T, Yujiri T, Papst PJ, Chan ED, Johnson GL, et al. (1999) MEKK1 suppresses oxidative stress-induced apoptosis of embryonic stem cell-derived cardiac myocytes. Proceedings of the National Academy of Sciences 96: 15127–15132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Butterfield DA, Drake J, Pocernich C, Castegna A (2001) Evidence of oxidative damage in Alzheimer's disease brain: central role for amyloid β-peptide. Trends in Molecular Medicine 7: 548–554. [DOI] [PubMed] [Google Scholar]
- 32. Castegna A, Thongboonkerd V, Klein JB, Lynn B, Markesbery WR, et al. (2003) Proteomic identification of nitrated proteins in Alzheimer's disease brain. Journal of Neurochemistry 85: 1394–1401. [DOI] [PubMed] [Google Scholar]
- 33. Markesbery WR (1997) Oxidative Stress Hypothesis in Alzheimer's Disease. Free Radical Biology and Medicine 23: 134–147. [DOI] [PubMed] [Google Scholar]
- 34. Drane P, Bravard A, Bouvard V, May E (2001) Reciprocal down-regulation of p53 and SOD2 gene expression-implication in p53 mediated apoptosis. Oncogene 20: 430–439. [DOI] [PubMed] [Google Scholar]
- 35. Dhar SK, Xu Y, Chen Y, St Clair DK (2006) Specificity protein 1-dependent p53-mediated suppression of human manganese superoxide dismutase gene expression. J Biol Chem 281: 21698–21709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sompol P, Ittarat W, Tangpong J, Chen Y, Doubinskaia I, et al. (2008) A neuronal model of Alzheimer's disease: An insight into the mechanisms of oxidative stress–mediated mitochondrial injury. Neuroscience 153: 120–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rhee SG, Kang SW, Netto LE, Seo MS, Stadtman ER (1999) A family of novel peroxidases, peroxiredoxins. Biofactors 10: 207–209. [DOI] [PubMed] [Google Scholar]
- 38. Budanov AV, Sablina AA, Feinstein E, Koonin EV, Chumakov PM (2004) Regeneration of peroxiredoxins by p53-regulated sestrins, homologs of bacterial AhpD. Science 304: 596–600. [DOI] [PubMed] [Google Scholar]
- 39. Budanov AV, Shoshani T, Faerman A, Zelin E, Kamer I, et al. (2002) Identification of a novel stress-responsive gene Hi95 involved in regulation of cell viability. Oncogene 21: 6017–6031. [DOI] [PubMed] [Google Scholar]
- 40. Velasco-Miguel S, Buckbinder L, Jean P, Gelbert L, Talbott R, et al. (1999) PA26, a novel target of the p53 tumor suppressor and member of the GADD family of DNA damage and growth arrest inducible genes. Oncogene 18: 127–137. [DOI] [PubMed] [Google Scholar]
- 41. Nonn L, Berggren M, Powis G (2003) Increased Expression of Mitochondrial Peroxiredoxin-3 (Thioredoxin Peroxidase-2) Protects Cancer Cells Against Hypoxia and Drug-Induced Hydrogen Peroxide-Dependent Apoptosis11CA52995 and CA772049. Molecular Cancer Research 1: 682–689. [PubMed] [Google Scholar]
- 42. Chen L, Na R, Gu M, Salmon AB, Liu Y, et al. (2008) Reduction of mitochondrial H2O2 by overexpressing peroxiredoxin 3 improves glucose tolerance in mice. Aging Cell 7: 866–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim SH, Fountoulakis M, Cairns N, Lubec G (2001) Protein levels of human peroxiredoxin subtypes in brains of patients with Alzheimer's disease and Down syndrome. J Neural Transm Suppl: 223–235. [DOI] [PubMed]
- 44. Macellaro A, Tujulin E, Hjalmarsson K, Norlander L (1998) Identification of a 71-kilodalton surface-associated Hsp70 homologue in Coxiella burnetii. Infect Immun 66: 5882–5888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Agoff SN, Hou J, Linzer DI, Wu B (1993) Regulation of the human hsp70 promoter by p53. Science 259: 84–87. [DOI] [PubMed] [Google Scholar]
- 46. Mosser DD, Caron AW, Bourget L, Meriin AB, Sherman MY, et al. (2000) The chaperone function of hsp70 is required for protection against stress-induced apoptosis. Mol Cell Biol 20: 7146–7159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kouchi Z, Sorimachi H, Suzuki K, Ishiura S (1999) Proteasome Inhibitors Induce the Association of Alzheimer's Amyloid Precursor Protein with Hsc73. Biochemical and Biophysical Research Communications 254: 804–810. [DOI] [PubMed] [Google Scholar]
- 48. Dworniczak B, Mirault ME (1987) Structure and expression of a human gene coding for a 71 kd heat shock ‘cognate’ protein. Nucleic Acids Res 15: 5181–5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Yoo BC, Kim SH, Cairns N, Fountoulakis M, Lubec G (2001) Deranged Expression of Molecular Chaperones in Brains of Patients with Alzheimer's Disease. Biochemical and Biophysical Research Communications 280: 249–258. [DOI] [PubMed] [Google Scholar]
- 50. Castegna A, Aksenov M, Thongboonkerd V, Klein JB, Pierce WM, et al. (2002) Proteomic identification of oxidatively modified proteins in Alzheimer's disease brain. Part II: dihydropyrimidinase-related protein 2, alpha-enolase and heat shock cognate 71. J Neurochem 82: 1524–1532. [DOI] [PubMed] [Google Scholar]
- 51. Holley AK, St Clair DK (2009) Watching the watcher: regulation of p53 by mitochondria. Future Oncology 5: 117–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Shoshan-Barmatz V, Israelson A, Brdiczka D, Sheu SS (2006) The voltage-dependent anion channel (VDAC): function in intracellular signalling, cell life and cell death. Curr Pharm Des 12: 2249–2270. [DOI] [PubMed] [Google Scholar]
- 53. Cheng EH, Sheiko TV, Fisher JK, Craigen WJ, Korsmeyer SJ (2003) VDAC2 inhibits BAK activation and mitochondrial apoptosis. Science 301: 513–517. [DOI] [PubMed] [Google Scholar]
- 54. Ferecatu I, Bergeaud M, Rodríguez-Enfedaque A, Le Floch N, Oliver L, et al. (2009) Mitochondrial localization of the low level p53 protein in proliferative cells. Biochemical and Biophysical Research Communications 387: 772–777. [DOI] [PubMed] [Google Scholar]
- 55. Olovnikov IA, Kravchenko JE, Chumakov PM (2009) Homeostatic functions of the p53 tumor suppressor: Regulation of energy metabolism and antioxidant defense. Seminars in Cancer Biology 19: 32–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Perluigi M, Di Domenico F, Giorgi A, Schininà ME, Coccia R, et al. (2010) Redox proteomics in aging rat brain: Involvement of mitochondrial reduced glutathione status and mitochondrial protein oxidation in the aging process. Journal of Neuroscience Research 88: 3498–3507. [DOI] [PubMed] [Google Scholar]
- 57. Schägger H, Ohm TG (1995) Human Diseases with Defects in Oxidative Phosphorylation. European Journal of Biochemistry 227: 916–921. [DOI] [PubMed] [Google Scholar]
- 58. Reddy PH (2009) Role of mitochondria in neurodegenerative diseases: mitochondria as a therapeutic target in Alzheimer's disease. CNS Spectr 14: 8–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Puzio-Kuter AM (2011) The Role of p53 in Metabolic Regulation. Genes Cancer 2: 385–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Maddocks OD, Vousden KH (2011) Metabolic regulation by p53. J Mol Med (Berl) 89: 237–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Boyd-Kimball D, Castegna A, Sultana R, Poon HF, Petroze R, et al. (2005) Proteomic identification of proteins oxidized by Aβ(1–42) in synaptosomes: Implications for Alzheimer's disease. Brain Research 1044: 206–215. [DOI] [PubMed] [Google Scholar]
- 62. Boyd-Kimball D, Poon HF, Lynn BC, Cai J, Pierce Jr WM, et al. (2006) Proteomic identification of proteins specifically oxidized in Caenorhabditis elegans expressing human Aβ(1–42): Implications for Alzheimer's disease. Neurobiology of Aging 27: 1239–1249. [DOI] [PubMed] [Google Scholar]
- 63. Perluigi M, Poon HF, Maragos W, Pierce WM, Klein JB, et al. (2005) Proteomic Analysis of Protein Expression and Oxidative Modification in R6/2 Transgenic Mice. Molecular & Cellular Proteomics 4: 1849–1861. [DOI] [PubMed] [Google Scholar]
- 64. Lindahl R (1992) Aldehyde dehydrogenases and their role in carcinogenesis. Crit rev Biochem Mol Biol 27: 283–335. [DOI] [PubMed] [Google Scholar]
- 65. Murphy TC, Amarnath V, Gibson KM, Picklo Sr MJ (2003) Oxidation of 4-hydroxy-2-nonenal by succinic semialdehyde dehydrogenase (ALDH5A). Journal of Neurochemistry 86: 298–305. [DOI] [PubMed] [Google Scholar]
- 66. Murphy TC, Amarnath V, Picklo Sr MJ (2003) Mitochondrial oxidation of 4-hydroxy-2-nonenal in rat cerebral cortex. Journal of Neurochemistry 84: 1313–1321. [DOI] [PubMed] [Google Scholar]
- 67. Butterfield DA, Bader Lange ML, Sultana R (2010) Involvements of the lipid peroxidation product, HNE, in the pathogenesis and progression of Alzheimer's disease. Biochimica et Biophysica Acta (BBA) – Molecular and Cell Biology of Lipids 1801: 924–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Yoon K-A, Nakamura Y, Arakawa H (2004) Identification of ALDH4 as a p53-inducible gene and its protective role in cellular stresses. J Hum Genet 49: 134–140. [DOI] [PubMed] [Google Scholar]
- 69. Papazoglu C, Mills AA (2007) p53: at the crossroad between cancer and ageing. The Journal of Pathology 211: 124–133. [DOI] [PubMed] [Google Scholar]