Abstract
Human embryonic stem cells (hESC) have a unique capacity to self-renew and differentiate into all the cell types found in human body. Although the transcriptional regulators of pluripotency are well studied, the role of cytoplasmic regulators is still poorly characterized. Here, we report a new stem cell-specific RNA-binding protein L1TD1 (ECAT11, FLJ10884) required for hESC self-renewal and cancer cell proliferation. Depletion of L1TD1 results in immediate downregulation of OCT4 and NANOG. Furthermore, we demonstrate that OCT4, SOX2, and NANOG all bind to the promoter of L1TD1. Moreover, L1TD1 is highly expressed in seminomas, and depletion of L1TD1 in these cancer cells influences self-renewal and proliferation. We show that L1TD1 colocalizes and interacts with LIN28 via RNA and directly with RNA helicase A (RHA). LIN28 has been reported to regulate translation of OCT4 in complex with RHA. Thus, we hypothesize that L1TD1 is part of the L1TD1-RHA-LIN28 complex that could influence levels of OCT4. Our results strongly suggest that L1TD1 has an important role in the regulation of stemness.
Keywords: L1TD1, Pluripotent stem cells, Embryonic stem cells, Embryonal carcinoma, Proliferation
INTRODUCTION
Human embryonic stem cells (hESCs) have a unique capacity to self-renew and differentiate into all the cell types found in human body. The transcription factors OCT4, NANOG, and SOX2 constitute a regulatory network required for the maintenance of pluripotency [1]. In addition, a variety of RNA-binding proteins (RBPs) and noncoding RNAs play key roles in the control of RNA stability, processing, and translation during embryonic development. Post-transcriptional control of maternal mRNAs is a predominant mechanism of regulation in early embryos [2]. The role of RBPs in this process is well studied in C. elegans [3]; however, their role in pluripotency and stem cell function is still poorly characterized.
To define the mechanisms regulating self-renewal and pluripotency of hESCs, we have analyzed the transcriptome data of 21 hESC lines and extracted the genes rapidly regulated during the early differentiation (R. Lund, N. Rahkonen et al., manuscript in preparation). One of the top genes selectively expressed by hESCs was L1TD1 encoding for an uncharacterized protein. The gene L1TD1, also known as ECAT11 or FLJ10884, was originally identified by Mitsui et al. [4] to be one of the ESC-associated transcripts (ECATs) in mouse ESCs. Recently, while this manuscript was in preparation Wong et al. [5] reported L1TD1 as a marker for undifferentiated hESCs. In addition, Iwabuchi et al. [6] reported L1TD1 to be enriched in mouse ESCs and rapidly activated during generation of induced pluripotent cells. Nonetheless, they showed that mouse L1TD1, that has low overall identity with human, rhesus macaques, rat, and dog orthologs, was dispensable for the maintenance of pluripotency.
In this study, we report that human L1TD1 (ECAT11, FLJ10884) is an RBP that, is localized in P-bodies, interacts with LIN28 via RNA and directly with RNA helicase A (RHA), and is required for hESC self-renewal and cancer cell proliferation.
MATERIALS AND METHODS
Cell Culture
Human ESC lines HS401 and HS360 were obtained from Outi Hovatta (Karolinska Institutet, Sweden) and H9 from WiCell Research Institute (Madison, WI). The cells were maintained on 0.1% gelatin-coated (Sigma-Aldrich, St. Louis, MO, www.sigmaaldrich.com)) plates on mitomycin C-inactivated human foreskin fibroblasts (ATCC, Manassas, VA, www.lgcstandards-atcc.org). The ES culture media consisted of Dulbecco’s modified Eagle’s medium (DMEM)-F12 (Stem Cell Technologies, Vancouver, Canada, www.stemcell.com) supplemented with 20% serum replacement, 2 mM glutamax, 1% nonessential amino acids, 50 U/ml penicillin–streptomycin, 0.1 mM 2-mercaptoethanol (all from Gibco [Life Technologies, Grand Island, NY, www.lifetechnologies.com]), and 4 ng/ml basic fibroblast growth factor (R&D Systems, Minneapolis, MN, www.rndsystems.com). In feeder-free culture conditions, the cells were cultured on Matrigel (BD Biosciences, San Jose, CA, www.bdbiosciences.com) and maintained in feeder cell conditioned ES culture media or mTeSR1 media (Stem Cell Technologies). Lines were passaged using type IV collagenase (Gibco).
Embryonal carcinoma (EC) cell lines 2102Ep, NT2D1, and NT2D1-TetR3 were obtained from Dr. Peter Andrews and Dr. Jianliang Li (University of Sheffield, U.K.) [7, 8] grown in DMEM (Sigma) supplemented with 10% fetal calf serum (FCS) (PromoCell, Heidelberg, Germany, www.promocell.com) and 2 mM L-glutamine (Sigma). TCam2 seminoma cell line was obtained from Dr. Jukka Westermarck (Turku Centre for Biotechnology, Finland). Cells were grown in RPMI 1640+GlutaMAX (Gibco) medium supplemented with 10% FCS (PromoCell) and 1% penicillin-streptomycin (Sigma). 2102Ep and TCam2 cells were passaged using 0.05% trypsin-EDTA and NT2D1 lines by scraping.
hESC Differentiation
hESCs were plated on Matrigel in feeder-free conditions, and the culture medium was supplemented with 13.7 µM of retinoic acid (Sigma). The media was changed daily. For spontaneous differentiation and embryonic body formation, hESCs were grown on uncoated plates without fibroblasts in normal ES culture medium. The medium was changed every 3 days.
Analysis of the Stem Cell Matrix Data
The expression of L1TD1 was analyzed in the Stem Cell Matrix data (http://www.ncbi.nlm.nih.gov/geo/): accession code GSE11508 [9]. Samples were preprocessed with the lumi-package of R [10] using the quantile normalization algorithm [11]. The probe values were linked to the Ensembl genes (NCBI 36) and in cases where several probes were detected within the region of the same gene, the probe values were mean centered.
Vectors
The open reading frame sequence of L1TD1 was polymerase chain reaction (PCR) amplified from cDNA prepared from hESC mRNA and cloned into the following plasmids and restriction sites: pET-20b(+), XhoI and NcoI (Novagen [Merck KGaA], Darmstadt, Germany, www.merck.de); pEF6/V5-His-TOPO, ligated by TA cloning (Invitrogen [Life Technologies]); pCAGG-EGFP, AgeI and XhoI [12] a gift from Dr. Peter Andrews (University of Sheffield, U.K.).
Antibodies
To generate antibodies, L1TD1 was overexpressed as a [His]6-tagged protein in pET20b vector in Escherichia coli strain BL21(DE3)C43 (Avidis [Imaxio], Saint Beauzire, France, www.imaxio.com). After induction with 0.4 mM isopropyl-β-d-thiogalactopyranoside (AppliChem, Darmstadt, Germany, www.applichem.com), expressed protein was isolated from inclusion bodies, solubilized and purified with Histag-based Talon metal affinity resin (Clontech, Mountain View, CA, www.clontech.com). Protein antigen was further purified by size separation on 10% sodium dodecyl sulfate (SDS) gel, extracted, and its identity was verified by liquid chromatography–tandem mass spectrometry. Using this purified L1TD1 as an antigen, a rabbit polyclonal antibody was produced by BioGenes (Berlin, Germany). Another antibody was produced by peptide immunization with custom-designed ISKERQRDIEERSR peptide and affinity purified by Bio-Genes. Details of other antibodies are provided in supporting information Table 1.
Real-Time PCR
Real-time PCR analysis was carried out as previously described [13]. The results were normalized with the expression values of housekeeping gene EF1α. The primer and probe sequences are given in supporting information Table 2.
RNA Interference and hESC Transfection
For transient knockdown, L1TD1 short hairpin RNA (shRNA) constructs were generated by cloning specific shRNA sequences (supporting information Table 2) into pSuper-green fluorescent protein (GFP)-Neo (Oligoengine, Seattle, WA, www.oligoengine.com) using BglII and XhoI cloning sites. Sequence for siL1TD1 #3 was selected from the RNAi Codex shRNA database, others from siRNA Target Findersoftware of GeneScript. ShRNA oligos were synthesized by DNA Technology A/S (Risskov, Denmark, www.dna-technology.dk). The L1TD1 shRNA sequences 1, 2, and 5 were selected for synthesizing siRNA oligonucleotides (Sigma) for the knockdown experiments. Transfections were performed according to the manufacturer’s protocols using Lipofectamine 2000 reagent (Invitrogen) with 150–200 nM concentration of siRNAs. hESC double transfections were performed 2 and 3 days after plating using cells cultured in feeder-free conditions. 2102ep cells were transfected 1 day after plating.
For inducible knockdown of L1TD1, shRNA sequences 1 and 5 were cloned into pSUPERIOR.neo plasmid (Oligoengine) between BglII and XhoI cloning sites. To establish clonal cell lines with doxycycline-dependent inducible expression of shRNAs, NT2D1 cells stably expressing TetR3 (a generous gift from Dr. Jianliang Li, University of Sheffield) were transfected with L1TD1 shRNA constructs using Lipofectamine 2000 reagent. EC medium containing puromycin (Sigma) at 3 µg/ml and G418 (Sigma) at 750 µg/ml was used for selecting the clones that express the transgene. From the emerging colonies, clonal sublines were expanded and screened for the most efficient inducible knockdown of the L1TD1 mRNA.
Chromatin Immunoprecipitation
Chromatin immunoprecipitation (ChIP) assays were performed as described [14]. Five hundred micrograms of sonicated chromatin, 10 µg of antibodies (supporting information Table 1), and anti-rabbit #112.04 or protein G #112.02 (Dynal Biotech [Life Technologies]) magnetic beads were used. Reverse crosslinking was done at 65°C for 12 hours, and DNA was treated sequentially with Proteinase K and RNase A, and purified (QIAquick PCR purification kit, #28706, Qiagen). Binding was confirmed by performing PCR with Phusion High-Fidelity DNA Polymerase (F-530L, Finnzymes [Thermo Fisher Scientific, Waltham, MA, www.thermofisher.com]) using primers upstream of L1TD1 (supporting information Table 2).
Transciptome Analysis with Illumina BeadChips
The shRNAs were induced using doxycycline (Sigma) at the concentration of 1 µg/ml. After 6 days, samples were collected from the control and L1TD1 knockdown. The experiment was carried out with three different NT2D1 cell clones expressing two different sequences of inducible shRNAs. These six pairs of samples were hybridized on Illumina Human HT-12 v.3 Expression BeadChip. The data were normalized by using quantile normalization. R-package Limma was used for statistics. Statistically significant differentially expressed genes were selected based on the filtering criteria for fold change ≥1.3 and for p value ≤0.05. The raw data are available in the Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo) with the accession code GSE21275.
Proliferation and Colony Forming Assays
The cell growth was monitored using CellTiter 96 Nonradioactive Cell Proliferation Assay (Promega, Madison, WI, www.promega.com), with 1 hour solubilization step. For colony formation assays, siRNA-transfected cells were plated, cultured for 12 days, and stained with crystal violet.
Western Blotting
Cells were lysed in lysis buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 0.5% TX-100, 5% glycerol, 1% SDS, 1 mM Na3VO4, 10 mM NaF, and 1 mM phenylmethanesulfonyl fluoride). Protein concentrations were determined with DC Protein Assay (Bio-Rad, Hercules, CA, www.bio-rad.com) and 6×SDS sample buffer (0.5 M Tris-HCl pH 6.8, 28% glycerol, 9% SDS, 5% 2-mercaptoethanol, 0.01% bromphenol blue) was added. Lysates were electrophoresed on a 10% SDS-PAGE gel and transferred to a nitrocellulose membrane. Membranes were incubated overnight at 4°C with primary antibodies. The details of the antibodies are given in supporting information Table 1. Secondary antibodies were horseradish peroxidase-conjugated, and enhanced chemiluminescence (Amersham Biosciences [GE Healthcare, Piscataway, NJ, www.gelifesciences.com]) reagent or Pierce developing solution (Pierce [Thermo Fisher Scientific]) was used for detection.
Immunofluorescence
Mitotically inactivated fibroblasts were plated on microscopy coverslips placed in cell culture dishes. The hESCs were plated on the following day. Transfections of the cells with Lipofectamine 2000 were performed 24 hours after plating. The immunocytochemistry was carried out 48 hours after transfections. For the staining, cells were fixed with 4% paraformaldehyde for 10–20 minutes at room temperature. The cells were stained for surface markers, after which the cells were permeabilized for intracellular stainings with 0.1%–1% Triton-X 100 for 20 minutes. Antibodies (supporting information Table 1) were used in dilutions 1:50–200. After intracellular staining, incubation with 40,6-diamidino-2-phenylindole (1 µg/ml; Invitrogen) was performed. Fluorescence images were captured with Zeiss AxioVert 200M or with Leica TCS Sp2 confocal microscope (Spectral Physics, Irvine, CA, www.newport.com). Colocation was analyzed with Leica TCS confocal software.
In Situ RNase A Treatment for Immunofluorescence
The hESCs were plated on the Matrigel-coated coverslips. Cells were allowed to proliferate on coverslips in conditioned media for 4 days. For the staining, cells were washed with 1× phosphate-buffered saline (PBS) and permeabilized in 0.5% Triton X-100 + protease inhibitors for 5 minutes. Cells were incubated in PBS with 10 µg/ml RNase A (Qiagen 1007885) at RT for 20 minutes. Control coverslips were treated similarly but without RNase A, as mentioned in [15].
Flow Cytometry
Cells were harvested from feeder-free plates with trypsin and washed twice with cold buffer (D-PBS, 2% FCS, 0.01% atzide). Primary antibodies were incubated for 30 minutes at 4°C, after which cells were washed. Secondary antibodies were incubated for 30 minutes at 4°C. The cells were washed and run with fluorescence-activated cell sorting (FACS) Calibur (BD) and analyzed with Cell Quest FACS diva.
Immunoprecipitation
Cells were washed twice with cold PBS and lysed into NP40-buffer (20–50 mM Tris, 150 mM NaCl, 0.5% sodium deoxylate, 0.5% NP-40) containing PhosSTOP (Roche, Espoo, Finland, www.roche.fi) and complete EDTA-free (Roche) inhibitors. Lysates were treated in the presence or absence of 20–1,000 µg/ml RNase A (Qiagen). Immunoprecipitation was carried out using Bio-Adembeads (PAG 0463) (Ademtech, Pessac, France, www.ademtech.com) or proteinG Dynabeads (Invitrogen) according to manufacturers’ protocol.
RNA-Binding Assays
Protocol was carried out as published by Hafner et al. [16]. Cells were incubated 16–20 hours with 100–500 µM 4-thiouridine (Sigma) before UV crosslinking and harvesting. For RTPCR analysis, cDNA synthesis from RNA was prepared using first strand cDNA synthesis kit (Roche) with random hexamer primers. Primers and probes are listed supporting information Table 2. Results are represented as ΔRn value (ΔRn = Rn – baseline) where Rn is the ratio of the fluorescence emission intensity of the reporter dye to the fluorescence emission intensity of the passive reference dye.
Ethical Consideration
Ethics Committee of South-West Finland Hospital District has given us the permission to grow human ESC lines. Research was carried out following the good scientific practice and guidelines of the National Advisory Board on Research Ethics.
RESULTS
L1TD1 Is Highly Expressed in hESCs and Rapidly Downregulated in Response to Differentiation
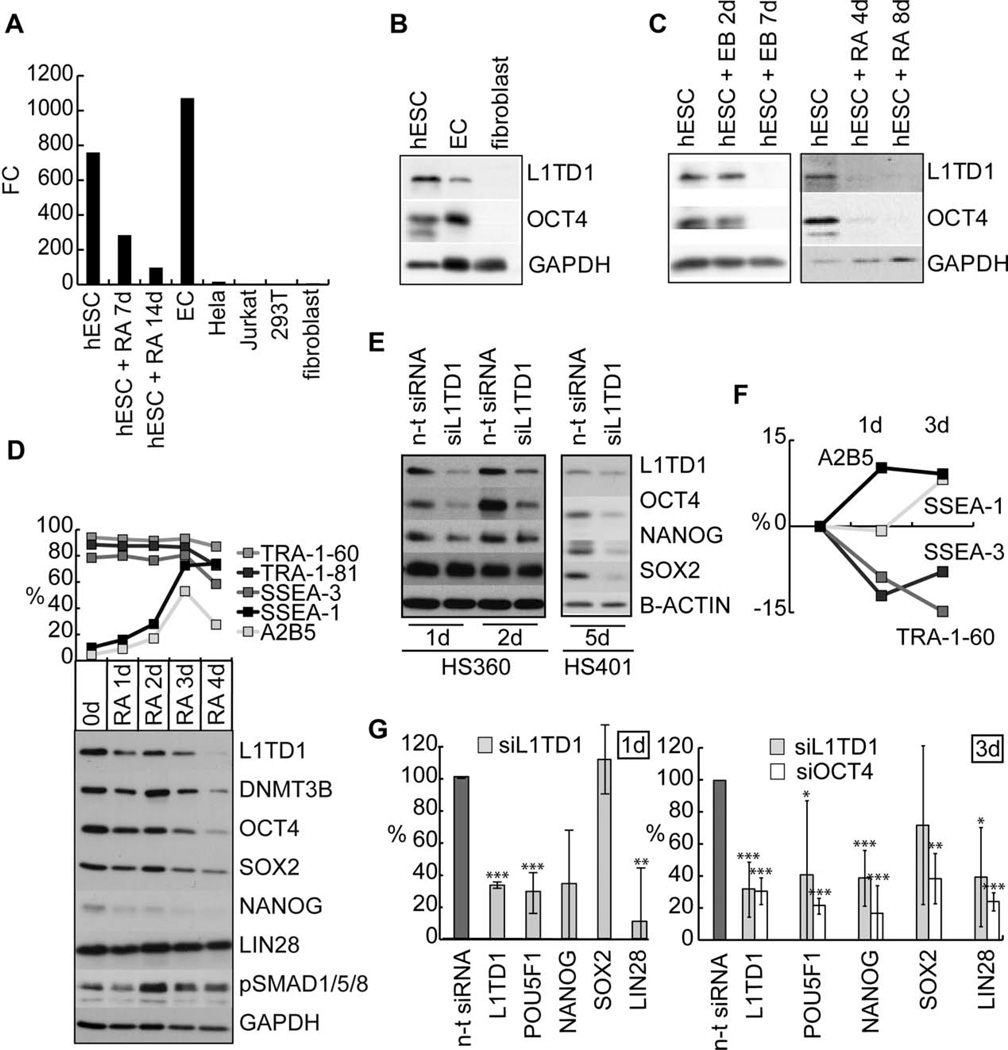
Based on our transcriptome data of 21 hESC lines (manuscript in preparation), we identified L1TD1 as a transcript selectively expressed by hESCs. Analysis of the expression profile of L1TD1 in a comprehensive Stem Cell Matrix data [9] revealed that compared with the known regulators of pluripotency OCT4 (POU5F1), SOX2, and NANOG, L1TD1 displayed the highest level of expression in hESCs, human induced pluripotent cells, and EC cells (supporting information Fig. 1A). We identified shRNAs that efficiently depleted epitope-tagged human L1TD1 (supporting information Fig. 1B) and used them to verify the specificity of antibodies against human L1TD1 (supporting information Fig. 1C). We then showed that at both the mRNA and protein levels L1TD1 was expressed in hESC and EC cells but not in fibroblasts (Fig. 1A, 1B) and that L1TD1 was downregulated in response to differentiation (Fig. 1C, 1D, supporting information Fig. 1D).
Figure 1.
L1TD1 is highly expressed in hESCs and required for self-renewal. (A): Real-time PCR analysis of L1TD1 expression in hESCs (HS401), RA-induced differentiation (HS401), EC cells (NTERA2), Hela, Jurkat, 293T, and human fibroblasts. (B): Western blot analysis of L1TD1 and OCT4 expression in hESCs (H9), ECs (2102Ep), and human fibroblasts. (C): Western blot analysis of L1TD1 and OCT4 expression in EB and RA-induced differentiation of hESC (H9 and HS401, respectively). (D): Flow cytometry and Western blot analysis of selected pluripotency and differentiation markers in RA-induced differentiation (hESC line H9). Average data from two replicate cultures. (E): Western blot analysis of the OCT4, SOX-2, and NANOG 1, 2 (oligo 5), and 5 (oligo 2) days after siRNA knockdown of L1TD1 (hESC lines HS360 and HS401). (F): Flow cytometry analysis of pluripotency (SSEA-3, TRA-1-60) and differentiation (A2B5 and SSEA-1) markers 1 and 3 days after knockdown of L1TD1 in relation to the n-t control (hESC line H9) (siRNA oligo 5). Average data from two replicate cultures. (G): Real-time PCR analysis of L1TD1 and pluripotency markers: POU5F1 (OCT4), NANOG, SOX2, and LIN28 1 and 3 days after knockdown of L1TD1. Average data of hESC lines: H9 and HS360 for day 1 (siRNA oligo 1 and 5), H9 and HS237 for day 3 (siRNA oligos 2 and 5). p values: *, ≤ 0.05; **, ≤ 0.01; ***, ≤ 0.001. Abbreviations: EB, embryoid body; EC, embryonal carcinoma; FC, fold change; hESC, human embryonic stem cell; n-t, nontargeting; PCR, polymerase chain reaction; RA, retinoic acid; siRNA, small interfering RNA.
L1TD1 Is Required for the Self-Renewal of hESCs
Depletion of L1TD1 with siRNAs induced morphological changes in hESC colonies within 4 days and resulted in downregulation of OCT4 and NANOG protein levels already on the 1st day after transfection (Fig. 1E) indicating immediate effect to the pluripotency of hESCs. At later time points all core factors OCT4, NANOG, and SOX2 were affected (Fig. 1E). Silencing of OCT4 has been reported to influence the expression of surface antigens SSEA-1, SSEA-3, and TRA-1-60 [8, 17]. To study if L1TD1 depletion has comparable effects to these antigens, we analyzed cell populations 1 and 3 days after transfections. We detected decrease of pluripotency antigens SSEA-3 and TRA-1-60 and increase of differentiation antigens SSEA-1 and A2B5 (Fig. 1F), indicating that the cells were committed for differentiation. Similar results were obtained with OCT4 knockdowns (supporting information Fig. 2).
To further analyze how L1TD1 depletion influences pluripotency of hESCs, we measured the expression of pluripotency genes 1 and 3 days after L1TD1 siRNA introduction. The expression of POU5F1 (OCT4), NANOG, and LIN28 were clearly decreased (Fig. 1G), indicating rapid loss of pluripotency. The expression of SOX2 was not directly influenced on the day 1 but started to decline on day 3. Silencing of OCT4 resulted in comparable effects analyzed on day 3 (Fig. 1G).
OCT4, SOX2, and NANOG Bind to Promoter of L1TD1
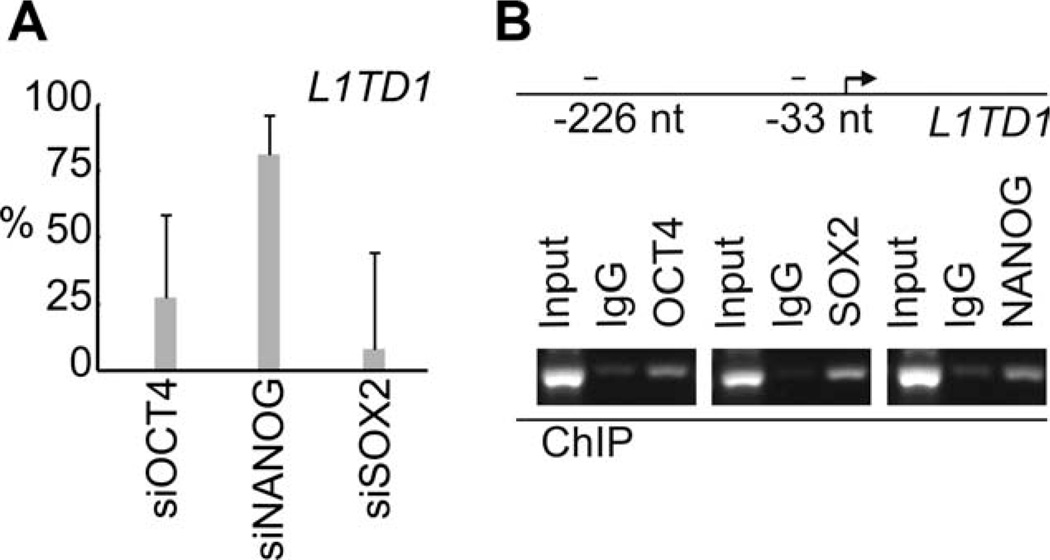
To test if expression of L1TD1 is driven by core pluripotency factors, we first silenced expression of OCT4, NANOG, and SOX2 and found decreased expression of L1TD1 mRNA, indicating that these factors could regulate expression of L1TD1 (Fig. 2A). Next, we used ChIP to demonstrate that OCT4, SOX2, and NANOG all bind to the L1TD1 promoter in hESCs (Fig. 2B).
Figure 2.
Expression of L1TD1 is regulated by core stem cell factors. (A): Expression of L1TD1 measured with real-time PCR after knockdown (KD) of OCT4 (KD-88%), NANOG (KD-75%), and SOX2 (KD-33%). Average data from days 1, 3, and 4 (human embryonic stem cells [hESC] line H9). (B): ChIP-PCR analysis of OCT4, SOX-2, and NANOG occupancy at L1TD1 locus (hESC line HS360). Abbreviations: ChIP, chromatin immunoprecipitation; PCR, polymerase chain reaction.
L1TD1 Supports Proliferation of the Seminoma and EC Cell Lines
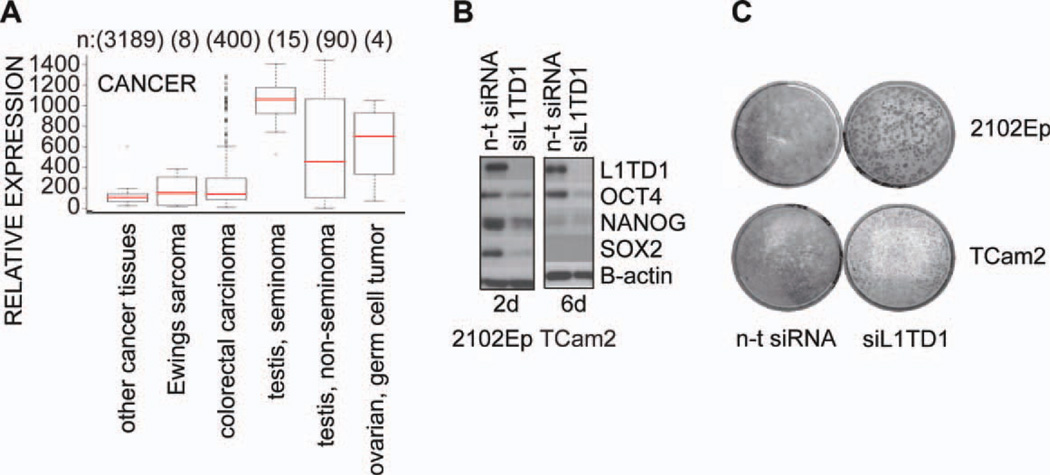
Because many stem cell-specific genes are expressed in malignant cancers and L1TD1 is highly expressed in EC cells, we analyzed L1TD1 expression profile in cancer tissues using In Silico Transcriptomics Database [18] (Fig. 3A). L1TD1 was most highly expressed in seminomas, testicular germ cell tumors that develop from pluripotent germ cells and are the most common malignancy in young men [19, 20]. Indeed, L1TD1 protein was highly expressed in seminoma (TCam2) and nonseminomatous EC cells (2102Ep) (Fig. 3B, left). Moreover, L1TD1 knockdown resulted in downregulation of the pluripotency factors OCT4, NANOG, and SOX2 in 2102Ep cells. Downregulation of OCT4 and NANOG was observed also in TCam2 cells which lack SOX2 expression (Fig. 3B, right), implying that observed effects of L1TD1 affecting stem cell core factors are not regulated through SOX2.
Figure 3.
L1TD1 is necessary for self-renewal of embryonal carcinoma (EC) cells. (A): Relative expression of L1TD1 in cancer tissues in in silico transcriptomics database [18]. (B): Western blot analysis of expression of indicated proteins and their response to L1TD1 depletion in 2102Ep and TCam2 cells (siRNA oligo 2). (C): Colony forming assay performed with nonseminoma EC cell line (2102Ep) and seminoma cell line (TCam2) after silencing with n-t siRNA or siL1TD1 (siRNA oligo 2). Cells were stained with crystal violet 12 days post-transfection. Abbreviations: n-t, nontargeting; siRNA, small interfering RNA.
We compared the transcriptional profile of the NT2D1 EC cell line to that of the same cell line where L1TD1 had been silenced in a stable fashion. Ingenuity Pathway Analysis of the transcriptome data indicated that genes involved in cancer and cell proliferation are influenced by L1TD1 (supporting information Fig. 1E). Consistent with these findings, we observed reduced colony formation capacity and reduced proliferation of these cancer cells in response to L1TD1 silencing (Fig. 3C, supporting information Fig. 1F). Taken together, our results suggest that L1TD1 may provide a useful diagnostic marker for certain cancers and its role in malignant transformation should be further investigated.
L1TD1 Is an RBP
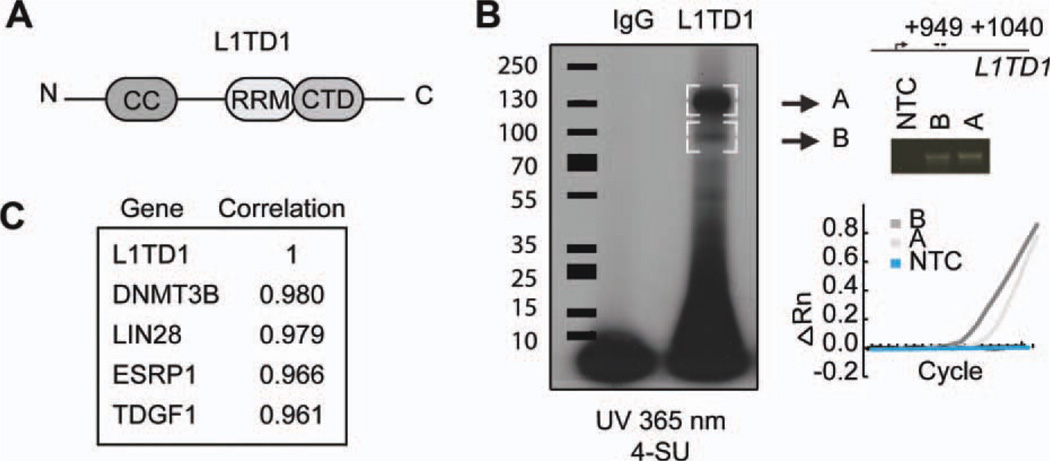
L1TD1 is composed of an N-terminal coiled-coil domain thought to be required for trimerization, an RNA recognition motif (RRM), and a C-terminal domain (CTD) (Fig. 4A) [21, 22]. L1TD1 belongs to the same family of RBPs as the mammalian LINE-1 element ORF1 protein (ORF1p). Alignment of the putative RNA-binding region of human L1TD1 with ORF1p revealed high homology with key residues known to mediate RNA binding (supporting information Fig. 3), suggesting that L1TD1 function is mediated through RNA binding. To study whether L1TD1 binds RNA, the RNA was crosslinked to protein complexes with UV and immunoprecipitated with L1TD1 antibody [16]. Two radioactive bands (A and B) were observed, whose sizes were consistent with the expected L1TD1 protein sizes detected in Western analysis of L1TD1, indicating that L1TD1 protein binds labeled RNA (Fig. 4B). As ORF proteins tend to bind their own transcripts [23], we tested if L1TD1 binds to its RNA. Indeed, L1TD1 mRNA was detected in RNA bands coimmunoprecipitated with L1TD1 protein (Fig. 4B).
Figure 4.
L1TD1 is an RNA-binding protein. (A): Structure of L1TD1 protein. N-terminal CC domain, an RRM, and a CTD. (B): SDS-PAGE autoradiography of 4-SU-labeled RNA after L1TD1 immunoprecipitation. Embryonic carcinoma cell line 2102Ep. Real-time PCR and gel analysis of L1TD1 bound L1TD1 messenger RNA extracted from gel bands A and B. (ΔRn = Rn − baseline), Rn = The ratio of the fluorescence emission intensity of the reporter dye to the fluorescence emission intensity of the passive reference dye. (C): The genes with the highest correlation of gene expression profiles with L1TD1 in Stem Cell Matrix data [9]. Abbreviations: CC, coiled coil; CTD, C-terminal domain; NTC, non-template control; RRM, RNA recognition motif; SDS-PAGE, sodium dodecyl sulfate polyacrylamide gel electrophoresis; SU, thiouridine.
L1TD1 Interacts with LIN28 in P-Bodies Through RNA
Further analysis of the Stem Cell Matrix data [9] showed that the genes with highest correlation of expression with L1TD1 were the known pluripotency-associated factors DNMT3B, LIN28, TDGF1 [24], and ESRP1 (Fig. 4C). Interestingly, ESRP1 and LIN28 are also RBPs [25, 26]. LIN28 localizes to stress granules (SGs) and processing bodies (P-bodies) [27]. Moreover, as we have shown here for L1TD1, LIN28 is also a marker for undifferentiated phenotype of hESCs [28], its knockdown has similar effect on cancer cell proliferation, and it is expressed in testicular and ovarian cancers [29, 30].
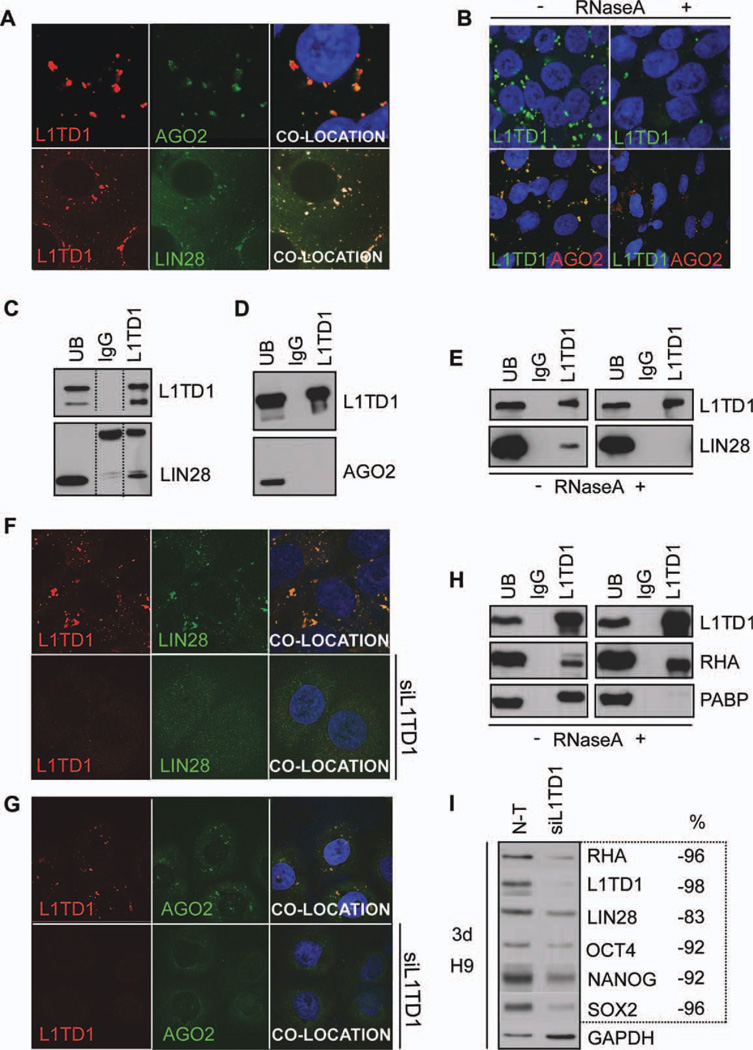
These observations prompted us to examine the intracellular location of L1TD1. Both endogenous and overexpressed L1TD1 were found in cytoplasmic structures that also contained LIN28 and the P-body marker, AGO2 [31] (supporting information Fig. 4, Fig. 5A). We also detected partial colocation with P-body markers DCP1A and GW182 in addition to SG marker TIA1 [32, 33] but not with endosome marker EEA1 [34] (supporting information Fig. 4). P-bodies are dynamic structures that are dependent on RNA for their formation [35] and are involved in mRNA and miRNA processing [36]. To verify that the cytoplasmic structures were P-bodies, we treated cells with RNase A. This resulted in loss of the cytoplasmic structures formed by L1TD1 and AGO2, as expected [31] (Fig. 5B), indicating that they corresponded to P-bodies.
Figure 5.
L1TD1 localizes in P-bodies and interacts with LIN28 and RHA. (A): L1TD1 (red) colocalization with AGO2 (green) or LIN28 (green) (cell line HS360). (B): Effect of RNase A treatment on the localization of L1TD1 and AGO2 (cell line HS360). (C): Coimmunoprecipitation analysis of L1TD1 and LIN28 (hESC line HS360). (D): Coimmunoprecipitation analysis of L1TD1 and AGO2 (NTERA2 cell line). (E): Effect of RNase A treatment on the coimmunoprecipitation of L1TD1 and LIN28 (2102Ep cell line). (F): Detection of L1TD1 (red) and LIN28 (green) in the presence or absence of siL1TD1 treatment (siRNA oligo 5) (hESC line H9). (G): Detection of L1TD1 (red) and AGO2 (green) in the presence or absence of siL1TD1 treatment (siRNA oligo 2) (TCam2 cell line). (H): Coimmunoprecipitation analysis of L1TD1 with RHA and PABP in the presence and absence of RNase A (2102Ep cell line). (I): Western blot analysis of the expression of indicated proteins and their response to L1TD1 depletion (siRNA oligo 5). Proportion (%) of the protein expression relative to that in the n-t siRNA. The values have been normalized with the housekeeping protein expression (GAPDH). Abbreviations: n-t, nontargeting; PABP, poly(A)-binding protein; RHA, RNA helicase A; UB, unbound fraction.
These results led us to ask whether LIN28 or AGO2 interacted physically or functionally with L1TD1. Coimmunoprecipitation experiments indicated that L1TD1 and LIN28 interact (Fig. 5C), in contrast, AGO2 did not coimmmunoprecipitate with L1TD1 (Fig. 5D). Because both L1TD1 and LIN28 are RBPs, we tested if these proteins interact via RNA. The RNase A treatment showed clearly that the interaction of L1TD1 and LIN28 was RNA dependent (Fig. 5E).
L1TD1 Depletion Leads to Reduction of P-Bodies
P-body assembly requires aggregation of mRNA and RBPs. As L1TD1 and LIN28 are both highly expressed stem cell-specific RBPs in P-bodies, we hypothesized that their loss could disturb P-body assembly. We were not able to detect either AGO2 or LIN28 by immunostaining after treatment of cells with siL1TD1. This indicates that the depletion of L1TD1 leads to a significant reduction in the number of P-bodies or disturbs LIN28 and AGO2 localization in these structures (Fig. 5F, 5G). These results suggest a key role for L1TD1 in the formation or maintenance of P-bodies in stem cells and especially in the localization of LIN28 to these structures.
L1TD1 Binds Directly to RHA
Human LIN28 has been shown to regulate OCT4 protein levels through binding to OCT4 mRNA [37]. To test if L1TD1 directly binds OCT4 mRNA, we probed L1TD1 immunoprecipitated RNA fraction for OCT4. The result was negative for the sequences analyzed, implying that L1TD1 does not bind OCT4 mRNA directly but possibly in complex with LIN28 and other RNA molecules.
LIN28 promotes translation of OCT4 by recruiting RHA into translation complex in hESCs [37–39]. To find out if L1TD1 is part of this complex, we tested if poly(A)-binding protein (PABP) and RHA coimmunoprecipitate with L1TD1. We demonstrated that L1TD1 directly interacts with RHA, whereas its interaction with PABP appears to be indirect (Fig. 5H), as reported for LIN28. Interestingly, depletion of L1TD1 resulted in a decrease of RHA and LIN28 levels (Fig. 5I).
DISCUSSION
We report here for the first time that human L1TD1 is an RBP that is localized in P-bodies, forms a complex with LIN28 and RHA, and is essential for the self-renewal of hESCs and embryonic carcinoma cells.
We demonstrate that OCT4, SOX2, and NANOG bind to promoter of L1TD1. Consistent with our data, binding of NANOG to the L1TD1 promoter was experimentally shown also by Wong et al. [5], and binding of SOX2 and NANOG could be detected in ChIP-on-Chip analysis [1]. Furthermore, a sequencing-based study demonstrated that the promoter region of L1TD1 appears to be active in an ESC-specific manner [40]. In addition, Iwabuchi et al. [6] showed that expression of mouse L1td1 is rapidly activated by Oct3/4 and Sox2 in the induction of pluripotency. These findings clearly indicate that L1TD1 is regulated and activated by core stem cell factors.
Several reports have indicated that the regulation of pluripotency of mouse and human differ in various aspects [41]. Based on our data L1TD1 has an important role in the regulation of pluripotency in human cells. However, it has been shown that despite the stem cell-specific expression of the mouse L1td1 ortholog, L1td1-deficient mouse ESCs are normally self-renewing [6]. This could be explained possibly by the difference of these proteins and their capability to bind RNA. The key residues, RRM and CTD domains, have 71% and 57% identical amino acids, respectively, between human and mouse L1TD1, while they share 45% identity in total sequence. Another possibility is, as discussed by Iwabuchi et al., that a lot of truncated sequences of L1 are dispersed in the mouse genome, which might work as complementary factors for L1TD1 in the knockin model. It has been also shown that LIN28, which we have shown here to interact via RNA with L1TD1, has distinct effects on pluripotency in mouse and human systems. Silencing of LIN28 in human, but not mouse, leads to reduced OCT4 protein levels [37, 42]. Hence there are important differences in the regulation of pluripotency between mouse and human ESCs.
In hESCs, LIN28 has been shown to regulate OCT4 protein translation through binding to OCT4 mRNA in complex with RHA in polysomes [37–39]. We show here that L1TD1 binds to RHA-LIN28 complex and that depletion of L1TD1 rapidly downregulates OCT4, RHA, and LIN28 levels. Our hypothesis is that the L1TD1-RHA-LIN28 complex may regulate OCT4 translation and protein levels. This would influence self-renewal as OCT4 is one of the key core regulators of pluripotency [1]. Moreover, in response to differentiation, L1TD1 and OCT4 decline with comparable kinetics, whereas level of LIN28 is reduced much later (Fig. 1D) implying that L1TD1 is an essential part of the complex regulating pluripotency in hESCs. Importantly, downregulation of L1TD1 immediately influences both mRNA and protein levels of OCT4, whereas LIN28 depletion results in the decrease of the OCT4 protein levels only [37]. We hypothesize that L1TD1 controls also levels of mRNAs to be processed for translation, as it has been reported for other RBPs whose downregulation lead to decreased levels of their targets on both RNA and protein level [43]. The subcellular location of L1TD1 also supports the idea, as one of the most important function of P-bodies is to store mRNA until needed for translation [44]. L1TD1 complex would control the stable translation of OCT4, as increase or depletion in the OCT4 levels leads to differentiation [45].
Our results strongly support the scheme that L1TD1 has an important role in the regulation of stemness in human cells. The ability of L1TD1 to bind RNA, interact with RHA, and its cytoplasmic localization, suggest a role for L1TD1 in the regulation of RNA processing. RBPs usually exert their function via binding to multiple targets that influence development in a concerted fashion [3]. As LIN28 has been shown to have a variety of functions including protein and miRNA regulation [46], L1TD1 may also have several ways to influence stem cell biology. The reduction of P-bodies can in part explain the wide effects of L1TD1 depletion on self-renewal, as P-bodies process multiple stem cell-specific mRNAs and miRNAs.
CONCLUSION
In summary, we report that L1TD1 is highly expressed in pluripotent cells. For the first time, we demonstrate that L1TD1 is an RNA-binding protein required for the self-renewal of hESCs, as depletion of L1TD1 leads to rapid reduction of OCT4 and NANOG levels and increased differentiation. Interestingly, L1TD1 interacts and colocalizes with LIN28. Moreover, L1TD1 supports proliferation of seminoma cell lines, suggesting a potential role in certain cancers. Taken together, our results propose that L1TD1 is one of the key core regulators of the stem cell fate.
Supplementary Material
ACKNOWLEDGMENTS
We thank Outi Hovatta (Karolinska Institute, Sweden) for the hESC lines; Peter Andrews (University of Sheffield, U.K.) for the EC cell lines, antibodies and plasmids; Jianliang Li (University of Sheffield, U.K.) for the NT2D1-TetR3 cell line; Anne-Mari Tveit, Joel Nyström, Subhanjan Bhowmik, Kirsi Rautajoki, and technicians Marjo Hakkarainen, Päivi Junni, Arja Reinikainen, and Sarita Heinonen for the assistance with the experiments and data mining; Cell Imaging Core and The Finnish Microarray and Sequencing Centre. This study was supported by funding for the ESTOOLS consortium under the Sixth Research Framework Programme of the European Union, JDRF, The Academy of Finland, the Finnish Cancer Organizations, Turku Graduate School of Biomedical Sciences, Ida Montin Foundation, and Finnish Cultural Foundation. A.R. is currently affiliated with La Jolla Institute for Allergy and Immunology, La Jolla, CA.
Footnotes
DISCLOSURE OF POTENTIAL CONFLICTS OF INTEREST
The authors indicate no potential conflicts of interest.
Author contributions: E.N. and N.R.: conception and design, collection and assembly of data, data analysis and interpretation, manuscript writing, and responsibility for the integrity of the work as a whole, from inception to published article; M.R.E. and J.-P.P.: conception and design, collection and assembly of data, data analysis and interpretation, and manuscript writing; R.Lu.: conception and design, data analysis and interpretation, and manuscript writing; J.N.: collection and assembly of data; R.A., K.D., and H.L.: data analysis and interpretation; O.R.: molecular biology and construct design; A.R.: conception and design, interpretation of results, and manuscript writing; R.L.: conception and design, interpretation of results, leader and supervision of the project, and manuscript writing. E.N., N.R., and M.R.E.
REFERENCES
- 1.Boyer LA, Lee TI, Cole MF, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kuersten S, Goodwin EB. The power of the 30 UTR: Translational control and development. Nat Rev Genet. 2003;4:626–637. doi: 10.1038/nrg1125. [DOI] [PubMed] [Google Scholar]
- 3.Lee MH, Schedl T. RNA-binding proteins. WormBook. 2006:1–13. doi: 10.1895/wormbook.1.79.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mitsui K, Tokuzawa Y, Itoh H, et al. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell. 2003;113:631–642. doi: 10.1016/s0092-8674(03)00393-3. [DOI] [PubMed] [Google Scholar]
- 5.Wong RC, Ibrahim A, Fong H, et al. L1TD1 is a marker for undifferentiated human embryonic stem cells. PLoS One. 2011;6:e19355. doi: 10.1371/journal.pone.0019355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iwabuchi KA, Yamakawa T, Sato Y, et al. ECAT11/L1td1 is enriched in ESCs and rapidly activated during iPSC generation, but it is dispensable for the maintenance and induction of pluripotency. PLoS One. 2011;6:e20461. doi: 10.1371/journal.pone.0020461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Andrews PW, Damjanov I, Simon D, et al. Pluripotent embryonal carcinoma clones derived from the human teratocarcinoma cell line Tera-2. Differentiation in vivo and in vitro. Lab Invest. 1984;50:147–162. [PubMed] [Google Scholar]
- 8.Zafarana G, Avery SR, Avery K, et al. Specific knockdown of OCT4 in human embryonic stem cells by inducible short hairpin RNA interference. Stem Cells. 2009;27:776–782. doi: 10.1002/stem.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Muller FJ, Laurent LC, Kostka D, et al. Regulatory networks define phenotypic classes of human stem cell lines. Nature. 2008;455:401–405. doi: 10.1038/nature07213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Du P, Kibbe WA, Lin SM. Lumi: A pipeline for processing Illumina microarray. Bioinformatics. 2008;24:1547–1548. doi: 10.1093/bioinformatics/btn224. [DOI] [PubMed] [Google Scholar]
- 11.Bolstad BM, Irizarry RA, Astrand M, et al. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 12.Liew CG, Draper JS, Walsh J, et al. Transient and stable transgene expression in human embryonic stem cells. Stem Cells. 2007;25:1521–1528. doi: 10.1634/stemcells.2006-0634. [DOI] [PubMed] [Google Scholar]
- 13.Narva E, Autio R, Rahkonen N, et al. High-resolution DNA analysis of human embryonic stem cell lines reveals culture-induced copy number changes and loss of heterozygosity. Nat Biotechnol. 2010;28:371–377. doi: 10.1038/nbt.1615. [DOI] [PubMed] [Google Scholar]
- 14.Li Z, Van Calcar S, Qu C, et al. A global transcriptional regulatory role for c-Myc in Burkitt’s lymphoma cells. Proc Natl Acad Sci USA. 2003;100:8164–8169. doi: 10.1073/pnas.1332764100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maison C, Bailly D, Peters AH, et al. Higher-order structure in pericentric heterochromatin involves a distinct pattern of histone modification and an RNA component. Nat Genet. 2002;30:329–334. doi: 10.1038/ng843. [DOI] [PubMed] [Google Scholar]
- 16.Hafner M, Landthaler M, Burger L, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matin MM, Walsh JR, Gokhale PJ, et al. Specific knockdown of Oct4 and beta2-microglobulin expression by RNA interference in human embryonic stem cells and embryonic carcinoma cells. Stem Cells. 2004;22:659–668. doi: 10.1634/stemcells.22-5-659. [DOI] [PubMed] [Google Scholar]
- 18.Kilpinen S, Autio R, Ojala K, et al. Systematic bioinformatic analysis of expression levels of 17,330 human genes across 9,783 samples from 175 types of healthy and pathological tissues. Genome Biol. 2008;9:R139. doi: 10.1186/gb-2008-9-9-r139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holmes L, Jr, Escalante C, Garrison O, et al. Testicular cancer incidence trends in the USA (1975–2004): Plateau or shifting racial paradigm? Public Health. 2008;122:862–872. doi: 10.1016/j.puhe.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bray F, Sankila R, Ferlay J, et al. Estimates of cancer incidence and mortality in Europe in 1995. Eur J Cancer. 2002;38:99–166. doi: 10.1016/s0959-8049(01)00350-1. [DOI] [PubMed] [Google Scholar]
- 21.Clery A, Blatter M, Allain FH. RNA recognition motifs: Boring? Not quite. Curr Opin Struct Biol. 2008;18:290–298. doi: 10.1016/j.sbi.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 22.Kaneko S, Manley JL. The mammalian RNA polymerase II C-terminal domain interacts with RNA to suppress transcription-coupled 30 end formation. Mol cell. 2005;20:91–103. doi: 10.1016/j.molcel.2005.08.033. [DOI] [PubMed] [Google Scholar]
- 23.Martin SL. Ribonucleoprotein particles with LINE-1 RNA in mouse embryonal carcinoma cells. Mol Cell Biol. 1991;11:4804–4807. doi: 10.1128/mcb.11.9.4804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Assou S, Cerecedo D, Tondeur S, et al. A gene expression signature shared by human mature oocytes and embryonic stem cells. BMC Genomics. 2009;10:10. doi: 10.1186/1471-2164-10-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Warzecha CC, Sato TK, Nabet B, et al. ESRP1 and ESRP2 are epithelial cell-type-specific regulators of FGFR2 splicing. Mol Cell. 2009;33:591–601. doi: 10.1016/j.molcel.2009.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moss EG, Tang L. Conservation of the heterochronic regulator Lin- 28, its developmental expression and microRNA complementary sites. Dev Biol. 2003;258:432–442. doi: 10.1016/s0012-1606(03)00126-x. [DOI] [PubMed] [Google Scholar]
- 27.Balzer E, Moss EG. Localization of the developmental timing regulator Lin28 to mRNP complexes, P-bodies and stress granules. RNA Biol. 2007;4:16–25. doi: 10.4161/rna.4.1.4364. [DOI] [PubMed] [Google Scholar]
- 28.Richards M, Tan SP, Tan JH, et al. The transcriptome profile of human embryonic stem cells as defined by SAGE. Stem Cells. 2004;22:51–64. doi: 10.1634/stemcells.22-1-51. [DOI] [PubMed] [Google Scholar]
- 29.Cao D, Allan RW, Cheng L, et al. RNA-binding protein LIN28 is a marker for testicular germ cell tumors. Hum Pathol. 2011;42:710–718. doi: 10.1016/j.humpath.2010.09.007. [DOI] [PubMed] [Google Scholar]
- 30.Peng S, Maihle NJ, Huang Y. Pluripotency factors Lin28 and Oct4 identify a sub-population of stem cell-like cells in ovarian cancer. Oncogene. 2010;29:2153–2159. doi: 10.1038/onc.2009.500. [DOI] [PubMed] [Google Scholar]
- 31.Sen GL, Blau HM. Argonaute 2/RISC resides in sites of mammalian mRNA decay known as cytoplasmic bodies. Nat Cell Biol. 2005;7:633–636. doi: 10.1038/ncb1265. [DOI] [PubMed] [Google Scholar]
- 32.Eystathioy T, Jakymiw A, Chan EK, et al. The GW182 protein colocalizes with mRNA degradation associated proteins hDcp1 and hLSm4 in cytoplasmic GW bodies. RNA. 2003;9:1171–1173. doi: 10.1261/rna.5810203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Anderson P, Kedersha N. Stressful initiations. J Cell Sci. 2002;115:3227–3234. doi: 10.1242/jcs.115.16.3227. [DOI] [PubMed] [Google Scholar]
- 34.Mu FT, Callaghan JM, Steele-Mortimer O, et al. EEA1, an early endosome- associated protein. EEA1 is a conserved alpha-helical peripheral membrane protein flanked by cysteine “fingers” and contains a calmodulin- binding Iq motif. J Biol Chem. 1995;270:13503–13511. doi: 10.1074/jbc.270.22.13503. [DOI] [PubMed] [Google Scholar]
- 35.Teixeira D, Sheth U, Valencia-Sanchez MA, et al. Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA. 2005;11:371–382. doi: 10.1261/rna.7258505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kulkarni M, Ozgur S, Stoecklin G. On track with P-bodies. Biochem Soc Trans. 2010;38:242–251. doi: 10.1042/BST0380242. [DOI] [PubMed] [Google Scholar]
- 37.Qiu C, Ma Y, Wang J, et al. Lin28-mediated post-transcriptional regulation of Oct4 expression in human embryonic stem cells. Nucleic Acids Res. 2010;38:1240–1248. doi: 10.1093/nar/gkp1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jin J, Jing W, Lei XX, et al. Evidence that Lin28 stimulates translation by recruiting RNA helicase A to polysomes. Nucleic Acids Res. 2011;39:3724–3734. doi: 10.1093/nar/gkq1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peng S, Chen LL, Lei XX, et al. Genome-wide studies reveal that Lin28 enhances the translation of genes important for growth and survival of human embryonic stem cells. Stem Cells. 2011;29:496–504. doi: 10.1002/stem.591. [DOI] [PubMed] [Google Scholar]
- 40.Zhao XD, Han X, Chew JL, et al. Whole-genome mapping of histone H3 Lys4 and 27 trimethylations reveals distinct genomic compartments in human embryonic stem cells. Cell Stem Cell. 2007;1:286–298. doi: 10.1016/j.stem.2007.08.004. [DOI] [PubMed] [Google Scholar]
- 41.Stewart R, Stojkovic M, Lako M. Mechanisms of self-renewal in human embryonic stem cells. Eur J Cancer. 2006;42:1257–1272. doi: 10.1016/j.ejca.2006.01.033. [DOI] [PubMed] [Google Scholar]
- 42.Xu B, Zhang K, Huang Y. Lin28 modulates cell growth and associates with a subset of cell cycle regulator mRNAs in mouse embryonic stem cells. RNA. 2009;15:357–361. doi: 10.1261/rna.1368009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weidensdorfer D, Stohr N, Baude A, et al. Control of c-myc mRNA stability by IGF2BP1-associated cytoplasmic RNPs. RNA. 2009;15:104–115. doi: 10.1261/rna.1175909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brengues M, Teixeira D, Parker R. Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science. 2005;310:486–489. doi: 10.1126/science.1115791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Niwa H, Miyazaki J, Smith AG. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat Genet. 2000;24:372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- 46.Viswanathan SR, Daley GQ. Lin28: A microRNA regulator with a macro role. Cell. 2010;140:445–449. doi: 10.1016/j.cell.2010.02.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.