Abstract
Mistic is a small Bacillus subtilis protein which is of current interest to the field of structural biology and biochemistry because of its unique ability to increase integral membrane protein yields in Escherichia coli expression. Using the osmo-sensing histidine kinase receptor, EnvZ, an E. coli two-component system, and its cytoplasmic cognate response regulator, OmpR, we provide the first evidence that a Mistic-fused integral membrane protein maintains functionality both in vitro and in vivo. When the purified and detergent-solubilized receptor EnvZ is fused to Mistic, it maintains the ability to autophosphorylate on residue His243 and phosphotransfers to residue Asp55 located on OmpR. Functionality was also observed in vivo by means of a β-galactosidase assay in which RU1012 [Φ(ompC-lacZ)10-15, ΔenvZ::Kmr] cells transformed with Mistic-fused EnvZ led to an increase in downstream signal transduction events detected by the activation of ompC gene expression. These findings illustrate that Mistic preserves the functionality of the Mistic-fused cargo protein and thus provides a beneficial alternate approach to study integral membrane proteins by not only improving expression levels but also for direct use in functional characterization.
Accounting for approximately 30% of all proteins in both prokaryotic and eukaryotic organisms are the integral membrane proteins. They are required for major cellular functions and are thus important pharmaceutical targets (1-4). Unfortunately, structural and biochemical studies of integral membrane proteins are hampered in part by low levels of expression. Therefore, a heterologous expression system is often employed to overcome this setback. Mistic is a 13 kDa, 110-amino acid Bacillus subtilis protein that has unique structural and functional properties. The NMR structure of Mistic has illustrated that it consists of a four α-helical bundle with a hydrophilic surface (5). Mistic differs from other membrane-integrated proteins in that it appears to interact with the lipid bilayer and can bypass the traditional cellular translocon machinery for membrane integration. Previous studies illustrate that both prokaryotic and eukaryotic membrane protein expression levels were boosted when target proteins are fused to Mistic (6, 7). Despite the utility of this Mistic-fusion system in improving expression levels of membrane proteins, the critical question still remains whether the overexpressed cargo protein remains functional as a fusion partner to Mistic. In this study, we chose to analyze the prokaryotic two-component signal transduction system EnvZ-OmpR to address this question.
Prokaryotic organisms utilize two-component signal transduction systems as their principal mode for adapting to various environmental stresses (8). One of the most widely studied and best characterized two-component systems involves the interaction between the osmo-sensing histidine kinase receptor, EnvZ and its cytoplasmic cognate response regulator, OmpR (8-10). EnvZ is a 450-amino acid inner membrane protein consisting of an NH2-terminal cytoplasmic tail, periplasmic sensor domain, two transmembrane domains, and a COOH-terminal cytoplasmic domain. The cytoplasmic domain is further divided into a HAMP-linker (histidine kinases, adenylyl cyclases, methyl-accepting chemotaxis proteins, and phosphatases), DHp (dimerization and histidine phosphotransfer) domain and the CA (catalytic and ATP-binding) domain (11-14) (Figure 1A). Upon activation EnvZ will autophosphorlyate on His243 (15) and phophotransfer the phosphoryl group to residue Asp55 on OmpR. Phosphorylated OmpR, OmpR-P, functions as a transcription factor and subsequently controls the expression of the genes for the two major outer membrane porins OmpF and OmpC (16-18). Like the majority of other histidine kinases, EnvZ has two major functions, possessing not only kinase activity but also the ability to act as a phosphatase when complexed with OmpR (16, 18), where it dephosphorylates OmpR-P and in turn regulates the concentration of OmpR-P in the cytoplasm (19).
Figure 1. Schematic Representation of EnvZ's Domain Organization.
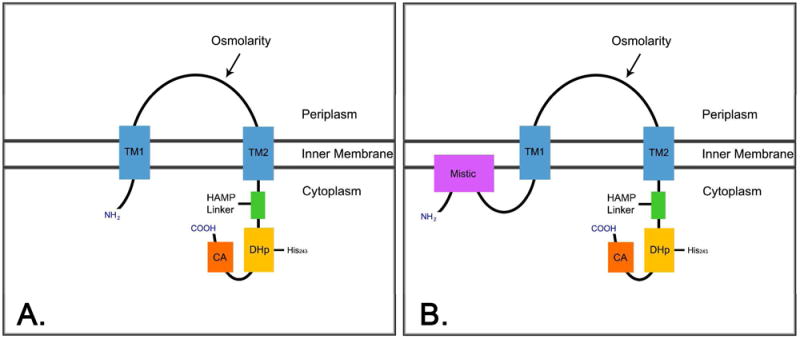
EnvZ is composed of two-transmembrane domains (blue), a periplasmic sensor domain, and a cytoplasmic domain composed of a HAMP Linker (green), DHp (gold), and CA domain (orange) (A). EnvZ domain organization when Mistic is NH2-terminally fused (B).
In addition to its use in studies involving two-component phosphorelays, EnvZ has also been exploited for various protein engineering purposes (20). In the past, EnvZ has been utilized to create many different chimeras which involve domain swapping with different chemoreceptors such as Tar (21, 22) and Trg (23) in order to study the signaling mechanisms behind two-component systems. More recently, EnvZ has been fused to the cyanobacterium light sensing phytochrome, Cph1, to create a chimera which functions in a unique system with an image processing role, thus permitting bacteria to exhibit properties like that of film (24). In this study we have used EnvZ to test if the Mistic fusion affects its catalytic and signaling capabilities (Figure 1B). Here we provide the first report which illustrates that the E. coli histidine kinase receptor, EnvZ, maintains its transmembrane signaling abilities when fused to Mistic, based on the data from both in vitro assays, through autophosphorylation and phosphotransfer to OmpR, and from in vivo assays through activation of ompC-lacZ gene expression.
Materials and Methods
Strains and Plasmids
All vector construction was Gateway (Invitrogen)- adapted. For the [γ-32P] ATP Kinase Assay, Mistic was fused to the NH2–terminus of all targets (referred to as ‘Misticated’), following an NH2-terminal octahistidine-tag in the Gateway-adapted vector, pMis3.0E (5). The genes that were not Misticated were placed in frame in a Gateway-adapted NH2-terminal nonylhistidine-tagged vector modified from pET28, (referred to as ‘pHis9’, non-Misticated). A thrombin cleavage site is present between the histidine tag and the target protein on each construct. The Gateway vector pDEST17 (Invitrogen) was used for expression of non-Misticated targets in the β-galactosidase assay to keep the antibiotic resistance (Amp) consistent between Misticated and non-Misticated samples.
E. coli BL21 (DE3) cells (Invitrogen) were used for expression of all samples. Experimental E. coli RU1012 [Φ(ompC-lacZ)10-15, ΔenvZ::Kmr] cells (courtesy of Dr. Masayori Inouye) and control E. coli MC4100 cells (courtesy of Dr. Kit Pogliano) were used for the β-galactosidase assay.
Expression and Membrane Isolation
Recombinant vectors were used to transform E. coli BL21 (DE3) cells (Invitrogen). A 5 ml overnight culture was used to inoculate 1 L of Terrific Broth (EMD) at a 1:1000 ratio. Cells were grown at 37°C to OD600= 1. Temperature was decreased to 18°C and 0.5 mM IPTG was added to induce expression of EnvZ constructs. For the soluble proteins OmpR and EnvZ cytoplasmic domain, cells were grown at 37°C to OD600= 0.4. 1 mM IPTG was added to induce expression, the temperature was kept at 37°C and cells were harvested 3 h later.
The cell pellet was weighed and the lysis buffer was added at 4X the weight of the cell pellet (20 mM Tris pH 8.0, 200 mM NaCl, 10 mM EDTA, 5 mM PMSF). The pellet was resuspended, then 5 mM β-Me and 1 mg/ml lysozyme was added and the sample was stirred at 4°C for 30 m. Cells were further lysed by sonication 3X on ice for a total of 1 m, pulses at 1 s on and 2 s off, in volumes of no more than 40 ml at a time. The sample was then centrifuged at 100,000 × g for 2 h. The pellet was resuspended in lysis buffer and centrifuged at 10,000 × g for 20 m. The supernatant was collected and centrifuged at 100,000 × g for 2 h. The membrane pellets were then re-suspended in cold salt wash buffer (20 mM Tris pH 8.0, 500 mM NaCl, 5 mM βMe, 5 mM PMSF, 10 mM EDTA) and stirred overnight at 4°C. The next day, the salt-washed membranes were centrifuged at 100,000 × g for 2 h, the pellet was then re-suspended in cold storage buffer (20mM Tris pH 8.0, 0.1 M NaCl, 5 mM βMe, 20 % v/v glycerol & protease inhibitor cocktail tablets (Roche)). The homogenous membrane mixture was then aliquoted and frozen at -80°C.
Purification
EnvZ Purification
Membranes were solubilized in solubilization buffer (20 mM Tris pH 8.0, 20 mM FC-12, 0.3 M NaCl, 1 mM MgCl2, 5 mM βMe) and stirred overnight at 4°C. Solubilized membranes were centrifuged 100,000 × g 2 h. The protein was purified on a Ni-NTA column (Qiagen) and the detergent was exchanged by washing with Wash buffer (20 mM Tris pH 8.0, 0.2 M NaCl, 4 mM FC-12, 10 mM imidazole, 3 mM βMe). The protein was eluted with Elution buffer (20 mM Tris pH 8.0, 0.2 M NaCl, 4 mM FC-12, 0.3 M imidazole, 3 mM βMe) and concentrated to 2 ml using a Vivaspin concentrator and injected on a S200 16/60 size exclusion column (Pharmacia) with FPLC buffer (20 mM Tris pH 8.0, 0.2 M NaCl, 1 mM DTT, 1 mM EDTA, 2 mM FC-12). To digest with thrombin, a 1:200 molar ratio of thrombin:protein was added and the sample was dialyzed overnight at 4°C, post Ni-NTA purification. Following cleavage on the next day, the thrombin and uncleaved protein were removed by purification on a benzamidine and Ni-NTA column, the protein was concentrated to 2 ml and purified via size exclusion chromatography as stated above.
OmpR and EnvZ Cytoplasmic Domain Purification
Cells were lysed as stated above using Lysis buffer (20 mM Tris pH 8.8, 0.3 M NaCl, 1 mM imidazole, 5 mM PMSF, 5 mM βMe). Protein was purified on a Ni-NTA column (Qiagen), washed with Wash buffer (20 mM Tris pH 8.8, 0.3 M NaCl, 20 mM imdazole), and eluted with Elution buffer (20 mM Tris pH 8.8, 0.3 M NaCl, 0.25 M imidazole, 2 mM CaCl2). Thrombin (Sigma) was added with a 1:2000 dilution, and dialyzed overnight 4°C, 3500 MWCO tubing in dialysis buffer (20 mM Tris pH 8.8, 0.3 M NaCl, 2.5 mM CaCl2). After thrombin cleavage, the protein was purified on Ni-NTA and benzamidine resin to remove uncleaved protein and thrombin. The sample was concentrated in a Vivaspin concentrator and injected on a S200 16/60 size exclusion column (Pharmacia) with FPLC buffer (20 mM Tris pH 8.8, 0.2 M NaCl, 5 mM DTT, 1 mM EDTA).
[γ-32P] ATP Kinase Assay
1 μM of protein was added to the reaction mixture (100 mM Tris pH 8.0, 100 mM KCl, 10 mM MgCl2, 10% glycerol) along with 20 μM cold ATP, 10 μCi [γ 32P] ATP in a total volume of 20 μl, and incubated at room temperature for 15 m. 10 μl of 2X SDS PAGE sample buffer was added to stop the reaction. Samples were heated in a 95°C water bath for 2 m and loaded on a 10% SDS polyacrylamide gel along with the Biorad precision plus pre-stained molecular weight marker. The gel was incubated with amberlite cation/anion exchange resin (Polysciences, Inc., polylite MB-3) to absorb free [γ 32P] ATP, dried, then exposed to KODAK BioMax XAR film for analysis.
β-galactosidase Assay
RU1012 cells or MC4100 cells were electroporated with the recombinant plasmid of choice and spread. The next day colonies were picked and a 5 ml overnight culture was made. Cells were diluted 1:100 into 150 ml Loria Broth (EMD). Cells were then grown to mid-log phase and induced with 0.5 mM IPTG. 10 ml aliquots were taken out before induction and 0.5 h, 1 h, 1.5 h, 2 h, 2.5 h, 3 h, and 20 h post induction, harvested and frozen in -80°C.
Cells were thawed and resuspended in chilled Z buffer (0.06 M Na2P04·7H2O, 0.04 M NaH2PO4·H20, 0.01 M KCl, 0.001 M MgSO4, 0.05 M βMe, pH 7.0) and normalized to 0.2 OD600. A fraction of cells were taken out and diluted to about 1:2 with Z buffer for a total of 1 ml, if this ratio did not yield a sufficient yellow color, the ratio was changed by the addition of more cells (or less cells if sample turned yellow too fast). The cells were permeabilized by the addition of 100 μl chloroform and 50 μl 0.1% SDS, then vortexed and equilibrated for 5 m in a 28°C water bath. The reaction was started by the addition of 0.2 ml 4 mg/ml ONPG, followed by incubation at 28°C. The reaction was terminated by the addition of 0.5 ml 1 M Na2CO3 once a sufficient yellow color developed. Cells were centrifuged at 17,000 × g for 5 m to remove chloroform and cell debris. OD420 and OD550 was recorded for all samples to calculate the units of activity (25).
Results
Expression and Purification of EnvZ, Misticated EnvZ and OmpR
One major challenge of studying membrane proteins is the ability to overexpress and isolate a pure homogeneous sample. With the use of a Mistic fusion (Figure 1B), approximately 19 mg of pure homogenous Misticated EnvZ was obtained by Ni-NTA affinity and size exclusion chromatography from a liter of cultured media. In contrast, EnvZ expressed without Mistic fusion yielded approximately 7 mg of pure homogeneous protein per liter of culture. The soluble response regulator OmpR also expressed to large quantities as well, yielding approximately 7 mg of pure homogenous protein by Ni-NTA affinity and size exclusion chromatography (Figure 2).
Figure 2. Purification of EnvZ, Misticated-EnvZ, and OmpR.
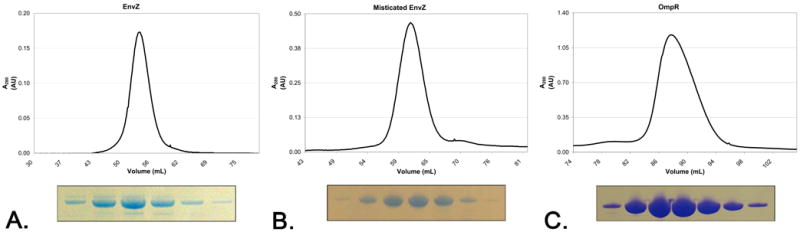
Size exclusion chromatogram profiles of EnvZ (A), Misticated EnvZ (B), and OmpR (C) from a FPLC run on a S200 16/60 column, post Ni-NTA affinity chromatography. Coomassie stained SDS-PAGE gels of the peak fractions are shown below each chromatogram.
Autophosphorylation and Phosphotransfer of EnvZ and Misticated EnvZ in vitro
[γ-32P] ATP kinase assay was used to test EnvZ and Misticated EnvZ's ability to autophosphorylate in vitro. Here purified EnvZ and Misticated EnvZ samples were incubated in the presence of Mg2+ and [γ-32P] ATP. Autoradiography was performed after running samples on a 10 % acrylamide gel. The soluble cytoplasmic domain demonstrated the ability to autophosphorylate in the absence of the sensor and transmembrane domains and confirmed the location of autophosphorylation as previously shown (17, 26), (Figure 3, lane 1).
Figure 3. [γ-32P] ATP Kinase Assay Detecting Autophosphorylation and Phosphotransfer of EnvZ and Misticated EnvZ.
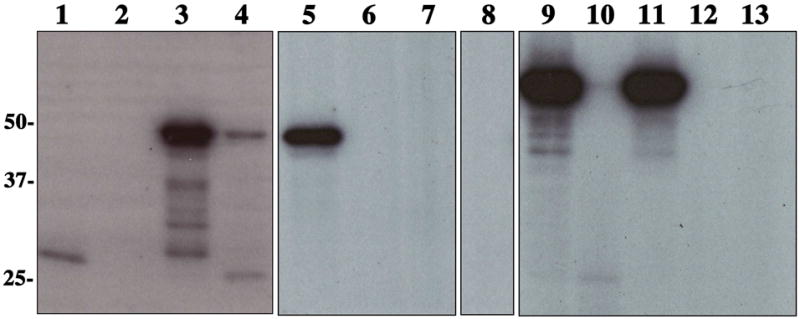
Autoradiogram of samples which were incubated in the presence of [γ-32P] ATP as described previously. Lane 1: EnvZ cytoplasmic domain; Lane 2: OmpR; Lane 3: EnvZ; Lane 4: EnvZ (upper band) and OmpR (lower band); Lane 5: EnvZ and OmpR D55Q; Lane 6: EnvZ H243V; Lane 7: EnvZ H243V and OmpR; Lane 8: OmpR D55Q; Lane 9: Misticated EnvZ; Lane 10: Misticated EnvZ (upper band) and OmpR (lower band); Lane 11: Misticated EnvZ and OmpR D55Q; Lane 12: Misticated EnvZ H243V; Lane 13: Misticated EnvZ H243V and OmpR.
Figure 3, lane 3 and 9, illustrate that both purified full-length EnvZ and Misticated EnvZ, when solubilized in FC-12, are also able to autophosphorylate. To demonstrate that this autophosphorylation event occurs on the predicted site of phosphorylation, His243, the point mutant H243V was created which has been previously shown to knock out kinase activity (17). Figure 3 (lanes 6 and 12) shows that no autophosphorylation takes place for EnvZ H243V, indicating that the autophosphorylation is dependent on residue His243.
EnvZ's ability to phosphotransfer to its cognate response regulator OmpR can also be detected using this method. When solubilized in FC-12, both EnvZ and Misticated EnvZ were incubated in the presence of OmpR and [γ-32P] ATP and exhibited the ability to autophosphorylate and phosphotransfer to OmpR (Figure 3, lanes 3-4, 9-10). When EnvZ H243V and Misticated EnvZ H243V were incubated in the presence of OmpR, no phosphotransfer (Figure 3, lanes 7 and 13) took place, demonstrating the dependence of these events on the initial autophosphorylation of EnvZ His243. To determine if phosphorylation of OmpR is dependent on EnvZ, we repeated the assay in the absence of EnvZ and Misticated EnvZ. No phosphorylation was dectected, thus illustrating its dependence on the histidine kinase (Figure 3, lane 2). To confirm the phosphorylation site, Asp55 of OmpR, we created the point mutant D55Q which was previously described to knock out phosphorylation (27). Figure 3 (lanes 5 and 11) shows that EnvZ and Misticated EnvZ are not able to phosphotransfer to OmpR D55Q, confirming its residue specificity for residue Asp55. The reaction mixture alone, in the absence of EnvZ, does not phosphorylate OmpR D55Q non-specifically (Figure 3, lane 8).
β-galactosidase Assay Illustrates EnvZ and Misticated EnvZ Signaling in vivo
The ability of EnvZ and Misticated EnvZ to autophosphorylate and phosphotransfer in vitro suggests that the CA domain and DHp domain are properly oriented allowing for such activities to take place. However, this does not give insight into the functionality of the periplasmic sensor domain, transmembrane domains, or the activity of all domains of EnvZ and Misticated EnvZ as a whole. In order to analyze the activity of all the domains of EnvZ and to look at EnvZ's ability to signal downstream to the level of inducing porin expression, an in vivo β-galactosidase assay was performed. Various EnvZ constructs were transformed into two different E. coli strains: RU1012 [Φ(ompC-lacZ)10-15, Δ envZ::Kmr] (21) and MC4100 (lac-). In this assay we electroporated cells with full-length EnvZ, Misticated EnvZ, EnvZ H243V, Misticated EnvZ H243V, and Misticated KvPae, a voltage gated K+ Channel-like protein from Pseudomonas aeruginosa, as a negative control. In addition, cells were tested in the absence of vector, as an additional negative control. Cells were grown to mid-log phase at 37°C. 10 ml aliquots were harvested before the cells reached mid-log phase, and 0.5, 1.0, 1.5, 2.0, 2.5, 3.0, and 20 h post mid-log phase. The samples were centrifuged, frozen, and the Miller Assay was completed on all samples.
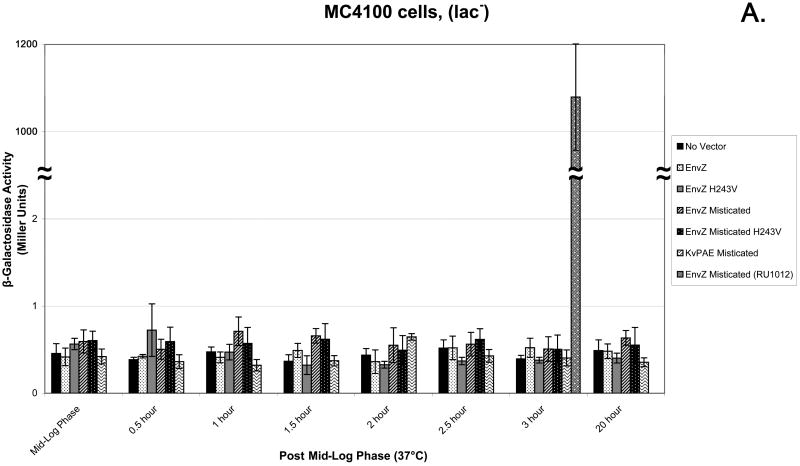
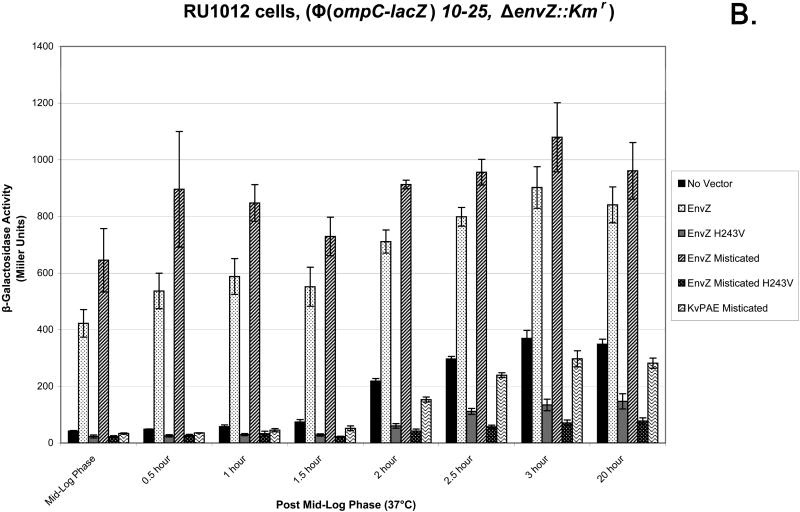
The MC4100 (lac-) E. coli cell strain was used as a control to ensure that any activity seen in the Miller Assay was due to the production of β-gal. When the Miller Assay was completed there was no significant β-gal activity, measured in Miller Units, from any of the six samples (Figure 4A). The use of the RU1012 [Φ(ompC-lacZ)10-15, Δ envZ::Kmr] (21) strain allows for the measurement of ompC gene expression as the consequence of the downstream signal transduction events of EnvZ and Misticated EnvZ. EnvZ- and Misticated EnvZ-transformed cells showed activity before mid-log phase with a rise in activity up to 3 h post mid-log phase and continuing up to 20 h post mid-log phase. The negative control which lacked a transformed vector exhibited a low level of β-gal activity starting at 2 h post mid-log phase in comparison to EnvZ and Misticated EnvZ samples. Null mutants EnvZ H243V and Misticated EnvZ H243V exhibited the lowest amount of activity than any other samples (Figure 4B).
Figure 4. β-Galactosidase Assay using the MC4100 and RU1012 E. coli strains illustrating EnvZ and Misticated EnvZ Activity.
(A) The MC4100 E. coli strain illustrating that any β-galactosidase activity (in Miller Units) is a result of the production of β-galactosidase produced by the lac-Z reporter gene. (B) The experimental RU1012 [Φ(ompC-lacZ)10-15, Δ envZ::Kmr] E. coli strain was tested to measure downstream signaling of EnvZ and Misticated-EnvZ as a result of β-galactosidase activity (in Miller Units). The following samples were tested: no vector control, EnvZ, EnvZ H243V mutant, Mistcated EnvZ, Misticated EnvZ H243V, and Misticated KvPAE. 10 ml aliquots of cells were harvested and frozen 0.5, 1.0, 1.5, 2.0, 2.5, 3.0, and 20 h post mid log phase. The Miller Assay was completed when all aliquots were collected.
Discussion
In this study, we provide direct evidence that Mistic-fused full-length EnvZ is active both in vitro and in vivo. By means of the in vitro [γ-32P] ATP kinase assay, we illustrate that EnvZ and Misticated EnvZ autophosphorylate residue His243, the conserved site of phosphorylation. In addition, we show that EnvZ and Misticated EnvZ are both able to phosphotransfer the phosphoryl group from EnvZ His243 to OmpR Asp55, in a site specific manner. Results from these assays demonstrate that both full-length constructs solubilized in the presence of the detergent FC-12 have a properly folded and functional cytoplasmic domain.
To further address the functionality of the whole receptor as a transmembrane signaling molecule, we performed an in vivo β-galactosidase assay using RU1012 E. coli cells, where EnvZ and Misticated EnvZ were tested to determine the activation of ompC-lacZ gene expression through the binding of OmpR to the ompC promoter. The results from this experiment illustrate that both EnvZ and Misticated EnvZ are active in their natural cell environment and are able to transduce a downstream signal such that the ompC promoter becomes activated. The lowest β-gal activity was found in the H243V samples and might be caused by the physical presence of the null EnvZ receptor. The expression of the non-functional EnvZ could possibly interfere or turn off alternative pathways subsequently inhibiting the otherwise recoverable ompC gene expression. When the negative control was tested in the absence of vector, a small rise in activity was seen 2 h post mid-log phase, which could be explained by the complexity of the gene regulation system of the major E. coli outer membrane porins.
Since OmpC is one of the major E. coli outer membrane porins under complex gene regulation (28), the slight rise in β-gal activity seen in our negative control of the in vivo β-gal assay, could be due to the interference of other pathways attempting to compensate for the absence of EnvZ. Both major E. coli outer membrane porins, OmpC and OmpF, are under the control of a very intricate regulatory system comprised of many components within the cell including sRNAs such as MicF (29-32), MicC (33), RseX (34, 35), RybB (36, 37), and Ipex (35, 38, 39) which function by forming base pairs with their target mRNAs in the translation start site region and thus prevent translation. There are also numerous indirect/direct protein regulators some of which include: Rob, SoxS, MarA, CpxR, Lrp, HU, IHF, and H-NS (Figure 5). Belonging to the AraC/XylS family of transcriptional regulators are Rob, SoxS, and MarA, and they function by repressing OmpF expression through activation of micF transcription (31, 40-49). The histidine kinase receptor CpxA responds to different stimuli some of which include misfolded proteins and alkaline pH and functions in conjunction with the response regulator CpxR to positively and negatively regulate ompC and ompF, respectively (50-52). The activity of the regulator Lrp increases when the cell is exposed to conditions of limited accessibility to nutrients like that of minimal medium, where this protein negatively regulates ompC and positively regulates ompF through repression of micF (53, 54). Histone-like proteins such as HU, IHF, and H-NS are also involved in the regulation of OmpC and OmpF. The nucleoid protein HU partakes in porin regulation through its involvement in a pathway which decreases OmpF levels through regulation of micF expression (55). IHF is a DNA-binding protein that functions by not only negatively regulating the ompR-envZ operon but also by negatively regulating both ompC and ompF by binding near their promoter region (56-59). The histone-like protein H-NS plays a role through repression of ompC and through the regulation of MicF resulting in a decreased level of OmpF (60, 61). Due to the complexity of porin regulation it is possible that one of these other pathways may take over the regulation of the ompC gene when EnvZ is absent.
Figure 5. The Complexity of OmpC and OmpF Porin Expression Regulation.
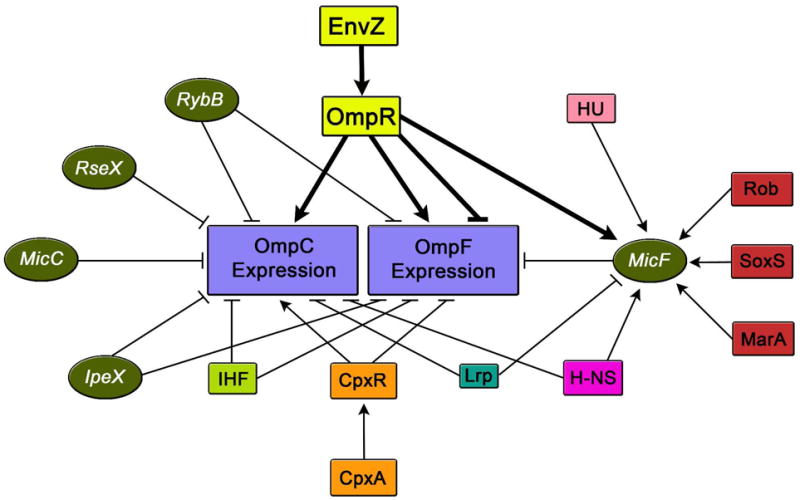
These are a number of the several different factors within the cell which are involved in the regulation of OmpC and OmpF porin expression. Various constituents illustrated include regulatory sRNAs (green circles), indirect and direct protein regulators (multi-colored boxes). The → symbol represents gene activation and ┤ represents gene repression.
Since there is an increasing demand to overcome the difficulties facing the structural studies of integral membrane proteins, biochemists and structural biologists have looked into alternative modes to not only increase the expression level of integral membrane proteins, but also to ensure that these proteins are functionally active. We describe in this study that the Mistic-fusion system provided one such alternative, by not only increasing the expression level of EnvZ but also preserving its functional activity both in vitro and in vivo.
Acknowledgments
This work was supported by grants from the National Institutes of Health GM74929 (S.C.). K.Y.B. acknowledges the fellowship support from the American Heart Association and from the H. A. and Mary K. Chapman Charitable Trust and the Mary K. Chapman Foundation. We would like to thank Innokentiy Maslennikov, Georgia Kefala, Mizuki Okamura, Luis Esquivies and Chris Dickson for their help during discussions. We thank M. Inouye for the gift of the RU1012 strain, and K. Pogliano for the gift of the MC4100 strain.
Abbreviations
- NMR
Nuclear Magnetic Resonance
- IPTG
Isopropyl β-D-1-thiogalactopyranoside
- EDTA
Ethylenediaminetetraacetic acid
- PMSF
phenylmethanesulfonyl fluoride
- B-Me
β-mercaptoethanol
- FC-12
FOS-Choline-12
- FPLC
Fast Protein Liquid Chromatography
- DTT
Dithiothreitol
- MWCO
Molecular weight cut-off
- SDS
Sodium dodecyl sulfate
- ONPG
o-Nitrophenyl-β-D-galactopyranoside
- β-gal
β-galactosidase
Footnotes
This work was supported by National Institutes of Health Grant GM74929 (S.C.), the American Heart Association Pre-doctoral Fellowship 0615005Y (K.Y.B.), and the H. A. and Mary K. Chapman Charitable Trust and the Mary K. Chapman Foundation (K.Y.B.)
References
- 1.Arkin IT, Brunger AT, Engelman DM. Are there dominant membrane protein families with a given number of helices? Proteins. 1997;28:465–466. doi: 10.1002/(sici)1097-0134(199708)28:4<465::aid-prot1>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 2.Wallin E, von Heijne G. Genome-wide analysis of integral membrane proteins from eubacterial, archaean, and eukaryotic organisms. Protein Sci. 1998;7:1029–1038. doi: 10.1002/pro.5560070420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.White SH, Wimley WC. Membrane protein folding and stability: physical principles. Annu Rev Biophys Biomol Struct. 1999;28:319–365. doi: 10.1146/annurev.biophys.28.1.319. [DOI] [PubMed] [Google Scholar]
- 4.Fraser CM, Gocayne JD, White O, Adams MD, Clayton RA, Fleischmann RD, Bult CJ, Kerlavage AR, Sutton G, Kelley JM, Fritchman RD, Weidman JF, Small KV, Sandusky M, Fuhrmann J, Nguyen D, Utterback TR, Saudek DM, Phillips CA, Merrick JM, Tomb JF, Dougherty BA, Bott KF, Hu PC, Lucier TS, Peterson SN, Smith HO, Hutchison CA, 3rd, Venter JC. The minimal gene complement of Mycoplasma genitalium. Science. 1995;270:397–403. doi: 10.1126/science.270.5235.397. [DOI] [PubMed] [Google Scholar]
- 5.Roosild TP, Greenwald J, Vega M, Castronovo S, Riek R, Choe S. NMR structure of Mistic, a membrane-integrating protein for membrane protein expression. Science. 2005;307:1317–1321. doi: 10.1126/science.1106392. [DOI] [PubMed] [Google Scholar]
- 6.Roosild TP, Vega M, Castronovo S, Choe S. Characterization of the family of Mistic homologues. BMC Struct Biol. 2006;6:10. doi: 10.1186/1472-6807-6-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kefala G, Kwiatkowski W, Esquivies L, Maslennikov I, Choe S. Application of Mistic to improving the expression and membrane integration of histidine kinase receptors from Escherichia coli. J Struct Funct Genomics. 2007;8:167–172. doi: 10.1007/s10969-007-9033-4. [DOI] [PubMed] [Google Scholar]
- 8.Hoch JA, Silhavy TJ. Two-Component Signal Transduction. ASM Press; Washington, DC: 1995. [Google Scholar]
- 9.Egger LA, Park H, Inouye M. Signal transduction via the histidyl-aspartyl phosphorelay. Genes Cells. 1997;2:167–184. doi: 10.1046/j.1365-2443.1997.d01-311.x. [DOI] [PubMed] [Google Scholar]
- 10.Forst SA, Roberts DL. Signal transduction by the EnvZ-OmpR phosphotransfer system in bacteria. Res Microbiol. 1994;145:363–373. doi: 10.1016/0923-2508(94)90083-3. [DOI] [PubMed] [Google Scholar]
- 11.Forst S, Comeau D, Norioka S, Inouye M. Localization and membrane topology of EnvZ, a protein involved in osmoregulation of OmpF and OmpC in Escherichia coli. J Biol Chem. 1987;262:16433–16438. [PubMed] [Google Scholar]
- 12.Park H, Inouye M. Mutational analysis of the linker region of EnvZ, an osmosensor in Escherichia coli. J Bacteriol. 1997;179:4382–4390. doi: 10.1128/jb.179.13.4382-4390.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Park H, Saha SK, Inouye M. Two-domain reconstitution of a functional protein histidine kinase. Proc Natl Acad Sci U S A. 1998;95:6728–6732. doi: 10.1073/pnas.95.12.6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dutta R, Qin L, Inouye M. Histidine kinases: diversity of domain organization. Mol Microbiol. 1999;34:633–640. doi: 10.1046/j.1365-2958.1999.01646.x. [DOI] [PubMed] [Google Scholar]
- 15.Roberts DL, Bennett DW, Forst SA. Identification of the site of phosphorylation on the osmosensor, EnvZ, of Escherichia coli. J Biol Chem. 1994;269:8728–8733. [PubMed] [Google Scholar]
- 16.Igo MM, Ninfa AJ, Stock JB, Silhavy TJ. Phosphorylation and dephosphorylation of a bacterial transcriptional activator by a transmembrane receptor. Genes Dev. 1989;3:1725–1734. doi: 10.1101/gad.3.11.1725. [DOI] [PubMed] [Google Scholar]
- 17.Forst S, Delgado J, Inouye M. Phosphorylation of OmpR by the osmosensor EnvZ modulates expression of the ompF and ompC genes in Escherichia coli. Proc Natl Acad Sci U S A. 1989;86:6052–6056. doi: 10.1073/pnas.86.16.6052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aiba H, Mizuno T, Mizushima S. Transfer of phosphoryl group between two regulatory proteins involved in osmoregulatory expression of the ompF and ompC genes in Escherichia coli. J Biol Chem. 1989;264:8563–8567. [PubMed] [Google Scholar]
- 19.Tokishita S, Yamada H, Aiba H, Mizuno T. Transmembrane signal transduction and osmoregulation in Escherichia coli: II. The osmotic sensor, EnvZ, located in the isolated cytoplasmic membrane displays its phosphorylation and dephosphorylation abilities as to the activator protein, OmpR. J Biochem. 1990;108:488–493. doi: 10.1093/oxfordjournals.jbchem.a123226. [DOI] [PubMed] [Google Scholar]
- 20.Xie W, Blain KY, Kuo MM, Choe S. Protein Engineering of Bacterial Histidine Kinase Receptor Systems. Protein Pept Lett. 2010;17:867–873. doi: 10.2174/092986610791306706. [DOI] [PubMed] [Google Scholar]
- 21.Utsumi R, Brissette RE, Rampersaud A, Forst SA, Oosawa K, Inouye M. Activation of bacterial porin gene expression by a chimeric signal transducer in response to aspartate. Science. 1989;245:1246–1249. doi: 10.1126/science.2476847. [DOI] [PubMed] [Google Scholar]
- 22.Zhu Y, Inouye M. Analysis of the role of the EnvZ linker region in signal transduction using a chimeric Tar/EnvZ receptor protein, Tez1. J Biol Chem. 2003;278:22812–22819. doi: 10.1074/jbc.M300916200. [DOI] [PubMed] [Google Scholar]
- 23.Baumgartner JW, Kim C, Brissette RE, Inouye M, Park C, Hazelbauer GL. Transmembrane signalling by a hybrid protein: communication from the domain of chemoreceptor Trg that recognizes sugar-binding proteins to the kinase/phosphatase domain of osmosensor EnvZ. J Bacteriol. 1994;176:1157–1163. doi: 10.1128/jb.176.4.1157-1163.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Levskaya A, Chevalier AA, Tabor JJ, Simpson ZB, Lavery LA, Levy M, Davidson EA, Scouras A, Ellington AD, Marcotte EM, Voigt CA. Synthetic biology: engineering Escherichia coli to see light. Nature. 2005;438:441–442. doi: 10.1038/nature04405. [DOI] [PubMed] [Google Scholar]
- 25.Miller JH. Experiments in Molecular Genetics. Cold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory; 1972. pp. 352–355. [Google Scholar]
- 26.Igo MM, Silhavy TJ. EnvZ, a transmembrane environmental sensor of Escherichia coli K-12, is phosphorylated in vitro. J Bacteriol. 1988;170:5971–5973. doi: 10.1128/jb.170.12.5971-5973.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kanamaru K, Aiba H, Mizuno T. Transmembrane signal transduction and osmoregulation in Escherichia coli: I. Analysis by site-directed mutagenesis of the amino acid residues involved in phosphotransfer between the two regulatory components, EnvZ and OmpR. J Biochem. 1990;108:483–487. doi: 10.1093/oxfordjournals.jbchem.a123225. [DOI] [PubMed] [Google Scholar]
- 28.Batchelor E, Goulian M. Imaging OmpR localization in Escherichia coli. Mol Microbiol. 2006;59:1767–1778. doi: 10.1111/j.1365-2958.2006.05048.x. [DOI] [PubMed] [Google Scholar]
- 29.Mizuno T, Chou MY, Inouye M. A unique mechanism regulating gene expression: translational inhibition by a complementary RNA transcript (micRNA) Proc Natl Acad Sci U S A. 1984;81:1966–1970. doi: 10.1073/pnas.81.7.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schmidt M, Zheng P, Delihas N. Secondary structures of Escherichia coli antisense micF RNA, the 5′-end of the target ompF mRNA, and the RNA/RNA duplex. Biochemistry. 1995;34:3621–3631. doi: 10.1021/bi00011a017. [DOI] [PubMed] [Google Scholar]
- 31.Delihas N, Forst S. MicF: an antisense RNA gene involved in response of Escherichia coli to global stress factors. J Mol Biol. 2001;313:1–12. doi: 10.1006/jmbi.2001.5029. [DOI] [PubMed] [Google Scholar]
- 32.Coyer J, Andersen J, Forst SA, Inouye M, Delihas N. micF RNA in ompB mutants of Escherichia coli: different pathways regulate micF RNA levels in response to osmolarity and temperature change. J Bacteriol. 1990;172:4143–4150. doi: 10.1128/jb.172.8.4143-4150.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen S, Zhang A, Blyn LB, Storz G. MicC, a second small-RNA regulator of Omp protein expression in Escherichia coli. J Bacteriol. 2004;186:6689–6697. doi: 10.1128/JB.186.20.6689-6697.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fernandez-Mora M, Oropeza R, Puente JL, Calva E. Isolation and characterization of ompS1, a novel Salmonella typhi outer membrane protein-encoding gene. Gene. 1995;158:67–72. doi: 10.1016/0378-1119(95)00171-2. [DOI] [PubMed] [Google Scholar]
- 35.Douchin V, Bohn C, Bouloc P. Down-regulation of porins by a small RNA bypasses the essentiality of the regulated intramembrane proteolysis protease RseP in Escherichia coli. J Biol Chem. 2006;281:12253–12259. doi: 10.1074/jbc.M600819200. [DOI] [PubMed] [Google Scholar]
- 36.Papenfort K, Pfeiffer V, Mika F, Lucchini S, Hinton JC, Vogel J. SigmaE-dependent small RNAs of Salmonella respond to membrane stress by accelerating global omp mRNA decay. Mol Microbiol. 2006;62:1674–1688. doi: 10.1111/j.1365-2958.2006.05524.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wassarman KM, Repoila F, Rosenow C, Storz G, Gottesman S. Identification of novel small RNAs using comparative genomics and microarrays. Genes Dev. 2001;15:1637–1651. doi: 10.1101/gad.901001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pugsley AP, Schnaitman CA. Identification of three genes controlling production of new outer membrane pore proteins in Escherichia coli K-12. J Bacteriol. 1978;135:1118–1129. doi: 10.1128/jb.135.3.1118-1129.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Castillo-Keller M, Vuong P, Misra R. Novel mechanism of Escherichia coli porin regulation. J Bacteriol. 2006;188:576–586. doi: 10.1128/JB.188.2.576-586.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rosenberg EY, Bertenthal D, Nilles ML, Bertrand KP, Nikaido H. Bile salts and fatty acids induce the expression of Escherichia coli AcrAB multidrug efflux pump through their interaction with Rob regulatory protein. Mol Microbiol. 2003;48:1609–1619. doi: 10.1046/j.1365-2958.2003.03531.x. [DOI] [PubMed] [Google Scholar]
- 41.Bennik MH, Pomposiello PJ, Thorne DF, Demple B. Defining a rob regulon in Escherichia coli by using transposon mutagenesis. J Bacteriol. 2000;182:3794–3801. doi: 10.1128/jb.182.13.3794-3801.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rosner JL, Dangi B, Gronenborn AM, Martin RG. Posttranscriptional activation of the transcriptional activator Rob by dipyridyl in Escherichia coli. J Bacteriol. 2002;184:1407–1416. doi: 10.1128/JB.184.5.1407-1416.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li Z, Demple B. SoxS, an activator of superoxide stress genes in Escherichia coli. Purification and interaction with DNA. J Biol Chem. 1994;269:18371–18377. [PubMed] [Google Scholar]
- 44.Gil F, Hernandez-Lucas I, Polanco R, Pacheco N, Collao B, Villarreal JM, Nardocci G, Calva E, Saavedra CP. SoxS regulates the expression of the Salmonella enterica serovar Typhimurium ompW gene. Microbiology. 2009;155:2490–2497. doi: 10.1099/mic.0.027433-0. [DOI] [PubMed] [Google Scholar]
- 45.Gallegos MT, Schleif R, Bairoch A, Hofmann K, Ramos JL. Arac/XylS family of transcriptional regulators. Microbiol Mol Biol Rev. 1997;61:393–410. doi: 10.1128/mmbr.61.4.393-410.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miller PF, Sulavik MC. Overlaps and parallels in the regulation of intrinsic multiple-antibiotic resistance in Escherichia coli. Mol Microbiol. 1996;21:441–448. doi: 10.1111/j.1365-2958.1996.tb02553.x. [DOI] [PubMed] [Google Scholar]
- 47.Balague C, Vescovi EG. Activation of multiple antibiotic resistance in uropathogenic Escherichia coli strains by aryloxoalcanoic acid compounds. Antimicrob Agents Chemother. 2001;45:1815–1822. doi: 10.1128/AAC.45.6.1815-1822.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cohen SP, Hachler H, Levy SB. Genetic and functional analysis of the multiple antibiotic resistance (mar) locus in Escherichia coli. J Bacteriol. 1993;175:1484–1492. doi: 10.1128/jb.175.5.1484-1492.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hachler H, Cohen SP, Levy SB. marA, a regulated locus which controls expression of chromosomal multiple antibiotic resistance in Escherichia coli. J Bacteriol. 1991;173:5532–5538. doi: 10.1128/jb.173.17.5532-5538.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Batchelor E, Walthers D, Kenney LJ, Goulian M. The Escherichia coli CpxA-CpxR envelope stress response system regulates expression of the porins ompF and ompC. J Bacteriol. 2005;187:5723–5731. doi: 10.1128/JB.187.16.5723-5731.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dorel C, Lejeune P, Rodrigue A. The Cpx system of Escherichia coli, a strategic signaling pathway for confronting adverse conditions and for settling biofilm communities? Res Microbiol. 2006;157:306–314. doi: 10.1016/j.resmic.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 52.Raivio TL. Envelope stress responses and Gram-negative bacterial pathogenesis. Mol Microbiol. 2005;56:1119–1128. doi: 10.1111/j.1365-2958.2005.04625.x. [DOI] [PubMed] [Google Scholar]
- 53.Ferrario M, Ernsting BR, Borst DW, Wiese DE, 2nd, Blumenthal RM, Matthews RG. The leucine-responsive regulatory protein of Escherichia coli negatively regulates transcription of ompC and micF and positively regulates translation of ompF. J Bacteriol. 1995;177:103–113. doi: 10.1128/jb.177.1.103-113.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Calvo JM, Matthews RG. The leucine-responsive regulatory protein, a global regulator of metabolism in Escherichia coli. Microbiol Rev. 1994;58:466–490. doi: 10.1128/mr.58.3.466-490.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Painbeni E, Caroff M, Rouviere-Yaniv J. Alterations of the outer membrane composition in Escherichia coli lacking the histone-like protein HU. Proc Natl Acad Sci U S A. 1997;94:6712–6717. doi: 10.1073/pnas.94.13.6712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tsui P, Helu V, Freundlich M. Altered osmoregulation of ompF in integration host factor mutants of Escherichia coli. J Bacteriol. 1988;170:4950–4953. doi: 10.1128/jb.170.10.4950-4953.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huang L, Tsui P, Freundlich M. Integration host factor is a negative effector of in vivo and in vitro expression of ompC in Escherichia coli. J Bacteriol. 1990;172:5293–5298. doi: 10.1128/jb.172.9.5293-5298.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ramani N, Huang L, Freundlich M. In vitro interactions of integration host factor with the ompF promoter-regulatory region of Escherichia coli. Mol Gen Genet. 1992;231:248–255. doi: 10.1007/BF00279798. [DOI] [PubMed] [Google Scholar]
- 59.Tsui P, Huang L, Freundlich M. Integration host factor binds specifically to multiple sites in the ompB promoter of Escherichia coli and inhibits transcription. J Bacteriol. 1991;173:5800–5807. doi: 10.1128/jb.173.18.5800-5807.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Deighan P, Free A, Dorman CJ. A role for the Escherichia coli H-NS-like protein StpA in OmpF porin expression through modulation of micF RNA stability. Mol Microbiol. 2000;38:126–139. doi: 10.1046/j.1365-2958.2000.02120.x. [DOI] [PubMed] [Google Scholar]
- 61.Suzuki T, Ueguchi C, Mizuno T. H-NS regulates OmpF expression through micF antisense RNA in Escherichia coli. J Bacteriol. 1996;178:3650–3653. doi: 10.1128/jb.178.12.3650-3653.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]