Abstract
Rapid, quantitative detection of cell-secreted biomarker proteins with a low sample volume holds great promise to advance cellular immunophenotyping techniques for personalized diagnosis and treatment of infectious diseases. Here we achieved such an assay with the THP-1 human acute moncytic leukemia cell line (a model for human monocyte) using a highly integrated microfluidic platform incorporating a no-wash bead-based chemiluminescence immunodetection scheme. Our microfluidic device allowed us to stimulate cells with lipopolysaccharide (LPS), which is an endotoxin causing septic shock due to severely pronounced immune response of the human body, under a well-controlled on-chip environment. Tumor necrosis factor-alpha (TNF-α) secreted from stimulated THP-1 cells was subsequently measured within the device with no flushing process required. Our study achieved high-sensitivity cellular immunophenotyping with 20-fold fewer cells than current cell-stimulation assay. The total assay time was also 7 times shorter than that of a conventional enzyme-linked immunosorbent assay (ELISA). Our strategy of monitoring immune cell functions in situ using a microfluidic platform could impact future medical treatments of acute infectious diseases and immune disorders by enabling a rapid, sample-efficient cellular immunophenotyping analysis.
Introduction
Cell-stimulation assays have provided critical means for determining functional immune responsiveness to a variety of stimuli in multiple clinical settings in order to provide diagnostic,1,2 prognostic,3–10 and therapeutic insight11,12 across a broad spectrum of patient cohorts. These assays involve stimulating white blood cells, either isolated or within whole blood, and quantitatively examining the amount of cell-secreted cytokines (i.e., cell-signaling protein molecules). At present, enzyme-linked immunosorbent assay/spot (ELISA/ELISpot)13,14 and intracellular cytokine staining (ICCS) assays are the most accepted methods to quantify cellular cytokine production while enabling multiplex, high-sensitivity (~1 pg mL−1) analyses.1,15 However, these techniques are laborious and time-consuming, prohibiting their utility in real-time clinical decision making. Both ELISA/ ELISpot and ICCS assays usually require numerous reagent manipulation processes that involve multiple staining, washing and blocking steps. Further, during these multiple sample processing steps, unpredictable signal variation and unintended analyte dilution are often induced, resulting in a narrow dynamic range, low screen throughput and compromised reproducibility. Even though ELISA is most commonly used for immunophenotyping analyses in a clinical setting, the ELISA-based approach usually requires the transfer of cell-secreted cytokine samples from culture petri dishes or centrifuge tubes to multi-well plates for signal reading in a plate reader. These steps can prove challenging, largely because edge effects and uncontrolled evaporation from very small wells can result in poor assay conditions.16
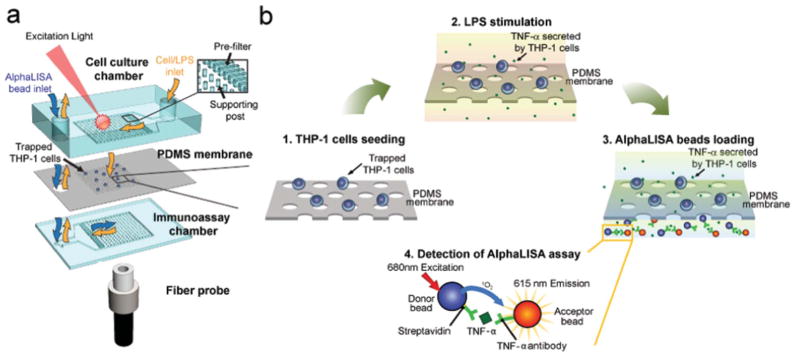
To overcome the current limitations of the aforementioned conventional functional immunophenotyping assay techniques, we have developed a polydimethylsiloxane (PDMS)-based microfluidic immunophenotyping assay (MIPA) device (Fig. 1a) capable of integrating all the assay operations on a single chip, including cell seeding, cell stimulation, and cell-secreted cytokine detection. The MIPA device incorporated a surface-micromachined PDMS microfiltration membrane (PMM). The MIPA device with the PMM provided a well-confined and miniaturized microenvironment for cell seeding and stimulation and permitted rapid diffusion of cell-secreted cytokine molecules from the cell culture chamber to the immunoassay chamber as illustrated in Fig. 1b. The miniaturized size of the MIPA device required less sample volume and shortened cytokine diffusion time. Our biomarker detection scheme further employed a bead-based chemiluminescence assay requiring no washing and lysing step while conjugating cell-secreted cytokines with assay beads, which enabled in situ cell-secreted cytokine detections with the MIPA device. We used a canonical stimulant, lipopolysaccharide (LPS), to trigger a human immune response that is routinely characterized by cytokine production.17 The detected cytokine is tumor necrosis factor-α (TNF-α), a proinflammatory cytokine and a key biomarker associated with host defense and immunosurveillance.18–20 TNF-α secretion from LPS-stimulated immune cells has been shown to reflect a functioning innate immune response.5,6,8,21
Fig. 1.
Functional immunophenotyping using the MIPA device (a) Schematic of the multi-layered MIPA device consisting of a cell culture chamber, a PDMS microfiltration membrane (PMM), and an immunoassay chamber. The size of both the cell culture and immunoassay chambers is 3.7 mm (L: length) × 3 mm (W: width) × 100 μm (H: height). The inset shows the pre-filter structure (300 μm L × 50 μm W × 100 μm H) to block particles larger than 50 μm in diameter (e.g. aggregated cells) and the supporting posts to prevent deformation of the PMM. The supporting post diameter is 50 μm with a center-to-center distance of 200 μm. A fiber probe was attached underneath the immunoassay chamber to collect AlphaLISA emission signal. (b) Schematic showing the immunophenotyping assay protocol used in this study: (1) Isolation and enrichment of THP-1 cells on the PMM; (2) LPS-stimulation of cells; (3) Loading and incubation of AlphaLISA beads in the immunoassay chamber; (4) TNF-α detection using the AlphaLISA assay, in which the streptavidin-coated donor (blue) and acceptor beads (orange) are both conjugated with TNF-α antibodies. The beads are brought into close proximity (<200 nm) through binding simultaneously to TNF-α. Using 680 nm laser for excitation, the singlet oxygen released by the donor bead diffuses to the nearby acceptor bead and triggers it to emit 615 nm fluorescent light.
Moreover, our MIPA device allowed simultaneous counting of the number of viable cells among those seeded and preserved the viability of these cells for downstream live-cell culture and analysis. The MIPA device demonstrated here eliminated a need for complex instrumental operations, prolonged sample pre-treatments, and protein surface immobilizations, which are required by other microfluidic approaches reported in previous studies.22–25 Using the MIPA device, we demonstrated a rapid, convenient, and reagent-saving functional immunophenotyping assay with only 1000 cells required, which is 20-fold less than required by current cell-stimulation assays. Owing to the miniaturized microenvironment coupled with no-wash bead-based homogenous immunoassay, the total assay time required for all of the cell loading, cell stimulation, reagent incubation, and detection processes in the MIPA device was only 3.5 h, 7 times faster than conventional ELISA-based assay. Given the shortened assay time and enhanced sample efficiencies of this approach, a microfluidic immunophenotyping technique using the MIPA device should realize a substantial number of applications in infectious and inflammatory diseases across a broad patient spectrum that includes neonates3 and pediatric patients2,5 whereby limited sample volume has precluded real-time, serial functional measurements. Thus, our microfluidic immunophenotyping technique promises to advance our understanding of immune dysfunctions, critical for developing effective interventions that will guide personalized therapy.
Methods and materials
MIPA device fabrication and materials
The PMM was fabricated using a PDMS surface icromachining technique described previously.26 Briefly, a silicon wafer was first activated using the O2 plasma (Plasma Cleaner PDC-001, Harrick Plasma) for 2 min and silanized with (tridecafluoro-1,1,2,2,-tetrahydrooctyl)-1-trichlorosilane vapour (United Chemical Technologies) for 1 h under vacuum to facilitate subsequent release of patterned PDMS layers. PDMS prepolymer (Sylgard-184, Dow Corning) was prepared by thoroughly mixing the PDMS curing agent with the PDMS base monomer (wt : wt = 1 : 10). PDMS prepolymer was then spin-coated on the silanized silicon wafer at a spin speed of 7000 rpm and completely cured after baking at 110 °C for 4 h. The PDMS surface was activated using the O2 plasma for 5 min to allow a uniform photoresist coating for photolithography. After the O2 plasma activation, photoresist (AZ 9260, AZ Electronic Materials) was spin-coated on PDMS, soft-baked at 90 °C for 10 min, and then patterned using contact photolithography. The silicon wafer was then processed with reactive ion etching (RIE; LAM 9400, Lam Research) using SF6 and O2 gas mixtures to transfer patterns from patterned photoresist to the underlying PDMS layer. During RIE, reactive gas ions etched exposed PDMS regions anisotropically. Photoresist was then stripped using organic solvents, leaving patterned PDMS thin films on the silicon wafer. Scanning electron microscopy (SEM) images were then taken for inspection of the geometrical features of the PMM, in which the PMM was mounted on stubs, sputtered with gold palladium, observed and photographed under a SEM machine (Hitachi SU8000 Ultra-High Resolution SEM).
The cell culture and immunoassay PDMS chambers were fabricated using soft lithography. Briefly, silicon molds were first fabricated using photolithography and deep reactive ion-etching (DRIE) (Deep Silicon Etcher, STS). The silicon molds were then silanized with (tridecafluoro-1,1,2,2,-tetrahydrooctyl)-1-trichlorosilane vapor for 4 h under vacuum to facilitate subsequent release of PDMS structures from the silicon molds. PDMS prepolymer with a 1 : 10 wt ratio of PDMS curing agent to base monomer was poured onto the silicon molds and cured at 110 °C for 4 h. Fully cured PDMS structures were peeled off from the silicon molds, and excessive PDMS was trimmed using a razor blade. O2 plasma-assisted PDMS–PDMS bonding process was then used to assemble the cell culture and immunoassay chambers with the PMM to form a completely sealed MIPA device. Assembly of the MIPA device was performed under eye inspection using alignment marks of the cell culture and immunoassay PDMS chambers.
Numerical simulation of flow field and cytokine diffusion in the MIPA device
The flow velocity field pattern in the MIPA device was calculated using the incompressible Navier-Stokes equations:
| (1) |
where μ denoted the dynamic viscosity (kg/(m s)), ρ represented the fluid density (kg m−3), u was the flow velocity (m s−1), p denoted the pressure (Pa), I was the inertia force, and F was the external body force. A velocity boundary condition was set to be u (=0.001 m s−1) and zero at the inlet and the outlet, respectively. To simplify our simulation, we developed a computational model with an array of 30 × 30 through holes to estimate the x–y and y–z direction flow pattern in the MIPA device. The hole diameter was 25 μm with the hole center-to-center distance of 100 μm. The total PMM area was 3 mm × 3 mm.
The cytokine diffusion profile was estimated using the transient convection and diffusion mass transfer equation:
| (2) |
where c was the concentration (mol m−3), D denoted the diffusion coefficient (m2 s−1), and u was the flow velocity (m s−1). In the cytokine diffusion model, LPS-stimulated cells were considered as the cytokine secretion source with the initial concentration of C0. To simplify our simulation, we assumed 25 single cytokine-secreting cells were uniformly distributed on the PMM.
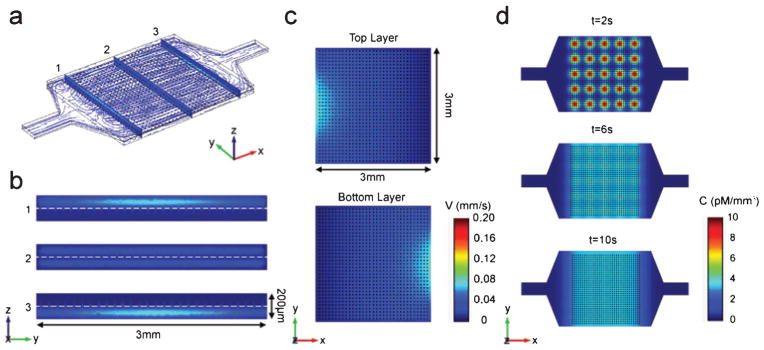
The commercial finite-element method (FEM) software (COMSOL 4.2a Multiphysics) was used to simulate the flow velocity field and cytokine diffusion in the MIPA device (Fig. 2). For the cytokine diffusion model, the size of the cytokine secreting cell was set to 25 μm. The time-dependant mass transfer equation was used with the diffusion coefficient (D) of TNF-α of 10−10 m2 s−1 and the initial TNF-α concentration (C0) of 1.0 nM mm−2; all values used in our simulation were from our experimental conditions or results reported in literature.27,28 In the time-dependant diffusion model, the velocity boundary condition at both the inlet and outlet of the cell culture and immunoassay chambers were set to zero, and secreted cytokines were homogenized over the MIPA device purely through diffusion.
Fig. 2.
Finite element simulation of MIPA device (a) Three dimensional velocity field and flow stream line profile of MIPA device. Slice figures show the detailed velocity field in the (b) y–z plane and (c) x–y plane. The model sets the inlet and outlet velocities to 0.001 m s−1 and zero, respectively. (d) Time lapse of the TNF-α diffused concentration in MIPA device. Diffusion coefficient D = 10−10 m2 s−1. Initial THP-1 cell secreted TNF-α concentration C0 = 1.0 nM mm−3. The model sets the both inlet and outlet velocities to zero.
Cell culture, deactivation, and viability text reagents
This study used the human acute monocytic leukemia cell line, THP-1, as a model for mimicking the functional immune responsiveness of human monocytes. THP-1 cells were cultured in a complete cell growth medium (RPMI-1640, ATCC) supplemented with 0.05 mM 2-mercaptoethanol (Invitrogen) and 10% (v/v) heat-inactivated fetal bovine serum (FBS, Gibco). Cells were maintained at 37 °C with 5% CO2 and 100% humidity.
To determine specificity of cellular activation, in some studies, THP-1 cells were “deactivated” or “reprogrammed” to a state of immune paralysis by culturing in the complete growth medium supplemented with 10 ng mL−1 LPS for 24 h before loading the cells into the MIPA device for immunophenotyping. This in vitro treatment has been commonly employed to induce endotoxin tolerance in THP-1 cells so that they are subsequently unresponsive to LPS.29
To examine viability of THP-1 cells after LPS stimulation, LIVE/DEAD® Viability/Cytotoxicity Kit for mammalian cells (L-3224, Invitrogen) was used. Specifically, calcein AM and ethidium homodimer-1 diluted in PBS to a final concentration of 1 μM and 2 μM, respectively, were added to THP-1 cells captured on the PMM for 30 min before the cells were examined under fluorescence microscopy for quantification of cell viability. A 130 W mercury lamp (Intensilight C-HGFIE, Nikon) was used for fluorescent illumination. Calcein AM was visualized with a FITC filter set (excitation, 498 nm; emission, 530 nm; Nikon), while ethidium homodimer-1 was visualized with a Texas Red filter set (excitation, 570 nm; emission, 625 nm; Nikon).
Immunophenotyping assay using the MIPA device
The overall immunophenotyping assay protocol using the MIPA device is shown in Fig. S1, ESI.† First, a 10 μL cell solution with various THP-1 cell concentrations was loaded into the MIPA device by a syringe pump (Fig. S1a, ESI†). After the cells were uniformly seeded on the PMM, 2 μL LPS (L5886, Sigma) solutions of different concentrations (10, 50, and 100 ng mL−1) were loaded into the MIPA device from the inlet of the cell culture chamber using pipette tips (Fig. S1b, ESI†). After LPS loading, two pipette tips were inserted into the inlets of both the cell culture and immunoassay chambers to prevent evaporation and provide a shear stress free microenvironment for cell stimulation. The MIPA device was incubated with the cells stimulated with LPS at 37 °C and 5% CO2 for 2 h. Then, the pipette tip inserted into the inlet of the immunoassay chamber was replaced by another pipette tip filled with 2 μL AlphaLISA acceptor beads (10 μg mL−1) mixed with 2 μL of 10 nM biotinylated TNF-α antibody. AlphaLISA acceptor beads in the MIPA device were incubated with the cells at 37 °C and 5% CO2 for 1 h, before another pipette tip filled with 2 μL AlphaLISA streptavidin-coated donor beads (400 μg mL−1) was loaded into the inlet of the immunoassay chamber. The whole MIPA setup was incubated at 37 °C and 5% CO2 for another 30 min (Fig. S1c, ESI†). During the whole 1.5 h bead incubation period, TNF-α secreted by LPS-stimulated THP-1 cells would diffuse from the top cell culture chamber through the PMM into the bottom immunoassay chamber to conjugate with antibody-coated donor and acceptor beads. After bead incubation, the MIPA device was placed into the customized optical setup for AlphaLISA signal detection.
No-wash, homogeneous bead-based sandwich immunoassay for immunophenotyping
AlphaLISA is a no-wash, homogeneous bead-based sandwich immunoassay technique well validated by the standard ELISA technique in previous studies.30,31 AlphaLISA eliminates washing and blocking steps required for ELISA that often result in analyte dilution and potential human contaminations. AlphaLISA is based on photo-induced chemiluminescence between pairs of antibody-conjugated donor and acceptor beads (250–350 nm in diameter) in close proximity to each other in the presence of a sandwiched analyte molecule (Fig. 1b, step 4).32 When the analyte is captured by sandwich antibodies, both antibody-conjugated donor and acceptor beads are brought into close proximity (<200 nm) to each other. Upon a laser excitation at 680 nm, donor beads generate singlet oxygen triggering a cascade of chemical events in the acceptor bead, resulting in a sharp chemiluminescent emission from the acceptor bead peaking at 615 nm. The emission signal from acceptor beads is only generated when the antibodies conjugated on both donor and acceptor beads capture analytes. Thus, AlphaLISA is highly specific and can preserve biological activities of immune cells that can be disrupted by blocking or washing steps required in ELISA.
Assay signal detection setup
The MIPA device was placed on a customized optical setup for detection of the AlphaLISA signal (Fig. S2a, ESI†). In this setup, a 500 mW 680 nm laser diode (S-67-500C-100-H, Coherent) was used to induce singlet oxygen from AlphaLISA donor beads. An optical fiber (A57-746, Edmund Optics) with a signal collection area of 1000 μm in diameter and N.A. of 0.22 was placed underneath the MIPA device to collect AlphaLISA emission signal, which was transmitted through the optical fiber and detected by a photomultiplier tube (PMT) (H9306-03, Hamamatsu). A 660 nm shortpass filter (ET660SP, Chroma) and an electronic shutter (DSS1033250A, Unibliz) were placed in front of the PMT to cut off undesired scattering light from the excitation laser. A function generator (Agilent) was used to control timing of triggering the laser pulse for excitation and of opening the shutter in front of PMT for collecting AlphaLISA signal. Both laser pulse and PMT signals were recorded by a multifunctional data acquisition card (NI PCI-6111, National Instruments). Signal analysis software custom-developed using LabVIEW 7.0 (National Instruments) program was used for simultaneous recording of time-sequenced shutter trigger and emission signal detected by the PMT.
To generate the TNF-α standard curve, known amounts of TNF-α were spiked in RPMI media (0–10 000 pg mL−1). Then, 10 μg mL−1 AlphaLISA acceptor beads, 10 nM biotinylated TNF-α antibody and 400 μg mL−1 streptavidin-coated donor beads were mixed with the TNF-α spiked solution. The AlphaLISA signal was measured using the same customized optical system as a function of the TNF-α concentration and fitted by a sigmoid dose–response curve using GraphPad Prism software.
Quantitative analysis of cell seeding in the MIPA device
Before seeding cells, the MIPA device was filled with 2% (w/w) Pluronics F127 (P2443-250G, Sigma) in PBS to remove air bubbles trapped in the device. The MIPA device was flushed twice with PBS, followed by loading a fresh cell growth medium. THP-1 cells were then seeded into the MIPA device in the complete cell medium using a syringe infusion pump (World Precision Instruments) at a flow rate of 5 μL min−1. Cell seeding was monitored under an inverted microscope (Nikon Eclipse TiS, Nikon) equipped with an electron multiplying charge-coupled device (EMCCD) camera (Photometrics). Sequential brightfield and fluorescent images were taken using 10× (Ph1 ADL, numerical aperture or N.A. = 0.25, Nikon) and 20× (CFI Plan Fluor ELWD, N.A. = 0.45, Nikon) objectives. A 130 W mercury lamp (Intensilight C-HGFIE, Nikon) was used for fluorescent illumination. To examine cell seeding uniformity, the whole PMM area was scanned on a motorized stage (ProScan III, Prior Scientific). The images obtained from scanning were stitched using microscopic analysis software (NIS-Element BR, Nikon). To count viable and dead cells, recorded FITC-Texas Red fluorescent images were processed using NIS-Element BR software. Specifically, the threshold function was applied to optimize image contrast. Then, the Canny edge detection method was used to identify cell boundary, after which certain measurement criterions including cell area (100–500 μm2) and circularity (0.5–1.0) were applied to identify THP-1 cells isolated on the PMM and perform cell counting.
Results and discussion
Device design and simulation
The structure of the MIPA device consisted of three different PDMS layers. The top and bottom PDMS layers were the cell culture and immunoassay chambers, respectively, and the middle layer was a PDMS microfiltration membrane (PMM). The top cell culture chamber of the MIPA device was designed for seeding and stimulation of THP-1 cells using LPS. The bottom immunoassay chamber of the MIPA device was designed for loading immunoassay beads and optical detection of AlphaLISA signals. Embedded between the top and bottom microfluidic layers was the PMM, which was designed (1) for isolation and enrichment of THP-1 cells and (2) for allowing cytokines secreted from LPS-stimulated cells to diffuse rapidly into the bottom immunoassay chamber for quantitative immunosensing. The PMM contained an array of closely packed through holes of 4 μm in diameter and with a center-to-center distance of 10 μm. The PMM had an effective filtration area of 7 mm2 and a thickness of 10 μm. The cell isolation and cytokine diffusion efficiency is critically dependent on the membrane porosity, which is defined as the ratio between the total pore area to the total membrane area. In this work, we successfully fabricated the PMM with 25% porosity. In comparison, conventional track-etched polycarbonate filters used for blood cell isolation has reported porosity of less than 2%.33,34 The other more recently developed Parylene-based micropore membrane has porosity of 7%–15%.35 Even with this high porosity of 25% in the PMM, we did not observe any deformation of the PMM during all cell loading experiments (with a flow rate from 1 to 10 μL min−1), suggesting the PMM structure has superior mechanical robustness owing to the supporting post structures integrated in both the cell culture and immunoassay chambers.
To reduce the computational load in our numerical simulation, we modelled the PMM as a membrane with holes of 25 μm in diameter and their center-to-center distance of 100 μm, which has a coarser distribution of holes than the real PMM design (4 μm hole diameter and 10 μm spacing). The membrane hole dimensions used in this model are expected to yield a worse cell flow condition than the actual design. Our simulation results in Fig. 2c still showed sufficiently uniform flow distributions in the middle x–y plane of both the cell culture and immunoassay chambers in the MIPA device. Therefore, we could ensure that the real PMM design, having a finer and more dense hole pattern, would enable us to obtain a flow velocity distribution and an analyte diffusion profile both at high uniformity. Fig. 2d plotted the spatial distribution of the TNF-α concentration over time. After diffusion for 10 s, the TNF-α concentration became largely homogeneous within both the cell culture and the immunoassay chambers of the MIPA device. Our simulation result suggested that TNF-α secreted by THP-1 cells in the MIPA device could rapidly become spatially homogeneous owing to the miniaturized microfluidic environment of the MIPA device.
Cell seeding and cell viability in the MIPA device
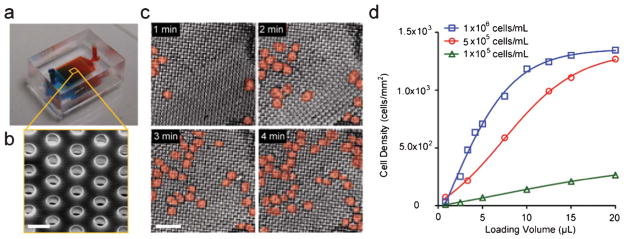
We characterized the cell seeding performance of the MIPA device for on-chip isolation and enrichment of THP-1 cells. THP-1 cells of 10–30 μm in diameter were loaded into the MIPA device in the complete cell medium at three different concentrations of 1 × 105, 5 × 105, 1 × 106 cells mL−1 under a flow rate of 5 μL min−1. Fig. 3a and b show a photograph of the MIPA device and a SEM image of the PMM. Fig. 3c represents a temporal sequence of false-colored brightfield images showing isolation and enrichment of THP-1 cells on the PMM. Using these brightfield images, we quantified the cell seeding density on the PMM as a function of the sample loading volume (Fig. 3d). Our results in Fig. 3 demonstrate that we could conveniently control the total number of THP-1 cells trapped on the PMM by modulating the sample injection time, necessary for normalizing the amount of TNF-α secreted by single THP-1 cells.
Fig. 3.
Isolation and enrichment of THP-1 cells using the MIPA device (a) A photograph of the MIPA device. The MIPA device was injected with dyed solutions for visualization of the cell culture chamber and the immunoassay chamber. The device dimension is 9 mm L × 7 mm W × 4 mm H. (b) SEM image showing the PMM. Scale bar, 10 μm. (c) Temporal sequence of false-colored brightfield images showing isolation and enrichment of THP-1 cells on the PMM. The cell loading concentration was 5 × 105 cells mL−1 at 5 μL min−1 flow rate. Scale bar, 100 μm. (d) Plot of density of trapped cells on the PMM as a function of injection volume, using three different cell loading concentrations as indicated.
Under optical microscopy, the top surface of the PMM as well as the immunoassay chamber could be monitored in real time during cell loading by vertically changing the focal plane of the microscope (Fig. S3b, ESI†). During the cell loading process, no cell was observed in the immunoassay chamber as well as at the outlet of the MIPA device, suggesting no cell could pass through the PMM and all the cells were captured and retained on the PMM. We confirmed the cell capture efficiency of the PMM for THP-1 cells by comparing the number of captured cells on the PMM to the number of cells injected into the MIPA device. Our results suggested a nearly 100% capture efficiency of the PMM for THP-1 cells.
We further examined the cell seeding uniformity by taking fluorescence images across the whole PMM area for calcein AM-labeled THP-1 cells. In this experiment, the MIPA device was loaded with a cell solution with the cell concentration of 5 × 105 cells mL−1 and the flow rate of 10 μL min−1 for 3 min. The cell density at three different locations uniformly distributed on the PMM was calculated and compared. Our result showed that the cell density at these different locations on the PMM were in the range of 1.20 × 103–1.32 × 103 cells mm−2 with a variation of 3–5% (Fig. S3c, ESI†), suggesting a good cell seeding uniformity for THP-1 cells on the PMM in the MIPA device.
We next verified cell viability of THP-1 cells under various levels of LPS stimulations. We loaded a 10 μL cell solution with the THP-1 cells concentration of 5 × 105 cells mL−1 into the MIPA device. The cells were stimulated and incubated with different concentrations of LPS (10, 50, 100 ng mL−1) for 2 h. THP-1 cells were then stained using the cell LIVE/DEAD® Viability/Cytotoxicity Kit. The cell viability rate after LPS stimulations was as high as 90–92%, regardless of the LPS concentration (Fig. S4b, ESI†). Compared to 96% viability rate in the control group without LPS stimulation, we concluded that our cell capture procedure using the PMM and the subsequent LPS stimulation had a minimum effect on viability of THP-1 cells.
Effect of cell population and endotoxin concentration on cytokine secretion
After validating the minimal cytotoxic effect of our immunophenotyping protocol on THP-1 cell viability, we systematically quantified the levels of TNF-α secreted by THP-1 cells as a function of the total cell population trapped on the PMM (n = 1000, 5000, and 20 000 cells) and the LPS concentration (10, 50, 100 ng mL−1). The AlphaLISA signal detected using the optical system was converted to the TNF-α concentration using a TNF-α standard curve (Fig. 4a) generated using AlphaLISA with samples spiked with known concentrations of TNF-α. This TNF-α standard curve provided a correlation between the TNF-α concentration in the MIPA device and the corresponding AlphaLISA signal intensity. We further compared this result with the TNF-α standard curve by ELISA using a commercial plate reader (SpectraMax M2e, Molecular Devices). The two curves showed comparable results (Fig. S5a, ESI†), suggesting AlphaLISA is indeed suitable for detection of TNF-α secreted from stimulated immune cells. The limit of detection (LOD), which is defined as 3 times the standard deviation of the blank (without spiked analyte condition) was 75 pg mL−1 and 10 pg mL−1 with our AlphaLISA method and the ELISA method, respectively, which are of the same order of magnitude.
Fig. 4.
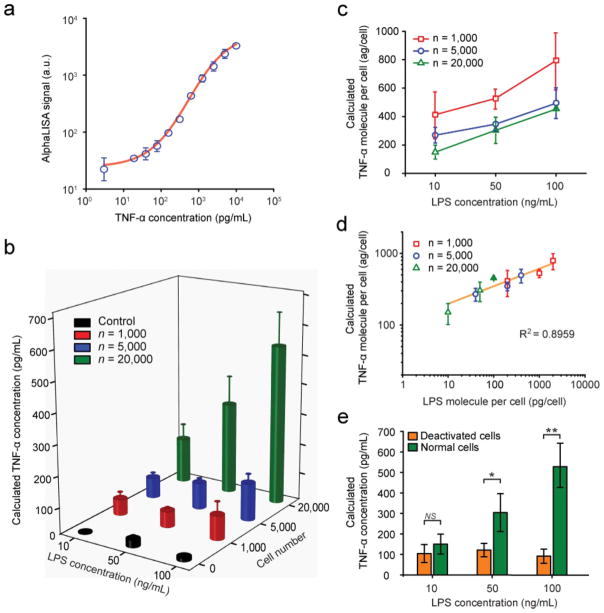
Detection of TNF-α secreted from LPS-stimulated THP-1 cells using the MIPA device (a) Standard curve for TNF-α detection. TNF-α with a known concentration (0–10 000 pg mL−1) was spiked in the complete cell growth medium and detected using AlphaLISA and the customized optical setup. (b) Plot of TNF-α concentration secreted by LPS-stimulated THP-1 cells as a function of cell number and LPS concentration. (c and d) Plots of average TNF-α concentration secreted by individual cells as a function of LPS concentration (c) or LPS concentration per cell (d). (e) Plot of TNF-α concentration secreted by normal and LPS-deactivated THP-1 cells trapped on the PMM (n = 20 000 cells). P-values calculated using the paired Student’s t-test are indicated for significant differences (P < 0.05 (*) and P < 0.005 (**)). NS, statistically not significant.
Fig. 4b plotted the TNF-α concentration secreted by THP-1 cells as a function of the total cell population and the LPS concentration. Our result demonstrated that, as expected, the concentration of TNF-α secreted by THP-1 cells increased according to both the cell number and the LPS concentration. When the LPS concentration increased from 10 to 100 ng mL−1, the TNF-α concentration secreted by 1000, 5000 and 20 000 THP-1 cells increased from 53 to 80 pg mL−1, 67 to 123 pg mL−1 and 150 to 528 pg mL−1, respectively. With the LPS concentration of 100 ng mL−1, the AlphaLISA signal-to-noise ratio was 6.97, 10.85 and 46.41 with 1000, 5000 and 20 000 THP-1 cells, respectively. We also used two commercial plate readers to compare, side by side, the ELISA (done by SpectraMax M2e, Molecular Devices) and AlphaLISA (done by PHERAstar MicroPlate Reader, BMG) signals from 20 000 THP-1 cells stimulated at various LPS concentrations (Fig. S5b, ESI†). The result also showed a highly linear correlation (R2 = 0.9889) between ELISA and AlphaLISA techniques. For 1000 THP-1 cells treated with 10 ng mL−1 LPS, the AlphaLISA signal was detected to be 2-fold greater than that for control samples generated by loading 10 ng mL−1 LPS in the MIPA without any THP-1 cells trapped on the PMM. The sensitive optical signal detection used in our study was susceptible to external noise, likely coming from the environmental background noise or the electronic noise of the PMT detector (e.g. dark current), which set up the lower limit for the detection sensitivity of our measurements.
Fig. 4c and d plotted the average amount of TNF-α secreted by single THP-1 cells as a function of LPS concentrations (Fig. 4c) or the amount of LPS molecules available to single THP-1 cells (Fig. 4d). As shown in Fig. 4c, the amount of TNF-α secreted by single THP-1 cells appeared to increase as the cell number decreased. More interestingly, as shown in Fig. 4d, the amount of TNF-α secreted by single THP-1 cells for different cell densities (n = 1000, 5000, and 20 000) collapsed and followed a single linear positive trend with the amount of LPS molecules available to single THP-1 cells, suggesting that TNF-α secretion process by single THP-1 cells might be dictated by the available LPS molecules independent of the cell population size.
Finally, we compared the levels of TNF-α secretion between normal and deactivated THP-1 cells that were both stimulated with LPS. Identifying deactivation of monocytes (also termed as immunoparalysis) can provide an effective means to predict health risks such as development of infectious complications.2–9 It is believed that real-time phenotypic identification of patients with immunoparalysis could be used to guide alternative care strategies, such as immune stimulation.5 To examine whether the MIPA device could distinguish normal THP-1 cells vs. immunoparalyzed immune cells, THP-1 cells were first treated with the complete cell growth medium supplemented with 10 ng mL−1 LPS for 24 h to deactivate them and attenuate the secretion of cytokines, including TNF-α, in response to a second stimulation with LPS.36 Deactivated THP-1 cells were then loaded into the MIPA device for TNF-α secretion measurements. Fig. 4e compared TNF-α concentrations secreted by normal and deactivated THP-1 cells trapped on the PMM for n = 20 000 cells. Consistent with prior in vitro models, TNF-α secretion by deactivated THP-1 cells was 2–4 times less than those of normal THP-1 cells, especially when LPS concentration was greater than 50 ng mL−1. More interestingly, deactivated THP-1 cells appeared to be not sensitive to changes of LPS concentration as compared to normal THP-1 cells, as concentrations of the TNF-α secreted by deactivated THP-1 cells remained roughly constant (105 ± 12 pg mL−1) as the LPS concentration increased from 10 to 100 ng mL−1. In distinct contrast, concentrations of TNF-α secreted by normal THP-1 cells increased from 150 to 528 pg mL−1 with the LPS concentration increasing from 10 to 100 ng mL−1.
Conclusion
We developed a microfluidic cellular immunophenotyping assay device capable of cell seeding, on-chip endotoxin stimulation, and in situ cell cytokine secretion detection. Our study demonstrated four important features of the MIPA device. First, the PMM integrated in the MIPA device enabled high-efficiency and uniform cell trapping/seeding with a cell population accurately adjustable by modulating the sample injection volume. Second, the MIPA device required a significantly reduced amount of sample volume (or cell population) as compared to conventional whole blood assays. The miniaturized microfluidic environment of the MIPA device permitted a spatial confinement of stimulated cells and their secreted cytokines, yielding significantly improved detection sensitivity with a much smaller cell population. More specifically, our assay using the MIPA device allowed reliable signal measurements with a signal-to-noise ratio of 2.2 for only 1000 THP-1 cells, while a conventional cell-stimulation assay typically requires 2 × 104 cells with whole human blood of 50 μL containing monocyte cells at a concentration of 4 × 105 cells mL−1.37 Thus, immunophenotyping of human monocytes using the MIPA device would only require whole blood of ~2.5 μL with a 20-fold cell number reduction. The detection sensitivity of the MIPA device could be further heightened by using smaller cell culture and immunoassay chambers.
Third, the MIPA device coupled with the optical detection system permitted cell stimulation assays and quantifications of cytokine secretions operated in the same microfluidic platform. Cytokine secretion from LPS-stimulated THP-1 cells could be quantified without any cell flushing, analyte dilution, cellular condition alterations, or potential human contaminations. Most importantly, stimulated THP-1 cells could remain alive after immunophenotyping with the MIPA device, permitting downstream cellular analysis that would require live cells. Such analysis includes, for example, examination of the proliferative potential of immune cells.
Fourth, by reducing the reagent diffusion distance and eliminating the need for multiple reagent loading, blocking, and washing steps, our immunophenotyping method could limit the LPS stimulation time to 2 h and the total assay time to 3.5 h. In contrast, conventional ELISA-based cell-stimulation assays would require a much longer assay time of >8–24 h. Previous studies22,24 based on microfluidics-based cellular immunophenotyping devices usually used heterogeneous immunoassay techniques (e.g., ELISA/ELISpot) and thus required a substantially longer assay time due to multiple surface immobilization processes and washing steps. For patients exhibiting acute immune responses, a rapid and accurate evaluation of their immune status is highly critical. Our cellular immunophenotyping assay with the MIPA device holds significant promise to open ways for rapid immune status determination in real clinical settings. Our future work will integrate multi-parallel cell culture chambers, each connected to sub-immunoassay chambers that allow for simultaneous detection of multiple cytokines. With such a device enabling multi-parallel loading of cells and reagents on the common microfluidic platform, the total multiplexed assay time would be the same with the singleplex analysis demonstrated by this study.
Supplementary Material
Acknowledgments
We acknowledge financial support from the National Science Foundation (CMMI 1129611 to J. Fu, ECCS-0601237 to K. Kurabayashi), the MICHR Pilot Program (CTSA UL1RR024986 to J. Fu, K. Kurabayashi, T. T. Cornell, and T. P. Shanley), the Coulter Foundation (to K. Kurabayashi), the National Institute of Health (R01HL097361 to T. P. Shanley), the University of Michigan Rackham Predoctoral Fellowship (to Nien-Tsu Huang) and the Department of Mechanical Engineering (J. Fu) at the University of Michigan, Ann Arbor. The Lurie Nanofabrication Facility at the University of Michigan, a member of the National Nanotechnology Infrastructure Network (NNIN) funded by the National Science Foundation, is acknowledged for support in microfabrication.
Footnotes
Electronic Supplementary Information (ESI) available. See DOI: 10.1039/c2lc40619e
Contributor Information
Jianping Fu, Email: jpfu@umich.edu.
Katsuo Kurabayashi, Email: katsuo@umich.edu.
References
- 1.Boomer JS, To K, Chang KC, Takasu O, Osborne DF, Walton AH, Bricker TL, Jarman SD. JAMA, J Am Med Assoc. 2011;306:2594–2605. doi: 10.1001/jama.2011.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cornell TT, Sun L, Hall MW, Gurney JG, Ashbrook MJ, Ohye RG, Shanley TP. J Thorac Cardiovasc Surg. 2012;143:1160–1166. e1161. doi: 10.1016/j.jtcvs.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Azizia M, Lloyd J, Allen M, Klein N, Peebles D. Pediatrics. 2012;129:e967–e974. doi: 10.1542/peds.2011-1579. [DOI] [PubMed] [Google Scholar]
- 4.Lee N, Wong CK, Chan PKS, 111C M, Chan W, Wong RYK, Lun SWM, Ngai KLK, Lui GCY, Wong BCK, Lee SKW, Choi KW, Hui DSC. PLoS One. 2011;6:e26050. doi: 10.1371/journal.pone.0026050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hall MW, Knatz NL, Vetterly C, Tomarello S, Wewers MD, Volk HD, Carcillo JA. Intensive Care Med. 2011;37:525–532. doi: 10.1007/s00134-010-2088-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frazier WJ, Hall MW. Pediatr Clin North Am. 2008;55:647–668xi. doi: 10.1016/j.pcl.2008.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Myrianthefs PM, Lazaris N, Venetsanou K, Smigadis N, Karabatsos E, Anastasiou-Nana MI, Baltopoulos GJ. Cytokine. 2007;37:150–154. doi: 10.1016/j.cyto.2007.03.007. [DOI] [PubMed] [Google Scholar]
- 8.Heagy W, Nieman K, Hansen C, Cohen M, Danielson D, West MA. Surg Infect. 2003;4:171–180. doi: 10.1089/109629603766956960. [DOI] [PubMed] [Google Scholar]
- 9.Kayakabe K, Kuroiwa T, Sakurai N, Ikeuchi H, Kadiombo AT, Sakairi T, Kaneko Y, Maeshima A, Hiromura K, Nojima Y. Rheumatology. 2012 doi: 10.1093/rheumatology/kes094. [DOI] [PubMed] [Google Scholar]
- 10.Araya AV, Pavez V, Perez C, Gonzalez F, Columbo A, Aguirre A, Schiattino I, Aguillon JC. Eur Cytokine Netw. 2003;14:128–133. [PubMed] [Google Scholar]
- 11.Hotchkiss RS, Opal S. N Engl J Med. 2010;363:87–89. doi: 10.1056/NEJMcibr1004371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bernard GR, Vincent JL, Laterre PF, LaRosa SP, Dhainaut JF, Lopez-Rodriguez A, Steingrub JS, Garber GE, Helterbrand JD, Ely EW, Fisher CJ., Jr N Engl J Med. 2001;344:699–709. doi: 10.1056/NEJM200103083441001. [DOI] [PubMed] [Google Scholar]
- 13.Cox JH, Ferrari G, Janetzki S. Methods. 2006;38:274–282. doi: 10.1016/j.ymeth.2005.11.006. [DOI] [PubMed] [Google Scholar]
- 14.Guerkov RE, Targoni OS, Kreher CR, Boehm BO, Herrera MT, Tary-Lehmann M, Lehmann PV, Schwander SK. J Immunol Methods. 2003;279:111–121. doi: 10.1016/s0022-1759(03)00240-0. [DOI] [PubMed] [Google Scholar]
- 15.Seder RA, Darrah PA, Roederer M. Nat Rev Immunol. 2008;8:247–258. doi: 10.1038/nri2274. [DOI] [PubMed] [Google Scholar]
- 16.El-Ali J, Sorger PK, Jensen KF. Nature. 2006;442:403–411. doi: 10.1038/nature05063. [DOI] [PubMed] [Google Scholar]
- 17.Louis E, Franchimont D, Piron A, Gevaert Y, Schaaf-Lafontaine N, Roland S, Mahieu P, Malaise M, De Groote D, Louis R, Belaiche J. Clin Exp Immunol. 1998;113:401–406. doi: 10.1046/j.1365-2249.1998.00662.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shurety W, Merino-Trigo A, Brown D, Hume DA, Stow JL. J Interferon Cytokine Res. 2000;20:427–438. doi: 10.1089/107999000312379. [DOI] [PubMed] [Google Scholar]
- 19.Aderka D. Cytokine Growth Factor Rev. 1996;7:231–240. doi: 10.1016/s1359-6101(96)00026-3. [DOI] [PubMed] [Google Scholar]
- 20.Aggarwal BB, Natarajan K. Eur Cytokine Netw. 1996;7:93–124. [PubMed] [Google Scholar]
- 21.Thurm CW, Halsey JF. Current Protocols in Immunology. John Wiley & Sons, Inc; 2001. [Google Scholar]
- 22.Ma C, Fan R, Ahmad H, Shi Q, Comin-Anduix B, Chodon T, Koya RC, Liu CC, Kwong GA, Radu CG, Ribas A, Heath JR. Nat Med. 2011;17:738–743. doi: 10.1038/nm.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Love JC, Ronan JL, Grotenbreg GM, van der Veen AG, Ploegh HL. Nat Biotechnol. 2006;24:703–707. doi: 10.1038/nbt1210. [DOI] [PubMed] [Google Scholar]
- 24.Zhu H, Stybayeva G, Macal M, Ramanculov E, George MD, Dandekar S, Revzin A. Lab Chip. 2008;8:2197–2205. doi: 10.1039/b810244a. [DOI] [PubMed] [Google Scholar]
- 25.Dimov IK, Kijanka G, Park Y, Ducree J, Kang T, Lee LP. Lab Chip. 2011;11:2701–2710. doi: 10.1039/c1lc20105k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen W, Lam RHW, Fu J. Lab Chip. 2012;12:391–395. doi: 10.1039/c1lc20721k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Han Q, Bradshaw EM, Nilsson B, Hafler DA, Love JC. Lab Chip. 2010;10:1391–1400. doi: 10.1039/b926849a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fiala M, Zhang L, Gan X, Sherry B, Taub D, Graves MC, Hama S, Way D, Weinand M, Witte M, Lorton D, Kuo YM, Roher AE. Mol Med. 1998;4:480–489. [PMC free article] [PubMed] [Google Scholar]
- 29.Nimah M, Zhao B, Denenberg AG, Bueno O, Molkentin J, Wong HR, Shanley TP. Shock. 2005;23:80–87. doi: 10.1097/01.shk.0000145206.28812.60. [DOI] [PubMed] [Google Scholar]
- 30.Bielefeld-Sevigny M. Assay Drug Dev Technol. 2009;7:90–92. doi: 10.1089/adt.2009.9996. [DOI] [PubMed] [Google Scholar]
- 31.Poulsen F, Jensen KB. J Biomol Screening. 2007;12:240–247. doi: 10.1177/1087057106297566. [DOI] [PubMed] [Google Scholar]
- 32.Leister KP, Huang R, Goodwin BL, Chen A, Austin CP, Xia M. Curr Chem Genomics. 2011;5:21–29. doi: 10.2174/1875397301105010021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hofman VJ, Ilie MI, Bonnetaud C, Selva E, Long E, Molina T, Vignaud JM, Flejou JF, Lantuejoul S, Piaton E, Butori C, Mourad N, Poudenx M, Bahadoran P, Sibon S, Guevara N, Santini J, Venissac N, Mouroux J, Vielh P, Hofman PM. Am J Clin Pathol. 2011;135:146–156. doi: 10.1309/AJCP9X8OZBEIQVVI. [DOI] [PubMed] [Google Scholar]
- 34.Vona G, Sabile A, Louha M, Sitruk V, Romana S, Schutze K, Capron F, Franco D, Pazzagli M, Vekemans M, Lacour B, Brechot C, Paterlini-Brechot P. Am J Pathol. 2000;156:57–63. doi: 10.1016/S0002-9440(10)64706-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zheng S, Lin HK, Lu B, Williams A, Datar R, Cote RJ, Tai YC. Biomed Microdevices. 2011;13:203–213. doi: 10.1007/s10544-010-9485-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cavaillon JM, Adib-Conquy M. Crit Care. 2006;10:233. doi: 10.1186/cc5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alberts B. Molecular Biology of the Cell. Garland Science; New York: 2008. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.