Abstract
OBJECTIVE
We sought to comprehensively evaluate the association of laminin gamma-1 (LAMC1) and advance pelvic organ prolapse.
STUDY DESIGN
We conducted a candidate gene association of patients (n =239) with stages III–IV prolapse and controls (n =197) with stages 0–I prolapse. We used a linkage disequilibrium (LD)–tagged approach to identify single-nucleotide polymorphisms (SNPs) in LAMC1 and focused on non-Hispanic white women to minimize population stratification. Additive and dominant multivariable logistic regression models were used to test for association between individual SNPs and advanced prolapse.
RESULTS
Fourteen SNPs representing 99% coverage of LAMC1 were genotyped. There was no association between SNP rs10911193 and advanced prolapse (P =.34). However, there was a trend toward significance for SNPs rs1413390 (P =.11), rs20563 (P =.11), and rs20558 (P =.12).
CONCLUSION
Although we found that the previously reported LAMC1 SNP rs10911193 was not associated with nonfamilial prolapse, our results support further investigation of this candidate gene in the pathophysiology of prolapse.
Keywords: candidate gene, genetic epidemiology, LAMC1, laminin gamma-1, pelvic organ prolapse
Pelvic organ prolapse is a major public health issue for women because 40% of postmenopausal women are affected.1 Symptomatic prolapse results in a marked reduction in quality of life because of pelvic discomfort and bladder and bowel dysfunction.2 Although prolapse can be managed surgically, the recurrence rate is as high as 30%,3 and the annual health care costs of surgery alone total more than a billion dollars in the United States.4 Given the public health impact of prolapse, it is critical to understand the underlying pathophysiology of this disease to develop effective prevention strategies and specific treatment recommendations for high-risk women.
Currently, the underlying mechanisms of prolapse are poorly understood; however, a growing body of evidence supports a genetic predisposition.5–9 Epidemiologic studies suggest that a family history of prolapse is a significant risk factor for disease.6,8 A pedigree analysis of familial prolapse found that inheritance occurs in an autosomal dominant fashion with incomplete penetrance,5 and a linkage analysis of affected sibling pairs identified a predisposition gene for pelvic floor disorders on chromosome 9q21.10
Hypotheses regarding the pathophysiology of prolapse include abnormal synthesis or degradation of collagen11 and elastin;12,13 thus, genetic mutations in extracellular matrix (ECM) pathways may result in the genesis and progression of prolapse. Nikolova et al9 identified a single-nucleotide polymorphism (SNP) (rs10911193) in the LAMC1 gene encoding laminin, an ECM component in a study of familial prolapse. However, when studying nonfamilial prolapse in a case-control association study of 265 Caucasians and 146 African Americans, Chen et al14 reported that this laminin mutation and 2 additional functional SNPs in LAMC1 (rs20563 and rs20558) were not associated with advanced prolapse. One limitation of the study by Chen et al may have been the smaller sample size.
Given the significant findings of LAMC1 in familial prolapse by Nikolova et al,9 we felt that further investigation of LAMC1 as a potential candidate gene for advanced pelvic organ prolapse was warranted in a larger cohort of unrelated individuals. In this study, we sought to perform a comprehensive analysis of the entire LAMC1 gene, using a linkage disequilibrium (LD)-tagged SNP approach, in a larger case-control association study of non-Hispanic white women.
Materials and Methods
Study population
After institutional review board approval, we recruited subjects from the Division of Urogynecology at Duke University Medical Center for this case-control association study from February 2009 to December 2010.15 Patients were defined as nonpregnant women with stages III–IV prolapse based on the Pelvic Organ Prolapse Quantification (POP-Q) examination. Controls were women with stages 0–I prolapse who had no prior history of prolapse surgery. Women with a prior hysterectomy for nonprolapse indications were eligible to be controls.
We excluded women with a gynecological malignancy or inherited or acquired connective tissue diseases, including rheumatoid arthritis, systemic lupus erythematosus, scleroderma, polymyositis, dermatomyositis, or Marfan or Ehlers-Danlos syndromes from both groups. Sociodemographic data, past medical and obstetric history, and physical examination information were collected, which included age, race, ethnicity, parity, body mass index (BMI), and POP-Q points.
Although we recruited subjects of all ages with advanced prolapse, one approach commonly used in genetic epidemiology studies is to preferentially enroll younger subjects with more advanced disease to select for a high-risk group with a presumably higher genetic load.16,17 In this study, we preferentially recruited younger women with advanced prolapse for our patients, and for our control group, we preferentially recruited older women. Because the prevalence of prolapse increases with age, older controls help to ensure that these individuals are not at risk for developing prolapse. By selecting these more extreme cases, we attempted to enhance our ability to detect genetic differences between our 2 groups.
To avoid issues of confounding by race, also known as population stratification, we focused on non-Hispanic white women in this study. By focusing on non-Hispanic white women, our goal was to further ensure that our cases and controls were derived from the same source population.18 In other words, we matched on race and ethnicity to minimize the risk of differences in the genetic background of our 2 cohorts.19 It is critically important to consider racial and ethnic groups in genetic epidemiologic studies because different racial/ethnic groups can have markedly different population frequencies of genetic variants as well as different linkage disequilibrium structures.
Genotyping and SNP selection
Genomic deoxyribonucleic acid was extracted from frozen stored peripheral venous whole-blood samples. To enable a more comprehensive evaluation of the LAMC1 gene, we utilized an LD tagged SNP approach. A tagged SNP approach is an efficient methodology for comprehensive evaluation of a gene because most SNPs are in LD (ie, genetically linked and thus correlated) with other SNPs.20–22 That is, within each gene, there are groups of SNPs that are highly correlated with each other. Representative SNPs from each group (ie, tagged SNPs) can be assessed, which represents a detailed evaluation of the gene without having to genotype every SNP. Thus, a tagged SNP approach is comprehensive as well as efficient, in terms of time and cost.
We utilized the Tagger subsection of Haploview 4.2 software (Broad Institute, Cambridge, MA)23 to identify tagged SNPs in LAMC1 as well as in the 5 kb upstream and downstream of LAMC1 and used a correlation (r2) of greater than 0.8 and SNPs with a minor allele frequency (MAF) of greater than 0.05. We also included known and putative functional coding SNPs with an MAF greater than 0.05 from any given LD bin.
Genotyping was performed using the 7900 HT TaqMan system, which incorporates a standard PCR-based, dual-fluorescence, allele discrimination assay with a dual-laser scanner. Assays by Design and Assays on Demand were purchased through Applied Biosystems (Applied Biosystems, Foster City, CA). Quality control (QC) samples, composed of 12 reference controls, were included in each quadrant of the plate. All SNPs examined were successfully genotyped for 98% or greater of the individuals in the study. Error rate estimates for SNPs meeting QC benchmarks were 0.2%.
Statistical analysis
Sociodemographic data and physical examination findings for cases and controls were compared using a χ2 test for discrete variables and a Student t test for continuous variables. A 2-sided alpha of 0.05 was considered significant. For all SNPs, deviation from Hardy-Weinberg equilibrium (HWE) was tested using a χ2 test. Genotype clusters and genotyping calling for SNPs that were significantly out of HWE were re-reviewed and genotyping accuracy confirmed. To test for association of single SNPs with pelvic organ prolapse, we constructed logistic regression models and evaluated both unadjusted and adjusted models, using both additive and dominant models of genetic risk. The multivariable models adjusted for age, vaginal parity (dichotomous variable regarding any prior vaginal deliveries), and BMI.
A full-tagged SNP approach was conducted in an initial cohort of 396 non-Hispanic white women. Because 3 SNPs showed a nominal P value of .10, we expanded the final cohort to 436 women to increase the power. Given these independent effects of several SNPs, we also performed a haplotype analysis of all 2-way SNP combinations using the tool set of PLINK24 to evaluate the possible association of a genetic background of more than 1 SNP for pelvic organ prolapse risk.
A power calculation was conducted for a nonfamilial case-control association using the QUANTO program (Gauderman and Morrison, http://hydra.usc.edu/gxe. Accessed Oct. 24, 2008.).25 A range of allele frequencies of 0.05–0.5 and varying effect sizes as well as dominant and additive models were considered. We recognize that these calculations are based on model parameters that we cannot know with certainty; however, we can derive a sense of the expected power for the proposed sample sizes. Given a sample size of 150 cases and 150 controls, we have greater than 80% power to detect an effect size of approximately 2.0 and higher for most allele frequencies and genetic models. We chose an effect size of 2.0 because we believed that this is a clinically meaningful difference to detect and represented a balance between clinical/biological significance and feasibility. All statistical analyses were conducted with SAS version 9.2 (SAS Institute, Cary, NC) unless otherwise noted.
Results
We enrolled a total of 436 women, of whom 239 were cases with advanced prolapse and 197 were controls with none to minimal prolapse. Cases were slightly younger (64.8 ±10.3 vs 69.0 ±10.2 years, P <.001) and more likely to have had 1 or more vaginal deliveries (96.6% vs 82.2%, P <.001) when compared with controls (Table 1). Cases and controls were similar in the Charlson co-morbidity index, smoking status, and BMI (27.8 ±5.1 vs 28.5 ±6.3 kg/m2, P =.18). Of controls, 57.4% had stage 0 prolapse and 42.6% had stage I prolapse. For cases, a majority had stage III prolapse (94.6%) and 5.4% had stage IV prolapse.
TABLE 1.
Sociodemographic characteristics and physical examination findings for cases and controls
| Characteristic | Controls (n =197) | Cases (n =239) | P value |
|---|---|---|---|
| Age, y | 69.0 ±10.2 | 64.8 ±10.3 | <.001 |
| Vaginal parity | 162 (82.2) | 230 (96.6) | <.001 |
| Charlson comorbidity index | 0.48 ±0.90 | 0.42 ±0.83 | .51 |
| Current smoker | 8 (4.1) | 16 (6.8) | .26 |
| Body mass index, kg/m2 | 28.5 ±6.3 | 27.8 ±5.1 | .18 |
| Prolapse stage | |||
| 0 | 113 (57.4) | 0 (0.0) | |
| I | 84 (42.6) | 0 (0.0) | |
| II | 0 (0.0) | 0 (0.0) | |
| III | 0 (0.0) | 226 (94.6) | |
| IV | 0 (0.0) | 13 (5.4) | |
Data presented as mean ±SD or n (%).
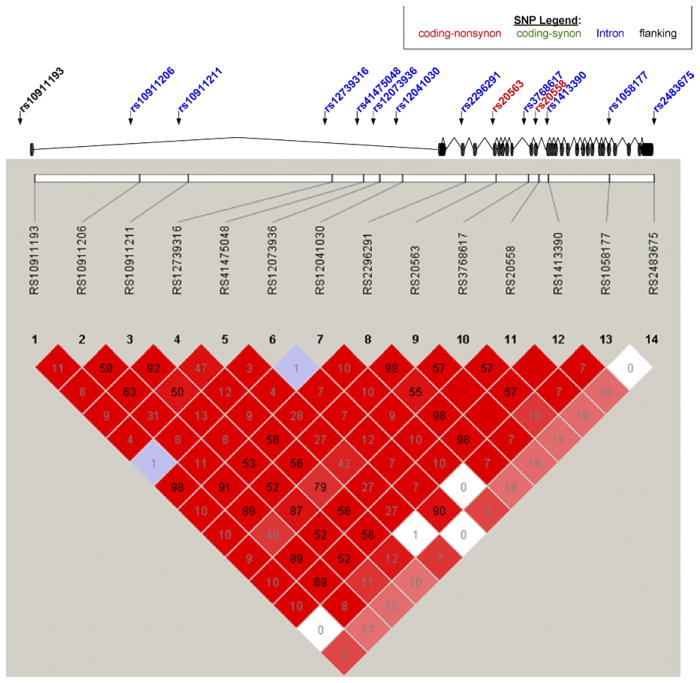
We genotyped 15 SNPs based on our LD tagging selection. The Figure is a schematic of the LAMC1 gene structure, which indicates exons, introns, and sites of translation initiation and termination. The genotyped SNPs are marked on the schematic with descriptions of the different types of SNPs, whether nonsynonymous coding, synonymous coding, or intronic. All SNPs were in Hardy-Weinberg equilibrium except rs17477864 (P <.001). This SNP was regenotyped and remained out of HWE and thus was not included in the final analysis. At an r2 of 0.80, the remaining 14 SNPs provided 99% coverage of the gene.
Figure 1.
The LAMC1 gene structure is displayed diagrammatically with exons represented as black discs. Genotyped SNPs are noted with arrows and are color coded as follows: coding-nonsynonymous (red); coding-synonymous (green); intronic (blue); and flanking (black). The bottom panel illustrates an LD graphic with r2 values and squares without any value indicates perfect LD.
LAMC1, laminin gamma-1; LD, linkage disequilibrium; SNP, single-nucleotide polymorphism.
In this cohort of non-Hispanic white women, there was no association between the previously reported LAMC1 SNP rs10911193 with advanced pelvic organ prolapse, based on both the dominant and additive adjusted models (odds ratio [OR], 1.29; 95% confidence interval [CI], 0.77–2.17; P =.34) and OR, 1.23; 95% CI, 0.77–1.97; P =.39, respectively) (Table 2). However, there was a trend toward significance for the following SNPs in the dominant model: rs1413390, an intronic SNP (adjusted OR, 1.45; 95% CI, 0.92–2.29; P =.11), rs20558; a nonsynonymous coding SNP (adjusted OR, 1.44; 95% CI, 0.91–2.26; P =.12); and rs20563, a nonsynonymous coding SNP (adjusted OR, 1.44; 95% CI, 0.92–2.28; P =.11) (Table 2). These 3 SNPs were in perfect linkage disequilibrium with each other (r2 =1.0) (Figure). No other SNPs in LAMC1 were associated with prolapse in either the dominant or additive adjusted models.
TABLE 2.
Logistic regression results for both the additive and dominant adjusted models for LAMC1 SNPs adjusted for age, BMI, and vaginal parity
| SNP | Minor allele (frequency) | Dominant adjusted model
|
Additive adjusted model
|
||
|---|---|---|---|---|---|
| OR (95% CI) | P value | OR (95% CI) | P value | ||
| rs10911193 | T (0.11) | 1.29 (0.77–2.17) | .34 | 1.23 (0.77–1.97) | .39 |
|
| |||||
| rs1413390 | G (0.45) | 1.45 (0.92–2.29) | .11 | 1.22 (0.90–1.65) | .21 |
|
| |||||
| rs20558 | T (0.45) | 1.44 (0.91–2.26) | .12 | 1.19 (0.88–1.61) | .25 |
|
| |||||
| rs20563 | A (0.46) | 1.44 (0.92–2.28) | .11 | 1.20 (0.89–1.63) | .23 |
|
| |||||
| rs10911206 | A (0.48) | 1.37 (0.85–2.20) | .19 | 1.16 (0.85–1.57) | .35 |
|
| |||||
| rs2296291 | C (0.45) | 1.36 (0.85–2.16) | .20 | 1.14 (0.84–1.56) | .39 |
|
| |||||
| rs12041030 | G (0.11) | 1.36 (0.80–2.29) | .25 | 1.28 (0.80–2.07) | .30 |
|
| |||||
| rs12739316 | T (0.41) | 0.79 (0.51–1.23) | .30 | 0.82 (0.60–1.11) | .20 |
|
| |||||
| rs3768617 | T (0.42) | 0.83 (0.53–1.31) | .42 | 0.84 (0.61–1.16) | .29 |
|
| |||||
| rs2483675 | A (0.22) | 1.15 (0.76–1.74) | .51 | 1.17 (0.82–1.68) | .38 |
|
| |||||
| rs10911211 | T (0.40) | 0.89 (0.57–1.39) | .62 | 0.90 (0.66–1.24) | .52 |
|
| |||||
| rs41475048 | T (0.25) | 1.04 (0.69–1.56) | .86 | 1.08 (0.77–1.50) | .66 |
|
| |||||
| rs1058177 | C (0.09) | 1.01 (0.58–1.76) | .97 | 1.01 (0.58–1.76) | .97 |
|
| |||||
| rs12073936 | G (0.08) | 1.00 (0.56–1.78) | .99 | 0.96 (0.55–1.68) | .88 |
BMI, body mass index; CI, confidence interval; LAMC1, laminin gamma-1; OR, odds ratio; SNP, single-nucleotide polymorphism.
In haplotype analyses of all 2-SNP combinations, only 2 haplotypes were statistically significantly associated with prolapse (rs12739316_rs3768617, omnibus, P =.005; and rs10911211_rs12739316, P =.04). Interestingly, 3 haplotypes including the previously reported rs10911193 and each of the borderline significant SNPs from our study also showed a trend for significance in haplotype analyses (rs10911193_rs20563, omnibus, P =.07; rs10911193_rs20558, omnibus, P =.07; and rs10911193_rs1413390, omnibus, P =.06). Although none of the haplotype analyses would meet statistical significance after adjustment for multiple comparisons, these results continue to corroborate the potential role of these SNPs in prolapse.
Comment
In this comprehensive analysis of LAMC1 genetic variants, we provide additional evidence that LAMC1 is a potential candidate gene for nonfamilial, advanced pelvic organ prolapse. Although we did not find an association between the previously reported SNP rs10911193 and advanced prolapse, we report on 3 LAMC1 SNPs that show a trend toward significance, although they were not strictly statistically significant.
LAMC1 encodes the γ1 chain of laminin. Laminins are a heterogeneous group of ECM glycoproteins that form the major noncollagenous portion of the basement membranes.26 There are at least 15 distinct forms of laminin, and these isoforms are comprised of 3 nonidentical chains: α, β, and γ chains. Nikovola et al9 first reported that the LAMC1 SNP rs10911193 minor T allele was more prevalent among probands in a highrisk family with multigenerational early-onset prolapse and hernias when compared with the general population. Subsequently, Chen et al14 evaluated 3 LAMC1 SNPs (rs10911192, rs20563, and rs20558) in 265 Caucasians (102 cases and 163 controls) and 146 African Americans(63 cases and 83 controls) and found no association with nonfamilial prolapse. However, their power to detect a significant association may have been limited by their sample size.
In our study, rs20563 and rs20558, both nonsynonymous, or missense, SNPs, showed a trend toward significance. The rs20563 G produces an amino acid substitution of isoleucine to valine, which is a fairly conservative change because both are amino acids with hydrophobic side chains. The rs20558 C results in a less conservative amino acid change from leucine to proline. The amino acid changes associated with rs20563 and rs20558 are predicted to have tolerated and benign effects on the protein function based on the Sorting Tolerant From Intolerant27 and the Polymorphism Phenotyping v228 predictions in the Ensembl genome browser.29,30
A major strength of this study is that we used a tagged SNP approach to comprehensively evaluate the entirety of the LAMC1 gene. We evaluated a total of 14 SNPs, which provided 99% coverage of the gene. In contrast, Chen et al14 evaluated only 3 SNPs in LAMC1. Another critical factor is that we phenotyped cases and controls based on a standardized objective assessment, the POP-Q examination, and we selected more extreme phenotypes to try to concentrate the genetic effect. Another important aspect of this study is that we conducted a case-control association study, which evaluates unrelated individuals. It is critical to distinguish between studying a high-risk family vs unrelated individuals because the disease phenotype may be quite different.
A limitation of our study is that we were powered to detect an effect size of 2.0, which represented a balance between clinical/biological significance and feasibility; however, we did not have sufficient power to detect more modest effects (ie, an OR of <2.0). Thus, although we showed a trend toward significance for 3 SNPs, we were likely underpowered to show a significant association. Of note, effect sizes less than 2.0 have been shown to be clinically significant in other phenotypes; for example, the most consistent and recognized genetic locus for susceptibility to coronary artery disease, chromosome 9p21, shows effect sizes of 1.3–1.6.31 In this study, we focused on non-Hispanic white women to avoid population stratification;19 however, future genetic epidemiological studies should evaluate other races/ethnicities.
Another important issue in a case-control study is careful selection of the control subjects. In this study, we defined our cases and controls based on stage of prolapse. However, at this point, we do not know the ideal method for phenotyping pelvic organ prolapse. Should we define cases and controls based on physical examination and stage of prolapse, as we did in this study? Should we define prolapse based on the degree of bothersome symptoms, age at onset, or history of treatment, whether with surgery or a pessary? Should family history of prolapse be included? Also, it is possible that different pelvic floor disorders may share the same genetic susceptibility, for example, stress urinary incontinence (SUI) and prolapse. If SUI and prolapse share the same genetic susceptibility, there is the potential to introduce misclassification bias if controls have SUI, even though they did not have prolapse. This point highlights the importance for future research into how best to phenotype prolapse, as well as the other pelvic floor disorders, and the importance of genetic and genetic epidemiological research of pelvic floor disorders.
Our current knowledge of the genetic susceptibility of pelvic organ prolapse is limited, although research in this field is growing. Pelvic organ prolapse is considered a common complex disease, for which there are likely multiple genetic susceptibility markers. Historically, the field of human genetics was focused on Mendelian disorders, in which single-gene mutations with a high penetrance and large effect size result in rare diseases in families. There has been a paradigm shift to study common, complex diseases, in which there is a polygenic model of genetic risk, with multiple, often common, genetic polymorphisms conferring moderate risk in a general population, with concomitant gene-gene and gene-environment interactions. For prolapse, additional candidate genes that have been investigated include collagen type III alpha 1 chain (COL3A1),32,33 collagen type I alpha 1 chain (COL1A1),34,35 and matrix metalloproteinase-9 (MMP9).13,36
Animal models have also highlighted the association of prolapse and defective ECM proteins, such as lysyl oxidase-like-1,12,37 fibulin-3,38 and fibulin-5.39 Although these findings highlight potential candidate genes for pelvic organ prolapse, we have yet to fully understand the pathophysiology of pelvicorgan prolapse.
It is critically important to pursue future investigations into the genetics of pelvic organ prolapse because this research may uncover the underlying mechanisms for this highly prevalent and burdensome disease. These data will not only provide invaluable information regarding the pathophysiology of disease but will also help to identify genetically high-risk women to target for prevention strategies or specific treatment recommendations. LAMC1 is an interesting candidate gene, and future large-scale investigations into this gene as well as other extracellular matrix genes are definitely warranted.
Acknowledgments
This study was supported by grant R03HD061766 from the, Eunice Kennedy Shriver National Institute of Child Health and Human Development. J.M.W. is supported by grant K23HD068404 from the Eunice Kennedy Shriver National Institute of Child Health and Human Development.
Footnotes
The authors report no conflict of interest.
Presented at the 32nd Annual Scientific Meeting of the American Urogynecologic Society, Providence, RI, Sept. 16, 2011.
Reprints not available from the authors.
References
- 1.Hendrix SL, Clark A, Nygaard I, Aragaki A, Barnabei V, McTiernan A. Pelvic organ prolapse in the Women’s Health Initiative: gravity and gravidity. Am J Obstet Gynecol. 2002;186:1160–6. doi: 10.1067/mob.2002.123819. [DOI] [PubMed] [Google Scholar]
- 2.Jelovsek JE, Maher C, Barber MD. Pelvic organ prolapse. Lancet. 2007;369:1027–38. doi: 10.1016/S0140-6736(07)60462-0. [DOI] [PubMed] [Google Scholar]
- 3.Olsen AL, Smith VJ, Bergstrom JO, Colling JC, Clark AL. Epidemiology of surgically managed pelvic organ prolapse and urinary incontinence. Obstet Gynecol. 1997;89:501–6. doi: 10.1016/S0029-7844(97)00058-6. [DOI] [PubMed] [Google Scholar]
- 4.Subak LL, Waetjen LE, van den Eeden S, Thom DH, Vittinghoff E, Brown JS. Cost of pelvic organ prolapse surgery in the United States. Obstet Gynecol. 2001;98:646–51. doi: 10.1016/s0029-7844(01)01472-7. [DOI] [PubMed] [Google Scholar]
- 5.Jack GS, Nikolova G, Vilain E, Raz S, Rodriguez LV. Familial transmission of genitovaginal prolapse. Int Urogynecol J Pelvic Floor Dysfunct. 2006;17:498–501. doi: 10.1007/s00192-005-0054-x. [DOI] [PubMed] [Google Scholar]
- 6.Chiaffarino F, Chatenoud L, Dindelli M, et al. Reproductive factors, family history, occupation and risk of urogenital prolapse. Eur J Obstet Gynecol Reprod Biol. 1999;82:63–7. doi: 10.1016/s0301-2115(98)00175-4. [DOI] [PubMed] [Google Scholar]
- 7.Buchsbaum GM, Duecy EE, Kerr LA, Huang LS, Perevich M, Guzick DS. Pelvic organ prolapse in nulliparous women and their parous sisters. Obstet Gynecol. 2006;108:1388–93. doi: 10.1097/01.AOG.0000245784.31082.ed. [DOI] [PubMed] [Google Scholar]
- 8.McLennan MT, Harris JK, Kariuki B, Meyer S. Family history as a risk factor for pelvic organ prolapse. Int Urogynecol J Pelvic Floor Dysfunct. 2008;19:1063–9. doi: 10.1007/s00192-008-0591-1. [DOI] [PubMed] [Google Scholar]
- 9.Nikolova G, Lee H, Berkovitz S, et al. Sequence variant in the laminin gamma1 (LAMC1) gene associated with familial pelvic organ prolapse. Hum Genet. 2007;120:847–56. doi: 10.1007/s00439-006-0267-1. [DOI] [PubMed] [Google Scholar]
- 10.Norton P, Allen-Brady K, Cannon-Albright L. Significant linkage evidence of a predisposition gene for pelvic floor disorders on chromosome 9. J Pelvic Med Surg. 2008;14:219. doi: 10.1016/j.ajhg.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moalli PA, Shand SH, Zyczynski HM, Gordy SC, Meyn LA. Remodeling of vaginal connective tissue in patients with prolapse. Obstet Gynecol. 2005;106:953–63. doi: 10.1097/01.AOG.0000182584.15087.dd. [DOI] [PubMed] [Google Scholar]
- 12.Liu X, Zhao Y, Pawlyk B, Damaser M, Li T. Failure of elastic fiber homeostasis leads to pelvic floor disorders. Am J Pathol. 2006;168:519–28. doi: 10.2353/ajpath.2006.050399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Budatha M, Roshanravan S, Zheng Q, et al. Extracellular matrix proteases contribute to progression of pelvic organ prolapse in mice and humans. J Clin Invest. 2011;121:2048–59. doi: 10.1172/JCI45636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen C, Hill LD, Schubert CM, Strauss JF, 3rd, Matthews CA. Is laminin gamma-1 a candidate gene for advanced pelvic organ prolapse? Am J Obstet Gynecol. 2010;202:505. e1–5. doi: 10.1016/j.ajog.2010.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hattersley AT, McCarthy MI. What makes a good genetic association study? Lancet. 2005;366:1315–23. doi: 10.1016/S0140-6736(05)67531-9. [DOI] [PubMed] [Google Scholar]
- 16.Hauser ER, Crossman DC, Granger CB, et al. A genomewide scan for early-onset coronary artery disease in 438 families: the GENECARD Study. Am J Hum Genet. 2004;75:436–47. doi: 10.1086/423900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shah SH, Kraus WE, Crossman DC, et al. Serum lipids in the GENECARD study of coronary artery disease identify quantitative trait loci and phenotypic subsets on chromosomes 3q and 5q. Ann Hum Genet. 2006;70:738–48. doi: 10.1111/j.1469-1809.2006.00288.x. [DOI] [PubMed] [Google Scholar]
- 18.Cardon LR, Palmer LJ. Population stratification and spurious allelic association. Lancet. 2003;361:598–604. doi: 10.1016/S0140-6736(03)12520-2. [DOI] [PubMed] [Google Scholar]
- 19.Lee WC, Wang LY. Reducing population stratification bias: stratum matching is better than exposure. J Clin Epidemiol. 2009;62:62–6. doi: 10.1016/j.jclinepi.2008.02.016. [DOI] [PubMed] [Google Scholar]
- 20.Carlson CS, Eberle MA, Rieder MJ, Yi Q, Kruglyak L, Nickerson DA. Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. Am J Hum Genet. 2004;74:106–20. doi: 10.1086/381000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu H, Gregory SG, Hauser ER, et al. SNPselector: a web tool for selecting SNPs for genetic association studies. Bioinformatics. 2005;21:4181–6. doi: 10.1093/bioinformatics/bti682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meng Z, Zaykin DV, Xu CF, Wagner M, Ehm MG. Selection of genetic markers for association analyses, using linkage disequilibrium and haplotypes. Am J Hum Genet. 2003;73:115–30. doi: 10.1086/376561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Bakker PI, Yelensky R, Pe’er I, Gabriel SB, Daly MJ, Altshuler D. Efficiency and power in genetic association studies. Nat Genet. 2005;37:1217–23. doi: 10.1038/ng1669. [DOI] [PubMed] [Google Scholar]
- 24.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cohen J, Pertsemlidis A, Kotowski IK, Graham R, Garcia CK, Hobbs HH. Low LDL cholesterol in individuals of African descent resulting from frequent nonsense mutations in PCSK9. Nat Genet. 2005;37:161–5. doi: 10.1038/ng1509. [DOI] [PubMed] [Google Scholar]
- 26.National Center for Biotechnology Information. [Accessed July 5, 2011.];LAMC1 laminin, gamma 1. Available at: www.ncbi.nlm.nih.gov/gene/3915.
- 27.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4:1073–81. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 28.Adzhubei IA, Schmidt S, Peshkin L, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–9. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. [Accessed Jan. 9, 2012.];Ensembl. Variation rs20558, Genomic context, gene/transcript. Available at: http://useast.ensembl.org/Homo_sapiens/Variation/Mappings?db=core;g=ENSG00000135862;r=1:182992597-183114727;t=ENST00000258341;v=rs20558;vdb=variation;vf=22315591.
- 30. [Accessed Jan. 12, 2012.];Ensembl. Variation rs20563, Genomic context, gene/transcript. Available at: http://useast.ensembl.org/Homo_sapiens/Variation/Mappings?db=core;r=1:183085255-183086255;v=rs20563;vdb=variation;vf=22315596.
- 31.Helgadottir A, Thorleifsson G, Manolescu A, et al. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science. 2007;316:1491–3. doi: 10.1126/science.1142842. [DOI] [PubMed] [Google Scholar]
- 32.Kluivers KB, Dijkstra JR, Hendriks JC, Lince SL, Vierhout ME, van Kempen LC. COL3A1 2209G>A is a predictor of pelvic organ prolapse. Int Urogynecol J Pelvic Floor Dysfunct. 2009;20:1113–8. doi: 10.1007/s00192-009-0913-y. [DOI] [PubMed] [Google Scholar]
- 33.Chen HY, Chung YW, Lin WY, Wang JC, Tsai FJ, Tsai CH. Collagen type 3 alpha 1 polymorphism and risk of pelvic organ prolapse. Int J Gynaecol Obstet. 2008;103:55–8. doi: 10.1016/j.ijgo.2008.05.031. [DOI] [PubMed] [Google Scholar]
- 34.Feiner B, Fares F, Azam N, Auslender R, David M, Abramov Y. Does COLIA1 SP1-binding site polymorphism predispose women to pelvic organ prolapse? Int Urogynecol J Pelvic Floor Dysfunct. 2009;20:1061–5. doi: 10.1007/s00192-009-0895-9. [DOI] [PubMed] [Google Scholar]
- 35.Rodrigues AM, Girao MJ, da Silva ID, Sartori MG, Martins KD, Castro RD. COL1A1 Sp1-binding site polymorphism as a risk factor for genital prolapse. Int Urogynecol J Pelvic Floor Dysfunct. 2008;19:1471–5. doi: 10.1007/s00192-008-0662-3. [DOI] [PubMed] [Google Scholar]
- 36.Chen HY, Lin WY, Chen YH, Chen WC, Tsai FJ, Tsai CH. Matrix metalloproteinase-9 polymorphism and risk of pelvic organ prolapse in Taiwanese women. Eur J Obstet Gynecol Reprod Biol. 2010;149:222–4. doi: 10.1016/j.ejogrb.2009.12.014. [DOI] [PubMed] [Google Scholar]
- 37.Liu X, Zhao Y, Gao J, et al. Elastic fiber homeostasis requires lysyl oxidase-like 1 protein. Nat Genet. 2004;36:178–82. doi: 10.1038/ng1297. [DOI] [PubMed] [Google Scholar]
- 38.Rahn DD, Acevedo JF, Roshanravan S, et al. Failure of pelvic organ support in mice deficient in fibulin-3. Am J Pathol. 2009;174:206–15. doi: 10.2353/ajpath.2009.080212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Drewes PG, Yanagisawa H, Starcher B, et al. Pelvic organ prolapse in fibulin-5 knockout mice: pregnancy-induced changes in elastic fiber homeostasis in mouse vagina. Am J Pathol. 2007;170:578–89. doi: 10.2353/ajpath.2007.060662. [DOI] [PMC free article] [PubMed] [Google Scholar]