Abstract
Non-Watson-Crick base pairing provides an in situ approach for actuation of DNA nanostructures through responses to solution conditions. Here we demonstrate this concept by using physiologically-relevant changes in pH to regulate DNA pyramid assembly/disassembly and to control the release of protein cargo.
The ability of DNA to form predictable nanostructures through sequence-directed hybridization has allowed the design of complex supramolecular materials.1, 2 Such scaffolds can organize functional proteins and bioactive molecules in well-designed patterns and are therefore of great interest to biosensing and drug delivery.3, 4 To date, the mechanical actuation of DNA nanostructures has been largely achieved through the addition of thermodynamically more favorable DNA partners and the resulting rearrangement of interactions between DNA strands.5 However, these approaches require the addition of new DNA strands and generate waste DNA with each cycle. Non-canonical Watson-Crick base pairing provides an in situ approach for actuation through sensitivity to solution conditions. Here we demonstrate this concept with DNA pyramids containing i-motifs along one face. We show that i-motifs can regulate pyramid assembly/disassembly and control the release of protein cargo. Importantly, disassembly is triggered with physiologically-relevant acidification, a key criteria for drug and gene delivery vehicles.
An i-motif refers to a quadruplex structure that forms due to base pairing between protonated and unprotonated cytosines.6 A number of pH-responsive DNA devices based on i-motifs have been reported,7–11 although these i-motifs were typically applied to contexts outside of drug delivery. We are interested in using physiological changes in pH to induce conformational changes and release cargo, inspired by the endosomal acidification that triggers viruses, pH differences across compartments within the body (e.g., blood-brain-barrier), and the acidity of tumor micro-environments.12, 13 To our knowledge, there have not yet been attempts to use i-motifs within DNA nanostructures to achieve such triggered changes. Conventional pH-responsive materials produce physicochemical changes including swelling, dissociation, and surface charge switching in order to favor drug release at target sites.13, 14 However, synthetic polymers often present concerns regarding biocompatibility, biodegradation, and immunogenicity.
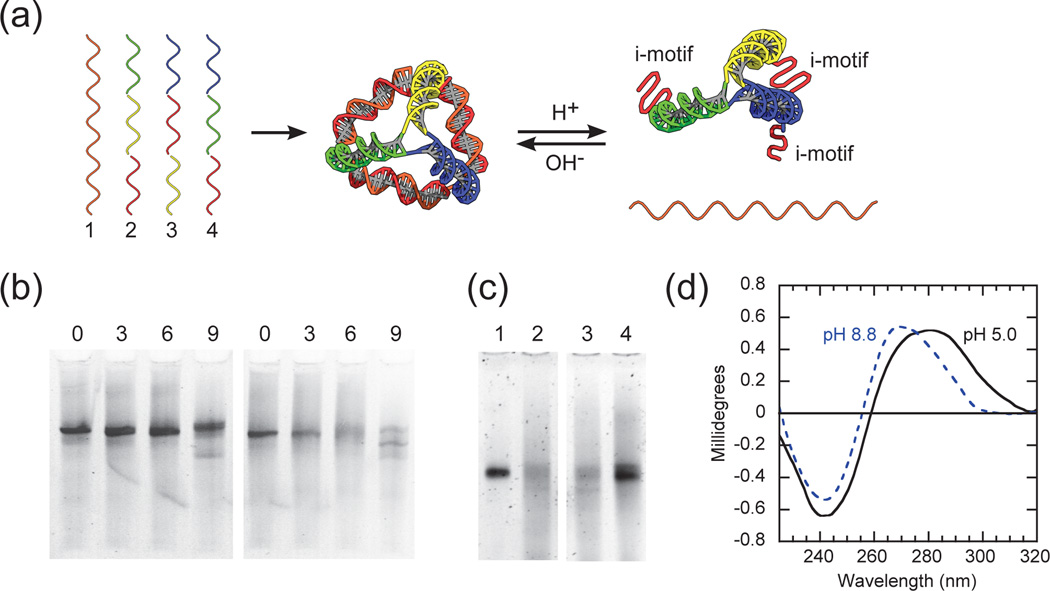
As illustrated in Fig. 1a, the DNA pyramids are composed of four oligonucleotides. Similar to our previous work and others, each strand runs around one face of the pyramid and hybridizes to form double-helical edges.15–19 Cytosine-rich stretches are distributed into strands 2–4 so that three edges of the pyramid exhibit identical i-motif sequences along one face. Strand 1 is composed of a repetitive sequence, complementary to the cytosine-rich stretches, such that it is hybridized in the pyramid under alkaline conditions (pH 8.8, unless otherwise stated). In our design, the cytosine-rich stretches in strands 2–4 become partially protonated under acidic conditions (pH 5.0, unless otherwise stated), leading to the formation of intramolecular i-motifs (Fig. 1a). These i-motifs are intended to promote an “open” state (i.e., partial pyramid disassembly). Once the overall distribution of i-motifs within the DNA pyramid was decided, the DNA sequences in the remaining regions were generated using the Tiamat software.20
Fig. 1.
(a) Schematic of pH-triggered conformational changes of an i-motif DNA pyramid. The i-motif sequence is colored red and the complementary sequence is colored orange. Otherwise, identical colored regions are complementary to each other. (b) Mismatch-controlled conformational changes of i-motif DNA pyramids at different pH. The left and right gel are at alkaline (pH 8.8, unless otherwise stated) and acidic conditions (pH 5.0, unless otherwise stated), respectively. In both gels the lane numbers indicate the number of mismatches in strand 1. (c) pH-controlled assembly of i-motif DNA pyramids. Switching from alkaline to acidic conditions (lanes 1 and 2), and vice versa (lanes 3 and 4); (d) Circular dichroism (CD) of i-motif DNA pyramids at different pH.
DNA pyramids, or tetrahedrons, are now well-known and characterized systems.15–19 Successful assembly of our DNA pyramids was first verified using native PAGE (Fig. S1†). To achieve efficient and reversible transitions upon pH changes, the pyramid stability was modulated by the introduction of mismatches into strand 1. These mismatches reduce the effective melting temperature under alkaline conditions, and thereby facilitate the transition to intramolecular i-motifs. We varied the number of mismatches in strand 1 and examined the effect on pyramid assembly at different pH values. For 0 – 6 mismatches, pyramid assembly always remains efficient under alkaline conditions (Fig. 1b, left). In the case of 9 mismatches, a new structure is formed (note the distinct band and intensity). Under acidic conditions, for 0 and 3 mismatches, pyramids did not undergo disassembly whereas 9 mismatches induced nearly complete disassembly (Fig. 1b, right). The case of 6 mismatches results in a diffuse band, indicative of an “open” structure, and it is likely that strand 1 has not fully dissociated from all pyramids. Nevertheless, 6 mismatches appears to give an acceptable assembly/disassembly response to pH, and the sequence of strand 1 was fixed as such for all further experiments. As controls, we verified that the assembly of a DNA pyramid and hybridization of linear dsDNA, both lacking i-motifs, were unaffected by pH (Fig. S2†).
To further demonstrate pH-controlled assembly/ disassembly, structures were switched from alkaline to acidic conditions, or vice versa, and were consistent with the presumed role of i-motifs (Fig. 1c). The structures associated with “open” and “closed” states (i.e., partial and complete assembly) were further confirmed by circular dichroism (CD) (Fig. 1d). Under alkaline conditions, the DNA pyramid has a CD spectrum which shows the characteristics of B-form double-helical DNA with positive band near 275 nm and a negative band near 240 nm.7 Under acidic conditions, the appearance of i-motifs are reflected in a red-shift of the CD spectrum, with a strong positive band near 285 nm and a smaller negative band near 260 nm.21 These results are consistent with our design, where acidic conditions will lead to three i-motifs and three double-helical edges (Fig. 1a). Assembly and conformational changes of i-motif pyramids were further confirmed using fluorescence quenching (Fig. S3†) and FRET (Fig. 2). We conclude that changes in the assembly/disassembly of our DNA pyramid due to pH arise solely from formation of i-motifs.
Fig. 2.
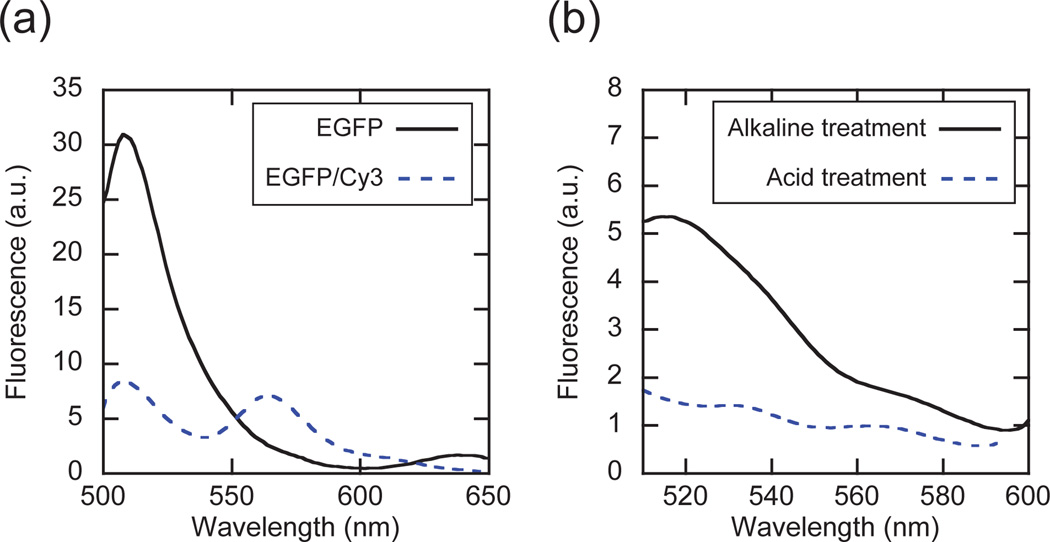
EGFP incorporation into, and release from, i-motif pyramids. (a) FRET analysis of singly-labeled (EGFP) and dual-labeled (EGFP/Cy3) pyramids under alkaline conditions. (b) Triggered release of EGFP via pH changes. EGFP i-motif pyramids were incubated under alkaline or acidic conditions and the released EGFP was removed by Ni2+-NTA agarose beads, followed by fluorescence analysis of the flow-through solution.
Next, we were motivated to demonstrate the potential of our pH-responsive DNA pyramids for triggered cargo delivery. While we previously showed (non-triggered) delivery of therapeutic nucleic acid,18 protein delivery has not yet been achieved with these types of vehicles. As a model, we attached enhanced green fluorescent protein (EGFP) to DNA via nickel-mediated interactions between nitrilotriacetic acid (NTA) and the protein’s hexa-histidine tag.22 This strategy possesses several attractive features: (i) any recombinant protein with hexa-histidine tag can be attached; (ii) the interaction is site-specific and (iii) reversible. In order to demonstrate the specificity of the NTA-modified strand 1 with EGFP, several control experiments were performed (Fig. S4†).
Based on their geometry and dimensions, DNA pyramids should be able to accommodate globular proteins with diameters of about 2.8–4.8 nm. By comparison, GFP is reported to have a hydrodynamic diameter of about 3.2 nm.23 According to Erben et al.,24 the position and orientation of an attached protein relative to the DNA scaffold can be controlled by the location of the attachment point. This control relies on the fact that with increasing linear distance along the pyramid edge, there is a corresponding clockwise rotation of 34.3° per base, due to the helical nature of dsDNA. When the attachment point is 8 bases away from the pyramid vertex, an attached protein should be oriented towards the interior of the structure.
With the above considerations in mind, EGFP-labeled pyramids were assembled using NTA-modified strand 1. Separately, strand 3 was Cy3-labeled so as to assess intra-pyramid FRET between EGFP and Cy3. The fluorescence of pyramids with EGFP alone was compared with that of pyramids with both EGFP and Cy3 (Fig. 2a). To exclusively assess FRET-induced Cy3 excitation, the fluorescence of Cy3-pyramids was subtracted from the fluorescence of dual-labeled pyramids. This corrected fluorescence of dual-labeled pyramids was then compared to that of EGFP-labeled pyramids. As shown in Fig. 2a, the fluorescence emission of EGFP was decreased, while the emission from Cy3 at 565 nm was increased. Given a FRET efficiency E = 72% and a Förster distance for the EGFP/Cy3 pair R0 = 6.3 nm,25 the distance between the two dyes in the pyramid is calculated to be R = 5.4 nm. If we take into consideration the additional length due to linker units, the calculated value is reasonably close to the value of 6.8 nm that is expected from the design. Moreover, when pyramids bearing Cy3 but lacking NTA were incubated with free EGFP, no FRET signal was observed (Fig. S5†), further confirming that the intended conjugation was successful.
EGFP-incorporated i-motif pyramids were then exposed to acidic conditions to demonstrate pH-responsive behavior (Fig. 2b). Since the NTA-Ni2+-His linkage connecting EGFP to the DNA pyramid is also pH sensitive, acidification will facilitate both pyramid disassembly and EGFP release. Even though the fluorescence of EGFP is quenched at low pH (Fig. S6†),26 its release can still be determined, albeit in a slightly complex manner. Following a 10 minute incubation under acidic conditions, the EGFP-labeled i-motif pyramids were quickly re-adjusted to alkaline conditions, with the released EGFP captured using Ni2+-NTA agarose beads. Fluorescence measurements show that under acidic conditions, Ni2+-NTA beads effectively captured the released EGFP since the flow-through did not exhibit the characteristic 510 nm emission peak of EGFP (Fig. 2b). Under alkaline conditions, Ni2+- NTA beads were unable to capture EGFP since it is presumably still pyramid-bound, consistent with the observed fluorescence. We emphasize that according to Erben et al.,24 the EGFP should be oriented toward the interior of the pyramid and therefore release will be most efficient upon i-motif switching. Although a crude estimate of 33% release can be calculated from the 510 nm signal, we plan to study questions regarding both loading/release kinetics, and efficiency. Here our main intent is to demonstrate the proof-of-concept.
The linkage of EGFP to DNA pyramids can also be regulated by the addition of a competitive ligand (e.g., imidazole) or by the removal of nickel ions with chelating agents (e.g., EDTA). Treatment of EGFP-incorporated pyramids with either 25 µM of EDTA or 100 µM imidazole showed higher levels of released EGFP as compared to controls, indicating the ability to trigger release with orthogonal cues (Fig. S7†). Of course, the kinetics of cargo release with imidazole or EDTA triggers are likely to be much slower than pH triggers.
In conclusion, we have demonstrated pH-dependent conformational changes of DNA pyramids made possible by the introduction of i-motif sequences. Structural changes were verified using electrophoresis, circular dichroism, and FRET. Triggered release of a protein cargo (EGFP) from pH-responsive pyramids highlights the potential of DNA nanostructures in drug delivery and biosensing applications. Towards more closely mimicking the efficiency of viruses, future work will entail pH-triggered delivery of fusogenic cargo and selective targeting via aptamer motifs.
Supplementary Material
Acknowledgments
We thank P. Charoenphol, S. Dawn, D. Frisardi and L. Ramos-Mucci for their assistance. This work was supported by NIH 1R21CA158977-01, NSF DMR-0847558, and partially by NSF CMMI-0531171.
Footnotes
Electronic Supplementary Information (ESI) available: DNA sequences, details of assembly and analysis including gel electrophoresis, circular dichroism, and fluorescence measurements. See DOI: 10.1039/b000000x/
References
- 1.Chhabra R, Sharma J, Ke Y, Liu Y, Rinker S, Lindsay S, Yan H. J. Am. Chem. Soc. 2007;129:10304–10305. doi: 10.1021/ja072410u. [DOI] [PubMed] [Google Scholar]
- 2.Park N, Um SH, Funabashi H, Xu J, Luo D. Nat. Mater. 2009;8:432–437. doi: 10.1038/nmat2419. [DOI] [PubMed] [Google Scholar]
- 3.Krishnan Y, Simmel FC. Angew. Chem. Int. Ed. Engl. 2011;50:3124–3156. doi: 10.1002/anie.200907223. [DOI] [PubMed] [Google Scholar]
- 4.Lo PK, Metera KL, Sleiman HF. Curr. Opin. Chem. Biol. 2010;14:597–607. doi: 10.1016/j.cbpa.2010.08.002. [DOI] [PubMed] [Google Scholar]
- 5.Goodman RP, Heilemannt M, Dooset S, Erben CM, Kapanidis AN, Turberfield AJ. Nat. Nanotechnol. 2008;3:93–96. doi: 10.1038/nnano.2008.3. [DOI] [PubMed] [Google Scholar]
- 6.Guéron M, Leroy JL. Curr. Opin. Struct. Biol. 2000;10:326–331. doi: 10.1016/s0959-440x(00)00091-9. [DOI] [PubMed] [Google Scholar]
- 7.Liu D, Balasubramanian S. Angew. Chem. Int. Ed. Engl. 2003;42:5734–5736. doi: 10.1002/anie.200352402. [DOI] [PubMed] [Google Scholar]
- 8.Cheng E, Xing Y, Chen P, Yang Y, Sun Y, Zhou D, Xu L, Fan Q, Liu D. Angew. Chem. Int. Ed. Engl. 2009;48:7660–7663. doi: 10.1002/anie.200902538. [DOI] [PubMed] [Google Scholar]
- 9.Modi S, MG S, Goswami D, Gupta GD, Mayor S, Krishnan Y. Nat. Nanotechnol. 2009;4:325–330. doi: 10.1038/nnano.2009.83. [DOI] [PubMed] [Google Scholar]
- 10.Chen L, Di J, Cao C, Zhao Y, Ma Y, Luo J, Wen Y, Song W, Song Y, Jiang L. Chem. Commun. 2011;47:2850–2852. doi: 10.1039/c0cc04765a. [DOI] [PubMed] [Google Scholar]
- 11.Shimron S, Magen N, Elbaz J, Willner I. Chem. Commun. 2011;47:8787–8789. doi: 10.1039/c1cc12795k. [DOI] [PubMed] [Google Scholar]
- 12.Torchilin VP. Annu Rev Biomed Eng. 2006;8:343–375. doi: 10.1146/annurev.bioeng.8.061505.095735. [DOI] [PubMed] [Google Scholar]
- 13.Gao W, Chan JM, Farokhzad OC. Mol. Pharm. 2010;7:1913–1920. doi: 10.1021/mp100253e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ulbrich K, Subr V. Adv Drug Deliv Rev. 2004;56:1023–1050. doi: 10.1016/j.addr.2003.10.040. [DOI] [PubMed] [Google Scholar]
- 15.Goodman RP, Berry RM, Turberfield AJ. Chem. Commun. 2004:1372–1373. doi: 10.1039/b402293a. [DOI] [PubMed] [Google Scholar]
- 16.Ozhalici-Unal H, Armitage BA. ACS Nano. 2009;3:425–433. doi: 10.1021/nn800727x. [DOI] [PubMed] [Google Scholar]
- 17.Li J, Pei H, Zhu B, Liang L, Wei M, He Y, Chen N, Li D, Huang Q, Fan C. ACS Nano. 2011;5:8783–8789. doi: 10.1021/nn202774x. [DOI] [PubMed] [Google Scholar]
- 18.Keum J-W, Ahn J-H, Bermudez H. Small. 2011;7:3529–3535. doi: 10.1002/smll.201101804. [DOI] [PubMed] [Google Scholar]
- 19.Greene DG, Keum J-W, Bermudez H. Small. 2012;8:1320–1325. doi: 10.1002/smll.201102221. [DOI] [PubMed] [Google Scholar]
- 20.Williams S, Lund K, Lin C, Wonka P, Lindsay S, Yan H. Tiamat: A Three-Dimensional Editing Tool for Complex DNA Structures. 1st edn. vol. 5347. New York: Springer; 2009. [Google Scholar]
- 21.Li W, Wu P, Ohmichi T, Sugimoto N. FEBS Lett. 2002;526:77–81. doi: 10.1016/s0014-5793(02)03118-6. [DOI] [PubMed] [Google Scholar]
- 22.Shen W, Zhong H, Neff D, Norton ML. J. Am. Chem. Soc. 2009;131:6660–6661. doi: 10.1021/ja901407j. [DOI] [PubMed] [Google Scholar]
- 23.Busch N, Kim T, Bloomfield V. Macromolecules. 2000;33:5932–5937. [Google Scholar]
- 24.Erben CM, Goodman RP, Turberfield AJ. Angew. Chem. Int. Ed. Engl. 2006;45:7414–7417. doi: 10.1002/anie.200603392. [DOI] [PubMed] [Google Scholar]
- 25.Fessenden JD. PLoS One. 2009;4:e7338. doi: 10.1371/journal.pone.0007338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ward WW, Prentice HJ, Roth AF, Cody CW, Reeves SC. Photochem. Photobiol. 1982;35:803–808. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.