Abstract
Despite the high frequency of HCV and HIV coinfection, little is known about HCV quasispecies in HIV-positive patients. The current analysis included 236 HIV+/anti-HCV+ women enrolled in the Women’s Interagency HIV Study (WIHS). Hypervariable region 1 of the second envelope gene was analyzed by single-strand conformation polymorphism (SSCP). The relationship between the HCV quasispecies and clinical and demographic features were analyzed in multivariate models. Age over 40 years and high HCV RNA load were the only factors significantly associated with quasispecies complexity, assessed as the number of SSCP bands. High HIV and HCV plasma loads were associated with quasispecies stability over time, as reflected by stable SSCP band patterns. However, women who were actively injecting drugs were 3 times more likely to experience quasispecies changes than their noninjecting counterparts. No affect on HCV quasi-species dynamics was noted in relation to CD4 count or highly active antiretroviral therapy (HAART). Conclusion: among HIV/HCV coinfected patients, HCV quasispecies complexity and dynamics correlate more closely with HIV and HCV plasma loads than with CD4+ cell counts. Active drug use is associated with quasispecies changes probably due to repeated superinfections with new HCV strains. This needs to be considered when planning treatment and prevention strategies for HCV in coinfected individuals.
Hepatitis C virus (HCV) coinfection is common among HIV-infected patients. In the United States and Europe 13% to 43% of HIV-positive persons are coinfected with HCV1 and this proportion can be as high as 85% among intravenous drug users (IDU).2 HIV coinfection increases the risk of HCV-related cirrhosis3–6 and because of highly active antiretroviral therapy (HAART)-related reduction in mortality, HCV-associated liver disease has become the primary cause of death among HIV-positive patients.7 This deleterious effect of HIV coinfection could be conveniently explained by increased HCV replication secondary to immunodeficiency. However, there is some emerging evidence that HCV replication may be enhanced by HIV infection more directly, although the mechanism behind this interaction remains unclear. Thus, plasma HCV RNA levels have been found to be more closely associated with HIV RNA levels than with CD4+ T cell count,8 and HIV seroconversion in HCV-infected patients has been associated with a rapid increase in HCV replication.9 It has also been reported that HIV infection may facilitate HCV infection/replication in in vitro cell culture systems.10 Interestingly, there is some emerging evidence that HCV infection may negatively affect HIV disease as well as response to HAART.11,12 The detrimental effects of HIV on the course of HCV infection are further compounded by its effect on treatment outcome. In 2 large clinical trials, the overall rate of sustained virological response among HIV/HCV coinfected patients treated with pegylated interferon plus ribavirin ranged from 27% to 40%, but the response rate was only 14% to 29% among those infected with the most common genotype 1, which is a markedly lower success rate than in HIV-negative patients.13,14
HCV is genetically heterogeneous because of low viral RNA polymerase fidelity resulting from lack of proofreading 3′-5′ exonuclease activity within the enzyme.15,16 Because of high mutation rates, HCV circulates as a population of closely related but nonidentical genomes, referred to as quasispecies.16–18 The N terminus of the E2 protein gene, which is the most variable part of the entire HCV genome, is called hypervariable region 1 (HVR1).19,20 This region encodes for a prominent B cell epitope and its high mutation rate could be important for viral evasion of the host immune system.19,21–23
HCV quasispecies heterogeneity and dynamics has been extensively studied in various settings, including in those with acute and chronic hepatitis C, those receiving anti-HCV therapy, and following liver transplantation for HCV-related liver disease.24–28 However, despite the high frequency of HCV/HIV coinfection, HCV quasi-species have rarely been analyzed in HIV-positive patients. In the few studies that were published, the number of analyzed patients was small and the role of associated factors could not be properly analyzed.29–31 The current study is the first systematic analysis of quasispecies complexity and dynamics in a large cohort of HIV-positive women taking into account the impact of multiple epidemiological factors and HAART.
Patients and Methods
Study Design and Classifications
The current analysis includes a subset of women enrolled in the Women’s Interagency HIV Study (WIHS), a prospective, multi-center cohort study established in 1993 to conduct comprehensive investigations of HIV infection in women. A detailed description of the WIHS has been published elsewhere.32 Participants are seen every 6 months and undergo an extensive interview, physical, and gynecological examinations, as well as multiple laboratory evaluations at each visit, including CD4 counts and plasma viral load measurements. We conducted the study in accordance with institutional review board guidelines for human research at each of the participating sites and we obtained informed consent from all study participants or their guardians.
Women chosen for this study were anti-HCV-positive, AIDS-free and hepatitis B surface antigen-negative at baseline and initiated HAART during follow-up. Among the 236 women evaluated, 97 had sequential evaluations pre-HAART and post-HAART with 1 to 2 pre-HAART and 1 post-HAART follow-up visit (at 6 to 12 months). WIHS used a standard definition of antiretroviral therapy: (1) untreated; (2) monotherapy (any single antiretroviral therapy within the preceding 6 months); (3) combination therapy (all combination therapies except HAART within the preceding 6 months); and (4) HAART within the preceding 6 months. HAART was defined as: (1) 2 nucleoside reverse-transcriptase inhibitors (NRTIs) in combination with at least 1 protease inhibitor (PI) or 1 nonnucleoside reverse-transcriptase inhibitor (NNRTI; 82% of regimens classified as HAART); (2) 1 NRTI in combination with at least 1 PI and at least 1 NNRTI (14% of regimens); (3) a regimen that contained PIs ritonavir and saquinavir in combination with 1 NRTI and no NNRTIs (2% of regimens); and (4) a regimen of 3 NRTIs including abacavir or tenofovir, in the absence of both PIs and NNRTIs (2% of regimens). We did not consider combinations of nucleoside reverse-transcriptase inhibitors zidovudine and stavudine with either a PI or NNRTI to constitute HAART. We initiated HIV treatment from 1996 to 2001. None of the participants received treatment for HCV for the duration of the study.
Virologic Evaluations
For viral load measurements and cell and plasma storage, we collected blood in sodium citrate cell-preparation tubes (Vacutainer tubes; Becton-Dickinson), which we either processed within 6 hours or centrifuged at 1500g and then processed them as directed by the manufacturer. We placed the majority of samples at −80°C within 6 hours from drawing and none later than after 24 hours. However, we previously found that delaying freezing by up to 48 hours has little effect on HIV RNA.33 We measured plasma HIV RNA levels using the NASBA/NucliSens HIV RNA assay (bioMe’rieux), in accordance with the manufacturer’s recommendations in laboratories that participate in and are certified by the National Institutes of Health, National Institute of Allergy and Infectious Diseases, Virology Quality Assurance certification program. We measured HCV RNA using a COBAS Amplicor Monitor 2.0 (Roche Diagnostics) with a linear range of 600 to 500,000 IU/mL. Because we anticipated that coinfected women would have HCV RNA levels higher than the upper limit of the assay, we initially diluted all samples 1:10; if we found them to be negative for HCV RNA, we retested them in undiluted form using a qualitative Amplicor HCV assay (Roche Diagnostics), which has a lower detection limit of 50 IU/mL; if these were positive, we retested them in undiluted form with the quantitative assay. We considered all specimens that were nonreactive in both HCV quantitative and qualitative PCR assays to be HCV RNA negative. We determined HCV serologic results at entry by the contemporary HCV commercial EIAs (Abbott EIA 2.0 and 3.0). Additionally, we retested all women with undetectable HCV RNA by HCV 3.0 EIA (Ortho Diagnostic). For women found to be viremic, we determined HCV genotype using the NC TRUGENE HCV 5′NC Genotyping Kit (Bayer HealthCare LLC, Tarrytown, NY) as recommended by the manufacturer and as described.34
Reverse-Transcription PCR
We extracted RNA from 100 μL serum by means of a modified guanidinium thiocyanatephenol/chloroform technique using a commercially available kit (Triazol, Gibco) and dissolved it in 20 μL water. We reverse transcribed 5 μL of this RNA solution with MMLV, and E2/HVR1 sequences were PCR amplified as described.35 We employed extensive measures to prevent and detect carryover contamination.36 Positive controls consisted of endpoint dilutions of synthetic RNA strand and negative controls included un-infected sera.
Analysis of Quasispecies by Single-Strand Conformation Polymorphism (SSCP)
We ran the assay as described.35 In brief, we purified PCR products with a DNA binding resin system (Wizard PCR, Promega, WI) and resuspended them in 50 μL water. Next, we diluted 2 to 4 μL purified product in 15 μL low ionic strength solution (10% sucrose, 0.5% bromophenol blue, 0.5% xylene cyanol), denatured it by heating at 97°C for 3 minutes, immediately cooled it on ice and subjected it to nondenaturing 8% PAGE in 1× Tris-borate-ethylene diamine tetraacetic acid buffer with 400 V applied for 4 to 5 hours at a constant temperature of 25°C. We visualized the bands with silver staining (Silver Stain, Promega). Controls included samples with known polymorphism and we tested many reverse transcription PCR (RT-PCR) products repeatedly.
We have previously shown that SSCP can reliably distinguish between amplified PCR products differing by a single nucleotide and can detect minor viral variants representing as little as 1.5% to 3% of the whole population.26 The technique is immune to cloning artifacts, which are due to preferential selection of genomes by E. coli.37,38 Analysis of cloned RT-PCR is also prone to sampling errors related to the typically small number of studied clones.37,38 Consequently, attaining the theoretical sensitivity limit of 3% for the detection of minor viral variants, which is expected for SSCP assays, would require analyzing 99 clones per each sample (confidence 95%).
However, SSCP does not measure genetic distance among viral variants and may underestimate complexity due to possible band comigration.
Genetic Characterization of HCV Evolution by Sequencing
We cloned the RT-PCR products amplified from the E2/HVR1 into the TA cloning vector (Invitrogen, Carlsbad, CA) and we sequenced clones using a PerkinElmer ABI 377 automatic sequencer. We sequenced 14 to 20 clones for each time point. We did multiple sequence alignments with MegAlign (DNAS-TAR, Inc.; Madison, WI). We constructed phylogenetic trees with the MEGA 3.0 computer package,39 using the neighbor-joining method and the Tamura-Nei distance measure, which corrects for base composition and transition/transversion bias. We discounted positions where gaps were inserted to preserve alignment. We did a bootstrap analysis using 1000 bootstrap replicates to assess the reliability at each of the internal nodes of the trees.
We calculated mean distances within viral sequences at each visit (diversity) and mean distances from the first analyzed visit consensus sequence (divergence) according to the Kimura 2-parameter model with a transition-to-transversion ratio of 2. We calculated the average number of synonymous substitutions per synonymous site and the number of nonsynonymous substitutions per nonsynonymous site by the method of Nei and Gojobori, using a Jukes-Cantor correction to account for multiple substitutions at the same site.39 We used the consensus sequence from the first time point as a reference.
Study Definitions
We measured 2 measures of HCV quasispecies variability in relation to various clinical and virologic features in this study: complexity and “shift” (change in the pattern of major SSCP bands).
We measured quasispecies complexity by calculating the number of bands on the SSCP gels as described previously.26 To prevent bias from manual counting, we recorded the images as high-resolution TIFF files (Epson Expression 1680; Epson) and analyzed them by Quantity One software (Bio-Rad Laboratories, Hercules, CA). We utilized women with single evaluations for analyses of quasispecies complexity only.
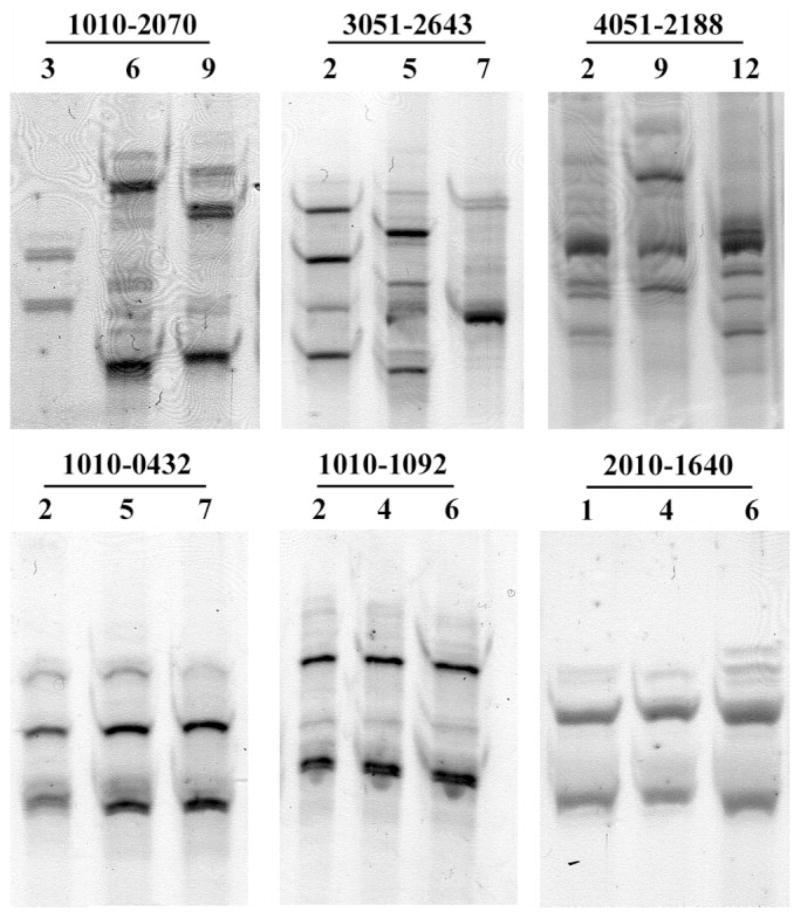
To assess for changes in quasispecies, we included only women who had 2 or more evaluations. For the purpose of this study, we defined any change in the position of major SSCP bands on sequential samples from the same patient as “shift” and presumed it to be indicative of sequence change. We assessed shift visually by 2 independent readers. Because of assay sensitivity, a “shift” could represent anything from single nucleotide substitution to a new infecting HCV strain. Conversely, unchanged SSCP band pattern would indicate complete stability of viral quasispecies sequence. Examples of SSCP analysis of stable (cases 1010-0432, 1010-1092, 2010-1640) and changing quasispecies patterns (cases 1010-2070, 3051-2643, 4051-2188) are presented in (Fig. 1).
Fig. 1.
SSCP analysis of hypervariable region 1 HCV RNA amplified from sequential plasma samples of 6 different WIHS patients. Three patients in the upper row showed changes in the position of SSCP bands (“shift”), indicating viral sequence changes. In 3 patients in the lower row the position of bands did not change, indicating stability of viral quasi-species. In patient 2010-1640 (lower row) new minor bands appeared at visit 6 (last lane); however, since position of the major bands did not change, for the purpose of this study patient’s quasispecies was classified as stable. Patients’ ID and visit numbers are shown above each gel.
Data Analysis
To accommodate analysis of the samples from multiple visits within subjects, we used a generalized estimating equations approach with a linear regression model to determine the factors associated with the number of SSCP bands (viral complexity). In this analysis, the dependent variable was the number of SSCP bands. We used data from all subject-visits, and we assumed an exchangeable correlation matrix between multiple measures from each subject.
We used a generalized estimating equations approach with a logistic regression model to determine the correlates of SSCP band “shift.” The dependent variable was the presence or absence of “shift” over 2 visits. Because the detection of a “shift” required repeated samples within subjects, we included only subjects who contributed samples from at least 2 visits in this analysis. The covariates of interest were measures from the visit prior to the assessment for band “shift” to be certain that the exposures occurred before the outcome was assessed (presence or absence of band “shift”). We defined measures of antiretroviral therapy and HAART use as use in the past 6 months (i.e., since the last study visit); to assess the association of band “shift” with initiation of HAART, we pulled this exposure variable from the visit at which the presence or absence of band “shift” was determined.
In both analyses, covariates tested for association with the outcomes (number of bands or the presence of a band “shift”) included current age (categorized as <35, 35 to 40, or >40 years), race, HCV-RNA level (≤13.1 million, >13.1 million to 27.4 million, >27.4 million to 39.4 million, or >39.4 million), HIV-RNA level (<5000, 5000 to 9999, 10,000 to 49,000, 50,000 to 99,999, or 100,000 or more), CD4 cell count (<200, 200 to 349, 350 to 500, or >500), CD8 cell count (<500, 500 to 1199, or 1200 or more), IDU in the past 6 months, current smoking, antiretroviral therapy (none, monotherapy, combination therapy, or HAART), and current HAART use. Additional independent variables evaluated in the band “shift” analyses were noninjection drug use in the past 6 months, (heroin, cocaine, or methadone) and number of sex partners in the past 6 months. We tested all independent variables first for univariate (unadjusted) association with the outcome (number of bands or the presence of a band “shift”). We further evaluated the factors that were at least marginally significant (P < 0.1) in the unadjusted analysis in the multivariate regression model that adjusted for each of these parameters.
Results
We presented the baseline demographic and clinical information for all 236 HIV+/anti-HCV+ women participating in the study in Table 1. The vast majority of women were black or Hispanic and although the majority had a history of injection drug use, only 17% were actively injecting and less than one-third were cocaine, methadone, or heroine users. Women had high HCV RNA levels (median: 2,370,000 IU/mL) and 91% were infected with genotype 1.
Table 1.
Baseline Characteristics of the Women Participating in the Study (n = 236)
| Characteristic | Value* |
|---|---|
| Age (years) | 39.7 (23.6–54.3) |
| Race (%) | |
| White | 18 |
| Black | 60 |
| Hispanic | 21 |
| CD4 cell count (μ/L)* | 367 (4–1386) |
| CD8 cell count (μ/L)* | 829 (145–3478) |
| HIV-viral load (copies/mL)* | 20,500 (80–1,700,000) |
| HCV-viral load (IU/mL)* | 2,370,000 (0–12,300,000) |
| Intravenous drug use (%) | 17 |
| Heroin use (%) | 20 |
| Cocaine use (%) | 33 |
| Methadone use (%) | 4 |
| Number of sex partners | 1 (0–60) |
| HCV genotype (%) | |
| 2 and 3 | 9 |
| 1a and 1b | 91 |
Continuous variables are reported as median (range).
Altogether, we amplified HCV RNA from plasma samples collected at 410 patient visits. In 139 out of the 236 women we analyzed only 1 sample from a single visit, while in the remaining 97 women we assessed 1 to 2 pre-HAART and 1 post-HAART visits. Among the latter women, we drew the final sample at the second visit after HAART initiation. The mean ± SD number of SSCP bands among all 410 samples was 10.8 ± 4.7.
We first analyzed the relationship between the number of SSCP bands and demographic characteristics, HCV and HIV viral loads, smoking, active IDU (injecting within the last 6 months), HIV therapy, and CD4+ and CD8+ cell counts in a univariate model (Table 2). In these analyses, age over 40 years and high HIV RNA load were associated with lower number of SSCP bands, indicating lower HCV complexity, while high HCV RNA load was associated with increased complexity. Patients on HAART were also more likely to have higher number of viral variants in circulation, but this difference did not reach statistical significance. In a subsequent multivariate model analysis, which adjusted for age, HIV and HCV viral load, CD4 and CD8 counts, and treatment, the only factors significantly correlated with quasispecies complexity were age and high HCV RNA load (Table 3).
Table 2.
Univariate Analysis of Characteristics Associated with the Number of SSCP Bands Among 410 Specimens from 236 Women
| Variables | Mean Number of Bands (SD) | P Value for Mean Difference From Reference Group | Trend P Value |
|---|---|---|---|
| Age (years) | 0.005 | ||
| <35 | 11.39 (0.55) | Reference group | |
| 35–40 | 10.97 (0.57) | 0.60 | |
| >40 | 9.57 (0.42) | 0.01 | |
| Race | |||
| White | 10.56 (0.72) | Reference group | |
| Black | 10.01 (0.38) | 0.43 | |
| Hispanic | 11.35 (0.60) | 0.44 | |
| HCV-RNA (IU/mL) | 0.04 | ||
| <1,310,000 | 9.67 (0.43) | Reference group | |
| 1,310,001–2,740,000 | 10.52 (0.43) | 0.09 | |
| 2,740,001–3,940,000 | 10.71 (0.47) | 0.07 | |
| >3,940,000 | 10.99 (0.48) | 0.02 | |
| HIV-viral load (copies/mL) | 0.01 | ||
| <5,000 | 10.92 (0.38) | Reference group | |
| 5000–9999 | 11.37 (0.73) | 0.52 | |
| 10,000–49,000 | 9.96 (0.48) | 0.05 | |
| 50,000–99,999 | 10.22 (0.53) | 0.20 | |
| 100,000+ | 9.75 (0.46) | 0.03 | |
| Intravenous drug use (in the past 6 months) | |||
| No | 10.44 (0.32) | Reference group | |
| Yes | 10.32 (0.63) | 0.87 | |
| Current smoking | |||
| No | 10.41 (0.52) | Reference group | |
| Yes | 10.42 (0.33) | 0.98 | |
| Antiretroviral therapy | |||
| None | 10.13 (0.37) | Reference group | |
| Monotherapy | 10.53 (0.49) | 0.44 | |
| Combination therapy | 10.64 (0.38) | 0.23 | |
| HAART | 10.99 (0.47) | 0.07 | |
| Current HAART use | |||
| No | 10.35 (0.30) | Reference group | |
| Yes | 10.94 (0.47) | 0.16 | |
| CD4 cell count/mm3 | 0.27 | ||
| >500 | 10.76 (0.43) | Reference group | |
| 350–500 | 10.55 (0.49) | 0.69 | |
| 200–349 | 10.16 (0.43) | 0.29 | |
| >200 | 10.17 (0.46) | 0.32 | |
| CD8 cell count/mm3 | 0.09 | ||
| <500 | 9.59 (0.60) | Reference group | |
| 500–1199 | 10.51 (0.36) | 0.18 | |
| 1200 or more | 10.76 (0.41) | 0.08 |
Table 3.
Multivariate Analysis of Characteristics Associated with the Number of SSCP Bands Among 410 Specimens from 236 Women
| Variables | Mean (SD) Number of Bands | P-value for Mean Difference From Reference Group |
|---|---|---|
| Age (years) | ||
| <35 | 11.53 (0.57) | |
| 35–40 | 11.07 (0.57) | 0.68 |
| >40 | 9.76 (0.46) | 0.01 |
| HCV-RNA (IU/mL) | ||
| <1,310,000 | 10.12 (0.47) | |
| 1,310,001–2,740,000 | 11.0 (0.42) | 0.04 |
| 2,740,001–3,940,000 | 10.79 (0.48) | 0.14 |
| >3,940,000 | 11.24 (0.53) | 0.04 |
| HIV-viral load (copies/mL) | ||
| <5000 | 11.14 (0.39) | |
| 5000–9999 | 11.71 (0.72) | 0.55 |
| 10,000–49,000 | 10.16 (0.47) | 0.04 |
| 50,000–99,999 | 10.50 (0.57) | 0.20 |
| 100,000+ | 10.41 (0.50) | 0.18 |
| Antiretroviral therapy | ||
| None | 10.29 (0.41) | |
| Monotherapy | 10.78 (0.49) | 0.55 |
| Combination therapy | 10.74 (0.42) | 0.56 |
| HAART | 11.33 (0.48) | 0.12 |
| CD8 cell count | ||
| <500 | 10.38 (0.55) | |
| 500–1199 | 10.95 (0.38) | 0.39 |
| 1200 or more | 11.03 (0.47) | 0.30 |
We evaluated a total of 97 women on multiple occasions, of whom 77 had 3 evaluations: 2 before HAART and 1 after initiation of HAART, and the remaining 20 women had 2 evaluations with 1 before HAART and 1 after initiation of HAART. Analyzed visits were a median 1 to 1.5 years apart. A total of 70 (72%) women showed a shift in the SSCP band pattern at some point during follow-up and 39 of these women had SSCP band shift both between the first and second, and between the second and third of the analyzed visits. In 27 women, the quasispecies pattern remained stable throughout the follow-up.
We first did the investigations of the relationship between the presence or absence of SSCP “shift” and demographic characteristics, HCV and HIV viral loads, smoking, active IDU and noninjection drug use, HIV therapy, number of sex partners, and CD4+ and CD8+ cell counts in an unadjusted model (Table 4). We further evaluated the relationship between the presence/absence of SSCP band “shift” and the above factors in the multivariate logistic regression model that adjusted for each of the parameters that were at least borderline significant (P < 0.1) in the univariate analysis. The only statistically significant correlates of the occurrence of quasispecies “shift” were HIV RNA and HCV RNA plasma levels and active IDU (Table 5). The association with HAART was of borderline statistical significance (P = 0.09). While high HCV and HIV loads were associated with quasispecies stability, actively injecting women were over 3 times more likely to experience SSCP band pattern changes than their noninjecting counterparts.
Table 4.
Univariate Analysis of the Presence of SSCP Band “Shift” in Sequential Plasma Samples from 97 Women*
| Variables | Odds Ratio (95% Confidence Interval) | P Value | Trend P Value |
|---|---|---|---|
| Age (years) | |||
| <35 | Reference group | ||
| 35–40 | 1.06 (0.58–1.93) | 0.85 | |
| >40 | 0.75 (0.40–1.38) | 0.35 | |
| Race | |||
| White | Reference group | ||
| Black | 1.38 (0.64–2.98) | 0.41 | |
| Hispanic | 0.95 (0.40–2.26) | 0.91 | |
| HCV-RNA (IU/mL) | |||
| <1,310,000 | Reference group | ||
| 1,310,001–2,740,000 | 0.66 (0.29–1.54) | 0.34 | |
| 2,740,001–3,940,000 | 0.67 (0.28–1.63) | 0.38 | 0.10 |
| >3,940,000 | 0.44 (0.18–1.09) | 0.07 | |
| HIV-viral load (copies/mL) | |||
| <5000 | Reference group | ||
| 5000–9999 | 0.79 (0.23–2.76) | 0.71 | |
| 10,000–49,000 | 0.63 (0.29–1.40) | 0.26 | 0.03 |
| 50,000–99,999 | 0.63 (0.25–1.55) | 0.31 | |
| 100,000+ | 0.30 (0.11–0.84) | 0.02 | |
| Intravenous drug use (in the past 6 months) | |||
| No | Reference group | ||
| Yes | 2.67 (0.90–7.75) | 0.07 | |
| Current smoking | |||
| No | Reference group | ||
| Yes | 1.93 (0.83–4.49) | 0.13 | |
| Antiretroviral therapy | |||
| None | Reference group | ||
| Monotherapy | 0.99 (0.41–2.41) | 0.99 | |
| Combination therapy | 1.47 (0.75–2.89) | 0.26 | |
| Current HAART use | |||
| No | Reference group | ||
| Yes | 4.28 (2.53–7.22) | <0.0001 | |
| CD4 cell count/mm3 | |||
| >500 | Reference group | ||
| 350–500 | 0.88 (0.39–1.98) | 0.76 | |
| 200–349 | 0.37 (0.16–0.86) | 0.02 | 0.08 |
| <200 | 0.60 (0.25–1.43) | 0.25 | |
| CD8 cell count/mm3 | |||
| <500 | Reference group | ||
| 500–1199 | 0.66 (0.30–1.47) | 0.31 | 0.13 |
| 1200 or more | 1.48 (0.56–3.94) | 0.43 | |
| Heroin use | |||
| No | Reference group | ||
| Yes | 2.33 (0.85–6.40) | 0.52 | |
| Cocaine use | |||
| No | Reference group | ||
| Yes | 1.12 (0.52–2.44) | 0.77 | |
| Methadone use | |||
| No | Reference group | ||
| Yes | 2.89 (0.44–18.9) | 0.27 | |
| Number of sex partners | |||
| None | Reference group | ||
| 1 | 0.71 (0.34–1.47) | 0.35 | 0.30 |
| 2 or more | 0.64 (0.25–1.60) | 0.34 | |
All the exposures are from previous visit, except for HAART, which is from the current visit.
Table 5.
Multivariate Analysis of the Presence of SSCP Band “Shift” in Sequential Plasma Samples from 97 Women
| Variables | Odds Ratio (95% Confidence Interval) | P Value |
|---|---|---|
| HCV-RNA (IU/mL) | ||
| <1,310,000 | Reference group | |
| 1,310,001–2,740,000 | 0.54 (0.23–1.28) | 0.16 |
| 2,740,001–3,940,000 | 0.49 (0.19–1.26) | 0.14 |
| >3,940,000 | 0.37 (0.14–0.95) | 0.04 |
| HIV-viral load (copies/mL) | ||
| <5000 | Reference group | |
| 5000–9999 | 0.79 (0.22–2.86) | 0.72 |
| 10,000–49,000 | 0.81 (0.35–1.91) | 0.63 |
| 50,000–99,999 | 0.86 (0.33–2.26) | 0.76 |
| 100,000+ | 0.28 (0.95–0.85) | 0.02 |
| Intravenous drug use (in the past 6 months) | ||
| No | Reference group | |
| Yes | 3.26 (1.02–10.41) | 0.04 |
| Current HAART use | ||
| No | Reference group | |
| Yes | 1.74 (0.92–3.27) | 0.09 |
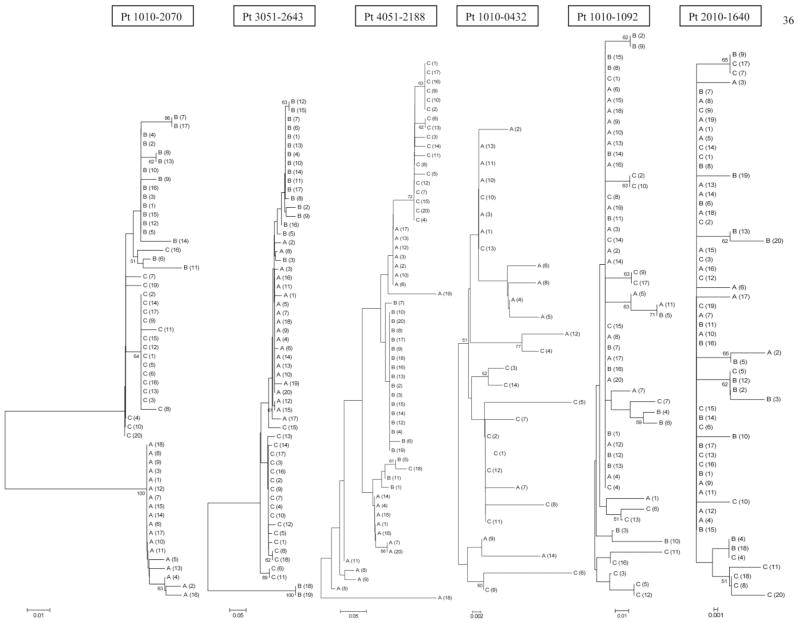
To validate and extend findings based on SSCP analysis, we cloned 17 RT-PCR products derived from 6 patients shown in Figure 1 and sequenced them. Altogether, we analyzed 312 clones (14 to 20 per sample). Phylogenetic analysis of E2/HVR1 sequences obtained from each of the 6 patients showed 2 different topological patterns. In patients 1010-0432, 1010-1092, and 2010-1640, who were characterized by stable SSCP band patterns, there was a monophyletic population with intermingling of sequences from different times. In patients 010-2070, 3051-2643, and 4051-2188, who were characterized by changing SSCP patterns, phylogenetic reconstruction was compatible with sequential shifts in the viral population, and sequences derived from individual visits tended to form clusters (Fig. 2).
Fig. 2.
Phylogenetic reconstruction of evolutionary relationship of HVR1/E2 sequences in 6 patients. Bootstrap proportions of greater than 50% are shown at branch points. The taxa are labeled A, B, and C indicating sequential samples from each patient, which correspond to visit numbers shown in Fig. 1. In patients 1010-0432, 1010-1092, and 2010-1640 viral population was relatively monophyletic with intermingling of sequences from different visits, while in patients 1010-2070, 3051-2643, and 4051-2188 quasispecies from sequential visits tended to group separately.
Quasispecies diversity and divergence measurements in the above 6 patients are shown in Table 6. Whereas the 3 patients with stable SSCP patterns showed little genetic evolution over time, patients with “shift” were characterized by much higher divergence. Furthermore, the latter patients were characterized by a high number of nonsynonymous substitutions, which suggests the presence of positive immune selection. The above findings support the validity of SSCP analysis in differentiating between stable and changing quasispecies.
Table 6.
Analysis of HCV E2/HVR1 Genetic Diversity and Divergence in Sequential Plasma Samples from 6 HIV/HCV Coinfected Patients*
| Patient ID | Genotype* | SSCP Pattern | Diversity†
|
Divergence‡
|
dS§
|
dN|
|
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| A | B | C | B | C | B | C | B | C | |||
| 1010-2070 | 1a | Shift | 0.005 | 0.010 | 0.007 | 0.133 | 0.126 | 0.182 | 0.184 | 0.105 | 0.098 |
| 3051-2643 | 1b | 0.015 | 0.137 | 0.036 | 0.093 | 0.065 | 0.050 | 0.057 | 0.097 | 0.067 | |
| 4051-2188 | 1a | 0.107 | 0.040 | 0.037 | 0.101 | 0.097 | 0.000 | 0.004 | 0.143 | 0.139 | |
| 1010-0432 | 1a | Stable | 0.016 | ND | 0.018 | ND | 0.013 | ND | 0.031 | ND | 0.007 |
| 1010-1092 | 1a | 0.011 | 0.027 | 0.036 | 0.014 | 0.020 | 0.020 | 0.030 | 0.012 | 0.016 | |
| 2010-1640 | 1a | 0.004 | 0.011 | 0.009 | 0.013 | 0.013 | 0.000 | 0.007 | 0.008 | 0.004 | |
NOTE. A, B, C refers to sequential plasma samples; ND, not done.
Genotypes were tested at each of the 3 time points.
Mean Kimura 2-parameter distance within all clones derived from a single visit.
Mean Kimura 2-parameter distance between all clones from a follow-up visit and the consensus sequence from the initial visit.
Synonymous mutations per synonymous site compared to consensus sequence from the first time point.
Nonsynonymous mutations per nonsynonymous site compared to consensus sequence from the first time point.
Discussion
Our study is the first systematic analysis of quasispecies complexity and dynamics in a large cohort of HIV-infected patients taking into consideration the impact of multiple epidemiological factors and HAART. We found that extremely high HIV RNA and HCV RNA levels were associated with quasispecies stability over time, while active injection drug use was associated with changing quasispecies patterns. Level of CD4 did not independently contribute to HCV quasispecies complexity or changes.
It is generally assumed that the immune response acts as a major selective force on the HVR1 region during HCV infection. This view is supported by the observation that in immunosuppressed patients, including patients with hypogammaglobulinemia40 and those undergoing bone marrow transplantation,41 variability of HVR1 quasispecies is low. Analysis of HCV quasispecies provides therefore an insight into the strength of the immune pressure against HCV. Not surprisingly, quasispecies dynamics were found to be highly predictive of clinical outcomes. In 2 extensive studies of acute hepatitis, resolving infection was characterized by a homogeneous viral population and evolutionary stasis, while chronic infection was associated with genetic evolution within the first months of infection.25,26 Interestingly, the timing of viral clearance coincided with the advent of quasispecies changes in those who remained chronically infected, suggesting that the mechanisms responsible for viral clearance and those driving HVR1 quasispecies changes are the same. A reduction in genetic diversity leading to an increasingly homogeneous viral population was also associated with viral clearance in patients undergoing therapy with interferon.27 While in the above 2 settings lack of escape mutations is indicative of strong immune pressure leading to infection clearance, stability or reduction of quasispecies diversity and complexity in the natural course of chronic infection implies a weak immune response and may entail poor prognosis with respect to liver disease.28,42,43 Similarly, narrowing of HVR1 complexity and diversity was associated with progressive liver disease in HCV/HIV coinfected hemophiliacs.44
Despite the fact that HIV/HCV coinfection is common and may have grave clinical consequences, few studies have analyzed factors affecting HCV quasispecies in this setting. Two different studies found correlation between CD4+ cell count and HCV diversity, assessed as genetic distance,29,30 but in a third study correlation was statistically not significant.31 No relationship was found between CD4+ cell count and quasispecies complexity in any of these 3 reports. However the number of studied patients was small, ranging from 10 to 27, which yielded low statistical power and did not allow for multivariate analysis. Furthermore, the technique used— cloning of PCR products—is prone to artificial polymorphism introduced during cloning and is influenced by sampling errors related to the typically small number of analyzed clones.37,38 In 1 study using SSCP, which is more conducive to large-scale analysis, CD4+ count emerged as an independent predictor of the number of circulating viral variants among 56 patients.45 However, a recent report using clonal frequency analysis through heteroduplex mobility assay did not find any independent association between CD4 count and quasispecies complexity or diversity when analyzing 28 HCV/HIV coinfected patients.46
In our current study neither quasispecies complexity nor dynamics correlated with CD4+ cell count. However, very high HIV RNA plasma levels of >100,000 copies/mL were associated with lack of escape mutations, as suggested by the stability of bands over the time. Such high HIV RNA levels are prognostic of AIDS development47 and perhaps they could identify patients with severely compromised immune system, who are unable to exert immune pressure against HCV infection. An alternate explanation is that HIV selectively facilitates growth of some HCV variants, to the extent that variants of lower abundance fall below the level of detection. This hypothesis is compatible with observations that HCV replication may be directly enhanced by HIV coinfection both in vivo and in vitro.8–10
Interestingly, we found a positive association between HCV load and viral complexity. The reason for this phenomenon is unclear, but could be related to high-replication fitness of the infecting strain, which would be conducive for the emergence of new mutations and quasispecies variants. Although similar findings were reported by others,30,48 the association was invariably weak and some studies did not find any association between viral load and quasispecies complexity.25,49
HIV-related immunodeficiency facilitates progression of liver disease and several studies found a close association between low CD4 cell count and the development of fibrosis and cirrhosis among HIV/HCV coinfected patients.50,51 Not surprisingly, HAART was found to reduce long-term liver-related mortality in HIV/HCV coinfected patients.52 However, successful immune restoration may facilitate immune-mediated hepatocyte destruction resulting in increased liver damage, increased serum aminotransferase levels, and higher HCV RNA serum viral loads.53 There may be a relationship between low CD4 cell counts and the magnitude and duration of HCV RNA increase. Chung et al.54 reported an increase of HCV load at 16 and 48 weeks of HAART in patients with entry CD4 cell counts <350 cells/mm3, while in those with higher CD4 cell counts the viral load increase was small and temporary. Nevertheless, in our analysis HAART did not have significant effect on HCV quasi-species complexity or dynamics. This could have been due to the relatively short (mean 9.8 months; range 6.3 to 17.5 months) duration of therapy among our patients. In their study on the effect of HAART on HCV quasispecies, Babik and Holodniy30 analyzed 7 patients treated for 7 to 10 months and 6 patients who were treated longer. They found increased viral diversity and complexity in patients on long-term HAART but no changes were evident among patients treated short-term. Similarly, Blackard et al.55 observed increase in HVR1 diversity in 11 patients treated for 48 weeks. However, no adjustments for covariates were made in either of these 2 studies. In our sample, HAART was highly significantly associated with a higher likelihood of band “shift” on univariate analysis (odds ratio [OR] =4.28; 95% confidence interval [CI] = 2.53 to 7.22; Table 4), but this association was reduced (OR = 1.74, 95% CI = 0.92 to 3.27; Table 5) in a multivariate model. In a recently published study comparing 11 HAART-naive and 17 HAART-treated patients, HAART was associated with increased HCV complexity and diversity in a multivariate analysis. However, the median duration of HAART in that study was 46 months.46
An intriguing finding in our study was a relationship between changes in SSCP band positions, indicating quasispecies sequence changes, and recent IDU. This is compatible with superinfection with a new virus. It was demonstrated in chimpanzees that the presence of persistent infection does not provide protection against subsequent infection with heterologous and even with homologous strains.56–58 As the neutralizing antibodies are isolate-specific, they may be ineffective against other variants present in the complex of quasispecies: in a chimpanzee challenge experiment a hyperimmune anti-HVR1 serum raised against the predominant strain present in the inoculum was able to neutralize the predominant clone but was ineffective against minor variants.59 However, there is clear evidence for the presence of some cross-reactive immunity as convalescent chimpanzees and humans usually do not develop chronic infection upon reexposure.60–62 Similarly, immunization of chimpanzees with recombinant envelope glycoproteins gpE1 and gpE2 significantly reduced chronic infection when the animals were experimentally challenged with homologous as well as heterologous strains.63
Nevertheless, chronically infected humans remain susceptible to new infection. Eyster et al.64 documented a common change of infecting HCV genotypes over time in high-risk patients with hemophilia and recently Herring et al.65 identified superinfection in 20% of active IDUs over a period of 12 months. Similarly, it was shown that chronically infected patients can be superinfected with new HCV strains in the setting of blood transfusion, where HCV-positive patients were exposed to HCV-positive blood,66 and in the setting of liver transplantation, where patients with HCV-related liver disease receive organs from HCV-infected donors.36 Interestingly, the superinfecting strain often became dominant, eliminating or suppressing replication of the original virus below detection level. It can be argued that the superinfecting strains are immunologically favored, as it represents, to a certain extent, an “escape mutant.” However, all 3 patients with SSCP defined “shift” who were analyzed by cloning and sequencing remained infected by the same genotype strain. While the possibility of infection with multiple genotypes presenting a multiple quasispecies variants can not be excluded, mixed genotype infections appear to be rare.67,68
An alternate explanation for HCV quasispecies changes is that they are directly related to IDU itself and such a possibility is supported by a reported association between active IDU and increased diversity in the HIV env region.69 HIV superinfection is unlikely to affect HIV quasispecies dynamics among IDUs, as it is a relatively rare event.70 However, drugs of abuse were reported to enhance HIV replication and this process is likely to be mediated by cytokines.71 Whether drugs could affect HCV infection is currently unclear.
In summary, among HIV/HCV coinfected patients, HCV quasispecies complexity and dynamics correlate more closely with HIV and HCV plasma loads than with CD4+ cell counts. Active drug use is associated with quasispecies changes probably as the result of repeated superinfections with new HCV strains. HAART did not have a significant effect on HCV quasispecies within the 6 to 12 months after its initiation.
Acknowledgments
We thank the study participants for their cooperation. Data were collected by the WIHS Collaborative Study Group, with centers (Principal Investigators) at New York City/Bronx Consortium, Bronx, NY (Kathryn Anastos); Brooklyn, New York (Howard Minkoff); Washington, DC, Metropolitan Consortium (Mary Young); The Connie Wofsy Study Consortium of Northern California, San Francisco (Ruth Greenblatt and Herminia Palacio); Los Angeles County/Southern California Consortium, Los Angeles, CA (Alexandra Levine); Chicago Consortium, Chicago, IL (Mardge Cohen); and Data Coordinating Center, Boston, MA (Alvaro Munoz and Stephen J. Gange).
Supported by the National Institutes of Health, National Institute of Allergy and Infectious Diseases (NIAID; grant R01 AI 052065-04 to A.K.); National Cancer Institute, National Institute of Child Health and Human Development, National Institute on Drug Abuse, National Institute of Craniofacial and Dental Research (grants U01-AI-35004, U01-AI-31834, U01-AI-34994, U01-AI-34989, U01-HD-32632, U01-AI-34993, U01-AI-42590, M01-RR00079, M01-RR00083, and U01-HD-3-2632-11 to the Women’s Interagency HIV Study); Institute of Mental Health (grant 1R21 MH073422-01A1 to T.L.).
Abbreviations
- anti-HCV
antibody to HCV
- HAART
highly active antiretroviral therapy
- HVR1
hypervariable region 1
- IDU
injection drug use(rs)
- NNRTI
non-nucleoside reverse transcriptase inhibitor
- NRTI
nucleoside reverse transcriptase inhibitor(s)
- PI
protease inhibitor
- RT-PCR
reverse transcription PCR
- SSCP
single-strand conformation polymorphism
- WIHS
Women’s Interagency HIV Study
Footnotes
Potential conflict of interest: Nothing to report.
References
- 1.Winnock M, Salmon-Ceron D, Dabis F, Chene G. Interaction between HIV-1 and HCV infections: towards a new entity? J Antimicrob Chemother. 2004;53:936–946. doi: 10.1093/jac/dkh200. [DOI] [PubMed] [Google Scholar]
- 2.Thomas DL, Vlahov D, Solomon L, Cohn S, Taylor E, Garfein R, et al. Correlates of hepatitis C virus infections among injection drug users. Medicine (Baltimore) 1995;74:212–220. doi: 10.1097/00005792-199507000-00005. [DOI] [PubMed] [Google Scholar]
- 3.Ridzon R, Gallagher K, Ciesielski C, Ginsberg MB, Robertson BJ, Luo CC, et al. Simultaneous transmission of human immunodeficiency virus and hepatitis C virus from a needle-stick injury [see comments] N Engl J Med. 1997;336:919–922. doi: 10.1056/NEJM199703273361304. [DOI] [PubMed] [Google Scholar]
- 4.Eyster ME, Diamondstone LS, Lien JM, Ehmann WC, Quan S, Goedert JJ. Natural history of hepatitis C virus infection in multitransfused hemophiliacs: effect of coinfection with human immunodeficiency virus. The Multicenter Hemophilia Cohort Study. J Acquir Immune Defic Syndr. 1993;6:602–610. [PubMed] [Google Scholar]
- 5.Garcia-Samaniego J, Soriano V, Castilla J, Bravo R, Moreno A, Carbo J, et al. Influence of hepatitis C virus genotypes and HIV infection on histological severity of chronic hepatitis C. The Hepatitis/HIV Spanish Study Group. Am J Gastroenterol. 1997;92:1130–1134. [PubMed] [Google Scholar]
- 6.Hanley JP, Jarvis LM, Andrews J, Dennis R, Lee R, Simmonds P, et al. Investigation of chronic hepatitis C infection in individuals with haemophilia: assessment of invasive and non-invasive methods. Br J Haematol. 1996;94:159–165. doi: 10.1046/j.1365-2141.1996.6192064.x. [DOI] [PubMed] [Google Scholar]
- 7.Monga HK, Breauz K, Rodrigues-Barradas MC, Yoffe B. Increased HCV-related morbidity and mortality in HIV patients. Hepatology. 1998;28:565A. [Google Scholar]
- 8.Thomas DL, Rich JD, Schuman P, Smith DK, Astemborski JA, Nolt KR, et al. Multicenter evaluation of hepatitis C RNA levels among female injection drug users. J Infect Dis. 2001;183:973–976. doi: 10.1086/319256. [DOI] [PubMed] [Google Scholar]
- 9.Beld M, Penning M, Lukashov V, McMorrow M, Roos M, Pakker N, et al. Evidence that both HIV and HIV-induced immunodeficiency enhance HCV replication among HCV seroconverters. Virology. 1998;244:504–512. doi: 10.1006/viro.1998.9130. [DOI] [PubMed] [Google Scholar]
- 10.Laskus T, Radkowski M, Jablonska J, Kibler K, Wilkinson J, Adair D, et al. Human immunodeficiency virus facilitates infection/replication of hepatitis C virus in native human macrophages. Blood. 2004;103:3854–3859. doi: 10.1182/blood-2003-08-2923. [DOI] [PubMed] [Google Scholar]
- 11.Greub G, Ledergerber B, Battegay M, Grob P, Perrin L, Furrer H, et al. Clinical progression, survival, and immune recovery during antiretroviral therapy in patients with HIV-1 and hepatitis C virus coinfection: the Swiss HIV Cohort Study. Lancet. 2000;356:1800–1805. doi: 10.1016/s0140-6736(00)03232-3. [DOI] [PubMed] [Google Scholar]
- 12.De Luca A, Bugarini R, Lepri AC, Puoti M, Girardi E, Antinori A, et al. Coinfection with hepatitis viruses and outcome of initial antiretroviral regimens in previously naive HIV-infected subjects. Arch Intern Med. 2002;162:2125–2132. doi: 10.1001/archinte.162.18.2125. [DOI] [PubMed] [Google Scholar]
- 13.Torriani FJ, Rodriguez-Torres M, Rockstroh JK, Lissen E, Gonzalez-Garcia J, Lazzarin A, et al. Peginterferon Alfa-2a plus ribavirin for chronic hepatitis C virus infection in HIV-infected patients. N Engl J Med. 2004;351:438–450. doi: 10.1056/NEJMoa040842. [DOI] [PubMed] [Google Scholar]
- 14.Chung RT, Andersen J, Volberding P, Robbins GK, Liu T, Sherman KE, et al. Peginterferon Alfa-2a plus ribavirin versus interferon alfa-2a plus ribavirin for chronic hepatitis C in HIV-coinfected persons. N Engl J Med. 2004;351:451–459. doi: 10.1056/NEJMoa032653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Choo QL, Richman KH, Han JH, Berger K, Lee C, Dong C, et al. Genetic organization and diversity of the hepatitis C virus. Proc Natl Acad Sci U S A. 1991;88:2451–2455. doi: 10.1073/pnas.88.6.2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Domingo E, Martinez-Salas E, Sobrino F, de la Torre JC, Portela A, Ortin J, et al. The quasispecies (extremely heterogeneous) nature of viral RNA genome populations: biological relevance–a review. Gene. 1985;40:1–8. doi: 10.1016/0378-1119(85)90017-4. [DOI] [PubMed] [Google Scholar]
- 17.Martell M, Esteban JI, Quer J, Genesca J, Weiner A, Esteban R, et al. Hepatitis C virus (HCV) circulates as a population of different but closely related genomes: quasispecies nature of HCV genome distribution. J Virol. 1992;66:3225–3229. doi: 10.1128/jvi.66.5.3225-3229.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Steinhauer DA, Holland JJ. Rapid evolution of RNA viruses. Annu Rev Microbiol. 1987;41:409–433. doi: 10.1146/annurev.mi.41.100187.002205. [DOI] [PubMed] [Google Scholar]
- 19.Weiner AJ, Brauer MJ, Rosenblatt J, Richman KH, Tung J, Crawford K, et al. Variable and hypervariable domains are found in the regions of HCV corresponding to the flavivirus envelope and NS1 proteins and the pestivirus envelope glycoproteins. Virology. 1991;180:842–848. doi: 10.1016/0042-6822(91)90104-j. [DOI] [PubMed] [Google Scholar]
- 20.Hijikata M, Kato N, Ootsuyama Y, Nakagawa M, Ohkoshi S, Shimotohno K. Hypervariable regions in the putative glycoprotein of hepatitis C virus. Biochem Biophys Res Commun. 1991;175:220–228. doi: 10.1016/s0006-291x(05)81223-9. [DOI] [PubMed] [Google Scholar]
- 21.Shirai M, Arichi T, Chen M, Masaki T, Nishioka M, Ikeda K, et al. T cell recognition of hypervariable region-1 from hepatitis C virus envelope protein with multiple class II MHC molecules in mice and humans: preferential help for induction of antibodies to the hypervariable region. J Immunol. 1999;162:568–576. [PubMed] [Google Scholar]
- 22.Scarselli E, Cerino A, Esposito G, Silini E, Mondelli MU, Traboni C. Occurrence of antibodies reactive with more than one variant of the putative envelope glycoprotein (gp70) hypervariable region 1 in viremic hepatitis C virus-infected patients. J Virol. 1995;69:4407–4412. doi: 10.1128/jvi.69.7.4407-4412.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weiner AJ, Geysen HM, Christopherson C, Hall JE, Mason TJ, Saracco G, et al. Evidence for immune selection of hepatitis C virus (HCV) putative envelope glycoprotein variants: potential role in chronic HCV infections. Proc Natl Acad Sci U S A. 1992;89:3468–3472. doi: 10.1073/pnas.89.8.3468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Honda M, Kaneko S, Sakai A, Unoura M, Murakami S, Kobayashi K. Degree of diversity of hepatitis C virus quasispecies and progression of liver disease. Hepatology. 1994;20:1144–1151. [PubMed] [Google Scholar]
- 25.Farci P, Shimoda A, Coiana A, Diaz G, Peddis G, Melpolder JC, et al. The outcome of acute hepatitis C predicted by the evolution of the viral quasispecies. Science. 2000;288:339–344. doi: 10.1126/science.288.5464.339. [DOI] [PubMed] [Google Scholar]
- 26.Laskus T, Wilkinson J, Gallegos-Orozco JF, Radkowski M, Adair DM, Nowicki M, et al. Analysis of hepatitis C virus quasispecies transmission and evolution in patients infected through blood transfusion. Gastroenterology. 2004;127:764–776. doi: 10.1053/j.gastro.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 27.Farci P, Strazzera R, Alter HJ, Farci S, Degioannis D, Coiana A, et al. Early changes in hepatitis C viral quasispecies during interferon therapy predict the therapeutic outcome. Proc Natl Acad Sci U S A. 2002;99:3081–3086. doi: 10.1073/pnas.052712599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arenas JI, Gallegos-Orozco JF, Laskus T, Wilkinson J, Khatib A, Fasola C, et al. Hepatitis C virus quasi-species dynamics predict progression of fibrosis after liver transplantation. J Infect Dis. 2004;189:2037–2046. doi: 10.1086/386338. [DOI] [PubMed] [Google Scholar]
- 29.Toyoda H, Fukuda Y, Koyama Y, Takamatsu J, Saito H, Hayakawa T. Effect of immunosuppression on composition of quasispecies population of hepatitis C virus in patients with chronic hepatitis C coinfected with human immunodeficiency virus. J Hepatol. 1997;26:975–982. doi: 10.1016/s0168-8278(97)80105-5. [DOI] [PubMed] [Google Scholar]
- 30.Babik JM, Holodniy M. Impact of highly active antiretroviral therapy and immunologic status on hepatitis C virus quasispecies diversity in human immunodeficiency virus/hepatitis C virus-coinfected patients. J Virol. 2003;77:1940–1950. doi: 10.1128/JVI.77.3.1940-1950.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mao Q, Ray SC, Laeyendecker O, Ticehurst JR, Strathdee SA, Vlahov D, et al. Human immunodeficiency virus seroconversion and evolution of the hepatitis c virus quasispecies. J Virol. 2001;75:3259–3267. doi: 10.1128/JVI.75.7.3259-3267.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barkan SE, Melnick SL, Preston-Martin S, Weber K, Kalish LA, Miotti P, et al. The Women’s Interagency HIV Study. WIHS Collaborative Study Group. Epidemiology. 1998;9:117–125. [PubMed] [Google Scholar]
- 33.Lew J, Reichelderfer P, Fowler M, Bremer J, Carrol R, Cassol S, et al. Determinations of levels of human immunodeficiency virus type 1 RNA in plasma: reassessment of parameters affecting assay outcome. TUBE Meeting Workshop Attendees. Technology Utilization for HIV-1 Blood Evaluation and Standardization in Pediatrics. J Clin Microbiol. 1998;36:1471–1479. doi: 10.1128/jcm.36.6.1471-1479.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Germer JJ, Majewski DW, Rosser M, Thompson A, Mitchell PS, Smith TF, et al. Evaluation of the TRUGENE HCV 5′NC genotyping kit with the new GeneLibrarian module 3.1. 2 for genotyping of hepatitis C virus from clinical specimens. J Clin Microbiol. 2003;41:4855–4857. doi: 10.1128/JCM.41.10.4855-4857.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Laskus T, Radkowski M, Wang LF, Nowicki M, Rakela J. Uneven distribution of hepatitis C virus quasispecies in tissues from subjects with end-stage liver disease: confounding effect of viral adsorption and mounting evidence for the presence of low-level extrahepatic replication. J Virol. 2000;74:1014–1017. doi: 10.1128/jvi.74.2.1014-1017.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Laskus T, Wang LF, Rakela J, Vargas H, Pinna AD, Tsamandas AC, et al. Dynamic behavior of hepatitis C virus in chronically infected patients receiving liver graft from infected donors. Virology. 1996;220:171–176. doi: 10.1006/viro.1996.0297. [DOI] [PubMed] [Google Scholar]
- 37.Smith DB, McAllister J, Casino C, Simmonds P. Virus ‘quasispecies’: making a mountain out of a molehill? J Gen Virol. 1997;78:1511–1519. doi: 10.1099/0022-1317-78-7-1511. [DOI] [PubMed] [Google Scholar]
- 38.Forns X, Bukh J, Purcell RH, Emerson SU. How Escherichia coli can bias the results of molecular cloning: preferential selection of defective genomes of hepatitis C virus during the cloning procedure. Proc Natl Acad Sci U S A. 1997;94:13909–13914. doi: 10.1073/pnas.94.25.13909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kumar S, Tamura K, Nei M. MEGA3: Integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- 40.Booth JC, Kumar U, Webster D, Monjardino J, Thomas HC. Comparison of the rate of sequence variation in the hypervariable region of E2/NS1 region of hepatitis C virus in normal and hypogammaglobulinemic patients. Hepatology. 1998;27:223–227. doi: 10.1002/hep.510270134. [DOI] [PubMed] [Google Scholar]
- 41.Ni YH, Chang MH, Chen PJ, Hsu HY, Lu TW, Lin KH, et al. Decreased diversity of hepatitis C virus quasispecies during bone marrow transplantation. J Med Virol. 1999;58:132–138. doi: 10.1002/(sici)1096-9071(199906)58:2<132::aid-jmv6>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 42.Doughty AL, Painter DM, McCaughan GW. Post-transplant quasispecies pattern remains stable over time in patients with recurrent cholestatic hepatitis due to hepatitis C virus. J Hepatol. 2000;32:126–134. doi: 10.1016/s0168-8278(00)80199-3. [DOI] [PubMed] [Google Scholar]
- 43.Izopet J, Rostaing L, Sandres K, Cisterne JM, Pasquier C, Rumeau JL, et al. Longitudinal analysis of hepatitis C virus replication and liver fibrosis progression in renal transplant recipients. J Infect Dis. 2000;181:852–858. doi: 10.1086/315355. [DOI] [PubMed] [Google Scholar]
- 44.Qin H, Shire NJ, Keenan ED, Rouster SD, Eyster ME, Goedert JJ, et al. HCV quasispecies evolution: association with progression to end-stage liver disease in hemophiliacs infected with HCV or HCV/HIV. Blood. 2005;105:533–541. doi: 10.1182/blood-2004-04-1452. [DOI] [PubMed] [Google Scholar]
- 45.Roque-Afonso AM, Robain M, Simoneau D, Rodriguez-Mathieu P, Gigou M, Meyer L, et al. Influence of CD4 cell counts on the genetic heterogeneity of hepatitis C virus in patients coinfected with human immunodeficiency virus. J Infect Dis. 2002;185:728–733. doi: 10.1086/339297. [DOI] [PubMed] [Google Scholar]
- 46.Shuhart MC, Sullivan DG, Bekele K, Harrington RD, Kitahata MM, Mathisen TL, et al. HIV infection and antiretroviral therapy: effect on hepatitis C virus quasispecies variability. J Infect Dis. 2006;193:1211–1218. doi: 10.1086/502974. [DOI] [PubMed] [Google Scholar]
- 47.Mellors JW, Rinaldo CR, Jr, Gupta P, White RM, Todd JA, Kingsley LA. Prognosis in HIV-1 infection predicted by the quantity of virus in plasma. Science. 1996;272:1167–1170. doi: 10.1126/science.272.5265.1167. [DOI] [PubMed] [Google Scholar]
- 48.Gonzalez-Peralta RP, Qian K, She JY, Davis GL, Ohno T, Mizokami M, et al. Clinical implications of viral quasispecies heterogeneity in chronic hepatitis C. J Med Virol. 1996;49:242–247. doi: 10.1002/(SICI)1096-9071(199607)49:3<242::AID-JMV14>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 49.Naito M, Hayashi N, Moribe T, Hagiwara H, Mita E, Kanazawa Y, et al. Hepatitis C viral quasispecies in hepatitis C virus carriers with normal liver enzymes and patients with type C chronic liver disease. Hepatology. 1995;22:407–412. [PubMed] [Google Scholar]
- 50.Benhamou Y, Bochet M, Di Martino V, Charlotte F, Azria F, Coutellier A, et al. Liver fibrosis progression in human immunodeficiency virus and hepatitis C virus coinfected patients. The Multivirc Group. Hepatology. 1999;30:1054–1058. doi: 10.1002/hep.510300409. [DOI] [PubMed] [Google Scholar]
- 51.Di Martino V, Rufat P, Boyer N, Renard P, Degos F, Martinot-Peignoux M, et al. The influence of human immunodeficiency virus coinfection on chronic hepatitis C in injection drug users: A long-term retrospective cohort study. Hepatology. 2001;34:1193–1199. doi: 10.1053/jhep.2001.29201. [DOI] [PubMed] [Google Scholar]
- 52.Qurishi N, Kreuzberg C, Luchters G, Effenberger W, Kupfer B, Sauerbruch T, et al. Effect of antiretroviral therapy on liver-related mortality in patients with HIV and hepatitis C virus coinfection. Lancet. 2003;362:1708–1713. doi: 10.1016/S0140-6736(03)14844-1. [DOI] [PubMed] [Google Scholar]
- 53.Vento S, Garofano T, Renzini C, Casali F, Ferraro T, Concia E. Enhancement of hepatitis C virus replication and liver damage in HIV-coinfected patients on antiretroviral combination therapy. AIDS. 1998;12:116–117. [PubMed] [Google Scholar]
- 54.Chung RT, Evans SR, Yang Y, Theodore D, Valdez H, Clark R, et al. Immune recovery is associated with persistent rise in hepatitis C virus RNA, infrequent liver test flares, and is not impaired by hepatitis C virus in co-infected subjects. AIDS. 2002;16:1915–1923. doi: 10.1097/00002030-200209270-00008. [DOI] [PubMed] [Google Scholar]
- 55.Blackard JT, Yang Y, Bordoni P, Sherman KE, Chung RT. Hepatitis C virus (HCV) diversity in HIV-HCV-coinfected subjects initiating highly active antiretroviral therapy. J Infect Dis. 2004;189:1472–1481. doi: 10.1086/382959. [DOI] [PubMed] [Google Scholar]
- 56.Farci P, Alter HJ, Govindarajan S, Wong DC, Engle R, Lesniewski RR, et al. Lack of protective immunity against reinfection with hepatitis C virus. Science. 1992;258:135–140. doi: 10.1126/science.1279801. [DOI] [PubMed] [Google Scholar]
- 57.Okamoto H, Mishiro S, Tokita H, Tsuda F, Miyakawa Y, Mayumi M. Superinfection of chimpanzees carrying hepatitis C virus of genotype II/1b with that of genotype III/2a or I/1a. Hepatology. 1994;20:1131–1136. [PubMed] [Google Scholar]
- 58.Prince AM, Brotman B, Huima T, Pascual D, Jaffery M, Inchauspe G. Immunity in hepatitis C infection. J Infect Dis. 1992;165:438–443. doi: 10.1093/infdis/165.3.438. [DOI] [PubMed] [Google Scholar]
- 59.Farci P, Alter HJ, Wong DC, Miller RH, Govindarajan S, Engle R, et al. Prevention of hepatitis C virus infection in chimpanzees after antibody-mediated in vitro neutralization. Proc Natl Acad Sci U S A. 1994;91:7792–7796. doi: 10.1073/pnas.91.16.7792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mehta SH, Cox A, Hoover DR, Wang XH, Mao Q, Ray S, et al. Protection against persistence of hepatitis C. Lancet. 2002;359:1478–1483. doi: 10.1016/S0140-6736(02)08435-0. [DOI] [PubMed] [Google Scholar]
- 61.Lanford RE, Guerra B, Chavez D, Bigger C, Brasky KM, Wang XH, et al. Cross-genotype immunity to hepatitis C virus. J Virol. 2004;78:1575–1581. doi: 10.1128/JVI.78.3.1575-1581.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bassett SE, Guerra B, Brasky K, Miskovsky E, Houghton M, Klimpel GR, et al. Protective immune response to hepatitis C virus in chimpanzees rechallenged following clearance of primary infection. Hepatology. 2001;33:1479–1487. doi: 10.1053/jhep.2001.24371. [DOI] [PubMed] [Google Scholar]
- 63.Houghton M, Abrignani S. Prospects for a vaccine against the hepatitis C virus. Nature. 2005;436:961–966. doi: 10.1038/nature04081. [DOI] [PubMed] [Google Scholar]
- 64.Eyster ME, Sherman KE, Goedert JJ, Katsoulidou A, Hatzakis A. Prevalence and changes in hepatitis C virus genotypes among multitransfused persons with hemophilia. The Multicenter Hemophilia Cohort Study. J Infect Dis. 1999;179:1062–1069. doi: 10.1086/314708. [DOI] [PubMed] [Google Scholar]
- 65.Herring BL, Page-Shafer K, Tobler LH, Delwart EL. Frequent hepatitis C virus superinfection in injection drug users. J Infect Dis. 2004;190:1396–1403. doi: 10.1086/424491. [DOI] [PubMed] [Google Scholar]
- 66.Laskus T, Wang LF, Radkowski M, Vargas H, Nowicki M, Wilkinson J, et al. Exposure of hepatitis C virus (HCV) RNA-positive recipients to HCV RNA-positive blood donors results in rapid predominance of a single donor strain and exclusion and/or suppression of the recipient strain. J Virol. 2001;75:2059–2066. doi: 10.1128/JVI.75.5.2059-2066.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Di Liberto G, Roque-Afonso AM, Kara R, Ducoulombier D, Fallot G, Samuel D, et al. Clinical and therapeutic implications of hepatitis C virus compartmentalization. Gastroenterology. 2006;131:76–84. doi: 10.1053/j.gastro.2006.04.016. [DOI] [PubMed] [Google Scholar]
- 68.Guadagnino V, Stroffolini T, Rapicetta M, Costantino A, Kondili LA, Menniti-Ippolito F, et al. Prevalence, risk factors, and genotype distribution of hepatitis C virus infection in the general population: a community-based survey in southern Italy. Hepatology. 1997;26:1006–1011. doi: 10.1002/hep.510260431. [DOI] [PubMed] [Google Scholar]
- 69.Carneiro M, Yu XF, Lyles C, Templeton A, Weisstein AE, Safaeian M, et al. The effect of drug-injection behavior on genetic evolution of HIV-1. J Infect Dis. 1999;180:1025–1032. doi: 10.1086/315044. [DOI] [PubMed] [Google Scholar]
- 70.Tsui R, Herring BL, Barbour JD, Grant RM, Bacchetti P, Kral A, et al. Human immunodeficiency virus type 1 superinfection was not detected following 215 years of injection drug user exposure. J Virol. 2004;78:94–103. doi: 10.1128/JVI.78.1.94-103.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Peterson PK, Gekker G, Schut R, Hu S, Balfour HH, Jr, Chao CC. Enhancement of HIV-1 replication by opiates and cocaine: the cytokine connection. Adv Exp Med Biol. 1993;335:181–188. doi: 10.1007/978-1-4615-2980-4_26. [DOI] [PubMed] [Google Scholar]