Abstract
Mutation of staphylococcal accessory regulator (sarA) results in increased production of extracellular proteases in Staphylococcus aureus, which has been correlated with decreased biofilm formation and decreased accumulation of extracellular toxins. We used murine models of implant-associated biofilm infection and S. aureus bacteremia (SAB) to compare virulence of USA300 strain LAC, its isogenic sarA mutant, and derivatives of each of these strains with mutations in all 10 of the genes encoding recognized extracellular proteases. The sarA mutant was attenuated in both models, and this was reversed by eliminating production of extracellular proteases. To examine the mechanistic basis, we identified proteins impacted by sarA in a protease-dependent manner. We identified 253 proteins where accumulation was reduced in the sarA mutant compared to the parent strain, and was restored in the sarA/protease mutant. Additionally, in SAB, the LAC protease mutant exhibited a hypervirulent phenotype by comparison to the isogenic parent strain, demonstrating that sarA also positively regulates production of virulence factors, some of which are subject to protease-mediated degradation. We propose a model in which attenuation of sarA mutants is defined by their inability to produce critical factors and simultaneously repress production of extracellular proteases that would otherwise limit accumulation of virulence factors.
AUTHOR SUMMARY
Staphylococcus aureus infections associated with biofilms are difficult to treat without surgery. Mutation of the staphylococcal accessory regulator (sarA) limits biofilm formation to an extent that enhances the ability to treat these infections using antibiotics alone. However, it also has other consequences including the increased transcription of genes encoding toxins. In the laboratory, this is not apparent because the toxins are degraded by extracellular proteases. Thus, the critical question is whether the toxin phenotype of sarA mutants in vivo is defined by increased transcription or decreased accumulation, with the first potentially compromising therapeutic strategies targeting sarA to limit biofilm formation. We demonstrate that this is not the case, in that mutation of sarA reduced virulence in animal models of bacteremia and biofilm-associated infection. We also demonstrate that the increased production of proteases in sarA mutants limits the accumulation many factors known to contribute to virulence. Thus, we propose a model in which sarA plays an important role in acute and chronic forms of S. aureus infection owing to its ability to simultaneously promote the production of important virulence factors and repress the production of proteases that would otherwise limit the accumulation of these factors.
INTRODUCTION
Regulatory circuits in Staphylococcus aureus are complex and highly interactive. Given our interest in biofilm-associated musculoskeletal infections, we have focused much of our effort on the staphylococcal accessory regulator (sarA), mutation of which limits biofilm formation to a degree that can be correlated with increased antibiotic susceptibility under in vitro and in vivo conditions (Weiss et al., 2009; Weiss et al., 2009). Mutation of sarA has also been shown to limit virulence in animal models of endocarditis, endopthalmitis, and septic arthritis (Blevins et al., 2003; Booth et al., 1997; Cheung et al., 1994; Nilsson et al., 1996). The regulatory functions of sarA are generally attributed to transcriptional changes mediated by the binding of SarA to cis elements associated with the promoters of its target genes, one of which is the accessory gene regulator (agr) (Blevins et al., 2002; Didier et al., 2010; Rechtin et al., 1999; Reyes et al., 2011). Efforts to define a consensus SarA binding site have met with limited success (Chien et al., 1999; Sterba et al., 2003), and it has been suggested that SarA functions as an architectural protein with broad binding specificity (Fujimoto et al., 2009). However, mutation of sarA has also been associated with changes in mRNA stability, particularly during the post-exponential and stationary growth phases (Roberts et al., 2006; Morrison et al., 2012). Thus, SarA modulates the production of S. aureus proteins at the mRNA level via both transcriptional and post-transcriptional pathways.
The relative impact of these two pathways remains unclear, but it is clear that mutation of sarA has a global effect on the abundance of many S. aureus transcripts (Cassat et al., 2006; Dunman et al., 2001). Included among these are the transcripts encoding multiple extracellular proteases, all of which are present in increased amounts in sarA mutants (Cassat et al., 2006). This is reflected in the increased accumulation of the corresponding proteases, with the amounts of the metalloproteinase aureolysin (Aur), the staphylococcal serine glutamyl endopeptidase A (SspA) and its co-transcribed cysteine protease (SspB), and the cysteine protease staphopain (ScpA) all being elevated in sarA mutants (Jones et al., 2008). The increased production of these proteases has been correlated with a reduced capacity to form a biofilm (Beenken et al., 2010), and reduced accumulation of critical extracellular toxins including alpha toxin and phenol-soluble modulins (Zielinska et al., 2011), thus suggesting that the impact of sarA on protease production also makes an important contribution to multiple phenotypes in S. aureus.
The phenotypes that would be predicted based on the accumulation of mRNA vs. the accumulation of the corresponding proteins in sarA mutants are sometimes opposite with respect to each other. For example, in S. aureus isolates of the USA300 clonal lineage, mutation of sarA results in increased accumulation of the hla transcript but decreased accumulation of alpha toxin owing to protease-mediated degradation (Zielinska et al., 2011). Alpha toxin plays an important role in the virulence of USA300 isolates (Li et al., 2010), and this contrast makes it impossible to predict the impact of mutating sarA on the virulence of such isolates. This is particularly true since many reports concluding that sarA mutants are attenuated in animal models of S. aureus infection were based on studies done with the 8325-4 strain RN6390 (Booth et al., 1997; Cheung et al., 1994; Kielian et al., 2001), a strain in which mutation of sarA results in decreased rather than increased hla transcription (Blevins et al., 2002; Cheung et al., 1994). This has been attributed to the rsbU and tcaR mutations present in 8325 strains or, more precisely, the impact of these mutations on the regulatory functions of sigB and sarS (Oscarsson et al., 2006). In this report, we investigated both of these issues with a specific focus on isolates of the USA300 clonal lineage.
RESULTS AND DISCUSSION
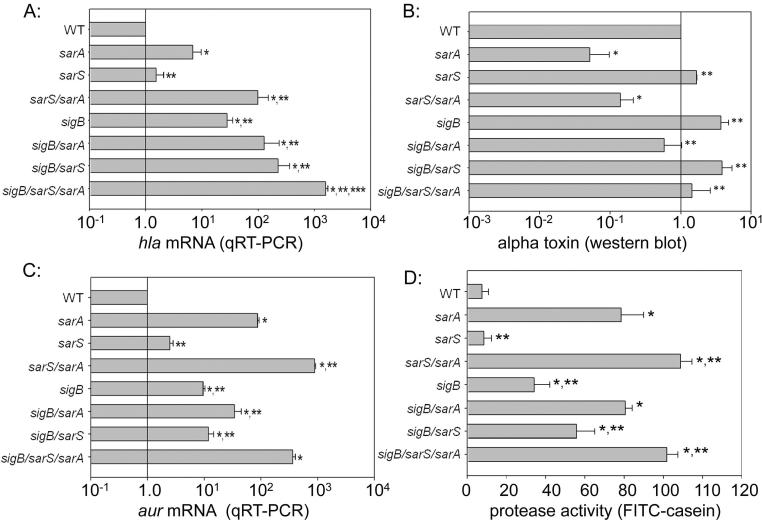
We first investigated the role of sigB and sarS in defining the impact of sarA on the accumulation of the hla transcript. In the USA300 strain FPR3757, mutation of either or both of these loci did impact the overall levels of hla mRNA, but neither mutation, either alone or in combination with each other, reversed the increase in hla transcription observed in an FPR3757 sarA mutant (Fig. 1A). Additionally, mutation of sarA resulted in decreased accumulation of alpha toxin irrespective of the functional status of sarS and/or sigB (Fig. 1B). The decreased accumulation of alpha toxin was correlated with the level of aur transcription (Fig. 1C) and overall protease activity in all strains (Fig. 1D), thus confirming the potential phenotypic contrast in the impact of sarA on the alpha toxin phenotype.
Figure 1. Impact of sarA, sarS, and sigB on hla expression and accumulation of alpha toxin.
A: Relative levels of hla transcript were assessed by qRT-PCR in stationary phase (16 hr) cultures of FPR3757 and derivatives with mutations in the indicated genes. Levels in the parent strain (WT) were set to 1.0, with the levels observed in each of the other strains shown relative to this standard. B: Relative amounts of alpha toxin were assessed by western blot of conditioned medium from stationary phase cultures of the indicated strains. C: Relative levels of aur transcript were assessed by qRT-PCR in stationary phase (16 hr) cultures FPR3757 and derivatives with mutations in the indicated genes. Levels observed in the parent strain (WT) were set to 1.0, with levels in the other strains shown relative to this standard. D: Overall protease activity was assessed in conditioned medium from stationary phase cultures. Single asterisk indicates a statistically significant difference by comparison to the parent strain. Double asterisk indicates statistical significance by comparison to the isogenic sarA mutant. Triple asterisk indicates significance by comparison to the sarS/sarA mutant.
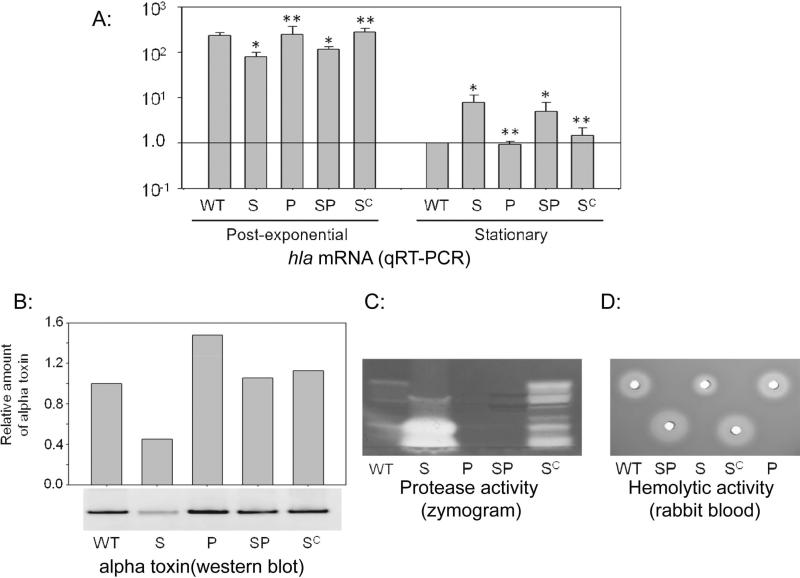
At the same time, these results are based on in vitro experiments, and under these conditions it would be anticipated that the impact of extracellular proteases would be maximized owing to the physical proximity of the proteases and their targets. Thus, the critical question is whether the in vivo alpha toxin phenotype of a sarA mutant is defined by the impact of sarA on hla transcription or by its impact on protease production. To investigate this, we employed the USA300 strain LAC and its isogenic sarA mutant with mutations inactivating all 10 of the genes encoding extracellular proteases (aur, scpA, splABCDEF, and sspAB) (Wörman et al., 2011); these strains are referred to hereinafter as protease and sarA/protease mutants respectively. It is perhaps important to note in this regard that FPR3757 and LAC are essentially identical at both the genotypic and phenotypic levels (Kennedy et al., 2008). Subsequent studies confirmed all of the expected phenotypes in these strains with respect to hla transcription (Fig. 2A), accumulation of alpha toxin (Fig. 2B), protease production (Fig. 2C), and overall hemolytic activity as assessed using rabbit blood agar (Fig. 2D). They also confirmed the reduced accumulation of alpha toxin and reduced hemolytic activity observed in the sarA mutant were reversed in the sarA/protease mutant and that all of the sarA-defined phenotypes could be complemented with a functional copy of sarA.
Figure 2. Correlation between sarA, extracellular proteases, and hemolytic activity.
A: Relative levels of hla transcript were assessed by qRT-PCR in LAC, its sarA mutant (S), protease-deficient derivatives of each of these strains (P, SP), and the sarA mutant complemented with a functional copy of sarA (SC). RNA was isolated from the post-exponential (OD560 = 3.0) and stationary (16 hr) growth phases. Levels observed in the parent strain (WT) during the stationary growth phase were set to 1.0, with the levels observed in all other samples shown relative to this standard. Single asterisk indicates a statistically significant difference by comparison to the parent strain. Double asterisk indicates statistical significance by comparison to the isogenic sarA mutant. Accumulation of alpha toxin (B), protease activity (C), and hemolytic activity (D) was assessed in stationary phase samples obtained from the same strains.
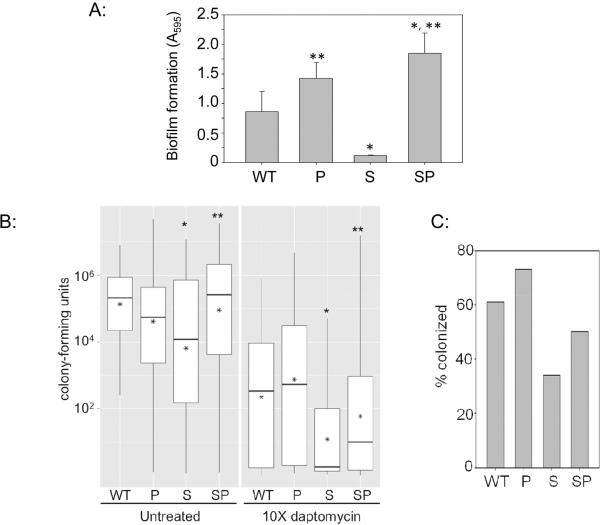
Under in vitro conditions, eliminating production of all 10 extracellular proteases also fully restored biofilm formation in the LAC sarA mutant (Fig. 3A). To assess the in vivo relevance of these observations, we first examined the relative capacity of LAC, its sarA mutant, and their protease-deficient derivatives to form a biofilm in vivo using a murine model of catheter-associated biofilm formation (Thurlow et al., 2011). Comparisons were done with and without daptomycin treatment as previously described in the context of sarA itself (Weiss et al., 2009). Mutation of sarA was found to limit biofilm formation by comparison to LAC, and this could be correlated with increased daptomycin susceptibility (Fig. 3B). Additionally, while the effect was not absolute, both the reduced capacity to form a biofilm, and the increased susceptibility of the sarA mutant, were reversed to a statistically significant degree in the LAC sarA/protease mutant. This increased susceptibility was reflected in both average colony counts per catheter (Fig. 3B) and in the overall percentage of catheters that remained colonized at any level after exposure to daptomycin, with this percentage being lowest with catheters colonized with the sarA mutant and highest with those colonized with the LAC protease mutant (Fig. 3C), the latter suggesting that biofilm formation is limited by the production of extracellular proteases even in LAC. To the extent that this reflects a reduced capacity to form a biofilm, which we believe to be the case given that mutation of sarA does not alter daptomycin susceptibility as assessed under in vitro conditions (Beenken et al., 2012), this confirms that the increased production of extracellular proteases plays an important role defining the biofilm-deficient phenotype of sarA mutants even under in vivo conditions. At the same time, eliminating protease production did not fully restore biofilm formation in the sarA mutant as assessed either directly or in the context of daptomycin exposure, thus suggesting that mutation of sarA also impacts the accumulation of relevant virulence factors in a protease-independent manner.
Figure 3. Impact of sarA and proteases on biofilm formation in vitro and in vivo.
A: Biofilm formation was assessed in vitro in LAC (WT), its sarA mutant (S), and protease-deficient derivatives of each of these strains (P, SP) using a microtiter plate assay (Beenken et al., 2010). B: A murine model of implant-associated infection (Weiss et al., 2009) was used to assess the capacity of the same strains to form a biofilm and relative susceptibility to daptomycin in the context of biofilm-associated infection. Results are shown as colony counts per catheter. Boxes indicate the 25th and 75th percentiles for each experimental group, with the asterisk within each box indicating the mean and the horizontal line indicating the median. Single asterisk above a box indicates statistical significance by comparison to the parent strain (WT). Double asterisk indicates significance by comparison to the sarA (S) mutant. C: Percentage of catheters that remained colonized at any level following exposure to daptomycin.
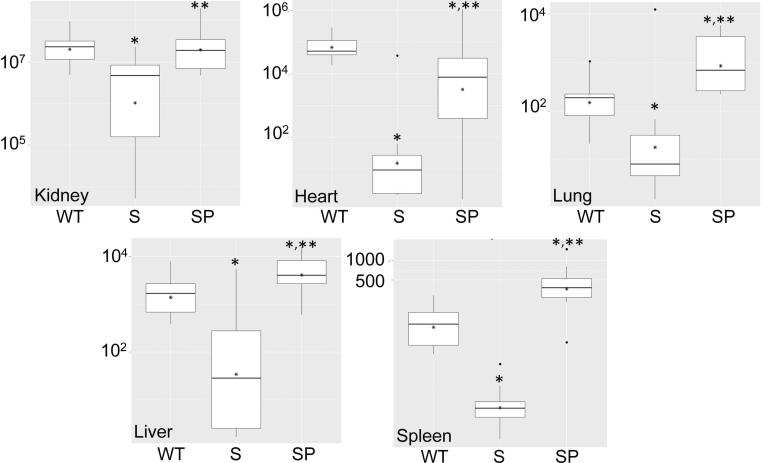
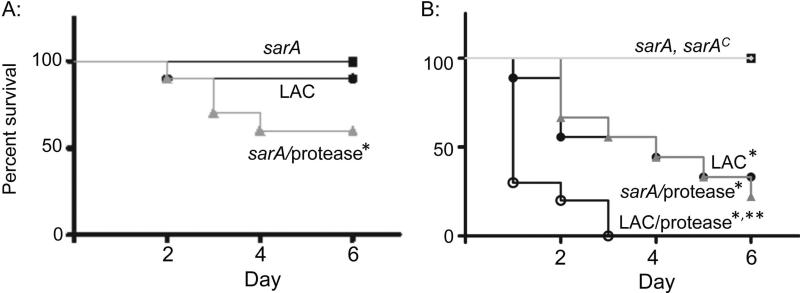
We also assessed the impact of extracellular proteases on the phenotype of sarA mutants using a murine model of S. aureus bacteremia (SAB) (Blevins et al., 2003). This was done in two independent experiments, with the first limited to LAC, its sarA mutant, and the protease-deficient sarA mutant. Results were assessed 6 days post-infection or, in those cases in which compassionate euthanasia was required before this time, immediately upon death. Based on bacterial counts in liver, spleen, kidney, heart, and lungs, the results confirmed that, by comparison to LAC, the sarA mutant was attenuated to a significant degree and that this was reversed, in most cases to a statistically-significant degree, by eliminating the production of extracellular proteases (Fig. 4). Also, all of the mice infected with the sarA mutant survived, while 40% of those infected with the sarA/protease mutant died, a proportion that exceeded even that observed with the LAC parent strain (Fig. 5A).
Figure 4. Relative virulence of sarA and sarA/protease mutants in SAB.
Mice were infected by tail vein injection of 108 cfu of LAC (WT), its sarA mutant (S), or its protease-deficient sarA mutant (SP). Tissues were harvested after 6 days or immediately after compassionate euthanasia. Analysis of soft tissue samples was done by colony count. Boxes indicate the 25th and 75th percentiles for each experimental group, with the asterisk within each box indicating the mean and the horizontal line indicating the median. Single asterisk above a box indicates statistical significance by comparison to the parent strain (WT). Double asterisk indicates significance by comparison to the sarA (S) mutant.
Figure 5. Impact of sarA and protease production on virulence as assessed by lethality.
Mice were infected by tail vein injection of 108 cfu of LAC (WT), its sarA mutant (S), or its protease-deficient sarA mutant (SP). Results shown in panels A and B are from two independent experiments. Single asterisk indicates statistical significance by comparison to the sarA mutant. Double asterisk indicates statistical significance by comparison to LAC.
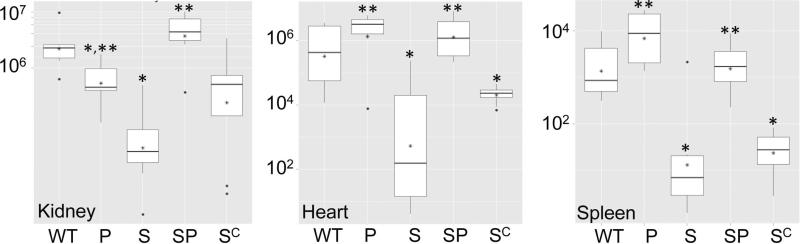
When this experiment was repeated using a protease-deficient derivative of LAC and the complemented sarA mutant, only 30% of mice infected with the LAC parent strain survived after 6 days, while 100% infected with the isogenic sarA mutant survived (Fig. 5B). Moreover, the virulence of the sarA mutant as assessed based on lethality was fully restored by eliminating the production of extracellular proteases. The attenuation of the sarA mutant, and its reversal in the sarA/protease mutant, were also evident in colony counts of kidney, spleen, and heart (Fig. 6), which were the only tissues examined in this 2nd experiment. The only inconsistency was that virulence was not restored in the complemented sarA mutant as assessed based on lethality (Fig. 5B). Although incomplete, complementation was observed based on soft tissue colony counts (Fig. 6). Characterization of individual colonies isolated from infected mice confirmed that, when averaged across all tissues, >90% of the recovered colonies had lost the complementing plasmid (data not shown), and this presumably accounts for this partial complementation. In this respect it is important to note that, as was observed with this same plasmid construct (pSarA) in our previous studies (Blevins et al., 1999), complementation of the LAC sarA mutation was achieved in vitro (Fig. 2), thus confirming that this partial complementation is due to plasmid instability in vivo rather than an undetected defect in the LAC sarA mutant.
Figure 6. Impact of protease production in LAC.
Mice were infected by tail vein injection of 108 cfu of LAC (WT), its sarA mutant (S), protease-deficient derivatives of each (P and SP respectively), and the complemented sarA mutant (SC). Tissues were harvested after 6 days or immediately after compassionate euthanasia. Analysis of soft tissue samples was done by colony count. Boxes indicate the 25th and 75th percentiles for each experimental group, with the asterisk within each box indicating the mean and the horizontal line indicating the median. Single asterisk above a box indicates statistical significance by comparison to the parent strain (WT). Double asterisk indicates significance by comparison to the sarA (S) mutant.
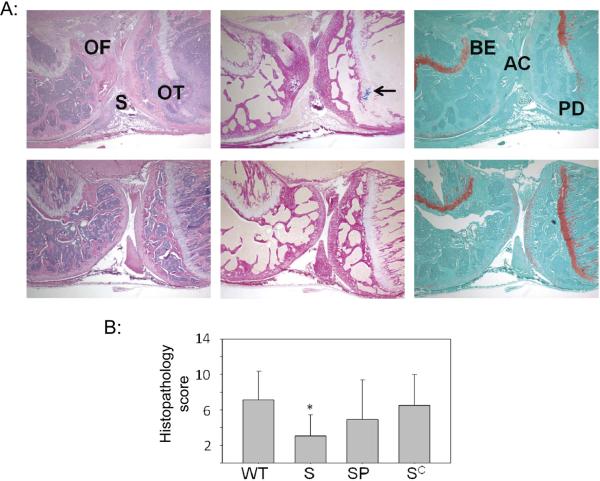
The impact of sarA and the increased production of extracellular proteases was also assessed based on development of morphologic features of septic arthritis and osteomyelitis (Fig. 7A). As assessed based on overall histopathological scores based on inflammatory changes in the synovial space and bone (see Materials and Methods), mutation of sarA was also associated with reduced virulence in this context. Additionally, both complementation of the sarA defect and elimination of extracellular protease production reversed this attenuation (Fig. 7B). Thus, when taken together, these results demonstrate that it is possible to phenotypically complement the sarA defect in LAC to a significant degree in murine models of implant-associated infection and SAB by eliminating its ability to produce extracellular proteases.
Figure 7. Histological evidence of bone and joint involvement.
A: Representative images illustrating histopathological changes in bone and joint infection following bacteremia. Top panel is from a mouse exhibiting a high pathological score defined by acute inflammation (2+) in the synovium (S) and severe osteomyelitis (3+) in both femur (OF) and tibia (OT). The bottom panel of photomicrographs depicts low pathologic score with no acute inflammation, Gram-positive microorganisms or bone/cartilage erosion seen. Stains employed in both panels are (left to right): Hematoxylin and eosin (H&E) to assess overall pathology, Gram-stain to detect intraosseous Gram-positive cocci (arrow, top panel), and Safranin O, which exhibits orange staining of proteoglycan and highlights the loss of articular cartilage (AC), bone erosion (BE) and physis destruction (PD). Magnification = 40×. B: Comparison of overall histopathological scores in mice infected with LAC (WT), its isogenic sarA mutant (S), a protease-deficient sarA mutant (SP), and the complemented sarA mutants (SC). No results are shown for mice infected with the LAC protease mutant because none survived long enough to develop histopathological evidence of disease.
At the same time, if the increased production of extracellular proteases were solely responsible for the attenuation of sarA mutants, then LAC protease and sarA/protease mutants would be expected to exhibit the same in vivo phenotypes, and this was not the case. This was most evident in the observation that none of the mice infected with the LAC protease mutant survived the 6-day post-infection period (Fig. 5). This hypervirulence was not evident when assessed based on colony counts, and in one case (kidney), the LAC protease mutant was significantly attenuated by comparison to its isogenic parent strain (Fig. 6). However, these results must be interpreted with caution in that all mice infected with the LAC protease mutant died by 3 days post-infection, thus compromising the ability to make direct comparisons between the results obtained with these strains beyond those associated with lethality itself.
Thus, while we placed a primary emphasis in this study on determining the extent to which we could “complement” the sarA defect by eliminating protease production, we also found that we could, in effect, “complement” the hypervirulence phenotype of the LAC protease mutant by eliminating sarA. To the extent that this hypervirulent phenotype required a functional sarA locus, this presumably reflects the fact that sarA is known to modulate the production of important virulence factors owing to its impact on transcription and/or transcript stabilization. In the context of this report, these results also suggest that at least a subset of these virulence factors is relatively insensitive to protease-mediated degradation. However, this too must be interpreted with caution in that it is also possible that the hypervirulence of the LAC protease mutant merely reflects a protease-defined gradient in the accumulation of specific virulence factors. Support for this hypothesis comes from the observation that the accumulation of alpha toxin was found to increase progressively in a fashion that paralleled the “virulence gradient” observed in our SAB experiments, with the amount present being highest in the LAC protease mutant and lowest in the sarA mutant (Fig. 2).
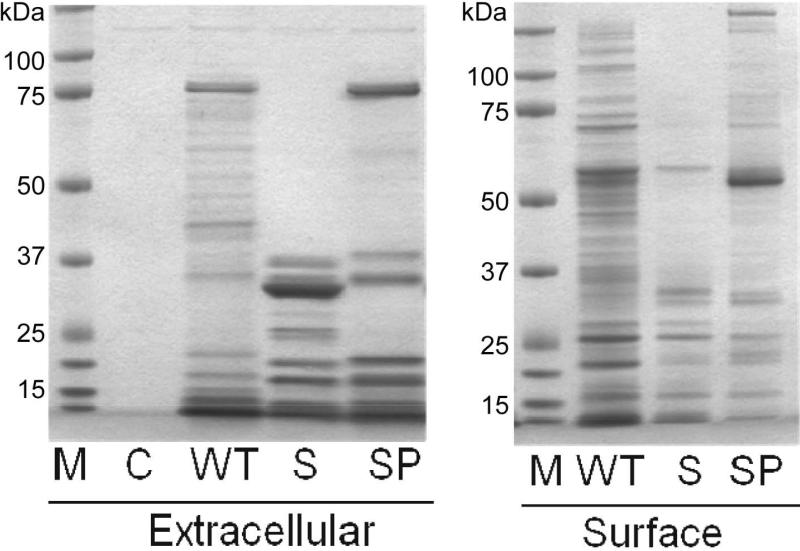
When taken together, these results suggest that the regulatory functions of sarA as defined under in vivo conditions is not limited to its impact on the production of important virulence factors but requires the simultaneous repression of extracellular protease production. This emphasizes the need to understand both sides of this regulatory equation. To this end, we used a GeLC-MS/MS proteomics approach to compare overall protein profiles of LAC, its sarA mutant, and its sarA/protease mutant. In an effort to be as comprehensive as possible, we combined the results obtained with samples from conditioned medium and cellular extracts enriched for surface proteins, both of which were prepared from stationary phase cultures and both of which were first resolved by SDS-PAGE prior to quantitative analysis (Fig. 8). Nine of the 10 extracellular proteases were detected in these studies, the only exception being SplF, and all that were detected were present in increased amounts in the sarA mutant by comparison to its LAC parent strain (Fig. 9). Some were also detected in samples prepared from the sarA/protease mutant, but in all cases the total spectral counts were very low (≤4). Based on this, we defined this as background signal, with any protein with a spectral count ≤4 in all 3 strains being excluded from further analysis. We then defined a protein as being impacted by mutation of sarA in a protease-dependent manner based on its abundance in the sarA mutant being ≤50% of that observed in the parent strain and its abundance in the sarA/protease mutant being ≥200% of that observed in the sarA mutant or ≥95% of that observed in the parent strain.
Figure 8. Correlations between sarA, extracellular proteases, and protein profiles.
Comparisons were made by SDS-PAGE using conditioned medium (extracellular) or samples enriched for surface-assciated proteins (right). Strain examined included LAC (WT), its sarA mutant (S), and its sarA/protease mutant (SP). All samples were prepared from stationary phase (16 hr) cultures. M refers to molecular size standards, while C refers to sterile, non-conditioned medium (C).
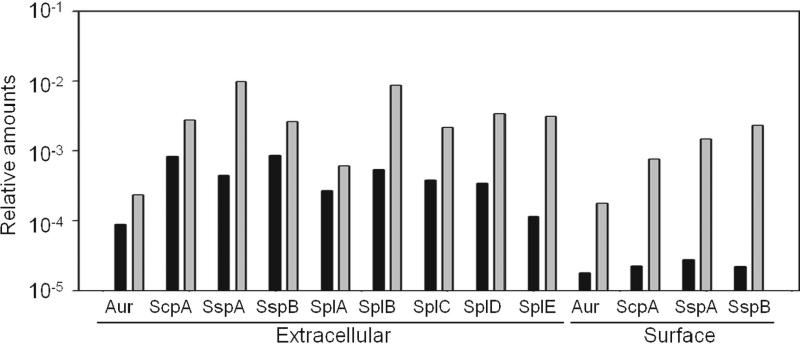
Figure 9. Impact of sarA on extracellular proteases.
Results illustrate relative amounts of the indicated proteases as determined by LC-MS/MS in LAC (black) and its isogenic sarA mutant (gray). Results are shown for conditioned medium (extracellular) and samples enriched for surface-associated proteins (surface). Proteases not listed were present in amounts below our experimental definition of background (see text). All proteases where present in amounts below this background level in samples prepared from the isogenic LAC sarA/protease mutant (data not shown).
A total of 404 proteins were identified in conditioned medium, with 565 identified in preparations enriched for surface proteins (Tables S1 and S2 respectively). When overlap between the two datasets was taken into account, we could identify and quantify 640 different S. aureus proteins. In some cases, spectral counts were below the cut-off in one but not all samples; we did not exclude these proteins from further analysis, but we would note that this precludes the ability to obtain an accurate assessment of the impact of sarA and/or protease production on their accumulation. After removing overlap between the two datasets, we identified 253 proteins that were present in decreased amounts in sarA mutants owing at least partially to protease-mediated degradation. Included among these were protein A, multiple phenol-soluble modulins (PSMs), and alpha toxin, the identification of which is consistent with our earlier, more targeted studies (Zielinska et al., 2011; Mrak et al., 2012), thus providing validation of our experimental approach.
Included among these 253 proteins were a large number with presumed intracellular functions, an observation that is consistent with previous reports describing global proteomics comparisons Jones et al., 2008) and perhaps reflects our use of samples from stationary phase cultures. None of these should be excluded from further consideration, particularly given the growing number of reports implicating proteins involved in central metabolic processes in the virulence of S. aureus (Somerville et al., 2009). However, we focused our analysis on the production of extracellular and surface-associated proteins previously associated with virulence properties including biofilm formation.
Included among these was gamma hemolysin component A, which is potentially significant in that gamma hemolysin has been shown to function in conjunction with alpha toxin to promote the development of septic arthritis (Nilsson et al., 1999). Also included were two different lipases, staphylokinase, and catalase (Table 1), all of which have also been previously associated with virulence either directly or indirectly via their impact on colonization (Dinges et al., 2000; Hu et al., 2012; Jacobsson et al., 2010; Kwiecinski et al., 2010; Park et al., 1996; Schlievert et al., 2000). The IgG binding proteins Sbi and Spa, which are produced in both surface-associated and extracellular forms (Merino et al., 2009; Haupt et al., 2008; Smith et al., 2012), were also identified in our experiments (Table 1). Spa met the parameters used to define a protein as modulated by sarA in a protease-dependent manner only in our conditioned medium samples, but even in samples enriched for surface-associated proteins, its abundance was increased almost 2-fold in the sarA/protease mutant by comparison to the isogenic sarA mutant (Table S2). This suggests that surface-anchored Spa is more resistant to protease-mediated degradation than extracellular Spa (eSpa), but given that both forms contribute to biofilm formation (Merino et al., 2009), it is likely that the increased production of extracellular proteases has a significant impact on the Spa phenotype of S. aureus sarA mutants in the context of biofilm formation. Sbi has not been previously associated with biofilm formation, but it has been shown to bind human complement Factor H and C3b and act as a complement inhibitor (Haupt et al., 2008).
Table 1.
Proteins defined as impacted by sarA in a protease-dependent manner.
| Accession Number | Molecular Weight | Gene | Protein name | WT | S | SP |
|---|---|---|---|---|---|---|
| Extracellular | ||||||
| Q2FJU4 | 76 kDa | Triacylglycerol lipase | 1317 | 20 | 1088 | |
| Q2FDJ1 | 77 kDa | lip | Triacylglycerol lipase | 555 | 2 | 420 |
| Q2FHS2 | 36 kDa | hla | Alpha-hemolysin | 206 | 36 | 340 |
| Q2FI25 | 137 kDa | atl | Autolysin | 148 | 23 | 136 |
| Q2FIL7 | 47 kDa | eno | Enolase | 122 | 1 | 90 |
| Q2FIK2 | 25 kDa | nuc | Thermonuclease | 96 | 40 | 744 |
| P0C7Y0 | 2 kDa | psmA1 | Phenol-soluble modulin alpha 1 peptide | 80 | 1 | 39 |
| Q2FH99 | 58 kDa | katA | Catalase | 63 | 9 | 43 |
| Q2FKE8 | 56 kDa | spa | Immunoglobulin G binding protein A | 49 | 23 | 47 |
| P0C817 | 2 kDa | psmA4 | Phenol-soluble modulin alpha 4 peptide | 43 | 0 | 24 |
| Q2FE78 | 35 kDa | hlgA | Gamma-hemolysin component A | 30 | 8 | 27 |
| Q2FE79 | 50 kDa | sbi | Immunoglobulin-binding protein sbi | 30 | 1 | 8 |
| Q2FGW1 | 53 kDa | ebpS | Elastin-binding protein ebpS | 24 | 2 | 10 |
| Q2FIH7 | 28 kDa | seq | Staphylococcal enterotoxin Q | 10 | 0 | 10 |
| Q2FFF5 | 18 kDa | sak | Staphylokinase | 7 | 0 | 21 |
| Surface | ||||||
| Q2FIK6 | 97 kDa | clfA | Clumping factor A | 214 | 52 | 155 |
| Q2FGW1 | 53 kDa | ebpS | Elastin-binding protein ebpS | 52 | 10 | 22 |
| Q2FDM9 | 96 kDa | clfB | Clumping factor B | 31 | 6 | 98 |
| Q2FJ78 | 149 kDa | sdrD | Serine-aspartate repeat-containing protein D | 31 | 2 | 87 |
| Q2FG07 | 101 kDa | isdH | Iron-regulated surface determinant protein H | 17 | 2 | 14 |
| Q2FHV2 | 72 kDa | isdB | Iron-regulated surface determinant protein B | 8 | 0 | 35 |
Results represent total spectral count in the parent strain (WT), their sarA mutants (S), and protease deficient sarA mutants (SP).
The abundance of the major S. aureus autolysin Atl was also impacted by mutation of sarA in a protease-dependent manner (Table 1). Atl was recently shown to contribute to the early stages of biofilm formation, and it was suggested that this is due to its impact on the release of extracellular DNA and/or PIA production, with the fibronectin-binding proteins being required for biofilm maturation (Houston et al., 2011). However, Atl has also been shown to bind extracellular matrix components including fibrinogen, fibronectin, and vitronectin (Heilmann et al., 2005). Like Spa, Atl met our criteria for a protein impacted by sarA in a protease-dependent manner only in extracellular samples, but its abundance in surface protein preparations was also reduced in the LAC sarA mutant (~55%) and partially restored in the sarA/protease mutant (~82% of WT).
Another protein identified in these comparisons was alpha enolase. While not directly implicated in biofilm formation, alpha enolase, along with Atl, was identified as a target protein in a recent study examining the use of a Serratia marcescens extracellular protease (serratiopeptidade) as a means of limiting S. aureus biofilm formation (Artini et al., 2011). As with Spa and Atl, this was true only in conditioned medium, but even in our surface protein samples it was present in reduced amounts in the sarA mutant by comparison to the parent strain (39%), and its abundance was increased in the sarA/protease mutant by comparison to the sarA mutant (160% of that observed in the parent strain) (Table S2). Alpha enolase has also been shown to bind laminin and promote staphylokinase-mediated plasminogen activation (Carneiro et al., 2004; Molkanen et al., 2002).
Proteins that did meet both of our criteria in samples enriched for surface proteins included EbpS, IsdB, IsdH, and SdrD. EbpS is an elastin-binding protein that has been well-characterized biochemically (Parket et al., 1996; Nakakido et al., 2007) but to our knowledge has not been directly implicated in any aspect of virulence. A recent study found that biofilm formation is enhanced in response to iron limitation and that this can be correlated with increased production of IsdA and IsdB (Lin et al., 2012). Atl, FnbA, and Sbi were also identified in this same study. SdrD promotes adherence to dequamated nasal cells, thus suggesting that it plays a role in colonization (Corrigan et al., 2009), an important contributing factor in the development of all forms of S. aureus infection. SdrD, IsdA, IsdB, Spa, ClfA and ClfB also contribute to abscess formation (Cheng et al., 2009), with Spa, ClfA and ClfB also associated with activation of platelet aggregation (O'Brien et al., 2002). ClfA and ClfB are also of interest in that USA300 isolates exhibit a high binding capacity for fibrinogen (Cheung et al., 2011), which could contribute to their ability to form a biofilm despite their high level of agr expression. Indeed, mutation of clfB has been shown to limit biofilm formation under certain in vitro conditions (Abraham et al., 2012).
Although LAC encodes both fnbA and fnbB (Diep et al., 2006), neither of the fibronectin-binding proteins was identified in any samples included in our experiments. One possible explanation for this is that USA300 isolates do express the accessory gene regulator (agr) at high levels (Cheng et al., 2009), and this is associated with reduced expression of these genes and a reduced capacity to bind fibronectin (Novick et al., 2003). Additionally, previous studies employing other strains demonstrated that these proteins are subject to protease-mediated degradation and that this is enhanced in sarA mutants (Karlsson et al., 2001). Moreover, we recently demonstrated that, in the commonly-studied strain Newman, this can be correlated with increased production of SspA and/or SspB and a decreased capacity to form a biofilm (Mrak et al., 2012).
The experiments we report also shed light on sarA-mediated phenotypes in the context of its impact as a modulator of mRNA production and/or stability. For instance, while accumulation of all of the proteins discussed above was restored to some degree in the sarA/protease mutant vs. the sarA mutant, in many cases it was not fully restored. This implies that sarA also negatively regulates the production of these proteins. We also found that the amounts of LukF-PV and LukS-PV, as well as a number of other S. aureus proteins, were comparable in sarA and sarA//protease mutants and elevated over 10-fold by comparison to the parent strain (Table 2). While not the primary focus of the experiments we report, the fact that eliminating protease production had no impact on this phenotype suggest that sarA represses the production of these exotoxins at the level of transcription and/or mRNA stability. Identifying such targets has the potential to significantly enhance efforts to define the interaction between SarA and its DNA and/or RNA targets. Nevertheless, the results we present collectively demonstrate that sarA plays a critical role in defining the virulence of USA300 isolates in multiple forms of infection and suggest that a primary reason for this is its ability to repress the production of extracellular proteases, thereby allowing for the accumulation of critical virulence factors, the phenotypic impact of which would otherwise be limited if not reversed.
Table 2.
Proteins impacted by sarA in a protease-independent manner.
| Accession Number | Molecular Weight | Gene | Protein name | WT | S | SP |
|---|---|---|---|---|---|---|
| Extracellular | ||||||
| Q2FEQ4 | 37 kDa | lukF-PV | Panton-Valentine leukocidin, | 41 | 586 | 591 |
| Q2FIH8 | 35 kDa | lukS-PV | Panton-Valentine leukocidin, | 31 | 471 | 468 |
| Q2FJP8 | 37 kDa | hlgB | Gamma-hemolysin component B | 15 | 63 | 62 |
| Q2FKN3 | 19 kDa | isaB | Immunodominant staphylococcal antigen B | 6 | 28 | 39 |
| Q2FHP4 | 37 kDa | lukD | Leukotoxin LukD | 3 | 17 | 32 |
| Surface | ||||||
| Q2FDM1 | 19 kDa | isaB | Immunodominant staphylococcal antigen B | 24 | 33 | 52 |
| Q2FHS2 | 36 kDa | hla | Alpha-hemolysin | 6 | 24 | 48 |
| Q2FGU9 | 35 kDa | lukS-PV | Panton-Valentine leukocidin, | 0 | 37 | 74 |
| Q2FGV0 | 37 kDa | lukF-PV | Panton-Valentine leukocidin, | 0 | 16 | 34 |
| Q2FIK2 | 25 kDa | nuc | Thermonuclease | 0 | 16 | 28 |
| Q2FE76 | 37 kDa | hlgB | Gamma-hemolysin component B | 0 | 1 | 6 |
Results represent total spectral count in the parent strain (WT), their sarA mutants (S), and protease deficient sarA mutants (SP).
MATERIALS AND METHODS
Ethics Statement
All animal experiments were done in accordance with the policies of the Public Health Service (PHS) policy in Care and Use of Laboratory Animals, the Animal Welfare Act, and the NIH Guide for the Care and Use of Laboratory Animals in an AAALAC (Association for Assessment and Accreditation of Laboratory Animal Care) International accredited facility. All animal procedures were reviewed and approved by the Institutional Animal Care and Use Committee of the University of Arkansas for Medical Sciences.
Bacterial strains and growth conditions
The S. aureus strains used in this study included a plasmid-cured, erythromycin-sensitive derivative of the USA300 strains LAC (Wörman et al., 2011) and FPR3757 (Diep et al., 2006). Isogenic sarA, sarS, and sigB mutants were generated by Φ11-mediated transduction from existing mutants (Zielinska et al., 2011; Blevins et al., 1999; Hsieh et al., 2008; Bischoff et al., 2001). Where appropriate, the sarA mutation was complemented using a plasmid (pSarA) previously shown to be sufficient for this purpose (Blevins et al., 1999). Strains were maintained at −80°C in tryptic soy broth (TSB) containing 25% (vol/vol) glycerol. For each experiment, strains were retrieved from cold storage by plating on tryptic soy agar (TSA) with appropriate antibiotic selection. Antibiotics were used at the following concentrations: erythromycin (Erm; 5 μg per ml), kanamycin (Kan; 50 μg per ml), and neomycin (Neo; 50 μg per ml), with kanamycin and neomycin being used together to avoid selection for spontaneous mutants. For phenotypic assays, strains were inoculated into TSB at an initial optical density at 560 nm (OD560) of 0.05 and to the post-exponential (OD560 = 3.0) or stationary (16 hr) growth phases. All cultures employed for phenotypic assays were grown without antibiotic selection at 37°C with constant aeration and a medium-to-flask volume ratio of 0.40.
Hemolytic activity
Hemolytic activity was assessed by adding 10 μl of standardized, sterile conditioned medium from stationary phase cultures to wells formed in rabbit blood agar plates (Remel, Lenexa, KS). Plates were incubated overnight at 37°C followed by 1 hr at 4°C. The production of alpha-toxin was also assessed directly by Western blotting of standardized cell-free supernatants using rabbit polyclonal anti-alpha-toxin IgG antibody (Sigma Chemical Co., St. Louis, MO) as previously described (Zielinska et al., 2011). For quantitative comparisons, signal intensity was assessed with an Alpha Innotech Flourochem FC2 gel documentation system (Cell Biosciences, Santa Clara, CA) and Image J software analysis, with the amount of signal observed in the wild-type strain set to 1.0 and all other results shown relative to this value. Western blots were blocked with 0.5% skim milk containing 0.1 mg/ml human IgG (Sigma Chemical Co., St. Louis, MO).
Production of extracellular proteases
Protease activity in standardized samples of stationary phase conditioned medium was assessed using the Protease Flourescent Detection Kit (Sigma, St. Louis, MO) with a 1 hr incubation period and by zymogram using 10% Zymogram (Gelatin) Gels (Life Technologies, Grand Island, NY). Samples were first concentrated 15-fold using Centricon YM-3 filter units (Millipore, Bedford, MA) before loading equivalent samples using a buffer containing DTT but not β-mercaptoethanol. After electrophoresis, gels were first incubated for 30 min at room temperature (RT) in renaturing buffer (2.5% TritonX-100) and then overnight at 37°C in developing buffer (0.2 M Tris, 5 mM CaCl2, 1 mM DTT). Gels were then stained with SimplyBlue SafeStain (Life Technologies, Grand Island, NY) at RT for 2 hrs before destaining overnight in distilled water.
Transcriptional analysis
To assess the levels of hla, aur and RNAIII accumulation, total bacterial RNA was isolated using the Qiagen RNeasy Mini Kit. Quantitative, real-time RT-PCR (qRT-PCR) was performed using primers and TaqMan probes corresponding to RNAIII or hla as previously described (Zielinska et al., 2011). Primers and probe used to assess the relative abundance of aur mRNA were: 5'-TATGGTGATGGTGATGGTCGCACA-3', 5'-ATTTAGAGCGCCTGACTGGTCCTT-3', 5'-/FAM/TCGGGTGCAAATGACGTAGTAGCACA/BHQ/-3'. Results were calibrated by comparison to those obtained with the same RNA samples for a 16S ribosomal RNA gene (Zielinska et al., 2011). Results are reported as relative units by comparison to the results observed with the parent strain in stationary phase samples, the value for which was set to 1.0.
Assessment of biofilm formation in vitro
Biofilm formation was assessed in vitro using a microtiter plate assay in which the wells were first coated with human plasma proteins and the medium (tryptic soy broth) was supplemented with both salt and glucose (Beenken et al., 2010).
Assessment of biofilm formation and relative daptomycin susceptibility in vivo
Biofilm formation was assessed in vivo using a murine model of catheter-associated biofilm formation (Weiss et al., 2009). Briefly, uncoated catheters were implanted into each flank of NIH Swiss mice and inoculated by direct injection into the lumen of each catheter with 105 colony-forming units (cfu) of the test strain in a total volume of 100 μl of phosphate-buffered saline (PBS). After 24 h, mice were randomly divided into experimental groups (n = 5). Because each mouse had two catheters implanted and because previous experiments have confirmed the absence of cross-contamination between catheters in opposite flanks of the same mouse (Weiss et al., 2009), each catheter was treated as an independent data point (n = 10). In untreated mice, 100 μl of sterile PBS was injected in the lumen of each catheter daily for 5 days. In the treated groups, 100 μl of sterile PBS containing daptomycin was injected into the lumen at daily intervals. The concentration of daptomycin used (10 μg/ml) corresponds to 10 times (10X) the concentration defined by the Clinical and Laboratory Standards Institute (CLSI) as the breakpoint MIC for S. aureus (≤1.0 μg/ml) (Weiss et al., 2009). All experiments were done three times and the results combined for statistical analysis.
Bacteremia model
5–8 week old female, outbred NIH-Swiss mice (Harlan, Indianapolis, Ind.) were infected via tail vein injection with 108 cfu (Blevins et al., 2003). Two independent trials with at least 8 mice per experimental group were carried out. After 6 days, mice were euthanized and tissues harvested for analysis; in those cases in which compassionate euthanasia was required, tissues were harvested immediately after death. For analysis of soft tissue samples, the targeted organ was removed aseptically and homogenized on ice. Dilutions of each homogenate were then plated on CHROMagar (Blevins et al., 2003) for quantitative analysis. The number of cfu per organ was then determined following overnight incubation at 37°C. To assess retention of pSarA, replicate samples were also plated on TSA containing chloramphenicol.
Additionally, the left hind limb was removed and prepared for histological analysis. Briefly, formalin fixed samples were decalcified in 5% formic acid until completely decalcified as determined by ammonium oxalate test (Skinner et al., 1997). Decalcified samples were washed in running water to remove any residual decalcifier, dehydrated through graded ethanol washes, cleared through multiple changes of methyl salicylate, and infiltrated with and embedded in paraffin as previously described (Skinner et al., 1997). Paraffin embedded blocks were sectioned at 4 microns and stained with Hematoxylin & eosin to assess general cellular and architectural detail. Sequential sections were also stained to detect intraosseous Gram positive cocci (Skinner et al., 1992) and with Saffranin O-Fast Green to visualize changes in cartilage components (Kalscheur et al. 2001). All tissue sections were evaluated microscopically in a blinded fashion and scored for the degree of inflammation (0–3) based on the extent of acute inflammatory cells seen in the synovial space and bone. The sections were also scored for the absence (0) or presence (1) of Gram-positive cocci/abscess formation, articular cartilage erosion, cortical bone erosion and physis destruction. A total score was generated for each animal based on these parameters (range: 0–13), with statistical comparisons between experimental groups made based on these scores.
Preparation of samples for analysis of extracellular proteins
After overnight incubation, cultures were standardized with respect to each other by the addition of sterile TSB. Standardized samples were then centrifuged and the supernatants filtered through 0.2 μm syringe filters to remove residual bacterial cells. 10 μl was then mixed with sample loading buffer and resolved by SDS-PAGE using 4–12% gradient NuPAGE Novex Bis-Tris Gels (Life Technologies, Grand Island, NY).
Preparation of samples for analysis of surface-associated proteins
S. aureus cells were harvested from stationary phase. After standardization, volumes calculated to contain 109 cells were centrifuged and the cell pellets washed with water. Pellets were then suspended in 200 μl of a digestion buffer composed of 40mM Tris-HCl, pH 7.5, 100 mM NaCl, 27% sucrose (w/v), 20 mM MgCl2, lysostaphin (100 mg/mL), mutanolysin (50 mg/mL), DNase I (20 mg/mL), and the protease inhibitors PMSF (2 mM), benzamidine (1 mM), TAME (0.2 mM), leupeptin (10 mg/mL), and pepstatin (5 mg/mL). Cells suspensions were incubated at 37°C for 4 hr, with 10 μl aliquots removed before and after the incubation period to monitor the decrease in absorption at 540 nm. Absorption values were typically 10-fold lower after incubation indicating extensive cell wall digestion. Preparations were centrifuged at 6000 × g for 20 min at 4°C. Proteins were then precipitated from the supernatants by adding TCA to a final concentration of 10% and incubating overnight at 4°C. The precipitate was harvested by centrifugation at 7000 × g for 20 min at 4°C, washed with ice-cold 96% (w/v) ethanol, and dried. Proteins were then resuspended in sample loading buffer (Life Technologies, Grand Island, NY) and resolved by SDS-PAGE using 4–12% NuPAGE Novex Bis-Tris Gels.
MS analysis
SDS-PAGE lanes were divided into 20 slices and subjected to in-gel trypsin digestion. Briefly, gel slices were destained in 50% methanol, 100 mM ammonium bicarbonate, followed by reduction in 10 mM Tris[2-carboxyethyl]phosphine and alkylation in 50 mM iodoacetamide. Gel slices were then dehydrated in acetonitrile, followed by addition of 100 ng sequencing grade porcine trypsin (Promega, Madison, WI) in 100 mM ammonium bicarbonate and incubation at 37°C for 12–16 hours. Peptide products were then acidified in 0.1% formic acid (Fluka, Milwaukee, WI). Tryptic peptides were separated by reverse phase HPLC on a 10 cm C18 column using a NanoLC 2D system (Eksigent, Dublin, CA) and ionized by electrospray upon elution, followed by MS/MS analysis using an LTQ XL mass spectrometer (Thermo Scientific, Waltham, MA). Proteins were identified from MS/MS spectra by database searching using the Mascot search engine (Matrix Science, Boston, MA).
Statistical analysis
Bacterial count data from harvested catheters were logarithmically transformed and analyzed using analysis of variance (ANOVA) models to evaluate the effect of each mutation. Using these ANOVA models, we were able to assess not only the effect of any given mutation but also any interaction between the effect of the mutation and daptomycin exposure, when applicable. Pair-wise testing was performed using t tests on the logarithmically transformed data. The significance of the ANOVA and t-test analyses were calculated using permutation tests. Wilcoxon rank-sum tests were used to analyze hemolytic activity, protease activity and histopathology data, while Kaplan-Meier methods were used to calculate survival estimates for lethality studies. Statistical analyses were performed using R (version 2.7; The Foundation for Statistical Computing) and Mann-Whitney Rank Sum Test and SigmaPlot. P-values ≤0.05 considered to be statistically significant. Statistical analysis of survival curves was performed using GraphPad Prism 5.0 and the log-rank test (Mantel-Cox test).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants AI069087 and AI093126 (MSS) from the National Institute of Allergy and Infectious Diseases and grant 10PRE3220017 (AKZ) from the American Heart Association. Support was also obtained from resources provided through the Clinical and Translational Sciences Award (RR0298884) to the University of Arkansas for Medical Sciences. We would also like to acknowledge the UAMS Proteomics Facility for mass spectrometric support and support from NIH grants R01DA025755, P30GM103450, and P20GM103429.
LITERATURE CITED
- Abraham NM, Jefferson KK. Staphylococcus aureus clumping factor B mediates biofilm formation in the absence of calcium. Microbiology. 2012 Mar 22; doi: 10.1099/mic.0.057018-0. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artini M, Scoarughi GL, Papa R, Cellini A, Carpentieri A, et al. A new anti-infective strategy to reduce adhesion-mediated virulence in Staphylococcus aureus affecting surface proteins. Int J Immunopathol Pharmacol. 2011;24:661–672. doi: 10.1177/039463201102400312. [DOI] [PubMed] [Google Scholar]
- Beenken KE, Mrak LN, Griffin LM, Zielinska AK, Shaw LN, et al. Epistatic relationships between sarA and agr in Staphylococcus aureus biofilm formation. PLoS One. 2010;5:e10790. doi: 10.1371/journal.pone.0010790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beenken KE, Spencer H, Griffin LM, Smeltzer MS. Impact of extracellular nuclease production on the biofilm phenotype of Staphylococcus aureus under in vitro and in vivo conditions. Infect Immun. 2012;80(5):1634–8. doi: 10.1128/IAI.06134-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff M, Berger-Bächi B. Teicoplanin stress-selected mutations increasing sigma(B) activity in Staphylococcus aureus. Antimicrob Agents Chemother. 2001;45(6):1714–20. doi: 10.1128/AAC.45.6.1714-1720.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blevins JS, Beenken KE, Elasri MO, Hurlburt BK, Smeltzer MS. Strain-dependent differences in the regulatory roles of sarA and agr in Staphylococcus aureus. Infect Immun. 2002;70:470–480. doi: 10.1128/IAI.70.2.470-480.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blevins JS, Elasri MO, Allmendinger SD, Beenken KE, Skinner RA, et al. Role of sarA in the pathogenesis of Staphylococcus aureus musculoskeletal infection. Infect Immun. 2003;71:516–523. doi: 10.1128/IAI.71.1.516-523.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blevins JS, Gillaspy AF, Rechtin TM, Hurlburt BK, Smeltzer MS. The staphylococcal accessory regulator (sar) represses transcription of the Staphylococcus aureus collagen adhesion gene (cna) in an agr-independent manner. Mol Microbiol. 1999;33:317–326. doi: 10.1046/j.1365-2958.1999.01475.x. [DOI] [PubMed] [Google Scholar]
- Booth MC, Cheung AL, Hatter KL, Jett BD, Callegan MC, et al. Staphylococcal accessory regulator (sar) in conjunction with agr contributes to Staphylococcus aureus virulence in endophthalmitis. Infect Immun. 1997;65(4):1550–56. doi: 10.1128/iai.65.4.1550-1556.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carneiro CR, Postol E, Nomizo R, Reis LF, Brentani RR. Identification of enolase as a laminin-binding protein on the surface of Staphylococcus aureus. Microbes Infect. 2004;6:604–608. doi: 10.1016/j.micinf.2004.02.003. [DOI] [PubMed] [Google Scholar]
- Cassat J, Dunman PM, Murphy E, Projan SJ, Beenken KE, et al. Transcriptional profiling of a Staphylococcus aureus clinical isolate and its isogenic agr and sarA mutants reveals global differences in comparison to the laboratory strain RN6390. Microbiology. 2006;152:3075–3090. doi: 10.1099/mic.0.29033-0. [DOI] [PubMed] [Google Scholar]
- Cheng AG, Kim HK, Burts ML, Krausz T, Schneewind O, et al. Genetic requirements for Staphylococcus aureus abscess formation and persistence in host tissues. FASEB J. 2009;23:3393–3404. doi: 10.1096/fj.09-135467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung AL, Eberhardt KJ, Chung E, Yeaman MR, Sullam PM, et al. Diminished virulence of a sar-/agr- mutant of Staphylococcus aureus in the rabbit model of endocarditis. J Clin Invest. 1994;94(4):1815–22. doi: 10.1172/JCI117530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung AL, Ying P. Regulation of alpha- and beta-hemolysins by the sar locus of Staphylococcus aureus. J Bacteriol. 1994;176(3):580–5. doi: 10.1128/jb.176.3.580-585.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung GY, Wang R, Khan BA, Sturdevant DE, Otto M. Role of the accessory gene regulator agr in community-associated methicillin-resistant Staphylococcus aureus pathogenesis. Infect Immun. 2011;79:1927–1935. doi: 10.1128/IAI.00046-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien Y, Manna AC, Projan SJ, Cheung AL. SarA, a global regulator of virulence determinants in Staphylococcus aureus, binds to a conserved motif essential for sar-dependent gene regulation. J Biol Chem. 1999;274:37169–37176. doi: 10.1074/jbc.274.52.37169. [DOI] [PubMed] [Google Scholar]
- Corrigan RM, Miajlovic H, Foster TJ. Surface proteins that promote adherence of Staphylococcus aureus to human desquamated nasal epithelial cells. BMC Microbiol. 2009;9:22. doi: 10.1186/1471-2180-9-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didier JP, Cozzone AJ, Duclos B. Phosphorylation of the virulence regulator SarA modulates its ability to bind DNA in Staphylococcus aureus. FEMS Microbiol Lett. 2010;306(1):30–6. doi: 10.1111/j.1574-6968.2010.01930.x. [DOI] [PubMed] [Google Scholar]
- Diep BA, Gill SR, Chang RF, Phan TH, Chen JH, et al. Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. Lancet. 2006;367:731–739. doi: 10.1016/S0140-6736(06)68231-7. [DOI] [PubMed] [Google Scholar]
- Dinges MM, Orwin PM, Schlievert PM. Exotoxins of Staphylococcus aureus. Clin Microbiol Rev. 2000;13:16–34. doi: 10.1128/cmr.13.1.16-34.2000. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunman PM, Murphy E, Haney S, Palacios D, Tucker-Kellogg G, et al. Transcription profiling-based identification of Staphylococcus aureus genes regulated by the agr and/or sarA loci. J Bacteriol. 2001;183:7341–7353. doi: 10.1128/JB.183.24.7341-7353.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto DF, Higginbotham RH, Sterba KM, Maleki SJ, Segall AM, et al. Staphylococcus aureus SarA is a regulatory protein responsive to redox and pH that can support bacteriophage lambda integrase-mediated excision/recombination. Mol Microbiol. 2009;74:1445–1458. doi: 10.1111/j.1365-2958.2009.06942.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haupt K, Reuter M, van den Elsen J, Burman J, Halbich S, et al. The Staphylococcus aureus protein Sbi acts as a complement inhibitor and forms a tripartite complex with host complement Factor H and C3b. PLoS Pathog. 2008;4:e1000250. doi: 10.1371/journal.ppat.1000250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heilmann C, Hartleib J, Hussain MS, Peters G. The multifunctional Staphylococcus aureus autolysin AAA mediates adherence to immobilized fibrinogen and fibronectin. Infect Immun. 2005;73:4793–4802. doi: 10.1128/IAI.73.8.4793-4802.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houston P, Rowe SE, Pozzi C, Waters EM, O'Gara JP. Essential role for the major autolysin in the fibronectin-binding protein-mediated Staphylococcus aureus biofilm phenotype. Infect Immun. 2011;79:1153–1165. doi: 10.1128/IAI.00364-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh HY, Tseng CW, Stewart GC. Regulation of Rot expression in Staphylococcus aureus. J Bacteriol. 2008;190(2):546–54. doi: 10.1128/JB.00536-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu C, Xiong N, Zhang Y, Rayner S, Chen S. Functional characterization of lipase in the pathogenesis of Staphylococcus aureus. Biochem Biophys Res Commun. 2012 doi: 10.1016/j.bbrc.2012.02.057. [DOI] [PubMed] [Google Scholar]
- Jacobsson G, Colque-Navarro P, Gustafsson E, Andersson R, Mollby R. Antibody responses in patients with invasive Staphylococcus aureus infections. Eur J Clin Microbiol Infect Dis. 2010;29:715–725. doi: 10.1007/s10096-010-0919-x. [DOI] [PubMed] [Google Scholar]
- Jones RC, Deck J, Edmondson RD, Hart ME. Relative quantitative comparisons of the extracellular protein profiles of Staphylococcus aureus UAMS-1 and its sarA, agr, and sarA agr regulatory mutants using one-dimensional polyacrylamide gel electrophoresis and nanocapillary liquid chromatography coupled with tandem mass spectrometry. J Bacteriol. 2008;190:5265–5278. doi: 10.1128/JB.00383-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalscheur VL. Bone and cartilage changes in osteoarthritis: a proteoglycan stain. Histologic. 2001;34:14–15. [Google Scholar]
- Karlsson A, Saravia-Otten P, Tegmark K, Morfeldt E, Arvidson S. Decreased amounts of cell wall-associated protein A and fibronectin-binding proteins in Staphylococcus aureus sarA mutants due to up-regulation of extracellular proteases. Infect Immun. 2001;69:4742–48. doi: 10.1128/IAI.69.8.4742-4748.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy AD, Otto M, Braughton KR, Whitney AR, Chen L, Mathema B, Mediavilla JR, Byrne KA, Parkins LD, Tenover FC, Kreiswirth BN, Musser JM, DeLeo FR. Epidemic community-associated methicillin-resistant Staphylococcus aureus: Recent clonal expansion and diversification. Proc Nat Acad Sci. 2008;105:1327–1232. doi: 10.1073/pnas.0710217105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kielian T, Cheung A, Hickey WF. Diminished virulence of an alpha-toxin mutant of Staphylococcus aureus in experimental brain abscesses. Infect Immun. 2001;69(11):6902–11. doi: 10.1128/IAI.69.11.6902-6911.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiecinski J, Josefsson E, Mitchell J, Higgins J, Magnusson M, et al. Activation of plasminogen by staphylokinase reduces the severity of Staphylococcus aureus systemic infection. J Infect Dis. 2010;202:1041–1049. doi: 10.1086/656140. [DOI] [PubMed] [Google Scholar]
- Li M, Cheung GY, Hu J, Wang D, Joo HS. Comparative analysis of virulence and toxin expression of global community-associated methicillin-resistant Staphylococcus aureus strains. J Infect Dis. 2010;202(12):1866–76. doi: 10.1086/657419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin MH, Shu JC, Huang HY, Cheng YC. Involvement of iron in biofilm formation by Staphylococcus aureus. PLoS ONE. 2012;7:e34388. doi: 10.1371/journal.pone.0034388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merino N, Toledo-Arana A, Vergara-Irigaray M, Valle J, Solano C, et al. Protein A-mediated multicellular behavior in Staphylococcus aureus. J Bacteriol. 2009;191:832–843. doi: 10.1128/JB.01222-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molkanen T, Tyynela J, Helin J, Kalkkinen N, Kuusela P. Enhanced activation of bound plasminogen on Staphylococcus aureus by staphylokinase. FEBS Lett. 2002;517:72–78. doi: 10.1016/s0014-5793(02)02580-2. [DOI] [PubMed] [Google Scholar]
- Morrison JMAK, Beenken KE, Smeltzer MS, Dunman PM. The staphylococcal accessory regulator, SarA, is an RNA-binding protein that modulates the mRNA turnover properties of late-exponential and stationary phase Staphylococcus aureus cells. Front Cell Inf Microbio. 2012;2 doi: 10.3389/fcimb.2012.00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mrak LN, Zielinska AK, Beenken KE, Mrak IN, Atwood DN. saeRS and sarA act synergistically to repress protease production and promote biofilm formation in Staphylococcus aureus. PLoS One. 2012;7(6):e38453. doi: 10.1371/journal.pone.0038453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakakido M, Tanaka Y, Tsumoto K. The N-terminal domain of elastin-binding protein of Staphylococcus aureus changes its secondary structure in a membrane-mimetic environment. J Biochem. 2007;142:131–134. doi: 10.1093/jb/mvm131. [DOI] [PubMed] [Google Scholar]
- Nilsson IM, Bremell T, Rydén C, Cheung AL, Tarkowski A. Role of the staphylococcal accessory gene regulator (sar) in septic arthritis. Infect Immun. 1996;64(11):4438–43. doi: 10.1128/iai.64.11.4438-4443.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson IM, Hartford O, Foster T, Tarkowski A. Alpha-toxin and gamma-toxin jointly promote Staphylococcus aureus virulence in murine septic arthritis. Infect Immun. 1999;67:1045–1049. doi: 10.1128/iai.67.3.1045-1049.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick RP. Mobile genetic elements and bacterial toxinoses: the superantigen-encoding pathogenicity islands of Staphylococcus aureus. Plasmid. 2003;49:93–105. doi: 10.1016/s0147-619x(02)00157-9. [DOI] [PubMed] [Google Scholar]
- O'Brien L, Kerrigan SW, Kaw G, Hogan M, Penades J, et al. Multiple mechanisms for the activation of human platelet aggregation by Staphylococcus aureus: Roles for the clumping factors ClfA and ClfB, the serine-aspartate repeat protein SdrE and protein A. Mol Microbiol. 2002;44:1033–1044. doi: 10.1046/j.1365-2958.2002.02935.x. [DOI] [PubMed] [Google Scholar]
- Oscarsson J, Kanth A, Tegmark-Wisell K, Arvidson S. SarA is a repressor of hla (alpha-hemolysin) transcription in Staphylococcus aureus: Its apparent role as an activator of hla in the prototype strain NCTC 8325 depends on reduced expression of sarS. J Bacteriol. 2006;188:8526–8533. doi: 10.1128/JB.00866-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park PW, Rosenbloom J, Abrams WR, Mecham RP. Molecular cloning and expression of the gene for elastin-binding protein (ebpS) in Staphylococcus aureus. J Biol Chem. 1996;271:15803–15809. doi: 10.1074/jbc.271.26.15803. [DOI] [PubMed] [Google Scholar]
- Rechtin TM, Gillaspy AF, Schumacher MA, Brennan RG, Smeltzer MS, et al. Characterization of the SarA virulence gene regulator of Staphylococcus aureus. Mol Microbiol. 1999;33(2):307–16. doi: 10.1046/j.1365-2958.1999.01474.x. [DOI] [PubMed] [Google Scholar]
- Reyes D, Andrey DO, Monod A, Kelley WL, Zhang G, et al. Coordinated regulation by AgrA, SarA, and SarR to control agr expression in Staphylococcus aureus. J Bacteriol. 2011;193(21):6020–31. doi: 10.1128/JB.05436-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts C, Anderson KL, Murphy E, Projan SJ, Mounts W, et al. Characterizing the effect of the Staphylococcus aureus virulence factor regulator, SarA, on log-phase mRNA half-lives. J Bacteriol. 2006;188:2593–2603. doi: 10.1128/JB.188.7.2593-2603.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlievert PM, Jablonski LM, Roggiani M, Sadler I, Callantine S, et al. Pyrogenic toxin superantigen site specificity in toxic shock syndrome and food poisoning in animals. Infect Immun. 2000;68:3630–3634. doi: 10.1128/iai.68.6.3630-3634.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner RA, Hickmon SG, Lumpkin CK, Aronson JA, Nicholas RW. Decalcified bone: Twenty years of successful specimen management. J Histotechnology. 1997;20(3):267–277. [Google Scholar]
- Skinner RA, Hickmon SG, Nelson CL, Germer RA. Modified stain for identification of Staphylococcus aureus in osteomyelitis. J Histotech. 1992;15:303–306. [Google Scholar]
- Smith EJ, Corrigan RM, van der Sluis T, Grundling A, Speziale P, et al. The immune evasion protein Sbi of Staphylococcus aureus occurs both extracellularly and anchored to the cell envelope by binding lipoteichoic acid. Mol Microbiol. 2012;83:789–804. doi: 10.1111/j.1365-2958.2011.07966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somerville GA, Proctor RA. At the crossroads of bacterial metabolism and virulence factor synthesis in staphylococci. Microbiol Mol Biol Rev. 2009;73(2):233–48. doi: 10.1128/MMBR.00005-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterba KM, Mackintosh SG, Blevins JS, Hurlburt BK, Smeltzer MS. Characterization of Staphylococcus aureus SarA binding sites. J Bacteriol. 2003;185:4410–4417. doi: 10.1128/JB.185.15.4410-4417.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurlow LR, Hanke ML, Fritz T, Angle A, Aldrich A, Williams SH, Engebretsen IL, Bayles KW, Horswill AR, Kielian T. Staphylococcus aureus biofilms prevent macrophage phagocytosis and attenuate inflammation in vivo. J Immunol. 2011;186(11):6585–96. doi: 10.4049/jimmunol.1002794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss EC, Spencer HJ, Daily SJ, Weiss BD, Smeltzer MS. Impact of sarA on antibiotic susceptibility of Staphylococcus aureus in a catheter-associated in vitro model of biofilm formation. Antimicrob Agents Chemother. 2009;53:2475–2482. doi: 10.1128/AAC.01432-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss EC, Zielinska A, Beenken KE, Spencer HJ, Daily SJ, et al. Impact of sarA on daptomycin susceptibility of Staphylococcus aureus biofilms in vivo. Antimicrob Agents Chemother. 2009;53:4096–4102. doi: 10.1128/AAC.00484-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wörmann ME, Reichmann NT, Malone CL, Horswill AR, Gründling A. Proteolytic cleavage inactivates the Staphylococcus aureus lipoteichoic acid synthase. J Bacteriol. 2011;193(19):5279–91. doi: 10.1128/JB.00369-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielinska AK, Beenken KE, Joo HS, Mrak LN, Griffin LM, et al. Defining the strain-dependent impact of the staphylococcal accessory regulator (sarA) on the alpha-toxin phenotype of Staphylococcus aureus. J Bacteriol. 2011;193:2948–2958. doi: 10.1128/JB.01517-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.