Abstract
Host range factors, expressed by the poxvirus family, determine the host tropism of species, tissue, and cell specificity. C7L family members exist in the genomes of most sequenced mammalian poxviruses, suggesting an evolutionarily conserved effort adapting to the hosts. In general, C7L orthologs influence the host tropism in mammalian cell culture, and for some poxviruses it is essential for the complete viral life cycle in vitro and in vivo. The C7L family members lack obvious sequence homology with any other known viral or cellular proteins. Here we review recent findings from an evolutionary perspective and summarize recent progress that broadens our view on the role of C7L family members in mediating poxvirus host range and antagonizing the host defense system.
Introduction
Members of Poxviridae are large complex DNA viruses encoding a plethora of viral factors to manipulate host defense mechanisms in addition to a more conserved set of essential genes necessary for the viral cytoplasmic life cycle. Host range factors are a group of virus-encoded proteins that are essential for the biologic tropism features. Phenotypically, the deletion of specific host range factor genes leads to the inability of the resulting virus to infect cells of certain species, tissues, and/or certain cell types for which the parental virus is permissive. Although, at the molecular level, the direct mechanistic cause of these phenotypes varies in different model systems, the crucial roles of these factors in direct engagement of the various host defense systems highlights the constant pressures exerted by evolutionary host-pathogen interactions. The poxvirus C7L family, named after the prototypical gene from vaccinia virus (VACV), is a perfect example of related host range factors that have uniquely evolved to orchestrate the tropism specificities of individual virus isolates. The majority of sequenced mammalian poxviruses contain one or more C7L family members in their genomes, with rare exceptions, such as molluscum contagiosum virus and parapoxvirus [1,2]. Members of C7L family share a sequence homology (Figure 1) that is unique among poxviruses and no conserved motif of any other viral or cellular protein can be found. Through the study of evolutionary divergence of this gene family, we can gain insights into how poxviruses compete for survival within various host species, each with a diverse repertoire of anti-viral response pathways. With recent progress in the identification of novel host cell targets and signaling effectors that interact with specific viral host range factors of the C7L family, we review here what has been learned to date about the mechanisms governing virus-host tropism at the cellular, tissue and organismal levels.
Figure 1.
Multiple sequence alignment of poxvirus C7L family members. A multiple sequence alignment of poxvirus C7L orthologs (see Table 1 for abbreviations) that show less than 93% amino acid sequence identity with one another by pairwise comparison was carried out using MAFT. Residues that are 100% conserved are shown in red, residues that are conserved in ≥90% of sequences are shown in blue. Residues 134 (N = Asparagine) and 135 (F = Phenylalanine) of sheeppox virus (SPPV) 063 are highlighted in green, and corresponding residues Y (Tyrosine) and I (Isoleucine) that were introduced into SPPV 063 and rescued virus replication in mouse 3T3 cells [19**] are shown in purple.
Overview-the Perspective of Evolutionary Biology
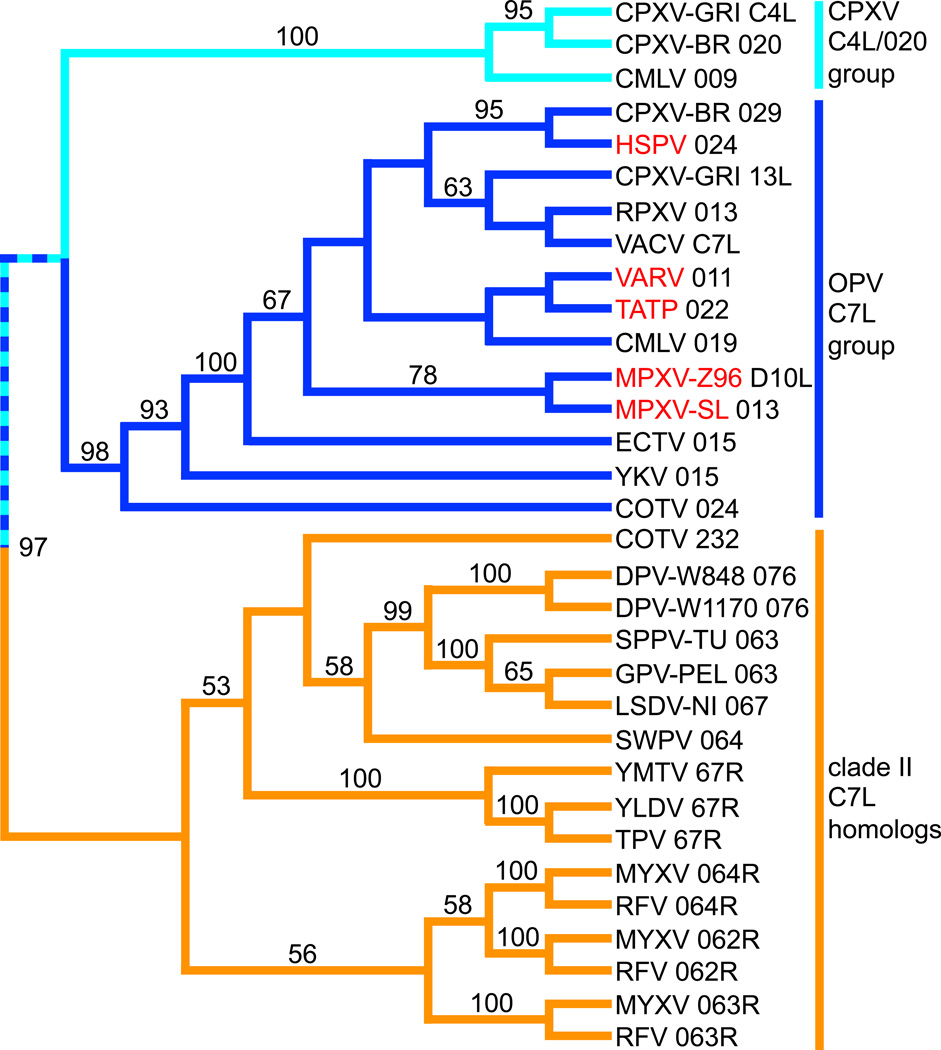
C7L gene family orthologs can be found in all completely sequenced orthopoxviruses (OPV) as well as in leporipoxviruses, suipoxviruses, carpipoxviruses, yatapoxviruses, cervidpoxviruses and Cotia poxvirus, the latter six of which can be referred to as clade II poxviruses because they form an independent clade in phylogenetic analyses [3,4]. Viruses discussed in this review and their abbreviations are listed in Table 1. Some OPV members, including each of the 10 sequenced cowpox viruses, contain two genes with sequence homology to the canonical C7L gene of VACV: a direct C7L ortholog and a more distantly related gene, which is called C4L and 020 in cowpox virus strains GRI-90 and Brighton Red (CPXV-GRI and CPXV-BR), respectively. The leporipoxviruses, including MYXV and RFV, each contain three copies of tandemly arranged C7L-related genes. In a recent phylogenetic analysis (Bratke, McLysaght and Rothenburg, submitted) that included 28 C7L homologs from 21 poxviruses revealed the existence of 3 major clades, which were separated by high bootstrap values. We extended this analysis by including C7L homologs from Yoka and Cotia poxviruses and found a comparable phylogenetic relationship of the three major clades (Figure. 2): one clade contains C7L members from clade II poxviruses (orange branches), which forms a sister clade to the one that contains two subclades: One clade contains OPV C7L orthologs (dark blue) and one clade contains the CPXV C4L/020 group (azure branches). The OPV C7L clade includes Yoka poxvirus 015 and Cotia poxvirus 024. Whereas Yoka poxvirus is most closely related to extant OPVs [5], previous phylogenetic analyses showed that Cotia poxvirus was nested within the clade II poxviruses [6], in which only the second Cotia poxvirus C7L homolog 232 was included. The presence of Cotia poxvirus 024 within the OPV clade indicates a putative recombination event. In agreement with this, the genes neighboring 024 show highest sequence identity with OPV genes [6]. The analysis indicates that one C7L homolog was present in a poxvirus that was ancestral to both clade II poxviruses and OPVs, and that two gene duplication events led to the emergence of three C7L-like gene copies in the leporipoxviruses. Another duplication event in an OPV ancestor led to the emergence of both the C7L and CPXV C4L/020 clades. In addition to the phylogenetic analysis, a closer relationship between the latter clades is also supported by higher sequence identities of CPXV C4L/020 and OPV C7L orthologs (table 2, grey background). Taking the low sequence identities of about 32% (table 2) between C7L orthologs and the CPXV C4L/020 group and the higher sequence identities between OPV C7L and YKV 015 (57%) and COTV 024 (51–54%) into account, it appears that the split into C7L and CPXV C4L/020 occurred early during the evolution of an OPV ancestor.
Table 1.
Poxviruses discussed in this review are listed.
| Abbreviation | Full name | Genus |
|---|---|---|
| CMLV | Camelpox virus | Orthopoxvirus (OPV) |
| CPXV | Cowpox virus | |
| ECTV | Ectromelia virus | |
| HSPV | Horsepox virus | |
| MPXV | Monkeypox virus | |
| RPXV | Rabbitpox virus | |
| TATP | Taterapox virus | |
| VACV | Vaccinia virus | |
| VARV | Variola virus | |
| YKV | Yoka virus | Unclassified |
| COTV | Cotia virus | |
| GPV | Goatpox virus | Capripoxvirus |
| LSDV | Lumpy skin disease virus | |
| SPPV | Sheeppox virus | |
| DPV | Deerpox virus | Cervidpoxvirus |
| MYXV | Myxoma virus | Leporipoxvirus |
| RFV | Rabbit fibroma virus | |
| SWPV | Swinepox virus | Suipoxvirus |
| TPV | Tanapox virus | Yatapoxvirus |
| YLDV | Yaba-like disease virus | |
| YMTV | Yata monkey tumor virus |
Figure 2.
Phylogenetic relationship between poxvirus C7L family members. A multiple sequence alignment of poxvirus C7L orthologs (see Table 1 for abbreviations) was carried out using MAFT [36]. Midpoint-rooted phylogenetic tree was generated using the neighbor-joining method with nodal support assessed via bootstrapping (1000 replicates) as implemented in PAUP[37]. Bootstrap values above 50 are shown on branches. Three major clades are colored as follows: dark blue = C7L orthologs of orthopoxviruses (OPV) and related sequences; azure blue = CPXV C4L/020 clade; orange = C7L homologs from clade II poxviruses. OPV that contain putatively inactivated or truncated CPXV C4L/020 orthologs in their genomes are highlighted in red.
Table 2.
Identities between C7 orthologs. The gaps and poorly aligned parts of the multiple sequence alignment shown in Figure 1 were removed using Gblocks (http://molevol.cmima.csic.es/castresana/Gblocks_server.html). Sequence identities of the resulting alignment were calculated with MegAlign (Lasergene 10). Sequence identities between CPXV-BR020 and CMLV 009 with the other sequences are highlighted with grey background.
| VACV_C7L | ECTV_015 | YKV_171 | COTV_024 | CPXV_BR020 | CMLV_009 | COTV_232 | YMTV_67R | YLDV_67R | DPV_076 | SPPV_063 | SWPV_064 | MYXV_64R | RFV_064R | MYXV_62R | RFV_062R | MYXV_63R | RFV_063R | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| *** | 94 | 57.9 | 54.1 | 32.3 | 30.8 | 27.8 | 33.8 | 34.6 | 33.1 | 32.3 | 37.6 | 24.8 | 27.8 | 31.6 | 30.8 | 19.5 | 24.1 | VACV_C7L |
| *** | 57.1 | 51.1 | 32.3 | 30.8 | 24.8 | 32.3 | 33.1 | 31.6 | 31.6 | 33.8 | 24.8 | 27.1 | 30.1 | 29.3 | 19.5 | 24.1 | ECTV_015 | |
| *** | 47.4 | 32.3 | 32.3 | 27.8 | 31.6 | 32.3 | 32.3 | 28.6 | 26.3 | 24.1 | 29.3 | 28.6 | 26.3 | 18 | 19.5 | YKV_171 | ||
| *** | 35.3 | 33.8 | 30.1 | 33.1 | 34.6 | 30.8 | 30.8 | 29.3 | 21.8 | 26.3 | 29.3 | 26.3 | 22.6 | 21.1 | COTV_024 | |||
| *** | 94 | 20.3 | 22.6 | 21.8 | 17.3 | 20.3 | 21.1 | 18.8 | 22.6 | 22.6 | 24.1 | 18.8 | 20.3 | CPXV_BR020 | ||||
| *** | 20.3 | 21.8 | 21.1 | 18 | 20.3 | 21.8 | 18.8 | 21.8 | 22.6 | 22.6 | 18.8 | 19.5 | CMLV_009 | |||||
| *** | 42.9 | 44.4 | 42.9 | 40.6 | 42.9 | 33.1 | 37.6 | 41.4 | 39.8 | 28.6 | 29.3 | COTV_232 | ||||||
| *** | 85.7 | 46.6 | 45.1 | 49.6 | 37.6 | 40.6 | 42.1 | 36.8 | 30.8 | 27.1 | YMTV_67R | |||||||
| *** | 47.4 | 46.6 | 51.9 | 36.8 | 41.4 | 44.4 | 39.1 | 32.3 | 27.8 | YLDV_67R | ||||||||
| *** | 66.9 | 50.4 | 34.6 | 37.6 | 41.4 | 36.1 | 27.1 | 26.3 | DPV_076 | |||||||||
| *** | 49.6 | 37.6 | 39.8 | 42.1 | 39.8 | 25.6 | 28.6 | SPPV_063 | ||||||||||
| *** | 38.3 | 38.3 | 45.1 | 40.6 | 24.8 | 27.8 | SWPV_064 | |||||||||||
| *** | 69.2 | 40.6 | 42.1 | 24.8 | 27.8 | MYXV_64R | ||||||||||||
| *** | 39.1 | 38.3 | 25.6 | 26.3 | RFV_064R | |||||||||||||
| *** | 76.7 | 30.1 | 32.3 | MYXV_62R | ||||||||||||||
| *** | 33.1 | 37.6 | RFV_062R | |||||||||||||||
| *** | 70.7 | MYXV_63R | ||||||||||||||||
| *** | RFV_063R |
Interestingly, inactivated CPXV C4L/020 orthologs can be found in the following OPVs: VARV, TATP, HSPV and MPXV. A close inspection of those genes reveals that independent gene-inactivating mutations are found in each of those viruses (Figure. 3). In HSPV and TATP the open reading frames (ORFs) are interrupted by short insertions/deletions (indels) that lead to premature stop-codons. In all VARV and MPXV strains the putative start codon is mutated from an ATG to TTG. Alternative downstream ATGs are out-of frame and are not predicted to lead to functional proteins. VARV strains can be grouped into 3 major clades: West African, South American and Asian clades, where the first two share a common ancestor [7]. The ORFs corresponding to the C4L/020 orthologs in all three South American strains as well as in strain NIG69_001 (West Africa) are intact and could potentially lead to a full-length product if translation was initiated at the non-cognate start-codon TTG, which can serve as a start-codon for some cellular genes [8]. The other West African VARV strains contain a 1 bp insertion at position 48, while all Asian strains contain an independent 1 bp deletion at position 140. All MPXV strains contain five gene-inactivating indels, three of which are shared between West African and Central African strains. The 3’ region of the C4L/020 ortholog in the West African clade is deleted at the genomic level. The presence of inactivated genes indicates that inactivation events occurred relatively recently or possibly that the inactivated/truncated genes, might have retained some function.
Figure 3.
Gene-inactivating mutations in orthopoxvirus CPXV-C4L/BR20 orthologs. Horizontal lines represent CPXV-C4L/BR20 orthologs of poxviruses that contain predicted gene-inactivating mutations. Mutation of start-codons that are followed by out-of-frame start-codons are indicated by red circles (stop codons). Deletions and insertions are indicated by blue triangles and inverse green triangles, respectively. Numbers above triangles indicate deleted or inserted nucleotides, which lead to frame shifts and premature stop-codons. Nucleotide identity (nt) in % of C4L/020 orthologs compared to CPXV-BR020 is shown on the right. Abbreviations of viruses are shown in Table 1.
C4L/020 orthologs are absent in the genomes of VACV, RPXV, ECTV and YKV
In Clade II poxviruses, at least one copy of a C7L ortholog is present. One unique example constitutes the three related C7L-like members in Leporipoxviruses. Phylogenetic analysis indicates that these three C7L-related genes (M062R, M063R and M064R) arose during Leporipoxvirus evolution through two distinct duplication events (Figure 2). MYXV, a rabbit specific pathogen, is the most extensively studied representative of Leporipoxvirus genus. MYXV is pathogenic only in European rabbits (O. cuniculus), and this in vivo model has been an instructive experimental system to study viral host range gene functions in pathogenesis and virulence [9–11**]. Targeted gene knockouts have been created for each of the M062R, M063R, and M064R genes, and their respective biological properties were characterized in vitro and in vivo. Interestingly, the M064R knockout virus does not exhibit a host range defect in any cell line tested to date, but instead is a virulence factor that controls the kinetics of MYXV infection in vitro and in vivo [10]. In contrast, M063R is a classic host range factor, in that the M063R-knockout virus cannot replicate in any tested cells from rabbit species [9,12]. Finally, M062R expression determines the host tropism in cells from almost all species tested, and only a few primate and human cancer cell lines were found to be permissive for the M062R-knockout virus [11**]. M062R has been shown to be able to compliment the functions of VACV-C7L in vitro and in vivo, at least when expressed in the genetic background of VACV without K1L and C7L, while neither M063R nor M064R can rescue this host range function [13**] (Table 3.). In addition, during MYXV infection, M062 and M063 proteins bind to each other to form a complex [11**]. Interestingly, in rabbit cells such as RK13, transiently transfected VACV-C7L cannot rescue the abortive infection by the M062R knockout MYXV [11**]. It suggests that the emergence of M063 and its interaction to M062 protein during evolution may well be a part of the unique adaption of the leporipoxviruses to the rabbit host.
Table 3.
A summary of the functional tests for C7L homologs.
| C7L homologs | Viral background tested |
Cell line tested for its Functions |
Animal model tested and apparent functions | |
|---|---|---|---|---|
| VACV-C7L | VACV-K1L−C7L− | Host range function: (human) A-431 and HeLa, (mouse) NIH3T3 and LA-4; Antagonize IFN effector: (human) MCF-7, Huh7; (mouse) P815 |
Balb/c mouse (intranasal inoculation) [13**] |
Virulent with high viral yield in lung and spleen |
| MYXV-M062R | VACV-K1L−C7L− | Host range function: A-431, HeLa, NIH3T3; Antagonize IFN effector: Huh7, P815 |
Balb/c mouse (intranasal inoculation) [13**] |
Virulent with high viral yield in lung and spleen |
| MYXV-M063R | VACV-K1L−C7L− | No host range function: A-431, HeLa, NIH3T3; Not antagonize IFN effector: Huh7 and P815 |
Not tested. | |
| MYXV-M064R | VACV-K1L−C7L− | No host range function: A-431, HeLa, NIH3T3; Not antagonize IFN effector: Huh7 and P815 |
Balb/c mouse (intranasal inoculation) [13**] |
Not virulent. |
| MYXV-M062R | MYXV | [11**] | European rabbit (intradermal inoculation) [11**] |
A determinant of pathogenesis |
| MYXV-M063R | MYXV | [9,12] | European rabbit (subcutaneous/intrader mal inoculation) [9] |
A determinant of pathogenesis |
| MYXV-M064R | MYXV | [10] | European rabbit (intradermal inoculation) [10] |
A virulence factor |
| YDLV-67R | VACV-K1L−C7L− | Host range function: A-431, HeLa, NIH3T3; Antagonize IFN effector: Huh7 and P815 |
Not tested. | |
| CPXV-BR020 | VACV-K1L−C7L− | No host range function: A-431, HeLa, NIH3T3; Not antagonize IFN effector: Huh7 and P815 |
Not tested. | |
| SWPV-064 | VACV-K1L−C7L− | Host range function: HeLa, NIH3T3, LA-4; Antagonize IFN effector: Huh7 and P815 |
Not tested. | |
| SPPV-063 | VACV-K1L−C7L− |
|
Not tested. | |
| VACV-C7L | modified vaccinia virus Ankara (MVA) (K1L fragmented) |
Antagonize eIF2α phosphorylation in a PKR dependent manner: HeLa, NIH3T3, MRC-5 |
Not tested. | |
The presence of multiple copies of C7L homologs in some poxviruses indicates distinct roles in the life cycle of the respective viruses. They may target different host factors for a given species or may have evolved to inhibit anti-viral host proteins from different species and therefore increase the host range of these viruses. In that respect it is interesting to note that two C7L-like genes are found in all 10 completely sequences cowpox viruses, which have the broadest host range amongst all poxviruses. The poxvirus C7L family represents an example of the model of birth-and-death evolution of multigene family members derived from common ancestral gene precursors [14]. New genes derive through duplication, such as C4L/BR020 vs. C7L, and M062R/M064R vs. M063R. Some genes are probably maintained through selective pressure during the viral replication cycle, while others become dysfunctional through deleterious mutations, as evidenced in the C4L/BR020 clade. In other words, the status of the host range gene family members can reveal novel features of the evolutionary divergence between related viruses.
Functional Analyses of C7L Family Members
In VACV, the functions of C7L and K1L, a member of another poxvirus host range gene family, have been known to complement each other, at least in terms of host restriction in human cells. In contrast to all of the C7L orthologs, K1L contains multiple copies of ankyrin repeats, motifs important for protein-protein interactions. It has been shown that both C7L and K1L can antagonize the dsRNA-activated protein kinase (PKR) pathway by inhibiting the phosphorylation of eIF2α [13**,15,16]. However, when the role of C7L was specifically investigated, various conditions used in these studies, such as virus genetic backgrounds, species of cell lines tested, and readout approaches, have led to sometimes conflicting conclusions. Given the complexity of the PKR pathway, an indirect effect of C7L and K1L on eIF2α phosphorylation may be mediated through allowing E3L and/or K3L to antagonize different aspects of PKR pathway. For example, murine PKR is more sensitive to inhibition by VACV-K3 than is the human PKR [17], and this might explain why ectopic E3 alone cannot rescue the defect of the lack of both C7L and K1L in inducing PKR dependent phosphorylation of eIF2α in murine cells [15]. The role of various C7L members at regulating the PKR pathway will rely on further investigation.
Within the VACV background with K1L knockout, it has been shown that C7 can also antagonize interferon (IFN) induced anti-viral effects, not at the step of IFN signaling, but the establishment of the anti-viral state induced by type I IFN in two human cell lines, Huh7 and MCF-7 [18*]. This effect is possibly mediated via specific IFN inducible host factors that are inhibitory targets of C7L family members. By sequential screening, the IFN regulatory factor 1 (IRF1) was identified to be responsible for mediating the anti-viral effects against C7L-/K1L-null VACV (VACV-C7L−K1L−) in Huh7 cells [19**]. However, in the same study it was also shown that, at least in murine embryonic fibroblasts, IRF1 is not the factor accountable for the host restriction against VACV-C7L− K1L−. Several questions remain to be answered, for example: 1) whether IRF1 is responsible for IFN-induced anti-viral effects against VACVC7L −K1L− in other cell lines than Huh7 and P815 that are permissive to its infection; 2) what is the mechanistic link between IFN/IRF1 mediated antiviral activities and C7L’s host range function in overcoming host restriction; 3) what is the host pathway that C7L family directly targets. The IFN-induced anti-viral effects antagonized by VACV-C7 were apparently PKR independent, at least in human Huh7 cells, but it is entirely possible that the role of PKR activation in mediating the anti-viral state may vary greatly between cell lines. So far, human and murine cancer cells have also been useful model systems to investigate of the role of C7L family member functions. Caution needs to be taken, because of the myriad of mutations occurring in different cancer cells may allow phenotypes that are not permitted in normal cells [20,21].
It has also been shown that VACV C7 can inhibit apoptosis when the gene is inserted into the genome of NYVAC, another attenuated VACV strain in which 18 viral ORFs (including C7L and K1L) were deleted [22]. The application in vaccine development would be to insert both C7L and K1L into the NYVAC genetic background to improve the priming potential of host adaptive immunity while maintaining the vector safety profile [23,24].
Poxvirus host range factors can be powerful tools to uncover novel functions of uncharacterized host factors and signaling pathways. For example, CPXV-CP77, another ankyrin-repeat host range factor unique to OPVs, was shown to interact with a host factor HMG20A [25]. Functionally, CPXV-CP77 can complement K1L in overcoming host restriction of VACV in RK-13 cells whereas VACV-C7L cannot. However, CPXV-CP77 is not 100% efficient in complementing VACV-K1L, suggesting its host targets may not be exactly the same as K1L or alternatively might not be as efficiently targeted in the cell line tested [26]. Similarly, the efficiency in mediating host range function may well not be equivalent between VACV-C7L and K1L. It was shown that VACV-K1 binds to ACAP2, a GTPase activating protein in rabbits, but in humans the interaction between VACV-K1 and human ACAP2 is not apparently associated with its host range function [27]. A definitive test is still needed to confirm whether the interaction between rabbit ACAP2 and VACV-K1 is essential to VACV host range in rabbit cells. Finally, these somewhat interchangeable viral factors are probably not functionally equal in terms of their specific molecular targets, suggesting their interactions with host countermeasure pathways are not identical. So far, the reported host-virus protein interactions that have been implicated in mediating viral tropism to date only paint an incomplete view.
Host-Pathogen Interactions Mediated by C7L Host Range Factors
Poxvirus immunoregulatory factors have been instrumental in uncovering players of host antiviral immunity, such as the discovery of the PKR [28,29], and 2’-5’OAS/RNase L pathways [30]. The conserved presence of C7L-like family members among poxviruses suggests an evolutionary advantage to maintain this gene family during the adaptation to new host species. Among poxviruses that belong to Clade II, the corresponding C7L-like genes map to the conserved region of viral genome between the Thymidine kinase (TK) and poly (A) polymerase small subunit. It is thus expected that these C7L members play an essential role in the viral life cycle. For example, the only functional homolog of C7L in MYXV is M062R, whose deletion from the MYXV genome completely abrogates the ability of the resulting virus to replicate within not only cells from its pathogenic host (European rabbit) but most mammalian cells tested from other species as well [11**]. However, the OPV members that are categorized in Clade I possess C7L-like genes that reside in the terminal regions of the viral genome. These members are not necessarily essential to viral replication in cultured cells but instead play a role as immunoregulators that counteract host defenses in vivo. Each individual poxvirus has evolved in a unique series of host species. Thus when the functions of C7L family members are surveyed, it should not be surprising that they have adapted to attack common defense pathways in each host but in different ways.
When Clade II C7L orthologs were investigated in the VACV background without K1L, they are not all equal in terms of their ability to functionally replace C7L. Even though all functional homologs tested can compensate the lack of C7L in antagonizing IFN effectors in Huh7 cells, SPPV-063, specifically, was not able to overcome the host restriction of murine cells [19**]. It is noteworthy that the replacement of amino acids at 134 and 135 residues from Asparagine and Phenylalanine to Tyrosine (Tyr) and Isoleucine (Ile), respectively, corrected this defect in murine cells, such as NIH3T3. It is possible these corresponding residues in SPPV-063 are customized for its interaction with their cognate host targets in cells from susceptible sheep but are not designed to do the same in murine system. On the other hand, these same two amino acids (Tyr and Ile) in the corresponding location of VACV-C7L are likely essential for its host range function in both human and murine cells [19**].
There is evidence that related C7L family member proteins may interact with divergent host targets. A cellular factor, sterile α motif domain containing 9 (SAMD9), has been identified to be the binding partner of M062, in human cells and possibly rabbit cells [11**]. This protein interaction prevents anti-viral functions of SAMD9 in inhibiting MYXV replication. However, the related VACV-C7 and M067 of Yaba-like disease virus (YLDV) do not directly bind to SAMD9, at least in the context of MYXV infection in the absence of M062. On the other hand, both VACV-C7 and YLDV-M067 can compensate for the function of MYXV-M062, but with less efficiency, in the MYXV background when infecting human cells. A conclusion can be drawn that a direct binding to SAMD9 is not the only mechanism for the function of all members of C7L family, although SAMD9 may be a component of a larger pathway that is targeted elsewhere by the other C7L family members. The biological function of SAMD9 remains unknown, but it has an essential role in the inflammatory response associated with calcified tumors [31–33]. SAMD9 is also localized at a central node of the signaling network, regulated by IFNs, TNF, and cellular stress signals, and the gene possesses promoter elements responsive to IFNγ and the binding domain corresponding to IRF1 [32]. Interestingly, VACV-C7 was found to antagonize IRF-1 induced anti-viral effectors in some cells [19**] suggesting the potential link between other functional members of C7L family and the SAMD9 pathway. However, in mice the homologous gene of SAMD9 has been lost through chromosomal rearrangement during evolution [34]. It is known that compensatory evolution allows the tolerance of the loss of an important gene to restore the phenotype [35]. A paralog of SAMD9, named SAMD9-like (SAMD9L), which was derived from the same ancestral gene as SAMD9 during mammalian evolution, is kept intact in mice and is a potential candidate for mediating the equivalent anti-viral pathway in mice. The differences between the human and murine SAMD9/SAMD9L systems in relation to the host range functions of C7L family members is currently under investigation.
Conclusions and Future Directions
The quest to understand the host range functions of C7L family members so far has reminded us that viruses manipulate host defenses at multiple levels, each with different goals, and these manipulations can be remarkably host-specific. The presence of members of multiple host range families within a single virus may seem redundant at first glance, but it is exactly what makes individual viruses successful during evolution as they subvert host defenses in a potentially progressive series of host species. This virus-host co-evolution has played a considerable part in shaping the architecture of the C7L gene family. Thus, the cellular countermeasure pathway(s) targeted by each C7L member may well be different, depending on the evolutionary past of each individual virus. For some poxviruses, the exact co-evolutionary host remains to be identified, such as VACV and CPXV. To begin the investigation on viruses with the known natural host and with a well characterized model system is most appropriate. A good candidate virus in this regard is ECTV, whose natural reservoir is the mouse species and experimental mouse model systems have been well developed. It is noteworthy that C7L family members from OPV may well have a role in the immunoregulatory process, and murine models of ECTV infection allow the further investigation in this aspect due to the abundant availability of material, such as antibodies, necessary to probe host immune responses. To identify host factors interacting with C7L family members, proteomic analyses, such as co-immunoprecipitation (co-IP) and mass spectrometry have been instrumental. However, as some interactions can be transient or cellular localizationdependent, this co-IP directed approach may not always be satisfying. Nevertheless, the identification of the interaction between SAMD9 and MYXV M062 has been the only clue so far that a C7L family member can directly interact with a host antiviral factor. Further investigation of SAMD9’s cellular functions and the identification of the pathway involved with SAMD9 will allow us to understand which aspect of the host defense system C7L family member has evolved to antagonize. The complex nature of the functions of C7L family members provides an excellent opportunity of discovery of uncharacterized cellular pathways that are comprised of diverse host factors in a species-specific manner.
Highlights.
C7L family members have evolved to adapt to their corresponding hosts.
Not all C7L family members function via the same mechanism.
Some C7L members can overcome host restriction and inhibit type I IFN induced antiviral activities.
MYXV-M062, binds to antagonize host SAMD9, providing clues on other C7L orthologs’ functions.
Acknowledgements
This work was supported by grant RO1 AI080607 from the National Institute of Health to GM and NIH grant P20 RR016475 from the INBRE Program of the National Center for Research Resources to SR. We thank Loubna Tazi for helpful discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Francis RD, Bradford HB., Jr Some biological and physical properties of molluscum contagiosum virus propagated in cell culture. J Virol. 1976;19:382–388. doi: 10.1128/jvi.19.2.382-388.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McInnes CJ, Wood AR, Nettleton PE, Gilray JA. Genomic comparison of an avirulent strain of Orf virus with that of a virulent wild type isolate reveals that the Orf virus G2L gene is non-essential for replication. Virus Genes. 2001;22:141–150. doi: 10.1023/a:1008117127729. [DOI] [PubMed] [Google Scholar]
- 3.Hughes AL, Friedman R. Poxvirus genome evolution by gene gain and loss. Mol Phylogenet Evol. 2005;35:186–195. doi: 10.1016/j.ympev.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 4.Bratke KA, McLysaght A. Identification of multiple independent horizontal gene transfers into poxviruses using a comparative genomics approach. BMC Evol Biol. 2008;8:67. doi: 10.1186/1471-2148-8-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhao G, Droit L, Tesh RB, Popov VL, Little NS, Upton C, Virgin HW, Wang D. The genome of Yoka poxvirus. J Virol. 2011;85:10230–10238. doi: 10.1128/JVI.00637-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Afonso PP, Silva PM, Schnellrath LC, Jesus DM, Hu J, Yang Y, Renne R, Attias M, Condit RC, Moussatche N, et al. Biological characterization and next-generation genome sequencing of the unclassified Cotia virus SPAn232 (Poxviridae) J Virol. 2012;86:5039–5054. doi: 10.1128/JVI.07162-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Esposito JJ, Sammons SA, Frace AM, Osborne JD, Olsen-Rasmussen M, Zhang M, Govil D, Damon IK, Kline R, Laker M, et al. Genome sequence diversity and clues to the evolution of variola (smallpox) virus. Science. 2006;313:807–812. doi: 10.1126/science.1125134. [DOI] [PubMed] [Google Scholar]
- 8.Peabody DS. Translation initiation at non-AUG triplets in mammalian cells. J Biol Chem. 1989;264:5031–5035. [PubMed] [Google Scholar]
- 9.Barrett JW, Shun Chang C, Wang G, Werden SJ, Shao Z, Barrett C, Gao X, Belsito TA, Villenevue D, McFadden G. Myxoma virus M063R is a host range gene essential for virus replication in rabbit cells. Virology. 2007;361:123–132. doi: 10.1016/j.virol.2006.11.015. [DOI] [PubMed] [Google Scholar]
- 10.Liu J, Wennier S, Moussatche N, Reinhard M, Condit R, McFadden G. Myxoma virus M064 is a novel member of the poxvirus C7L superfamily of host range factors that controls the kinetics of myxomatosis in European rabbits. J Virol. 2012;86:5371–5375. doi: 10.1128/JVI.06933-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liu J, Wennier S, Zhang L, McFadden G. M062 is a host range factor essential for myxoma virus pathogenesis and functions as an antagonist of host SAMD9 in human cells. J Virol. 2011;85:3270–3282. doi: 10.1128/JVI.02243-10. This study is the first to identify the direct and functional interaction between a C7L family member and a host antiviral factor.
- 12.Barrett JW, Alston LR, Wang F, Stanford MM, Gilbert PA, Gao X, Jimenez J, Villeneuve D, Forsyth P, McFadden G. Identification of host range mutants of myxoma virus with altered oncolytic potential in human glioma cells. J Neurovirol. 2007;13:549–560. doi: 10.1080/13550280701591526. [DOI] [PubMed] [Google Scholar]
- 13. Meng X, Chao J, Xiang Y. Identification from diverse mammalian poxviruses of host-range regulatory genes functioning equivalently to vaccinia virus C7L. Virology. 2008;372:372–383. doi: 10.1016/j.virol.2007.10.023. This study tested several members of C7L superfamily on the ability to compensate the loss of VACV C7L in vitro and in vivo. It was shown that MYXV M062R, but not M063R or M064R, can fully compliment the function of VACV C7L. In addition, it was shown that CPXV-BR- 020 cannot compliment the function of VACV-C7L, at least in the VACV background.
- 14.Nei M, Rooney AP. Concerted and birth-and-death evolution of multigene families. Annu Rev Genet. 2005;39:121–152. doi: 10.1146/annurev.genet.39.073003.112240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Backes S, Sperling KM, Zwilling J, Gasteiger G, Ludwig H, Kremmer E, Schwantes A, Staib C, Sutter G. Viral host-range factor C7 or K1 is essential for modified vaccinia virus Ankara late gene expression in human and murine cells, irrespective of their capacity to inhibit protein kinase R-mediated phosphorylation of eukaryotic translation initiation factor 2alpha. J Gen Virol. 2010;91:470–482. doi: 10.1099/vir.0.015347-0. [DOI] [PubMed] [Google Scholar]
- 16.Willis KL, Patel S, Xiang Y, Shisler JL. The effect of the vaccinia K1 protein on the PKR-eIF2alpha pathway in RK13 and HeLa cells. Virology. 2009;394:73–81. doi: 10.1016/j.virol.2009.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rothenburg S, Seo EJ, Gibbs JS, Dever TE, Dittmar K. Rapid evolution of protein kinase PKR alters sensitivity to viral inhibitors. Nat Struct Mol Biol. 2009;16:63–70. doi: 10.1038/nsmb.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Meng X, Jiang C, Arsenio J, Dick K, Cao J, Xiang Y. Vaccinia virus K1L and C7L inhibit antiviral activities induced by type I interferons. J Virol. 2009;83:10627–10636. doi: 10.1128/JVI.01260-09. Authors used two unique human cancer cell lines as the model system to examine the functions of VACV-K1L and C7L. They showed that both viral factors can antagonize the antiviral activities induced by type I interferon.
- 19. Meng X, Schoggins J, Rose L, Cao J, Ploss A, Rice CM, Xiang Y. C7L family of poxvirus host range genes inhibits antiviral activities induced by type I interferons and interferon regulatory factor-1. J Virol. 2012;86:4538–4547. doi: 10.1128/JVI.06140-11. This study confirmed that poxvirus C7L orthologs are all capable of antagonizing the antiviral effect induced by type I interferon against the recombinant VACV without both C7L and K1L. They also identified IRF-1 to be responsible for the inhibitory effect against this double knockout VACV in Huh7 and P815 cells. Finally, the authors identified the residues of Asn134 and Phe135 to be responsible for the defect in SPPV-063 in complimenting VACV-C7L by overcoming host range restriction in mouse cells.
- 20.Bartee E, McFadden G. Human cancer cells have specifically lost the ability to induce the synergistic state caused by tumor necrosis factor plus interferon-beta. Cytokine. 2009;47:199–205. doi: 10.1016/j.cyto.2009.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McFadden G, Mohamed MR, Rahman MM, Bartee E. Cytokine determinants of viral tropism. Nat Rev Immunol. 2009;9:645–655. doi: 10.1038/nri2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Najera JL, Gomez CE, Domingo-Gil E, Gherardi MM, Esteban M. Cellular and biochemical differences between two attenuated poxvirus vaccine candidates (MVA and NYVAC) and role of the C7L gene. J Virol. 2006;80:6033–6047. doi: 10.1128/JVI.02108-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kibler KV, Gomez CE, Perdiguero B, Wong S, Huynh T, Holechek S, Arndt W, Jimenez V, Gonzalez-Sanz R, Denzler K, et al. Improved NYVAC-based vaccine vectors. PLoS One. 2011;6:e25674. doi: 10.1371/journal.pone.0025674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Najera JL, Gomez CE, Garcia-Arriaza J, Sorzano CO, Esteban M. Insertion of vaccinia virus C7L host range gene into NYVAC-B genome potentiates immune responses against HIV-1 antigens. PLoS One. 2010;5:e11406. doi: 10.1371/journal.pone.0011406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hsiao JC, Chao CC, Young MJ, Chang YT, Cho EC, Chang W. A poxvirus host range protein, CP77, binds to a cellular protein, HMG20A, and regulates its dissociation from the vaccinia virus genome in CHO-K1 cells. J Virol. 2006;80:7714–7728. doi: 10.1128/JVI.00207-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ramsey-Ewing AL, Moss B. Complementation of a vaccinia virus host-range K1L gene deletion by the nonhomologous CP77 gene. Virology. 1996;222:75–86. doi: 10.1006/viro.1996.0399. [DOI] [PubMed] [Google Scholar]
- 27.Meng X, Xiang Y. Vaccinia virus K1L protein supports viral replication in human and rabbit cells through a cell-type-specific set of its ankyrin repeat residues that are distinct from its binding site for ACAP2. Virology. 2006;353:220–233. doi: 10.1016/j.virol.2006.05.032. [DOI] [PubMed] [Google Scholar]
- 28.Garcia MA, Meurs EF, Esteban M. The dsRNA protein kinase PKR: virus and cell control. Biochimie. 2007;89:799–811. doi: 10.1016/j.biochi.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 29.Metz DH, Esteban M. Interferon inhibits viral protein synthesis in L cells infected with vaccinia virus. Nature. 1972;238:385–388. doi: 10.1038/238385a0. [DOI] [PubMed] [Google Scholar]
- 30.Hovanessian AG, Justesen J. The human 2'-5'oligoadenylate synthetase family: unique interferon-inducible enzymes catalyzing 2'-5' instead of 3'-5' phosphodiester bond formation. Biochimie. 2007;89:779–788. doi: 10.1016/j.biochi.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 31.Chefetz I, Ben Amitai D, Browning S, Skorecki K, Adir N, Thomas MG, Kogleck L, Topaz O, Indelman M, Uitto J, et al. Normophosphatemic familial tumoral calcinosis is caused by deleterious mutations in SAMD9, encoding a TNF-alpha responsive protein. J Invest Dermatol. 2008;128:1423–1429. doi: 10.1038/sj.jid.5701203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hershkovitz D, Gross Y, Nahum S, Yehezkel S, Sarig O, Uitto J, Sprecher E. Functional Characterization of SAMD9, a Protein Deficient in Normophosphatemic Familial Tumoral Calcinosis. J Invest Dermatol. 2010 doi: 10.1038/jid.2010.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Topaz O, Indelman M, Chefetz I, Geiger D, Metzker A, Altschuler Y, Choder M, Bercovich D, Uitto J, Bergman R, et al. A deleterious mutation in SAMD9 causes normophosphatemic familial tumoral calcinosis. Am J Hum Genet. 2006;79:759–764. doi: 10.1086/508069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li CF, MacDonald JR, Wei RY, Ray J, Lau K, Kandel C, Koffman R, Bell S, Scherer SW, Alman BA. Human sterile alpha motif domain 9, a novel gene identified as down-regulated in aggressive fibromatosis, is absent in the mouse. BMC Genomics. 2007;8:92. doi: 10.1186/1471-2164-8-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harcombe WR, Springman R, Bull JJ. Compensatory evolution for a gene deletion is not limited to its immediate functional network. BMC Evol Biol. 2009;9:106. doi: 10.1186/1471-2148-9-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Katoh K, Kuma K, Toh H, Miyata T. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Swofford DL. PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods). Version 4. edn 4. Sunderland, Massachusetts: Sinauer Associates; 2003. [Google Scholar]