Abstract
As a relatively simple virus, hepatitis C virus (HCV) depends extensively on its host to infect, replicate and disseminate. HCV has evolved host interactions that result in a restricted tropism, both in terms of cell type and species. Efforts into identifying and validating HCV-host interactions have been hampered by a limited number of infectious virus clones and cell lines that support HCV infection. Despite these limitations, consensus HCV-host interactions have emerged that help define the entry, replication, assembly, and tropism of HCV. This has had important implications in expanding our in vitro and in vivo systems to study HCV replication and pathogenesis. Additionally, a number of these host factors are being targeted for therapeutic development. In this review, we focus on medically relevant pro-viral host factors, their role in HCV biology, and their importance in expanding our model systems.
Introduction
HCV is a major cause of chronic hepatitis leading to liver cirrhosis and hepatocellular carcinoma [1]. An estimated 130 million people worldwide are persistently infected with HCV [2]. The available therapy, a combination of pegylated interferon-α (pegIFN-α) and ribavarin, is efficacious in only 50% of the infected individuals and associated with side effects [3,4]. Direct acting antivirals (DAAs), such as boceprevir and telaprevir, which inhibit the viral NS3/4A protease, will cure ~70% of patients when combined with pegIFN-α/ribavirin, although they are associated with side effects [5,6]. In addition to DAAs, a number of drugs targeting HCV host factors are in development. These may have utility in reducing resistance in combination with DAAs.
Expanding technologies, such as RNA interference (RNAi) screens, have identified literally hundreds of putative HCV cofactors that expand beyond the possible focus of this review. Additionally, HCV infection of Huh7 cell derivatives produces numerous cellular changes in signaling and metabolism that we unfortunately omit due to space considerations. We will instead highlight consensus host cofactors and their roles in expanding HCV model systems and therapeutic options.
HCV Entry
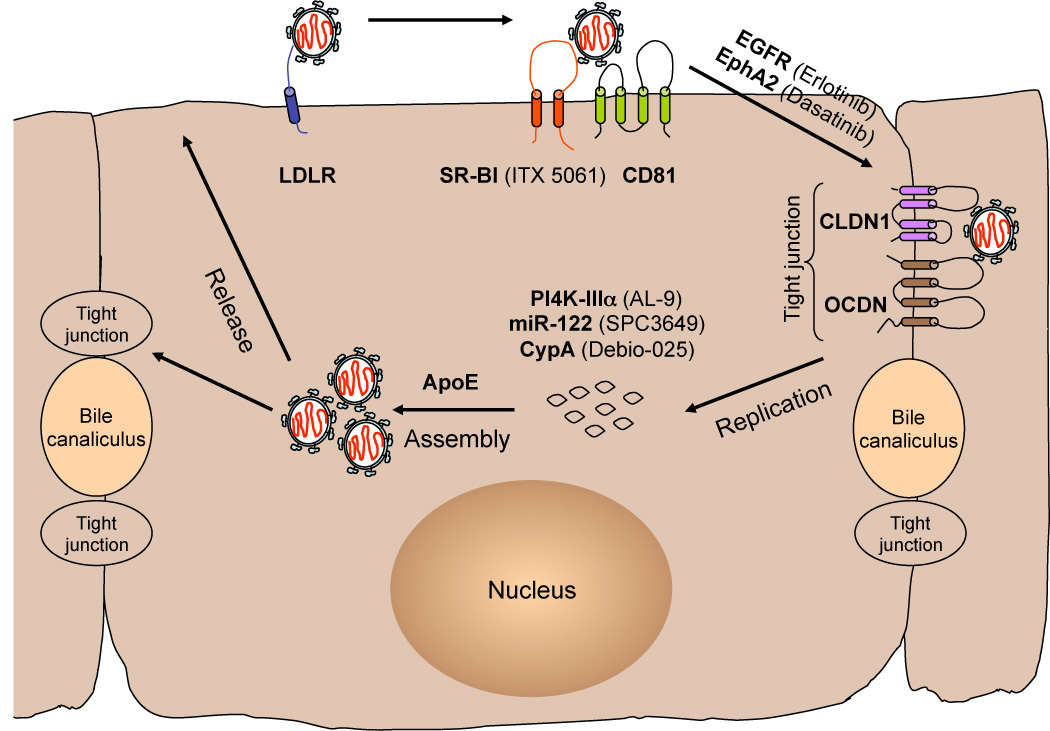
HCV entry into hepatocytes is a complex process that engages several cellular proteins such as the low density lipoprotein receptor (LDLR) [7,8], the tetraspanin CD81[9–11], the scavenger receptor class B type I (SR-BI) [12,13] and the tight junction (TJ) proteins, claudin-1 (CLDN1) [14] and occludin (OCLN) [15,16]. The in vitro data so far suggest that these host factors are used in a sequential manner, with virus particles attaching to hepatic cells via a lower affinity LDLR interaction and a higher affinity binding to SR-BI. Soluble E2 proteins, but not HCV particles, bind CD81 [9,10], suggesting that HCV-SRBI interactions alter HCV virion conformation [17,18], thus enabling E2-CD81 binding and subsequent priming of E2 for pH-dependent fusion [19*]. Following engagement of CD81, signaling events are necessary for recruitment of CLDN1. The receptor tyrosine kinases, epidermal growth factor receptor (EGFR) and ephrin receptor A2 (EphA2) modulate CD81-CLDN1 association, likely by influencing tyrosine kinase signaling [20**]. Following CD81-CLDN1 binding, the HCV-receptor complex is proposed to interact with OCLN and internalize at cellular TJs. HCV internalizes via clathrin-mediated endocytosis and uncoats from acidified endosomes [21,22]. This has led to a model, with similarities to group B coxsackieviruses [23], of initial binding of HCV to SR-BI/CD81 co-receptors followed by signaling and migration to TJs where TJ co-receptors mediate late stages of virion internalization. Numerous steps of this model remain untested for HCV.
In addition to direct virus interactions, cellular receptors that modulate lipid/cholesterol uptake influence HCV entry. Mutations in SR-BI that influence cholesterol uptake, but not E2 binding inhibit HCV entry [24] as does inhibition of the cholesterol uptake receptor Niemann Pick C1 like 1 (NPC1L1) [25*]. Inhibitors of EGFR (erlotinib), EphA2 (dasatinib) and NPC1L1 (ezetimibe) [26] are already licensed molecules and shown to inhibit HCV entry in vitro [20,25]. Additionally, there is a small molecule inhibitor of SR-BI (ITX 5061) which is the most advanced HCV entry inhibitor in clinical trials [27].
HCV Replication
As do all other positive-strand RNA viruses, HCV replicates its genome in association with virally-induced membrane structures. RNAi screens have uncovered numerous host cofactors of HCV replication, with the most consistently identified being phosphatidylinositol 4-kinase III α (PI4K-IIIα) [28–35**]. This enzyme phosphorylates phosphatidylinositol (PI) in the 4 position of the inositol ring to generate PI4P. During HCV infection, PI4K-IIIα interacts with the viral NS5A protein resulting in increased local levels of PI4P. PI4K-IIIα kinase activity is required for the fidelity of membranous web formation, as silencing PI4K-IIIα results in an aggregation of double membrane vesicles and HCV replication complexes [35**,36*,37]. The exact function of PI4K-IIIα, PI4P, or further modifications of PI4P in HCV replication are unclear. AL-9, a member of the 4-anilino quinazoline-containing kinase inhibitor family, was recently shown to inhibit HCV replication in vitro by direct inhibition of PI4K-IIIα [38*]. Additionally, resistance mutations to a class of NS5A DAAs map to NS5A domain I, which also interacts with PI4K-IIIα [34**], suggesting that these drugs may impact NS5A activation of PI4K-IIIα [39].
The liver-specific micro-RNA 122 (miR-122), which regulates cholesterol biosynthesis, binds to two closely spaced sites on the 5’ noncoding region of the HCV genome and facilitates viral replication [40–42]. miR-122 enhances HCV translational initiation [43,44], however, this is not sufficient to explain the full effect of miR-122 on HCV replication [45]. Recent data suggest that miR-122 binding to the HCV genome has a protective and stabilizing role. Indeed, miR-122 binds HCV RNA with 3’ overhanging nucleotides that seem to mask the 5’ terminal sequences from nucleolytic degradation or innate immunity cytoplasmic sensors of viral RNA [46*]. miR-122 binds HCV RNA in association with Argonaute 2 (Ago2) protein resulting in slower decay of the viral genome and protection from the cellular exonuclease decay machinery [47*]. In support of the stability hypothesis, HCV deleted in part of the miR-122 binding sites can be partially rescued by recombination with stable viral or cellular RNA structures [48*]. Miravirsen (SPC3649/Santaris Pharma) is a locked nucleic −acid modified oligonucleotide complimentary to miR-122, currently in Phase II clinical trial. When administered in chronically infected chimpanzees, this inhibitor resulted in long-lasting suppression of HCV viremia with no evidence of viral resistance and minimal side effects [49**]. Additionally, miravirsen is capable of potently antagonizing multiple HCV genotypes in vitro [48*].
Cyclophilin A (CypA) is a cellular chaperone with peptidyl-prolyl cis-trans isomerase (PPiase) activity that functions in protein folding and trafficking and plays an essential role in HCV replication and particle production [50–53]. CypA may play a role in the correct folding of several viral proteins, given that its PPiase activity is required for HCV replication in vitro [53,54]. Initially, CypA was found to associate with the viral NS5B replicase and modulate its RNA binding and/or synthesis capability [55–57]. More recently, CypA has been shown to bind specific motifs in domain II and III of the viral NS5A protein catalyzing cis/trans isomerization and thus stimulating the RNA binding activity and dimerization of NS5A [58, 59*–62]. Additionally, CypA may affect HCV polypeptide processing at the NS5A-B junction [52]. The original CypA inhibitor was cyclosporine A (CsA), an immunosuppressive drug used routinely in organ transplantation [63]. Chemical modification of CsA resulted in non-immunosuppressive analogs of the inhibitor that potently suppress HCV replication in cell culture and have clinical efficacy in HCV patients [64–66*]. The most promising of the cyclophilin inhibitors is alisporivir (Debio-025/Novartis) which is currently in Phase III clinical trials. Debio-025 given in combination with pegIFN-α/ribavirin has superior efficacy compared to standard of care treatment [52].
HCV Assembly and Release
The molecular events that take place during assembly of infectious HCV particles and the interplay between viral and host factors, are just being uncovered. It is hypothesized that HCV assembly is initiated in close proximity to intracellular lipid droplet (LD) structures on the surface of which the viral core (capsid) protein accumulates [67,68]. In addition to viral factors, several host factors likely participate in HCV particle assembly and envelopment, including the clathrin adaptor AP2M1 [69] and group IVA phospholipase A2 (PLA2G4A) [70*]. Components of the very-low-density lipoprotein (VLDL) synthesis and secretion pathway such as microsomal triglyceride transfer protein [71], apolipoprotein B (apoB) [72] and apolipoprotein E (apoE) have been implicated in HCV assembly [73–75]. Of these, substantial evidence supports a role for apoE as an HCV infectivity factor. HCV infectious virions purified either from cell culture supernatants [73] or infected patients can be specifically immuno-precipitated by anti-apoE monoclonal antibodies [76]. Furthermore, apoE was found to be part of affinity purified infectiousHCV particles [77*]. Live cell imaging of single HCV particles indicates that mature infectious particles containing apoE are transported along the secretory pathway [78**]. It has been suggested that apoE may promote HCV infectivity during entry by virtue of its interaction with LDLR [8] or heparan sulfate [79].
Another host factor implicated in HCV assembly is diacylglycerol acyltransferase-1 (DGAT1) [80*]. DGAT1 induces LD formation in cells and its interaction with viral core protein was identified to be essential in proper localization of core around LDs. Of note, DGAT1 inhibitors currently in clinical trials for obesity-related diseases could be potential candidates against HCV [81].
HCV tropism
HCV has a restricted tropism, both in terms of species and cell type. Only humans and chimpanzees are naturally infected, with the primary target cell being the hepatocyte. Extra-hepatic target sites, such as B lymphocytes, neuro-epithelioma cells and endothelial cells of the blood-brain barrier, can support virus entry but not a productive infection by cell culture derived HCV [82–84]. While host restriction factors could influence HCV tropism in vivo, the experimental data point to a primary contribution by pro-viral host factors [85]. The approach of screening for cDNAs that enable HCV pseudoparticle infection of non-permissive cells identified the HCV entry factors CLDN1 and OCDN [14,15]. OCDN in conjunction with CD81 define the species barrier to entry [15]. Expression of human OCDN and CD81 in mice, either by adenoviral delivery or transgenic approaches allows infection of mice, albeit without subsequent replication [86**]. This validates the significance of the entry factors in HCV infection, in addition to being an important step in developing a small animal model for HCV.
The main host factors for HCV replication appear to be conserved in multiple cell types and species. HCV JFH1 replicons can replicate in non-hepatic cells, including HeLa and 293T cells, albeit at lower levels than the hepatic Huh-7 cells [87,88]. One host factor that can enhance the tropism of HCV is miR-122. Ectopic expression of miR-122 in HepG2, Hec1b, 293T-CLDN, Hep3B and mouse fibroblasts enhances HCV replication [89–93]. Another tropism enhancing host cofactor is apoE. Over-expression of either mouse or human apoE isoforms increases the production of infectious HCV in mouse cells [92,94] and may be used as a tool to expand the cell types that support a complete infectious HCV cycle, in addition to improving the current small animal models. Notably, expression of the four entry receptor molecules in combination with miR-122 and apoE is sufficient to reconstitute the entire HCV life cycle in 293T cells [95*].
New in vitro models to validate and identify HCV-host interactions
A major limitation of studying HCV-host interactions is the limited number of HCV strains and cell types used to characterize them. There are seven known HCV phylogenetic groups (genotypes) with a nucleotide sequence diversity of around 30–35% [96], however viral infectious clones are available only for genotypes 1a, 1b, 2a, 3a and 4a [97–99]. Of these, only genotype 2a JFH1 and adapted genotypes 2a J6, 2b J8, and 1a H77S complete the viral life cycle in cell culture. An advance is the generation of chimeric HCV containing the JFH1 nonstructural genes fused to structural genes from all 7 HCV genotypes [100]. This will enable cross-genotypic comparisons of virus-host interactions with the HCV core, E1, E2, p7 and NS2; for example, infection with all 7 HCV genotype chimeras can be neutralized with antibodies to CD81 and SR-BI [100].
The in vitro studies validating host cell factors important for HCV replication were carried out predominantly in a single human hepatoma cell line, Huh-7 and other sublines termed Huh-7.5 [101], Huh-7.5.1 [102] and Huh-7 Lunet cells [103]. Human hepatocytes possess a unique complex polarity that is finely keened for uptake of nutrients at the basolateral surface (sinusoidal pole) and secretion of bile at the apical surface (bile canaliculus). The role of TJs in HCV entry is not clear yet, since these cell structures do not seem to be accessible to virions in vivo and entry into non-polarized Huh-7.5 cells does not preferentially occur at inter-cellular junctions, likely because these cells do not form TJs under normal culturing conditions [21]. Some progress has been made in developing polarized cell models for HCV entry, such as the Caco-2 colorectal adenocarcinoma cells that develop columnar polarity and the HepG2 hepatoma cells that develop complex hepatic polarity [104,105]. HepG2-CD81 cells expressing miR-122 have particular promise [89]. It is also likely that culturing conditions of existing HCV cell culture models can impact polarity. Recently, it shown that Huh-7 hepatocytes embedded in matrigel 3-dimensional cultures do polarize and support the entire HCV life cycle thus creating a system to validate host cell factors with respect to cell polarity [106].
Primary hepatocytes from patients can be productively infected with HCV; however, there is high variability between patients and access to fresh hepatocytes is limited [107,108]. An alternative approach is the development of induced pluripotent stem (iPS) cells. Multiple groups have successfully developed iPS cell culture systems that can be differentiated into human hepatocyte-like cells and infected with HCV [109*–111*]. Interestingly, the stage of cell differentiation susceptible to HCV infection correlates with expression of miR-122 and PI4K-IIIα [111]. Thus far, these cultures have (at least) two intriguing properties: (i) they produce an inflammatory response to infections and (ii) they can be infected with HCV1a and 1b patient isolates, suggesting that they may support the replication of a broad range of HCV genotypes [109*,111*].
Conclusions
Despite significant experimental hurdles, numerous bona fide HCV-host interactions have been defined. The newly developed model systems can begin to address questions that remain for these host factors. For instance, polarized hepatocytes are crucial to understanding HCV entry and trafficking. These model systems will also be important in defining the significance and function of the hundreds of less characterized putative host cofactors of HCV infection. Are there requirements conserved for multiple HCV genotypes in multiple cell lines? Are the HCV-induced changes in Huh7 cell physiology conserved in other hepatocyte models? How does the inflammatory signaling in response to HCV infection of iPS cells alter HCV replication and spread? Can driving over-expression of apoE in current HCV mouse models produce a robust small animal model? Finally, what will be the impact of therapeutics targeting host factors? Given the number of DAAs in advanced clinical trials, host targets are unlikely to be a primary therapeutic in the short term. However, they may emerge as useful tools in an arsenal to treat difficult populations. In any case, HCV should prove to be an important test model for targeting the host in emerging viral infections.
Figure 1. Model of hepatitis C virus life cycle in polarized hepatocytes.
Pro-viral host factors are highlighted in bold and their respective inhibitors in parenthesis. Abbreviations: LDLR, lipoprotein receptor; SR-BI, scavenger receptor type B1; EGFR, epidermal growth factor receptor; EphA2, ephrin receptor A2; CLDN-1, claudin-1; OCDN, occludin; PI4K-IIIa, phosphatidylinositol 4-kinase III α; miR-122, microRNA-122; CypA, cyclophilin A; ApoE, apolipoprotein E.
Highlights.
-
➢
Consensus HCV-host interactions have been identified at all stages of the viral life cycle.
-
➢
Many of these interactions are targets of drug development.
-
➢
These interactions define HCV tropism and are being used to expand our model systems.
Acknowledgements
We apologize to researchers whose relevant papers were omitted due to space restrictions. We would like to thank Vineela Chukkapalli and Yasmine Baktash for critical reading of the manuscript. G.R. acknowledges support from the National Institute of Allergy and Infectious Disease (AI080703), the American Cancer Society (118676-RSG-10-059-01-MPC), and Susan and David Sherman.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Shepard CW, Finelli L, Alter MJ. Global epidemiology of hepatitis C virus infection. Lancet Infect Dis. 2005;5:558–567. doi: 10.1016/S1473-3099(05)70216-4. [DOI] [PubMed] [Google Scholar]
- 2.Alter MJ. Epidemiology of hepatitis C virus infection. World J Gastroenterol. 2007;13:2436–2441. doi: 10.3748/wjg.v13.i17.2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Patel K, McHutchison JG. Initial treatment for chronic hepatitis C: current therapies and their optimal dosing and duration. Cleve Clin J Med. 2004;71(Suppl 3):S8–S12. doi: 10.3949/ccjm.71.suppl_3.s8. [DOI] [PubMed] [Google Scholar]
- 4.Manns MP, Wedemeyer H, Cornberg M. Treating viral hepatitis C: efficacy, side effects, and complications. Gut. 2006;55:1350–1359. doi: 10.1136/gut.2005.076646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kwo PY, Lawitz EJ, McCone J, Schiff ER, Vierling JM, Pound D, Davis MN, Galati JS, Gordon SC, Ravendhran N, et al. Efficacy of boceprevir, an NS3 protease inhibitor, in combination with peginterferon alfa-2b and ribavirin in treatment-naive patients with genotype 1 hepatitis C infection (SPRINT-1): an open-label, randomised, multicentre phase 2 trial. Lancet. 2010;376:705–716. doi: 10.1016/S0140-6736(10)60934-8. [DOI] [PubMed] [Google Scholar]
- 6.Zeuzem S, Andreone P, Pol S, Lawitz E, Diago M, Roberts S, Focaccia R, Younossi Z, Foster GR, Horban A, et al. Telaprevir for retreatment of HCV infection. N Engl J Med. 2011;364:2417–2428. doi: 10.1056/NEJMoa1013086. [DOI] [PubMed] [Google Scholar]
- 7.Molina S, Castet V, Fournier-Wirth C, Pichard-Garcia L, Avner R, Harats D, Roitelman J, Barbaras R, Graber P, Ghersa P, et al. The low-density lipoprotein receptor plays a role in the infection of primary human hepatocytes by hepatitis C virus. J Hepatol. 2007;46:411–419. doi: 10.1016/j.jhep.2006.09.024. [DOI] [PubMed] [Google Scholar]
- 8.Owen DM, Huang H, Ye J, Gale M., Jr Apolipoprotein E on hepatitis C virion facilitates infection through interaction with low-density lipoprotein receptor. Virology. 2009;394:99–108. doi: 10.1016/j.virol.2009.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pileri P, Uematsu Y, Campagnoli S, Galli G, Falugi F, Petracca R, Weiner AJ, Houghton M, Rosa D, Grandi G, et al. Binding of hepatitis C virus to CD81. Science. 1998;282:938–941. doi: 10.1126/science.282.5390.938. [DOI] [PubMed] [Google Scholar]
- 10.Cormier EG, Tsamis F, Kajumo F, Durso RJ, Gardner JP, Dragic T. CD81 is an entry coreceptor for hepatitis C virus. Proc Natl Acad Sci U S A. 2004;101:7270–7274. doi: 10.1073/pnas.0402253101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang J, Randall G, Higginbottom A, Monk P, Rice CM, McKeating JA. CD81 is required for hepatitis C virus glycoprotein-mediated viral infection. J Virol. 2004;78:1448–1455. doi: 10.1128/JVI.78.3.1448-1455.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scarselli E, Ansuini H, Cerino R, Roccasecca RM, Acali S, Filocamo G, Traboni C, Nicosia A, Cortese R, Vitelli A. The human scavenger receptor class B type I is a novel candidate receptor for the hepatitis C virus. Embo J. 2002;21:5017–5025. doi: 10.1093/emboj/cdf529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bartosch B, Vitelli A, Granier C, Goujon C, Dubuisson J, Pascale S, Scarselli E, Cortese R, Nicosia A, Cosset FL. Cell entry of hepatitis C virus requires a set of co-receptors that include the CD81 tetraspanin and the SR-B1 scavenger receptor. J Biol Chem. 2003;278:41624–41630. doi: 10.1074/jbc.M305289200. [DOI] [PubMed] [Google Scholar]
- 14.Evans MJ, von Hahn T, Tscherne DM, Syder AJ, Panis M, Wolk B, Hatziioannou T, McKeating JA, Bieniasz PD, Rice CM. Claudin-1 is a hepatitis C virus co-receptor required for a late step in entry. Nature. 2007;446:801–805. doi: 10.1038/nature05654. [DOI] [PubMed] [Google Scholar]
- 15.Ploss A, Evans MJ, Gaysinskaya VA, Panis M, You H, de Jong YP, Rice CM. Human occludin is a hepatitis C virus entry factor required for infection of mouse cells. Nature. 2009;457:882–886. doi: 10.1038/nature07684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu S, Yang W, Shen L, Turner JR, Coyne CB, Wang T. Tight junction proteins claudin-1 and occludin control hepatitis C virus entry and are downregulated during infection to prevent superinfection. J Virol. 2009;83:2011–2014. doi: 10.1128/JVI.01888-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Catanese MT, Ansuini H, Graziani R, Huby T, Moreau M, Ball JK, Paonessa G, Rice CM, Cortese R, Vitelli A, et al. Role of scavenger receptor class B type I in hepatitis C virus entry: kinetics and molecular determinants. J Virol. 2010;84:34–43. doi: 10.1128/JVI.02199-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zeisel MB, Koutsoudakis G, Schnober EK, Haberstroh A, Blum HE, Cosset FL, Wakita T, Jaeck D, Doffoel M, Royer C, et al. Scavenger receptor class B type I is a key host factor for hepatitis C virus infection required for an entry step closely linked to CD81. Hepatology. 2007;46:1722–1731. doi: 10.1002/hep.21994. [DOI] [PubMed] [Google Scholar]
- 19. Sharma NR, Mateu G, Dreux M, Grakoui A, Cosset FL, Melikyan GB. Hepatitis C virus is primed by CD81 protein for low pH-dependent fusion. J Biol Chem. 2011;286:30361–30376. doi: 10.1074/jbc.M111.263350. *The authors show that CD81 acts to prime the fusogenic activity of the HCV glycoproteins. In addition they show that cells lacking CD81 do not support cell surface fusion and this block can be rescued by adding soluble CD81 protein
- 20. Lupberger J, Zeisel MB, Xiao F, Thumann C, Fofana I, Zona L, Davis C, Mee CJ, Turek M, Gorke S, et al. EGFR and EphA2 are host factors for hepatitis C virus entry and possible targets for antiviral therapy. Nat Med. 2011;17:589–595. doi: 10.1038/nm.2341. **This study identifies two receptor tyrosine kinases (EGFR and EphA2) as important for HCV entry. The data implicate the signaling pathways downstream of these receptors in coordinating interactions between CD81 and CLDN1. Licensed inhibitors of EGFR (erlotinib) and EphA2 (dasatinib) have antiviral activity both in vitro and in vivo.
- 21.Coller KE, Berger KL, Heaton NS, Cooper JD, Yoon R, Randall G. RNA interference and single particle tracking analysis of hepatitis C virus endocytosis. PLoS Pathog. 2009;5:e1000702. doi: 10.1371/journal.ppat.1000702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blanchard E, Belouzard S, Goueslain L, Wakita T, Dubuisson J, Wychowski C, Rouille Y. Hepatitis C virus entry depends on clathrin-mediated endocytosis. J Virol. 2006;80:6964–6972. doi: 10.1128/JVI.00024-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Coyne CB, Bergelson JM. Virus-induced Abl and Fyn kinase signals permit coxsackievirus entry through epithelial tight junctions. Cell. 2006;124:119–131. doi: 10.1016/j.cell.2005.10.035. [DOI] [PubMed] [Google Scholar]
- 24.Dreux M, Dao Thi VL, Fresquet J, Guerin M, Julia Z, Verney G, Durantel D, Zoulim F, Lavillette D, Cosset FL, et al. Receptor complementation and mutagenesis reveal SR-BI as an essential HCV entry factor and functionally imply its intra- and extra-cellular domains. PLoS Pathog. 2009;5:e1000310. doi: 10.1371/journal.ppat.1000310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sainz B, Jr, Barretto N, Martin DN, Hiraga N, Imamura M, Hussain S, Marsh KA, Yu X, Chayama K, Alrefai WA, et al. Identification of the Niemann-Pick C1-like 1 cholesterol absorption receptor as a new hepatitis C virus entry factor. Nat Med. 2012;18:281–285. doi: 10.1038/nm.2581. *The authors identify the cholesterol uptake receptor NPC1L1 as influencing entry of HCV in hepatic cells. The FDA-approved inhibitor of NPC1L1, ezetimibe, inhibits HCV infection in vitro and delays the establishment of HCV genotype 1b infection in vivo.
- 26.Garcia-Calvo M, Lisnock J, Bull HG, Hawes BE, Burnett DA, Braun MP, Crona JH, Davis HR, Jr, Dean DC, Detmers PA, et al. The target of ezetimibe is Niemann-Pick C1-Like 1 (NPC1L1) Proc Natl Acad Sci U S A. 2005;102:8132–8137. doi: 10.1073/pnas.0500269102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Syder AJ, Lee H, Zeisel MB, Grove J, Soulier E, Macdonald J, Chow S, Chang J, Baumert TF, McKeating JA, et al. Small molecule scavenger receptor BI antagonists are potent HCV entry inhibitors. J Hepatol. 2011;54:48–55. doi: 10.1016/j.jhep.2010.06.024. [DOI] [PubMed] [Google Scholar]
- 28.Berger KL, Cooper JD, Heaton NS, Yoon R, Oakland TE, Jordan TX, Mateu G, Grakoui A, Randall G. Roles for endocytic trafficking and phosphatidylinositol 4-kinase III alpha in hepatitis C virus replication. Proc Natl Acad Sci U S A. 2009;106:7577–7582. doi: 10.1073/pnas.0902693106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tai AW, Benita Y, Peng LF, Kim SS, Sakamoto N, Xavier RJ, Chung RT. A functional genomic screen identifies cellular cofactors of hepatitis C virus replication. Cell Host Microbe. 2009;5:298–307. doi: 10.1016/j.chom.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vaillancourt FH, Pilote L, Cartier M, Lippens J, Liuzzi M, Bethell RC, Cordingley MG, Kukolj G. Identification of a lipid kinase as a host factor involved in hepatitis C virus RNA replication. Virology. 2009;387:5–10. doi: 10.1016/j.virol.2009.02.039. [DOI] [PubMed] [Google Scholar]
- 31.Borawski J, Troke P, Puyang X, Gibaja V, Zhao S, Mickanin C, Leighton-Davies J, Wilson CJ, Myer V, Cornellataracido I, et al. Class III phosphatidylinositol 4-kinase alpha and beta are novel host factor regulators of hepatitis C virus replication. J Virol. 2009;83:10058–10074. doi: 10.1128/JVI.02418-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trotard M, Lepere-Douard C, Regeard M, Piquet-Pellorce C, Lavillette D, Cosset FL, Gripon P, Le Seyec J. Kinases required in hepatitis C virus entry and replication highlighted by small interference RNA screening. Faseb J. 2009;23:3780–3789. doi: 10.1096/fj.09-131920. [DOI] [PubMed] [Google Scholar]
- 33.Li Q, Brass AL, Ng A, Hu Z, Xavier RJ, Liang TJ, Elledge SJ. A genome-wide genetic screen for host factors required for hepatitis C virus propagation. Proc Natl Acad Sci U S A. 2009;106:16410–16415. doi: 10.1073/pnas.0907439106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hsu NY, Ilnytska O, Belov G, Santiana M, Chen YH, Takvorian PM, Pau C, van der Schaar H, Kaushik-Basu N, Balla T, et al. Viral reorganization of the secretory pathway generates distinct organelles for RNA replication. Cell. 2010;141:799–811. doi: 10.1016/j.cell.2010.03.050. **This study shows that enteroviruses use PI4K-IIIβ for efficient replication and that the viral polymerase activity is enhanced by PI4P binding. Furthermore, replication complexes of enteroviruses and HCV are enriched in PI4P
- 35. Reiss S, Rebhan I, Backes P, Romero-Brey I, Erfle H, Matula P, Kaderali L, Poenisch M, Blankenburg H, Hiet MS, et al. Recruitment and activation of a lipid kinase by hepatitis C virus NS5A is essential for integrity of the membranous replication compartment. Cell Host Microbe. 2011;9:32–45. doi: 10.1016/j.chom.2010.12.002. ** The authors show that HCV NS5A stimulates PI4K-IIIα and that PI4P is increased in hepatocytes of HCV infected patients. Silencing PI4K-IIIα in cells expressing the HCV replicase results in the accumulation of double membrane vesicles
- 36. Berger KL, Kelly SM, Jordan TX, Tartell MA, Randall G. Hepatitis C virus stimulates the phosphatidylinositol 4-kinase III alpha-dependent phosphatidylinositol 4-phosphate production that is essential for its replication. J Virol. 2011;85:8870–8883. doi: 10.1128/JVI.00059-11. *The authors show that HCV NS5A stimulates PI4K-IIIα and that impairing PI4K-IIIα leads to formation of aggregated viral replication complex proteins
- 37.Tai AW, Salloum S. The role of the phosphatidylinositol 4-kinase PI4KA in hepatitis C virus-induced host membrane rearrangement. PLoS One. 2011;6:e26300. doi: 10.1371/journal.pone.0026300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bianco A, Reghellin V, Donnici L, Fenu S, Alvarez R, Baruffa C, Peri F, Pagani M, Abrignani S, Neddermann P, et al. Metabolism of phosphatidylinositol 4-kinase IIIalpha-dependent PI4P Is subverted by HCV and is targeted by a 4-anilino quinazoline with antiviral activity. PLoS Pathog. 2012;8:e1002576. doi: 10.1371/journal.ppat.1002576. *The authors show that 4-anilino quinazoline compounds that were previously thought to inhibit HCV NS5A are in fact inhibitors of cellular PI4K-IIIα and act by reducing PI4P synthesis required for HCV membranous web formation
- 39.Gao M, Nettles RE, Belema M, Snyder LB, Nguyen VN, Fridell RA, Serrano-Wu MH, Langley DR, Sun JH, O'Boyle DR, 2nd, et al. Chemical genetics strategy identifies an HCV NS5A inhibitor with a potent clinical effect. Nature. 2010;465:96–100. doi: 10.1038/nature08960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309:1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- 41.Randall G, Panis M, Cooper JD, Tellinghuisen TL, Sukhodolets KE, Pfeffer S, Landthaler M, Landgraf P, Kan S, Lindenbach BD, et al. Cellular cofactors affecting hepatitis C virus infection and replication. Proc Natl Acad Sci U S A. 2007;104:12884–12889. doi: 10.1073/pnas.0704894104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jopling CL, Schutz S, Sarnow P. Position-dependent function for a tandem microRNA miR-122-binding site located in the hepatitis C virus RNA genome. Cell Host Microbe. 2008;4:77–85. doi: 10.1016/j.chom.2008.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Henke JI, Goergen D, Zheng J, Song Y, Schuttler CG, Fehr C, Junemann C, Niepmann M. microRNA-122 stimulates translation of hepatitis C virus RNA. Embo J. 2008;27:3300–3310. doi: 10.1038/emboj.2008.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roberts AP, Lewis AP, Jopling CL. miR-122 activates hepatitis C virus translation by a specialized mechanism requiring particular RNA components. Nucleic Acids Res. 2011;39:7716–7729. doi: 10.1093/nar/gkr426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jangra RK, Yi M, Lemon SM. Regulation of hepatitis C virus translation and infectious virus production by the microRNA miR-122. J Virol. 2010;84:6615–6625. doi: 10.1128/JVI.00417-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Machlin ES, Sarnow P, Sagan SM. Masking the 5' terminal nucleotides of the hepatitis C virus genome by an unconventional microRNA-target RNA complex. Proc Natl Acad Sci U S A. 2011;108:3193–3198. doi: 10.1073/pnas.1012464108. *This study shows that miR-122 binds to the 5' terminus of the HCV RNA with 3' overhanging nucleotides that mask the 5' terminal sequences of the viral genome. In addition, the 3' overhanging nucleotides are required for maintaining HCV RNA abundance
- 47. Shimakami T, Yamane D, Jangra RK, Kempf BJ, Spaniel C, Barton DJ, Lemon SM. Stabilization of hepatitis C virus RNA by an Ago2-miR-122 complex. Proc Natl Acad Sci U S A. 2012;109:941–946. doi: 10.1073/pnas.1112263109. *The authors show that miR-122 slows decay of the HCV genome by recruiting an Ago2 RISC-like complex to the 5' end of the HCV RNA
- 48. Li YP, Gottwein JM, Scheel TK, Jensen TB, Bukh J. MicroRNA-122 antagonism against hepatitis C virus genotypes 1–6 and reduced efficacy by host RNA insertion or mutations in the HCV 5' UTR. Proc Natl Acad Sci U S A. 2011;108:4991–4996. doi: 10.1073/pnas.1016606108. *The authors develop efficient JFH1-based HCV genotypes 1a, 1b, 2a, 2b, 3a, 4a, 5a, and 6a with genotype specific 5'UTR-NS2 and use these culture systems for functional analysis of miR-122 binding to the 5'UTR
- 49. Lanford RE, Hildebrandt-Eriksen ES, Petri A, Persson R, Lindow M, Munk ME, Kauppinen S, Orum H. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science. 2010;327:198–201. doi: 10.1126/science.1178178. **The authors demonstrate that SPC3649 leads to long-lasting suppression of HCV in chronically infected chimpanzees
- 50.Wang P, Heitman J. The cyclophilins. Genome Biol. 2005;6:226. doi: 10.1186/gb-2005-6-7-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang F, Robotham JM, Nelson HB, Irsigler A, Kenworthy R, Tang H. Cyclophilin A is an essential cofactor for hepatitis C virus infection and the principal mediator of cyclosporine resistance in vitro. J Virol. 2008;82:5269–5278. doi: 10.1128/JVI.02614-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kaul A, Stauffer S, Berger C, Pertel T, Schmitt J, Kallis S, Zayas M, Lohmann V, Luban J, Bartenschlager R. Essential role of cyclophilin A for hepatitis C virus replication and virus production and possible link to polyprotein cleavage kinetics. PLoS Pathog. 2009;5:e1000546. doi: 10.1371/journal.ppat.1000546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chatterji U, Bobardt M, Selvarajah S, Yang F, Tang H, Sakamoto N, Vuagniaux G, Parkinson T, Gallay P. The isomerase active site of cyclophilin A is critical for hepatitis C virus replication. J Biol Chem. 2009;284:16998–17005. doi: 10.1074/jbc.M109.007625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.von Hahn T, Schiene-Fischer C, Van ND, Pfaender S, Karavul B, Steinmann E, Potthoff A, Strassburg C, Hamdi N, Abdelaziz AI, et al. Hepatocytes that express variants of cyclophilin a are resistant to HCV infection and replication. Gastroenterology. 2012;143:439–447. e431. doi: 10.1053/j.gastro.2012.04.053. [DOI] [PubMed] [Google Scholar]
- 55.Liu Z, Yang F, Robotham JM, Tang H. Critical role of cyclophilin A and its prolyl-peptidyl isomerase activity in the structure and function of the hepatitis C virus replication complex. J Virol. 2009;83:6554–6565. doi: 10.1128/JVI.02550-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Watashi K, Ishii N, Hijikata M, Inoue D, Murata T, Miyanari Y, Shimotohno K. Cyclophilin B is a functional regulator of hepatitis C virus RNA polymerase. Mol Cell. 2005;19:111–122. doi: 10.1016/j.molcel.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 57.Robida JM, Nelson HB, Liu Z, Tang H. Characterization of hepatitis C virus subgenomic replicon resistance to cyclosporine in vitro. J Virol. 2007;81:5829–5840. doi: 10.1128/JVI.02524-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hanoulle X, Badillo A, Wieruszeski JM, Verdegem D, Landrieu I, Bartenschlager R, Penin F, Lippens G. Hepatitis C virus NS5A protein is a substrate for the peptidyl-prolyl cis/trans isomerase activity of cyclophilins A and B. J Biol Chem. 2009;284:13589–13601. doi: 10.1074/jbc.M809244200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Yang F, Robotham JM, Grise H, Frausto S, Madan V, Zayas M, Bartenschlager R, Robinson M, Greenstein AE, Nag A, et al. A major determinant of cyclophilin dependence and cyclosporine susceptibility of hepatitis C virus identified by a genetic approach. PLoS Pathog. 2010;6:e1001118. doi: 10.1371/journal.ppat.1001118. * The authors use a genetic approach to identify a dipeptide motif (D316 and Y317) in domain II of NS5A as being important in the dependence of HCV on CypA. The authors show that wild-type HCV carrying these mutations can efficiently replicate in CypA knockdown cells and is resistant to cyclosporine A
- 60.Verdegem D, Badillo A, Wieruszeski JM, Landrieu I, Leroy A, Bartenschlager R, Penin F, Lippens G, Hanoulle X. Domain 3 of NS5A protein from the hepatitis C virus has intrinsic alpha-helical propensity and is a substrate of cyclophilin A. J Biol Chem. 2011;286:20441–20454. doi: 10.1074/jbc.M110.182436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Foster TL, Gallay P, Stonehouse NJ, Harris M. Cyclophilin A interacts with domain II of hepatitis C virus NS5A and stimulates RNA binding in an isomerase-dependent manner. J Virol. 2011;85:7460–7464. doi: 10.1128/JVI.00393-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Grise H, Frausto S, Logan T, Tang H. A conserved tandem cyclophilin-binding site in hepatitis C virus nonstructural protein 5A regulates Alisporivir susceptibility. J Virol. 2012;86:4811–4822. doi: 10.1128/JVI.06641-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Borel JF, Feurer C, Gubler HU, Stahelin H. Biological effects of cyclosporin A: a new antilymphocytic agent. 1976. Agents Actions. 1994;43:179–186. doi: 10.1007/BF01986686. [DOI] [PubMed] [Google Scholar]
- 64.Flisiak R, Feinman SV, Jablkowski M, Horban A, Kryczka W, Pawlowska M, Heathcote JE, Mazzella G, Vandelli C, Nicolas-Metral V, et al. The cyclophilin inhibitor Debio 025 combined with PEG IFNalpha2a significantly reduces viral load in treatment-naive hepatitis C patients. Hepatology. 2009;49:1460–1468. doi: 10.1002/hep.22835. [DOI] [PubMed] [Google Scholar]
- 65.Hopkins S, Dimassimo B, Rusnak P, Heuman D, Lalezari J, Sluder A, Scorneaux B, Mosier S, Kowalczyk P, Ribeill Y, et al. The cyclophilin inhibitor SCY-635 suppresses viral replication and induces endogenous interferons in patients with chronic HCV genotype 1 infection. J Hepatol. 2012;57:47–54. doi: 10.1016/j.jhep.2012.02.024. [DOI] [PubMed] [Google Scholar]
- 66.Lawitz E, Godofsky E, Rouzier R, Marbury T, Nguyen T, Ke J, Huang M, Praestgaard J, Serra D, Evans TG. Safety, pharmacokinetics, and antiviral activity of the cyclophilin inhibitor NIM811 alone or in combination with pegylated interferon in HCV-infected patients receiving 14 days of therapy. Antiviral Res. 2011;89:238–245. doi: 10.1016/j.antiviral.2011.01.003. [DOI] [PubMed] [Google Scholar]
- 67.Boulant S, Targett-Adams P, McLauchlan J. Disrupting the association of hepatitis C virus core protein with lipid droplets correlates with a loss in production of infectious virus. J Gen Virol. 2007;88:2204–2213. doi: 10.1099/vir.0.82898-0. [DOI] [PubMed] [Google Scholar]
- 68.Miyanari Y, Atsuzawa K, Usuda N, Watashi K, Hishiki T, Zayas M, Bartenschlager R, Wakita T, Hijikata M, Shimotohno K. The lipid droplet is an important organelle for hepatitis C virus production. Nat Cell Biol. 2007;9:1089–1097. doi: 10.1038/ncb1631. [DOI] [PubMed] [Google Scholar]
- 69. Neveu G, Barouch-Bentov R, Ziv-Aviv A, Gerber D, Jacob Y, Einav S. Identification and Targeting of an Interaction between a Tyrosine Motif within Hepatitis C Virus Core Protein and AP2M1 Essential for Virus Assembly. PLoS Pathog. 2012;8:e1002845. doi: 10.1371/journal.ppat.1002845. * The authors identify a motif within the HCV core that binds AP2M1, the µ subunit of clathrin adaptor complex 2, and is essential for virion assembly
- 70. Menzel N, Fischl W, Hueging K, Bankwitz D, Frentzen A, Haid S, Gentzch J, Kaderali L, Bartenschlager R, Pietschmann T. MAP-Kinase Regulated Cytosolic Phospholipase A2 Activity is Essential for Production of Infectious Hepatitis C Virus Particles. PLoS Pathog. 2012;8:e1002829. doi: 10.1371/journal.ppat.1002829. *By using chemical inhibitors of the MAPK/ERK signaling pathway, the authors identify PLA2G4A as an important host factor of HCV assembly and show that inhibition of PLA2G4A specifically reduces core protein abundance on lipid droplets as well as capsid envelopment
- 71.Huang H, Sun F, Owen DM, Li W, Chen Y, Gale M, Jr, Ye J. Hepatitis C virus production by human hepatocytes dependent on assembly and secretion of very low-density lipoproteins. Proc Natl Acad Sci U S A. 2007;104:5848–5853. doi: 10.1073/pnas.0700760104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Icard V, Diaz O, Scholtes C, Perrin-Cocon L, Ramiere C, Bartenschlager R, Penin F, Lotteau V, Andre P. Secretion of hepatitis C virus envelope glycoproteins depends on assembly of apolipoprotein B positive lipoproteins. PLoS One. 2009;4:e4233. doi: 10.1371/journal.pone.0004233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chang KS, Jiang J, Cai Z, Luo G. Human apolipoprotein e is required for infectivity and production of hepatitis C virus in cell culture. J Virol. 2007;81:13783–13793. doi: 10.1128/JVI.01091-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jiang J, Luo G. Apolipoprotein E but not B is required for the formation of infectious hepatitis C virus particles. J Virol. 2009;83:12680–12691. doi: 10.1128/JVI.01476-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Benga WJ, Krieger SE, Dimitrova M, Zeisel MB, Parnot M, Lupberger J, Hildt E, Luo G, McLauchlan J, Baumert TF, et al. Apolipoprotein E interacts with hepatitis C virus nonstructural protein 5A and determines assembly of infectious particles. Hepatology. 2010;51:43–53. doi: 10.1002/hep.23278. [DOI] [PubMed] [Google Scholar]
- 76.Nielsen SU, Bassendine MF, Burt AD, Martin C, Pumeechockchai W, Toms GL. Association between hepatitis C virus and very-low-density lipoprotein (VLDL)/LDL analyzed in iodixanol density gradients. J Virol. 2006;80:2418–2428. doi: 10.1128/JVI.80.5.2418-2428.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Merz A, Long G, Hiet MS, Brugger B, Chlanda P, Andre P, Wieland F, Krijnse-Locker J, Bartenschlager R. Biochemical and morphological properties of hepatitis C virus particles and determination of their lipidome. J Biol Chem. 2011;286:3018–3032. doi: 10.1074/jbc.M110.175018. *By using immuno-electron microscopy the authors show that affinity purified HCV particles contain apoE. They then determine the morphology and lipidome profile of HCV virions
- 78. Coller KE, Heaton NS, Berger KL, Cooper JD, Saunders JL, Randall G. Molecular determinants and dynamics of hepatitis C virus secretion. PLoS Pathog. 2012;8:e1002466. doi: 10.1371/journal.ppat.1002466. **The authors combine the results of an siRNA screen with a live cell imaging approach to visualize the secretion of HCV core with host factors during virus egress
- 79.Jiang J, Cun W, Wu X, Shi Q, Tang H, Luo G. Hepatitis C virus attachment mediated by apolipoprotein E binding to cell surface heparan sulfate. J Virol. 2012;86:7256–7267. doi: 10.1128/JVI.07222-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Herker E, Harris C, Hernandez C, Carpentier A, Kaehlcke K, Rosenberg AR, Farese RV, Jr, Ott M. Efficient hepatitis C virus particle formation requires diacylglycerol acyltransferase-1. Nat Med. 2010;16:1295–1298. doi: 10.1038/nm.2238. *The authors show that DGAT1, a critical enzyme in lipid droplet formation, associates with HCV core on lipid droplets and is required for virus assembly and release
- 81.Yen CL, Stone SJ, Koliwad S, Harris C, Farese RV., Jr Thematic review series: glycerolipids. DGAT enzymes and triacylglycerol biosynthesis. J Lipid Res. 2008;49:2283–2301. doi: 10.1194/jlr.R800018-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Morsica G, Tambussi G, Sitia G, Novati R, Lazzarin A, Lopalco L, Mukenge S. Replication of hepatitis C virus in B lymphocytes (CD19+) Blood. 1999;94:1138–1139. [PubMed] [Google Scholar]
- 83.Fletcher NF, Yang JP, Farquhar MJ, Hu K, Davis C, He Q, Dowd K, Ray SC, Krieger SE, Neyts J, et al. Hepatitis C virus infection of neuroepithelioma cell lines. Gastroenterology. 2010;139:1365–1374. doi: 10.1053/j.gastro.2010.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fletcher NF, Wilson GK, Murray J, Hu K, Lewis A, Reynolds GM, Stamataki Z, Meredith LW, Rowe IA, Luo G, et al. Hepatitis C virus infects the endothelial cells of the blood-brain barrier. Gastroenterology. 2012;142:634–643. e636. doi: 10.1053/j.gastro.2011.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Frentzen A, Huging K, Bitzegeio J, Friesland M, Haid S, Gentzch J, Hoffman M, Lindemann D, Zimmer G, Zielecki F, et al. Completion of hepatitis C virus replication cycle in heterokaryons excludes dominant restrictions in human non-liver and mouse liver cell lines. PLoS Pathog. 2011;7:e1002029. doi: 10.1371/journal.ppat.1002029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Dorner M, Horwitz JA, Robbins JB, Barry WT, Feng Q, Mu K, Jones CT, Schoggins JW, Catanese MT, Burton DR, et al. A genetically humanized mouse model for hepatitis C virus infection. Nature. 2011;474:208–211. doi: 10.1038/nature10168. ** This study reports a genetically humanized mouse model supporting HCV entry
- 87.Zhu Q, Guo J, Seeger C. Replication of hepatitis C virus subgenomes in nonhepatic epithelial and mouse hepatoma cells. J Virol. 2003;77:9204–9210. doi: 10.1128/JVI.77.17.9204-9210.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Uprichard SL, Chung J, Chisari F, Wakita T. Replication of a hepatitis C virus replication clone in mouse cells. Virol J. 2006;3:89. doi: 10.1186/1743-422X-3-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Narbus C, Israelow B, Sourisseau M, Michta M, Hopcraft S, Zeiner G, Evans MJ. HepG2 cells expressing microRNA miR-122 support the entire hepatitis C virus life cycle. J Virol. 2011;85:12087–12092. doi: 10.1128/JVI.05843-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Fukuhara T, Kambara H, Shiokawa M, Ono C, Katoh H, Morita E, Okuzaki D, Maehara Y, Koike K, Matsuura Y. Expression of MicroRNA miR-122 Facilitates an Efficient Replication in Nonhepatic Cells upon Infection with Hepatitis C Virus. J Virol. 2012;86:7918–7933. doi: 10.1128/JVI.00567-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kambara H, Fukuhara T, Shiokawa M, Ono C, Ohara Y, Kamitani W, Matsuura Y. Establishment of a novel permissive cell line for the propagation of hepatitis C virus by expression of microRNA miR122. J Virol. 2012;86:1382–1393. doi: 10.1128/JVI.06242-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Long G, Hiet MS, Windisch MP, Lee JY, Lohmann V, Bartenschlager R. Mouse hepatic cells support assembly of infectious hepatitis C virus particles. Gastroenterology. 2011;141:1057–1066. doi: 10.1053/j.gastro.2011.06.010. [DOI] [PubMed] [Google Scholar]
- 93.Chang J, Guo JT, Jiang D, Guo H, Taylor JM, Block TM. Liver-specific microRNA miR-122 enhances the replication of hepatitis C virus in nonhepatic cells. J Virol. 2008;82:8215–8223. doi: 10.1128/JVI.02575-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lin LT, Noyce RS, Pham TN, Wilson JA, Sisson GR, Michalak TI, Mossman KL, Richardson CD. Replication of subgenomic hepatitis C virus replicons in mouse fibroblasts is facilitated by deletion of interferon regulatory factor 3 and expression of liver-specific microRNA 122. J Virol. 2010;84:9170–9180. doi: 10.1128/JVI.00559-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Da Costa D, Turek M, Felmlee DJ, Girardi E, Pfeffer S, Long G, Bartenschlager R, Zeisel MB, Baumert TF. Reconstitution of the entire hepatitis C virus life cycle in non-hepatic cells. J Virol. 2012 doi: 10.1128/JVI.01066-12. *The authors use lentiviral vectors to exogenously express CLDN1, CD81, OCLN and SR-BI in combination with miR-122 and apoE3 isoform in 293T cells. This supports the entire HCV life cycle
- 96.Simmonds P. Genetic diversity and evolution of hepatitis C virus-15 years on. J Gen Virol. 2004;85:3173–3188. doi: 10.1099/vir.0.80401-0. [DOI] [PubMed] [Google Scholar]
- 97.Wakita T, Pietschmann T, Kato T, Date T, Miyamoto M, Zhao Z, Murthy K, Habermann A, Krausslich HG, Mizokami M, et al. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med. 2005;11:791–796. doi: 10.1038/nm1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Scheel TK, Gottwein JM, Jensen TB, Prentoe JC, Hoegh AM, Alter MJ, Eugen-Olsen J, Bukh J. Development of JFH1-based cell culture systems for hepatitis C virus genotype 4a and evidence for cross-genotype neutralization. Proc Natl Acad Sci U S A. 2008;105:997–1002. doi: 10.1073/pnas.0711044105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gottwein JM, Scheel TK, Callendret B, Li YP, Eccleston HB, Engle RE, Govindarajan S, Satterfield W, Purcell RH, Walker CM, et al. Novel infectious cDNA clones of hepatitis C virus genotype 3a (strain S52) and 4a (strain ED43): genetic analyses and in vivo pathogenesis studies. J Virol. 2010;84:5277–5293. doi: 10.1128/JVI.02667-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gottwein JM, Scheel TK, Jensen TB, Lademann JB, Prentoe JC, Knudsen ML, Hoegh AM, Bukh J. Development and characterization of hepatitis C virus genotype 1–7 cell culture systems: role of CD81 and scavenger receptor class B type I and effect of antiviral drugs. Hepatology. 2009;49:364–377. doi: 10.1002/hep.22673. [DOI] [PubMed] [Google Scholar]
- 101.Blight KJ, McKeating JA, Rice CM. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J Virol. 2002;76:13001–13014. doi: 10.1128/JVI.76.24.13001-13014.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zhong J, Gastaminza P, Cheng G, Kapadia S, Kato T, Burton DR, Wieland SF, Uprichard SL, Wakita T, Chisari FV. Robust hepatitis C virus infection in vitro. Proc Natl Acad Sci U S A. 2005;102:9294–9299. doi: 10.1073/pnas.0503596102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Koutsoudakis G, Herrmann E, Kallis S, Bartenschlager R, Pietschmann T. The level of CD81 cell surface expression is a key determinant for productive entry of hepatitis C virus into host cells. J Virol. 2007;81:588–598. doi: 10.1128/JVI.01534-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Mee CJ, Grove J, Harris HJ, Hu K, Balfe P, McKeating JA. Effect of cell polarization on hepatitis C virus entry. J Virol. 2008;82:461–470. doi: 10.1128/JVI.01894-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Mee CJ, Harris HJ, Farquhar MJ, Wilson G, Reynolds G, Davis C, van ISC, Balfe P, McKeating JA. Polarization restricts hepatitis C virus entry into HepG2 hepatoma cells. J Virol. 2009;83:6211–6221. doi: 10.1128/JVI.00246-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Molina-Jimenez F, Benedicto I, Dao Thi VL, Gondar V, Lavillette D, Marin JJ, Briz O, Moreno-Otero R, Aldabe R, Baumert TF, et al. Matrigel-embedded 3D culture of Huh-7 cells as a hepatocyte-like polarized system to study hepatitis C virus cycle. Virology. 2012;425:31–39. doi: 10.1016/j.virol.2011.12.021. [DOI] [PubMed] [Google Scholar]
- 107.Ploss A, Khetani SR, Jones CT, Syder AJ, Trehan K, Gaysinskaya VA, Mu K, Ritola K, Rice CM, Bhatia SN. Persistent hepatitis C virus infection in microscale primary human hepatocyte cultures. Proc Natl Acad Sci U S A. 2010;107:3141–3145. doi: 10.1073/pnas.0915130107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Podevin P, Carpentier A, Pene V, Aoudjehane L, Carriere M, Zaidi S, Hernandez C, Calle V, Meritet JF, Scatton O, et al. Production of infectious hepatitis C virus in primary cultures of human adult hepatocytes. Gastroenterology. 2010;139:1355–1364. doi: 10.1053/j.gastro.2010.06.058. [DOI] [PubMed] [Google Scholar]
- 109. Schwartz RE, Trehan K, Andrus L, Sheahan TP, Ploss A, Duncan SA, Rice CM, Bhatia SN. Modeling hepatitis C virus infection using human induced pluripotent stem cells. Proc Natl Acad Sci U S A. 2012;109:2544–2548. doi: 10.1073/pnas.1121400109. * This study shows that induced pluripotent stem cells can be differentiated into hepatocyte-like cells that are permissive to HCV infection. Upon infection these cells mount an appropriate inflammatory antiviral response
- 110.Roelandt P, Obeid S, Paeshuyse J, Vanhove J, Van Lommel A, Nahmias Y, Nevens F, Neyts J, Verfaillie CM. Human pluripotent stem-cell derived hepatocytes support complete replication of hepatitis C virus. J Hepatol. 2012;57:246–251. doi: 10.1016/j.jhep.2012.03.030. [DOI] [PubMed] [Google Scholar]
- 111. Wu X, Robotham JM, Lee E, Dalton S, Kneteman NM, Gilbert DM, Tang H. Productive hepatitis C virus infection of stem-cell derived hepatocytes reveals a critical transition to viral permissiveness during differentiation. PLoS Pathog. 2012;8:e1002617. doi: 10.1371/journal.ppat.1002617. * This study shows that human embryonic and induced pluripotent stem cells can be differentiated into hepatocyte-like cells that are permissive to both cell culture- and patient-derived HCV.