Abstract
Biotin-dependent carboxylases include acetyl-CoA carboxylase (ACC), propionyl-CoA carboxylase (PCC), 3-methylcrotonyl-CoA carboxylase (MCC), geranyl-CoA carboxylase, pyruvate carboxylase (PC), and urea carboxylase (UC). They contain biotin carboxylase (BC), carboxyltransferase (CT), and biotin-carboxyl carrier protein components. These enzymes are widely distributed in nature and have important functions in fatty acid metabolism, amino acid metabolism, carbohydrate metabolism, polyketide biosynthesis, urea utilization, and other cellular processes. ACCs are also attractive targets for drug discovery against type 2 diabetes, obesity, cancer, microbial infections, and other diseases, and the plastid ACC of grasses is the target of action of three classes of commercial herbicides. Deficiencies in the activities of PCC, MCC, or PC are linked to serious diseases in humans. Our understanding of these enzymes has been greatly enhanced over the past few years by the crystal structures of the holoenzymes of PCC, MCC, PC, and UC. The structures reveal unanticipated features in the architectures of the holoenzymes, including the presence of previously unrecognized domains, and provide a molecular basis for understanding their catalytic mechanism as well as the large collection of disease-causing mutations in PCC, MCC, and PC. This review will summarize the recent advances in our knowledge on the structure and function of these important metabolic enzymes.
Keywords: Fatty acid metabolism, Carbohydrate metabolism, Amino acid metabolism, Metabolic syndrome, Obesity, Diabetes, Cancer, Drug discovery, Antibiotics, Propionic acidemia, 3-methylcrotonylglycinuria, Lactic acidemia
Introduction
Biotin-dependent carboxylases are widely distributed in nature and can be found in bacteria, archaea, fungi, algae, plants, and animals. They have crucial roles in the metabolism of fatty acids, amino acids, and carbohydrates [1–4]. In some microorganisms, these enzymes also have important functions in CO2 fixation [5, 6], methanol assimilation [7], acetyl-CoA assimilation [8–10], 3-hydroxypropionate assimilation [11], mycolic acid and methyl-branched fatty acid biosynthesis [12], polyketide biosynthesis [13], metabolism of terpenoids [14], and the utilization of urea as a nitrogen source [15, 16].
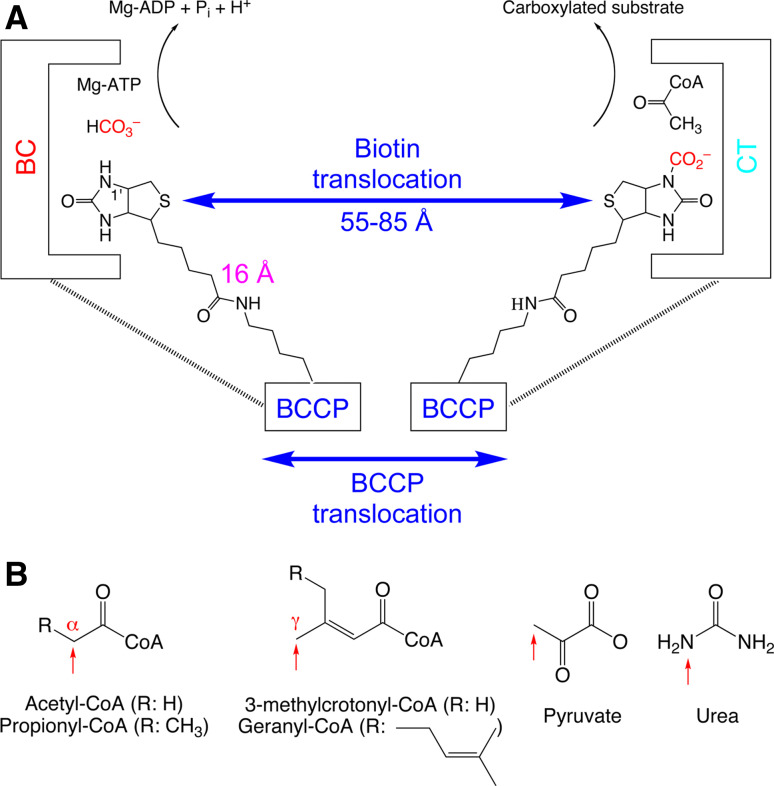
Biotin-dependent carboxylases have two distinct enzymatic activities and catalyze their reactions in two steps [17, 18]. First, a biotin carboxylase (BC) component catalyzes the MgATP-dependent carboxylation of the N1′ atom of the biotin cofactor, using bicarbonate as the CO2 donor (Fig. 1a). Biotin is covalently linked through an amide bond to a lysine side chain in the biotin carboxyl carrier protein (BCCP) component. In the second step of the reaction, a carboxyltransferase (CT) component catalyzes the CO2 transfer from carboxybiotin to the acceptor of the carboxyl group (referred to as the substrate here). Most of the substrates are coenzyme A (CoA) esters of organic acids, and the site of carboxylation is on the α carbon of a saturated acid (acetyl-CoA, propionyl-CoA) or the γ carbon of an α-β unsaturated acid (3-methylcrotonyl-CoA, geranyl-CoA) (Fig. 1b). In addition, small compounds can also serve directly as the substrate, such as pyruvate and urea (Fig. 1b). Especially, the urea substrate is unique in that carboxylation occurs on a nitrogen atom.
Fig. 1.
The biochemical activity of biotin-dependent carboxylases. a Biotin is carboxylated in the active site of the biotin carboxylase (BC) component, using bicarbonate as the CO2 donor with concomitant ATP hydrolysis. Biotin then translocates to the carboxyltransferase (CT) active site, where the CO2 is transferred to the acceptor (substrate, acetyl-CoA is shown as an example). In the swinging-arm model, biotin itself translocates between the BC and CT active sites, while the biotin-carboxyl carrier protein (BCCP) component remains stationary. The longest distance between the N1′ of biotin and the Cα atom of the covalently linked lysine residue is ~16 Å, giving the swinging arm a maximal reach of ~30 Å. This is significantly shorter than the distances observed in the holoenzymes so far, between 55 and 80 Å. Therefore, the BCCP domain must also translocate during catalysis, and this is known as the swinging-domain model. b The substrates of biotin-dependent carboxylases. The sites of carboxylation are indicated with the red arrows
Depending on the organism and the enzyme, the BC, BCCP, and CT components can be separate subunits or part of a multi-domain protein (Fig. 2). In addition, while the BC and BCCP components are conserved among these enzymes, the sequence and structure of the CT component depend on the chemical nature of the substrate. The CT components of enzymes that carboxylate CoA esters share recognizable sequence conservation because they recognize the CoA segment (Fig. 2). On the other hand, the CT component that carboxylates pyruvate or urea is entirely different.
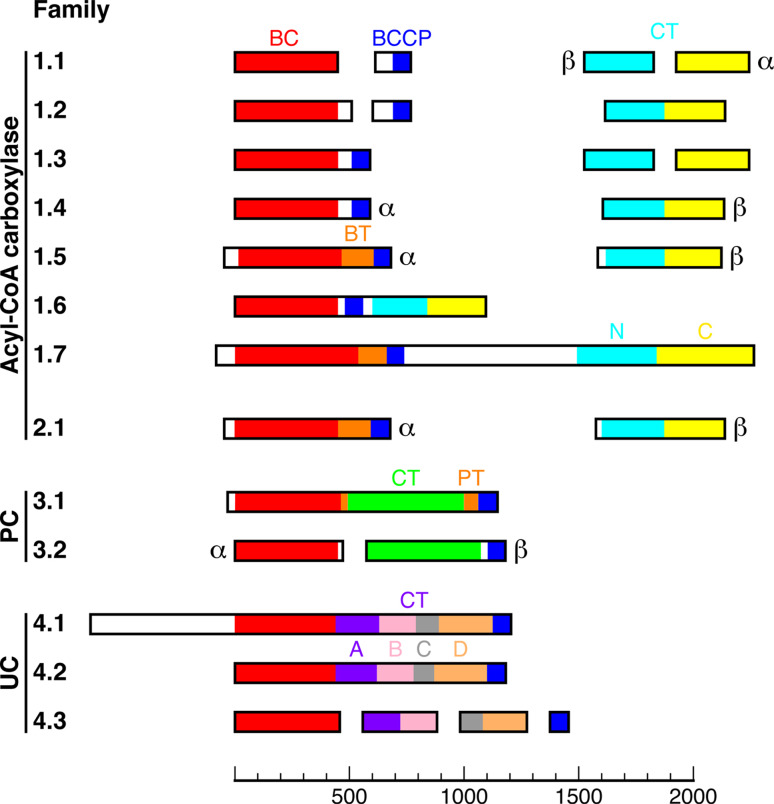
Fig. 2.
Classification of biotin-dependent carboxylases. These enzymes are classified into four major collections and 13 different families. Domains with sequence homology to each other are shown with the same color. More detailed descriptions of the families can be found in the text, and common examples of acyl-CoA carboxylase family members include E. coli ACC (family 1.1), eukaryotic ACC (family 1.7), PCC (family 1.5), and MCC (family 2.1). The proteins are drawn to size, which is indicated with the scale bar at the bottom (in number of residues)
Biotin must visit both the BC and CT active sites during catalysis by biotin-dependent carboxylases. A swinging-arm model had been the accepted mechanism for this translocation. The connection between biotin and BCCP contains eight methylene groups and ten rotatable single bonds, and is likely to be rather flexible (Fig. 1a). When fully extended, this flexible arm can approach a length of ~16 Å (the distance from the N1′ atom of biotin to the Cα atom of the lysine). Therefore, it may be expected that biotin can translocate by up to ~30 Å on this swinging arm [19]. However, recent structures on the holoenzymes of pyruvate carboxylase [20, 21], propionyl-CoA carboxylase [22], 3-methylcrotonyl-CoA carboxylase [23], and urea carboxylase [24] showed that the distance between the BC and CT active sites ranges between 55 and 85 Å (Fig. 1a). Therefore, the swinging-arm model is not sufficient for biotin to reach both active sites, and hence the BCCP domain must also translocate during catalysis. This is referred to as the “swinging-domain” model (Fig. 1a).
Besides biotin-dependent carboxylases, two other classes of biotin-dependent enzymes exist in nature. Biotin-dependent decarboxylases couple the decarboxylation of organic acids (possibly as CoA esters) to sodium ion transport in anaerobes [25–30], while the biotin-dependent transcarboxylase of Propionibacterium shermanii transfers the carboxyl group from methylmalonyl-CoA to pyruvate [31–33]. These enzymes are distinct from the biotin-dependent carboxylases in that they lack the BC component. They will not be specifically described further here.
Biotin-dependent carboxylases were first discovered more than 50 years ago. They have been studied intensively due to their important metabolic functions, and also feature prominently in most biochemistry textbooks. Over the past few years, there have been significant advances in our understanding of these enzymes, especially from the first structural information on several of the holoenzymes [20–24]. This review summarizes our current knowledge on the structure and function of biotin-dependent carboxylases, with emphasis on recent studies (over the past 5 years). Space limitations unfortunately prevent detailed descriptions of results from earlier studies or the citation of those primary publications. These results are summarized in the many reviews that have been published in the previous years, which are cited throughout this manuscript.
Classification of biotin-dependent carboxylases
Biotin-dependent carboxylases can be classified first based on the identity of the substrate that becomes carboxylated. This is dictated by the CT component, which can be highly distinct among these enzymes (Fig. 2). These enzymes can then be further classified by how the BC, BCCP, and CT components are organized in them (Fig. 2). The different components may exist as separate subunits, often in bacteria, or they can be fused together into a large, multi-domain enzyme in eukaryotes (Fig. 2). Various intermediates between these two extremes have also been observed in nature (Fig. 2).
The largest collection of biotin-dependent carboxylases uses CoA esters of (small) organic acids as the substrate; hence they are acyl-CoA carboxylases (YCCs) in general. These enzymes can have distinct substrate preferences, such as acetyl-CoA carboxylase (ACC), propionyl-CoA carboxylase (PCC), 3-methylcrotonyl-CoA carboxylase (MCC), and geranyl-CoA carboxylase (GCC), although some of them may have a wider collection of substrates, for example enzymes that are active toward both acetyl- and propionyl-CoA (ACC/PCC). In addition, some of these enzymes can be identified based on genome sequences but have not been studied in detail biochemically, and their substrate preference is currently not known. They are referred to generically as YCCs here. Acyl-CoA carboxylases have also been referred to as ACCases [12], although ACCs are sometimes called ACCases as well.
The CT components of the acyl-CoA carboxylases share readily detectable amino acid sequence conservation, because they all recognize CoA esters. It was generally believed that these enzymes have the same organization of their components. However, the recent structure of the MCC holoenzyme indicates that there may be two distinct lineages of these carboxylases [23]. Therefore, the acyl-CoA carboxylases have been divided into two separate collections, one including ACC, PCC, ACC/PCC, and most of the other YCCs (families 1.1 through 1.7), while the other consists of MCC and GCC (family 2.1) (Fig. 2). The different families of biotin-dependent carboxylases are described briefly next and in more detail in the later sections.
Family 1.1 includes the bacterial ACC enzymes (Fig. 2). They contain four subunits, with a BC subunit (~50 kD), a BCCP subunit (~17 kD), and two subunits (α and β, ~33 kD each) for the CT activity. These enzymes are also referred to as the multi-subunit ACCs [2]. The holoenzyme has the stoichiometry (BC)2(BCCP)4(CTα, CTβ)2, but it is generally unstable and readily dissociates during purification. These enzymes are also found in the chloroplasts of many plants, reflecting the evolutionary origin of this organelle.
Family 1.2 includes the ACC/PCC enzymes from M. sedula [34], A. fulgidus, and other archaeal organisms, but it appears to be absent in M. jannaschii. Compared to the multi-subunit bacterial ACCs, the two CT subunits are fused into a single protein (~60 kD), while the BC and CT components remain as separate subunits. The holoenzyme has the stoichiometry (BC)4(BCCP)4(CT)4.
Family 1.3 represents another possible way of fusing the different components of the multi-subunit bacterial ACCs, where the BC and BCCP subunits are fused together but the CT subunits remain separate. An actual example of such an enzyme has not been identified as yet, but its existence may be expected.
Family 1.4 includes the ACC/PCC/acyl-CoA carboxylases (YCCs) in S. coelicolor, M. tuberculosis, and other organisms [12]. The BC and BCCP components are fused into a single protein (α subunit, ~65 kD), and the two CT subunits are also fused (β subunit, ~60 kD). The holoenzyme has the stoichiometry α6β6 or α2β2 [12]. Another subunit (ε) is required for some of these enzymes to achieve maximal activity [12].
Family 1.5 includes the PCCs from various organisms. The difference to family 1.4 is that the BC-BCCP fusion (the α subunit, ~73 kD) also contains a BT domain, which mediates the interactions between the BC (in the α subunit) and CT (the β subunit, ~55 kD) domains [22]. This domain is likely absent in family 1.4, as the linker between BC and BCCP is too short to accommodate the residues of the BT domain in that family. However, it is possible that family 1.4 contains a modified version of the BT domain, for example a structure similar to that of the PT domain in pyruvate carboxylase (family 3.1) [21]. PCC holoenzyme is a 750 kD α6β6 dodecamer.
Family 1.6 includes the acyl-CoA carboxylases from P. aeruginosa and some other bacterial organisms. These enzymes can be readily identified based on sequence searches, but they have not been characterized biochemically (hence they are called YCCs here). All four subunits of the multi-subunit bacterial ACCs are fused together in these enzymes, giving rise to a multi-domain protein of ~120-kD molecular weight.
Family 1.7 includes the ACCs from most eukaryotic organisms. These proteins can be thought of as being made of three parts of equal lengths. The N-terminal one-third of the proteins contains the BC, BCCP, and possibly a BT domain, and the C-terminal one-third contains the CT activity. The middle one-third is unique to eukaryotic ACCs and has no other close sequence homologs in the database. The structure and function of this part of eukaryotic ACCs is currently not known. These enzymes are generally referred to as the multi-domain ACCs, with ~250-kD molecular weight, and they function as 500-kD homodimers and possibly higher oligomers. In light of the multi-domain bacterial YCCs of family 1.6, it is probably more appropriate to refer to family 1.7 as the multi-domain eukaryotic ACCs. The multi-domain bacterial YCCs lack the middle one-third of the multi-domain eukaryotic ACCs, and they may lack the BT domain as well (Fig. 2).
Family 2.1 includes the MCCs and GCCs from various organisms. The overall domain organization of these enzymes appears quite similar to that of PCCs (family 1.5). However, the crystal structure of MCC indicates a large difference in the architectures of the β subunit and the holoenzyme [23], and therefore it has been placed into a separate family. GCC is assigned to this family based on sequence conservation and the similarity of its substrate to that of MCC (Fig. 1b). These holoenzymes are also 750-kD α6β6 dodecamers.
Besides the acyl-CoA carboxylases, two other major collections of biotin-dependent carboxylases use small organic compounds as substrates, specifically pyruvate and urea (Fig. 1b). Family 3.1 includes the pyruvate carboxylases (PCs) that are present in most organisms, from bacteria to humans. It is a single-chain, multi-domain enzyme of ~130 kD and functions only as a 500-kD tetramer. Besides the BC, CT, and BCCP domains, structural studies have revealed the presence of another domain, known as the PT (PC tetramerization) or the allosteric domain, in these enzymes [20, 21].
Family 3.2 includes the two-subunit form of PCs. They are found in some bacteria (such as P. aeruginosa) as well as archaea (M. jannaschii). The α subunit contains the BC component (~52 kD), and the β subunit contains CT and BCCP (~65 kD). The stoichiometry of the holoenzyme is α4β4. Whether the PT domain also exists in these enzymes is currently not known.
Family 4.1 includes the urea carboxylase (UC) found in yeast and some other fungal organisms. It is a single-chain, multi-domain enzyme of ~200 kD, with an allophanate hydrolase (also known as the amidase) domain fused at the N-terminus. The entire enzyme is known as the urea amidolyase (UA). The CT component consists of four sub-domains (A, B, C, D) and is distinct from that in acyl-CoA carboxylase and PC [24]. UC functions as a monomer.
Family 4.2 includes the UCs found in many bacterial organisms, some fungal species, and green algae. They are different from family 4.1 in that they lack the allophanate hydrolase domain, and hence they are somewhat smaller, ~130 kD.
Family 4.3 includes the UC found in P. aeruginosa and possibly other bacterial organisms. It is a multi-subunit form of the enzyme, with a BC subunit (~50 kD), BCCP subunit (~10 kD), and two subunits for the CT activity (~35 kD, each containing two domains of the multi-domain form of the enzyme).
Overall, four major collections of biotin-dependent carboxylases can be identified based on current biochemical, sequence, and structural information, which can be further divided into 13 families. The availability of genome sequences has enabled the identification of all biotin-dependent enzymes in many organisms, which also provides insight into the evolution of these enzymes [35, 36]. An inventory of such enzymes can now be compiled for these organisms (Table 1). E. coli has only one biotin-dependent enzyme, a multi-subunit bacterial ACC (family 1.1, Fig. 2). The yeast S. cerevisiae has two multi-domain eukaryotic ACCs (family 1.7), two PCs (family 3.1), and one urea amidolyase (family 4.1), while most other fungal species has only one ACC, one PC, and one UA. There are five biotin-dependent carboxylases in humans, ACC1, ACC2, PCC, MCC, and PC. The structure and function of representative enzymes in these different families will be described in more detail below.
Table 1.
Inventory of biotin-dependent carboxylases in some common organisms
| Organism | Biotin-dependent carboxylase (family number) |
|---|---|
| Bacteria | |
| Escherichia coli K-12 | Multi-subunit ACC (1.1) |
| Pseudomonas aeruginosa PAO1 | Multi-subunit ACC (1.1), multi-domain YCC (1.6), MCC (2.1), GCC (2.1), two-subunit PC (3.2), multi-subunit UC (4.3) |
| Ruegeria pomeroyi DSS-3 | Multi-subunit ACC (1.1), PCC (1.5), MCC (2.1), GCC (2.1), PC (3.1) |
| Cupriavidus metallidurans CH34 | Multi-subunit ACC (1.1), multi-domain YCC (1.6, 2 copies), MCC (2.1), GCC (2.1), PC (3.1) |
| Deinococcus radiodurans R1 | Multi-subunit ACC (1.1), multi-domain YCC (1.6), PCC (1.5), MCC (2.1) |
| Streptomyces coelicolor A3(2) | Two-subunit YCC (1.4), PCC (1.5, 2 copies), MCC (2.1, 2 copies), PC (3.1) |
| Staphylococcus aureus | Multi-subunit ACC (1.1), PC (3.1), multi-subunit UC (4.3) |
| Bacillus subtilis | Multi-subunit ACC (1.1), three-subunit YCC (1.4), PC (3.1) |
| Archaea | |
| Methanocauldococcus jannaschii | Two-subunit PC (3.2) |
| Metallosphaera sedula | Two subunit YCC (1.4) |
| Fungi | |
| Saccharomyces cerevisiae | Eukaryotic ACC (1.7, 2 copies), PC (3.1, 2 copies), UC (4.1) |
| Kluyveromyces lactis | Eukaryotic ACC (1.7), PC (3.1), UC (4.1) |
| Schizosaccharomyces pombe | Eukaryotic ACC (1.7), PC (3.1) |
| Animals | |
| Xenopus laevis | Eukaryotic ACC (1.7), PCC (1.5), MCC (2.1), PC (3.1) |
| Danio rerio | Eukaryotic ACC (1.7), PCC (1.5), MCC (2.1), PC (3.1) |
| Mus musculus | Eukaryotic ACC (1.7, 2 copies), PCC (1.5), MCC (2.1), PC (3.1) |
| Homo sapiens | Eukaryotic ACC (1.7, 2 copies), PCC (1.5), MCC (2.1), PC (3.1) |
Acetyl-CoA carboxylase (ACC)
Biological functions of ACC
ACC catalyzes the conversion of acetyl-CoA to malonyl-CoA, which serves many functions [37]. It provides the two-carbon building block for fatty acid biosynthesis in most living organisms, and is also used for polyketide biosynthesis in some organisms (Fig. 3). In mammals, ACC1 (also known as ACCα) is cytoplasmic and catalyzes the rate-limiting and committed step of long-chain fatty acid biosynthesis in liver, adipose, and other lipogenic tissues. In the yeast S. cerevisiae, the cytosolic ACC1 is essential for viability. There is also a mitochondrial form of the enzyme (known as HFA1), which is important for growth on lactate or glycerol as carbon source and for fatty acid (especially lipoic acid) biosynthesis in this organelle [38–40]. However, this isoform is unique to S. cerevisiae and is not present in other fungal organisms, although ACC activity has been reported in the mitochondria of some plants. Malonyl-CoA for fatty acid biosynthesis in the mitochondria of other organisms may be produced by malonyl-CoA synthetase [41, 42] and/or PCC working on acetyl-CoA as the substrate.
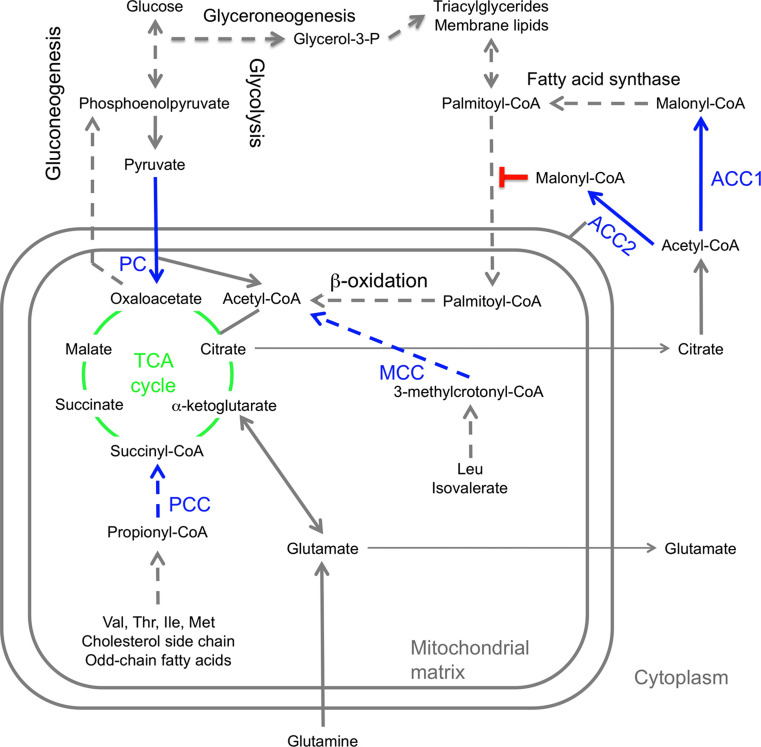
Fig. 3.
Functions of biotin-dependent carboxylases in mammals. Reactions involving the five biotin-dependent carboxylases are shown in blue. Selected intermediates of the TCA cycle (green) are shown. Glutamine can also be used for anaplerosis, especially in some cancer cells. Dashed arrows indicate pathways with more than one step. In other organisms, the biotin-dependent carboxylases have similar functions (with the exception of ACC2), and they are also involved in additional cellular processes
Mammals carry a second isoform of ACC, ACC2 (also known as ACCβ), which is highly conserved with ACC1, with 73 % amino acid sequence identity between human ACC1 and ACC2. ACC2 is associated with the outer mitochondrial membrane through a 140-residue segment at the N-terminus that is absent in ACC1, the first 20 residues of which are highly hydrophobic [43]. However, most of the ACC2 protein faces the cytosol (Fig. 3). This isoform is primarily expressed in heart and muscle tissues, as well as liver. The malonyl-CoA product is a potent inhibitor of carnitine palmitoyltransferase I (CPT-I), the crucial enzyme for the transport of long-chain fatty acyl-CoAs into the mitochondria for β-oxidation (Fig. 3) [44, 45]. There are differences in the expression patterns of human and rodent ACCs [46]. In addition, a form of ACC2 lacking the N-terminal mitochondrial targeting segment is expressed at high levels in white adipose tissue in humans and can participate in de novo lipogenesis [47].
Single-nucleotide polymorphisms (SNPs) in ACC1 and ACC2 are associated with hypertriglyceridemia and hypercholesterolemia, respectively, in patients taking antipsychotic drugs [48]. Polymorphisms in the promoter or the coding region of the ACC1 gene is linked to fatty acid composition in porcine meat [49], fatness traits in chickens [50], fatty acid composition in beef [51], and milk production traits in goats [52].
The activity of both ACC1 and ACC2 in mammals are stimulated by citrate, inhibited by long-chain saturated acyl-CoA, and inactivated by phosphorylation [53], especially AMP-activated protein kinase (AMPK, at Ser80 in ACC1, Ser222 in ACC2) and cAMP-dependent protein kinase (protein kinase A, at Ser1201 in ACC1). Steady-state kinetic studies show that citrate increases the k cat (threefold) and k cat/K m (tenfold) of the ATP and acetyl-CoA substrates for human ACC2, with a K a of ~0.5 mM [54]. In comparison, another report shows that human ACC2 is activated 1,000-fold by citrate, while ACC1 is activated only fourfold [55]. The data also suggest two binding sites for citrate, a higher-affinity (K d ~ 1 mM) activating site and a lower-affinity (K i ~ 30 mM) inhibitory site. A cytosolic protein MIG12 (~20 kD) promotes the polymerization and stimulation of ACC1 by citrate, and is incorporated into the ACC1 polymer [56]. The activity of MIG12 is negatively regulated by forming a complex with another protein, Spot 14 (~17 kD) [57].
Human ACC1 interacts with BRCA1 in a phosphorylation-dependent manner, which reduces the activity of ACC1 [58, 59]. The tandem BRCT domains of BRCA1 recognize the phosphorylated Ser1263 residue of ACC1, in a (pS)PTF motif, and the structure of this complex has been reported [60]. The phosphorylation of Ser1263 is regulated by the cell cycle, possibly through a proline-directed protein kinase such as the cyclin-dependent kinase (CDK).
In plants, plastid ACC (ACC1) produces malonyl-CoA for the biosynthesis of long-chain fatty acids, while cytoplasmic ACC (ACC2) is important for secondary metabolism, including the synthesis of very long-chain fatty acids, flavonoids, cuticular waxes, and other compounds, and for proper embryonic development [61–63]. Plastid ACC in dicots is a multi-subunit enzyme, similar to the multi-subunit bacterial ACCs [64, 65]. On the other hand, the plastid ACC in grasses is a single-chain, multi-domain enzyme, similar to the multi-domain eukaryotic ACCs and arising through gene duplication of the cytosolic ACC. This multi-domain plastid ACC is the target of three classes of herbicides (see below). The BCCP subunit of the multi-subunit plastid ACC of Arabidopsis thaliana forms a complex with the signaling protein PII, which reduces the V max of ACC but does not affect its K m for acetyl-CoA [66]. 2-oxoglutarate, pyruvate or oxaloacetate can disrupt this inhibition. A second isoform of BCCP (BCCP2) in A. thaliana plastids is expressed at much lower levels and cannot rescue the lethal phenotype of BCCP1 null mutants [67]. Two additional BCCP-like proteins lack the biotinylation motif (MKM) but co-purify with the 1–2 MDa ACC complex from A. thaliana chloroplasts [68].
ACC has important functions in other organisms as well. In the apicomplexan pathogen Toxoplasma gondii, ACC1 in the apicoplast is required for fatty acid biosynthesis [69]. In Trypanosoma brucei, RNAi knockdown of ACC results in reduced virulence of the pathogen, and lower fatty acid elongation in procyclic forms [70]. Aedes aegypti mosquitos with deficient ACC activity have reduced lipogenesis and produce significantly fewer eggs [71]. Human cytomegalovirus (HCMV) upregulates the expression and activity of ACC1 in infected cells, and inhibition of ACC1 attenuates viral replication [72]. In archaea and some bacteria, ACC (and PCC) activity is important for CO2 fixation (possibly coupled with ammonia oxidation) [5, 6].
ACCs as drug discovery targets
The crucial roles of ACCs in fatty acid metabolism make them attractive targets for drug discovery against a variety of human diseases, including bacterial infections, fungal infections, type 2 diabetes, cancer, artherosclerosis, and others [2, 3, 73–77]. Besides microbial infections, many of the other diseases are also manifestations of the metabolic syndrome, which is linked to the current obesity epidemic [78–80]. It has been projected that ~50 % of the adult population in the US will be obese by the year 2030 [81], indicating a pressing need for new therapeutic agents and modalities in this area.
The importance of ACCs as targets for drug discovery against the metabolic syndrome was first validated by observations on the ACC2 knockout mice [82–86]. These mice have elevated fatty acid oxidation, increased energy expenditure, reduced body fat and body weight, improved insulin sensitivity, smaller heart size but with normal function, and normal life span and fertility. The animals have increased food intake (hyperphagia), due to reduced malonyl-CoA levels in the brain [87, 88]. In comparison, a null mutation in the ACC1 gene causes embryonic lethality in mice [89]. ACC1 knockout in the liver in mice reduced hepatic lipid accumulation but did not disturb glucose homeostasis [90]. ACC1 knockout in the adipose tissues in mice reduced lipid accumulation in these tissues, but also caused prenatal growth retardation and impaired bone development [91].
Knockdown of ACC1 and ACC2 expression with antisense oligonucleotides confirms the beneficial effects of inhibiting these enzymes [92]. Mice on a high-fat diet show improved peripheral insulin sensitivity after treatment with the potent ACC inhibitor soraphen A [93]. Downregulation of ACC2 activity by the adipokine CTRP1, through AMPK-mediated phosphorylation, leads to elevated fatty acid oxidation and reduced adiposity in mice [94]. On the other hand, overexpression of ACC2 is associated with increased production of proinflammatory cytokines in a human renal cell, which can be reversed by inhibition of p38 MAP kinase [95]. This may be a mechanism for diabetic nephropathy development. An SNP in ACC2 is associated with type 2 diabetic nephropathy and proteinuria [96–98], while another SNP in ACC2 is linked to increased risk for metabolic syndrome [99].
Two recent reports have failed to observe the beneficial effects of ACC2 knock out or down regulation, as elevated fatty acid oxidation did not change energy expenditure or adiposity [100, 101]. Another study failed to demonstrate a correlation between ACC2 phosphorylation (down regulation) and fatty acid oxidation in skeletal muscle [102]. Some of these differences could be due to variations in the genetic background of the mice or the strategies that were used to create the knockout mice [86, 103, 104].
A large number of potent (nanomolar) inhibitors have been reported against human ACCs [105–116]. Some of these compounds have nearly equal activity against both isoforms (isoform nonselective), while others are more selective toward ACC2. They can reduce tissue malonyl-CoA levels, inhibit fatty acid biosynthesis, enhance fatty acid oxidation, reduce plasma triglyceride levels, improve insulin sensitivity in cells or in rodent models. However, long-term treatment with an inhibitor of both ACC1 and ACC2 did not lead to sustained reduction in hepatic triglyceride levels or body weight in rodent models, although the compound was able to stimulate fatty acid oxidation [117]. Long-term down-regulation of ACC1 activity in β-cells leads to reduced glucose-stimulated insulin secretion [118], indicating that ACC2 inhibition may be the more desirable approach for diabetes therapy.
Recent studies suggest that ACCs are also attractive targets for anti-cancer agents. ACC is over-expressed in liver, prostate, breast, and other cancers [119–122], and RNAi knockdown or chemical inhibition of ACC1 leads to growth inhibition and apoptosis [111, 123–125] as well as reduced tumor cell invasion [126]. AKR1B10 (aldo–keto reductase family 1 B10) is overexpressed in some cancer cells and can form a complex with ACC1 and stabilize its cellular levels [127], consistent with the importance of ACC1 in cancer. As discussed earlier, ACC1 activity is downregulated through a phosphorylation-dependent interaction with BRCA1 [58, 59].
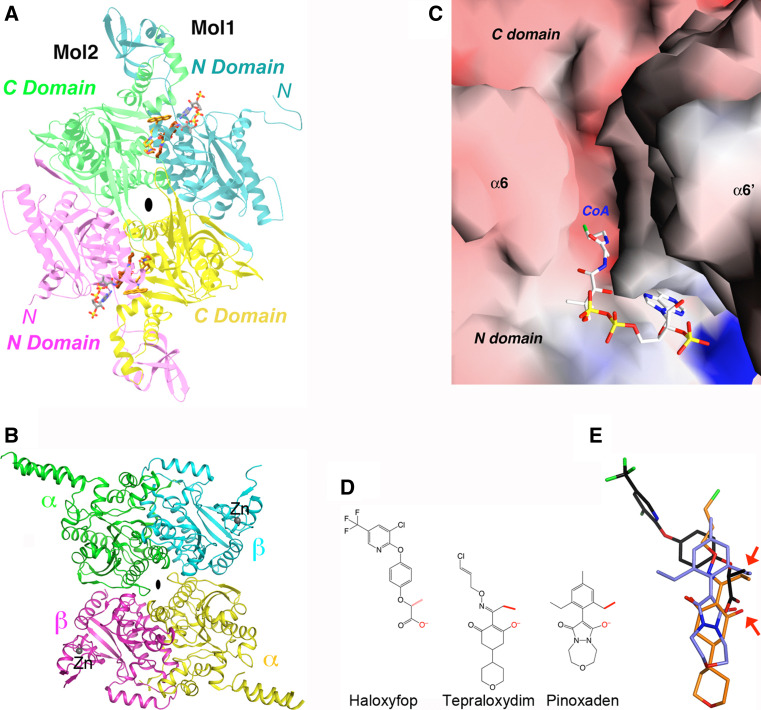
Plastid multi-domain eukaryotic ACCs from grasses are the targets of three classes of commercial herbicides, aryloxyphenoxypropionates (APPs or FOPs), cyclohexanediones (CHDs or DIMs), and pinoxaden [128–131]. FOP and DIM inhibitors have been used in the field for more than 30 years, and resistance mutations have been reported against them [132–136]. Pinoxaden was introduced in 2006, but resistance grasses were already present, likely due to cross resistance to the FOPs and/or DIMs [137, 138]. Additional pinoxaden resistance mutations have also been reported [136, 139]. A herbicide-resistant green foxtail plant has higher fitness than the wild-type [140]. Dimeric cyclohexanedione compounds also have activity against the malaria parasite Plasmodium falciparum [141].
Potent inhibitors that target the BC activity of bacterial ACCs have been developed, with potential use as antibiotics [142–146]. These compounds bind selectively to the ATP binding site of BC, and the initial leads were identified by directed high-throughput screening, virtual screening, and fragment-based approaches. For the CT activity, the natural products moiramide and andrimid are known inhibitors [146]. A single-site mutation (M203L) in the β subunit of E. coli CT produces a fivefold resistance to andrimid [147]. Components of cinnamon oil are also inhibitors of CT activity, partly explaining the antibacterial effect of cinnamon bark [148].
Structures of ACC BC component
The ACC holoenzymes have been difficult to study at the structural level. The multi-subunit bacterial ACCs dissociate readily during purification, while the multi-domain eukaryotic ACCs are exceptionally large (~250-kD monomers). On the other hand, using the divide-and-conquer approach, structural information has been obtained for the BC, BCCP, and CT components of both bacterial and eukaryotic ACCs. The structure of a bacterial BC subunit (from E. coli ACC) was first reported in 1994 [149], and the structure of a eukaryotic BC domain (from yeast ACC) was first reported in 2004 [150]. Currently, a large number of structures are available for bacterial and eukaryotic BC components [143, 151–157].
The BC structure contains three domains, A, B, and C domains (Fig. 4a). Residues in domains A and C form the active site. Domain B undergoes a large conformational change to close over the active site during catalysis. Eukaryotic BC domain contains several inserted segments, especially at the N-terminus and between domains A and B (the AB linker), which explains its larger size (~550 residues) compared to bacterial BC subunits (~450 residues) (Fig. 4b).
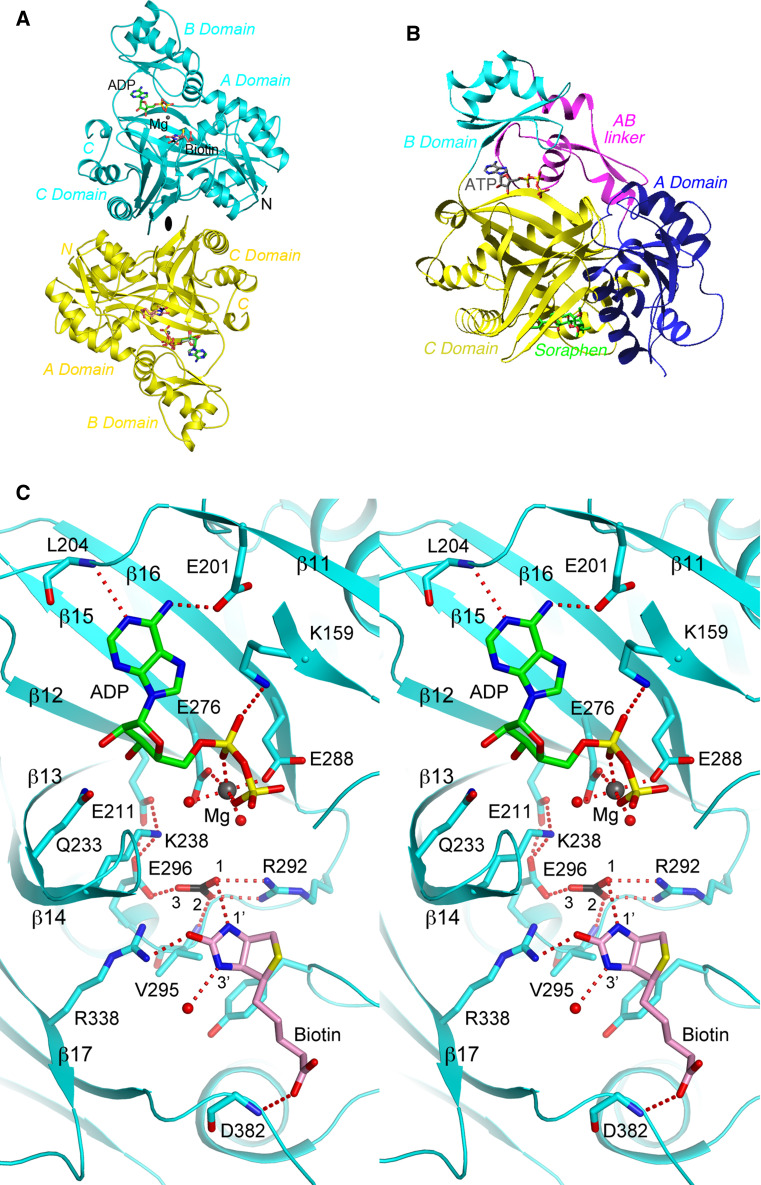
Fig. 4.
Structural information on the BC component. a Structure of the BC subunit dimer (cyan and yellow) of E. coli ACC in complex with MgADP, bicarbonate, and biotin [154]. The twofold axis of the dimer is indicated with the black oval. b Structure of the BC domain of yeast ACC in complex with the inhibitor soraphen A (green) [161]. The sub-domains of BC are given different colors. The bound position of ATP is also shown to indicate the location of the active site. The view is similar to that for the top monomer in a. c Stereo figure showing detailed interactions between MgADP, bicarbonate, and biotin with the active site of the E. coli BC subunit. Several segments of the protein, including the glycine-rich loop, are omitted for clarity. The structure figures were produced with the program PyMOL (www.pymol.org)
The structure of E. coli BC in a pentary complex with the substrates MgADP, bicarbonate, and biotin provides detailed insights in the catalysis by this enzyme (Fig. 4a) [154]. The substrates are recognized by an intricate network of ionic, hydrogen-bonding and van der Waals interactions (Fig. 4c). One of the oxygen atoms of bicarbonate is poised to initiate the nucleophilic attack on ATP to form the carboxyphosphate intermediate, and Glu296 is the general base that extracts the proton from bicarbonate. The orthophosphate (PO4 3–) derived from the decomposition of carboxyphosphate is the general base that extracts the proton on the N1′ atom of biotin [17], and Arg338 stabilizes the resulting enolate biotin intermediate for the carboxylation reaction.
Mutation of residues in the BC active site (including Glu296 and Arg338) confirms their importance for catalysis. Mutation of Gly165 and/or Gly166 in the glycine-rich loop in domain B and near the phosphates of ATP has no effect on the k cat but leads to a 40-fold increase in K m for ATP [158]. The catalytic activity of E. coli BC can be inhibited by ATP at high concentrations (substrate inhibition) and other nucleotides [157]. A second ADP molecule can be bound in the BC active site, with its phosphate groups occupying the binding sites of bicarbonate and biotin.
Escherichia coli BC is a dimer (Fig. 4a), and studies suggest that catalysis at the two active sites of the dimer may be coupled [159]. A chimeric mutant where one of the active sites of the dimer is knocked out is essentially inactive. However, each active site of the dimer is located ~25 Å away from the dimer interface, with no contributions from residues in the other monomer (Fig. 4a). It is postulated that there may be long-range communications between the two active sites, through the dimer interface. The two monomers may undergo alternating catalysis, with one monomer binding substrates and catalyzing turnover and the other releasing products. Half-site reactivity of the BC subunit has also been proposed [152, 160], although no cooperative behavior is observed from kinetic studies.
Mutations in the dimer interface of E. coli BC can produce mutants that are monomeric in solution [161]. These mutants are still catalytically active, indicating that dimerization is not absolutely required for BC catalysis in vitro. On the other hand, dimerization of BC is required in vivo, possibly for the assembly of the ACC holoenzyme [162].
The polyketide natural product soraphen A is an allosteric inhibitor of eukaryotic ACC, binding to the BC domain using the equivalent surface area as that for dimerization of E. coli BC (Fig. 4b) [150]. Conformational differences in this region of eukaryotic BC allow soraphen A binding but disallow dimerization. The BC domain of eukaryotic ACC is monomeric in solution and is catalytically inactive [163]. A BODIPY-labeled soraphen analog has been developed and can be useful for screening for new compounds and characterizing inhibitor binding to the BC domain [155].
The AMPK phosphorylation sites in mammalian ACCs are located in the N-terminal extension of the BC domain. The structure of human ACC2 BC domain phosphorylated by AMPK shows that the segment enclosing the phosphorylated Ser222 residue is located in the soraphen-binding site [156]. Therefore, structural information on soraphen A binding and the phosphorylated BC domain supports the hypothesis that this region of BC can allosterically regulate catalysis, although the exact molecular mechanism for this long-range communication is currently not known.
Structures of ACC BCCP component
The BCCP domain of E. coli ACC, including the “thumb” feature, is more flexible in the apo protein [164]. Upon biotinylation, the thumb interacts with biotin [165], leading to its stabilization as well as that of BCCP domain overall [164, 166]. Solution structure of the BCCP domain of human ACC2 is similar to that of BCCP subunit of E. coli ACC [165], except that the human protein does not have the thumb feature [167]. As a result, the covalently attached biotin is flexible in human BCCP, while it interacts with the thumb in E. coli BCCP. This thumb structure interferes with biotinylation by the human holocarboxylase synthase (HCS), though it has no effect on biotinylation by the E. coli BirA enzyme [168]. In comparison, an engineered E. coli BCCP lacking the thumb feature, and the BCCP domain of human PCC (which also lacks the thumb feature) can be readily biotinylated by HCS [168].
Structures of ACC CT component
The structure of a eukaryotic CT domain (from yeast ACC) was first reported in 2003 [169], and the structure of bacterial CT subunit was first reported in 2006 [170]. The structure of the CT domain contains two sub-domains, N and C domains (Fig. 5a), which are equivalent to the β and α subunits of bacterial CT (Figs. 2, 5b). Each domain/subunit has the crotonase fold (a β–β–α superhelix). The active site of CT is located at the bottom of a “canyon” in the interface of a dimer of the CT domains (Fig. 5c) or an α2β2 heterotetramer of the bacterial CT subunits. Therefore, CT must dimerize to be catalytically active.
Fig. 5.
Structural information on the CT component of acyl-CoA carboxylases. a Structure of the CT domain dimer of yeast ACC in complex with tepraloxydim (brown) [172]. The N and C domains for monomer 1 are colored in cyan and yellow, respectively, and those for monomer 2 in magenta and green. The bound positions of CoA (gray) [169] and CP-640186 (gold) [174] are also shown. b Structure of the CT subunit of S. aureus ACC [170]. The zinc ions are shown as spheres (dark gray). The view is similar to that for a. c Molecular surface showing the canyon in the active site region of the CT dimer. CoA is recognized by the N domain of one monomer, in the bottom half of the canyon. Biotin is recognized by the C domain of the other monomer, in the top half of the canyon. The side chain of Lys1764 has been omitted in this figure. d Chemical structures of the herbicides haloxyfop (FOP), tepraloxydim (DIM), and pinoxaden. The two anchoring points of interaction with the CT domain are highlighted by the red arrows. e Overlay of the binding modes of haloxyfop (black), tepraloxydim (brown), and pinoxaden (light blue)
Using the yeast CT domain as a surrogate since crystals of the CT domain of grass plastid ACCs are not yet available, the binding modes of all three classes of herbicides, haloxyfop, tepraloxydim, and pinoxaden, have been determined [171–173]. The compounds inhibit ACC by competing against the binding of the acetyl-CoA substrate. Despite their chemical diversity, the three compounds share two common anchoring points of interactions with the CT domain—a negatively charged oxygen atom that likely mimics the oxyanion in the substrate during catalysis, and a small hydrophobic group (methyl or ethyl) that is probably located in the binding site for the acetyl group. Each compound also establishes unique interactions with the CT domain. Haloxyfop binding requires a large conformational change in the active site, in the dimer interface of the CT domain, while tepraloxydim and especially pinoxaden require much smaller changes. Most of the herbicide resistance mutations are located in or near the binding sites, consistent with their effects on herbicide binding although the exact molecular mechanism is still not fully understood.
Studies using the yeast CT domain also reveal that a class of inhibitors of mammalian ACCs, as exemplified by the CP-640186 compound, block the binding of (carboxy)biotin to the CT-active site (Fig. 5a) [108, 174]. Structures of the human and bovine ACC2 CT domain have been reported at 3.2 Å and 2.4 Å resolution, respectively [112, 175]. In a different approach, nine mutations are introduced into the active site of yeast ACC CT domain to “humanize” this protein [176]. The resulting mutant binds human ACC inhibitors with similar potency as the human CT domain and produces crystals that diffract up to 2.4 Å resolution.
A unique feature of bacterial CT is the presence of a zinc finger in the β subunit (Fig. 5b) [170], which binds the CT mRNA and inhibits its translation, and this inhibition can be reversed by the substrate acetyl-CoA [177]. E. coli CT also binds and is inhibited by single-stranded, double-stranded, and hairpin DNA [178].
Propionyl-CoA carboxylase (PCC)
Biological functions of PCC
PCC catalyzes the conversion of propionyl-CoA to D-methylmalonyl-CoA. In most organisms, it is crucial for the catabolism of β-branched amino acids (Thr, Val, Ile) and Met, cholesterol side chain, and fatty acids with an odd number of carbon atoms (Fig. 3). Mammalian PCC is localized in the mitochondrial matrix, and is activated by mono-valent cations (K+, NH4 +, Cs+) [179–181]. PCC activity is also important for CO2 fixation in some archaeal organisms [5, 6], methanol assimilation in Methylobacterium extorquens [7], acetyl-CoA assimilation in α-proteobacteria [10], 3-hydroxypropionate assimilation in Rhodobacter sphaeroides [11], mycolic acid and methyl-branched long-chain fatty acid biosynthesis in M. tuberculosis [12], and polyketide biosynthesis in Streptomyces and other organisms [13].
Inherited deficiencies in PCC activity in humans are linked to propionic acidemia (PA), with symptoms usually first appearing during the neonatal period, including vomiting, lethargy, ketoacidosis, delayed growth, cardiomyopathy, mental retardation, and death in severe cases. A large number of autosomal recessive mutations in both subunits of PCC have been identified, including missense mutations, a few nonsense mutations, insertions/deletions, and splicing mutations [182–189]. PCC is expressed in the brain and may be important for neurodevelopment, which may be related to the neurological effects of PCC deficiency [190], including epilepsy [191] and seizure [192].
PCCα−/− mice die of propionic acidemia within 2 days after birth, but they can be rescued by a liver-specific transgene [193]. Anti-sense morpholino oligonucleotides have been used to correct splicing defects in PCC, which restored normal PCC activity in patients’ fibroblasts [194]. In another study, a modified U1 snRNA was used to correct a splicing defect, but it did not restore PCC enzymatic activity [195]. Successful rescue of PCC deficiency in mouse models by gene therapy has been demonstrated [196, 197], indicating a potential treatment strategy for PCC deficiency in humans.
Structure of PCC
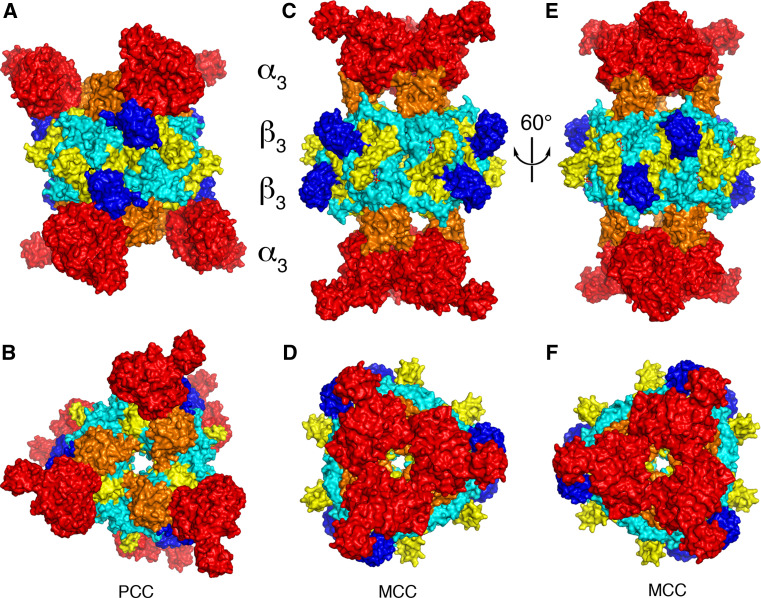
The crystal structure of the 750-kD α6β6 holoenzyme of a bacterial PCC was reported recently, and a similar structure was observed for human PCC based on a cryo-electron microscopy reconstruction [22]. The holoenzyme obeys 32-point group symmetry, with a central β6 hexamer core (Fig. 6a) and three α subunits at each end of the β6 core (Fig. 6b). Therefore, the structure of the PCC holoenzyme consists of four layers, with three subunits in each layer, α3–β3–β3–α3.
Fig. 6.
Striking differences in the overall architecture of the holoenzymes of PCC and MCC. a Crystal structure of the bacterial PCC holoenzyme [22], viewed down the twofold symmetry axis within a β2 dimer. The domains are colored as in Fig. 2. The four layers of the structure are indicated. b Structure of the PCC holoenzyme, viewed down the threefold symmetry axis. c Crystal structure of the P. aeruginosa MCC holoenzyme [23], viewed down the twofold axis within a β2 dimer. d Structure of the MCC holoenzyme, viewed down the threefold axis. e Structure of the MCC holoenzyme, after a ~60° rotation around the vertical axis from c. The view is down the twofold axis relating two β2 dimers. f Structure of the MCC holoenzyme, after a ~60° counterclockwise rotation from panel D
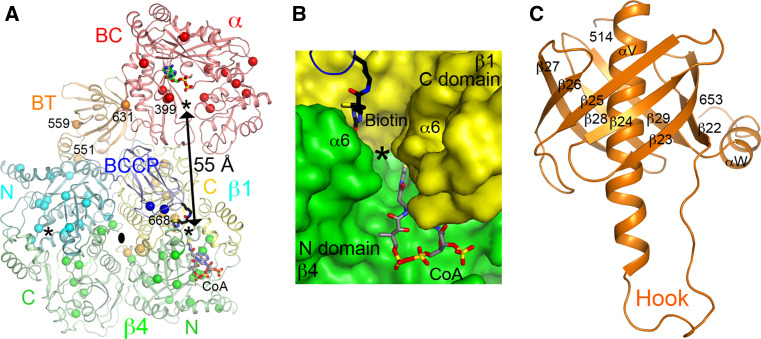
The β6 hexamer core is in the shape of a short cylinder and there are intimate contacts among the six monomers. The overall structure of this hexamer core is similar to the 12S subunit of transcarboxylase [31] as well as the β subunit of S. coelicolor and M. tuberculosis acyl-CoA carboxylases [198–200]. In addition, the β2 dimer of PCC, with one monomer from each layer of the β6 hexamer (Fig. 6a), is similar to the dimer of the CT component of ACC (Fig. 5a). The active site is located at the interface of this dimer (Fig. 7a), at the bottom of a deep canyon (Fig. 7b). In the PCC crystal, BCCP-biotin (from the α subunit) is bound in the CT active site, interacting with residues in the C domain of one β subunit (Fig. 7b). The propionyl-CoA substrate is recognized by the N domain of the other subunit (Fig. 7b).
Fig. 7.
The active sites of PCC. a Relationship between the BC and CT active sites (indicated with the asterisks) in the PCC holoenzyme. The CT active site is located at the interface of a β2 dimer, with the β subunit from the bottom layer colored in green. Sites of disease-causing missense mutations are indicated with the spheres. The third asterisk indicates the other active site of the CT dimer. b Molecular surface of the active site region of CT. The observed position of biotin is shown (stick model in black). The position of CoA (gray) is modeled based on that in the structure of the yeast ACC CT domain [169]. c Structure of the BT domain of the bacterial PCC α subunit. The hook region is labeled
In comparison, the α subunits are arranged as monomers, splayed far apart from each other, and there are essentially no contacts among them in the PCC holoenzyme (Fig. 6b). This is in sharp contrast to the dimeric organization of the BC subunit of bacterial ACCs (Fig. 4a) and the BC domain of pyruvate carboxylase (see below). While the overall structure of the BC domain of the PCC α subunit is similar to that of the bacterial BC subunit, sequence, and conformational differences for those residues in the putative dimer interface in PCC precludes the formation of a similar dimer.
There are few contacts between the BC domain in the α subunit and the β subunit (CT component) (Fig. 6a). Instead, interactions between the α and β subunits in PCC is mediated by a previously unrecognized domain, located between the BC and BCCP domains in the primary sequence of the α subunit (Fig. 2). It has been named the BT domain [22], for its role in mediating BC-CT interactions. The structure of this domain has a novel backbone fold, with a long helix surrounded by an eight-stranded anti-parallel β-barrel (Fig. 7c). A “hook”, comprising the C-terminal part of the helix and the loop connecting it to the first strand of the β-barrel, has a central role in the interactions between the α and β subunits in the PCC holoenzyme.
The distance between the BC and CT active sites in the PCC holoenzyme is ~55 Å (Fig. 7a). This cannot be reached by biotin in the swinging-arm model. Therefore, the BCCP domain (in addition to its attached biotin) must translocate during PCC catalysis, hence the swinging-domain model (Fig. 1a).
The structure of the PCC holoenzyme also provides a framework for understanding the large collection of disease-causing mutations. The missense mutations are distributed throughout the entire enzyme (Fig. 7a). Some of them affect substrate binding and/or catalysis, while others affect protein stability [201–207]. At the same time, few of the mutations are located directly in the interface between the subunits of the holoenzyme, consistent with the extensive nature of the interface. Single-site mutations in this interface may not be sufficient to disrupt the holoenzyme. In fact, five mutations in the hook region are needed to disrupt the interactions between the α and β subunits [22].
3-Methylcrotonyl-CoA carboxylase (MCC)
Biological functions of MCC
MCC catalyzes the conversion of 3-methylcrotonyl-CoA to 3-methylglutaconyl-CoA (Fig. 1b). It is essential for the catabolism of leucine and isovalerate in most living systems (Fig. 3) [61]. In Pseudomonas organisms, MCC is required for the metabolism of acyclic terpenoids [14, 208–212]. In Arabidopsis, MCC activity is required for seed development and germination [213]. MCC expression level is higher in male zebra finches, which may be important for the development of their singing behavior [214].
MCC shares good sequence conservation with PCC and its holoenzyme is also a 750-kD α6β6 dodecamer. On the other hand, while PCC carboxylates the α carbon of the substrate, MCC carboxylates the γ carbon of an α–β unsaturated acid (Fig. 1b). MCC is located in the mitochondrial matrix in eukaryotes, and mutations in the N-terminal mitochondrial targeting sequence of both subunits can affect their localization [215]. Human MCC has been expressed and purified from the baculovirus system in the active form [216]. It demonstrated hyperbolic kinetics with ATP and 3-methylcrotonyl-CoA, while the Pseudomonas MCC showed sigmoidal kinetics toward ATP [212].
Deficiencies in MCC activity in humans are linked to 3-methylcrotonylglycinuria (MCG), which constitute one of the most frequently observed inborn errors of metabolism [217–226]. The clinical manifestations of MCG are highly variable, from asymptomatic individuals to neonatal onset, severe cases that can result in death. Neurological symptoms such as psychomotor retardation, seizure and coma have also been observed. Like PCC, a large number of autosomal recessive mutations in both subunits of MCC have been identified.
Structure of MCC
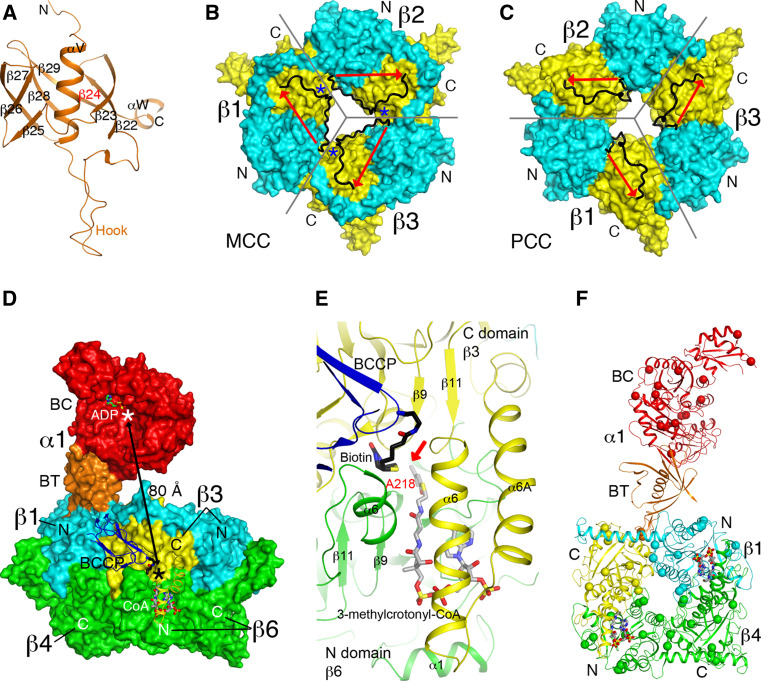
The structure of the P. aeruginosa MCC holoenzyme also obeys 32-point group symmetry, with three subunits in each of four layers (Fig. 6c) [23]. However, despite its sequence conservation with PCC, the overall architecture of the MCC holoenzyme is strikingly different from that of PCC. This is especially apparent for the locations of the α subunit relative to the β subunit, the organization of the N and C domains in the β subunit, and the structure and placement of the BT domain. As a result of these differences, the overall shape of the MCC holoenzyme is highly distinct from that of PCC (Fig. 6a, c).
Rather than being splayed away from each other as in PCC, the three α subunits at each end of the central β6 core of MCC are located close to each other, in fact showing trimeric association (Fig. 6d). In addition, the BC domain in the α subunit is located ~20 Å above the β subunit, and there is no contact between them. The BT domain bridges the BC domain and the β subunit. While the overall structure of this domain is similar to that in PCC, there are also important differences. Especially, the hook in the MCC BT domain has a different conformation (Fig. 8a), contributing to the different architecture of the MCC holoenzyme compared to PCC. The Pseudomonas MCC BT domain lacks the third strand of the eight-stranded β-barrel (Fig. 8a), although this strand is likely present in human and most other MCCs [23].
Fig. 8.
Structure of P. aeruginosa MCC. a Structure of the BT domain of the P. aeruginosa MCC α subunit. The missing third strand is indicated in red. b Structure of the β6 hexamer of MCC. The subunit boundaries are indicated with the gray lines. The N (cyan) and C (yellow) domains are labeled. The linker between the two domains is shown in black, with the direction given by the red arrow. The linkers from neighboring subunits come very close to each other at one point (blue asterisk), and therefore only a small change is needed to switch to the connectivity seen in the PCC β subunit. c Structure of the β6 hexamer of PCC, shown in the same scheme as panel B. The linker in PCC β runs in the opposite direction compared to MCC β. d Relationship between the BC and CT active sites (indicated with the asterisks) in the MCC holoenzyme. While the BT domain of the α subunit contacts one β2 dimer (β1 and β4), the BCCP domain of that α subunit is actually located in the active site of a different β2 dimer (β3 and β6). e Binding modes of biotin (black) and 3-methylcrotonyl-CoA (gray) to the CT active site of MCC. The position of 3-methylcrotonyl-CoA is modeled based on that of CoA in MCC [23] and crotonyl-CoA in GCDα [30]. The carbon atom to be carboxylated is indicated with the red arrow. f Locations of disease-causing missense mutations in the structure of MCC (indicated with the spheres). Only the β1–β4 dimer is shown
In the β subunit, the positions of its N and C domains in MCC (Fig. 8b) are swapped relative to those in PCC (Fig. 8c). This difference is not due to a swap of the two domains in the primary sequences of the two β subunits. Instead, it is due to a different connectivity between the N and C domains. In essence, each layer of the β6 hexamer contains alternating N and C domains of the three subunits. While PCC uses one way of connecting neighboring N and C domains to make a β subunit (Fig. 8c), MCC uses the alternative way, such that the linker between the two domains runs in the opposite direction as compared to PCC (Fig. 8b).
The N and C domains of the β subunits in PCC and MCC have the same backbone fold. The pseudo symmetry operation relating the two domains is a rotation of ~60° along the threefold axis of the β6 hexamer. Therefore, the β6 hexamers of MCC and PCC have pseudo sixfold symmetry. The swapping of the N and C domains in MCC follows this pseudo symmetry, equivalent to a 60° rotation around the threefold axis of the hexamer (Fig. 8b, c), and therefore does not lead to a large change in the overall shape of the hexamer. Such a rotation also places the BCCP domain in the same position in the MCC holoenzyme compared to PCC (Fig. 6e, f).
The connectivity between the N and C domains observed in MCC β subunit is the same as that in the α subunit of the Na+-transporting glutaconyl-CoA decarboxylase (GCDα) [27, 30]. However, GCDα is a dimer only, and therefore the different connectivity leads to a distinct organization of the active site in GCDα dimer compared to the CT dimer of ACC (Fig. 5a). Like MCC and GCC, GCDα is also active on the γ carbon of an α-β unsaturated acid. It is possible that these three enzymes share a common evolutionary origin, representing a second lineage of biotin-dependent carboxylases that act on CoA esters. Another common feature among MCC, GCDα and likely GCC is the extended N-terminal segment of the β subunit, which has extensive interactions with the C domain of the same subunit (Fig. 8b). In comparison, the N-terminal segment of PCC β subunit is much shorter (Fig. 8c). These additional interactions may be the molecular basis for the swapping of the positions of the N and C domains [23].
The distance between the BC and CT active sites in the MCC holoenzyme is ~80 Å (Fig. 8d), indicating that the BCCP domain must translocate during catalysis (the swinging-domain model). BCCP-biotin is located in the CT active site in the MCC structures (Fig. 8d). Compared to the structure of PCC, biotin is bound deeper into the CT active site, consistent with its carboxylation of the γ carbon of the substrate. The N1′ atom of biotin is ~6 Å from the γ carbon of the 3-methylcrotonyl-CoA substrate (Fig. 8e), modeled based on the observed positions of CoA bound to MCC [23] and crotonyl-CoA bound to GCDα [30]. A conformational change for two helices in the CT active site (α6 and α6A, Fig. 8e) is observed upon CoA binding in MCC [23]. A similar conformational change is also observed in GCDα [30].
The disease-causing mutations are distributed over the entire structure of the holoenzyme (Fig. 8f). Some of them are located in the BC or CT active site. For example, the R385S mutation in the α subunit abolishes the stabilization of the biotin enolate during BC catalysis. The A214T mutation in the β subunit is centrally located between biotin and 3-methylcrotonyl-CoA in the CT active site (Fig. 8e) and may block binding of either or both substrates. Some of the mutations may also disturb the interaction between the BCCP or the BT domain and the β subunit [23].
Geranyl-CoA carboxylase (GCC)
GCC is classified into the same family as MCC since its site of carboxylation in the substrate is also on the γ carbon of an α-β unsaturated acid (Fig. 1b). The amino acid sequences of bacterial GCC are highly conserved with those of MCC, and the holoenzyme of GCC is likely also an α6β6 dodecamer. At the same time, the geranyl group is much larger than the 3-methylcrotonyl group, and therefore the CT component (β subunit) of GCC is expected to have a larger pocket in the active site to accommodate this group. In fact, P. aeruginosa MCC cannot carboxylate geranyl-CoA, while GCC has activity toward both geranyl-CoA and 3-methylcrotonyl-CoA [212].
GCC is found in Pseudomonas organisms and several other bacteria (Table 1) [14, 208–212, 227]. It is important for the metabolism of the geranyl group and other acyclic terpenes. In fact, some Pseudomonas organisms can use these compounds as the sole carbon source. Through reactions that are similar to β-oxidation, γ-carboxygeranyl-CoA is converted to 3-methylcrotonyl-CoA (releasing two acetyl-CoAs and one acetate), and MCC facilitates the further degradation of this compound.
GCC activity has been purified from maize leaves, and it is probably also important for the metabolism of acyclic terpenes in plants [61, 228]. The biotin-containing component of this enzyme was found to have a molecular weight of ~122 kD, which is significantly larger than the ~75-kD α subunit of bacterial GCC (and MCC). Plant GCCs may constitute another family of biotin-dependent carboxylases, although a sequence for this protein is currently not available.
Other acyl-CoA carboxylases
Streptomyces, Mycobacterium, and Corynebacterium organisms have a collection of α6β6 or α2β2 acyl-CoA carboxylases (family 1.4, Fig. 2) [12, 229]. These enzymes are different from PCC (family 1.5) in that they probably lack the BT domain in the α subunit. In addition, they have more diverse substrate preferences, being active toward acetyl-, propionyl-, and/or butyryl-CoA [230, 231]. Moreover, some of these enzymes require the presence of a third subunit, ε subunit (~7 kD in S. coelicolor and ~25 kD in M. tuberculosis), for maximal activity, although the exact stoichiometry of this subunit is currently not known [12, 232, 233]. These acyl-CoA carboxylases are important for the biosynthesis of fatty acids, branched-chain fatty acids and other compounds, such as mycolic acid [234, 235]. An acyl-CoA carboxylase in Mycobacterium tuberculosis is crucial for mycolic acid biosynthesis and pathogenesis [236].
The structures of the β6 hexamer of these enzymes have 32 symmetry [198–200, 237], and are similar to that of PCC [22] and the 12S subunit of transcarboxylase [31]. An Asp residue near the CT active site may be important for substrate preference for propionyl-CoA, while mutating this residue to Ile changes the preference to acetyl-CoA [198, 200]. On the other hand, an equivalent mutation in PCC is not able to change the substrate preference of that enzyme [22].
A Rhizobium etli acyl-CoA carboxylase has ~15-fold higher activity toward propionyl-CoA than acetyl-CoA, and it is activated substantially by mono-valent cations, K+, NH4 + and Cs+ [238].
Pyruvate carboxylase (PC)
Biological functions of PC
PC catalyzes the conversion of pyruvate to oxaloacetate (Fig. 1b) and it is an important enzyme in intermediary metabolism [4, 239–243]. In mammals, PC is localized in the mitochondrial matrix and has crucial roles in gluconeogenesis in liver and kidney (being the first enzyme in the gluconeogenesis pathway), lipogenesis and glyceroneogenesis in adipocytes, and biosynthesis of the excitatory neurotransmitter glutamate in astrocytes (Fig. 3). PC has an important anaplerotic role, replenishing the intermediates of the tricarboxylic acid (TCA) cycle that have been withdrawn for the biosynthesis of glucose, fatty acids, amino acids and other molecules [244]. Tumor cells depend on PC for anaplerosis in the absence of glutamine (Fig. 3), and cells with high PC activity may be resistant to inhibition of glutamine metabolism [245].
PC is important for glucose-induced insulin secretion by pancreatic β-cells in rats [246], and knock-down of PC activity by shRNA in rat insulinoma cells leads to impaired insulin secretion [247]. However, PC protein level and enzymatic activity are low in human pancreatic β-cells, and these cells may use a different pathway for stimulating insulin secretion [248].
Fungal PC is localized in the cytoplasm, in contrast to other eukaryotes. S. cerevisiae is unique in that it carries two PC enzymes, and the expression levels of the two enzymes are regulated independently in response to different nutrients [4]. PC is required for the assembly, import, and/or activation of some peroxisomal enzymes in several fungi, including alcohol oxidase in Hansenula polymorpha [249] and D-amino acid oxidase in Pichia pastoris [250]. This function is mediated by the CT domain of PC [251], but involves residues on the opposite face of the CT domain from the active site and is independent of its catalytic activity [252]. PC activity is important for carbon metabolism and virulence of the human pathogen Listeria monocytogenes [253].
PC is a single-chain, multi-domain enzyme in most organisms (family 3.1, Fig. 2), and is active only as a 500-kD tetramer. In archaea and some bacteria, such as P. aeruginosa [254], PC is in the two-chain form (family 3.2, Fig. 2), but it is also active as a tetramer of the two chains. Acetyl-CoA stimulates single-chain form of PC, by activating the BC activity, but it has no effect on the two-chain form of PC [255, 256]. An ATP analog, MgTNP-ATP, also activates PC activity and can allosterically compete with acetyl-CoA [257]. On the other hand, aspartate, glutamate and α-ketogluratate are allosteric, feedback inhibitors of the CT activity of PC [258].
Deficiency in PC activity is a rare, autosomal recessive metabolic disorder in humans, and has been associated with three forms of clinical manifestations [259–264]. Patients with form A PC deficiency have chronic, mild to moderate lactic acidemia, psychomotor retardation and hypotonia, and many patients die within a few years of birth. Form B deficiency is associated with the most serious symptoms, with severe lactic acidemia, hypoglycemia, hyperammonemia, anorexia, convulsions, and the patients generally die in the first months of life. Form C deficiency shows occasional mild lactic acidemia and may also show mild neurological symptoms. A collection of missense and insertion/deletion mutations has been identified in patients suffering from PC deficiency.
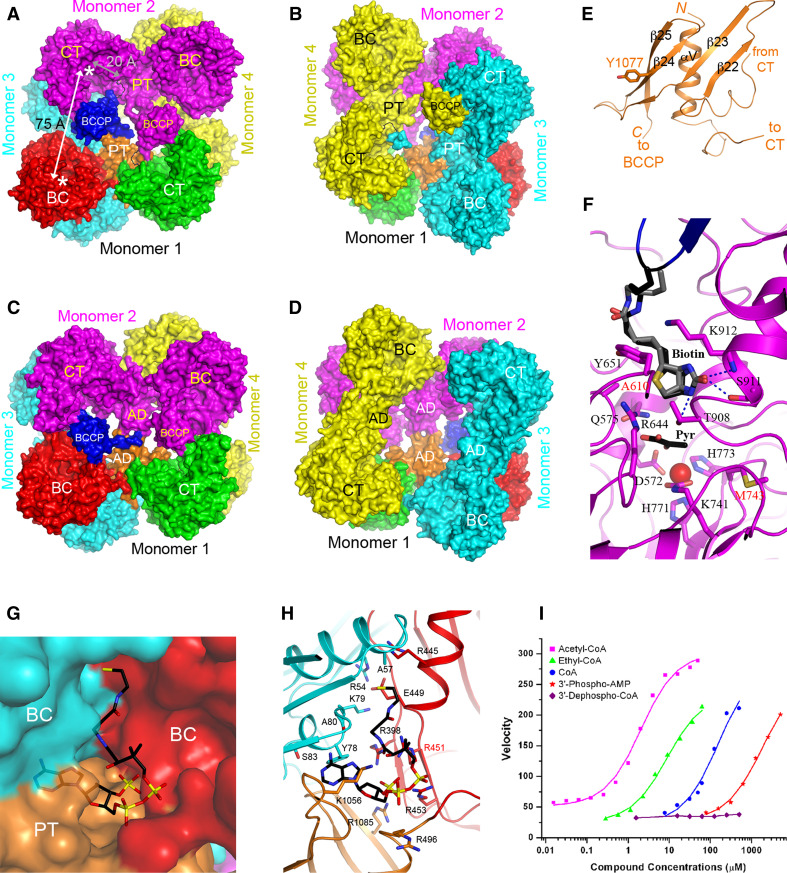
Structure of PC
Crystal structures of the full-length Rhizobium etli PC (RePC) [20, 265], Staphyloccocus aures PC (SaPC) [21, 266], and human PC (HsPC) lacking only the BC domain [21] are currently available. The overall structure of the tetramer is in the shape of a square (or a diamond), with approximate 222 symmetry for SaPC (Fig. 9a). The structure consists of two layers, with two monomers in each layer (Fig. 9b). BC and CT dimers are located at alternate corners of the square, creating an extensive interface between the two layers. On the other hand, there are few contacts between the two monomers in the same layer (Fig. 9a). The overall structures of the four monomers in the tetramer are similar to each other, but there are differences in the relative positions of their BC and CT domains. This is especially true for RePC, where the two monomers in one layer show large differences to those in the other layer. As a result, the RePC tetramer shows significant asymmetry between the two layers (Fig. 9c, d) [20, 265].
Fig. 9.
Structure of PC. a Crystal structure of SaPC tetramer [21]. The domains of monomer 1 are colored as in Fig. 2, and the other three monomers are in magenta, cyan and yellow. The BC and CT active sites are indicated with the asterisks. The distance between the exo site and the CT active site is also labeled (gray). b Structure of SaPC tetramer, viewed from the bottom layer. c Crystal structure of RePC tetramer [20]. d Structure of RePC tetramer, viewed from the bottom layer. e Structure of the PT domain of SaPC [21]. f Structure of the active site region of the CT domain, in complex with BCCP (blue). The position of biotin in the SaPC structure is shown in black, and that in the HsPC structure in gray [21]. g Molecular surface of the binding site of CoA in SaPC [266]. h Detailed interactions between CoA and the binding site in SaPC. i The activity of various acetyl-CoA analogs in stimulating the catalysis by SaPC [266]
Structural information on these PC enzymes reveals the presence another domain, named as the PT (PC tetramerization) [21] or allosteric [20] domain. The structure of this domain contains a helix (formed by 30 residues between the BC and CT domains, Fig. 2) surrounded by a highly twisted, four-stranded anti-parallel β-sheet (formed by 60 residues between the CT and BCCP domains) (Fig. 9e). In HsPC and SaPC, the PT domain is important for tetramerization, hence its name [21]. A PT domain in one layer interacts with a PT domain in the other layer, and mutations in this PT–PT interface can disrupt PC tetramerization and catalytic activity. In contrast, the equivalent allosteric domain in RePC does not appear to be important for tetramerization (Fig. 9c) [20]. RePC lacking the BC domain is a dimer in solution [265], while the same construct for HsPC is a tetramer [21].
The structure of the PT domain shares remote similarity to that of the BT domain in PCC and MCC. The eight-stranded β-barrel in the BT domain (Fig. 7c) is replaced by a four-stranded anti-parallel β-sheet in the PT domain (Fig. 9e). However, while the helix is connected directly to the β-barrel in the BT domain, the CT domain of PC is inserted between the helix and the β-sheet in the PT domain.
The structure of the CT domain of PC contains a triosephosphate isomerase (TIM) barrel with a long C-terminal extension, and has similarity to that of the 5S (CT) subunit of transcarboxylase [32] and the CT domain of oxaloacetate decarboxylase [29, 267]. In SaPC and HsPC structures [21], one BCCP and its covalently attached biotin is bound in the CT active site of the other monomer in the same layer (Fig. 9b), providing direct insight into the molecular mechanism of the CT reaction. In contrast, the BCCP domains are in a non-productive location and their attached biotins are disordered in the RePC structure (Fig. 9c) [20].
In the CT active site, Thr908 is hydrogen-bonded to the N1′ atom of biotin (Fig. 9f), indicating that it may play an important role in catalysis. The structure of the T908A mutant of SaPC is essentially identical to that of the wild-type enzyme, but the mutant is inactive [266]. Kinetic studies with the equivalent mutant of RePC also confirm the importance of this residue in the CT reaction [268]. The disease-causing A610T and M743I mutations are located in the active site of the CT domain, and are expected to disrupt catalysis by interfering with the binding of biotin (A610T mutation) [21, 266] or pyruvate (M743I mutation) (Fig. 9f) [21, 32]. The importance of other residues in the CT active site has also been assessed by mutagenesis studies in SaPC [266] and RePC [269].
The structural information defines the molecular basis for why PC is only active in the tetrameric form [20, 21]. BCCP-biotin is carboxylated in the BC domain of its own monomer, but it then transfers the CO2 to pyruvate in the CT domain of the other monomer in the same layer (Fig. 9a). Therefore, both monomers in each layer are required for catalysis, but their conformation is stabilized only in the context of the entire tetramer because there are few interactions between two monomers of the same layer.
The distance between the BC and CT active sites in PC is ~75 Å (Fig. 9a), supporting the swinging-domain model for catalysis (Fig. 1a). Binding of BCCP-biotin in the CT active site is observed in SaPC and HsPC [21]. Recently, BCCP-biotin in a nonproductive binding mode in the BC domain of RePC was reported [265].
The overall structures of the BC domain monomer and dimer of PC are similar to those of the BC subunit of E. coli ACC. Mutation of residues in RePC equivalent to those important for substrate binding and catalysis in E. coli BC [154] confirms their importance for PC catalysis as well [270].
The activator CoA is bound at the interface of the BC and PT domains of one monomer and the BC domain of another monomer (Fig. 9g) [20, 266]. A change in the organization of the dimer of the two BC domains is observed upon CoA binding [266], consistent with the fact that acetyl-CoA primarily activates the BC activity. The phosphate groups of CoA interact with a collection of Arg and Lys residues (Fig. 9h). Especially, Arg541 recognizes the 3′-phosphate group of CoA, which is critical for the activating effect of CoA (Fig. 9i), and the R541C disease-causing mutation abolishes acetyl-CoA activation [21]. In SaPC, CoA binding made the four monomers of the PC tetramer more similar to each other, essentially making the tetramer more symmetric. The symmetrical structure of SaPC is also supported by cryo-EM studies [266, 271]. In contrast, a highly asymmetric tetramer is observed in the RePC structure, and only two CoA molecules (the 3′-phospho-ADP portion) are bound to the tetramer [20]. However, this asymmetry may be an inherent property of RePC and does not depend on CoA binding, as the structure without CoA shows similar asymmetry [265].
In the SaPC structure, the other three biotin groups are located in another binding site, at the interface between the PT and CT domains of the other monomer in the same layer, ~20 Å from the CT active site (Fig. 9a) [21]. This exo site does not exist in HsPC due to sequence and structural differences. The functional importance of this binding site is currently unknown.
Urea carboxylase (UC)
Biological functions of UC
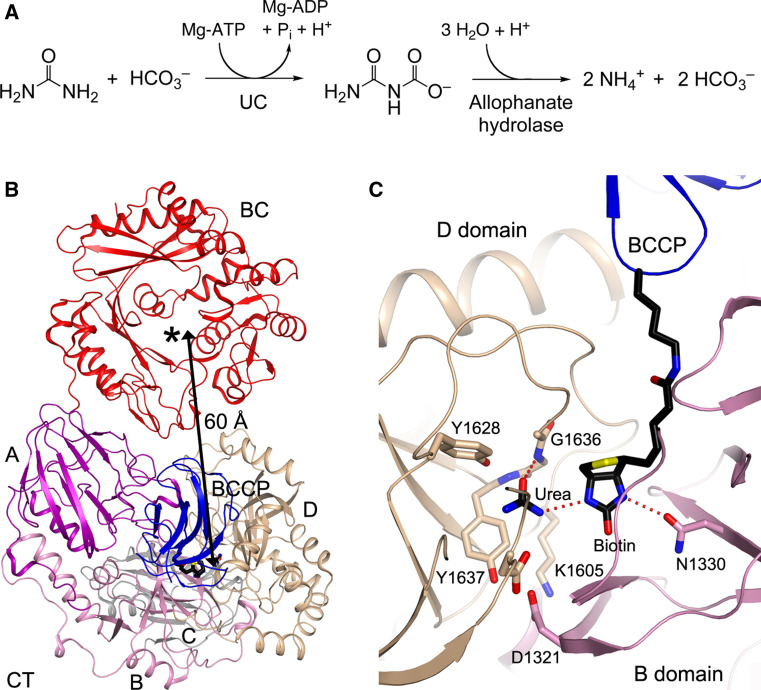
UC catalyzes the carboxylation of urea to produce allophanate, which is then hydrolyzed to generate ammonia and CO2 (Fig. 10a) [15, 16, 272]. Therefore, urea amidolyase (UA, family 4.1) can catalyze both steps of this conversion, while a separate allophanate hydrolase activity is needed for UC (families 4.2 and 4.3).
Fig. 10.
Structure of the urea carboxylase (UC) domain of K. lactis urea amidolyase. a The chemical reaction catalyzed by UC, converting urea to allophanate, which is then hydrolyzed to ammonia and bicarbonate. b Crystal structure of K. lactis UC [24]. The domains are labeled, and the active site of BC is indicated with the asterisk. c Interactions of biotin and urea with the CT active site of UC, located at the interface between the B and D domains
UC is found in some bacteria, fungi, and green algae, and its activity is important for the utilization of urea as a nitrogen source in these organisms [15, 16, 272]. A second pathway for urea utilization is catalyzed by the nickel-dependent enzyme urease, which can convert urea to ammonia in a single step. Some of the fungal organisms contain both pathways, although only the UA pathway is present in S. cerevisiae. In fact, the UA form of this enzyme is found primarily in fungal species. In Saccharomyces kluyveri, UA is part of a pyrimidine degradation pathway, converting the urea product of uracil degradation to ammonia and enabling this organism to use uracil as the sole nitrogen source [273]. This is a distinct pathway for uracil catabolism, and it may also be present in other eukaryotes.
In Candida albicans, UA activity is important for the virulence of this important human pathogen. The CO2 product of UA is a signal for hyphal switching [274], while the ammonia product can be secreted to alkalinize the extracellular environment, which also induces the yeast-hyphal transition. The hyphal form is important for C. albicans to escape phagocytosis by macrophages, thereby evading the host immune system [275].
Structure of UC
The structure of the UC component of K. lactis urea amidolyase was reported recently (Fig. 10b) [24]. In contrast to the other biotin-dependent carboxylases, the UC component is catalytically active as a monomer. The CT domain of UC consists of four sub-domains, named A, B, C and D (Fig. 10b), with many structural and sequence homologs. Especially, domains A and B together are similar to the KipA subunit, and domains C and D the KipI subunit of the B. subtilis KipA-KipI complex [276, 277]. KipI binds and inhibits the autophosphorylation of the histidine kinase KinA, thereby inhibiting sporulation. On the other hand, KipA can sequester KipI and prevent it from inhibiting KinA activation. CT and KipA-KipI have ~20–35 % sequence identity, but residues that are important for CT catalysis are not conserved in KipA and KipI. Therefore, the KipA-KipI complex is unlikely to have CT activity. In addition, both domains B and D have the cyclophilin fold, although they do not share any recognizable sequence homology with cyclophilin.
The active site of the CT domain is located in a cleft at the interface between the B and D domains (Fig. 10c) [24]. The biotin head group is fully extended away from the BCCP domain and interacts with highly conserved residues in the active site. The urea substrate is recognized by hydrogen-bonding interactions with the active site. Especially, one of the urea nitrogen atoms is directly hydrogen-bonded to the N1′ atom of biotin. Mutations of residues in the active site region have detrimental effects on the catalysis by UC.
The overall structure of the BC domain of UC is similar to that of other BC components. However, the BC domain is monomeric in UC, as the interface that mediates the dimerization of the BC subunit of bacterial ACC is not conserved in UC. The BC domain interacts with domains A and D of CT in the UC holoenzyme. The distance between the BC and CT active sites in UC is ~60 Å (Fig. 10b), indicating that the BCCP domain must translocate during catalysis, consistent with the swinging-domain model (Fig. 1).
Common features in the catalysis by biotin-dependent carboxylases
The BC component has strong sequence conservation among the biotin-dependent carboxylases. Insights into the BC reaction are obtained from the structure of the E. coli BC subunit in complex with substrates [154], which indicates the crucial importance of two residues, Glu296 as a general base to extract the proton from bicarbonate and Arg338 to stabilize the biotin enolate oxyanion. The hydrolysis of ATP, via the formation of a carboxyphosphate intermediate, serves at least two functions: (1) the dehydration of bicarbonate to CO2, which is a stronger electrophile; (2) the production of orthophosphate (PO4 3−), which is a strong base that is capable of extracting the proton on the N1′ atom of biotin [17]. This mechanism is likely shared among all biotin-dependent carboxylases, and Glu296 and Arg338 of E. coli BC are conserved among these enzymes.
The CT components of the acyl-CoA carboxylases show sequence and structural conservation. The acyl-CoA substrate is recognized by a domain/subunit with the crotonase fold, and it may be speculated that this part of the CT component evolved from a primordial crotonase. This fold is capable of binding CoA esters, and it also provides an oxyanion hole, in the form of two main-chain amides, that stabilizes the enolate oxyanion of the acyl group during catalysis. Remarkably, the main-chain carbonyl of one of the oxyanion hole residues recognizes the N6 amino group, and the main-chain amide of the following residue is hydrogen-bonded to the N1 atom of the adenine base of CoA. Biotin is recognized by a separate domain/subunit, but also with the crotonase fold. The equivalent oxyanion hole in this domain/subunit stabilizes the biotin enolate oxyanion during the CT reaction. It might be possible that the biotin-binding domain/subunit of CT arose through gene duplication of a crotonase enzyme and then evolved its specificity toward biotin.
The CT components of PC and UC are entirely different in sequence and structure from those of acyl-CoA carboxylases. However, they share the common feature of providing two oxyanion holes to stabilize the intermediates during the CT reaction. On the other hand, while acyl-CoA carboxylases use primarily (carboxy)biotin for catalysis [17, 169], side chains of conserved amino acids are important for the CT reaction in PC and UC, for example Thr908 in SaPC [21, 266, 268] and Lys1605 in KlUC [24].
Summary and perspective
Biotin-dependent carboxylases are crucial enzymes for metabolism and many other cellular processes. They produce compounds that are important for the biosynthesis of many molecules and/or help degrade potentially toxic metabolic intermediates. ACCs are drug discovery targets against type 2 diabetes, cancer, microbial infections, and other diseases in humans. Three classes of herbicides function by inhibiting the plastid ACC of grasses, demonstrating the feasibility of small-molecule ACC antagonists. On the other hand, deficiencies in PCC, MCC and PC activity are linked to serious diseases in humans, especially infants. Development of agents that can restore the activity of the deficient enzyme, possibly through gene therapy, will be of great benefits to these patients.
Structural information on the holoenzymes of biotin-dependent carboxylases demonstrates the remarkable degree of variability in these enzymes. They share conserved domains, which have similar structures on their own. However, the arrangement of these domains in the holoenzyme monomers and especially the architecture of the holoenzyme oligomers can be dramatically different, for example the comparison between PCC and MCC or between SaPC and RePC. These observations also serve as a cautionary tale for the limitations of homology modeling on such large, multi-domain systems.
The past few years have seen great progress in the studies on biotin-dependent caboxylases, especially with the first structural information on most of the holoenzymes (PCC, MCC, PC, UC). At the same time, many important questions remain to be answered. For example, structural information on the holoenzymes of multi-subunit and multi-domain ACCs is still not available. In addition, while the recognition of BCCP by the CT component has been observed in most of the holoenzyme structures, it is not clear how BCCP binds to the BC component for catalysis. Moreover, the molecular basis for substrate selectivity of the acyl-CoA carboxylases is still poorly understood, and presently one cannot readily determine the substrate preference(s) of a given acyl-CoA carboxylase based on a sequence analysis of its CT component. While substitutions of amino acid side chains are likely to play a major role in determining substrate selectivity (for example the size and/or shape of the acyl-CoA binding pocket), conformational differences in the main chain of residues in the active site may also be important and further structural information is needed to illuminate such differences. Therefore, the coming years should continue to be highly exciting in the studies of biotin-dependent carboxylases.
Acknowledgments
I thank Matthew Callaghan, Chi-Yuan Chou, Daniel Floyd, Christine Huang, Yi Seul Kim, Jiang Li, Svetlana Novoseletskaya, Kianoush Sadre-Bazzaz, Yang Shen, Timothy Tran, Benjamin Tweel, Jia Wei, Song Xiang, Zhiru Yang, Linda Yu, and Hailong Zhang for contributions to this project and for helpful discussions. I apologize for not being able to cite many of the earlier publications due to space limitations. This research was supported in part by a grant from the NIH to LT (DK067238).
Abbreviations
- ACC
Acetyl-CoA carboxylase
- BC
Biotin carboxylase
- BCCP
Biotin carboxyl carrier protein
- BT
BC-CT interaction
- CT
Carboxyltransferase
- GCC
Geranyl-CoA carboxylase
- GCD
Glutaconyl-CoA decarboxylase
- HCS
Holocarboxylase synthase
- MCC
3-methylcrotonyl-CoA carboxylase
- MCG
3-methylcrotonylglycinuria
- PA
Propionic acidemia
- PC
Pyruvate carboxylase
- PCC
Propionyl-CoA carboxylase
- PT
PC tetramerization
- UA
Urea amidolyase
- UC
Urea carboxylase
- YCC
Acyl-CoA carboxylase (generic name)
References
- 1.Wakil SJ, Stoops JK, Joshi VC. Fatty acid synthesis and its regulation. Ann. Rev. Biochem. 1983;52:537–579. doi: 10.1146/annurev.bi.52.070183.002541. [DOI] [PubMed] [Google Scholar]
- 2.Cronan JE, Jr, Waldrop GL. Multi-subunit acetyl-CoA carboxylases. Prog Lipid Res. 2002;41:407–435. doi: 10.1016/S0163-7827(02)00007-3. [DOI] [PubMed] [Google Scholar]
- 3.Tong L. Acetyl-coenzyme A carboxylase: crucial metabolic enzyme and attractive target for drug discovery. Cell Mol Life Sci. 2005;62:1784–1803. doi: 10.1007/s00018-005-5121-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jitrapakdee S, St. Maurice M, Rayment I, Cleland WW, Wallace JC, Attwood PV. Structure, mechanism and regulation of pyruvate carboxylase. Biochem J. 2008;413:369–387. doi: 10.1042/BJ20080709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berg IA, Kockelkorn D, Ramos-Vera WH, Say RF, Zarzycki J, Hugler M, Alber BE, Fuchs G. Autotrophic carbon fixation in archaea. Nat Rev Microbiol. 2010;8:447–460. doi: 10.1038/nrmicro2365. [DOI] [PubMed] [Google Scholar]
- 6.Pratscher J, Dumont MG, Conrad R. Ammonia oxidation coupled to CO2 fixation by archaea and bacteria in an agricultural soil. Proc Natl Acad Sci USA. 2011;108:4170–4175. doi: 10.1073/pnas.1010981108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smejkalova H, Erb TJ, Fuchs G. Methanol assimilation in Methylobacterium extorquens AM1: demonstration of all enzymes and their regulation. PLoS One. 2010;5:e13001. doi: 10.1371/journal.pone.0013001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Erb TJ, Berg IA, Brecht V, Muller M, Fuchs G, Alber BE. Synthesis of C5-dicarboxylic acids from C2-units involving crotonyl-CoA carboxylase/reductase: the ethylmalonyl-CoA pathway. Proc Natl Acad Sci USA. 2007;104:10631–10636. doi: 10.1073/pnas.0702791104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Khomyakova M, Bukmez O, Thomas LK, Erb TJ, Berg IA. A methylaspartate cycle in haloarchaea. Science. 2011;331:334–337. doi: 10.1126/science.1196544. [DOI] [PubMed] [Google Scholar]
- 10.Alber BE. Biotechnological potential of the ethylmalonyl-CoA pathway. Appl Microbiol Biotechnol. 2011;89:17–25. doi: 10.1007/s00253-010-2873-z. [DOI] [PubMed] [Google Scholar]
- 11.Schneider K, Asao M, Carter MS, Alber BE. Rhodobacter sphaeroides uses a reductive route via propionyl coenzyme A to assimilate 3-hydroxypropionate. J Bacteriol. 2012;194:225–232. doi: 10.1128/JB.05959-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gago G, Diacovich L, Arabolaza A, Tsai S-C, Gramajo H. Fatty acid biosynthesis in actinomycetes. FEMS Microbiol Rev. 2011;35:475–497. doi: 10.1111/j.1574-6976.2010.00259.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang H, Boghigian BA, Pfeifer BA. Investigating the role of native propionyl-CoA and methylmalonyl-CoA metabolism on heterologous polyketide production in Escherichia coli . Biotechnol Bioeng. 2010;105:567–573. doi: 10.1002/bit.22560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Forster-Fromme K, Jendrossek D. Catabolism of citronellol and related acyclic terpenoids in pseudomonads . Appl Microbiol Biotechnol. 2010;87:859–869. doi: 10.1007/s00253-010-2644-x. [DOI] [PubMed] [Google Scholar]
- 15.Navarathna DHMLP, Harris SD, Roberts DD, Nickerson KW. Evolutionary aspects of urea utilization by fungi. FEMS Yeast Res. 2010;10:209–213. doi: 10.1111/j.1567-1364.2009.00602.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Strope PK, Nickerson KW, Harris SD, Moriyama EN. Molecular evolution of urea amidolyase and urea carboxylase in fungi. BMC Evol Biol. 2011;11:80. doi: 10.1186/1471-2148-11-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Knowles JR. The mechanism of biotin-dependent enzymes. Ann Rev Biochem. 1989;58:195–221. doi: 10.1146/annurev.bi.58.070189.001211. [DOI] [PubMed] [Google Scholar]
- 18.Attwood PV, Wallace JC. Chemical and catalytic mechanisms of carboxyl transfer reactions in biotin-dependent enzymes. Acc Chem Res. 2002;35:113–120. doi: 10.1021/ar000049+. [DOI] [PubMed] [Google Scholar]
- 19.Perham RN. Swinging arms and swinging domains in multifunctional enzymes: catalytic machines for multistep reactions. Ann Rev Biochem. 2000;69:961–1004. doi: 10.1146/annurev.biochem.69.1.961. [DOI] [PubMed] [Google Scholar]
- 20.St. Maurice M, Reinhardt L, Surinya KH, Attwood PV, Wallace JC, Cleland WW, Rayment I. Domain architecture of pyruvate carboxylase, a biotin-dependent multifunctional enzyme. Science. 2007;317:1076–1079. doi: 10.1126/science.1144504. [DOI] [PubMed] [Google Scholar]
- 21.Xiang S, Tong L. Crystal structures of human and Staphylococcus aureus pyruvate carboxylase and molecular insights into the carboxyltransfer reaction. Nat Struct Mol Biol. 2008;15:295–302. doi: 10.1038/nsmb.1393. [DOI] [PubMed] [Google Scholar]
- 22.Huang CS, Sadre-Bazzaz K, Shen Y, Deng B, Zhou ZH, Tong L. Crystal structure of the a6b6 holoenzyme of propionyl-coenzyme A carboxylase. Nature. 2010;466:1001–1005. doi: 10.1038/nature09302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang CS, Ge P, Zhou ZH, Tong L. An unanticipated architecture of the 750-kDa a6b6 holoezyme of 3-methylcrotonyl-CoA carboxylase. Nature. 2012;481:219–223. doi: 10.1038/nature10691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fan C, Chou C-Y, Tong L, Xiang S. Crystal structure of urea carboxylase provides insights into the carboxyltransfer reaction. J Biol Chem. 2012;287:9389–9398. doi: 10.1074/jbc.M111.319475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Benning MM, Haller T, Gerlt JA, Holden HM. New reactions in the crotonase superfamily: structure of methylmalonyl CoA decarboxylase from Escherichia coli . Biochem. 2000;39:4630–4639. doi: 10.1021/bi9928896. [DOI] [PubMed] [Google Scholar]
- 26.Dimroth P, Jockel P, Schmid M. Coupling mechanism of the oxaloacetate decarboxylase Na+ pump. Biochim Biophys Acta. 2001;1505:1–14. doi: 10.1016/S0005-2728(00)00272-3. [DOI] [PubMed] [Google Scholar]
- 27.Wendt KS, Schall I, Huber R, Buckel W, Jacob U. Crystal structure of the carboxyltransferase subunit of the bacterial sodium ion pump glutaconyl-coenzyme A decarboxylase. EMBO J. 2003;22:3493–3502. doi: 10.1093/emboj/cdg358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boiangiu CD, Jayamani E, Brugel D, Herrmann G, Kim J, Forzi L, Hedderich R, Vgenopoulou I, Pierik AJ, Steuber J, Buckel W. Sodium ion pumps and hydrogen production in glutamate fermenting anaerobic bacteria. J Mol Microbiol Biotechnol. 2005;10:105–119. doi: 10.1159/000091558. [DOI] [PubMed] [Google Scholar]
- 29.Studer R, Dahinden P, Wang W-W, Auchli Y, Li X-D, Dimroth P. Crystal structure of the carboxyltransferase domain of the oxaloacetate decarboxylase Na+ pump from Vibrio cholerae. J Mol Biol. 2007;367:547–557. doi: 10.1016/j.jmb.2006.12.035. [DOI] [PubMed] [Google Scholar]
- 30.Kress D, Brugel D, Schall I, Linder D, Buckel W, Essen L-O. An asymmetric model for Na+-translocating glutaconyl-CoA decarboxylase. J Biol Chem. 2009;284:28401–28409. doi: 10.1074/jbc.M109.037762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hall PR, Wang Y-F, Rivera-Hainaj RE, Zheng X, Pustai-Carey M, Carey PR, Yee VC. Transcarboxylase 12S crystal structure: hexamer assembly and substrate binding to a multienzyme core. EMBO J. 2003;22:2334–2347. doi: 10.1093/emboj/cdg244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hall PR, Zheng R, Antony L, Pustai-Carey M, Carey PR, Yee VC. Transcarboxylase 5S structures: assembly and catalytic mechanism of a multienzyme complex subunit. EMBO J. 2004;23:3621–3631. doi: 10.1038/sj.emboj.7600373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carey PR, Sonnichsen FD, Yee VC. Transcarboxylase: one of nature’s early nanomachines. IUBMB Life. 2004;56:575–583. doi: 10.1080/15216540400022417. [DOI] [PubMed] [Google Scholar]
- 34.Hugler M, Krieger RS, Jahn M, Fuchs G. Characterization of acetyl-CoA/propionyl-CoA carboxylase in Metallosphaera sedula. Eur J Biochem. 2003;270:736–744. doi: 10.1046/j.1432-1033.2003.03434.x. [DOI] [PubMed] [Google Scholar]
- 35.Jordan IK, Henze K, Fedorova ND, Koonin EV, Galperin MY. Phylogenomic analysis of the Giardia intestinalis transcarboxylase reveals multiple instances of domain fusion and fission in the evolution of biotin-dependent enzymes. J Mol Microbiol Biotechnol. 2003;5:172–189. doi: 10.1159/000070268. [DOI] [PubMed] [Google Scholar]
- 36.Lombard J, Moreira D. Early evolution of the biotin-dependent carboxylase family. BMC Evol Biol. 2011;11:232. doi: 10.1186/1471-2148-11-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saggerson D. Malonyl-CoA, a key signaling molecule in mammalian cells. Annu Rev Nutr. 2008;28:253–272. doi: 10.1146/annurev.nutr.28.061807.155434. [DOI] [PubMed] [Google Scholar]
- 38.Hoja U, Marthol S, Hofmann J, Stegner S, Schulz R, Meier S, Greiner E, Schweizer E. HFA1 encoding an organelle-specific acetyl-CoA carboxylase controls mitochondrial fatty acid synthesis in Saccharomyces cerevisiae. J Biol Chem. 2004;279:21779–21786. doi: 10.1074/jbc.M401071200. [DOI] [PubMed] [Google Scholar]
- 39.Tehlivets O, Scheuringer K, Kohlwein SD. Fatty acid synthesis and elongation in yeast. Biochim Biophys Acta. 2007;1771:255–270. doi: 10.1016/j.bbalip.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 40.Hiltunen JK, Chen Z, Haapalainen AM, Wierenga RK, Kastaniotis AJ. Mitochondrial fatty acid synthesis—an adopted set of enzymes making a pathway of major importance for the cellular metabolism. Prog Lipid Res. 2010;49:27–45. doi: 10.1016/j.plipres.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 41.Witkowski A, Thweatt J, Smith S. Mammalian ACSF3 protein is a malonyl-CoA synthetase that supplies the chain extender units for mitochondrial fatty acid synthesis. J Biol Chem. 2011;286:33729–33736. doi: 10.1074/jbc.M111.291591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen H, Kim HU, Weng H, Browse J. Malonyl-CoA synthetase, encoded by ACYL ACTIVATING ENZYME 13, is essential for growth and development of Arabidopsis . Plant Cell. 2011;23:2247–2262. doi: 10.1105/tpc.111.086140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Abu-Elheiga L, Brinkley WR, Zhong L, Chirala SS, Woldegiorgis G, Wakil SJ. The subcellular localization of acetyl-CoA carboxylase 2. Proc Natl Acad Sci USA. 2000;97:1444–1449. doi: 10.1073/pnas.97.4.1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McGarry JD, Brown NF. The mitochondrial carnitine palmitoyl transferase system: from concept to molecular analysis. Eur J Biochem. 1997;244:1–14. doi: 10.1111/j.1432-1033.1997.00001.x. [DOI] [PubMed] [Google Scholar]
- 45.Ramsay RR, Gandour RD, van der Leij FR. Molecular enzymology of carnitine transfer and transport. Biochim Biophys Acta. 2001;1546:21–43. doi: 10.1016/S0167-4838(01)00147-9. [DOI] [PubMed] [Google Scholar]
- 46.Kreuz S, Schoelch C, Thomas L, Rist W, Rippmann JF, Neubauer H. Acetyl-CoA carboxylases 1 and 2 show distinct expression patterns in rats and humans and alterations in obesity and diabetes. Diabetes Metab Res Rev. 2009;25:577–586. doi: 10.1002/dmrr.997. [DOI] [PubMed] [Google Scholar]
- 47.Castle JC, Hara Y, Raymond CK, Garrett-Engele P, Ohwaki K, Kan Z, Kusunoki J, Johnson JM. ACC2 is expressed at high levels human white adipose and has an isoform with a novel N-terminus. PLoS One. 2009;4:e4369. doi: 10.1371/journal.pone.0004369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Diaz FJ, Meary A, Arranz MJ, Ruano G, Windermuth A, de Leon J. Acetyl-coenzyme A carboxylase alpha gene variations may be associated with the direct effects of some antipsychotics on triglyceride levels. Schizophr Res. 2009;115:136–140. doi: 10.1016/j.schres.2009.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gallardo D, Quintanilla R, Varona L, Diaz I, Ramirez O, Pena RN, Amills M. Polymorphism of the pig acetyl-coenzyme A carboxylase alpha gene is associated with fatty acid composition in a Duroc commercial line. Anim Genet. 2009;40:410–417. doi: 10.1111/j.1365-2052.2009.01854.x. [DOI] [PubMed] [Google Scholar]
- 50.Tian J, Wang S, Wang Q, Leng L, Hu X, Li H. A single nucleotide polymorphism of chicken acetyl-CoA carboxylase A gene associated with fatness traits. Anim Biotechnol. 2010;21:42–50. doi: 10.1080/10495390903347009. [DOI] [PubMed] [Google Scholar]
- 51.Zhang S, Knight TJ, Reecy JM, Wheeler TL, Shackelford SD, Cundiff LV, Beitz DC. Associations of polymorphisms in the promoter I of bovine acetyl-CoA carboxylase-alpha gene with beef fatty acid composition. Anim Genet. 2009;41:417–420. doi: 10.1111/j.1365-2052.2009.02006.x. [DOI] [PubMed] [Google Scholar]
- 52.Federica S, Francesco N, Giovanna DM, Carmela SM, Gennaro C, Carmela T, Bianca M. Identification of novel single nucleotide polymorphisms in promoter III of the acetyl-CoA carboxylase-alpha gene in goats affecting milk production traits. J Heredity. 2009;100:386–389. doi: 10.1093/jhered/esn098. [DOI] [PubMed] [Google Scholar]
- 53.Brownsey RW, Boone AN, Elliot JE, Kulpa JE, Lee WM. Regulation of acetyl-CoA carboxylase. Biochem Soc Trans. 2006;34:223–227. doi: 10.1042/BST20060223. [DOI] [PubMed] [Google Scholar]
- 54.Kaushik VK, Kavana M, Volz JM, Weldon SC, Hanrahan S, Xu J, Caplan SL, Hubbard BK. Characterization of recombinant human acetyl-CoA carboxylase-2 steady-state kinetics. Biochim Biophys Acta. 2009;1794:961–967. doi: 10.1016/j.bbapap.2009.02.004. [DOI] [PubMed] [Google Scholar]
- 55.Locke GA, Cheng D, Witmer MR, Tamura JK, Haque T, Carney RF, Rendina AR, Marcinkeviciene J. Differential activation of recombinant human acetyl-CoA carboxylases 1 and 2 by citrate. Arch Biochem Biophys. 2008;475:72–79. doi: 10.1016/j.abb.2008.04.011. [DOI] [PubMed] [Google Scholar]
- 56.Kim CW, Moon Y-A, Park SW, Cheng D, Kwon HJ, Horton JD. Induced polymerization of mammalian acetyl-CoA carboxylase by MIG12 provides a tertiary level of regulation of fatty acid biosynthesis. Proc Natl Acad Sci USA. 2010;107:9626–9631. doi: 10.1073/pnas.1001292107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Colbert CL, Kim CW, Moon Y-A, Henry L, Palnitkar M, McKean WB, Fitzgerald K, Deisenhofer J, Horton JD, Kwon HJ. Crystal structure of Spot 14, a modulator of fatty acid synthesis. Proc Natl Acad Sci USA. 2010;107:18820–18825. doi: 10.1073/pnas.1012736107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ray H, Moreau K, Dizin E, Callebaut I, Venezia ND. ACCA phosphopeptide recognition by the BRCT repeats of BRCA1. J Mol Biol. 2006;359:973–982. doi: 10.1016/j.jmb.2006.04.010. [DOI] [PubMed] [Google Scholar]
- 59.Ray H, Suau F, Vincent A, Venezia ND. Cell cycle regulation of the BRCA1/acetyl-CoA-carboxylase complex. Biochem Biophys Res Commun. 2009;378:615–619. doi: 10.1016/j.bbrc.2008.11.090. [DOI] [PubMed] [Google Scholar]
- 60.Shen Y, Tong L. Structural evidence for direct interactions between the BRCT domains of human BRCA1 and a phospho-peptide from human ACC1. Biochemical. 2008;47:5767–5773. doi: 10.1021/bi800314m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nikolau BJ, Ohlrogge JB, Wurtele ES. Plant biotin-containing carboxylases. Arch Biochem Biophys. 2003;414:211–222. doi: 10.1016/S0003-9861(03)00156-5. [DOI] [PubMed] [Google Scholar]
- 62.Chalupska D, Lee HY, Faris JD, Evrard A, Chalhoub B, Haselkorn R, Gornicki P. Acc homoeoloci and the evolution of wheat genomes. Proc Natl Acad Sci USA. 2008;105:9691–9696. doi: 10.1073/pnas.0803981105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lu S, Zhao H, Parsons EP, Xu C, Kosma DK, Xu X, Chao D, Lohrey G, Bangarusamy DK, Wang G, Bressan RA, Jenks MA. The glossyhead1 allele of ACC1 reveals a principal role for multidomain acetyl-coenzyme A carboxylase in the biosynthesis of cuticular waxes by Arabidopsis . Plant Physiol. 2011;157:1079–1092. doi: 10.1104/pp.111.185132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li ZG, Yin WB, Guo H, Song LY, Chen YH, Guan RZ, Wang JQ, Wang RRC, Hu ZM. Genes encoding the alpha-carboxyltransferase subunit of acetyl-CoA carboxylase from Brassica napus and parental species: cloning, expression patterns, and evolution. Genome. 2010;53:360–370. doi: 10.1139/G10-011. [DOI] [PubMed] [Google Scholar]
- 65.Li ZG, Yin WB, Song LY, Chen YH, Guan RZ, Wang JQ, Wang RRC, Hu ZM. Genes encoding the biotin carboxylase subunit of acetyl-CoA carboxylase from Brassica napus and parental species: cloning, expression patterns, and evolution. Genome. 2011;54:202–211. doi: 10.1139/G10-110. [DOI] [PubMed] [Google Scholar]
- 66.Bourrellier ABF, Valot B, Guillot A, Ambard-Bretteville F, Vidal J, Hodges M. Chloroplast acetyl-CoA carboxylase activity is 2-oxoglutarate-regulated by interaction of PII with the biotin carboxyl carrier subunit. Proc Natl Acad Sci USA. 2010;107:502–507. doi: 10.1073/pnas.0910097107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li X, Ilarslan H, Brachova L, Qian HR, Li L, Che P, Wurtele ES, Nikolau BJ. Reverse-genetic analysis of the two biotin-containing subunit genes of the heteromeric acetyl-coenzyme A carboxylase in Arabidopsis indicates a unidirectional functional redundancy. Plant Physiol. 2011;155:293–314. doi: 10.1104/pp.110.165910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Olinares PDB, Ponnala L, van Wijk KJ. Megadalton complexes in the chroloplast stroma of Arabidopsis thaliana characterized by size exclusion chromatography, mass spectrometry, and hierarchical clustering. Mol Cell Proteomics. 2010;9:1594–1615. doi: 10.1074/mcp.M000038-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gornicki P. Apicoplast fatty acid biosynthesis as a target for medical intervention in apicoplexan parasites. Int J Parasitol. 2003;33:885–896. doi: 10.1016/S0020-7519(03)00133-4. [DOI] [PubMed] [Google Scholar]
- 70.Vigueira PA, Paul KS. Requirement for acetyl-CoA carboxylase in Trypanosoma brucei is dependent upon the growth environment. Mol Microbiol. 2011;80:117–132. doi: 10.1111/j.1365-2958.2011.07563.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Alabaster A, Isoe J, Zhou G, Lee A, Murphy A, Day WA, Miesfeld RL. Deficiencies in acetyl-CoA carboxylase and fatty acid synthase 1 differentially affect eggshell formation and blood meal digestion in Aedes aegypti . Insect Biochem Mol Biol. 2011;41:946–955. doi: 10.1016/j.ibmb.2011.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Spencer CM, Schafer XL, Moorman NJ, Munger J. Human cytomegalovirus induces the activity and expression of acetyl-coenzyme A carboxylase, a fatty acid biosynthetic enzyme whose inhibition attenuates viral replication. J Virol. 2011;85:5814–5824. doi: 10.1128/JVI.02630-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Campbell JW, Cronan JE., Jr Bacterial fatty acid biosynthesis: targets for antibacterial drug discovery. Ann Rev Microbiol. 2001;55:305–332. doi: 10.1146/annurev.micro.55.1.305. [DOI] [PubMed] [Google Scholar]
- 74.Tong L, Harwood HJ., Jr Acetyl-coenzyme A carboxylases: versatile targets for drug discovery. J Cell Biochem. 2006;99:1476–1488. doi: 10.1002/jcb.21077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wakil SJ, Abu-Elheiga LA. Fatty acid metabolism: target for metabolic syndrome. J Lipid Res. 2009;50:S138–S143. doi: 10.1194/jlr.R800079-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schreurs M, Kuipers F, van der Leij FR. Regulatory enzymes of mitochondrial beta-oxidation as targets for treatment of the metabolic syndrome. Obes Rev. 2010;11:380–388. doi: 10.1111/j.1467-789X.2009.00642.x. [DOI] [PubMed] [Google Scholar]
- 77.Lenhard JM. Lipogenic enzymes as therapeutic targets for obesity and diabetes. Curr Pharm Des. 2011;17:325–331. doi: 10.2174/138161211795164185. [DOI] [PubMed] [Google Scholar]
- 78.Harwood HJ., Jr Acetyl-CoA carboxylase inhibition for the treatment of metabolic syndrome. Curr Opin Investig Drugs. 2004;5:283–289. [PubMed] [Google Scholar]
- 79.Grundy SM. Drug therapy of the metabolic syndrome: minimizing the emerging crisis in polypharmacy. Nat Rev Drug Discov. 2006;5:295–309. doi: 10.1038/nrd2005. [DOI] [PubMed] [Google Scholar]
- 80.Reaven GM. Insulin resistance: the link between obesity and cardiovascular disease. Med Clin North Amer. 2011;95:875–892. doi: 10.1016/j.mcna.2011.06.002. [DOI] [PubMed] [Google Scholar]
- 81.Wang YC, McPherson K, Marsh T, Gortmaker SI, Brown M. Health and economic burden of the projected obesity trends in the USA and the UK. Lancet. 2011;378:815–825. doi: 10.1016/S0140-6736(11)60814-3. [DOI] [PubMed] [Google Scholar]
- 82.Abu-Elheiga L, Matzuk MM, Abo-Hashema KAH, Wakil SJ. Continuous fatty acid oxidation and reduced fat storage in mice lacking acetyl-CoA carboxylase 2. Science. 2001;291:2613–2616. doi: 10.1126/science.1056843. [DOI] [PubMed] [Google Scholar]
- 83.Abu-Elheiga L, Oh W, Kordari P, Wakil SJ. Acetyl-CoA carboxylase 2 mutant mice are protected against obesity and diabetes induced by high-fat/high-carbohydrate diets. Proc Natl Acad Sci USA. 2003;100:10207–10212. doi: 10.1073/pnas.1733877100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Choi CS, Savage DB, Abu-Elheiga L, Liu Z-X, Kim S, Kulkarni A, Distefano A, Hwang Y-J, Reznick RM, Codella R, Zhang D, Cline GW, Wakil SJ, Shulman GI. Continuous fat oxidation in acetyl-CoA carboxylase 2 knockout mice increases total energy expenditure, reduces fat mass, and improves insulin sensitivity. Proc Natl Acad Sci USA. 2007;104:16480–16485. doi: 10.1073/pnas.0706794104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Essop MF, Camp HS, Choi CS, Sharma S, Fryer RM, Reinhart GA, Guthrie PH, Bentebibel A, Gu Z, Shulman GI, Taegtmeyer H, Wakil SJ, Abu-Elheiga L. Reduced heart size and increased myocardial fuel substrate oxidation in ACC2 mutant mice. Am J Physiol Heart Cir Physiol. 2008;295:H256–H265. doi: 10.1152/ajpheart.91489.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Abu-Elheiga L, Wu H, Gu Z, Bressler R, Wakil SJ. Acetyl-CoA carboxylase 2−/− protects mutant mice against fatty liver under high-fat, high-carbohydrate dietary and de novo lipogenic conditions. J Biol Chem. 2012;287:12578–12588. doi: 10.1074/jbc.M111.309559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lane MD, Wolfgang M, Cha SH, Dai Y. Regulation of food intake and energy expenditure by hypothalamic malonyl-CoA. Int J Obesity. 2008;32:S49–S54. doi: 10.1038/ijo.2008.123. [DOI] [PubMed] [Google Scholar]
- 88.Fantino M. Role of lipids in the control of food intake. Curr Opin Clin Nutr Metab Care. 2011;14:138–144. doi: 10.1097/MCO.0b013e3283437b78. [DOI] [PubMed] [Google Scholar]
- 89.Abu-Elheiga L, Matzuk MM, Kordari P, Oh W, Shaikenov T, Gu Z, Wakil SJ. Mutant mice lacking acetyl-CoA carboxylase 1 are embryonically lethal. Proc Natl Acad Sci USA. 2005;102:12011–12016. doi: 10.1073/pnas.0505714102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mao J, Demayo FJ, Li H, Abu-Elheiga L, Gu Z, Shaikenov T, Kordari P, Chirala SS, Heird WC, Wakil SJ. Liver-specific deletion of acetyl-CoA carboxylase 1 reduces hepatic triglyceride accumulation without affecting glucose homeostasis. Proc Natl Acad Sci USA. 2006;103:8552–8557. doi: 10.1073/pnas.0603115103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mao J, Yang T, Gu Z, Heird WC, Finegold MJ, Lee B, Wakil SJ. aP2-Cre-mediated inactivation of acetyl-CoA carboxylase 1 causes growth retardation and reduced lipid accumulation in adipose tissues. Proc Natl Acad Sci USA. 2009;106:17576–17581. doi: 10.1073/pnas.0909055106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Savage DB, Choi CS, Samuel VT, Liu ZX, Zhang D, Wang A, Zhang XM, Cline GW, Yu XX, Geisler JG, Bhanot S, Monia BP, Shulman GI. Reversal of diet-induced hepatic steatosis and hepatic insulin resistance by antisense oligonucleotide inhibitors of acetyl-CoA carboxylases 1 and 2. J Clin Investig. 2006;116:817–824. doi: 10.1172/JCI27300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Schreurs M, van Dijk TH, Gerding A, Havinga R, Reijngoud DJ, Kuipers F. Soraphen, an inhibitor of the acetyl-CoA carboxylase system, improves peripheral insulin sensitivity in mice fed a high-fat diet. Diabetes Obesity Metab. 2009;11:987–991. doi: 10.1111/j.1463-1326.2009.01078.x. [DOI] [PubMed] [Google Scholar]
- 94.Peterson JM, Aja S, Wei Z, Wong GW. CTRP1 protein enhances fatty acid oxidation via AMP-activated protein kinase (AMPK) activation and acetyl-CoA carboxylase (ACC) inhibition. J Biol Chem. 2012;287:1576–1587. doi: 10.1074/jbc.M111.278333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kobayashi MA, Watada H, Kawamori R, Maeda S. Overexpression of acetyl-coenzyme A carboxylase beta increases proinflammatory cytokines in cultured human renal proximal tubular epithelial cells. Clin Exp Nephrol. 2010;14:315–324. doi: 10.1007/s10157-010-0296-x. [DOI] [PubMed] [Google Scholar]
- 96.Tang SCW, Leung VTM, Chan LYY, Wong SSH, Chu DWS, Leung JCK, Ho YW, Lai KN, Ma L, Elbein SC, Bowden DW, Hicks PJ, Comeau ME, Langefeld CD, Freedman BI. The acetyl-coenzyme A carboxylase beta (ACACB) gene is associated with nephropathy in Chinese patients with type 2 diabetes. Nephrol Dial Transplant. 2010;25:3931–3934. doi: 10.1093/ndt/gfq303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Maeda S, Kobayashi MA, Araki SI, Babazono T, Freedman BI, Bostrom MA, Cooke JN, Toyoda M, Umezono T, Tarnow L, Hansen T, Gaede P, Jorsal A, Ng DPK, Ikeda M, Yanagimoto T, Tsunoda T, Unoki H, Kawai K, Imanishi M, Suzuki D, Shi HD, Park KS, Kashiwagi A, Iwamoto Y, Kaku K, Kawamori R, Parving HH, Bowden DW, Pedersen O, Nakamura Y. A single nucleotide polymorphism within the acetyl-coenzyme A carboxylase beta gene is associated with proteinuria in patients with type 2 diabetes. PLoS Genet. 2010;6:e1000842. doi: 10.1371/journal.pgen.1000842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Riancho JA, Vazquez L, Garcia-Perez MA, Sainz J, Olmos JM, Hernandez JL, Perez-Lopez J, Amado JA, Zarrabeitia MT, Cano A, Rodriguez-Rey JC. Association of ACACB polymorphisms with obesity and diabetes. Mol Gen Metab. 2011;104:670–676. doi: 10.1016/j.ymgme.2011.08.013. [DOI] [PubMed] [Google Scholar]
- 99.Phillips CM, Goumidi L, Bertrais S, Field MR, Cupples LA, Ordovas JM, McMonagle J, Defoort C, Lovegrove JA, Drevon CA, Blaak EE, Kiec-Wilk B, Riserus U, Lopez-Miranda J, McManus R, Hercberg S, Lairon D, Planells R, Roche HM. ACC2 gene polymorphisms, metabolic syndrome, and gene-nutrient interactions with dietary fat. J Lipid Res. 2010;51:3500–3507. doi: 10.1194/jlr.M008474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hoehn KL, Turner N, Swarbrick MM, Wilks D, Preston E, Phua Y, Joshi H, Furler SM, Larance M, Hegarty BD, Leslie SJ, Pickford R, Hoy AJ, Kraegen EW, James DE, Cooney GJ. Acute or chronic upregulation of mitochondrial fatty acid oxidation has no net effect on whole-body energy expenditure or adiposity. Cell Metab. 2010;11:70–76. doi: 10.1016/j.cmet.2009.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Olson DP, Pulinilkunnil T, Cline GW, Shulman GI, Lowell BB. Gene knockout of Acc2 has little effect on body weight, fat mass, or food intake. Proc Natl Acad Sci USA. 2010;107(16):7598–7603. doi: 10.1073/pnas.0913492107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Alkhateeb H, Holloway GP, Bonen A. Skeletal muscle fatty acid oxidation is not directly associated with AMPK or ACC2 phosphorylation. Appl Physiol Nutr Metab. 2011;36:361–367. doi: 10.1139/h11-024. [DOI] [PubMed] [Google Scholar]
- 103.Hoehn KL, Turner N, Cooney GJ, James DE. Phenotypic discrepancies in acetyl-CoA carboxylase 2-deficient mice. J Biol Chem. 2012;287:15801. doi: 10.1074/jbc.O112.356915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Abu-Elheiga L, Wu H, Gu Z, Wakil SJ. Reply to Hoehn et al.: phenotypic discrepancies in acetyl-CoA carboxylase 2-deficient mice. J Biol Chem. 2012;287:15802. doi: 10.1074/jbc.O112.362939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Harwood HJ, Jr, Petras SF, Shelly LD, Zaccaro LM, Perry DA, Makowski MR, Hargrove DM, Martin KA, Tracey WR, Chapman JG, Magee WP, Dalvie DK, Soliman VF, Martin WH, Mularski CJ, Eisenbeis SA. Isozyme-nonselective N-substituted bipiperidylcarboxamide acetyl-CoA carboxylase inhibitors reduce tissue malonyl-CoA concentrations, inhibit fatty acid synthesis, and increase fatty acid oxidation in cultured cells and in experimental animals. J Biol Chem. 2003;278:37099–37111. doi: 10.1074/jbc.M304481200. [DOI] [PubMed] [Google Scholar]
- 106.Corbett JW. Review of recent acetyl-CoA carboxylase inhibitor patents: mid-2007-2008. Expert Opin Ther Pat. 2009;19:943–956. doi: 10.1517/13543770902862180. [DOI] [PubMed] [Google Scholar]
- 107.Jonas M, LaMarr WA, Ozbal C. Mass spectrometry in high-throughput screening: a case study on acetyl-coenzyme A carboxylase using RapidFire-mass spectrometry (RF-MS) Comb Chem High Throughput Screen. 2009;12:752–759. doi: 10.2174/138620709789104924. [DOI] [PubMed] [Google Scholar]
- 108.Corbett JW, Freeman-Cook KD, Elliott R, Vajdos F, Rajamohan F, Kohls D, Marr E, Zhang H, Tong L, Tu M, Murdande S, Doran SD, Houser JA, Song W, Jones CJ, Coffey SB, Buzon L, Minich ML, Dirico KJ, Tapley S, McPherson RK, Sugarman E, Harwood HJ, Jr, Esler W. Discovery of small molecule isozyme non-specific inhibitors of mammalian acetyl-CoA carboxylase 1 and 2. Bioorg Med Chem Lett. 2010;20:2383–2388. doi: 10.1016/j.bmcl.2009.04.091. [DOI] [PubMed] [Google Scholar]
- 109.Keil S, Muller M, Zoller G, Haschke G, Schroeter K, Glien M, Ruf S, Focken I, Herling AW, Schmoll D. Identification and synthesis of novel inhibitors of acetyl-CoA carboxylase with in vitro and in vivo efficacy on fat oxidation. J Med Chem. 2010;53:8679–8687. doi: 10.1021/jm101179e. [DOI] [PubMed] [Google Scholar]
- 110.Marjanovic J, Chalupska D, Patenode C, Coster A, Arnold E, Ye A, Anesi G, Lu Y, Okun I, Tkachenko S, Haselkorn R, Gornicki P. Recombinant yeast screen for new inhibitors of human acetyl-CoA carboxylase 2 identifies potential drugs to treat obesity. Proc Natl Acad Sci USA. 2010;107:9093–9098. doi: 10.1073/pnas.1003721107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Abramson HN. The lipogenesis pathway as a cancer target. J Med Chem. 2011;54:5615–5638. doi: 10.1021/jm2005805. [DOI] [PubMed] [Google Scholar]
- 112.Bengtsson C, Blaho S, Saitton DB, Brickmann K, Broddefalk J, Dadidsson O, Drmota T, Folmer R, Hallberg K, Hallen S, Hovland R, Isin E, Johannesson P, Kull B, Larsson LO, Lofgren L, Nilsson KE, Noeske T, Oakes N, Plowright AT, Schnecke V, Stahlberg P, Sorme P, Wan H, Wellner E, Oster L. Design of small molecule inhibitors of acetyl-CoA carboxylase 1 and 2 showing reduction of hepatic malonyl-CoA levels in vivo in obese Zucker rats. Bioorg Med Chem. 2011;19:3039–3053. doi: 10.1016/j.bmc.2011.04.014. [DOI] [PubMed] [Google Scholar]
- 113.Jump DB, Torres-Gonzalez M, Olson LK. Soraphen A, an inhibitor of acetyl CoA carboxylase activity, interferes with fatty acid elongation. Biochem Pharmacol. 2011;81:649–660. doi: 10.1016/j.bcp.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Yamashita T, Kamata M, Endo S, Yamamoto M, Kakegawa K, Watanabe H, Miwa K, Yamano T, Funata M, Sakamoto JI, Tani A, Mol CD, Zou H, Dougan DR, Sang B, Snell G, Fukatsu K. Design, synthesis, and structure-activity relationships of spirolactones bearing 2-ureidobenzothiophene a acetyl-CoA carboxylase inhibitors. Bioorg Med Chem Lett. 2011;21:6314–6318. doi: 10.1016/j.bmcl.2011.08.117. [DOI] [PubMed] [Google Scholar]
- 115.Bagley SW, Southers JA, Cabral S, Rose CR, Bernhardson DJ, Edmonds DJ, Polivkova J, Yang X, Kung DW, Griffith DA, Bader SJ. Synthesis of 7-oxo-dihydrospiro [indazole-5,4′-piperidine] acetyl-CoA carboxylase inhibitors. J Org Chem. 2012;77:1497–1506. doi: 10.1021/jo202377g. [DOI] [PubMed] [Google Scholar]
- 116.Freeman-Cook KD, Amor P, Bader S, Buzon LM, Coffey SB, Corbett JW, Dirico KJ, Doran SD, Elliott RL, Esler W, Guzman-Perez A, Henegar KE, Houser JA, Jones CJ, Limberakis C, Loomis K, McPherson K, Murdande S, Nelson KL, Phillion D, Pierce BS, Song W, Sugarman E, Tapley S, Tu M, Zhao Z. Maximizing lipophilic efficiency: the use of free-Wilson analysis in the design of inhibitors of acetyl-CoA carboxylase. J Med Chem. 2012;55:935–942. doi: 10.1021/jm201503u. [DOI] [PubMed] [Google Scholar]
- 117.Glien M, Haschke G, Schroeter K, Pfenninger A, Zoller G, Keil S, Muller M, Herling AW, Schmoll D. Stimulation of fat oxidation, but no sustained reduction of hepatic lipids by prolonged pharmacological inhibition of acetyl CoA carboxylase. Horm Metab Res. 2011;43:601–606. doi: 10.1055/s-0031-1283138. [DOI] [PubMed] [Google Scholar]
- 118.Ronnebaum SM, Joseph JW, Ilkayeva O, Burgess SC, Lu D, Becker TC, Sherry AD, Newgard CB. Chronic suppression of acetyl-CoA carboxylase 1 in beta-cells impairs insulin secretion via inhibition of glucose rather than lipid metabolism. J Biol Chem. 2008;283:14248–14256. doi: 10.1074/jbc.M800119200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Swinnen JV, Brusselmans K, Verhoeven G. Increased lipogenesis in cancer cells: new players, novel targets. Curr Opin Clin Nutr Metab Care. 2006;9:358–365. doi: 10.1097/01.mco.0000232894.28674.30. [DOI] [PubMed] [Google Scholar]
- 120.Chajes V, Cambot M, Moreau K, Lenoir GM, Joulin V. Acetyl-CoA carboxylase alpha is essential to breast cancer cell survival. Cancer Res. 2006;66:5287–5294. doi: 10.1158/0008-5472.CAN-05-1489. [DOI] [PubMed] [Google Scholar]
- 121.Yoon S, Lee M-Y, Park SW, Moon J-S, Koh YK, Ahn Y-H, Park BW, Kim KS. Up-regulation of acetyl-CoA carboxylase alpha and fatty acid synthase by human epidermal growth factor receptor 2 at the translational level in breast cancer cells. J Biol Chem. 2007;282:26122–26131. doi: 10.1074/jbc.M702854200. [DOI] [PubMed] [Google Scholar]
- 122.Hilvo M, Denkert C, Lehtinen L, Muller B, Brockmoller S, Seppanen-Laakso T, Budczies J, Bucher E, Yetukuri L, Castillo S, Berg E, Nygren H, Sysi-Aho M, Griffin JL, Fiehn O, Loibl S, Richter-Ehrenstein C, Radke C, Hyotylainen T, Kallioniemi O, Iljin K, Oresic M. Novel theranostic opportunities offered by characterization of altered membrane lipid metabolism in breast cancer progression. Mol Cell Pathobiol. 2011;71:3236–3245. doi: 10.1158/0008-5472.CAN-10-3894. [DOI] [PubMed] [Google Scholar]
- 123.Beckers A, Organe S, Timmermans L, Scheys K, Peeters A, Brusselmans K, Verhoeven G, Swinnen JV. Chemical inhibition of acetyl-CoA carboxylase induces growth arrest and cytotoxicity selectively in cancer cells. Cancer Res. 2007;67:8180–8187. doi: 10.1158/0008-5472.CAN-07-0389. [DOI] [PubMed] [Google Scholar]
- 124.Wang C, Rajput S, Watabe K, Liao DF, Cao D. Acetyl-CoA carboxylase-alpha as a novel target for cancer therapy. Front Biosci. 2010;2:15–26. doi: 10.2741/s82. [DOI] [PubMed] [Google Scholar]
- 125.Olsen AM, Eisenberg BL, Kuemmerle NB, Flanagan AJ, Morganelli PM, Lombardo PS, Swinnen JV, Kinlaw WB. Fatty acid synthesis is a therapeutic target in human liposarcoma. Int J Oncology. 2010;36:1309–1314. doi: 10.3892/ijo_00000616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Scott KEN, Wheeler FB, Davis AL, Thomas MJ, Ntambi JM, Seals DF, Kridel SJ. Metabolic regulation of invadopodia and invasion by acetyl-CoA carboxylase 1 and de novo lipogenesis. PLoS One. 2012;7:e29761. doi: 10.1371/journal.pone.0029761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ma J, Yan R, Zu X, Cheng JM, Rao K, Liao DF, Cao D. Aldo-keto reductase family 1 B10 affects fatty acid synthesis by regulating the stability of acetyl-CoA carboxylase-alpha in breast cancer cells. J Biol Chem. 2008;283:3418–3423. doi: 10.1074/jbc.M707650200. [DOI] [PubMed] [Google Scholar]
- 128.Gronwald JW. Lipid biosynthesis inhibitors. Weed Sci. 1991;39:435–449. [Google Scholar]
- 129.Hofer U, Muehlebach M, Hole S, Zoschke A. Pinoxaden—for broad spectrum grass weed management in cereal crops. J Plant Dis Protection. 2006;20:989–995. [Google Scholar]
- 130.Muehlebach M, Boeger M, Cederbaum F, Cornes D, Friedmann AA, Glock J, Niderman T, Stoller A, Wagner T. Aryldiones incorporating a [1,4,5]oxadiazepane ring. Part I: discovery of the novel cereal herbicide pinoxaden. Bioorg Med Chem. 2009;17:4241–4256. doi: 10.1016/j.bmc.2008.12.062. [DOI] [PubMed] [Google Scholar]
- 131.Yang X, Guschina IA, Hurst S, Wood S, Langford M, Hawkes T, Harwood JL. The action of herbicides on fatty acid biosynthesis and elongation in barley and cucumber. Pest Manag Sci. 2010;66:794–800. doi: 10.1002/ps.1944. [DOI] [PubMed] [Google Scholar]
- 132.Devine MD, Shukla A. Altered target sites as a mechanism of herbicide resistance. Crop Protection. 2000;19:881–889. doi: 10.1016/S0261-2194(00)00123-X. [DOI] [Google Scholar]
- 133.Yu Q, Collavo A, Zheng MQ, Owen M, Sattin M, Powles SB. Diversity of acetyl-coenzyme A carboxylase mutations in resistant Lolium populations: evaluation using clethodim. Plant Physiol. 2007;145:547–558. doi: 10.1104/pp.107.105262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Liu W, Harrison DK, Chalupska D, Gornicki P, O’donnell CC, Adkins SW, Haselkorn R, Williams RR. Single-site mutations in the carboxyltransferase domain of plastid acetyl-CoA carboxylase confer resistance to grass-specific herbicides. Proc Natl Acad Sci USA. 2007;104:3627–3632. doi: 10.1073/pnas.0611572104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Delye C, Matejicek A, Michel S. Cross-resistance patterns to ACCase-inhibiting herbicides conferred by mutant ACCase isoforms in Alopecurus myosuroides Huds. (black-grass), re-examined at the recommended herbicide field rate. Pest Manag Sci. 2008;64:1179–1186. doi: 10.1002/ps.1614. [DOI] [PubMed] [Google Scholar]
- 136.Cruz-Hipolito H, Osuna MD, Dominguez-Valenzuela JA, Espinoza N, de Prado R. Mechanism of resistance to ACCase-inhibiting herbicides in wild oat (Avena fatua) from Latin America. J Agric Food Chem. 2011;59:7261–7267. doi: 10.1021/jf201074k. [DOI] [PubMed] [Google Scholar]
- 137.Petit C, Bay G, Pernin F, Delye C. Prevalence of cross- or multiple resistance to the acetyl-coenzyme A carboxylase inhibitors fenoxaprop, clodinafop and pinoxaden in black-grass (Alopecurus myosuroides Huds.) in France. Pest Manag Sci. 2010;66:168–177. doi: 10.1002/ps.1851. [DOI] [PubMed] [Google Scholar]
- 138.Scarabel L, Panozzo S, Varotto S, Sattin M. Allelic variation of the ACCase gene and response to ACCase-inhibiting herbicides in pinoxaden-resistant Lolium spp. Pest Manag Sci. 2011;67:932–941. doi: 10.1002/ps.2133. [DOI] [PubMed] [Google Scholar]
- 139.Kaundun SS. An aspartate to glycine change in the carboxyl transferase domain of acetyl CoA carboxylase and non-target-site mechanism(s) confer resistance to ACCase inhibitor herbicides in a Lolium multiflorum population. Pest Manag Sci. 2010;66:1249–1256. doi: 10.1002/ps.2003. [DOI] [PubMed] [Google Scholar]
- 140.Wang T, Picard JC, Tian X, Darmency H. A herbicide-resistant ACCase 1781 Setaria mutant shows higher fitness than wild type. Heredity. 2010;105:394–400. doi: 10.1038/hdy.2009.183. [DOI] [PubMed] [Google Scholar]
- 141.Louie T, Goodman CD, Holloway GA, McFadden GI, Mollard V, Watson KG. Dimeric cyclohexane-1,3-dione oximes inhibit wheat acetyl-CoA carboxylase and show anti-malarial activity. Bioorg Med Chem Lett. 2010;20:4611–4613. doi: 10.1016/j.bmcl.2010.06.007. [DOI] [PubMed] [Google Scholar]
- 142.Miller JR, Dunham S, Mochalkin I, Banotai C, Bowman M, Buist S, Dunkle B, Hanna D, Harwood HJ, Jr, Huband MD, Karnovsky A, Kuhn M, Limberakis C, Liu JY, Mehrens S, Mueller WT, Narasimhan L, Ogden A, Ohren J, Prasad JVNV, Shelly JA, Skerlos L, Sulavik M, Thomas VH, VanderRoest S, Wang L, Wang Z, Whitton A, Zhu T, Stover CK. A class of selective antibacterials derived from a protein kinase inhibitor pharmacophore. Proc Natl Acad Sci USA. 2009;106:1737–1742. doi: 10.1073/pnas.0811275106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Mochalkin I, Miller JR, Narasimhan L, Thanabal V, Erdman P, Cox PB, Prasad JVNV, Lightle S, Huband MD, Stover CK. Discovery of antibacterial biotin carboxylase inhibitors by virtual screening and fragment-based approaches. ACS Chem Biol. 2009;4:473–483. doi: 10.1021/cb9000102. [DOI] [PubMed] [Google Scholar]
- 144.Cheng CC, Shipps GW, Jr, Yang Z, Sun B, Kawahata N, Soucy KA, Soriano A, Orth P, Xiao L, Mann P, Black T. Discovery and optimization of antibacterial AccC inhibitors. Bioorg Med Chem Lett. 2009;19:6507–6514. doi: 10.1016/j.bmcl.2009.10.057. [DOI] [PubMed] [Google Scholar]
- 145.Waldrop GL. Smaller is better for antibiotic discovery. ACS Chem Biol. 2009;4:397–399. doi: 10.1021/cb900122j. [DOI] [PubMed] [Google Scholar]
- 146.Polyak SW, Abell AD, Wilce MCJ, Zhang L, Booker GW. Structure, function and selective inhibition of bacterial acetyl-coa carboxylase. Appl Microbiol Biotechnol. 2012;93:983–992. doi: 10.1007/s00253-011-3796-z. [DOI] [PubMed] [Google Scholar]
- 147.Liu X, Fortin PD, Walsh CT. Andrimid producers encode an acetyl-CoA carboxyltransferase subunit resistant to the action of the antibiotic. Proc Natl Acad Sci USA. 2008;105:13321–13326. doi: 10.1073/pnas.0806873105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Meades G, Jr, Henken RL, Waldrop GL, Rahman MM, Gilman SD, Kamatou GPP, Viljoen AM, Gibbons S. Constituents of cinnamon inhibit bacterial acetyl CoA carboxylase. Planta Med. 2010;76:1570–1575. doi: 10.1055/s-0030-1249778. [DOI] [PubMed] [Google Scholar]
- 149.Waldrop GL, Rayment I, Holden HM. Three-dimensional structure of the biotin carboxylase subunit of acetyl-CoA carboxylase. Biochem. 1994;33:10249–10256. doi: 10.1021/bi00200a004. [DOI] [PubMed] [Google Scholar]
- 150.Shen Y, Volrath SL, Weatherly SC, Elich TD, Tong L. A mechanism for the potent inhibition of eukaryotic acetyl-coenzyme A carboxylase by soraphen A, a macrocyclic polyketide natural product. Mol Cell. 2004;16:881–891. doi: 10.1016/j.molcel.2004.11.034. [DOI] [PubMed] [Google Scholar]
- 151.Thoden JB, Blanchard CZ, Holden HM, Waldrop GL. Movement of the biotin carboxylase B-domain as a result of ATP binding. J Biol Chem. 2000;275:16183–16190. doi: 10.1074/jbc.275.21.16183. [DOI] [PubMed] [Google Scholar]
- 152.Mochalkin I, Miller JR, Evdokimov A, Lightle S, Yan C, Stover CK, Waldrop GL. Structural evidence for substrate-induced synergism and half-sites reactivity in biotin carboxylase. Prot Sci. 2008;17:1706–1718. doi: 10.1110/ps.035584.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Cho YS, Lee JI, Shin D, Kim HT, Cheon YH, Seo CI, Kim YE, Hyun YL, Lee YS, Sugiyama K, Park SY, Ro S, Cho JM, Lee TG, Heo YS. Crystal structure of the biotin carboxylase domain of human acetyl-CoA carboxylase 2. Proteins. 2008;70:268–272. doi: 10.1002/prot.21611. [DOI] [PubMed] [Google Scholar]
- 154.Chou C-Y, Yu LPC, Tong L. Crystal structure of biotin carboxylase in complex with substrates and implications for its catalytic mechanism. J Biol Chem. 2009;284:11690–11697. doi: 10.1074/jbc.M805783200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Raymer B, Kavana M, Price A, Wang B, Corcoran L, Kulathila R, Groarke J, Mann T. Synthesis and characterization of a BODIPY-labeled derivative of soraphen A that binds to acetyl-CoA carboxylase. Bioorg Med Chem Lett. 2009;19:2804–2807. doi: 10.1016/j.bmcl.2009.03.107. [DOI] [PubMed] [Google Scholar]
- 156.Cho YS, Lee JI, Shin D, Kim HT, Jung HY, Lee TG, Kang LW, Ahn YJ, Cho HS, Heo YS. Molecular mechanism for the regulation of human ACC2 through phosphorylation by AMPK. Biochem Biophys Res Commun. 2010;391:187–192. doi: 10.1016/j.bbrc.2009.11.029. [DOI] [PubMed] [Google Scholar]
- 157.Chou C-Y, Tong L. Structural and biochemical studies on the regulation of biotin carboxylase by substrate inhibition and dimerization. J Biol Chem. 2011;286:24417–24425. doi: 10.1074/jbc.M111.220517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Bordelon T, Lill SON, Waldrop GL. The utility of molecular dynamics simulations for understanding site-directed mutagenesis of gycine residues in biotin carboxylase. Proteins. 2009;74:808–819. doi: 10.1002/prot.22190. [DOI] [PubMed] [Google Scholar]
- 159.Janiyani K, Bordelon T, Waldrop GL, Cronan JE., Jr Function of Escherichia coli biotin carboxylase requires catalytic activity of both subunits of the homodimer. J Biol Chem. 2001;276:29864–29870. doi: 10.1074/jbc.M104102200. [DOI] [PubMed] [Google Scholar]
- 160.de Queiroz MS, Waldrop GL. Modeling and numerical simulation of biotin carboxylase kinetics: implications for half-sites reactivity. J Theoretical Biol. 2007;246:167–175. doi: 10.1016/j.jtbi.2006.12.025. [DOI] [PubMed] [Google Scholar]
- 161.Shen Y, Chou C-Y, Chang G-G, Tong L. Is dimerization required for the catalytic activity of bacterial biotin carboxylase? Mol . Cell. 2006;22:807–818. doi: 10.1016/j.molcel.2006.04.026. [DOI] [PubMed] [Google Scholar]
- 162.Smith AC, Cronan JE. Dimerization of the bacterial biotin carboxylase subunit is required for acetyl coenzyme A carboxylase activity in vivo. J Bacteriol. 2012;194:72–78. doi: 10.1128/JB.06309-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Weatherly SC, Volrath SL, Elich TD. Expression and characterization of recombinant fungal acetyl-CoA carboxylase and isolation of a soraphen-binding domain. Biochem J. 2004;380:105–110. doi: 10.1042/BJ20031960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Yao X, Soden C, Jr, Summers MF, Beckett D. Comparison of the backbone dynamics of the apo- and holo-carboxy-terminal domain of the biotin carboxyl carrier subunit of Escherichia coli acetyl-CoA carboxylase. Prot Sci. 1999;8:307–317. doi: 10.1110/ps.8.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Athappilly FK, Hendrickson WA. Structure of the biotinyl domain of acetyl-coenzyme A carboxylase determined by MAD phasing. Structure. 1995;3:1407–1419. doi: 10.1016/S0969-2126(01)00277-5. [DOI] [PubMed] [Google Scholar]
- 166.Solbiati J, Chapman-Smith A, Cronan JE., Jr Stabilization of the biotinoyl domain of Escherichia coli acetyl-CoA carboxylase by interactions between the attached biotin and the protruding “thumb” structure. J Biol Chem. 2002;277:21604–21609. doi: 10.1074/jbc.M201928200. [DOI] [PubMed] [Google Scholar]
- 167.Lee CK, Cheong HK, Ryu KS, Lee JI, Lee W, Jeon YH, Cheong C. Biotinoyl domain of human acetyl-CoA carboxylase: structural insights into the carboxyl transfer mechanism. Proteins. 2008;72:613–624. doi: 10.1002/prot.21952. [DOI] [PubMed] [Google Scholar]
- 168.Healy S, McDonald MK, Wu X, Yue WW, Kochan G, Oppermann U, Gravel RA. Structural impact of human and Escherichia coli biotin carboxyl carrier proteins on biotin attachment. Biochem. 2010;49:4687–4694. doi: 10.1021/bi901612y. [DOI] [PubMed] [Google Scholar]
- 169.Zhang H, Yang Z, Shen Y, Tong L. Crystal structure of the carboxyltransferase domain of acetyl-coenzyme A carboxylase. Science. 2003;299:2064–2067. doi: 10.1126/science.1081366. [DOI] [PubMed] [Google Scholar]
- 170.Bilder P, Lightle S, Bainbridge G, Ohren J, Finzel B, Sun F, Holley S, Al-Kassim L, Spessard C, Melnick M, Newcomer M, Waldrop GL. The structure of the carboxyltransferase component of acetyl-CoA carboxylase reveals a zinc-binding motif unique to the bacterial enzyme. Biochem. 2006;45:1712–1722. doi: 10.1021/bi0520479. [DOI] [PubMed] [Google Scholar]
- 171.Zhang H, Tweel B, Tong L. Molecular basis for the inhibition of the carboxyltransferase domain of acetyl-coenzyme A carboxylase by haloxyfop and diclofop. Proc Natl Acad Sci USA. 2004;101:5910–5915. doi: 10.1073/pnas.0400891101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Xiang S, Callaghan MM, Watson KG, Tong L. A different mechanism for the inhibition of the carboxyltransferase domain of acetyl-coenzyme A carboxylase by tepraloxydim. Proc Natl Acad Sci USA. 2009;106:20723–20727. doi: 10.1073/pnas.0908431106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Yu LPC, Kim YS, Tong L. Mechanism for the inhibition of the carboxyltransferase domain of acetyl-coenzyme A carboxylase by pinoxaden. Proc Natl Acad Sci USA. 2010;107:22072–22077. doi: 10.1073/pnas.1012039107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Zhang H, Tweel B, Li J, Tong L. Crystal structure of the carboxyltransferase domain of acetyl-coenzyme A carboxylase in complex with CP-640186. Structure. 2004;12:1683–1691. doi: 10.1016/j.str.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 175.Madauss KP, Burkhart WA, Consler TG, Cowan DJ, Gottschalk WK, Miller AB, Short SA, Tran TB, Williams SP. The human ACC2 CT-domain C-terminus is required for full functionality and has a novel twist. Acta Cryst. 2009;D65:449–461. doi: 10.1107/S0907444909008014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Rajamohan F, Marr E, Reyes AR, Landro JA, Anderson MD, Corbett JW, Dirico KJ, Harwood HJ, Jr, Tu M, Vajdos FF. Structure-guided inhibitor design for human acetyl-coenzyme A carboxylase by interspecies active site conversion. J Biol Chem. 2011;286:41510–41519. doi: 10.1074/jbc.M111.275396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Meades G, Jr, Benson BK, Grove A, Waldrop GL. A tale of two functions: enzymatic activity and translational repression by carboxyltransferase. Nucl Acids Res. 2010;38:1217–1227. doi: 10.1093/nar/gkp1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Benson BK, Meades G, Jr, Grove A, Waldrop GL. DNA inhibits catalysis by the carboxyltransferase subunit of acetyl-CoA carboxylase: implications for active site communication. Prot Sci. 2008;17:34–42. doi: 10.1110/ps.073186408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Giorgio AJ, Plaut GWE. The effect of univalent cations on activities catalyzed by bovine-liver propionyl-CoA carboxylase. Biochim Biophys Acta. 1967;139:487–501. doi: 10.1016/0005-2744(67)90052-6. [DOI] [PubMed] [Google Scholar]
- 180.Edwards JB, Keech DB. Activation of pig heart propionyl-CoA carboxylase by potassium ions. Biochim Biophys Acta. 1968;159:167–175. doi: 10.1016/0005-2744(68)90255-6. [DOI] [PubMed] [Google Scholar]
- 181.Kalousek F, Darigo MD, Rosenberg LE. Isolation and characterization of propionyl-CoA carboxylase from normal human liver. Evidence for a protomeric tetramer of nonidentical subunits. J Biol Chem. 1980;255:60–65. [PubMed] [Google Scholar]
- 182.Ugarte M, Perez-Cerda C, Rodriguez-Pombo P, Desviat LR, Perez B, Richard E, Muro S, Campeau E, Ohura T, Gravel RA. Overview of mutations in the PCCA and PCCB genes causing propionic acidemia. Hum Mutat. 1999;14:275–282. doi: 10.1002/(SICI)1098-1004(199910)14:4<275::AID-HUMU1>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 183.Yang X, Sakamoto O, Matsubara Y, Kure S, Suzuki Y, Aoki Y, Yamaguchi S, Takahashi Y, Nishikubo T, Kawaguchi C, Yoshioka A, Kimura T, Hayasaka K, Kohno Y, Iinuma K, Ohura T. Mutation spectrum of the PCCA and PCCB genes in Japanese patients with propionic acidemia. Mol Gen Metab. 2004;81:335–342. doi: 10.1016/j.ymgme.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 184.Desviat LR, Perez B, Perez-Cerda C, Rodriguez-Pombo P, Clavero S, Ugarte M. Propionic acidemia: mutation update and functional and structural effects of the variant alleles. Mol Gen Metab. 2004;83:28–37. doi: 10.1016/j.ymgme.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 185.Desviat LR, Clavero S, Perez-Cerda C, Navarrete R, Ugarte M, Perez B. New splicing mutations in propionic acidemia. J Hum Genet. 2006;51:992–997. doi: 10.1007/s10038-006-0068-3. [DOI] [PubMed] [Google Scholar]
- 186.Desviat LR, Sanchez-Alcudia R, Perez B, Perez-Cerda C, Navarrete R, Vijzelaar R, Ugarte M. High frequency of large genomic deletions in the PCCA gene causing propionic acidemia. Mol Gen Metab. 2009;96:171–176. doi: 10.1016/j.ymgme.2008.12.009. [DOI] [PubMed] [Google Scholar]
- 187.Perez B, Angaroni C, Sanchez-Alcudia R, Merinero B, Perez-Cerda C, Specola N, Rodriguez-Pombo P, Wajner M, de Kremer RD, Cornejo V, Desviat LR, Ugarte M. The molecular landscape of propionic acidemia and methylmalonic aciduria in Latin America. J Inher Metab Dis. 2010;33:S307–S314. doi: 10.1007/s10545-010-9116-4. [DOI] [PubMed] [Google Scholar]
- 188.Scholl-Burgi S, Sass JO, Zschocke J, Karall D. Amino acid metabolism in patients with propionic acidemia. J Inher Metab Dis. 2012;35:65–70. doi: 10.1007/s10545-010-9245-9. [DOI] [PubMed] [Google Scholar]
- 189.Kraus JP, Spector E, Venezia S, Estes P, Chiang PW, Creadon-Swindell G, Mullerleile S, de Silva L, Barth M, Walter M, Walter K, Meissner T, Lindner M, Ensenauer R, Santer R, Bodamer OA, Baumgartner MR, Brunner-Krainz M, Karall D, Haase C, Knerr I, Marquardt T, Hennermann JB, Steinfeld R, Beblo S, Koch H-G, Konstantopoulou V, Scholl-Burgi S, van Teeffelen-Heithoff A, Suormala T, Ugarte M, Sperl W, Superti-Furga A, Schwab KO, Grunert SC, Sass JO. Mutation analysis in 54 propionic acidemia patients. J Inher Metab Dis. 2012;35:51–63. doi: 10.1007/s10545-011-9399-0. [DOI] [PubMed] [Google Scholar]
- 190.Ballhausen D, Mittaz L, Boulat O, Bonafe L, Braissant O. Evidence for catabolic pathway of propionate metabolism in CNS: expression pattern of methylmalonyl-CoA mutase and propionyl-CoA carboxylase alpha-subunit in developing and adult rat brain. Neurosci. 2009;164:578–587. doi: 10.1016/j.neuroscience.2009.08.028. [DOI] [PubMed] [Google Scholar]
- 191.Haberlandt E, Canestrini C, Brunner-Krainz M, Moslinger D, Mussner K, Plecko B, Scholl-Burgi S, Sperl W, Rostasy K, Karall D. Epilepsy in patients with propionic acidemia. Neuropediatrics. 2009;40:120–125. doi: 10.1055/s-0029-1243167. [DOI] [PubMed] [Google Scholar]
- 192.Rigo FK, Pasquetti L, Malfatti CRM, Fighera MR, Coelho RC, Petri CZ, Mello CF. Propionic acid induces convulsions and protein carboxylation in rats. Neurosci Lett. 2006;408:151–154. doi: 10.1016/j.neulet.2006.08.075. [DOI] [PubMed] [Google Scholar]
- 193.Miyazaki T, Ohura T, Kobayashi M, Shigematsu Y, Yamaguchi S, Suzuki Y, Hata I, Aoki Y, Yang X, Minjares C, Haruta I, Uto H, Ito Y, Muller U. Fatal propionic acidemia in mice lacking propionyl-CoA carboxylase and its rescue by postnatal, liver-specific supplementation via a transgene. J Biol Chem. 2001;276:35995–35999. doi: 10.1074/jbc.M105467200. [DOI] [PubMed] [Google Scholar]
- 194.Rincon A, Aguado C, Desviat LR, Sanchez-Alcudia R, Ugarte M, Perez B. Propionic and methylmalonic acidemia: antisense therapeutics for intronic variations causing aberrantly spliced messenger RNA. Am J Hum Genet. 2007;81:1262–1270. doi: 10.1086/522376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Sanchez-Alcudia R, Perez B, Perez-Cerda C, Ugarte M, Desviat LR. Overexpression of adapted U1snRNA in patients’ cells to correct a 5′ splice site mutation in propionic acidemia. Mol Gen Metab. 2011;102:134–138. doi: 10.1016/j.ymgme.2010.10.013. [DOI] [PubMed] [Google Scholar]
- 196.Hofherr SE, Senac JS, Chen CY, Palmer DJ, Ng P, Barry MA. Short-term rescue of neonatal lethality in a mouse model of propionic acidemia by gene therapy. Hum Gene Ther. 2009;20:169–180. doi: 10.1089/hum.2008.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Chandler RJ, Chandrasekaran S, Carrillo-Carrasco N, Senac JS, Hofherr SE, Barry MA, Venditti CP. Adeno-associated virus serotype 8 gene transfer rescues a neonatal lethal murine model of propionic acidemia. Hum Gene Ther. 2011;22:477–481. doi: 10.1089/hum.2010.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Diacovich L, Mitchell DL, Pham H, Gago G, Melgar MM, Khosla C, Gramajo H, Tsai S-C. Crystal structure of the b-subunit of acyl-CoA carboxylase: structure-based engineering of substrate specificity. Biochem. 2004;43:14027–14036. doi: 10.1021/bi049065v. [DOI] [PubMed] [Google Scholar]
- 199.Lin TW, Melgar MM, Kurth D, Swamidass SJ, Purdon J, Tseng T, Gago G, Baldi P, Gramajo H, Tsai S-C. Structure-based inhibitor design of AccD5, an essential acyl-CoA carboxylase carboxyltransferase domain of Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 2006;103:3072–3077. doi: 10.1073/pnas.0510580103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Arabolaza A, Shillito ME, Lin TW, Diacovich L, Melgar MM, Pham H, Amick D, Gramajo H, Tsai S-C. Crystal structures and mutational analyses of acyl-CoA carboxylase b subunit of Streptomyces coelicolor. Biochemical. 2010;49:7367–7376. doi: 10.1021/bi1005305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Muro S, Perez B, Desviat LR, Rodriguez-Pombo P, Perez-Cerda C, Clavero S, Ugarte M. Effect of PCCB gene mutations on the heteromeric and homomeric assembly of propionyl-CoA carboxylase. Mol Gen Metab. 2001;74:476–483. doi: 10.1006/mgme.2001.3254. [DOI] [PubMed] [Google Scholar]
- 202.Chloupkova M, Maclean KN, Alkhateeb A, Kraus JP. Propionic acidemia: analysis of mutant propionyl-CoA carboxylase enzymes expressed in Escherichia coli. Hum Mutat. 2002;19:629–640. doi: 10.1002/humu.10085. [DOI] [PubMed] [Google Scholar]
- 203.Clavero S, Martinez MA, Perez B, Perez-Cerda C, Ugarte M, Desviat LR. Functional characterization of PCCA mutations causing propionic acidemia. Biochim Biophys Acta. 2002;1588:119–125. doi: 10.1016/S0925-4439(02)00155-2. [DOI] [PubMed] [Google Scholar]
- 204.Perez-Cerda C, Clavero S, Perez B, Rodriguez-Pombo P, Desviat LR, Ugarte M. Functional analysis of PCCB mutations causing propionic acidemia based on expression studies in deficient human skin fibroblasts. Biochim Biophys Acta. 2003;1638:43–49. doi: 10.1016/S0925-4439(03)00039-5. [DOI] [PubMed] [Google Scholar]
- 205.Sloane V, Waldrop GL. Kinetic characterization of mutations found in propionic acidemia and methylcrotonylglycinuria. J Biol Chem. 2004;279:15772–15778. doi: 10.1074/jbc.M311982200. [DOI] [PubMed] [Google Scholar]
- 206.Jiang H, Rao KS, Yee VC, Kraus JP. Characterization of four variant forms of human propionyl-CoA carboxylase expressed in Escherichia coli. J Biol Chem. 2005;280:27719–27727. doi: 10.1074/jbc.M413281200. [DOI] [PubMed] [Google Scholar]
- 207.Rodriguez-Pombo P, Perez-Cerda C, Perez B, Desviat LR, Sanchez-Pulido L, Ugarte M. Towards a model to explain the intragenic complementation in the heteromultimeric protein propionyl-CoA carboxylase. Biochim Biophys Acta. 2005;1740:489–498. doi: 10.1016/j.bbadis.2004.10.009. [DOI] [PubMed] [Google Scholar]
- 208.Diaz-Perez AL, Zavala-Hernandez AN, Cervantes C, Campos-Garcia J. The gny RDBHAL cluster is involved in acyclic isoprenoid degradation in Pseudomonas aeruginosa . Appl Environ Microbiol. 2004;70:5102–5110. doi: 10.1128/AEM.70.9.5102-5110.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Hoschle B, Gnau V, Jendrossek D. Methylcrotonyl-CoA and geranyl-CoA carboxylases are involved in leucine/isovalerate utilization (Liu) and acyclic terpene utilization (Atu), and are encoded by liuB/liuD and atuC/atuF, in Pseudomonas aeruginosa . Microbiol. 2005;151:3649–3656. doi: 10.1099/mic.0.28260-0. [DOI] [PubMed] [Google Scholar]
- 210.Forster-Fromme K, Hoschle B, Mack C, Bott M, Armbruster W, Jendrossek D. Identification of genes and proteins necessary for catabolism of acyclic terpenes and leucine/isovalerate in Pseudomonas aeruginosa . Appl Environmental Microbiol. 2006;72:4819–4828. doi: 10.1128/AEM.00853-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Aguilar JA, Zavala AN, Diaz-Perez C, Cervantes C, Diaz-Perez AL, Campos-Garcia J. The atu and liu clusters are involved in the catabolic pathways for acyclic monoterpenes and leucine in Pseudomonas aeruginosa . Appl Environmental Microbiol. 2006;72:2070–2079. doi: 10.1128/AEM.72.3.2070-2079.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Aguilar JA, Diaz-Perez C, Diaz-Perez AL, Rodriguez-Zavala JS, Nikolau BJ, Campos-Garcia J. Substrate specificity of the 3-methylcrotonyl coenzyme A (CoA) and geranyl-CoA carboxylases from Pseudomonas aeruginosa . J Bacteriol. 2008;190:4888–4893. doi: 10.1128/JB.00454-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Ding G, Che P, Ilarslan H, Wurtele ES, Nikolau BJ. Genetic dissection of methylcrotonyl CoA carboxylase indicates a complex role for mitochondrial leucine catabolism during seed development and germination. Plant J. 2012;70(4):562–577. doi: 10.1111/j.1365-313X.2011.04893.x. [DOI] [PubMed] [Google Scholar]
- 214.Tomaszycki ML, Peabody C, Replogle K, Clayton DF, Tempelman RJ, Wade J. Sexual differentiation of the zebra finch song system: potential roles for sex chromosome genes. BMC Neurosci. 2009;10:24. doi: 10.1186/1471-2202-10-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Stadler SC, Polanetz R, Meier S, Mayerhofer PU, Herrmann JM, Anslinger K, Roscher AA, Roschinger W, Holzinger A. Mitochondrial targeting signals and mature peptides of 3-methylcrotonyl-CoA carboxylase. Biochem Biophys Res Commun. 2005;334:939–946. doi: 10.1016/j.bbrc.2005.06.190. [DOI] [PubMed] [Google Scholar]
- 216.Chu C-H, Cheng D. Expression, purification, characterization of human 3-methylcrotonyl-CoA carboxylase (MCCC) Prot Expr Purif. 2007;53:421–427. doi: 10.1016/j.pep.2007.01.012. [DOI] [PubMed] [Google Scholar]
- 217.Baumgartner MR, Almashanu S, Suormala T, Obie C, Cole RN, Packman S, Baumgartner ER, Valle D. The molecular basis of human 3-methylcrotonyl-CoA carboxylase deficiency. J Clin Investig. 2001;107:495–504. doi: 10.1172/JCI11948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Gallardo ME, Desviat LR, Rodriguez JM, Esparza-Gordillo J, Perez-Cerda C, Perez B, Rodriguez-Pombo P, Criado O, Sanz R, Morton DH, Gibson KM, Le TP, Ribes A, Rodriguez de Cordoba S, Ugarte M, Penalva MA. The molecular basis of 3-methylcrotonylglycinuria, a disorder of leucine metabolism. Am J Hum Genet. 2001;68:334–346. doi: 10.1086/318202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Holzinger A, Roschinger W, Lagler F, Mayerhofer PU, Lichtner P, Kattenfeld T, Thuy LP, Nyhan WL, Koch H-G, Muntau AC, Roscher AA. Cloning of the human MCCA and MCCB genes and mutations therein reveal the molecular cause of 3-methylcrotonyl-CoA carboxylase deficiency. Human Mol Gen. 2001;10:1299–1306. doi: 10.1093/hmg/10.12.1299. [DOI] [PubMed] [Google Scholar]
- 220.Desviat LR, Perez-Cerda C, Perez B, Esparza-Gordillo J, Rodriguez-Pombo P, Penalva MA, Rodriguez de Cordoba S, Ugarte M. Functional analysis of MCCA and MCCB mutations causing methylcrotonylglycinuria. Mol Gen Metab. 2003;80:315–320. doi: 10.1016/S1096-7192(03)00130-6. [DOI] [PubMed] [Google Scholar]
- 221.Stadler SC, Polanetz R, Maier EM, Heidenreich SC, Niederer B, Mayerhofer PU, Lagler F, Koch H-G, Santer R, Fletcher JM, Ranieri E, Das AM, Spiekerkotter U, Schwab KO, Potzsch S, Marquardt I, Hennermann JB, Knerr I, Mercimek-Mahmutoglu S, Kohlschmidt N, Liebl B, Fingerhut R, Olgemoller B, Muntau AC, Roscher AA, Roschinger W. Newborn screening for 3-methylcrotonyl-CoA carboxylase deficiency: population heterogeneity of MCCA and MCCB mutations and impact on risk assessment. Hum Mutat. 2006;27:748–759. doi: 10.1002/humu.20349. [DOI] [PubMed] [Google Scholar]
- 222.Uematsu M, Sakamoto O, Sugawara N, Kumagai N, Morimoto T, Yamaguchi S, Hasegawa Y, Kobayashi H, Ihara K, Yoshino M, Watanabe Y, Inokuchi T, Yokoyama T, Kiwaki K, Nakamura K, Endo F, Tsuchiya S, Ohura T. Novel mutations in five Japanese patients with 3-methylcrotonyl-CoA carboxylase deficiency. J Hum Genet. 2007;52:1040–1043. doi: 10.1007/s10038-007-0211-9. [DOI] [PubMed] [Google Scholar]
- 223.Stucki M, Suormala T, Fowler B, Valle D, Baumgartner MR. Cryptic exon activation by disruption of exon splice enhancer. Novel mechanism causing 3-methylcrotonyl-CoA carboxylase deficiency. J Biol Chem. 2009;284:28953–28957. doi: 10.1074/jbc.M109.050674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Nguyen KV, Naviaux RK, Patra S, Barshop BA, Nyhan WL. Novel mutations in the human MCCA and MCCB gene causing methylcrotonylglycinuria. Mol Gen Metab. 2011;102:218–221. doi: 10.1016/j.ymgme.2010.10.008. [DOI] [PubMed] [Google Scholar]
- 225.Morscher RJ, Grunert SC, Burer C, Burda P, Suormala T, Fowler B, Baumgartner MR. A single mutation in MCCC1 or MCCC2 as a potential cause of positive screening for 3-methylcrotonyl-CoA carboxylase deficiency. Mol Gen Metab. 2012;105:602–606. doi: 10.1016/j.ymgme.2011.12.018. [DOI] [PubMed] [Google Scholar]
- 226.Jung CW, Lee BH, Kim JH, Kim GH, Lee J, Choi JH, Yoo HW. Uneventful clinical courses of Korean patients with methylcrotonylglycinuria and their common mutations. J Hum Genet. 2012;57:62–64. doi: 10.1038/jhg.2011.116. [DOI] [PubMed] [Google Scholar]
- 227.Forster-Fromme K, Jendrossek D. AtuR is a repressor of acyclic terpene utilization (Atu) gene cluster expression and specifically binds two 13 bp inverted repeat sequences of the atuA-atuR intergenic region. FEMS Microbiol Lett. 2010;308:166–174. doi: 10.1111/j.1574-6968.2010.02005.x. [DOI] [PubMed] [Google Scholar]
- 228.Guan X, Diez T, Prasad TK, Nikolau BJ, Wurtele ES. Geranoyl-CoA carboxylase: a novel biotin-containing enzyme in plants. Arch Biochem Biophys. 1999;362:12–21. doi: 10.1006/abbi.1998.1014. [DOI] [PubMed] [Google Scholar]
- 229.Demirev AV, Khanal A, Hanh NPK, Nam KT, Nam DH. Biochemical characterization of propionyl-coenzyme A carboxylase complex of Streptomyces toxytricini. J Microbiol. 2011;49:407–412. doi: 10.1007/s12275-011-1122-1. [DOI] [PubMed] [Google Scholar]
- 230.Gago G, Kurth D, Diacovich L, Tsai S-C, Gramajo H. Biochemical and structural characterization of an essential acyl-coenzyme A carboxylase from Mycobacterium tuberculosis. J Bacteriol. 2006;188:477–486. doi: 10.1128/JB.188.2.477-486.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Daniel J, Oh TJ, Lee CM, Kolattukudy PE. AccD6, a member of the Fas II locus, is a functional carboxyltransferase subunit of the acyl-coenzyme A carboxylase in Mycobacterium tuberculosis. J Bacteriol. 2007;189:911–917. doi: 10.1128/JB.01019-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Diacovich L, Peiru S, Kurth D, Rodriguez E, Podesta F, Khosla C, Gramajo H. Kinetic and structural analysis of a new group of acyl-CoA carboxylases found in Streptomyces coelicolor A3(2) J Biol Chem. 2002;277:31228–31236. doi: 10.1074/jbc.M203263200. [DOI] [PubMed] [Google Scholar]
- 233.Oh TJ, Daniel J, Kim HJ, Sirakova TD, Kolattukudy PE. Identification and characterization of Rv3281 as a novel subunit of a biotin-dependent acyl-CoA carboxylase in Mycobacterium tuberculosis H37Rv. J Biol Chem. 2006;281:3899–3908. doi: 10.1074/jbc.M511761200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Kurth DG, Gago GM, de la Iglesia A, Lyonnet BB, Lin TW, Morbidoni HR, Tsai S-C, Gramajo H. ACCase 6 is the essential acetyl-CoA carboxylase involved in fatty acid and mycolic acid biosynthesis in mycobacteria. Microbiol. 2009;155:2664–2675. doi: 10.1099/mic.0.027714-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Gande R, Dover LG, Krumbach K, Besra GS, Sahm H, Oikawa T, Eggeling L. The two carboxylases of Corynebacterium glutamicum essential for fatty acid and mycolic acid synthesis. J Bacteriol. 2007;189:5257–5264. doi: 10.1128/JB.00254-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Pawelczyk J, Brzostek A, Kremer L, Dziadek B, Rumijowska-Galewicz A, Fiolka M, Dziadek J. AccD6, a key carboxyltransferase essential for mycolic acid synthesis in Mycobacterium tuberculosis, is dispensable in a nonpathogenic strain. J Bacteriol. 2011;193:6960–6972. doi: 10.1128/JB.05638-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Holton SJ, King-Scott S, Eddine AN, Kaufmann SHE, Wilmanns M. Structural diversity in the six-fold redundant set of acyl-CoA carboxyltransferases in Mycobacterium tuberculosis. FEBS Lett. 2006;580:6898–6902. doi: 10.1016/j.febslet.2006.11.054. [DOI] [PubMed] [Google Scholar]
- 238.Dunn MF, Araiza G, Mora J. Biochemical characterization of a Rhizobium etli monovalent cation-stimulated acyl-coenzyme A carboxylase with a high substrate specficity constant for propionyl-coenzyme A. Microbiol. 2004;150:399–406. doi: 10.1099/mic.0.26779-0. [DOI] [PubMed] [Google Scholar]
- 239.Attwood PV. The structure and the mechanism of action of pyruvate carboxylase. Int J Biochem Cell Biol. 1995;27:231–249. doi: 10.1016/1357-2725(94)00087-R. [DOI] [PubMed] [Google Scholar]
- 240.Wallace JC, Jitrapakdee S, Chapman-Smith A. Pyruvate carboxylase. Int J Biochem Cell Biol. 1998;30:1–5. doi: 10.1016/S1357-2725(97)00147-7. [DOI] [PubMed] [Google Scholar]
- 241.Jitrapakdee S, Wallace JC. Structure, function and regulation of pyruvate carboxylase. Biochem J. 1999;340:1–16. doi: 10.1042/0264-6021:3400001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Jitrapakdee S, Vidal-Puig A, Wallace JC. Anaplerotic roles of pyruvate carboxylase in mammalian tissues. Cell Mol Life Sci. 2006;63:843–854. doi: 10.1007/s00018-005-5410-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Wallace JC. My favorite pyruvate carboxylase. IUBMB Life. 2010;62:535–538. doi: 10.1002/iub.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Owen OE, Kalhan SC, Hanson RW. The key role of anaplerosis and cataplerosis for citric acid cycle function. J Biol Chem. 2002;277:30409–30412. doi: 10.1074/jbc.R200006200. [DOI] [PubMed] [Google Scholar]
- 245.Cheng T, Sudderth J, Yang C, Mullen AR, Jin ES, Mates JM, DeBerardinis RJ. Pyruvate carboxylase is required for glutamine-independent growth of tumor cells. Proc Natl Acad Sci USA. 2011;108:8674–8679. doi: 10.1073/pnas.1016627108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Xu J, Han J, Long YS, Epstein PN, Liu YQ. The role of pyruvate carboxylase in insulin secretion and proliferation in rat pancreatic beta cells. Diabetologia. 2008;51:2022–2030. doi: 10.1007/s00125-008-1130-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Hasan NM, Longacre MJ, Stoker SW, Boonsaen T, Jitrapakdee S, Kendrick MA, Wallace JC, MacDonald MJ. Impaired anaplerosis and insulin secretion in insulinoma cells caused by small interfering RNA-mediated suppression of pyruvate carboxylase. J Biol Chem. 2008;283:28048–28059. doi: 10.1074/jbc.M804170200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.MacDonald MJ, Longacre MJ, Stoker SW, Kendrick M, Thonpho A, Brown LJ, Hasan NM, Jitrapakdee S, Fukao T, Hanson MS, Fernandez LA, Odorico J. Differences between human and rodent pancreatic islets. Low pyruvate carboxylase, ATP citrate lyase, and pyruvate carboxylation and high glucose-stimulated acetoacetate in human pancreatic islets. J Biol Chem. 2011;286:18383–18396. doi: 10.1074/jbc.M111.241182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Ozimek P, van Dijk R, Latchev K, Gancedo C, Wang DY, van der Klei IJ, Veenhuis M. Pyruvate carboxylase is an essential protein in the assembly of yeast peroximal oligomeric alcohol oxidase. Mol Biol Cell. 2003;14:786–797. doi: 10.1091/mbc.E02-07-0417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Klompmaker SH, Kilic A, Baerends RJ, Veenhuis M, van der Klei IJ. Activation of a peroximal Pichia pastoris D-amino acid oxidase, which uses d-alanine as a preferred substrate, depends on pyruvate carboxylase. FEMS Yeast Res. 2010;10:708–716. doi: 10.1111/j.1567-1364.2010.00647.x. [DOI] [PubMed] [Google Scholar]
- 251.Ozimek PZ, Klompmaker SH, Visser N, Veenhuis M, van der Klei IJ. The transcarboxylase domain of pyruvate carboxylase is essential for assembly of the peroxisomal flavoenzyme alcohol oxidase. FEMS Yeast Res. 2007;7:1082–1092. doi: 10.1111/j.1567-1364.2007.00214.x. [DOI] [PubMed] [Google Scholar]
- 252.Huberts DHEW, Venselaar H, Vriend G, Veenhuis M, van der Klei IJ. The moonlighting function of pyruvate carboxylase resides in the non-catalytic end of the TIM barrel. Biochim Biophys Acta. 2010;1803:1038–1042. doi: 10.1016/j.bbamcr.2010.03.018. [DOI] [PubMed] [Google Scholar]
- 253.Schar J, Stoll R, Schauer K, Loeffler DIM, Eylert E, Joseph B, Eisenreich W, Fuchs TM, Goebel W. Pyruvate carboxylase plays a crucial role in carbon metabolism of extra- and intracellularly replicating Listeria monocytogenes. J Bacteriol. 2010;192:1774–1784. doi: 10.1128/JB.01132-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Lai H, Kraszewski JL, Purwantini E, Mukhopadhyay B. Identification of pyruvate carboxylase genes in Pseudomonas aeruginosa PAO1 and development of a P. aeruginosa-based overexpression system for a4- and a4b4-type pyruvate carboxylases. Appl Environ Microbiol. 2006;72:7785–7792. doi: 10.1128/AEM.01564-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Zeczycki TN, Menefee AL, Jitrapakdee S, Wallace JC, Attwood PV, St. Maurice M, Cleland WW. Activation and inhibition of pyruvate carboxylase from Rhizobium etli. Biochemical. 2011;50:9694–9707. doi: 10.1021/bi201276r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Adina-Zada A, Zeczycki TN, Attwood PV. Regulation of the structure and activity of pyruvate carboxylase by acetyl CoA. Arch Biochem Biophys. 2012;519:118–130. doi: 10.1016/j.abb.2011.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Adina-Zada A, Hazra R, Sereeruk C, Jitrapakdee S, Zeczycki TN, St. Maurice M, Cleland WW, Wallace JC, Attwood PV. Probing the allosteric activation of pyruvate carboxylase using 2′,3′-O-(2,4,6-trinitrophenyl) adenosine 5′-triphosphate as a fluorescent mimic of the allosteric activator acetyl CoA. Arch Biochem Biophys. 2011;509:117–126. doi: 10.1016/j.abb.2011.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Zeczycki TN, St. Maurice M, Attwood PV. Inhibitors of pyruvate carboxylase. Open Enzyme Inhib J. 2010;3:8–26. doi: 10.2174/1874940201003010008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Carbone MA, MacKay N, Ling M, Cole DEC, Douglas C, Rigat B, Feigenbaum A, Clarke JTR, Haworth JC, Greenberg CR, Seargeant L, Robinson BH. Amerindian pyruvate carboxylase deficiency is associated with two distinct missense mutations. Am J Hum Genet. 1998;62:1312–1319. doi: 10.1086/301884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Wexler ID, Kerr DS, Du Y, Kaung MM, Stephenson W, Lusk MM, Wappner RS, Higgins JJ. Molecular characterization of pyruvate carboxylase deficiency in two consanguineous families. Pediatr Res. 1998;43:579–584. doi: 10.1203/00006450-199805000-00004. [DOI] [PubMed] [Google Scholar]
- 261.Robinson BH. Lactic acidemia and mitochondrial disease. Mol Gen Metab. 2006;89:3–13. doi: 10.1016/j.ymgme.2006.05.015. [DOI] [PubMed] [Google Scholar]
- 262.Wang D, Yang H, de Braganca KC, Lu J, Shih LY, Briones P, Lang T, de Vivo DC. The molecular basis of pyruvate carboxylase deficiency: mosaicism correlates with prolonged survival. Mol Gen Metab. 2008;95:31–38. doi: 10.1016/j.ymgme.2008.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Monnot S, Serre V, Chadefaux-Vekemans B, Aupetit J, Romano S, de Lonlay P, Rival JM, Munnich A, Steffann J, Bonnefont JP. Structural insights on pathogenic effects of novel mutations causing pyruvate carboxylase deficiency. Hum Mutat. 2009;30:734–740. doi: 10.1002/humu.20908. [DOI] [PubMed] [Google Scholar]
- 264.Marin-Valencia I, Roe CR, Pascual JM. Pyruvate carboxylase deficiency: mechanisms, mimics and anaplerosis. Mol Gen Metab. 2010;101:9–17. doi: 10.1016/j.ymgme.2010.05.004. [DOI] [PubMed] [Google Scholar]
- 265.Lietzan AD, Menefee AL, Zeczycki TN, Kumar S, Attwood PV, Wallace JC, Cleland WW, St. Maurice M. Interaction between the biotin carboxyl carrier domain and the biotin carboxylase domain in pyruvate carboxylase from Rhizobium etli . Biochemical. 2011;50:9708–9723. doi: 10.1021/bi201277j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Yu LPC, Xiang S, Lasso G, Gil D, Valle M, Tong L. A symmetrical tetramer for S. aureus pyruvate carboxylase in complex with coenzyme A. Structure. 2009;17:823–832. doi: 10.1016/j.str.2009.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Balsera M, Buey RM, Li X-D. Quaternary structure of the oxaloacetate decarboxylase membrane complex and mechanistic relationships to pyruvate carboxylase. J Biol Chem. 2011;286:9457–9467. doi: 10.1074/jbc.M110.197442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Zeczycki TN, St. Maurice M, Jitrapakdee S, Wallace JC, Attwood PV, Cleland WW. Insight into the carboxyl transferase domain mechanism of pyruvate carboxylase from Rhizobium etli . Biochemical. 2009;48:4305–4313. doi: 10.1021/bi9003759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Duangpan S, Jitrapakdee S, Adina-Zada A, Byrne L, Zeczycki TN, St. Maurice M, Cleland WW, Wallace JC, Attwood PV. Probing the catalytic roles of Arg548 and Gln552 in the carboxyl transferase domain of the Rhizobium etli pyruvate carboxylase by site-directed mutagenesis. Biochemical. 2010;49:3296–3304. doi: 10.1021/bi901894t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Zeczycki TN, Menefee AL, Adina-Zada A, Jitrapakdee S, Surinya KH, Wallace JC, Attwood PV, St Maurice M, Cleland WW. Novel insights into the biotin carboxylase domain reactions of pyruvate carboxylase from Rhizobium etli . Biochemical. 2011;50:9724–9737. doi: 10.1021/bi2012788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Lasso G, Yu LPC, Gil D, Xiang S, Tong L, Valle M. Cryo-EM analysis reveals new insights into the mechanism of action of pyruvate carboxylase. Structure. 2010;18:1300–1310. doi: 10.1016/j.str.2010.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Kanamori T, Kanou N, Atomi H, Imanaka T. Enzymatic characterization of a prokaryotic urea carboxylase. J Bacteriol. 2004;186:2532–2539. doi: 10.1128/JB.186.9.2532-2539.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Andersen G, Bjornberg O, Polakova S, Pynyaha Y, Rasmussen A, Moller K, Hofer A, Moritz T, Sandrini MPB, Merico A-M, Compagno C, Akerlund HE, Gojkovic Z, Piskur J. A second pathway to degrade pyrimidine nucleic acid precursors in eukaryotes. J Mol Biol. 2008;380:656–666. doi: 10.1016/j.jmb.2008.05.029. [DOI] [PubMed] [Google Scholar]
- 274.Ghosh S, Navarathna DHMLP, Roberts DD, Cooper JT, Atkin AL, Petro TM, Nickerson KW. Arginine-induced germ tube formation in Candida albicans is essential for escape from murine macrophage line RAW 264.7. Infection Immunity. 2009;77:1596–1605. doi: 10.1128/IAI.01452-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Collette JR, Lorenz MC. Mechanisms of immune evasion in fungal pathogens. Curr Opin Microbiol. 2011;14:668–675. doi: 10.1016/j.mib.2011.09.007. [DOI] [PubMed] [Google Scholar]
- 276.Jacques DA, Langley DB, Hynson RMG, Whitten AE, Kwan A, Guss JM, Trewhella J. A novel structure of an antikinase and its inhibitor. J Mol Biol. 2011;405:214–226. doi: 10.1016/j.jmb.2010.10.047. [DOI] [PubMed] [Google Scholar]
- 277.Jacques DA, Langley DB, Kuramitsu S, Yokoyama S, Trewhella J, Guss JM. The structure of TTHA0988 from Thermus thermophilus, a KipI-KipA homolgue incorrectly annotated as an allophanate hydrolase. Acta Cryst. 2011;D67:105–111. doi: 10.1107/S0907444910051127. [DOI] [PubMed] [Google Scholar]