Abstract
Small heat-shock protein chaperones are important players in the protein quality control system of the cell, because they can immediately respond to partially unfolded proteins, thereby protecting the cell from harmful aggregates. The small heat-shock proteins can form large polydisperse oligomers that are exceptionally dynamic, which is implicated in their function of protecting substrate proteins from aggregation. Yet the mechanism of substrate recognition remains poorly understood, and little is known about what parts of the small heat-shock proteins interact with substrates and what parts of a partially unfolded substrate protein interact with the small heat-shock proteins. The transient nature of the interactions that prevent substrate aggregation rationalize probing this interaction by crosslinking mass spectrometry. Here, we used a workflow with lysine-specific crosslinking and offline nano-liquid chromatography matrix-assisted laser desorption/ionization tandem time-of-flight mass spectrometry to explore the interaction between the plant small heat-shock protein Hsp21 and a thermosensitive model substrate protein, malate dehydrogenase. The identified crosslinks point at an interaction between the disordered N-terminal region of Hsp21 and the C-terminal presumably unfolding part of the substrate protein.
Electronic supplementary material
The online version of this article (doi:10.1007/s12192-012-0360-4) contains supplementary material, which is available to authorized users.
Introduction
Small heat-shock proteins (sHsps) can interact with unfolding proteins to prevent their irreversible aggregation, thereby protecting cells from toxic aggregates (Basha et al. 2011; Nakamoto and Vigh 2007). Together with other molecular chaperones, they are important players in the protein quality control system, which takes care of unfolded and misfolded proteins in order to maintain protein homeostasis (proteostasis) in cells (Hartl et al. 2011). The sHsps do not require ATP and can hold substrate proteins in a refolding-competent state until ATP-dependent chaperones such as the Hsp70 and Hsp100 family members can promote refolding. An alternative destination of unfolded proteins, which may be at least as important for proteostasis as refolding, is the degradation machinery (Tyedmers et al. 2010), although the molecular details of this route are less well known. Irrespective of the final destination of unfolded proteins, sHsps can act as a first line of defence against the threat of unfolded proteins, capable of sensing very subtle changes in the stability of proteins (McHaourab et al. 2009). Considering the presence of multiple sHsp genes in almost all organisms (Haslbeck et al. 2005), their implication in a variety of different diseases (Sun and MacRae 2005), and their dramatic upregulation into the most abundant proteins upon stress (Beck et al. 2009), it is obvious that sHsps play an important role in maintaining proteostasis in the cell.
The elucidation of sHsp structure and function has been challenging because of their polydispersity and dynamics, both with respect to oligomeric structure and substrate binding (Eyles and Gierasch 2010). The common feature of sHsps is the β-sandwich-forming α-crystallin domain, which consists of around 90 amino acid residues. Two such domains form the dimeric building blocks of higher-order oligomers. Dimer formation is mediated by either β-strand-exchange of a strand called β6 (in plant, yeast, and bacteria sHsps), or by the interaction of two extended β-strands called β6 + 7 (in metazoan sHsps) (Basha et al. 2011; Poulain et al. 2010). The α-crystallin domain is flanked by the N-terminal region and the C-terminal extension, both of which are important for oligomer formation. The N-terminal region is very diverse across the sHsp superfamily and often not well-defined in crystal structures of sHsps determined so far (McHaourab et al. 2009). The C-terminal extension has a highly conserved I/V/L-X-I/V/L motif, which was shown to bind a hydrophobic groove on neighboring dimers in the larger oligomers in crystal structures of sHsps (Kim et al. 1998; Laganowsky et al. 2010; Laganowsky and Eisenberg 2010; Van Montfort et al. 2001).
The only two sHsps that have been crystallized in their native oligomeric state are Methanocaldococcus jannaschii Hsp16.5 and wheat Hsp16.9 (Kim et al. 1998; Van Montfort et al. 2001). In both crystal structures, the C-terminal I/V/L-X-I/V/L motif binds neighbouring dimers, but M. jannaschii Hsp16.5 forms a 24-mer and wheat Hsp16.9 forms a 12-mer, which is illustrating how the same interaction can give rise to different oligomeric states. Whereas M. jannaschii Hsp16.5 and wheat Hsp16.9 form homogeneous oligomers, other sHsps form heterogeneous oligomers. Human αB-crystallin was shown to form an ensemble of differently sized oligomers ranging from 12- to 48-mers by native mass spectrometry (Baldwin et al. 2011c), explaining failing crystallization attempts of the full-length protein. The same C-terminal I/V/L-X-I/V/L motif interaction in all individual αB-crystallin monomers is responsible for oligomerization and governs the equilibrium dynamics of the heterogeneous ensemble (Baldwin et al. 2011a).
The dynamic nature of sHsp oligomers was long since established by monitoring the subunit exchange of sHsp heterooligomers by FRET (Bova et al. 1997; 2000) and more recently by native MS (Painter et al. 2008), and the dynamics and polydispersity of sHsps are clearly linked to their function of binding substrate proteins and protecting them from aggregation. Combining NMR, ion-mobility native mass spectrometry, and EM, structural models of the most abundant oligomeric states (24-, 26-, and 28-mers) and a model for the conversion between them have been proposed, providing a deeper understanding of the polydispersity and dynamics of sHsp structures (Baldwin et al. 2011b). Further increasing possible complexity, it has been found that various human sHsps may form mixed heterooligomers (Mymrikov et al. 2012). The stable protein complexes formed between sHsp and substrate proteins also appear to be polydisperse (Stengel et al. 2010), with complexes of pea Hsp18.1 and substrate protein luciferase comprised of variable numbers of both sHsp and substrate protein, covering over 300 different stoichiometries.
Despite the recent advancements in understanding the structure and dynamics of sHsps, the mechanism of substrate recognition, which is essential to their function, remains poorly understood. The sHsps prevent aggregation by stabilizing partially folded intermediates via weak and transient interactions, which eventually leads to the formation of stable protein complexes (Lindner et al. 2001). The folding pathway is a fast and reversible process, in which the natively folded form of a protein is in equilibrium with partially folded intermediates and unfolded forms. Under conditions in which the partially folded intermediates persist, such as cellular stress or when heat-sensitive model substrate proteins are exposed to elevated temperatures, the protein leaves the folding pathway and enters the protein off-folding pathway, which is relatively slow, in the order of seconds (Ecroyd and Carver 2008). Under these conditions, a kinetic competition exists between substrate protein aggregation and substrate protein interaction with sHsps (Lindner et al. 2001; McHaourab et al. 2009).
In this study, we aim to gain insight into such interactions that compete with and prevent aggregation. We have used crosslinking mass spectrometry, i.e., the mass spectrometric detection of crosslinked peptides, to investigate crosslinks between proteins. This approach (also called CXMS) is currently maturing into a useful approach to characterize protein–protein interactions, even though some technical obstacles still prevent its exploitation to its full potential (Leitner et al. 2010; Singh et al. 2010; Sinz 2006). Our previously developed workflow, using lysine-specific crosslinking, offline nano-liquid chromatography matrix-assisted laser desorption/ionization tandem time-of-flight (LC MALDI-TOF/TOF) mass spectrometry and an in-house developed program FINDX, was shown to allow the identification of true crosslinks that are all within crosslinking distance (<20 Å), mapping well into the structure of a protein oligomer (Lambert et al. 2011b; Söderberg et al. 2012). In CXMS, unmodified peptides are the most abundant, closely followed by peptides with a dead-end crosslink modification (type 0) and after this, intra-peptide crosslinks (type 1). Thereafter, inter-peptide crosslinks (type 2) within the same subunit (intra-monomeric) are still abundant enough to be fairly easily detected, whereas the more informative inter-peptide crosslinks (type 2) between two subunits (inter-monomeric) within a protein complex are very low in abundance and difficult to detect (Söderberg et al. 2012). To detect inter-peptide crosslinks (type 2) between two transiently interacting proteins is therefore challenging. Nevertheless, having this workflow at hand, we decided to see to what extent it is possible to explore the interaction between sHsps and a model substrate protein.
The chloroplast-localized sHsp, Hsp21, increases plant resistance to heat stress upon overexpression (Harndahl et al. 1999), and we have previously characterized its structure and function (Ahrman et al. 2007a, b; Lambert et al. 2011a). We found that Hsp21 has an even better chaperone activity compared with human αB-crystallin, the most well-characterized sHsp. Furthermore, Hsp21 has, compared with human αB-crystallin and cytosolic plant homologues, a longer N-terminal arm containing a conserved methionine-rich motif, which is believed to be important for substrate-binding. Here, we present data on the chaperone activity of Hsp21 towards the model substrate protein malate dehydrogenase (MDH), and the crosslinks identified for the Hsp21–substrate interaction under conditions where the initial interactions in the process of sHsp substrate recognition supposedly are captured. The identified crosslinks point at an interaction between the disordered N-terminal region of Hsp21 and the C-terminal presumably unfolding part of the substrate protein.
Materials and methods
Proteins and reagents
Recombinantly expressed Hsp21 from Arabidopsis thaliana (UniProtKB ID P31170) was obtained as previously described (Ahrman et al. 2007b). MDH (UniProtKB ID P00346) from Sus scrofa (pig) was purchased from Roche (Basel, Switzerland). For the crosslinking experiments, proteins were desalted and buffer exchanged into crosslinking buffer, consisting of 50 mM HEPES (4-(2-hydroxyethyl)-1-piperazine ethanesulfonic acid) at pH 8.0, 150 mM NaCl, and 5 mM MgCl2, using disposable PD-10 protein desalting columns (GE Healthcare, Little Chalfont, UK). Protein concentrations are those of monomeric Hsp21 (21 kDa) and MDH (33 kDa) and were determined with the Bradford assay (Stoscheck 1990), using bovine serum albumin (UniProtKB ID P02769) as a standard. The isotope-labelled crosslinking reagent BS3 (bis[sulfosuccinimidylsuberate]), consisting of a 1:1 molar ratio mixture of BS3-H12 and BS3-D12, was purchased from Creative Molecules Inc. (Victoria, Canada).
In vitro chaperone activity by light-scattering
MDH aggregation was evaluated by following the light-scattering changes at 360 nm. Samples in buffer (50 mM HEPES pH 8.0, 5 mM MgCl2, and 100 mM NaCl, or otherwise as stated) were mixed at various concentrations of Hsp21 and 10 or 5 μM of MDH, as indicated, to a final volume of 150 μl in a 96-well plate (Mikroplate Corning 3881, 96-well, low binding, half area, Corning Incorporated Life Sciences, Acton, MA, USA). The plate was sealed with a plastic film to avoid evaporation and inserted in the FLUOstar Omega microplate reader (BMG Labtech, Offenburg, Germany), which was set to 45 °C and continuous shaking. Absorbance measurements at 360 nm were recorded every 80 s for up to 24 h. In absence of Hsp21, light-scattering increased after a lag-phase of approximately 30 min and was completed after 2–3 h, depending on the salt conditions.
Non-denaturing electrophoresis
Samples were pre-incubated at 25 °C or 43 °C. Samples in buffer (50 mM HEPES pH 8.0, 150 mM NaCl, and 5 mM MgCl2) contained Hsp21 (25 μM) and model substrate protein (MDH, 25 μM) pre-incubated separately or mixed at a 1:1 molar ratio calculated on monomer to monomer basis. Non-denaturing polyacrylamide gel electrophoresis (Native-PAGE) was performed using NativePAGE™ Novex® Bis–Tris gels (3–12 % gradient, Bis–Tris buffer pH 7.6, Life Technologies Europe BV, Stockholm, Sweden). Approximately 5 μg was loaded of each protein. During the run (for 90 min), the gels were stained by CBB G-250, added according to the manufacturer’s instructions to the running buffer, in order to charge proteins negatively without denaturation. At pH 7.6 however, the intrinsic charge of MDH (pI ≈ 8.6) is positive and the negative charging from the CBB stain not enough to make it focus into sharp bands, but it was still possible to evaluate from the Native-PAGE when MDH was complexed to Hsp21 or not. A superior resolution is obtained for Hsp21 and most other proteins, by another commercially available Native-PAGE system (Native-PAGE Phastgel™, 8–25 % gradient, Tris–alanine buffer, pH 8.8, GE Healthcare, Uppsala, Sweden) (see Supplementary Fig. S-1, and comment therein); however, this system is not possible to use with MDH since, at pH 8.8, it is basically uncharged.
Denaturing electrophoresis
Denaturing gel electrophoresis (sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE)) was performed using precast 4–12 % NuPAGE® Bis–Tris Gels (Life Technologies Europe BV, Stockholm, Sweden) in MES buffer according to the manufacturer’s instructions. Samples were diluted with LDS sample buffer (4×) (Life Technologies Europe BV, Stockholm, Sweden) and solubilized for 10 min at 95 °C. Gels were stained with Coomassie Brilliant Blue G-250 according to the Quick-stain colloidal CBB protocol (Lawrence and Besir 2009).
Chemical crosslinking
The protein concentration was 50 μM for each protein, or otherwise as indicated. The crosslinker BS3 (bis[sulfosuccinimidylsuberate]) was dissolved in distilled water to a concentration of 30 mM immediately before use. Sample aliquots of 20 μl were pre-incubated at 45 °C for 30 min and then incubated with isotopically coded BS3 (final concentration 3 mM, or less where indicated) at 45 °C. After 20 min, the crosslinking reaction was quenched by adding 1 M Tris-(hydroxymethyl)-aminomethane (Tris) to a final concentration of 20 mM. To remove excess reagent and Tris and to concentrate the proteins, the samples were precipitated with freeze-cold acetone.
Trypsin digestion
The protein pellets from acetone-precipitation were carefully re-dissolved in 20 μl 25 mM NH4HCO3, pH 7.8. The samples were digested with sequencing-grade modified trypsin (Promega, Madison, WI, USA) at 37 °C at a protease/protein (w/w) ratio of 1:100 for 1 h, followed by a ratio of 1:50 overnight. All samples were acidified by adding 2 μl 10 % trifluoroacetic acid (TFA) and stored at −20 °C until further analysis.
Reversed-phase liquid chromatography prior to MALDI-TOF/TOF mass spectrometry
The samples were separated by reversed-phase liquid chromatography using an 1100 Series Nanoflow LC system (Agilent Technologies, Waldbronn, Germany). The mobile phases used for separation were composed of A—1 % (v/v) acetonitrile and 0.1 % (v/v) TFA and B—90 % (v/v) acetonitrile and 0.1 % (v/v) TFA. For each sample, 2, 8, or 20 μl was injected, which was estimated to contain approximately 50, 200, or 500 pmol peptides. Samples were loaded onto a Zorbax 300SB-C18 0.3 mm pre-column (Agilent Technologies) in buffer A at a flow rate of 0.040 ml/min, delivered by the isocratic pump. By switching the micro 6-port/2-position module, the nano-pump delivering a linear gradient from 0 % to 100 % buffer B (flow rate, 1.4 μl/min) was then connected to the pre-column to move the sample onward to the separation column, a PepSwift Monolithic Capillary Column (200 μm i.d. × 5 cm) (Dionex, Amsterdam, The Netherlands). Before starting to collect fractions for a sample, 2 μl was injected to saturate the column and eluted into waste. Peptides eluted between about 5 % and 45 % buffer B and were collected in 64 or 192 fractions on the matrix-assisted laser desorption/ionization target plate.
MALDI-TOF/TOF mass spectrometry
Matrix solution consisting of 5 mg/ml α-cyano-4-hydroxy cinnamic acid, 50 % acetonitrile, 0.1 % TFA, 25 mM citric acid, and standard peptides (angiotensin II, m/z 1046.541; neurotensin, m/z 1672.91 and ACTH 18–39, m/z 2465.199) for internal calibration was manually applied to the dried peptide fractions and allowed to dry. Mass spectrometric data were acquired using a 4700 Proteomics Analyzer (Applied Biosystems/MDS SCIEX, USA) in the positive reflector mode. The spectra were internally calibrated using the standard peptides. After MS-data analysis by FINDX, inclusion lists with peaks suggested to be crosslinked peptides were used for selection of crosslink precursors for tandem mass spectrometry (MS/MS) fragmentation. MS/MS spectra were recorded in the MS/MS 1 kV positive mode with 3,000 laser shots per spectrum, or manually with various settings to optimize the output.
Data analysis
Mass spectrometric data were analyzed with FINDX version BS3g_beta3_20ppm, using the following settings in MS mode: crosslinking reagent, BS3; isotope-labelled, yes (1:1 mixture of BS3-H12 and BS3-D12); enzyme used, trypsin; number of missed cleavages allowed, 4; included in theoretical digest: dead-end (both hydrolyzed and Tris), intra-peptide, and inter-peptide crosslinks, all with possibly one extra dead-end crosslinker modification, methionine oxidation allowed; precursor tolerance, 20 ppm; peaklist merging of different LC-separated fractions of the same sample at 5 ppm; mass accuracy of the mass difference between the isotope doublet peaks, 20 ppm; intensity ratio deviation from 1 between the isotope doublet peaks, 0.67 (allowed difference of 50 %); no reference sample. Crosslink candidates were discarded if their mass matched (within 60 ppm) one or several other peptides modified with the crosslinking reagent (ambiguity). Because the FINDX analysis did not consider a reference sample (non-crosslinked sample), all crosslink candidates presented here were compared with a theoretical digest of both Hsp21 and MDH, generated by GPMAW (Peri et al. 2001) and confirmed not to match any unmodified peptides. The following settings were used in FINDX in MS/MS mode: crosslinking reagent, BS3; isotope-labelled, yes (1:1 mixture of BS3-H12 and BS3-D12); enzyme used, trypsin; number of missed cleavages allowed, 4; included in theoretical digest: dead-end (both hydrolyzed and Tris), intra-peptide, and inter-peptide crosslinks, all with possibly one extra dead-end crosslinker modification, methionine oxidation not allowed; precursor tolerance, 50 ppm; MS/MS fragment tolerance, 400 ppm.
Results
Chaperone activity of Hsp21 toward the model substrate protein malate dehydrogenase (MDH)
Suppression of thermal-induced aggregation of heat-sensitive model substrate proteins is commonly used to assess in vitro chaperone activity (Ehrnsperger et al. 1997), even though this may not necessarily reflect the full activity in vivo (Giese et al. 2005). We measured the light-scattering increase at 45 °C, which is indicative of aggregation and precipitation of the heat-sensitive model substrate protein MDH. In absence of Hsp21, MDH started to form precipitates after about 30 min, with different salt conditions giving somewhat different lag phase periods but similar endpoints. At molar ratios of Hsp21/MDH of 1/1 or higher (Fig. 1a), the light-scattering increase was fully suppressed. As the Hsp21/MDH molar ratio was decreased to below 1/1, suppression was no longer complete (Fig. 1b). Thus, Hsp21 can protect MDH at a molar ratio >1. Since the monomeric masses of Hsp21 and MDH are rather similar (21 and 33 kDa), we can conclude that Hsp21 can protect an amount of substrate protein MDH of up to about its own mass, which is consistent to what has previously been reported for other sHsps and substrate proteins (Basha et al. 2011).
Fig. 1.
Hsp21 chaperone activity towards heat-treated MDH. The model substrate MDH was heat-treated by incubation at 45 °C in absence or presence of varying amounts of Hsp21. Thermal-induced precipitation of MDH was followed as light-scattering increase over time, and the suppression of light-scattering increase in presence of Hsp21 used to assess the chaperone activity. Samples assayed in duplicate or triplicate are marked with same color. The numbers of the ratios in the legend refer to concentrations (Hsp21/MDH) expressed in micromolars. a Hsp21/MSH molar ratios 1/1 and above, under different salt conditions (*): 1, low salt; 2, 0.1 M NaCl; 3, 0.2 M Na2SO4. b Hsp21/MDH molar ratios 1/1 and below, in 0.1 M NaCl
Crosslinking Hsp21 and MDH to capture the interactions protective in thermal-induced unfolding
We crosslinked Hsp21 and the model substrate protein MDH with the lysine-specific crosslinking reagent BS3 (bis[sulfosuccinimidylsuberate]), to characterize possible sites of interaction between the small heat-shock protein and the unfolding substrate protein. After prolonged incubation at high temperature, sHsps and substrates form irreversible stable protein complexes, a process that can only be reversed with the help of other chaperone proteins (Basha et al. 2011). However, the sHsp–substrate interaction is initially transient and reversible as shown, e.g., for αB-crystallin (Lindner et al. 2001). We have repeatedly observed that a heat-treatment of model substrates in presence of Hsp21 is sufficient to rescue the substrate protein from aggregation, although no stable Hsp21–substrate protein complexes form, as shown for Hsp21 and MDH (Fig. 2). To catch a glimpse of the transient and reversible interactions that protect against aggregation, we crosslinked samples where Hsp21 and MDH were pre-incubated for 30 min at 45 °C before adding the crosslinker.
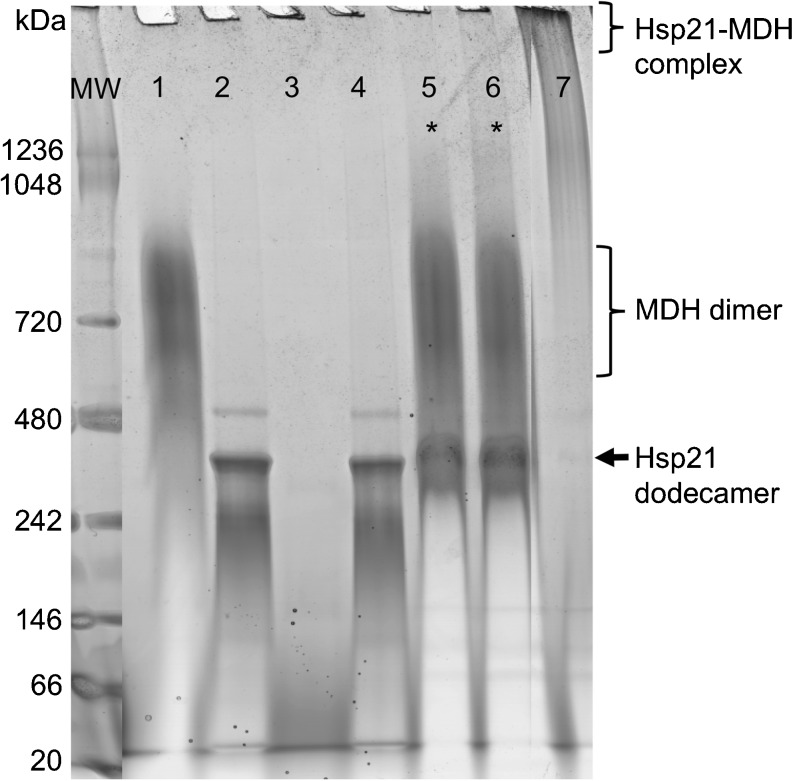
Fig. 2.
MDH heat-treated in absence and presence of Hsp21 analyzed by Native-PAGE. Samples were pre-incubated at 25 °C or 45 °C as indicated and directly loaded on Blue-Native PAGE (3–12 % gradient, Bis–Tris buffer pH 7.6). MW, molecular weight marker; lanes 1 and 2, MDH and Hsp21, respectively, pre-incubated at 25 °C; lanes 3 and 4, MDH and Hsp21, respectively, pre-incubated at 45 °C (note that aggregated MDH does not enter the gel); lane 5, Hsp21 and MDH mixed at a 1/1 molar ratio and pre-incubated for 30 min at 25 °C; lane 6, Hsp21 and MDH mixed at a 1/1 molar ratio and pre-incubated for 30 min at 45 °C; lane 7, Hsp21 and MDH mixed at a 1/1 molar ratio and incubated for 180 min at 45 °C. The large stable complexes formed between Hsp21 and MDH pre-incubated for 180 min at 45 °C cannot enter the gel and remain in the loading well (lane 7). Comparison of lane 5 and 6 (marked by asterisks) shows that no stable complexes are formed by pre-incubation for 30 min at 45 °C, since Hsp21 dodecamers and MDH dimers run separately (lane 6), just as in the non-heated sample (lane 5). The sub-optimal resolution of the Hsp21 dodecamer in this Native-PAGE, compared with the better performance shown in Supplementary Figure S-1 and the appearance of the MDH dimer as a smear at an apparent mass approximately 700 kDa instead of a band at 66 kDa are due to technical limitations imposed by the high pI-value of MDH as further explained in the “Supplement”
As revealed by SDS-PAGE, crosslinked species with a molecular weight of more than either monomeric Hsp21 or monomeric MDH were the result of this crosslinking reaction (Fig. 3). Without pre-incubation for 30 min at 45 °C (Fig. 3, lane 2), bands are visible at around 45 and 66 kDa, which presumably represent crosslinked Hsp21 dimer and MDH dimer, respectively. With pre-incubation for 30 min at 45 °C (Fig. 3, lanes 3–6), several additional bands are visible which may represent species of crosslinked Hsp21 and MDH. The most prominent of these, at around 55 kDa, matches the mass of one Hsp21 monomer and one MDH monomer. The bands with molecular weights of around 97 kDa and higher may be mixed Hsp21 and MDH crosslinked species of different stoichiometries. Samples pre-incubated at 45 °C contained high-molecular-weight species which had not entered the gel, material which we know is enriched in crosslinks (Söderberg et al. 2012). All crosslinks presented in this study were identified by analyzing the total crosslinked Hsp21-MDH sample (the unfractionated sample that was loaded to SDS-PAGE), because in our experience the crosslink yields are generally higher with in-solution digests as opposed to in-gel digests.
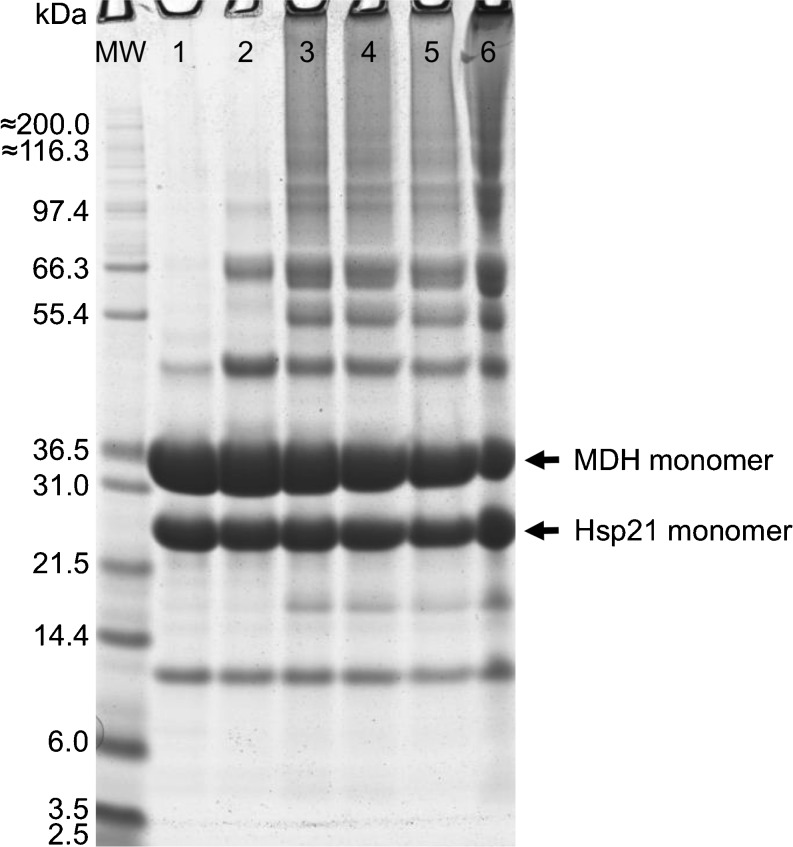
Fig. 3.
Crosslinked samples of Hsp21 and MDH analyzed by SDS-PAGE. Samples were pre-incubated at 20 °C or 45 °C M, as indicated, crosslinked, and then loaded to SDS-PAGE. MW, molecular weight marker; lane 1, Hsp21 + MDH, non-crosslinked reference sample; lane 2, Hsp21 + MDH, sample crosslinked after pre-incubation 30 min at 20 °C; lanes 3, 4, and 5 Hsp21 + MDH, triplicate samples crosslinked after pre-incubation 30 min at 45 °C; lane 6, Hsp21+MDH, sample crosslinked after pre-incubation 180 min at 45 °C. Extra bands are visible at 10 and 18 kDa which are degradation products of Hsp21 and at 42 kDa a dimer of Hsp21. The non-covalent bonds within the Hsp21 dimer are strong enough for a band to be visible in SDS-PAGE, more or less depending on the exact number of minutes and degrees in solubilization buffer. The Hsp21 dimer becomes stabilized by crosslinking and visible as an even stronger band (lanes 2–6)
Mass spectrometric analysis of crosslinked Hsp21 and MDH samples
By protease digestion of the crosslinked protein samples and subsequent mass spectrometric analysis of the resulting peptides, it is possible to identify peptide pairs that have been crosslinked after pre-incubation for 30 min at 45 °C, reflecting spatial proximity during the crosslinking reaction. We identified ten crosslinks between Hsp21 and MDH, representing ten pairs of amino acid residues crosslinked by BS3 (Table 1). Because BS3 is amine-reactive, potentially reactive amino acid residues include all lysine residues and the protein N termini of both Hsp21 (methionine) and MDH (alanine). We also analyzed Hsp21–Hsp21 crosslinks and MDH–MDH crosslinks (Supplementary Tables S1 and S2) and the presence of other types of crosslinks than type 2 (Supplementary Tables S3 and S4), for reasons discussed below.
Table 1.
Crosslinks identified between Hsp21 and MDH
| Theoretical (MH)+ (Da) | Matched Hsp21 sequence | Matched MDH sequence | Hsp21 res. | MDH res. | Observations | Ox. Met observationa | Fig. S-2b |
|---|---|---|---|---|---|---|---|
| 1503.7412 | M1QDQR5 | K81PGMTR86 | 1 | 81 | 3 | 2 | A-C |
| 1870.9234 | M1QDQR5 | K133HGVYNPNK141 | 1 | 133 | 5 | 1 | D, E |
| 2087.0807 | M1QDQR5 | I206QEAGTEVVKAK217 | 1 | 215 | 7 | 1 | F, G |
| 1388.7208 | M1QDQR5 | K273GIEK277 | 1 | 273 | 2 | 1 | H, I |
| 1842.9747 | M1QDQR5 | G274IEKNLGIGK283 | 1 | 277 | 11 | J | |
| 1360.7258 | M1QDQR5 | A301SIKK305 | 1 | 304 | 2 | 1 | K, L |
| 1650.8161 | M1QDQR5 | K305GEEFVK311 | 1 | 305 | 2 | M | |
| 1926.0157 | G19NQGSSVEKRPQQR32 | A1K2 | 27 | 1 | 3 | N | |
| 2544.3170 | G19NQGSSVEKRPQQR32 | K305GEEFVK311 | 27 | 305 | 3 | O | |
| 1605.8852 | T172KVER176 | K305GEEFVK311 | 173 | 305 | 4 | P |
aThe observation(s) of the same peptide pair but with one or two methionine residues oxidized strengthens the identification, and in this case, the MS spectrum of the oxidized version of the crosslink is also shown in Fig. S-2 (Supplementary information). This type of observation was only counted once, even if it was observed in different data sets
bThe capital letter refers to where the MS spectrum is shown in Fig. S-2 (Supplementary information). MS/MS spectra of precursor masses 2087.0807, 1842.9747, and 1360.7258 Da can be found in Fig. S-3 (Supplementary information)
The interaction sites in Hsp21 and MDH
The identified Hsp21–MDH crosslinks only involved three different amino acid residues of Hsp21 (Table 1), the protein N-terminal residues M1, K27, and K173 (Fig. 4a), whereas a range of different Hsp21 residues were involved in Hsp21–Hsp21 crosslinks (Supplementary Table S1), so it is noteworthy that the identified Hsp21–MDH exclusively involved these three Hsp21 residues. The identified Hsp21-MDH crosslinks (Table 1) involved eight amino acid residues of MDH: A1, K81, K133, K215, K273, K277, K304, and K305. The identified Hsp21–MDH crosslinks supposedly represent the interaction between Hsp21 and partially unfolded MDH. Partially unfolded MDH may rather be a collection of differently unfolded species than one defined structure. The structure is not known for such an intermediate MDH, so these eight crosslinked lysine residues were instead mapped into the crystal structure of folded MDH (Fig. 4b), to provide a hint as to which regions of MDH start to unfold first upon heat stress, as further commented in the “Discussion.”
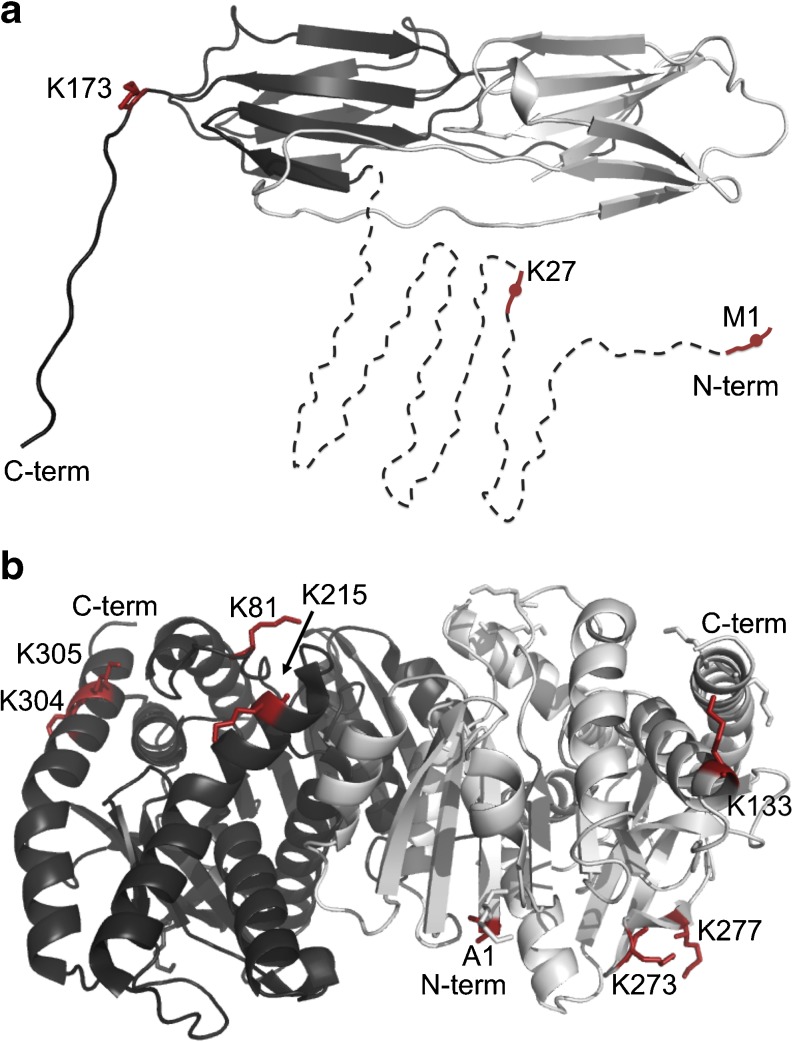
Fig. 4.
Residues involved in Hsp21–MDH crosslinks visualized in the 3D structures of Hsp21 and MDH. a Location of the three crosslinking Hsp21 residues M1, K27, and K173 in the Hsp21 dimeric subunit of the Hsp21 dodecamer. The crosslinking residues are only indicated on one of the two monomers shown. Residues M1 and K27 are within the predicted disordered N-terminal region (residues 1–84, dashed). b Location of the eight crosslinking MDH residues in the dimeric structure of MDH (PDB ID 1MLD). The dimer is made up of two monomers; one is colored dark gray, the other light gray. All eight residues crosslinking to Hsp21 are shown as sticks in both monomers. Residues 81, 215, 304, and 305 are only labelled in the dark gray monomer, and residues 1, 133, 273, and 277 are only labelled in the light gray monomer. a, b were prepared with PyMOL (www.pymol.org) (DeLano 2002)
Discussion
Crosslinks detected between Hsp21 and MDH
The main objective of this study was to explore by crosslinking mass spectrometry the transient interactions between an sHsp, Hsp21, and the model substrate protein MDH incubated 30 min at 45 °C, in order to investigate the interactions without which MDH aggregation would occur. Using this approach, we have identified ten Hsp21-MDH crosslinks (Table 1), which predominantly represent interactions between the N-terminal region of Hsp21 and the C-terminal part of MDH.
Three Hsp21 residues are involved in the Hsp21–MDH crosslinks: M1, K27, and K173. These three residues are also among the most abundant in Hsp21–Hsp21 crosslinks (Fig. 5), as further discussed below. Eight MDH residues are involved in the Hsp21–MDH crosslinks, five of which are located in the C-terminal half of MDH. These data support a view where, at 45 °C, the N-terminal domain of Hsp21 interacts with the model substrate protein MDH, and that these transient interactions with the unfolding C-terminal part of MDH prevent the unfolding intermediate MDH from aggregating.
Fig. 5.
Abundance of Hsp21 residues involved in crosslinks. Observations of Hsp21 residues involved in the identified crosslinks were calculated and shown here as bars for each residue that can possibly get modified by the crosslinking reagent. One or several observations of the methionine-oxidized version of a crosslink were counted as one extra observation
Apart from analyzing which Hsp21–MDH crosslinks that could be detected, we also analyzed the crosslinks detected within Hsp21 itself and within MDH itself, during the interactions between Hsp21 and MDH (Supplementary Tables S-1 and S-2, Figs. 5 and 6). Regarding the crosslinks within Hsp21, many of the crosslinks detected in presence of MDH are the same as detected without MDH (Lambert et al. 2011b; Söderberg et al. 2012). The difference is merely that the M1 amine, an efficient crosslinking probe as discussed below, interacts predominantly with MDH or Hsp21 itself, depending on presence or absence of substrate. This suggests that Hsp21 is not largely different in terms of structure when it is interacting with MDH. Contrastingly, the crosslinks detected within MDH during its interaction with Hsp21 do not fit the crystal structure of natively folded MDH, providing experimental support to the notion that the conformation of MDH that interacts with Hsp21 is an unfolding intermediate (Lindner et al. 2001; McHaourab et al. 2009).
Fig. 6.
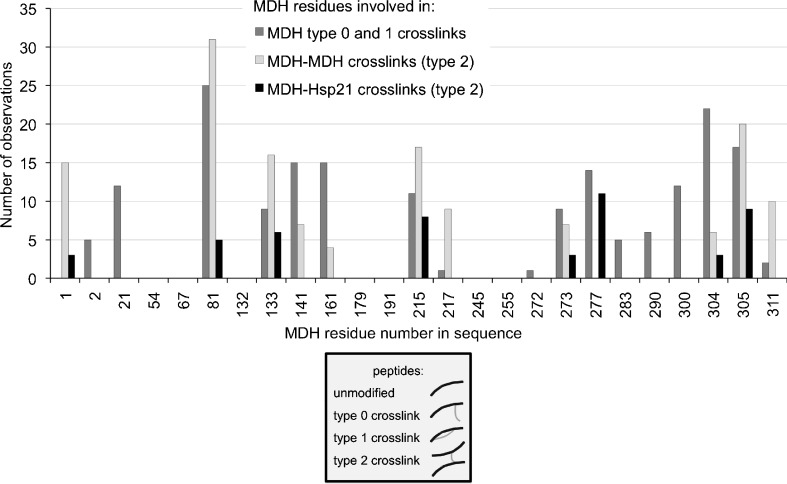
Abundance of MDH residues involved in crosslinks. Observations of MDH residues involved in the identified crosslinks were calculated and shown here as bars for each residue that can possibly get modified by the crosslinking reagent. One or several observations of the methionine-oxidized version of a crosslink were counted as one extra observation
Frequency of the different Hsp21 and MDH residues involved in crosslinks
Many different factors contribute to the final identification of crosslinks, which include technicalities such as the reactivity of the different lysine residues and the compatibility of different crosslinked peptides with detection by MALDI-TOF/TOF mass spectrometry. Comparing all lysine residues in both Hsp21 and MDH and the extent to which they were involved in various types of crossslinks (Figs. 5 and 6) gives an overview of the relative abundance of the different residues, taking the frequency of observation as a measure of abundance. There is a characteristic pattern of how often a certain residue is observed in crosslinks.
In case of Hsp21 (Fig. 5), the protein N-terminal methionine 1 (M1) accounts for most of the crosslinks within Hsp21. The M1 amine is particularly reactive because it has a lower pKa-value than lysine amine groups. That the different amines/lysine residues in proteins show different reactivity with amine-specific crosslinkers has previously been recognized, and acetylation of amine groups prior to crosslinking has been proposed to avoid exclusive identification of crosslinks involving the most reactive lysine residues only (Guo et al. 2008). Furthermore, the M1 amine is also located at the very end of a flexible N-terminal arm in a disordered region of Hsp21. The flexibility and reactivity of M1 may cause it to act as a very efficient crosslinking probe. In presence of MDH, the number of crosslinks M1 makes to Hsp21 itself is reduced compared with the situation where no MDH is present (Lambert et al. 2011b; Söderberg et al. 2012), and instead, the Hsp21 M1 amine crosslinks to MDH (Table 1). The residue most often observed after M1 is lysine 173 (K173). As well- known among protein mass spectrometrists, peptides are very differently ionized and detected in a mass spectrum, and both the tryptic peptides containing M1 (M1QDQR5) and K173 (T172KVER176) are relatively short and R-terminated, enhancing the likelihood of detection by MALDI-TOF/TOF mass spectrometry. Thus, two of the three lysine residues Hsp21 detected in crosslinks with MDH are located in the most abundantly detected Hsp21-peptides. For MDH (Fig. 6), it is seen that many, though not all, of the lysine residues detected in crosslinks with Hsp21 are actually the most abundantly detected.
The frequency of observation as a measure of abundance presented in the overviews in Figs. 5 and 6 shows that some lysine residues are more easily detected than others, in crosslinks within as well as between proteins. This abundance correlates also very well with the lysine residue distribution in detected dead-end crosslinks (Supplemental Tables S-3 and S-4). While the data need to be interpreted bearing this in mind, the crosslinks listed in Table 1 are representative of true contacts between Hsp21 and MDH.
Interactions between the N-terminal region of Hsp21 and the C-terminal region of MDH
Hsp21 residues M1 and K27, which are over-represented in Hsp21-MDH crosslinks, are part of the N-terminal region of Hsp21. This part of Hsp21 is a disordered, methionine-rich domain and has previously been suggested to be substrate-binding (Harndahl et al. 2001). For other sHsps, the N-terminal region has also previously been shown to be implicated in substrate-binding (Jaya et al. 2009; Sharma et al. 2000). Residues M1 and K27, and K18, are the only amine-group-containing amino acid residues out of the 81 amino acid residues in the disordered N-terminal region of wild-type Hsp21. By making R-to-K substitutions, it may be possible to further validate the importance of this region for substrate recognition.
Five of eight of the MDH residues involved in the identified Hsp21-MDH crosslinks (Table 1) are located in the C-terminal region of MDH. If we interpret the crosslinking residues to represent regions that had partially unfolded when interacting with Hsp21, the C-terminal regions of both monomers could be regions that start to unfold first upon heat stress. The absence of Hsp21-MDH crosslinks involving lysines in the proximity of the dimer interface (residues 21, 54, and 217) indicates that other regions of MDH may lose their tertiary structure before the monomer–monomer interaction is lost. The partial unfolding of the C-terminal regions while the MDH dimer is still intact makes sense with the N-terminal regions located closer to the dimer interface (Fig. 4b). Our results also compare well with a hydrogen/deuterium exchange study of sHsp–substrate complexes (Cheng et al. 2008). Partially unfolded MDH in complex with sHsp was characterized by measuring H/D exchange with mass spectrometry on tryptic peptides. Peptides with high H/D exchange levels imply unstructured protein regions, and six of the eight MDH residues involved in our identified Hsp21–MDH crosslinks are located in peptides with H/D exchange of >80 % after 5 s (MDH residues A1, K81, K273, K277, K304, and K305). MDH residues K133 and K215 are located in peptides with H/D exchange after 5 s of >40 %, and >60 %, resp.
The main problem with crosslinking mass spectrometry to study protein–protein interactions is the low yield of crosslinks. For the identification of the crosslinks listed in Table 1, we had to analyze a large number of datasets. Low yields of identified crosslinks with mass spectrometry can mainly be attributed to technical reasons, and the development of ways to tackle this problem is an intensive area of research in the field (Fritzsche et al. 2012; Leitner et al. 2012). Particularly, cation exchange chromatography (SCX) and affinity tags incorporated in the crosslinking reagent seem promising strategies to enrich crosslinks and will hopefully improve the yields and the intensity in the MS spectra also for crosslinks representing the sHsp–substrate interaction. Because the levels of Hsp21–MDH crosslinks appear to be only just above the lower limit of detection by the crosslinking mass spectrometry approach used here, we are most probably under-sampling the crosslinks present in the sample, and it cannot be excluded that there are also other interactions between Hsp21 and MDH than those detected.
Conclusion
To summarize, we have shown that Hsp21 can protect model substrate proteins at approximately 1:1 molar ratios. We investigated which crosslinks between Hsp21 and the model substrate protein MDH we could identify under conditions of transient Hsp21–MDH interactions after 30 min at 45 °C. Using the lysine-specific crosslinking reagent BS3, subsequent tryptic digestion, and LC-MALDI-TOF/TOF mass spectrometry, we could detect ten different sHsp–substrate crosslinks. Without excluding the possibility of additional interactions than those detected, we can conclude that the data points at a partially unfolded C-terminal region of substrate protein MDH, which is rescued from aggregation by the reversible interactions with the N-terminal region of Hsp21.
Electronic supplementary material
(DOCX 310 kb)
Acknowledgments
The Carl Tryggers Foundation (CTS 09:96) is thanked for financial support.
References
- Ahrman E, Gustavsson N, Hultschig C, Boelens WC, Emanuelsson CS. Small heat shock proteins prevent aggregation of citrate synthase and bind to the N-terminal region which is absent in thermostable forms of citrate synthase. Extremophiles. 2007;11:659–666. doi: 10.1007/s00792-007-0080-3. [DOI] [PubMed] [Google Scholar]
- Ahrman E, Lambert W, Aquilina JA, Robinson CV, Emanuelsson CS. Chemical cross-linking of the chloroplast localized small heat-shock protein, Hsp21, and the model substrate citrate synthase. Protein Sci. 2007;16:1464–1478. doi: 10.1110/ps.072831607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin AJ, Hilton GR, Lioe H, Bagneris C, Benesch JL, Kay LE. Quaternary dynamics of alphaB-crystallin as a direct consequence of localised tertiary fluctuations in the C-terminus. J Mol Biol. 2011;413:310–320. doi: 10.1016/j.jmb.2011.07.017. [DOI] [PubMed] [Google Scholar]
- Baldwin AJ, Lioe H, Hilton GR, Baker LA, Rubinstein JL, Kay LE, Benesch JL. The polydispersity of alphaB-crystallin is rationalized by an interconverting polyhedral architecture. Structure. 2011;19:1855–1863. doi: 10.1016/j.str.2011.09.015. [DOI] [PubMed] [Google Scholar]
- Baldwin AJ, Lioe H, Robinson CV, Kay LE, Benesch JL. alphaB-Crystallin polydispersity is a consequence of unbiased quaternary dynamics. J Mol Biol. 2011;413:297–309. doi: 10.1016/j.jmb.2011.07.016. [DOI] [PubMed] [Google Scholar]
- Basha E, O’Neill H, Vierling E (2011) Small heat shock proteins and alpha-crystallins: dynamic proteins with flexible functions. Trends Biochem Sci 37:106-117 [DOI] [PMC free article] [PubMed]
- Beck M, Malmstrom JA, Lange V, Schmidt A, Deutsch EW, Aebersold R. Visual proteomics of the human pathogen Leptospira interrogans. Nat Methods. 2009;6:817–823. doi: 10.1038/nmeth.1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bova MP, Ding LL, Horwitz J, Fung BK. Subunit exchange of alphaA-crystallin. J Biol Chem. 1997;272:29511–29517. doi: 10.1074/jbc.272.47.29511. [DOI] [PubMed] [Google Scholar]
- Bova MP, McHaourab HS, Han Y, Fung BK. Subunit exchange of small heat shock proteins. Analysis of oligomer formation of alphaA-crystallin and Hsp27 by fluorescence resonance energy transfer and site-directed truncations. J Biol Chem. 2000;275:1035–1042. doi: 10.1074/jbc.275.2.1035. [DOI] [PubMed] [Google Scholar]
- Cheng G, Basha E, Wysocki VH, Vierling E. Insights into small heat shock protein and substrate structure during chaperone action derived from hydrogen/deuterium exchange and mass spectrometry. J Biol Chem. 2008;283:26634–26642. doi: 10.1074/jbc.M802946200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano W (2002) The PyMOL molecular graphics system. DeLano Scientific, San Carlos, CA, USA
- Ecroyd H, Carver JA. Unraveling the mysteries of protein folding and misfolding. IUBMB Life. 2008;60:769–774. doi: 10.1002/iub.117. [DOI] [PubMed] [Google Scholar]
- Ehrnsperger M, Graber S, Gaestel M, Buchner J. Binding of non-native protein to Hsp25 during heat shock creates a reservoir of folding intermediates for reactivation. EMBO J. 1997;16:221–229. doi: 10.1093/emboj/16.2.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyles SJ, Gierasch LM. Nature’s molecular sponges: small heat shock proteins grow into their chaperone roles. Proc Natl Acad Sci USA. 2010;107:2727–2728. doi: 10.1073/pnas.0915160107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritzsche R, Ihling CH, Gotze M, Sinz A. Optimizing the enrichment of cross-linked products for mass spectrometric protein analysis. Rapid Commun Mass Spectrom. 2012;26:653–658. doi: 10.1002/rcm.6150. [DOI] [PubMed] [Google Scholar]
- Giese KC, Basha E, Catague BY, Vierling E. Evidence for an essential function of the N terminus of a small heat shock protein in vivo, independent of in vitro chaperone activity. Proc Natl Acad Sci USA. 2005;102:18896–18901. doi: 10.1073/pnas.0506169103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Bandyopadhyay P, Schilling B, Young MM, Fujii N, Aynechi T, Guy RK, Kuntz ID, Gibson BW. Partial acetylation of lysine residues improves intraprotein cross-linking. Anal Chem. 2008;80:951–960. doi: 10.1021/ac701636w. [DOI] [PubMed] [Google Scholar]
- Harndahl U, Hall RB, Osteryoung KW, Vierling E, Bornman JF, Sundby C. The chloroplast small heat shock protein undergoes oxidation-dependent conformational changes and may protect plants from oxidative stress. Cell Stress Chaperones. 1999;4:129–138. doi: 10.1379/1466-1268(1999)004<0129:TCSHSP>2.3.CO;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harndahl U, Kokke BP, Gustavsson N, Linse S, Berggren K, Tjerneld F, Boelens WC, Sundby C. The chaperone-like activity of a small heat shock protein is lost after sulfoxidation of conserved methionines in a surface-exposed amphipathic alpha-helix. Biochim Biophys Acta. 2001;1545:227–237. doi: 10.1016/S0167-4838(00)00280-6. [DOI] [PubMed] [Google Scholar]
- Hartl FU, Bracher A, Hayer-Hartl M. Molecular chaperones in protein folding and proteostasis. Nature. 2011;475:324–332. doi: 10.1038/nature10317. [DOI] [PubMed] [Google Scholar]
- Haslbeck M, Franzmann T, Weinfurtner D, Buchner J. Some like it hot: the structure and function of small heat-shock proteins. Nat Struct Mol Biol. 2005;12:842–846. doi: 10.1038/nsmb993. [DOI] [PubMed] [Google Scholar]
- Jaya N, Garcia V, Vierling E. Substrate binding site flexibility of the small heat shock protein molecular chaperones. Proc Natl Acad Sci U S A. 2009;106:15604–15609. doi: 10.1073/pnas.0902177106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KK, Kim R, Kim SH. Crystal structure of a small heat-shock protein. Nature. 1998;394:595–599. doi: 10.1038/29106. [DOI] [PubMed] [Google Scholar]
- Laganowsky A, Eisenberg D. Non-3D domain swapped crystal structure of truncated zebrafish alphaA crystallin. Protein Sci. 2010;19:1978–1984. doi: 10.1002/pro.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laganowsky A, Benesch JL, Landau M, Ding L, Sawaya MR, Cascio D, Huang Q, Robinson CV, Horwitz J, Eisenberg D. Crystal structures of truncated alphaA and alphaB crystallins reveal structural mechanisms of polydispersity important for eye lens function. Protein Sci. 2010;19:1031–1043. doi: 10.1002/pro.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert W, Koeck PJ, Ahrman E, Purhonen P, Cheng K, Elmlund D, Hebert H, Emanuelsson C. Subunit arrangement in the dodecameric chloroplast small heat shock protein Hsp21. Protein Sci. 2011;20:291–301. doi: 10.1002/pro.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert W, Soderberg CA, Rutsdottir G, Boelens WC, Emanuelsson C. Thiol-exchange in DTSSP crosslinked peptides is proportional to cysteine content and precisely controlled in crosslink detection by two-step LC-MALDI MSMS. Protein Sci. 2011;20:1682–1691. doi: 10.1002/pro.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence AM, Besir HU (2009) Staining of proteins in gels with Coomassie G-250 without organic solvent and acetic acid. J Vis Exp pii: 1350 [DOI] [PMC free article] [PubMed]
- Leitner A, Walzthoeni T, Kahraman A, Herzog F, Rinner O, Beck M, Aebersold R. Probing native protein structures by chemical cross-linking, mass spectrometry, and bioinformatics. Mol Cell Proteomics. 2010;9:1634–1649. doi: 10.1074/mcp.R000001-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitner A, Reischl R, Walzthoeni T, Herzog F, Bohn S, Foerster F, Aebersold R (2012) Expanding the chemical cross-linking toolbox by the use of multiple proteases and enrichment by size exclusion chromatography. Mol Cell Proteomics 11(3):M111.014126 [DOI] [PMC free article] [PubMed]
- Lindner RA, Treweek TM, Carver JA. The molecular chaperone alpha-crystallin is in kinetic competition with aggregation to stabilize a monomeric molten-globule form of alpha-lactalbumin. Biochem J. 2001;354:79–87. doi: 10.1042/0264-6021:3540079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHaourab HS, Godar JA, Stewart PL. Structure and mechanism of protein stability sensors: chaperone activity of small heat shock proteins. Biochemistry. 2009;48:3828–3837. doi: 10.1021/bi900212j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mymrikov EV, Seit-Nebi AS, Gusev NB. Heterooligomeric complexes of human small heat shock proteins. Cell Stress Chaperones. 2012;17:157–169. doi: 10.1007/s12192-011-0296-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamoto H, Vigh L. The small heat shock proteins and their clients. Cell Mol Life Sci. 2007;64:294–306. doi: 10.1007/s00018-006-6321-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Painter AJ, Jaya N, Basha E, Vierling E, Robinson CV, Benesch JL. Real-time monitoring of protein complexes reveals their quaternary organization and dynamics. Chem Biol. 2008;15:246–253. doi: 10.1016/j.chembiol.2008.01.009. [DOI] [PubMed] [Google Scholar]
- Peri S, Steen H, Pandey A. GPMAW—a software tool for analyzing proteins and peptides. Trends Biochem Sci. 2001;26:687–689. doi: 10.1016/S0968-0004(01)01954-5. [DOI] [PubMed] [Google Scholar]
- Poulain P, Gelly JC, Flatters D. Detection and architecture of small heat shock protein monomers. PLoS One. 2010;5:e9990. doi: 10.1371/journal.pone.0009990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma KK, Kumar RS, Kumar GS, Quinn PT. Synthesis and characterization of a peptide identified as a functional element in alphaA-crystallin. J Biol Chem. 2000;275:3767–3771. doi: 10.1074/jbc.275.6.3767. [DOI] [PubMed] [Google Scholar]
- Singh P, Panchaud A, Goodlett DR. Chemical cross-linking and mass spectrometry as a low-resolution protein structure determination technique. Anal Chem. 2010;82:2636–2642. doi: 10.1021/ac1000724. [DOI] [PubMed] [Google Scholar]
- Sinz A. Chemical cross-linking and mass spectrometry to map three-dimensional protein structures and protein–protein interactions. Mass Spectrom Rev. 2006;25:663–682. doi: 10.1002/mas.20082. [DOI] [PubMed] [Google Scholar]
- Söderberg CAG, Lambert W, Kjellström S, Wiegandt A, Peterson-Wulff R, Månsson C, Rutsdottir G, Emanuelsson C (2012) Detection of crosslinks within and between proteins by LC-MALDI-TOFTOF and the software FINDX to reduce the MSMS-data to acquire for validation. PLoS ONE 7(6):e38927 [DOI] [PMC free article] [PubMed]
- Stengel F, Baldwin AJ, Painter AJ, Jaya N, Basha E, Kay LE, Vierling E, Robinson CV, Benesch JL. Quaternary dynamics and plasticity underlie small heat shock protein chaperone function. Proc Natl Acad Sci USA. 2010;107:2007–2012. doi: 10.1073/pnas.0910126107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoscheck CM. Quantitation of protein. Methods Enzymol. 1990;182:50–68. doi: 10.1016/0076-6879(90)82008-P. [DOI] [PubMed] [Google Scholar]
- Sun Y, MacRae TH. The small heat shock proteins and their role in human disease. FEBS J. 2005;272:2613–2627. doi: 10.1111/j.1742-4658.2005.04708.x. [DOI] [PubMed] [Google Scholar]
- Tyedmers J, Mogk A, Bukau B. Cellular strategies for controlling protein aggregation. Nat Rev Mol Cell Biol. 2010;11:777–788. doi: 10.1038/nrm2993. [DOI] [PubMed] [Google Scholar]
- Montfort RL, Basha E, Friedrich KL, Slingsby C, Vierling E. Crystal structure and assembly of a eukaryotic small heat shock protein. Nat Struct Biol. 2001;8:1025–1030. doi: 10.1038/nsb722. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 310 kb)