Abstract
In the endoplasmic reticulum (ER), secretory and membrane proteins are properly folded and modified, and the failure of these processes leads to ER stress. At the same time, unfolded protein response (UPR) genes are activated to maintain homeostasis. Despite the thorough characterization of the individual gene regulation of UPR genes to date, further investigation of the mutual regulation among UPR genes is required to understand the complex mechanism underlying the ER stress response. In this study, we aimed to reveal a gene regulatory network formed by UPR genes, including immunoglobulin heavy chain-binding protein (BiP), X-box binding protein 1 (XBP1), C/EBP [CCAAT/enhancer-binding protein]-homologous protein (CHOP), PKR-like endoplasmic reticulum kinase (PERK), inositol-requiring 1 (IRE1), activating transcription factor 6 (ATF6), and ATF4. For this purpose, we focused on promoter-luciferase reporters for BiP, XBP1, and CHOP genes, which bear an ER stress response element (ERSE), and p5 × ATF6-GL3, which bears an unfolded protein response element (UPRE). We demonstrated that the luciferase activities of the BiP and CHOP promoters were upregulated by all the UPR genes, whereas those of the XBP1 promoter and p5 × ATF6-GL3 were upregulated by all the UPR genes except for BiP, CHOP, and ATF4 in HeLa cells. Therefore, an ERSE- and UPRE-centered gene regulatory network of UPR genes could be responsible for the robustness of the ER stress response. Finally, we revealed that BiP protein was degraded when cells were treated with DNA-damaging reagents, such as etoposide and doxorubicin; this finding suggests that the expression level of BiP is tightly regulated at the post-translational level, rather than at the transcriptional level, in the presence of DNA damage.
Keywords: ER stress, Unfolded protein response, ERSE, UPRE, BiP, Gene regulation
Introduction
The endoplasmic reticulum (ER) is responsible for the proper protein folding and modification of approximately one third of all cellular secretory and membrane proteins. Abnormalities such as the accumulation of unfolded proteins because of environmental deterioration, canceration, or senescence can interfere with these processes, leading to a dysfunctional situation known as ER stress. To avoid difficulties resulting from ER stress, the ER implements a protective response: the so-called unfolded protein response (UPR). The UPR is a system composed of multiple shields including the translational arrest of nascent protein, the degradation of misfolded proteins, and transcriptional induction of ER chaperones. Genes that are responsible for the UPR must be transcribed quickly and efficiently in response to abnormalities (for further details, please refer to previous reviews by Malhotra and Kaufman 2007; Parmar and Schröder 2012).
Among the UPR genes, PKR-like endoplasmic reticulum kinase (PERK; alias, EIF2AK3), inositol-requiring 1 (IRE1; alias, ERN1), and activating transcription factor 6 (ATF6) are membrane proteins that regularly form a heterodimer with BiP (immunoglobulin heavy chain-binding protein; alias, GRP78, or HSPA5A); however, these proteins separate from BiP immediately after ER stress is sensed. Subsequently, PERK promotes the translation of the transcription factor ATF4, IRE1 cleaves the x-box binding protein 1 (XBP1) pre-mRNA to produce mature XBP1 mRNA as an activated form, and ATF6 travels into the nucleus after cleavage. These XBP1, ATF4, and ATF6 proteins promote the transcription of molecular chaperons to cope with ER stress. On the other hand, apoptosis can be induced through the activation of c-Jun N-terminal kinase by IRE1 or the transcriptional induction of C/EBP [CCAAT/enhancer-binding protein]-homologous protein (CHOP; alias, DDIT3) by XBP1, ATF4, or ATF6 proteins (for further details, please refer to previous reviews by Malhotra and Kaufman 2007; Hetz 2012).
The importance of UPR gene regulation has been recognized not only in a physiological sense, but also as a basis for understanding the etiologies underlying serious human diseases such as neurodegenerative disorders and cancer (refer to reviews by Zhang and Kaufman 2006; Yoshida 2007). For instance, BiP plays a central role in the UPR used to avoid apoptosis and could be applied to protect neurons from abnormal death, attracting the attention of drug developers. Regarding BiP gene regulation, the transcriptional control of BiP has been well characterized (refer to reviews by Li and Lee 2006; Lee 2007); however, the proteolytic mechanism of BiP is poorly understood.
In this study, we investigated how the ER stress response element (ERSE) and the unfolded protein response element (UPRE) are regulated by UPR genes, including BiP, CHOP, PERK, IRE1, ATF6, and ATF4, through transcriptional analyses, confirming the existence of a regulatory network responsible for the robustness seen in the concurrent expression of UPR genes upon the sensing of ER stress. In addition, we found that topoisomerase II inhibitor affected the expression level of BiP through the proteasome-mediated proteolysis of BiP protein, rather than the transcriptional regulation of the BiP gene.
Materials and methods
Cells and reagents
HeLa cells were cultured in Minimum Essential Medium (cat. No. 11095, Life Technologies, Carlsbad, CA) supplemented with 10 % fetal bovine serum, 1 % non-essential amino acids (Life Technologies), and antibiotic-antimycotics (Life Technologies). Thapsigargin (Sigma, Saint Louis, MO), tunicamycin (Sigma), etoposide (Wako Pure Chemical Industries, Osaka, Japan), doxorubicin (Wako Pure Chemical Industries), 5-fluorouracil (5-FU, Sigma), proteasome inhibitor I, lactacystin, MG132, ALLN, clasto-lactacystin β-lactone, epoxomicin, ubiquitin aldehyde are purchased from Merck Biosciences (former Calbiochem, Darmstadt, Germany) and were dissolved in dimethyl sulfoxide (DMSO; Wako Pure Chemical Industries) before use.
Reverse-transcription polymerase chain reaction
Total RNA (500 ng) was extracted with a RNeasy Mini kit (Qiagen, Venlo, the Netherlands) and was treated with deoxyribonuclease I (Life Technologies), in accordance with the manufacturer’s instructions. The cDNA was synthesized with a High-capacity cDNA Reverse Transcription kit (Life Technologies). The PCR reaction mixture (20 μL) contained 1 × buffer, 200 μM of dNTPs, 400 nM of primers, 1 mM MgSO4, 1 unit of KOD plus DNA polymerase (Toyobo, Osaka, Japan), and twentieth part of synthesized cDNA. Primers used were listed in Table 1. These primers were designed to step over an exon. Refseq accession numbers and length of PCR products (annealing temperatures in degrees Celsius) are as follows, BiP, NM_005347 and 420-bp (55); XBP1, NM_005080 and 440-bp (54); CHOP, NM_004083 and 280-bp (57); PERK, NM_004836 and 520-bp (53); IRE1, NM_001433 and 403-bp (54); ATF6, NM_007348 and 380-bp (57); and ATF4, NM_001675 and 380-bp (54). PCR products were separated in 1.0–1.5 % agarose gel and visualized under ultraviolet irradiation. Gel images were quantified with Quantity One (Bio-Rad Laboratories, Hercules, CA). Values of the quantified band images were normalized with glyceraldehyde-3-phosphate dehydrogenase (GAPDH), and the background value was subtracted. For the cDNA panel analysis, 2.5 μL of cDNA purchased from Takara Bio (Otsu, Japan) was used (Human MTC panels I and II cDNA panel). GAPDH primer in the kit was used as a control.
Table 1.
List of primers used for RT-PCR of the UPR genes
| Gene | Sense primer | Antisense primer |
|---|---|---|
| BiP | GTATGGTGCTGCTGTCCAGG | GGTGTCAGGCGATTCTGGTC |
| XBP1 | GGATGGATGCCCTGGTTGCT | CTTGGCTCTCTGTCTCAGAG |
| CHOP | GCTTGGCTGACTGAGGAGGA | CCTCTACTTCCCTGGTCAGG |
| PERK | AGATCGCAGAGGCAGTGGAG | CGAGACCTCTGTCTGAGCAC |
| IRE1 | GGGTCTGAGGAAGGTGATGC | CAGTGGGGTTTCATGGTGTC |
| ATF6 | CATCCGCAGAAGGGGAGACA | CAGGGTCCCACGCTCAGTTT |
| ATF4 | AATGGCTGGCTGTGGATGGG | CACTTCACTGCCCAGCTCTA |
For a quantitative analysis, Taqman Gene Expression Assay was performed according to the manufacturer’s protocol (Life Technologies). The PCR mixtures included the Taqman Gene Expression Assay Primer for human BiP mRNA (assay ID: Hs99999174_m1) and human XBP1 mRNA (assay ID: Hs00964360_m1) and TaqMan Universal PCR Mix (Life Technologies) in a total reaction volume of 20 μL. Reactions were performed with the 7500 Standard program on a 7500 Fast Real-Time PCR System (Life Technologies). Cycling parameters were 95 °C for 10 min and then 40 cycles of 95 °C for 15 s, annealed/extended at 60 °C for 1 min. The cycle threshold (Ct) values, corresponding to the PCR cycle number at which fluorescence emission reached a threshold above baseline emission, were determined, and mRNA expression values were calculated using GAPDH as an endogenous control (Life Technologies) following the comparative Ct (ΔΔCt) method. All data represent mean and standard deviation of two experiments (in triplicate).
Plasmids
Plasmids were kindly provided as follows, pGL3-XBP1 and pGL3-CHOP (Dr. Hiderou Yoshida, Kyoto University), pcDNA3-BiP (Dr. Marina Gorbatyuk, University of North Texas Health Science Center), pcDNA-XBP1 (spliced; Dr. Kazutoshi Mori, Kyoto University) (Yoshida et al. 2001), pcDNA3.1-Myc-CHOP (Dr. Hidetoshi Hayashi, Nagoya City University), pcDNA-myc-mPERK wt and pcDNA-myc-mPERK K618A (Dr. Antonis E. Koromilas, McGill University), and pCMV-ATF4 (Dr. Guozhi Xiao, University of Pittsburgh). We obtained IRE1α-pcDNA3.EGFP and IRE1α KA-pcDNA3.EGFP from Addgene (plasmid 13009 and 13010; Cambridge, MA) deposited by Dr. Fumihiko Urano (University of Massachusetts Medical School) (Lipson et al. 2006), and also pCGN-ATF6, pCGN-ATF6(1-373), pCGN-ATF6(1-373)m1 and p5 × ATF6-GL3 from Addgene (plasmids 11974, 27173, 27174, and 11976) deposited by Dr. Ron Prywe (Columbia University) (Zhu et al. 1997; Wang et al. 2000).
To construct a pGL3 luciferase reporter plasmid that bears promoter region of each UPR genes upstream of luciferase gene, we amplified an assumed promoter region by PCR from human genomic DNA (Promega, Madison, WI) as the template DNA with primers listed in Table 2. 5′ end of sense and antisense primers were added with KpnI and BglII recognition sequence, respectively. Primers were designed with the GenBank accession number as follows, BiP (AL354710), PERK (AC062029), IRE1 (AC025362), ATF6 (AL359541), and ATF4 (AL022312). The amplified DNA was inserted into pGL3-Basic (Promega). We named each reporter plasmid for UPR genes based on the transcriptional start site as +1 as follows: pGL3-BiP, −761/+120; pGL3-PERK, −874/+26; pGL3-IRE1, −977/+23; pGL3-ATF6, −910/+60; and pGL3-ATF4, −723/+188. By sequencing of these plasmids, we found that T to A (−687) and A to G (−49) nucleotide substitution in pGL3-ATF6 (−910/+60) and C to T (−704) nucleotide substitution in pGL3-ATF4 (−723/+188).
Table 2.
List of primers used for the construction of promoter reporter plasmids for the UPR genes
| Plasmid | Sence primer (KpnI) | Antisense primer (BglII) |
|---|---|---|
| pGL3-BiP (−761/+120) | GGGGTACCACTGGAGTGGGTTGCCACAG | GAAGATCTAATACAGGCCGCGGCGCTTC |
| pGL3-PERK (−874/+26) | GGGGTACCGTCTTCTCCACTCTGCCCTT | GAAGATCTCTGAGTGACAGCCTATCTCG |
| pGL3-IRE1 (−977/+23) | GGGGTACCCTGGGCTCCAAATCTCAG | GAAGATCTGCGGACGCAGAACTGACTAG |
| pGL3-ATF6 (−910/+60) | GGGGTACCAGTTTGGAGGGTTCTGGGA | GAAGATCTGTTCCTTCTCCCTGGAACTC |
| pGL3-ATF4 (−723/+188) | GGGGTACCAACCGAAGGACGCGCAGGCT | GAAGATCTGCCATGGCTTAAGCCGCTGG |
The underlined sequences in the sense and antisense primers are the KpnI and BglII recognition sequences, respectively
Luciferase assay
Each 200 ng of the firefly luciferase reporter plasmid and/or expression vector and 0.6 ng of the Renilla luciferase reporter plasmid (pRL-TK, Promega) per 24-well dish were transfected with FuGENE6 (Roche, Basel, Switzerland), in accordance with the manufacturer’s instructions. Cells were lysed 24 h after transfection and light intensity was quantified in a GloMax 20/20n Luminometer (Promega) with Dual Luciferase Reporter Assay Kit (Promega). Experiments were performed at least in triplicate. As control for the transfection efficiency, the firefly luciferase activity values were normalized to the Renilla luciferase activity values. Statistical difference was determined by two-tailed Student’s t test (n = 3).
Caspase 3/7 assay
Cells were incubated with Caspase-Glo 3/7 substrate (Promega) for 1 h at room temperature, and the resulting activity was measured using a GloMax 20/20n Luminometer. The relative activity was presented by comparing relative light units obtained in the treated cells relative to those in the control value (treated with 0.1 % DMSO). Blanks were measured in wells containing 0.1 % DMSO or chemicals without cells. Statistical difference was determined by two-tailed Student’s t test (n = 4). P < 0.01 was considered as a statistical difference.
Western blot
Cells were harvested and lysed in RIPA lysis buffer on ice. The protein concentrations were determined using the Bio-Rad protein assay kit (Bio-Rad laboratories). The protein lysates were loaded onto each lane of a gel. Before performing sodium dodecyl sulfate polyacrylamide gel electrophoresis, the reaction was stopped by the addition of Laemmli sample buffer containing 100 mM of dithiothreitol. Cellular protein were electrophoresed on NuPAGE 4∼12 % Bis-Tris gel with MES running buffer (Life Technologies) and transferred to a Hybond-PVDF membrane (GE Healthcare, Piscataway, NJ). The membrane was first blocked using phosphate buffered saline containing 0.1 % Tween 20 and 5 % non-fat dried milk. The following antibodies were used: BiP (cat. No. 3177, Cell Signaling Technology, Beverley, MA), CHOP (cat. No. 2895, Cell Signaling Technology), p53 (cat. No. sc-6243, Santa Cruz Biotechnology, Santa Cruz, CA), calnexin (cat. No. M178-3, Medical and Biological Laboratories, Nagoya, Japan), protein disulfide isomerase (PDI; alias, P4HB; cat. No. ADI-SPA-891-D, Enzo Life Sciences, Exeter, UK), and GAPDH (cat. No. AM4300, Life Technologies). Alkaline phosphatase-labeled secondary antibodies were purchased from Promega. A Western Blue-stabilized substrate was used to detect the signals, according to the manufacturer’s protocol (Promega).
RNA interference
Stealth RNA interference (RNAi) for p53 (Life Technologies) and the Stealth RNAi Negative Control Kit with High GC (Life Technologies) was transfected into the cells using Lipofectamine 2000 (Life Technologies), according to the manufacturer’s protocol. Briefly, 125 pmol of RNA and 2.5 μL of transfection reagent were incubated in 0.5 ml of Opti-MEM I Reduced Serum Medium (Life Technologies) for 15 min to facilitate complex formation at room temperature. The resulting mixture was added to the cells cultured in 1.5 ml of Minimum Essential Medium in 6-well palate. After 24 h, RNA/transfection reagent mixture was re-added.
Results
Expression pattern of UPR genes
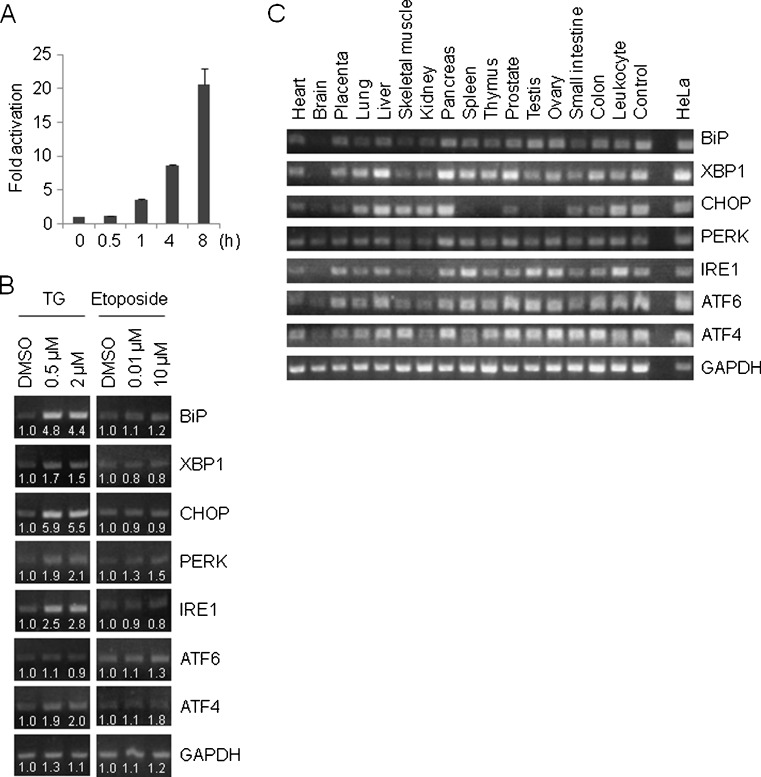
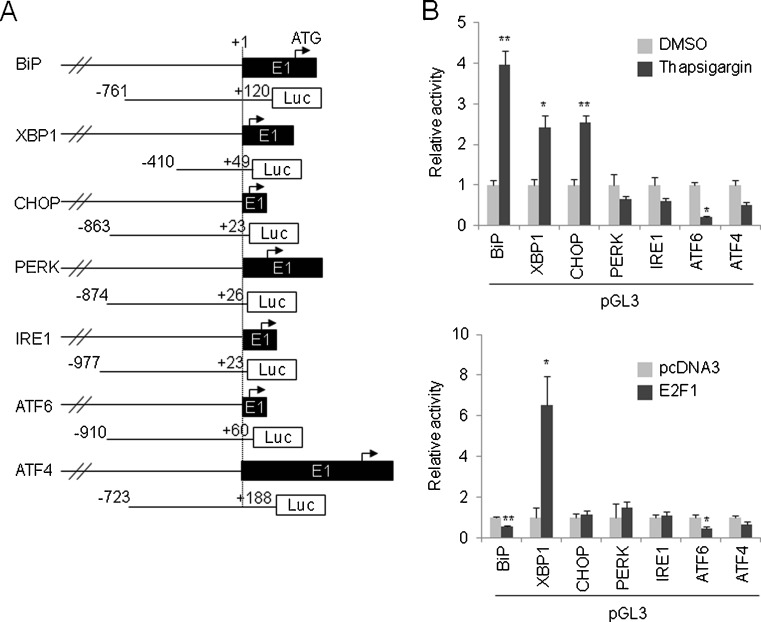
BiP, XBP1, and CHOP have been shown to be transcriptionally upregulated by thapsigargin and tunicamycin (Yoshida et al. 2000). We also confirmed that BiP mRNA was induced by thapsigargin shortly after thapsigargin treatment in HeLa cells using quantitative reverse-transcription (RT)-polymerase chain reaction (PCR) in a time-dependent manner (Fig. 1a). Similar to the case for BiP, UPR genes such as XBP1, CHOP, PERK, IRE1, and ATF4, but excluding ATF6, were also transcriptionally upregulated by thapsigargin (8-h treatment) but not by etoposide (48-h treatment) based on a semi-quantitative RT-PCR analysis (Fig. 1b). The ATF6 mRNA level has been shown to reach a relatively high level within a short time period (Namba et al. 2007). Indeed, ATF6 mRNA was induced to 1.5-fold and 1.6-fold levels by 0.5 and 2 μM of thapsigargin, respectively, compared with a DMSO-treated control, at 4 h after administration (data not shown). Similar to thapsigargin, the mRNA levels for UPR genes were also upregulated in tunicamycin-treated (2 μg/mL, 8 h) HeLa cells (data not shown). These observations suggest that the regulation of UPR genes could be inseparably connected. We further examined the mRNA expression patterns for UPR genes in human tissues and organs. As a result, out of the 17 tissues and organs that were examined, the UPR genes showed a tendency to be kept at a relatively low mRNA level in the brain, and CHOP transcription was shut down in the spleen, thymus, testis, and ovary (Fig. 1c). Taken together, these results suggest that uncovering the gene regulatory mechanisms of UPR genes would be of great value in understanding how UPR genes are deeply involved in pathophysiology. To this end, we established a promoter-luciferase reporter plasmids set for UPR genes by constructing pGL3-BiP, −761/+120; pGL3-PERK, −874/+26; pGL3-IRE1, −977/+23; pGL3-ATF6, −910/+60; and pGL3-ATF4, −723/+188 (see “Materials and methods”; Fig. 2a). The promoter regions were numbered relative to the transcriptional start site as +1. For XBP1 and CHOP promoter-luciferase reporter plasmids, we obtained pGL3-XBP1, −330/+129 and pGL3-CHOP, −870/+17 (Yoshida et al. 2000). These plasmids actually covered the −410/+49 and −863/+23 regions, respectively, based on the information of Refseq (NM_005080 and NM_004083, respectively) (Fig. 2a). Similar to BiP, the ERSE consensus (CCAAT-N9-CCACG) has been identified in XBP1 (reported as +33/+51 but changed to −49/−31 in this report) and CHOP (reported as −103/−85 and −93/−75 but changed to −97/−79 and −87/−69, respectively, in this report). We did not observe any ERSE in the vicinity of the transcriptional start site of the PERK, IRE1, ATF6, and ATF4 genes. In line with this observation, out of the seven pGL3 reporter plasmids, only ERSE-bearing reporter plasmids, such as pGL3-BiP, pGL3-XBP1, and pGL3-CHOP, resulted in the increased activity of luciferase at 8 h after thapsigargin exposure (0.5 μM) (Fig. 2b, top). This observation was unchanged when a higher concentration of thapsigargin was applied (2 μM) (data not shown). The decrease in the activity of PERK, IRE1, ATF6, and ATF4 promoter-reporters in thapsigargin-treated cells may be due to the PERK- or IRE1-mediated phosphorylation of eukaryotic translation initiation factor 2A, leading to translational attenuation.
Fig. 1.
Transcriptional regulation of UPR genes. a Time-dependent expression changes of BiP mRNA were detected using quantitative RT-PCR in thapsigargin (0.5 μM)-treated HeLa cells. GADH was used as an internal standard. The values represent the mean ± standard deviation (n = 2). b The changes in the mRNA expression level for UPR genes were determined using semi-quantitative RT-PCR in thapsigargin (TG; 8 h)- or etoposide (48 h)-treated HeLa cells. The band intensity was calculated using Quantity One. c mRNA expression pattern of UPR genes in human tissues and organs. HeLa cDNA was used as a positive control (rightmost lane)
Fig. 2.
Promoter analysis of UPR genes. a Schematic of promoter region of UPR genes and pGL3-based promoter-luciferase reporters. Exon 1 (black box with E1), the translation initiation codon (arrow), the position (number) relative to the start point of transcription (dotted line, designated as +1), and the luciferase gene (Luc within white box) are indicated. b Responsiveness of promoter region of UPR genes toward thapsigargin (top) and E2F1 (bottom). The indicated luciferase reporters were introduced into the cells, and 24 h later, the cells were treated with thapsigargin (0.5 μM) for 8 h (top), or the indicated luciferase reporters were co-introduced into the cells with the E2F1 expression vector for 24 h (bottom) and the luciferase activity was then measured. In each panel, pGL3-basic was used as an internal standard. The values represent the mean ± standard deviation (n = 3; *P < 0.05; **P < 0.01)
Recently, the negative regulation of the BiP promoter by E2F1 has been reported (Racek et al. 2008). In this previous report, pGL3-BiP (−371/+2) was used to analyze the BiP promoter. The −371/+2 should be −336/+41 based on Refseq NM_005347, and this region was fully covered by pGL3-BiP (−761/+120) in the present study. We coexpressed the E2F1 expression vector with the UPR gene promoter reporters and measured the luciferase activity. As a result, E2F1 suppressed the luciferase activity of pGL3-BiP, −761/+120 and pGL3-ATF6, −910/+60, whereas E2F1 approximately sextuplicated the luciferase activity of pGL3-XBP1, −410/+49 (Fig. 2b, bottom).
Considering the result obtained by RT-PCR, in which the mRNAs of UPR genes other than BiP, XBP1, and CHOP were induced by thapsigargin, using pGL3-based reporters for PERK, IRE1, ATF6, and ATF4 in subsequent assays seemed to be invalid. Therefore, we decided to employ pGL3-BiP, −761/+120; pGL3-XBP1, −410/+49; and pGL3-CHOP, −863/+23, all of which possess an ERSE and reacted to the ER stress-inducible chemical thapsigargin, in addition to the p5 × ATF6-GL3 reporter, which bears multiple UPREs, in subsequent assays.
ERSE- and UPRE-centered regulatory network among UPR genes
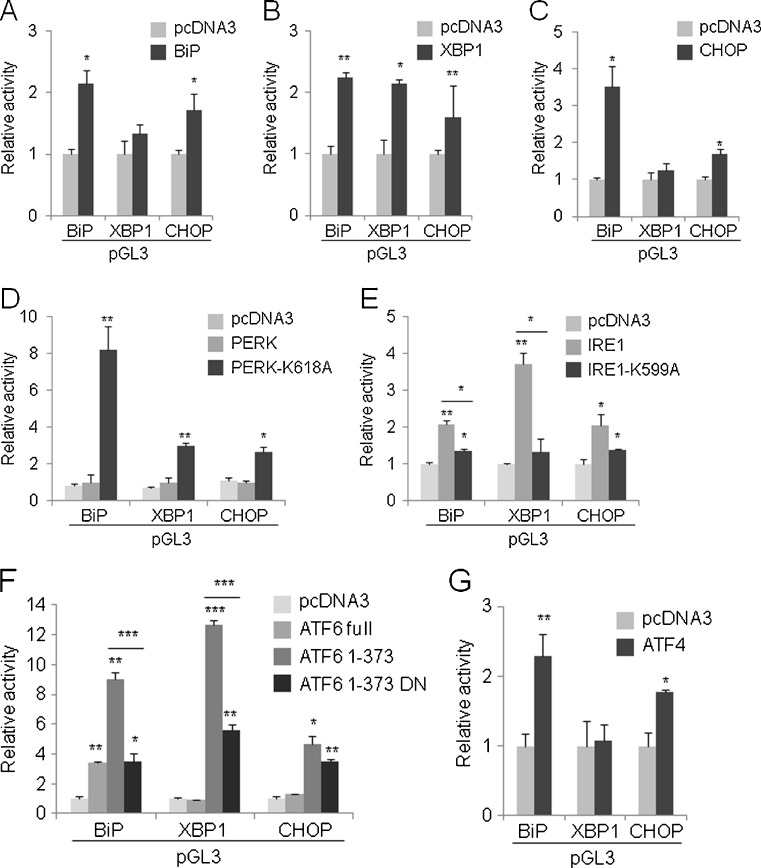
We continuously performed a luciferase assay in HeLa cells for pGL3-BiP, pGL3-XBP1, and pGL3-CHOP with expression vectors encoding seven UPR genes. Notably, BiP induced the luciferase activity driven by the BiP and CHOP promoters (Fig. 3a), XBP1 induced the luciferase activity driven by the BiP, XBP1, and CHOP promoters (Fig. 3b), and CHOP induced the luciferase activity driven by the BiP and CHOP promoters (Fig. 3c). These facts indicate that these three UPR genes have a complex regulatory loop; namely, BiP, XBP1, and CHOP have not only a positive feedback loop, but also form a triad structure where the three nodes are mutually facilitated, probably ensuring the concomitant expression of the three genes upon the sensing of ER stress.
Fig. 3.
Regulation of BiP, XBP1, and CHOP promoter by UPR genes. The expression vectors for the UPR genes were coexpressed with pGL3-BiP, pGL3-XBP1, or pGL3-CHOP for 24 h, and the luciferase activity was then measured. In each panel, pGL3-Basic was used as an internal standard. The values represent the mean ± standard deviation (n = 3; *P < 0.05; **P < 0.01; ***P < 0.001 versus pcDNA3). The asterisk above the line indicates a statistically significant difference between the below-the-line bar graph (e, f). a BiP, b spliced XBP1, c CHOP, d PERK and PERK-K618A, e IRE1 and IRE1-K599A, f ATF6 full-length, ATF6 1-373 and its dominant negative version (ATF6 1-373 DN), and g ATF4
A dominant negative mutant of PERK, PERK-K618A, exhibits a compensatory activation of XBP1 splicing and ATF6 cleavage (Yamaguchi et al. 2008). Actually, we demonstrated that wild-type PERK failed to affect the BiP, XBP1, and CHOP promoters, but PERK-K618A increased the luciferase activity driven by the BiP, XBP1, and CHOP promoters (Fig. 3d). On the other hand, IRE1 (or more accurately, IRE1α), showed a propensity to upregulate the luciferase activity driven by the BiP, XBP1, and CHOP promoters when compared with those obtained by the coexpression of a dominant negative form of IRE1, IRE1-K599A (Fig. 3e). In the case of ATF6, we first tested the ATF6 expression vector that can produce a full-length protein. Full-length ATF6 increased the luciferase activity driven by the BiP promoter only (Fig. 3f). Upon ER stress, ATF6 is cleaved to produce an N-terminal soluble form separate from the membrane region. We next tested a pCGN-ATF6(1-373) expression vector encoding the N-terminal active region of ATF6 and its mutant form pCGN-ATF6(1-373)m1, in which the 315-317 amino-acid residues were converted from KNR to TAA (Wang et al. 2000). Consequently, the N-terminal ATF6 increased the luciferase activity derived from all three promoter reporters, and the KNR-to-TAA mutant of ATF6 showed a relatively impaired activity level, compared with that obtained for the wild-type N-terminal ATF6 (Fig. 3f). ATF4 had a positive effect on the BiP and CHOP promoter reporters in the luciferase assay (Fig. 3g).
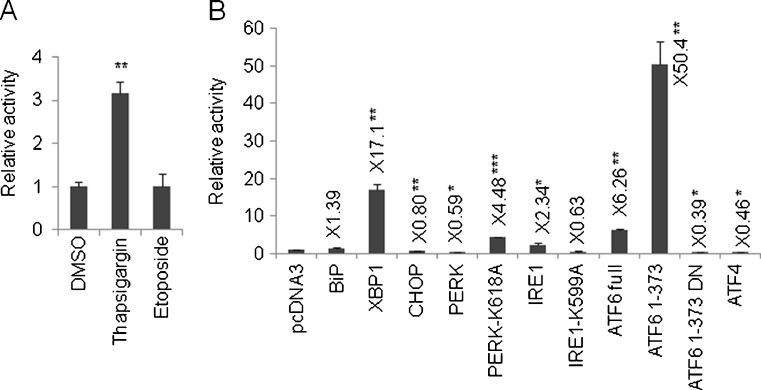
We then proceeded to analyze how the UPRE can be regulated by UPR genes. Prior to this analysis, we examined the responsiveness of p5 × ATF6-GL3, which possesses a UPRE consensus (TGACGTG(G/A)) upstream of the luciferase gene (Wang et al. 2000), to thapsigargin (0.5 μM, 8 h) and etoposide (10 μM, 48 h). The reporter plasmid was transfected into HeLa cells and 24 h later, the cells were treated with chemicals for the indicated time periods and the luciferase activity was measured. The luciferase activity of p5 × ATF6-GL3 was induced by thapsigargin, but not by etoposide (Fig. 4a). Next, the coexpression of p5 × ATF6-GL3 with UPR genes revealed that XBP1, PERK-K618A, and ATF6 were capable of augmenting UPRE-mediated reporter activity, compared with the value obtained using the pcDNA3 control (Fig. 4b). In particular, N-terminal soluble ATF6 enhanced the reporter activity to a remarkable extent (Fig. 4b). The ectopic overexpression of IRE1 reportedly promotes the reporter activity of p5 × ATF6-GL3 (Wang et al. 2000). In this experiment, IRE1 was also associated with an approximately two-fold increase in the luciferase activity of p5 × ATF6-GL3 (Fig. 4b). On the other hand, CHOP, PERK, ATF4, and the mutant form of ATF6 decreased the luciferase activity of p5 × ATF6-GL3, with statistically significant differences (Fig. 4b).
Fig. 4.
Regulation of UPRE by UPR genes. a p5 × ATF6-GL3 was transfected into the cells and 24 h later, thapsigargin (0.5 μM, 8 h) or etoposide (10 μM, 48 h) was added and the luciferase activity was measured. b p5 × ATF6-GL3 was cotransfected with the indicated expression vectors for 24 h, and the luciferase activity was measured. In each panel, pGL3-Basic was used as an internal standard. Values represent the mean ± standard deviation (n = 3; *P < 0.05; **P < 0.01; ***P < 0.001 versus pcDNA3). In (b), the average of the relative activity compared with pcDNA3 was inserted near the appropriate bar graph
Taken together, these results suggest that the robustness of the gene regulatory network arising from the ERSE-mediated gene expression of UPR genes could be expanded by including the UPRE-mediated gene expression of UPR genes. A peculiarly interesting point is that there are two patterns of regulation; basically, the BiP and CHOP promoters were responsive to all the UPR genes tested in this study, whereas the XBP1 promoter and p5 × ATF6-GL3 were similarly regulated by the UPR genes except for BiP, CHOP, and ATF4, albeit the BiP, CHOP, and XBP1 promoters share an ERSE. Obviously, this conclusion should be regarded with caution because of the limited regions that were tested for the BiP, XBP1, and CHOP promoters in the present study.
Changes in BiP expression upon DNA damage
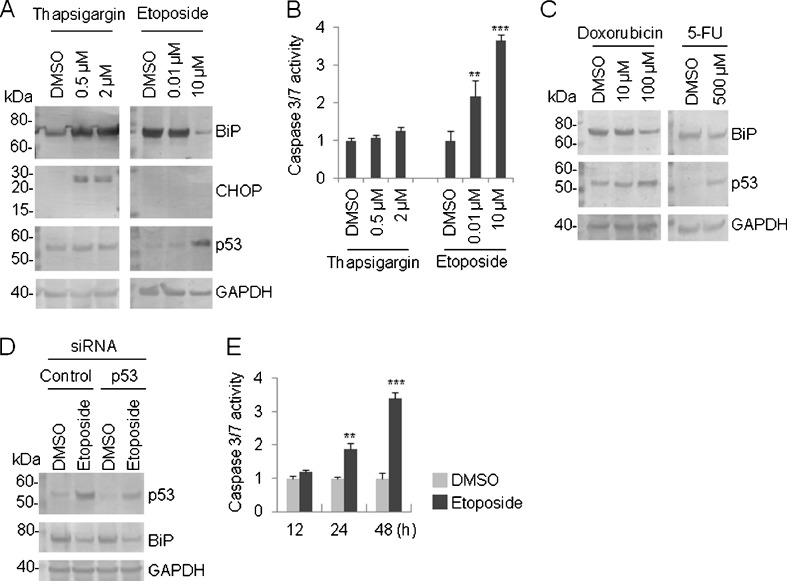
With an increase in BiP mRNA, the amount of BiP protein was increased at 8 h after exposure to thapsigargin in HeLa cells (Fig. 5a). In the same way, CHOP protein also accumulated after thapsigargin treatment (Fig. 5a). On the other hand, p53 protein level was unchanged by thapsigargin (Fig. 5a). Intriguingly, the BiP protein level dropped strikingly when the cells were treated with 10 μM of etoposide for 48 h, whereas p53 protein accumulated after etoposide treatment (Fig. 5a). As a loading control, GAPDH protein was equally detected in all the lanes (Fig. 5a). Caspase 3/7 activities, which are indicators of apoptosis, were strongly induced by etoposide treatment in conjunction with the enhancement of the p53 protein level, and the BiP protein level was also reduced (Fig. 5b, also see 5a). A phenomenon analogous to the BiP and p53 protein levels observed after etoposide treatment was reproducibly confirmed in doxorubicin-treated cells (Fig. 5c). To see if the downregulation of BiP was a specific response to the topoisomerase II inhibitors etoposide and doxorubicin, we also tested the cells using 5-FU. The BiP protein level was scarcely affected by 5-FU, whereas an accumulation of p53 was observed (Fig. 5c), implying that p53 is not a key regulator of BiP. To further examine the possibility of p53 involvement in the downregulation of BiP in response to DNA damage, we inhibited endogenous p53 expression using RNAi. To accomplish this goal, we introduced p53-specific short interference RNA (siRNA) into the cells on two successive days and then treated the cells with etoposide (10 μM) for 48 h. A western blot analysis showed that the elevated expression level of p53 protein in response to etoposide treatment (Fig. 5d, lane 1 versus lane 2) was clearly reduced by p53 siRNA (Fig. 5d, lane 2 versus lane 4). Most notably, the level of BiP protein hardly changed between the etoposide treatment and p53 siRNA transfection prior to etoposide treatment (Fig. 5d, lane 2 versus lane 4), indicating that p53 bears no relation to the regulation of the BiP protein level. GAPDH was constantly detected in all the lanes (Fig. 5d). Additionally, we examined whether the degradation of BiP protein was associated with the progress status of apoptosis. Based on the caspase 3/7 activities, etoposide strikingly enhanced apoptosis in a time-dependent manner (Fig. 5e).
Fig. 5.
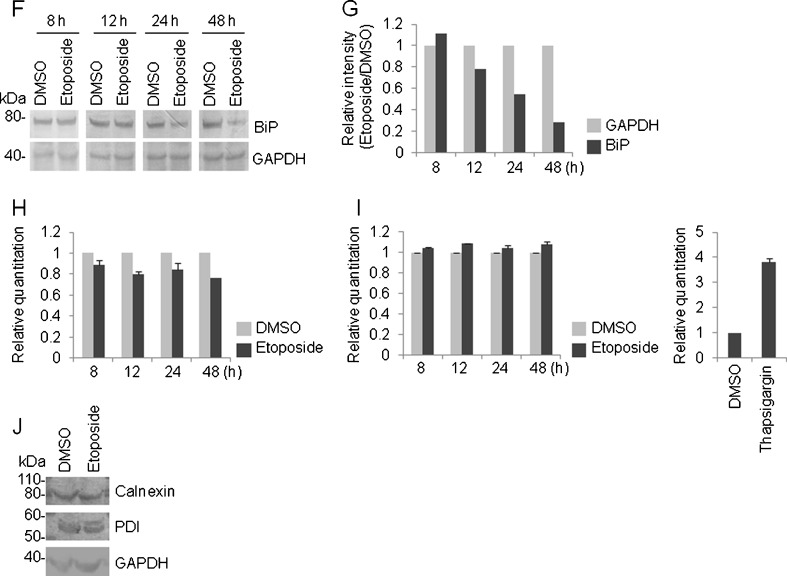
Alteration of BiP protein level in response to DNA damage. a HeLa cells were treated with the indicated concentration of thapsigargin (8 h) or etoposide (48 h), and a western blot analysis was performed using the indicated antibody. The position of the molecular marker (in kilodaltons) is indicated on the left side of the panel. b HeLa cells were treated with the indicated concentration of thapsigargin (8 h) or etoposide (48 h), and the caspase 3/7 activity was determined. The blank value was subtracted from the actual measured value. Values represent the mean ± standard deviation (n = 4; **P < 0.01; ***P < 0.001 versus DMSO). c HeLa cells were treated with the indicated concentration of doxorubicin or 5-FU for 48 h and a western blot analysis was performed using the indicated antibody. Left, the position of the molecular marker (in kilodaltons) is indicated. d Control or p53 siRNA was introduced into HeLa cells for two successive days and the cells were exposed to etoposide (10 μM) for 48 h; a western blot analysis was then performed using the indicated antibody. Left, the position of the molecular marker (in kilodaltons) is indicated. e HeLa cells were treated with etoposide (10 μM) for the indicated period, and a caspase 3/7 assay was performed to detect apoptosis. The blank value was subtracted from the actual measured value. Values represent the mean ± standard deviation (n = 4; **P < 0.01; ***P < 0.001 versus DMSO). f HeLa cells were treated with etoposide (10 μM) for the indicated period, and a western blot analysis was performed with the indicated antibody. Left, the position of the molecular marker (in kilodaltons) is indicated. g The blot image in (f) was processed using Quantity One. Briefly, each specific band was quantified by volume analysis using the background subtraction method and the resulting density was represented as the ratio of etoposide/DMSO. The resulting ratio was further normalized by the corresponding GAPDH band. h HeLa cells were treated with etoposide (10 μM) for the indicated period, and a TaqMan real-time RT-PCR was performed with the BiP primer. In each panel, GAPDH was used as an internal standard. Values represent the mean ± standard deviation (n = 2). i HeLa cells were treated with etoposide (10 μM) for the indicated period (left) and thapsigargin (0.5 μM) for 8 h (right), and a TaqMan real-time RT-PCR was performed with the XBP1 primer. In each panel, GAPDH was used as an internal standard. Values represent the mean ± standard deviation (n = 2). j HeLa cells were treated with etoposide (10 μM) for 48 h, and a western blot analysis was performed with the indicated antibody. Left, the position of the molecular marker (in kilodaltons) is indicated
Because the caspase 3/7 activities can be seen at a later stage of apoptosis, apoptosis is thought to occur after 12 h, but within 24 h at the latest, of etoposide treatment (Fig. 5e). To clarify the relationship between apoptosis induction and BiP protein level, we detected the BiP protein by western blot analysis (Fig. 5f). The BiP protein levels induced by etoposide were also shown as the relative intensity quantified by Quantity One software. The expression levels were divided by the DMSO-treated expression level and then normalized by GAPDH protein level. Subsequently, the BiP protein level initially decreased after 12 h of etoposide treatment (Fig. 5g). Together, these results suggest that the degradation of BiP protein occurred after apoptosis induction, but not prior to apoptosis induction.
To further illuminate the relationship between transcriptional regulation of BiP and BiP protein level regulated by etoposide, we performed a TaqMan real-time RT-PCR to detect BiP mRNA. As stated above, semi-quantitative RT-PCR revealed that BiP mRNA expression was unchanged by etoposide-induced apoptosis (Fig. 1b, also see Fig. 5b); however, apoptosis seems to happen when ER stress cannot be effectively relieved. Therefore, we tried to optimize the dose and treatment time for etoposide to draw a conclusion that UPR genes were not transcriptionally up-regulated by etoposide. As stated above, etoposide (10 μM) enhanced apoptosis after 24 h treatment but had no effect on the caspase 3/7 activities after 12 h treatment (Fig. 5e). We observed a slight decrease in BiP mRNA expression after etoposide treatment (12, 24, and 48 h) (Fig. 5h), suggesting that degradation of BiP protein by etoposide-induced apoptosis seems to be of little relevance to transcriptional regulation. Instead, etoposide treatment led to a decrease in the luciferase activity of pGL3-BiP (−761/+120) to ×0.449 ± 0.033 (mean ± standard deviation), which was significantly different from the activity in the DMSO-treated control (P < 0.01, n = 3). Given the above observations, etoposide likely suppresses the transcription of BiP as minimally as possible. As well BiP mRNA, XBP1 mRNA was unchanged in etoposide-treated HeLa cells, whereas XBP1 mRNA was clearly elevated in thapsigargin-treated HeLa cells (Fig. 5i).
Next, we performed a western blot using calnexin and PDI antibodies to check whether ER-associated degradation (ERAD) is intact or not in etoposide-treated HeLa cells. For this, HeLa cells were treated with etoposide (10 μM) for 48 h. As a result, calnexin and PDI proteins were almost equally detected in etoposide-treated HeLa cells compared with control DMSO-treated cells (Fig. 5j).
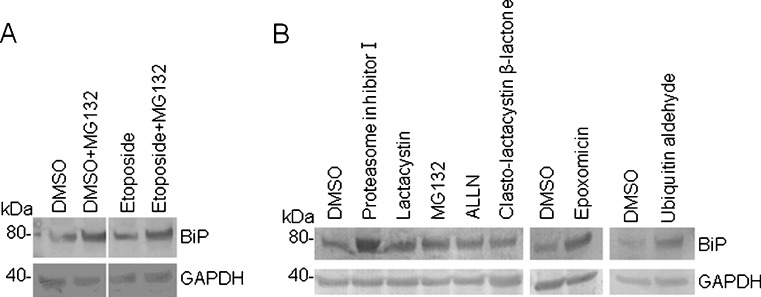
To validate the possible involvement of proteasome-mediated BiP protein degradation, the proteasome inhibitor MG132 was added to the culture medium of HeLa cells for 12 h. A western blot analysis showed the accumulation of BiP protein (Fig. 6a, lane 1 versus lane 2). In addition, MG132 also enhanced the BiP protein level even in the presence of etoposide (Fig. 6a, lane 3 versus lane 4). This result was achieved by treating the cells with MG132 for 12 h during the last half of the 24-h etoposide treatment. Aside from MG132, various proteasome inhibitors were tested to determine whether they could stabilize the BiP protein level. Surprisingly, the BiP protein level was significantly higher in proteasome inhibitor I-treated cellular lysates and moderately higher in lactacystin- and epoxomicin-treated cellular lysates (Fig. 6b). BiP protein was equally detected in ALLN and clasto-lactacystin β-lactone-treated cells, compared with in DMSO-treated cells (Fig. 6b). Finally, we confirmed the BiP protein level after adding a potent inhibitor of ubiquitin-hydrolyzing enzyme, ubiquitin aldehyde—which contributes to the stabilization of the ubiquitin-protein complex, to HeLa cells for 12 h; a western blot analysis was then performed to see whether the BiP protein had stabilized. As a result, BiP protein accumulated moderately in the ubiquitin aldehyde-treated cells, compared with the level in DMSO-treated cells (Fig. 6b). In both cells, GAPDH was almost equally detected (Fig. 6b). In conclusion, we favor the idea that proteasome-mediated degradation is more powerful than transcriptional regulation in the context of the expressional regulation of BiP in response to DNA damage based on the evidence obtained from the experiments using the proteasome inhibitor.
Fig. 6.
Effect of proteasomal inhibitor on BiP protein. a HeLa cells were treated with DMSO or etoposide (10 μM) for 24 h with or without MG132 for the latter 12 h, and a western blot analysis was performed using the indicated antibody. Left, the position of the molecular marker (in kilodaltons) is indicated. b HeLa cells were treated with the indicated proteasomal inhibitor (each 10 μM) or ubiquitin aldehyde (0.5 μM) for 12 h, and a western blot analysis was performed using the indicated antibody. Left, the position of the molecular marker (in kilodaltons) is indicated
Discussion
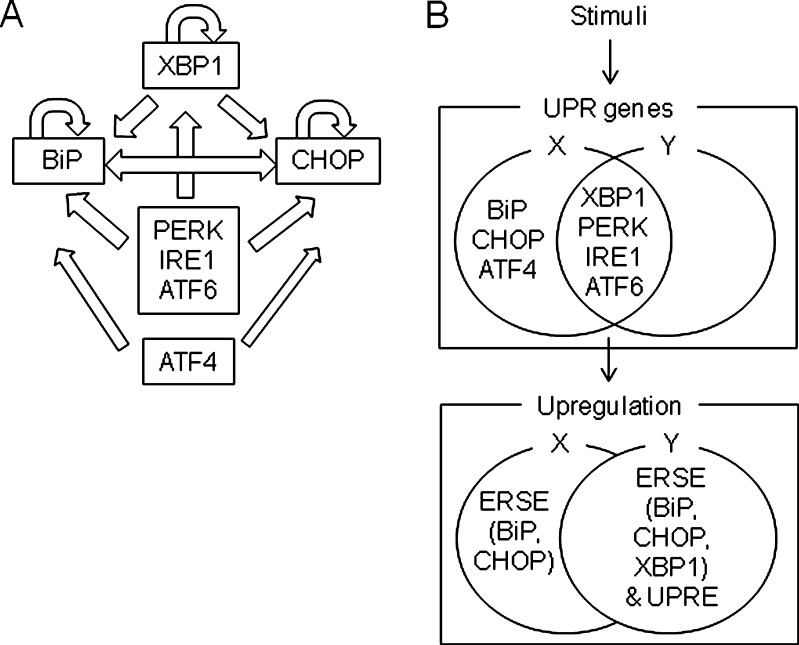
In this study, we investigated the gene regulatory network of UPR genes, focusing on ERSE- and UPRE-mediated gene expression. Among the promoter-reporter plasmids of seven UPR genes that were used, only pGL3-BiP, −761/+120; pGL3-XBP1, −410/+49; and pGL3-CHOP, −863/+23 (all of which possess an ERSE) and p5 × ATF6-GL3 (bearing a UPRE) were capable of increasing the luciferase activity upon thapsigargin-induced ER stress. We have successfully demonstrated that BiP, XBP1, and CHOP form unique self-regulatory loops, and BiP and CHOP, in particular, form a mutually facilitated structure (Fig. 7a; Table 3). These loops establish an infrastructure to ensure the robust and uniform regulation of BiP, XBP1, and CHOP genes during an ER stress response. Based on our findings, the infrastructure appears to be constructed from two components: namely, (1) all the UPR genes tested in this experiment positively regulated the BiP and CHOP promoters, and (2) all the UPR genes except for BiP, CHOP, and ATF4 positively regulated the XBP1 promoter and p5 × ATF6-GL3 (Fig. 7b; Table 3). Henceforth, we will call these two components patterns A and B, respectively (Table 3). Remarkably, pGL3-XBP1 (−410/+49) alone was differentially regulated by the UPR genes, even though the ERSE was shared with pGL3-BiP, −761/+120 and pGL3-CHOP, −863/+23 (Table 3, pattern B). Conversely, pGL3-XBP1 (−410/+49; characterized by the presence of an ERSE) and p5 × ATF6-GL3 (bearing an UPRE) were classified into the same pattern B (Table 3). Needless to say, some DNA element other than ERSE and UPRE might have a crucial role. Interestingly, all the UPR genes except for BiP, CHOP, and ATF4 were commonly responsible for both pattern A and pattern B (Fig. 7b; Table 3). In other words, the selection between pattern A and pattern B is performed by BiP, CHOP, and ATF4 (Fig. 7b and Table 3). The involvement of CHOP in the induction of apoptosis has been previously mentioned (see review by Oyadomari and Mori 2004). Therefore, we speculated that a dysregulation in the balance between patterns A and B might be responsible for determining the cell fate, such as apoptosis.
Fig. 7.
Diagrammatic illustration of the UPR gene regulatory network. a BiP, CHOP, and XBP1 have a positive feedback loop (indicated by the arrow) and are related to one another to secure robust and uniform gene expression as part of the ER stress response. For example, BiP and CHOP positively regulate each other. In addition, the BiP, CHOP, and XBP1 promoters have multiple layers of inputs consisting of UPR genes such as PERK, IRE1, ATF6, and ATF4. b Based on the results obtained in this study, the regulation of UPR genes centered on the ERSE (BiP, CHOP, and XBP1 genes) and the UPRE can be divided into two groups. As a matter of convenience, we call these groups X and Y in this figure. X contains all of the UPR genes, whereas Y consists of all the UPR genes except for BiP, CHOP, and ATF4. In other words, X and Y share XBP1, PERK, IRE1, and ATF6, and only these genes can induce both ERSE- and UPRE-dependent transcription. On the other hand, BiP, CHOP, and ATF4 exclusively belong to X and positively regulate ERSE, except for the XBP1 promoter. Taken together, our results suggest that cells might creatively use X (in other words, all the UPR genes, agreeing with the results obtained using RT-PCR in which all the UPR genes were transcriptionally activated by thapsigargin) and Y depending on the degree of ER stress. Alternatively, only BiP and CHOP can increase the amount of their expressions by changing from Y to X. This finding implies that cells resort to additional BiP expression to overcome unresolved and prolonged ER stress, and while simultaneous, a cellular strategy solicits the induction of apoptosis by CHOP
Table 3.
Summary of regulatory network identified for UPR genes pGL3-BiP, −761/+120; pGL3-XBP1, −410/+49; and pGL3-CHOP, −863/+23, which are characterized by the presence of an ERSE, and p5 × ATF6-GL3, which is characteristic by the presence of a UPRE, are shown in the columns, whereas the expression vectors of the UPR genes are shown in the rows
| Expression vector | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| BiP | CHOP | ATF4 | XBP1 | PERK | IRE1 | ATF6 | |||
| K618A | 1-373 | ||||||||
| ERSE | pGL3-BiP | ↑ | ↑ | ↑ | ↑ | ↑ | ↑ | ↑ | Pattern A |
| pGL3-CHOP | ↑ | ↑ | ↑ | ↑ | ↑ | ↑ | ↑ | ||
| pGL3-XBP1 | ↑ | ↑ | ↑ | ↑ | Pattern B | ||||
| UPRE | p5 × ATF6-GL3 | ↑ | ↑ | ↑ | ↑ | ||||
An arrow indicates the upregulation of reporter activity by co-expression with the expression vector. Patterns A and B were classified according to the appearance of the arrow
The transcription of ATF6 mRNA is activated by a calcium ion signal released from the ER into the cytoplasm by thapsigargin (Namba et al. 2007). In agreement with our findings, the activated soluble form of ATF6 has also been shown to regulate the BiP, XBP1, and CHOP promoters in a positive manner (Yoshida et al. 2000). Moreover, IRE1 as well as soluble ATF6 can induce BiP and ATF6 mRNA (Cao et al. 1995; Wang et al. 2000; Namba et al. 2007). Therefore, as proven by the experiment employing p5 × ATF6-GL3, de novo ATF6 immediately promotes the BiP, XBP1 and CHOP promoters; as an inevitable result, a robust infrastructure composed of BiP, XBP1, CHOP, and also ATF6 functions as a firm hub for the ER stress response.
Regarding the transcriptional regulation of BiP, ERSE-independent regulation by ATF4 as well as ERSE-dependent synergistic regulation by ATF6, NF-Y, and YY1 have been reported (Li et al. 2000; Luo et al. 2003). GATA4 and E2F1 have been shown to be involved in the transcriptional regulation of BiP; thus, the versatile properties of BiP other than the UPR are beginning to attract attention (Mao et al. 2006; Racek et al. 2008). As is the case with BiP, the transcriptional regulation of CHOP is also positively regulated by NF-Y through ERSE in the promoter, and its mRNA is known to be induced by ATF4, which acts downstream of PERK (Fawcett et al. 1999; Ubeda and Habener 2000; Ma et al. 2002). In addition, CHOP has been shown to be regulated by ATF5 through an amino acid response element located in the CHOP promoter and by Jun dimerizing protein 2 (Chérasse et al. 2008; Yamazaki et al. 2010). Taken together, this diversity in the responsiveness of BiP and CHOP promoters opens a new window for the identification of a hub centering on BiP, XBP1, CHOP, and ATF6, with inevitable adjustments enabling a variety of useful functions to be executed. Consequently, the regulatory network of UPR genes is both flexible and robust.
In human cell lines, a protein complex containing BiP, calnexin, and PDI has been identified to be required for dislocation or retrotranslocation of misfolded proteins from the ER lumen to the cytosolic proteasome to be degraded (Mueller et al. 2008). BiP is supposed to play a key role in the selection and targeting of misfolded proteins for the ERAD (Tamura et al. 2008). So far, BiP has been shown to be involved in ApoB degradation as a chaperon (Qiu et al. 2005); however, the degradation mechanism for BiP itself remains obscure. Interestingly, misfolded BiP has shown to be degraded in the ER lumen, suggesting that proteasome-independent ERAD machinery is likely existed (Donoso et al. 2005). We have found that the expression level of BiP is more tightly regulated at a post-translational degradation mechanism level, rather than at a transcriptional level, when confronted with DNA damage. ER stress is generally regarded as a protective cellular response against apoptosis. The loss of BiP, which means the lack of a key player in ER stress, triggered by DNA damage could accelerate cell death. The identification of an ubiquitinating enzyme specific to BiP protein is imperative. A comparison of the UPR gene expressional changes at both the mRNA and protein levels in ER stress versus apoptosis is also needed.
As we have discussed, the identification of a regulatory network of UPR genes is significant because the transcriptional regulation of UPR genes is strongly correlated with the functional aspects of UPR genes during the ER stress response. The whole picture of the ER stress response is likely to be clarified in the near future if the search for the molecular regulatory mechanism underlying the ER stress response is continued.
Acknowledgments
We would like to thank Moe Tategu for her technical assistance.
Glossary
- ATF
Activating transcription factor
- BiP
Immunoglobulin heavy chain-binding protein
- CHOP
C/EBP [CCAAT/enhancer-binding protein]-homologous protein
- DMSO
Dimethyl sulfoxide
- PDI
Protein disulfide isomerase
- ER
Endoplasmic reticulum
- ERAD
ER-associated degradation
- ERSE
ER stress response element
- 5-FU
5-fluorouracil
- GAPDH
Glyceraldehyde-3-phosphate dehydrogenase
- IRE1
Inositol-requiring 1
- PCR
Polymerase chain reaction
- PERK
PKR-like endoplasmic reticulum kinase
- RNAi
RNA interference
- RT
Reverse transcription
- siRNA
Short interference RNA
- UPR
Unfolded protein response
- UPRE
Unfolded protein response element
- XBP1
X-box binding protein 1
References
- Cao X, Zhou Y, Lee AS. Requirement of tyrosine- and serine/threonine kinases in the transcriptional activation of the mammalian grp78/BiP promoter by thapsigargin. J Biol Chem. 1995;270:494–502. doi: 10.1074/jbc.270.1.494. [DOI] [PubMed] [Google Scholar]
- Chérasse Y, Chaveroux C, Jousse C, Maurin AC, Carraro V, Parry L, Fafournoux P, Bruhat A. Role of the repressor JDP2 in the amino acid-regulated transcription of CHOP. FEBS Lett. 2008;582:1537–1541. doi: 10.1016/j.febslet.2008.03.050. [DOI] [PubMed] [Google Scholar]
- Donoso G, Herzog V, Schmitz A. Misfolded BiP is degraded by a proteasome-independent endoplasmic-reticulum-associated degradation pathway. Biochem J. 2005;387:897–903. doi: 10.1042/BJ20041312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawcett TW, Martindale JL, Guyton KZ, Hai T, Holbrook NJ. Complexes containing activating transcription factor (ATF)/cAMP-responsive-element-binding protein (CREB) interact with the CCAAT/enhancer-binding protein (C/EBP)-ATF composite site to regulate Gadd153 expression during the stress response. Biochem J. 1999;339:135–141. doi: 10.1042/0264-6021:3390135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetz C. The unfolded protein response: controlling cell fate decisions under ER stress and beyond. Nat Rev Mol Cell Biol. 2012;13:89–102. doi: 10.1038/nrm3270. [DOI] [PubMed] [Google Scholar]
- Lee AS. GRP78 induction in cancer: therapeutic and prognostic implications. Cancer Res. 2007;67:3496–3499. doi: 10.1158/0008-5472.CAN-07-0325. [DOI] [PubMed] [Google Scholar]
- Li J, Lee AS. Stress induction of GRP78/BiP and its role in cancer. Curr Mol Med. 2006;6:45–54. doi: 10.2174/156652406775574523. [DOI] [PubMed] [Google Scholar]
- Li M, Baumeister P, Roy B, Phan T, Foti D, Luo S, Lee AS. ATF6 as a transcription activator of the endoplasmic reticulum stress element: thapsigargin stress-induced changes and synergistic interactions with NF-Y and YY1. Mol Cell Biol. 2000;20:5096–5106. doi: 10.1128/MCB.20.14.5096-5106.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipson KL, Fonseca SG, Ishigaki S, Nguyen LX, Foss E, Bortell R, Rossini AA, Urano F. Regulation of insulin biosynthesis in pancreatic beta cells by an endoplasmic reticulum-resident protein kinase IRE1. Cell Metab. 2006;4:245–254. doi: 10.1016/j.cmet.2006.07.007. [DOI] [PubMed] [Google Scholar]
- Luo S, Baumeister P, Yang S, Abcouwer SF, Lee AS. Induction of Grp78/BiP by translational block: activation of the Grp78 promoter by ATF4 through and upstream ATF/CRE site independent of the endoplasmic reticulum stress elements. J Biol Chem. 2003;278:37375–37385. doi: 10.1074/jbc.M303619200. [DOI] [PubMed] [Google Scholar]
- Ma Y, Brewer JW, Diehl JA, Hendershot LM. Two distinct stress signaling pathways converge upon the CHOP promoter during the mammalian unfolded protein response. J Mol Biol. 2002;318:1351–1365. doi: 10.1016/S0022-2836(02)00234-6. [DOI] [PubMed] [Google Scholar]
- Malhotra JD, Kaufman RJ. The endoplasmic reticulum and the unfolded protein response. Semin Cell Dev Biol. 2007;18:716–731. doi: 10.1016/j.semcdb.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao C, Tai WC, Bai Y, Poizat C, Lee AS. In vivo regulation of Grp78/BiP transcription in the embryonic heart: role of the endoplasmic reticulum stress response element and GATA-4. J Biol Chem. 2006;281:8877–8887. doi: 10.1074/jbc.M505784200. [DOI] [PubMed] [Google Scholar]
- Mueller B, Klemm EJ, Spooner E, Claessen JH, Ploegh HL. SEL1L nucleates a protein complex required for dislocation of misfolded glycoproteins. Proc Nat Acad Sci U S A. 2008;105:12325–12330. doi: 10.1073/pnas.0805371105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namba T, Ishihara T, Tanaka K, Hoshino T, Mizushima T. Transcriptional activation of ATF6 by endoplasmic reticulum stressors. Biochem Biophys Res Commun. 2007;355:543–548. doi: 10.1016/j.bbrc.2007.02.004. [DOI] [PubMed] [Google Scholar]
- Oyadomari S, Mori M. Roles of CHOP/GADD153 in endoplasmic reticulum stress. Cell Death Differ. 2004;11:381–389. doi: 10.1038/sj.cdd.4401373. [DOI] [PubMed] [Google Scholar]
- Parmar VM, Schröder M. Sensing endoplasmic reticulum stress. Adv Exp Med Biol. 2012;738:153–168. doi: 10.1007/978-1-4614-1680-7_10. [DOI] [PubMed] [Google Scholar]
- Qiu W, Kohen-Avramoglu R, Mhapsekar S, Tsai J, Austin RC, Adeli K. Glucosamine-induced endoplasmic reticulum stress promotes ApoB100 degradation: evidence for Grp78-mediated targeting to proteasomal degradation. Arterioscler Thromb Vasc Biol. 2005;25:571–577. doi: 10.1161/01.ATV.0000154142.61859.94. [DOI] [PubMed] [Google Scholar]
- Racek T, Buhlmann S, Rüst F, Knoll S, Alla V, Pützer BM. Transcriptional repression of the prosurvival endoplasmic reticulum chaperone GRP78/BIP by E2F1. J Biol Chem. 2008;283:34305–34314. doi: 10.1074/jbc.M803925200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura T, Cormier JH, Hebert DN. Sweet bays of ERAD. Trends Biochem Sci. 2008;33:298–300. doi: 10.1016/j.tibs.2008.04.013. [DOI] [PubMed] [Google Scholar]
- Ubeda M, Habener JF. CHOP gene expression in response to endoplasmic-reticular stress requires NFY interaction with different domains of a conserved DNA-binding element. Nucleic Acids Res. 2000;28:4987–4997. doi: 10.1093/nar/28.24.4987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Shen J, Arenzana N, Tirasophon W, Kaufman RJ, Prywes R. Activation of ATF6 and an ATF6 DNA binding site by the endoplasmic reticulum stress response. J Biol Chem. 2000;275:27013–27020. doi: 10.1074/jbc.M003322200. [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y, Larkin D, Lara-Lemus R, Ramos-Castañeda J, Liu M, Arvan P. Endoplasmic reticulum (ER) chaperone regulation and survival of cells compensating for deficiency in the ER stress response kinase, PERK. J Biol Chem. 2008;283:17020–17029. doi: 10.1074/jbc.M802466200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki T, Ohmi A, Kurumaya H, Kato K, Abe T, Yamamoto H, Nakanishi N, Okuyama R, Umemura M, Kaise T, Watanabe R, Okawa Y, Takahashi S, Takahashi Y. Regulation of the human CHOP gene promoter by the stress response transcription factor ATF5 via the AARE1 site in human hepatoma HepG2 cells. Life Sci. 2010;87:294–301. doi: 10.1016/j.lfs.2010.07.006. [DOI] [PubMed] [Google Scholar]
- Yoshida H. ER stress and diseases. FEBS J. 2007;274:630–658. doi: 10.1111/j.1742-4658.2007.05639.x. [DOI] [PubMed] [Google Scholar]
- Yoshida H, Okada T, Haze K, Yanagi H, Yura T, Negishi M, Mori K. ATF6 activated by proteolysis binds in the presence of NF-Y (CBF) directly to the cis-acting element responsible for the mammalian unfolded protein response. Mol Cell Biol. 2000;20:6755–6767. doi: 10.1128/MCB.20.18.6755-6767.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001;107:881–891. doi: 10.1016/S0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
- Zhang K, Kaufman RJ. The unfolded protein response: a stress signaling pathway critical for health and disease. Neurology. 2006;66:S102–S109. doi: 10.1212/01.wnl.0000192306.98198.ec. [DOI] [PubMed] [Google Scholar]
- Zhu C, Johansen FE, Prywes R. Interaction of ATF6 and serum response factor. Mol Cell Biol. 1997;17:4957–4966. doi: 10.1128/mcb.17.9.4957. [DOI] [PMC free article] [PubMed] [Google Scholar]