SUMMARY
The ectopic expression of transcription factors can reprogram cell fate, yet it is unknown how the initial binding of factors to the genome relates functionally to the binding seen in the minority of cells that become reprogrammed. We report a map of Oct4, Sox2, Klf4, and c-Myc (O, S, K, and M) on the human genome during the first 48 hours of reprogramming fibroblasts to pluripotency. Three striking aspects of the initial chromatin binding events include: An unexpected role for c-Myc in facilitating OSK chromatin engagement, the primacy of O, S, and K as pioneer factors at enhancers of genes that promote reprogramming, and megabase-scale chromatin domains spanned by H3K9me3, including many genes required for pluripotency, that prevent initial OSKM binding and impede the efficiency of reprogramming. We find diverse aspects of initial factor binding that must be overcome in the minority of cells that become reprogrammed.
Keywords: Pluripotency, pioneer factors, reprogramming, chromatin, c-Myc, Oct4, Sox2, Klf4, H3K9me3
INTRODUCTION
Cellular reprogramming by transcription factors is remarkable for its potential to impact research and therapies, yet it remains inefficient in practice (Graf and Enver, 2009). One of the most striking examples is the ability of the transcription factors Oct4, Sox2, Klf4, and c-Myc (O, S, K, and M) to reprogram somatic cells to become induced pluripotent stem (iPS) cells (Takahashi and Yamanaka, 2006). Yet only a minority of cells in which the reprogramming factors are expressed will productively convert (Plath and Lowry, 2011). Little is known about the mechanisms by which reprogramming factors initially interact with the genome, how the initial binding network compares to that in later stages, and the chromatin structures that may guide or impede factor binding. By investigating the initial events in factor binding, we sought to shed light on mechanisms that may promote or limit reprogramming.
During embryonic development, a subset of transcription factors known as “pioneer factors” initially access silent chromatin and direct the binding of other transcription factors (Cirillo et al., 2002; Zaret and Carroll, 2011). Other transcription factors can access their chromatin target sites cooperatively (Adams and Workman, 1995; Filion et al., 2010). Reprogramming to pluripotency is stepwise, with morphological changes and apoptosis occurring in the first 48 hours (Samavarchi-Tehrani et al., 2010; Smith et al., 2010), changes in H3K4me2 modification occurring by one cell division (48 hr) (Koche et al., 2011), and transcriptional changes at genes whose promoters were marked by H3K4me3 (Koche et al., 2011; Mah et al., 2011). Early steps involve a mesenchymal-to-epithelial transition (MET) via silencing of Snail genes, suppression of TGF-β signaling, and up-regulating E-cadherin (Cdh1) (Li et al., 2010; Mah et al., 2011). The P53 pathway promotes apoptosis and senescence and its suppression enhances reprogramming (Hong et al., 2009; Kawamura et al., 2009; Marion et al., 2009). The timing and extent to which these genes and pathways are first targeted during reprogramming are unknown.
Genome-wide occupancy maps of ES and iPS cells have revealed that the OSK factors are central to the core pluripotency network, while c-Myc, bound to promoters, is part of a distinct network (Boyer et al., 2005; Chen et al., 2008; Kim et al., 2008; Sridharan et al., 2009). Mouse fibroblasts at a late stage of reprogramming (pre-iPS) contained c-Myc at promoters similar to that seen in iPS and ES cells (Sridharan et al., 2009). c-Myc can be replaced by Glis1 during reprogramming (Maekawa et al., 2011) or omitted, yet when combined with OSK, c-Myc greatly improves the efficiency and kinetics of the process (Nakagawa et al., 2008; Wernig et al., 2008). By promoting cell growth and survival, c-Myc is thought to indirectly enhance reprogramming by allowing more chances for stochastic events to occur (Hanna et al., 2009; Knoepfler, 2008). Whether c-Myc plays a direct role in global chromatin engagement is unclear.
Here we show that the O, S, K, and M factors initially occupy the genome in human fibroblasts in a markedly different way compared to that in pre-iPS, iPS, and ES cells. The factors bind together to distal elements of many genes required for reprogramming and there is minimal detectable effect of pre-existing histone modifications and DNaseI hypersensitivity in guiding most of the initial OSKM binding. We reveal a pioneer activity of Oct4, Sox2, and Klf4 in binding closed chromatin and a direct role for c-Myc in enhancing the binding of OSK to chromatin. We also discovered megabase-sized domains of the genome that are refractory to OSKM binding; most of these Differentially Bound Regions (OSKM-DBRs) will become bound in the minority of cells that become pluripotent. H3K9me3 is the dominant chromatin feature at the DBRs and knock-down of relevant methyltransferases allows OSKM binding and enhances reprogramming. Together, these studies reveal that progression to pluripotency requires re-organization of the initial network for c-Myc, a shift to promoter binding by OSKM, and overcoming broad heterochromatic features that occur in the somatic cell genome.
RESULTS
Extensive OSKM binding to chromatin in the first 48 hours of reprogramming
To investigate the initial engagement of O, S, K, and M with chromatin, we transcriptionally induced the factors together in human fibroblasts (hFib) using doxycyline (dox)-inducible lentiviral transduction (Brambrink et al., 2008) (Figure S1A). Cells were transferred to human ES (hES) culture conditions on the third day and gave rise to stable hFib-iPS lines that were maintained without dox for 10 passages; these expressed endogenous pluripotency marker proteins similar to the H1-hES control line (Figure S1B). As expected, the hFib-iPS cells can form embryoid bodies and differentiate into derivatives of the three germ layers (Figure S1C). The OSKM-infected human fibroblasts were reprogrammed at an efficiency of ~0.05% (Figure S1D), as seen by others (Hockemeyer et al., 2008; Takahashi et al., 2007).
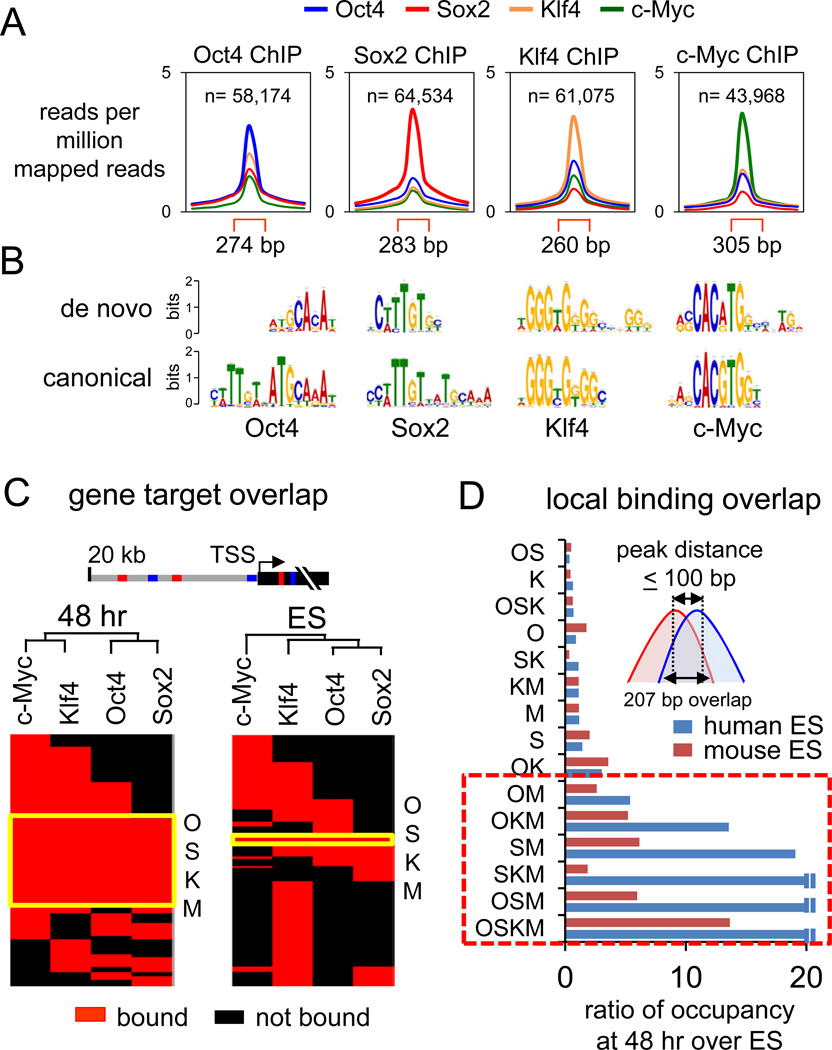
We used ChIP-seq to map the initial protein-DNA interactions of O, S, K, and M with the genome. A 48 hr dox induction point was chosen because it was the earliest for maximal OSKM expression (not shown), it preceded major transcriptional changes (Koche et al., 2011; Mah et al., 2011), and it preceded the time when the fibroblasts are changed to ES growth conditions. As a control, we probed non-infected fibroblasts that were treated with dox for 48 hours (Figures S1E, S1F). The DNA from eight ChIPs for each factor from infected cells, four ChIPs from non-infected control cells, and input DNA were sequenced. We applied MACS (Zhang et al., 2008) to identify bound regions containing unique-mapped reads from ChIP DNA over the input DNA and subtracted peaks from the control ChIP reactions from uninfected cells (Figure S2A, Table S1). Using qPCR, we validated all 65 of selected O, S, K, and/or M peaks that were FDR-controlled to 0.005 and confirmed poor or lack of enrichment in all of 30 regions that were distal to selected peaks (Figures S2B, Table S2). Our analysis identified 58174, 64534, 61075, and 43968 enriched peaks for O, S, K, and M, respectively, at 48 hr with an average peak width of ~280 bp (Figure 1A). De-novo motif discovery (Machanick and Bailey, 2011) showed that O, S, and K are able to access many of their specific DNA targets in early reprogramming, with M accessing a sequence related to its canonical target (p < 1e-05) (Figure 1B).
Figure 1. Extensive OSKM engagement with the genome at 48 hr post-induction.
(A) O, S, K, and M peak profiles showing average read counts, input subtracted, lane-normalized, and as reads per million mapped reads. Read counts within peak regions (red brackets) are compared to 1 kb flanking regions. See Figures S1, S2, and Table S1 for ChIP-seq details and peak examples.
(B) De novo sequence motifs overrepresented in the O, S, K, and M peaks compared to their canonical motifs found in the JASPAR database (Bryne et al., 2008).
(C) Hierarchical clustering of O, S, K, and/or M co-bound target genes in 48 hr (left) and in ES cells (right). Genes are called OSKM-targets if bound within the gene body or 20 kb upstream and are shown as red rectangles and non-targets as black rectangles. The genes co-targeted by all OSKM factors are indicated in the yellow boxed region, “OSKM”.
(D) OSKM co-bound regions at 48 hr with peak distances ≤100 bp are more frequent compared to those found in human ES (blue bars) (Lister et al., 2009) and mouse ES (red bars) (Chen et al., 2008). The O, S, K, and/or M co-bound regions are expressed as ratio of that seen at 48 hr to that seen in ES cells. Boxed area indicates extensive co-binding of c-Myc with O, S, and/or K.
See also Figures S1, S3, and Table S3.
Clustering of OSKM binding is a distinct feature of the initial reprogramming network
Genes bound by OSKM at 48 hr were compared to ChIP-Seq data from ES cells (Chen et al., 2008) (Table S3). By unsupervised hierarchical clustering, the initial regulatory network is extensively shared between the four factors, unlike that in ES cells (Figure 1C, red overlapping regions). Genes co-targeted by O, S, K, and M represent ~35% (11,702 out of 35,096) of all genes bound by the factors, while O, S, K, and M were only co-bound to ~3% (463 out of 15,062) of genes in ES cells (Figure 1C, yellow boxed region). At an FDR value of 0.005, our data has more peaks (188,595) than that of Chen et al. 2008 (19,208 at an FDR of 0.05), but the datasets have the same magnitude of expected error (~940 vs. ~960, respectively). The higher number of peaks in our dataset could reflect current ChIP-Seq methods and the level of OSKM expression, which is typical in the field and with typical levels of cellular conversion.
To gain a more mechanistic view of O, S, K, and/or M interactions with DNA at the nucleosome level at 48 hr, we identified binding peak centers that were no more than 100 bp apart; these overlapped by an average of 207 bp (Figure 1D, inset). We used the same method to analyze O, S, K, and M ChIP-seq datasets from human and mouse ES cells (Chen et al., 2008; Lister et al., 2009). Strikingly, we found that O, S, and K sites that were co-bound with c-Myc were more frequent at 48 hours with respect to those seen in ES cells (Figure 1D, red box). The DNA sequences of more than half (~56%) of the OSKM co-bound regions are conserved in mouse (p<0.0001), reflecting possible cis-regulatory function (Figures S3A, S3B). Overall, the OSKM factors bind together at conserved, closely spaced regions far more extensively during the initiation of reprogramming than in the partially reprogrammed and fully pluripotent states.
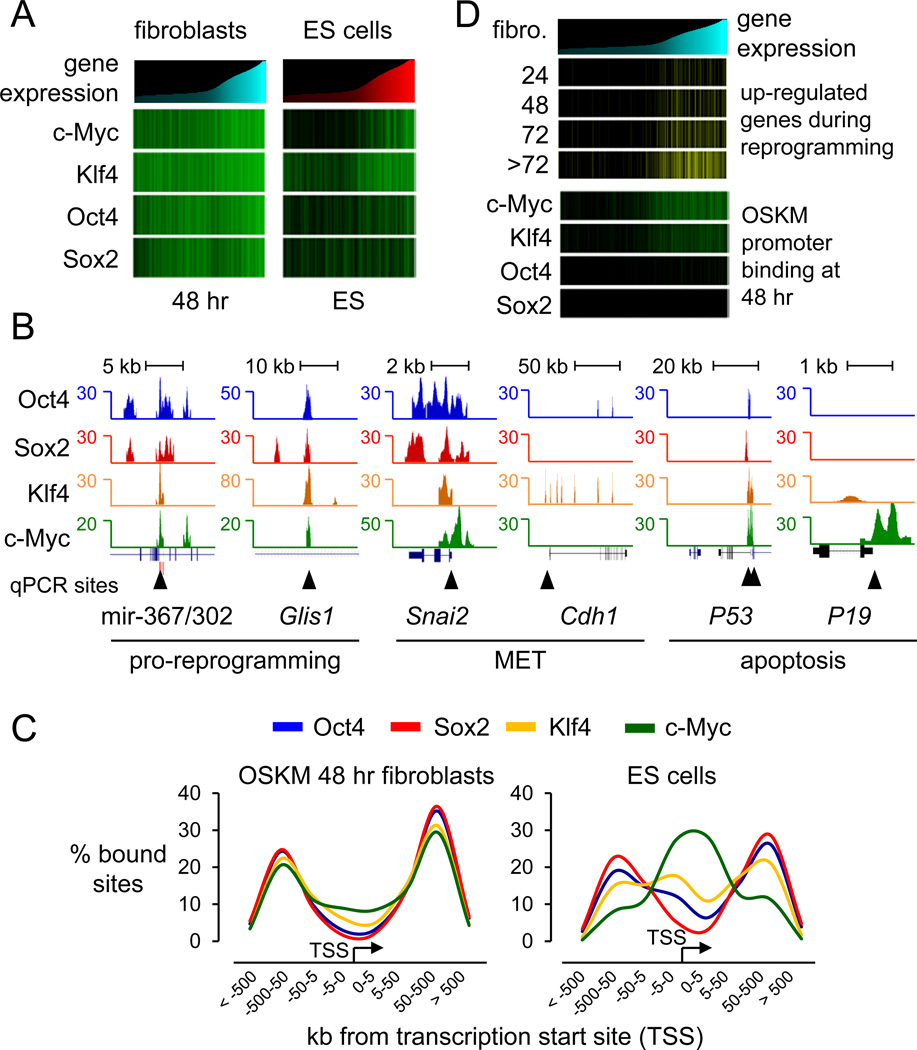
Using microarray expression datasets of human fibroblasts and ES cells (Lowry et al., 2008; Maherali et al., 2008), we found that silent genes were extensively targeted by the O, S, K, and M factors at 48 hours, in contrast to that seen in ES cells (Chen et al., 2008; Sridharan et al., 2009), where c-Myc and Klf4 were predominantly associated with active genes (Figure 2A, compare green bars at 48 hr with those in ES cells). About 90% of genes that change expression 2-fold or more between Fib and iPS (7,740 out of 8,587 transcripts) or ES (9,976 out of 11,216 transcripts) are targeted by O, S, K, and/or M at 48 hr, indicating that the initial bound genes will respond transcriptionally (Table S3).
Figure 2. OSKM initially targets genes for reprogramming and apoptosis.
(A) Heatmap analysis showing the correlation between O, S, K, and/or M binding in 48 hr and in ES cells, and gene expression in uninfected human fibroblasts and ES cells (Chen et al., 2008; Lowry et al., 2008; Sridharan et al., 2009). Genes were rank ordered according to their level of expression in fibroblast cells (cyan histogram, left panel) or in ES cells (red histogram, right panel) and correlated with their O, S, K, and/or M occupancy (green llines) at 48 hr (left panel) or in ES cells (right panel), respectively. Table S3 shows expression values in hFib, iPS, and ES cells.
(B) O, S, K, and M ChIP-seq profiles (blue, red, orange, and green respectively) at genes bound at 48 hr and important for reprogramming or apoptosis. OSKM peaks presented are normalized against input DNA sequenced tags. Black triangles indicate regions validated by ChIP-qPCR, as shown in Figure S3C.
(C) O, S, K, and M binding with respect to transcription start sites (TSS) are displayed with the frequency of total binding for Oct4 (blue), Sox2 (red), Klf4 (orange), and c-Myc (green) in 48 hr (left panel) and in ES (right panel, from Chen et al. 2008). See Figure S4 for detailed O, S, K, and/or M binding combination distributions and functional enhancer binding.
(D) Gene expression levels in fibroblasts are rank-ordered from low to high (top cyan histogram). Genes that change expression 2-fold or more over fibroblasts during 24, 48, 72, and >72 hr of reprogramming (Koche et al., 2011) are displayed as yellow lines and correlated with genes occupied by O, S, K, and/or M in their promoters (±1 kb of TSS) at 48 hr (green lines).
See also Figures S2–4.
The initial OSKM regulatory network promotes reprogramming and apoptosis
OSKM bind together at genes that promote reprogramming, including Glis1 (Maekawa et al., 2011), the mir-302/367 cluster (Anokye-Danso et al., 2011), and genes for MET such as Snai2 (Figures 2B, S3C). Gene Ontology analysis of O, S, K, and M co-targets revealed that pro-reprogramming pathways including MET are overrepresented at 48 hr (p<1.5e-7; Figure S3D). Klf4 targets Cdh1 at mulitple sites at 48 hr (Figures 2B, S3C), in concordance with ectopic Klf4 but not OSM potently inducing Cdh1 (Li et al., 2010). The factors did not target Nanog and Dppa4, which are activated late in reprogramming (Tables S2, S3) (Mikkelsen et al., 2008; Samavarchi-Tehrani et al., 2010; Sridharan et al., 2009).
Apoptotic genes were bound extensively by OSKM at 48 hr (GO targets p<2.8e-4; Figure S3D), including P53 being bound by all four factors and p19 (CDKN2D) by c-Myc (Figures 2B, S3C,D). These findings are consistent with c-Myc alone or OSK inducing P53, and c-Myc alone, but not OSK, inducing p19 (Kawamura et al., 2009). We conclude that OSKM together initially bind genes that are important for early stages of reprogramming as well as for apoptosis, with binding to the latter explaining how the cell death pathway is rapidly activated in reprogramming experiments.
OSKM predominantly bind to distal elements in early reprogramming
Using GREAT (McLean et al., 2010) to determine the distribution of O, S, K, and/or M-bound sites with respect to TSSs of RefSeq genes, we found that the factors bind distal to promoters at 48 hr of induction (Figure 2C). Apart from Klf4 and c-Myc co-bound regions, which showed a tendency to bind promoters, all the other O, S, K, and/or M combinations were primarily bound to distal elements at sites that were generally conserved between human and mouse (Figure S4A, compare magenta line to other lines). By contrast, c-Myc binds almost exclusively to promoters in ES cells (Figure 2C) (Chen et al., 2008; Kim et al., 2008; Sridharan et al., 2009). Most of the O, S, K, and/or M co-bound combinations shift from being predominantly distal to the TSS, at 48 hr, to being close to the TSS in ES cells, apart from Sox2-bound sites which remain distal (Figure S4A). The functionality of early distal binding sites was underscored by the marked binding of O, S, K, and M at 48 hr to validated enhancers (Visel et al., 2007), compared to flanking regions of similar size (Figure S4B). Our findings support a recent study showing that exogenous Oct4 first accesses the enhancer and later the promoter of MyoD1 gene during reprogramming (Taberlay et al., 2011). We further found that OSKM, not just Oct4, bind three of the four enhancers of MyoD1 and do not access the promoter at 48 hr (Figure S4C). Our observation of the vast majority (~85%) of initial OSKM binding events occur at distal sequences, prior to promoter occupancy, is consistent with only a fraction of the genes exhibiting transcriptional changes at 48 hr (Koche et al., 2011; Mah et al., 2011), and these largely comprising the subset that become bound by c-Myc and Klf4 at promoters (Figure 2D). Thus distal element binding is an early step in reprogramming, preceding promoter binding and the transcriptional activation of many target genes.
Oct4, Sox2 and Klf4 act as pioneer factors for c-Myc and c-Myc enhances chromatin binding by OSK
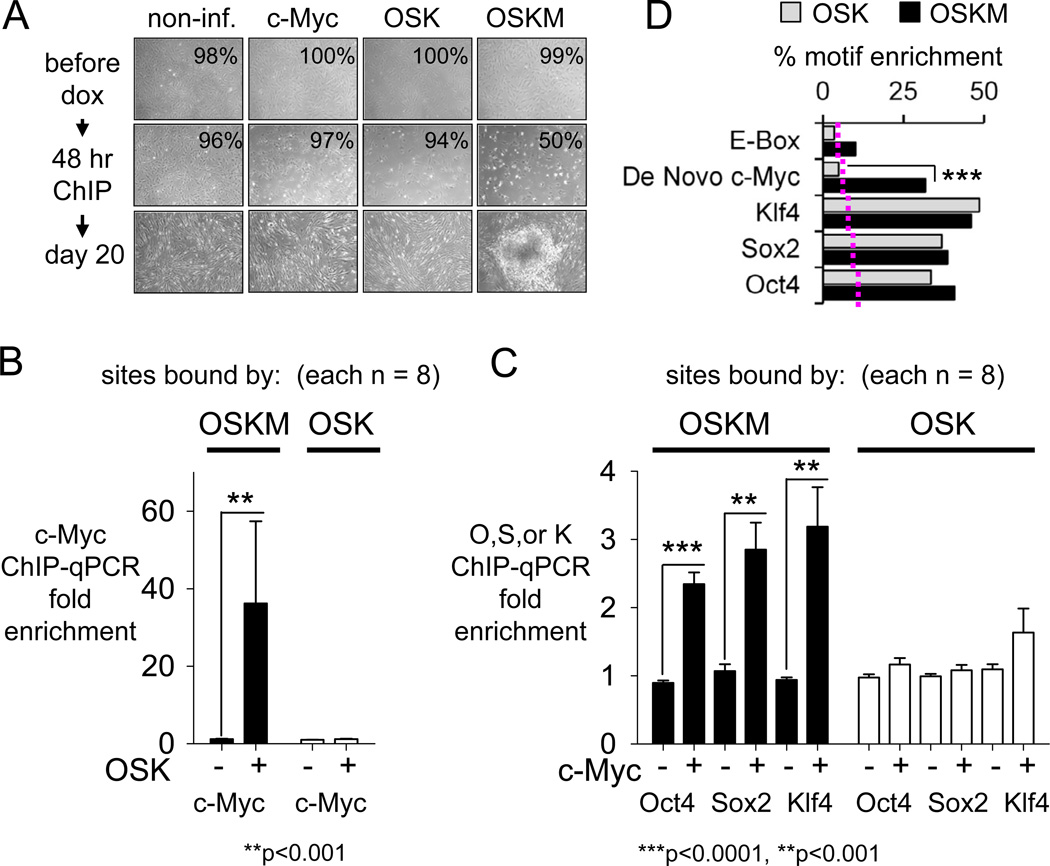
To investigate how c-Myc co-binds so extensively with OSK after 48 hr of co-expression in fibroblasts, we separately induced the expression of c-Myc alone, OSK, or OSKM in hFib cells. In the context of OSK, c-Myc promoted cell death in early reprogramming (Figures 3A). The OSKM-infected, but not the OSK- or the c-Myc only-infected fibroblasts, formed colonies after 20 days of expression (Figure 3A). We carried out ChIP-qPCR at 48 hr on 8 sites from the ChIP-Seq dataset where OSKM bound together and 8 sites where OSK bound together, but not with c-Myc (Figure S5).
Figure 3. OSK act as pioneer factors for c-Myc and c-Myc enhances OSK binding to chromatin.
(A) Non-infected and, c-Myc, OSK, or OSKM-infected fibroblasts at 0 hr, 48 hr and day 20 post-induction with dox. Viability is indicated at 0 and 48 hr post-induction with dox and shows marked apoptosis when c-Myc is expressed with OSK.
(B) ChIP-qPCR assays at 48 hr on 8 OSKM target sites (Mak10, Gins1, Jmjd2C, Jmjd1A, Smarcad1, Sox21, Lgr4, MIR-367; Figure S5B) and 8 OSK target sites (Pax6-1, Pax6-2, Pde4D, Gmpr, BckdhB, DdhD2, Astn2, Kcnip2; Figure S5B). Bars, means ± SEM of two biological replicates; each qPCR reaction with three technical replicates.
(C) As for (B).
(D) The enrichment of E-box, a de novo c-Myc (see Figure 1B), Oct4, Sox2, and Klf4 motifs within the OSK (gray bars) versus OSKM (black bars) are plotted against frequency of sites containing the motifs allowing no more than one bp mismatch. The threshold enrichment for each motif in random genomic sequences is displayed as dotted line. The data indicate that c-Myc is driven to OSKM sites in part by the novel, variant motif ***p<0.0001.
See also Figure S5.
Strikingly, we detected minimal or background signals of c-Myc at the OSKM “together” target sites in the absence of OSK and nearly a 40-fold increase in binding of c-Myc in the presence of OSK (Figures 3B, S5B). No c-Myc binding was observed at OSK “only” sites with or without OSK (Figure 3B, S5B) and OSK did not enhance c-Myc binding to sites where c-Myc binds by itself (not shown). We conclude that OSK together act as pioneer factors, directly enabling c-Myc binding at 48 hr.
We also found that c-Myc enhanced, by more than 2 fold (p<0.001–0.0001), the occupancy of O, S, and K to the OSKM “together” sites, while having no effect on the occupancy of O, S, and K to OSK “only” sites (Figure 3C, S5B), indicating cooperative binding of c-Myc with OSK. We used the MAST algorithm (Bailey and Gribskov, 1998) on our ChIP-Seq dataset to quantify the enrichment of the canonical E-box (Myc:Max) motif, the de novo motif we discovered in early reprogramming for c-Myc (Figure 1B), and the canonical motifs for O, S, and K. By allowing one mismatch for each consensus (p<10e-4), we found Oct4, Sox2, Klf4, and de novo c-Myc motifs, but not the E-box motif, to be overrepresented in OSKM co-bound regions compared to random DNA of similar sizes (Figure 3D, black bars; red dotted line indicates random DNA threshold). While Oct4, Sox2, and Klf4 binding motifs were equally enriched in OSK-versus OSKM-bound regions, the de novo c-Myc motif was underrepresented in OSK regions (Figure 3D, compare black to gray bars, p<0.0001). In conclusion, while the OSK factors allow c-Myc to access the OSKM sites, c-Myc also cooperatively enhances the initial OSK engagement with chromatin, driven in part by a variant c-Myc binding motif.
Oct4, Sox2, and Klf4 bind as pioneer factors to closed chromatin
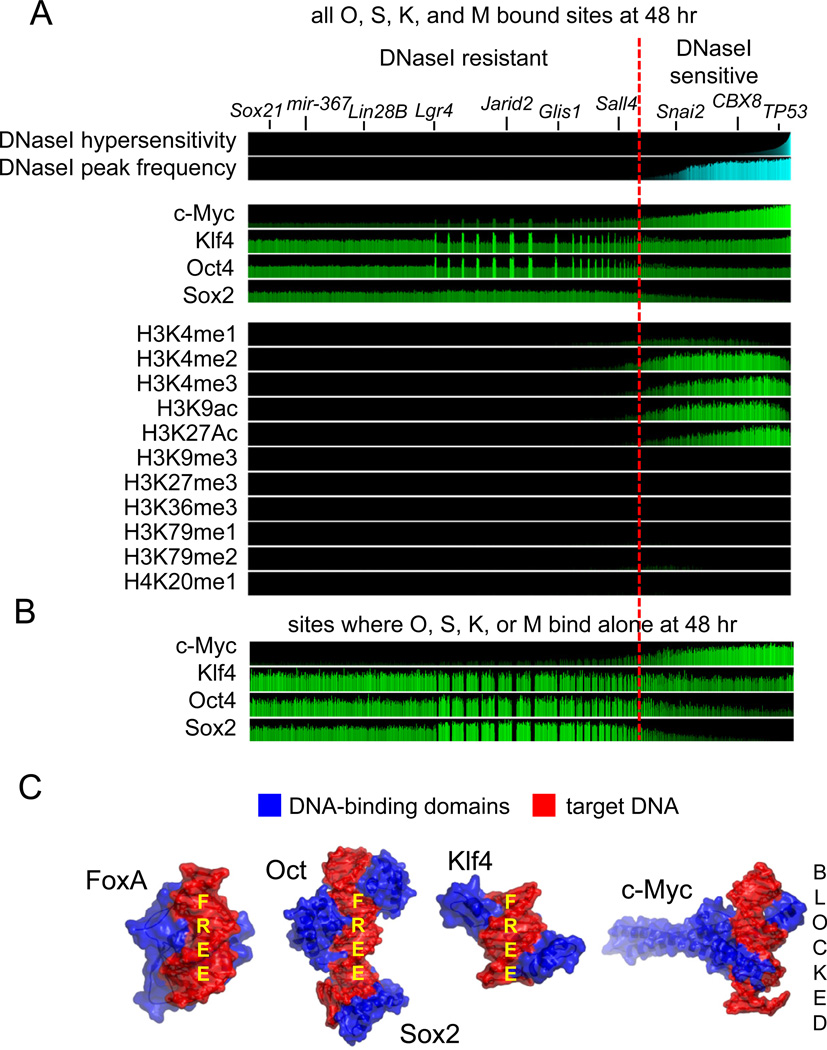
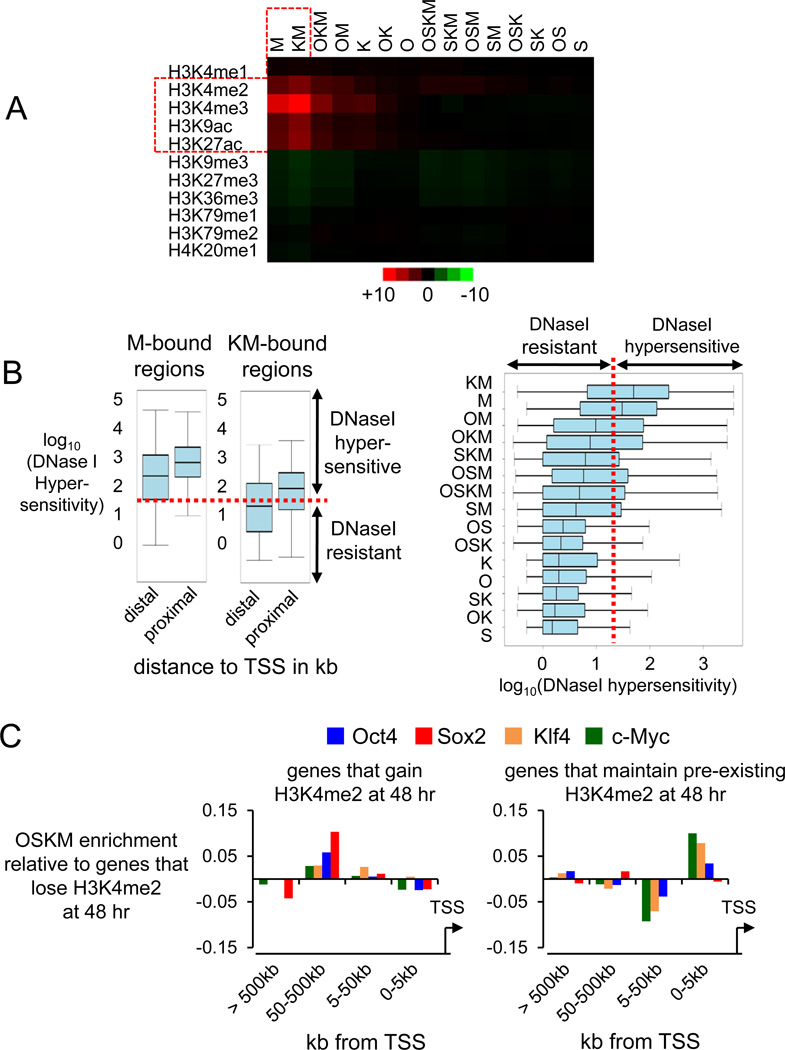
To further assess the pioneer activity of the pluripotency factors, we determined where they bound at 48 hr with regard to pre-existing DNaseI hypersensitive sites, using human BJ fibroblast data from the UW ENCODE project (Myers et al., 2011). Remarkably, we found that the majority (~70%, n=127,059) of all Oct4, Sox2, Klf4 and, to a much lesser extent, c-Myc initial binding events at 48 hr occur in DNaseI-resistant regions, representing closed chromatin (Figure 4A). Only 32% of OSKM binding events (n=61,536) occurred at pre-existing DNase-sensitive regions, mostly by c-Myc, Klf4, and Oct4. Within the DNaseI-resistant regions, sites that are most enriched for Oct4, Klf4, and c-Myc are co-bound by multiple factors (Figure 4, “spikes” in ChIP-Seq signals). Notably, many of the genes bound that promote reprogramming reside in the DNaseI-resistant portion of the genome (Figure 4A, top).
Figure 4. Oct4, Sox2, and Klf4 initially access closed chromatin extensively.
(A) For each of the 188,595 O, S, K, and/or M-bound sites at 48 hr (ave. peak width 208 bp), DNaseI sensitivity is displayed in pre-infected fibroblasts and the sites are rank ordered by degree of hypersensitivity (top cyan panel), with the DNaseI peaks occurrence shown in the second cyan panel. A red line separates the hypersensitive from the resistant regions. The top 4 green panels show read counts for O, S, K, and/or M within the bound sites at 48 hr, followed by read counts of ChIP-seq of designated histone modifications in uninfected fibroblasts (Lister et al., 2011). All read counts were input- and length-normalized and shown per million mapped tags. Genes at the top are relevant to reprogramming, validated for OSKM binding at 48 hr (Table S2), and reside in DNase-resistant chromatin. Spikes in the DNase-resistant regions for O,S,K,M are where the factors bind strongly together.
(B) As in A, but for the 156,401 sites where O, S, K, or M each bind alone at 48 hr.
(C) The three dimensional structures of the DNA-binding domains (blue) of FoxA, Oct-Sox2 (PDB-1GT0), Klf4 (PDB-2WBU), and cMyc (PDB-1NKP) in complex with their specific respective DNA sites (red). Free DNA surfaces are indicated, showing that OSK but not M could potentially bind DNA sites simultaneously with other proteins.
In asessing where the factors each bind alone at 48 hr, it is clear that c-Myc alone binds strictly to open chromatin regions, whereas Oct4, Sox2, and Klf4 each bind extensively, alone, to closed chromatin (Figure 4B). We note that these findings are consistent with the ability of O, S, and K, but not M, to bind on one side of the DNA helix (Nair and Burley, 2003; Remenyi et al., 2003; Schuetz et al., 2011), leaving the other side of DNA potentially bound to histone octamers or other chromatin complexes, as seen for FoxA (Cirillo et al., 1998; Clark et al., 1993) (Figure 4C).
Most initial Oct4, Sox2, and Klf4 binding sites lack an evident histone mark
We counted the ChIP-seq reads in uninfected human fibroblasts for pre-existing histone modifications (Lister et al., 2011) within each site where O, S, K, and M initially bind, focusing on the immediate environment of binding events. Put simply, ~70% of the sites where O, S, K, and/or M bound were not enriched for any histone modification tested (Figures 4A, 5A).
Figure 5. OSKM are not strongly directed by pre-existing histone modifications.
(A) The enrichment of the pre-existing histone modifications within the OSKM bound regions at 48 hr is displayed as a heatmap correlation. The counts of histone modifications ChIP-seq reads under each O, S, K, and/or M-bound region were input- and length-normalized and expressed as reads per million mapped tags. The color display ranges from +10 reads per million (red) to −10 reads per million (green). Red outlined regions denote marked correlations.
(B) Left. DNaseI hypersensitivity within regions co-bound by c-Myc alone and Klf4 and c-Myc (KM) were divided into groups based on their distance to TSS (distal, more than 1 kb away from TSS; proximal, within 1 kb of TSS) and assessed for DNaseI hypersensitivity, as shown in box and whisker plots on a log10 scale. The bottom and top of the box are the 25th and 75th percentiles and the middle band is the 50th percentile DNaseI hypersensitivity value; whisker ends are the min and max values. Right. As in the left panel but showing all O, S, K, and/or M combinations at 48 hr. The red dotted line represents the border between hypersensitive and resistant sites.
(C) O, S, K, and M binding enrichment at distal elements of genes that gain the H3K4me2 mark in 48 hr, relative to genes that lose the mark (left). Enrichment of c-Myc and Klf4 at proximal elements (promoters) of genes that maintain an H3K4me2 mark (right). H3K4me2 data of (Koche et al., 2011).
See also Figure S6.
We found that only a minority (~15%) of sites, mainly where c-Myc and Klf4 bind at promoters (Figures 5A, S6) were enriched for the open chromatin marks H3K4me2, H3K4me3, H3K9ac, and H3K27ac. c-Myc alone or in combination with Klf4 often binds promoters that are hypersensitive to DNaseI, consistent with their enrichment for the open histone marks (Figure 5B, left panel). However, c-Myc on its own can only access distal enhancers within open chromatin, unlike KM binding to enhancers within closed chromatin (Figure 5B, left panel). In virtually all cases, c-Myc binding to closed sites is dependent upon the pioneer activity of the OSK factors (Figure 5B, right panel).
As expected, the subset of sites that will be bound by O, S, K, and/or M and that were enriched for the open enhancer mark H3K4me1 (~17% of total sites) were located at distal elements, including functional enhancers (Figure S6B). OSKM binding to distal locations (50–500 kb) correlates with genes that gain the H3K4me2 mark de novo at 48 hr (Koche et al., 2011), relative to distal locations of genes that lose the mark (Figure 5C, left). The promoters of active genes in fibroblasts that maintain the H3K4me2 mark are bound by c-Myc and Klf4 and include genes that change expression at 48 hr (Figures 5C, right; S6C). In summary, we find minimal effects of pre-existing open histone modifications on guiding the initial binding of O, S, K, and M to distal elements; early chromatin changes occur at genes to which OSKM bind distally; and Oct4, Sox2, and Klf4 act as pioneer factors for c-Myc at distal elements and promoters.
Megabase-scale, H3K9me3-containing Differentially Bound Regions are refractory to initial OSKM binding
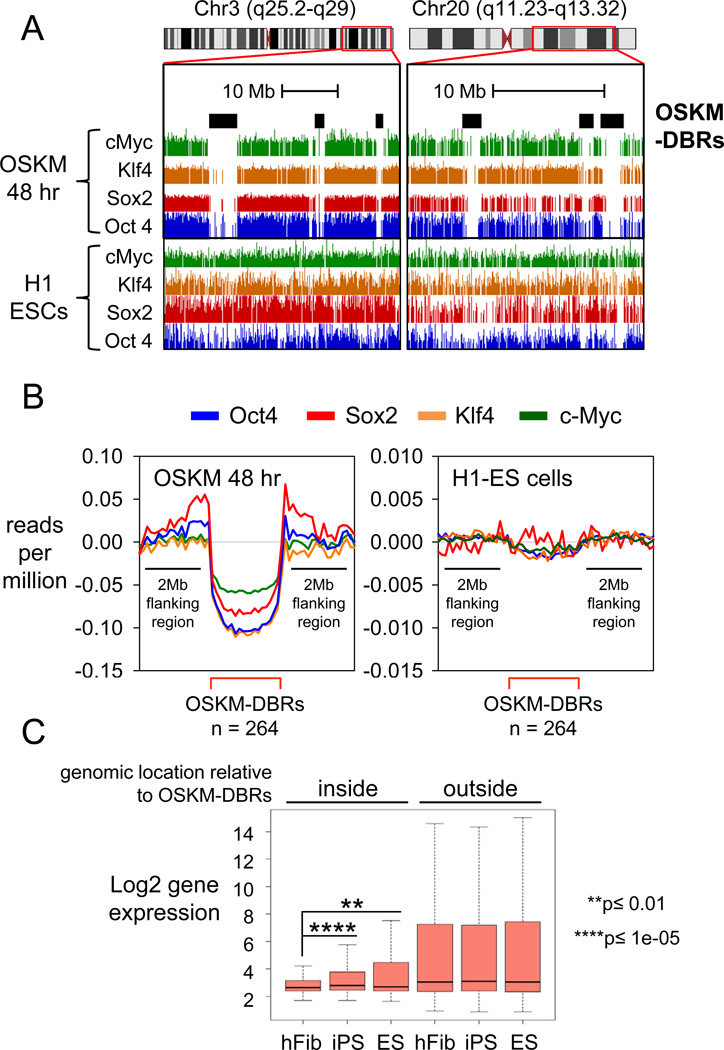
To identify impediments to the initial access of OSKM to chromatin, we searched for regions in the human genome that lacked OSKM binding at 48 hr and compared them to OSKM binding in H1-hES cells (Lister et al., 2009). We identified 264 contiguous swaths of the fibroblast genome of 2.2 megabase average size that were refractory to OSKM binding at 48 hr, compared to that in hES cells, which we call “OSKM Differentially Bound Regions” (OSKM-DBRs) (Figure 6A and 6B). Interestingly, Gene Ontology reveals late genes that are required for pluripotency, such as NANOG, DPPA4, and PRDM14 within OSKM-DBRs, along with many others required for cell differentiation (Table S4). Overall, the DBRs are gene-poor by about two-fold versus the rest of the genome and enriched for repeat elements (Table S4). While the expression of genes outside OSKM-DBRs is relatively similar between hFib, iPS, and ES cells, the expression of genes inside them is higher in ES and iPS compared to hFib cells (Figure 6C) and remains silent after 48 hr of ectopic OSKM expression (Figure S7A). Thus, despite the pioneer activity of the pluripotency factors at many genomic sites, large chromatin domains in differentiated cells are refractory to initial OSKM binding and the impediment is overcome during reprogramming.
Figure 6. Megabase-sized domains of the genome are refractory to the initial binding of OSKM.
(A) Examples of diminished OSKM ChIP-seq signals within OSKM-DBRs (black rectangles) at 48 hr after induction (top four tracks) compared to those in H1-ES (bottom four tracks).
(B) The average reads count for O, S, K, and M (blue, red, orange, and green lines respectively) within DBRs (red bracket) compared to 2 MB flanking regions at 48 hr (left) and H1-ES (right). Reads count is input subtracted, lane-normalized, and expressed as reads per million mapped reads. Size and number of OSKM-DBRs at 48 hr vs. ES cells are indicated.
(C) Genes are typically silent within OSKM-DBRs and exhibit activation in iPS and ES cells. Microarray gene expression in hFib, hFib-iPS and H1-ES is shown as box and whisker plots along a log2 scale.
See also Figure S7 and Table S4.
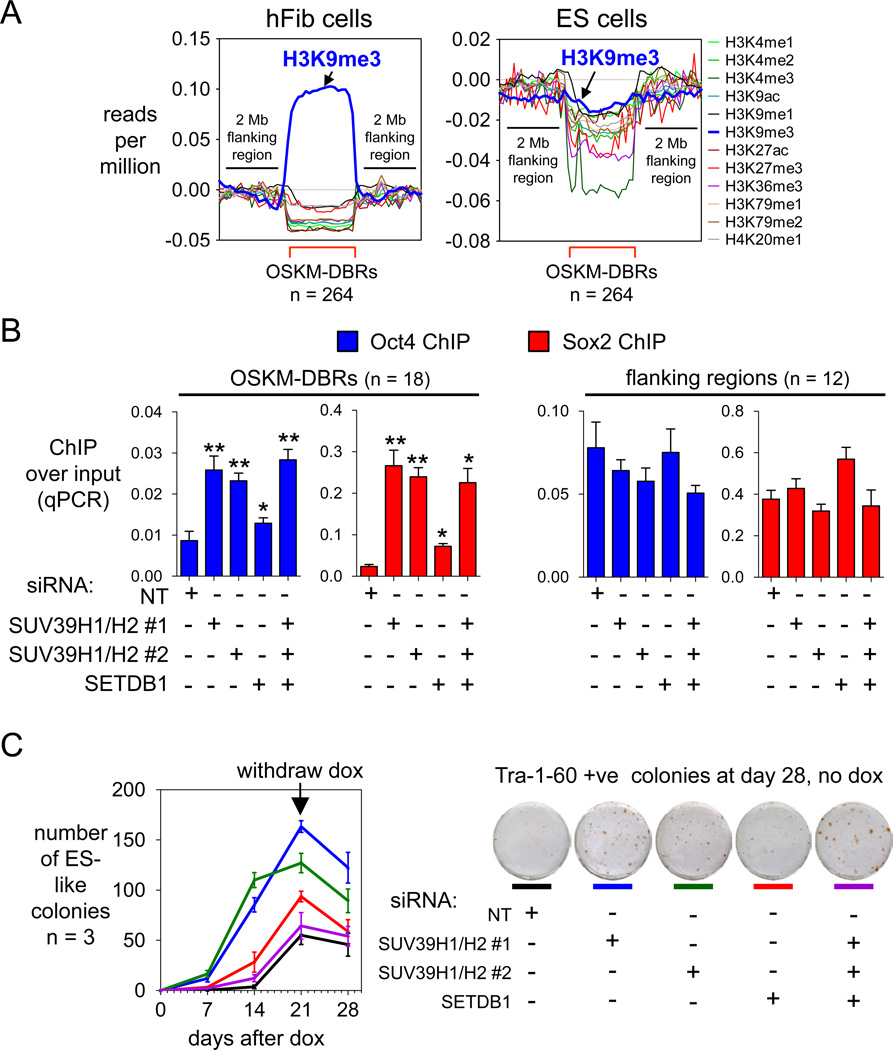
A recent screen of 21 histone modifiers showed that knocking down SUV39H1, which elicits H3K9me3 in heterochromatin, enhances reprogramming markedly (Onder et al., 2012), but the basis for the effect was not clear. Upon assessing the ChIP-seq tags of 12 different histone modifications, we observed a striking average enrichment for the H3K9me3 mark specifically at OSKM-DBRs in hFib cells, but not in H1-hES cells (Figure 7A). The OSKM-DBRs showed no such consistent enrichment for any other histone mark. H3K9me3 was about 10-fold more enriched, on average, than H3K9me2 at OSKM-DBRs (Figure S7B); this is like other studies showing only a partial concurrence of the two modifications (Chandra et al., 2012; Wen et al., 2009).
Figure 7. H3K9me3 is markedly enriched at OSKM-DBRs and is an impediment to initial OSKM binding.
(A) Read counts of ChIP-seq of histone modifications in uninfected human fibroblasts (left panel) and human ES cells (right panel) (Lister et al., 2011) are displayed within DBRs (red brackets) compared to 2 MB flanking regions. Read counts were input- and length-normalized and shown per million mapped tags.
(B) Fibroblasts were treated with either NT siRNA, SUV39H1/H2 siRNA, or SETDB1 siRNA twice before being infected with OSKM+rtTA lentiviruses, and OSKM expression was induced for 48 hr with dox (see Figure S7E,F). ChIP-qPCRs for Oct4 and Sox2 were carried out at 18 sites within DBRs and 12 sites flanking each DBR (Table S2) as controls. Assayed as in Figure 3B.
(C) Knock-down of H3K9 histone methyltransferases increases the speed with which colonies appear after inducing OSKM with dox (left). Knockdown of SUV39H1and SUV39H2 leads to an increase in Tra-1-60 positive colonies, indicating enhanced reprogramming (right).
See also Figure S7 and Table S4.
Further analysis of the OSKM-DBRs in “pre-infected” fibroblasts revealed them to be more DNase-resistant than flanking domains and to contain more highly demethylated DNA than in ES cells (Figure S7C). Strikingly, 75% of the OSKM-DBRs contain an overlap with lamin-associated domains (LADs) over 50% of the DBR sequence (Table S4), though we note that LADs are far more prevalent in the genome than the DBRs (Guelen et al., 2008). In summary, the OSKM-DBRs possess features of heterochromatin. Interestingly, the OSKM-DBR locations we mapped in fibroblasts exhibit varying H3K9me3 levels in different cell types (Figure S7D).
To investigate the role of H3K9me3 at OSKM-DBRs in fibroblasts, we transiently knocked down the related histone methyltransferases SUV39H1 and SUV39H2 together, or SETDB1 alone, or all three together with a two-stage siRNA knockdown strategy, compared to a non-targeting (NT) control siRNA. After 48 hr induction of OSKM in the knock-down cells, we performed ChIP-qPCR for Oct4 and Sox2 at 18 different sites inside OSKM-DBRs and 12 sites in flanking regions. Oct4 and Sox2 were significantly more enriched at sites inside OSKM-DBRs after inducing OSKM for 48 hr in the knock-down cells, compared to hFib cells treated with the NT siRNA (p<0.05) (Figure 7B). Importantly, we observed no difference in Oct4 and Sox2 enrichment at sites flanking OSKM-DBRs (Figure 7B). Also, knocking down SUV39H1/H2, which greatly diminished H3K9me3, allowed more access for Oct4 and Sox2 to DBRs with respect to the SETDB1 knock-down, which exhibited less diminution of H3K9me3 (Figure 7B, S7E,F). Thus, H3K9me3 impedes the initial access of OSKM to the DBRs. Overall, knocking down any of the H3K9 methyltransferases significantly accelerated the appearance of newly reprogrammed colonies (Figure 7C). More specifically, SUV39H1/H2 knockdown significantly enhanced the efficiency of reprogramming after dox withdrawal, as tested by Tra-1-60 staining (Figure 7C). Thus, H3K9me3 impedes reprogramming by blocking the initial engagement of OSKM at broad chromatin domains, some of which are necessary for promoting pluripotency.
DISCUSSION
Most studies of the ability of ectopic transcription factors to convert cells of one fate to another have largely been phenomenological; that is, seeing what works and how cells respond to the factors. Studies of OSKM being able to convert differentiated cells to pluripotency (Takahashi and Yamanaka, 2006) have focused on changes in gene expression and histone modification at early time points (Koche et al., 2011; Mah et al., 2011) and chromatin occupancy at the “pre-iPS” stage, well on the path to conversion (Sridharan et al., 2009). Our analysis reveals striking differences between the initial binding of the factors compared to later stages and unanticipated insights about what needs to be overcome for successful reprogramming. Notably, ectopically expressed factors extensively engage distal regulatory elements first; thus, subsequent events are required for the factors to engage promoters and activate transcription. O, S, and K act as pioneer factors at closed chromatin sites and their binding is not predominantly dictated by pre-existing histone modifications. c-Myc has a direct function, specified in part by a variant DNA binding motif, in enhancing the initial chromatin engagement of O, S, and K. And despite the pioneer activity of O, S, and K, we found megabase-sized regions of the genome that are refractory to OSKM binding and thereby impede the pace of reprogramming.
To what extent is the initial binding pattern in bulk fibroblast cells reflective of the minority of cells that will successfully reprogram? Three points argue that positive and negative features observed at 48 hr are informative in this regard. First, successfully reprogrammed human iPS cells do not appear for weeks; it is not the case that fully reprogrammed cells initially appear and then amplify their numbers (see Introduction). Many genes required for reprogramming are not expressed until late in the process, such as Nanog, which we find to be in a heterochromatic OSKM-DBR and not bound by OSKM in the first 48 hr. Indeed, we found that H3K9me3 at DBRs provides an impediment to OSKM binding and that knocking down enzymes that elicit this mark increases the pace of reprogramming. Further work is needed to understand the steps by which the OSKM-DBRs in fibroblasts lose H3K9me3 and allow factor binding in pluripotent cells, and the means by which heterochromatic DBR-like locations that resist reprogramming may differ across the genomes of different cell types. Along these lines, we note that 22 of the OSKM-DBRs correspond to “iPS-DMR” regions that fail to lose the H3K9me3 mark and fail to gain DNA methylation in human iPS cells, compared to ES cells (Lister et al., 2011) (Table S4). Thus a deficiency in converting a small subset of OSKM-DBRs may contribute to differences between iPS and ES cells.
A second point arguing that features of the bulk OSKM binding pattern at 48 hr are relevant to reprogramming stems from the binding of OSKM to many genes that are crucial for the process, including Glis1, mir-367/302, MET genes, Arid1B, Jarid2, Eed, Suz12, and Sall4 (Table S3). We find that such binding initially occurs at distal conserved elements or enhancers (Figure 2C, S4A,B), preceding marked transcriptional activation (Figure 2A,D), but correlating with where H3K4me2 changes occur at associated genes (Figure 5C). The binding of factors to enhancers first, prior to binding promoters and gene activity, is reminiscent of early developmental progenitor cells that are “marked” by pioneer factor binding at enhancers (Gualdi et al., 1996; Xu et al., 2009). Along these lines, we found that each of the Oct4, Sox2, and Klf4 factors, on their own, could extensively engage DNaseI-resistant chromatin lacking detectable histone marks and that the factors could permit c-Myc to gain access to its target sites where c-Myc could not do so on its own; these are all features expected of pioneer factors (Zaret and Carroll, 2011). Thus, early events in ectopic factor occupancy at reprogramming gene targets mimic regulatory mechanisms seen in natural progenitor cell contexts.
A third point arguing that studying early events in the bulk population is relevant to reprogramming is from novel insights gained about c-Myc. c-Myc is not absolutely required for reprogramming, yet c-Myc ectopic expression along with OSK dramatically enhances the efficiency and the kinetics of the early steps of the process, particularly in human cells (see Introduction). Our work provides a new explanation for the early role of c-Myc: We find that c-Myc facilitates, though is not absolutely required for, the initial engagement of Oct4, Sox2, and Klf4 with many chromatin sites, with no indirect effect on OSK engagement at sites lacking a c-Myc motif and lacking c-Myc binding (Figure 3C,D). Thus we suggest that c-Myc has a direct mechanistic role in facilitating the action of OSK.
A striking difference between the initial OSKM network and that seen in pluripotent cells is that initially, Oct4, Sox2, and Klf4 bind extensively with c-Myc at distal elements of silent genes, whereas in pre-iPS and ES cells, c-Myc targets a distinct network of mainly active genes (see Introduction). Indeed, many such targets at 48 hr are for genes that promote apoptosis and senescence (Figures 2B, S3D), which is prominent within 48 hr when c-Myc is used with OSK to reprogram cells (Figure 3A). Thus, in the bulk population, many of ectopic binding events contribute negatively to reprogramming. We suggest that the initial binding of ectopic transcription factors to such genes may serve as a protective mechanism to eliminate cells in which aberrant transcription factor expression has occurred, thereby preventing deleterious trans-differentiation and metaplasia.
In conclusion, the initial binding of OSKM to the fibroblast genome has provided diverse insights into the parameters that promote and impede the reprogramming process. By stepwise dissecting the mechanisms of reprogramming, starting from the initial binding of pioneer-like factors, we ultimately expect to improve the quality and efficiency of the process.
EXPERIMENTAL PROCEDURES
Cell Culture
Human foreskin fibroblast cells (BJ) were obtained from the ATCC (CRL-2522) at passage 6 and cultured in the ATCC-formulated Eagle's Minimum Essential Medium supplemented with 10% FBS at 37°C and 5% CO2. The human H1-ES line (Thomson et al., 1998) and our derived hFib-iPS cell lines were maintained as described (Lerou et al., 2008). Details on embryoid body cultures and other procedures below are in the Supplemental Experimental Procedures.
Viral Transduction and iPS Generation
We generated iPS cell lines as described (Hockemeyer et al., 2008; Park et al., 2008). Briefly, BJ cells at passage 10 were infected with lentiviruses encoding for dox-inducible Oct4, Sox2, Klf4, and c-Myc, along with lentiviruses expressing rtTA2M2 in the presence of 4.5 µg/ml polybrene. OSKM factors were induced by treating the cells with 1µg/ml dox for 48 hours. The cells were transferred onto irradiated MEFs and cultured in human ES media supplemented with 1µg/ml dox for 28 days. Dox was then withdrawn and the cells were cultured for 7 more days, when ES-like colonies were picked and expanded for 10 passages without dox.
Chromatin Immunoprecipitation (ChIP)
Chromatin fragments associated with Oct4, Sox2, Klf4, or c-Myc were prepared from 107 cells using 10 µg of antibodies for Oct4 (Abcam # ab19857), Sox2 (R&D # AF2018), Klf4 (R&D # AF3640), and c-Myc (R&D # AF3696). For ChIP of the OSKM factors in early reprogramming, the chromatin was obtained from BJ cells infected with OSKM plus rtTA2M2 lentiviruess and uninfected BJ control cells treated with 1 µg/ml dox for 48 hours, prior to ES culture. For ChIP of the OSKM factors in ES/iPS cells, the chromatin was obtained from hFib-iPS cells at passage 15 and human H1-ES cells (Thomson et al., 1998) at passage 35 that were expanded under feeder-free conditions for 3 passages (Ludwig et al., 2006).
Quantitative PCR (qPCR) Analysis
DNA from ChIP was amplified (ABI 7900HT Real-Time PCR System) with Power SYBR Green qPCR mix (ABI #4367659) as follows: 50° C for 4 min, 95° C for 10 min, then 45 cycles of 95° C for 15 sec, 54° C for 15 sec, and 72° C for 45 sec. Gene targets and oligonucleotides are in Table S2. PCR specificity for each primer pair was measured by gel electrophoresis and melting curve analysis. PCR efficiency for each primer pair was set between 90 and 100% by generating a standard curve from a 5 log dilution range of input DNA (slope of ~ 3.2 and R2 > 0.98). Threshold cycle values (Ct) used in our analysis were from 3 PCR replicates (S.D. ≤0.15) and fit within the ±8% dynamic range of the standard curve. The enrichment of ChIP DNA over input DNA was calculated by enrichment = 2(CtChIP DNA -Ctinput DNA).
Identification of OSKM Binding Sites and Computational Analysis
OSKM DNA libraries were sequenced using an Illumina GA2 Sequencer and aligned to the human genome (NCBI v36 assembly) using ELAND (default parameters). Non-unique sequence tags were removed from consideration. The remaining tags were used to call peaks with MACS (MFOLD=16) and were controlled for a False Discovery Rate (FDR) of 0.005 (0.5%). Computational analyses were performed as described by references in the text and in the Supplemental Experimental Procedures. See GEO repository GSE36570 for data.
Supplementary Material
Acknowledgements
We thank J. Kim for iPS reagents and G. Blobel, K. Blahnik, J. Caravaca, M. Iwafuchi-Doi, and J. Kim for comments on the manuscript and K. Kaestner and J. Schug for sequencing by the UPenn Diabetes Research Center Functional Genomics Gore (P30-DK19525). The work was supported by NIH grants R37GM36477 and P01GM099134 to K.S.Z.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Adams CC, Workman JL. Binding of disparate transcriptional activators to nucleosomal DNA is inherently cooperative. Mol Cell Biol. 1995;15:1405–1421. doi: 10.1128/mcb.15.3.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anokye-Danso F, Trivedi CM, Juhr D, Gupta M, Cui Z, Tian Y, Zhang Y, Yang W, Gruber PJ, Epstein JA, et al. Highly efficient miRNA-mediated reprogramming of mouse and human somatic cells to pluripotency. Cell Stem Cell. 2011;8:376–388. doi: 10.1016/j.stem.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Gribskov M. Combining evidence using p-values: application to sequence homology searches. Bioinformatics. 1998;14:48–54. doi: 10.1093/bioinformatics/14.1.48. [DOI] [PubMed] [Google Scholar]
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brambrink T, Foreman R, Welstead GG, Lengner CJ, Wernig M, Suh H, Jaenisch R. Sequential expression of pluripotency markers during direct reprogramming of mouse somatic cells. Cell Stem Cell. 2008;2:151–159. doi: 10.1016/j.stem.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryne JC, Valen E, Tang MH, Marstrand T, Winther O, da Piedade I, Krogh A, Lenhard B, Sandelin A. JASPAR, the open access database of transcription factor-binding profiles: new content and tools in the 2008 update. Nucleic Acids Res. 2008;36:D102–D106. doi: 10.1093/nar/gkm955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra T, Kirschner K, Thuret JY, Pope BD, Ryba T, Newman S, Ahmed K, Samarajiwa SA, Salama R, Carroll T, et al. Independence of Repressive Histone Marks and Chromatin Compaction during Senescent Heterochromatic Layer Formation. Mol Cell. 2012;47:203–214. doi: 10.1016/j.molcel.2012.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- Cirillo LA, Lin FR, Cuesta I, Friedman D, Jarnik M, Zaret KS. Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol Cell. 2002;9:279–289. doi: 10.1016/s1097-2765(02)00459-8. [DOI] [PubMed] [Google Scholar]
- Cirillo LA, McPherson CE, Bossard P, Stevens K, Cherian S, Shim EY, Clark KL, Burley SK, Zaret KS. Binding of the winged-helix transcription factor HNF3 to a linker histone site on the nucleosome. EMBO J. 1998;17:244–254. doi: 10.1093/emboj/17.1.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark KL, Halay ED, Lai E, Burley SK. Co-crystal structure of the HNF-3/fork head DNA-recognition motif resembles histone H5. Nature. 1993;364:412–420. doi: 10.1038/364412a0. [DOI] [PubMed] [Google Scholar]
- Filion GJ, van Bemmel JG, Braunschweig U, Talhout W, Kind J, Ward LD, Brugman W, de Castro IJ, Kerkhoven RM, Bussemaker HJ, et al. Systematic protein location mapping reveals five principal chromatin types in Drosophila cells. Cell. 2010;143:212–224. doi: 10.1016/j.cell.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf T, Enver T. Forcing cells to change lineages. Nature. 2009;462:587–594. doi: 10.1038/nature08533. [DOI] [PubMed] [Google Scholar]
- Gualdi R, Bossard P, Zheng M, Hamada Y, Coleman JR, Zaret KS. Hepatic specification of the gut endoderm in vitro: cell signaling and transcriptional control. Genes Dev. 1996;10:1670–1682. doi: 10.1101/gad.10.13.1670. [DOI] [PubMed] [Google Scholar]
- Guelen L, Pagie L, Brasset E, Meuleman W, Faza MB, Talhout W, Eussen BH, de Klein A, Wessels L, de Laat W, et al. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature. 2008;453:948–951. doi: 10.1038/nature06947. [DOI] [PubMed] [Google Scholar]
- Hanna J, Saha K, Pando B, van Zon J, Lengner CJ, Creyghton MP, van Oudenaarden A, Jaenisch R. Direct cell reprogramming is a stochastic process amenable to acceleration. Nature. 2009;462:595–601. doi: 10.1038/nature08592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockemeyer D, Soldner F, Cook EG, Gao Q, Mitalipova M, Jaenisch R. A drug-inducible system for direct reprogramming of human somatic cells to pluripotency. Cell Stem Cell. 2008;3:346–353. doi: 10.1016/j.stem.2008.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong H, Takahashi K, Ichisaka T, Aoi T, Kanagawa O, Nakagawa M, Okita K, Yamanaka S. Suppression of induced pluripotent stem cell generation by the p53-p21 pathway. Nature. 2009;460:1132–1135. doi: 10.1038/nature08235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura T, Suzuki J, Wang YV, Menendez S, Morera LB, Raya A, Wahl GM, Belmonte JC. Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature. 2009;460:1140–1144. doi: 10.1038/nature08311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Chu J, Shen X, Wang J, Orkin SH. An extended transcriptional network for pluripotency of embryonic stem cells. Cell. 2008;132:1049–1061. doi: 10.1016/j.cell.2008.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoepfler PS. Why myc? An unexpected ingredient in the stem cell cocktail. Cell Stem Cell. 2008;2:18–21. doi: 10.1016/j.stem.2007.12.004. [DOI] [PubMed] [Google Scholar]
- Koche RP, Smith ZD, Adli M, Gu H, Ku M, Gnirke A, Bernstein BE, Meissner A. Reprogramming factor expression initiates widespread targeted chromatin remodeling. Cell Stem Cell. 2011;8:96–105. doi: 10.1016/j.stem.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerou PH, Yabuuchi A, Huo H, Miller JD, Boyer LF, Schlaeger TM, Daley GQ. Derivation and maintenance of human embryonic stem cells from poor-quality in vitro fertilization embryos. Nat Protoc. 2008;3:923–933. doi: 10.1038/nprot.2008.60. [DOI] [PubMed] [Google Scholar]
- Li R, Liang J, Ni S, Zhou T, Qing X, Li H, He W, Chen J, Li F, Zhuang Q, et al. A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell. 2010;7:51–63. doi: 10.1016/j.stem.2010.04.014. [DOI] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Kida YS, Hawkins RD, Nery JR, Hon G, Antosiewicz-Bourget J, O'Malley R, Castanon R, Klugman S, et al. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature. 2011;471:68–73. doi: 10.1038/nature09798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry WE, Richter L, Yachechko R, Pyle AD, Tchieu J, Sridharan R, Clark AT, Plath K. Generation of human induced pluripotent stem cells from dermal fibroblasts. Proc Natl Acad Sci U S A. 2008;105:2883–2888. doi: 10.1073/pnas.0711983105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig TE, Bergendahl V, Levenstein ME, Yu J, Probasco MD, Thomson JA. Feeder-independent culture of human embryonic stem cells. Nat Methods. 2006;3:637–646. doi: 10.1038/nmeth902. [DOI] [PubMed] [Google Scholar]
- Machanick P, Bailey TL. MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics. 2011;27:1696–1697. doi: 10.1093/bioinformatics/btr189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa M, Yamaguchi K, Nakamura T, Shibukawa R, Kodanaka I, Ichisaka T, Kawamura Y, Mochizuki H, Goshima N, Yamanaka S. Direct reprogramming of somatic cells is promoted by maternal transcription factor Glis1. Nature. 2011;474:225–229. doi: 10.1038/nature10106. [DOI] [PubMed] [Google Scholar]
- Mah N, Wang Y, Liao MC, Prigione A, Jozefczuk J, Lichtner B, Wolfrum K, Haltmeier M, Flottmann M, Schaefer M, et al. Molecular insights into reprogramming-initiation events mediated by the OSKM gene regulatory network. PLoS One. 2011;6:e24351. doi: 10.1371/journal.pone.0024351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maherali N, Ahfeldt T, Rigamonti A, Utikal J, Cowan C, Hochedlinger K. A high-efficiency system for the generation and study of human induced pluripotent stem cells. Cell Stem Cell. 2008;3:340–345. doi: 10.1016/j.stem.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marion RM, Strati K, Li H, Murga M, Blanco R, Ortega S, Fernandez-Capetillo O, Serrano M, Blasco MA. A p53-mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity. Nature. 2009;460:1149–1153. doi: 10.1038/nature08287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, Bejerano G. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Hanna J, Zhang X, Ku M, Wernig M, Schorderet P, Bernstein BE, Jaenisch R, Lander ES, Meissner A. Dissecting direct reprogramming through integrative genomic analysis. Nature. 2008;454:49–55. doi: 10.1038/nature07056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers RM, Stamatoyannopoulos J, Snyder M, Dunham I, Hardison RC, Bernstein BE, Gingeras TR, Kent WJ, Birney E, Wold B, et al. A user's guide to the encyclopedia of DNA elements (ENCODE) PLoS Biol. 2011;9:e1001046. doi: 10.1371/journal.pbio.1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair SK, Burley SK. X-ray structures of Myc-Max and Mad-Max recognizing DNA. Molecular bases of regulation by proto-oncogenic transcription factors. Cell. 2003;112:193–205. doi: 10.1016/s0092-8674(02)01284-9. [DOI] [PubMed] [Google Scholar]
- Nakagawa M, Koyanagi M, Tanabe K, Takahashi K, Ichisaka T, Aoi T, Okita K, Mochiduki Y, Takizawa N, Yamanaka S. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat Biotechnol. 2008;26:101–106. doi: 10.1038/nbt1374. [DOI] [PubMed] [Google Scholar]
- Onder TT, Kara N, Cherry A, Sinha AU, Zhu N, Bernt KM, Cahan P, Mancarci OB, Unternaehrer J, Gupta PB, et al. Chromatin-modifying enzymes as modulators of reprogramming. Nature. 2012 doi: 10.1038/nature10953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park IH, Lerou PH, Zhao R, Huo H, Daley GQ. Generation of human-induced pluripotent stem cells. Nat Protoc. 2008;3:1180–1186. doi: 10.1038/nprot.2008.92. [DOI] [PubMed] [Google Scholar]
- Plath K, Lowry WE. Progress in understanding reprogramming to the induced pluripotent state. Nat Rev Genet. 2011;12:253–265. doi: 10.1038/nrg2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remenyi A, Lins K, Nissen LJ, Reinbold R, Scholer HR, Wilmanns M. Crystal structure of a POU/HMG/DNA ternary complex suggests differential assembly of Oct4 and Sox2 on two enhancers. Genes Dev. 2003;17:2048–2059. doi: 10.1101/gad.269303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samavarchi-Tehrani P, Golipour A, David L, Sung HK, Beyer TA, Datti A, Woltjen K, Nagy A, Wrana JL. Functional genomics reveals a BMP-driven mesenchymal-to-epithelial transition in the initiation of somatic cell reprogramming. Cell Stem Cell. 2010;7:64–77. doi: 10.1016/j.stem.2010.04.015. [DOI] [PubMed] [Google Scholar]
- Schuetz A, Nana D, Rose C, Zocher G, Milanovic M, Koenigsmann J, Blasig R, Heinemann U, Carstanjen D. The structure of the Klf4 DNA-binding domain links to self-renewal and macrophage differentiation. Cell Mol Life Sci. 2011;68:3121–3131. doi: 10.1007/s00018-010-0618-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ZD, Nachman I, Regev A, Meissner A. Dynamic single-cell imaging of direct reprogramming reveals an early specifying event. Nat Biotechnol. 2010;28:521–526. doi: 10.1038/nbt.1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridharan R, Tchieu J, Mason MJ, Yachechko R, Kuoy E, Horvath S, Zhou Q, Plath K. Role of the murine reprogramming factors in the induction of pluripotency. Cell. 2009;136:364–377. doi: 10.1016/j.cell.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taberlay PC, Kelly TK, Liu CC, You JS, De Carvalho DD, Miranda TB, Zhou XJ, Liang G, Jones PA. Polycomb-repressed genes have permissive enhancers that initiate reprogramming. Cell. 2011;147:1283–1294. doi: 10.1016/j.cell.2011.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- Visel A, Minovitsky S, Dubchak I, Pennacchio LA. VISTA Enhancer Browser--a database of tissue-specific human enhancers. Nucleic Acids Res. 2007;35:D88–D92. doi: 10.1093/nar/gkl822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen B, Wu H, Shinkai Y, Irizarry RA, Feinberg AP. Large histone H3 lysine 9 dimethylated chromatin blocks distinguish differentiated from embryonic stem cells. Nat Genet. 2009;41:246–250. doi: 10.1038/ng.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernig M, Meissner A, Cassady JP, Jaenisch R. c-Myc is dispensable for direct reprogramming of mouse fibroblasts. Cell Stem Cell. 2008;2:10–12. doi: 10.1016/j.stem.2007.12.001. [DOI] [PubMed] [Google Scholar]
- Xu J, Watts JA, Pope SD, Gadue P, Kamps M, Plath K, Zaret KS, Smale ST. Transcriptional competence and the active marking of tissue-specific enhancers by defined transcription factors in embryonic and induced pluripotent stem cells. Genes Dev. 2009;23:2824–2838. doi: 10.1101/gad.1861209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret KS, Carroll JS. Pioneer transcription factors: establishing competence for gene expression. Genes Dev. 2011;25:2227–2241. doi: 10.1101/gad.176826.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.