Abstract
Chromatin modifier Swi-independent 3a (SIN3A), together with associated histone deacetylases, influences gene expression during development and differentiation through a variety of transcription factors in a cell-specific manner. Sin3a is essential for the maintenance of inner cell mass cells of mouse blastocysts, embryonic fibroblasts, and myoblasts, but is not required for the survival of trophectoderm or Sertoli cells. To better understand how this transcriptional regulator modulates cells at different developmental stages within a single lineage, we used conditional gene targeting in mice to ablate Sin3a from perinatal quiescent male gonocytes and from postnatal differentiating spermatogonia. Mitotic germ cells expressing stimulated by retinoic acid gene 8 (Stra8) that lacked Sin3a exhibited increased DNA damage and apoptosis, yet collectively progressed through meiosis and spermiogenesis and generated epididymal sperm at approximately 50% of control levels, sufficient for normal fertility. In contrast, perinatal gonocytes lacking Sin3a underwent rapid depletion that coincided with cell cycle reentry, exhibiting 2.5-fold increased histone H3 phosphorylation upon cycling that suggested a prophase/metaphase block; germ cells were almost entirely absent two weeks after birth, resulting in sterility. Gene expression profiling of neonatal testes containing Sin3a-deleted gonocytes identified upregulated transcripts highly associated with developmental processes and pattern formation, and downregulated transcripts involved in nuclear receptor activity, including Nr4a1 (Nur77). Interestingly, Nr4a1 levels were elevated in testes containing Stra8-expressing, Sin3a-deleted spermatogonia. SIN3A directly binds to the Nr4a1 promoter, and Nr4a1 expression is diminished upon spermatogonial differentiation in vitro. We conclude that within the male germline, Sin3a is required for the mitotic reentry of gonocytes, but is dispensable for the maintenance of differentiating spermatogonia and subsequent spermatogenic processes.
Keywords: Sin3a, spermatogonia, DNA damage, cell proliferation, apoptosis, differentiation
Introduction
The mammalian SIN3 protein complex performs multiple functions throughout development by modulating gene expression, influencing nucleosome remodeling, and mediating DNA damage repair and replication timing (Silverstein and Ekwall, 2005). With a core comprised of SIN3A, SIN3B, histone deacetylases (HDACs) 1 and 2, retinoblastoma binding proteins (Rbbps) 1, 4, and 7, suppressor of defective silencing 3 homolog (Suds3), and SIN3A-associated proteins of 18 kDa, 30 kDa, and 130 kDa (SAP18, -30, and -130), the SIN3 complex transiently associates with transcription factors like p53, Rb, E2f4, Rest, and Mxd1 to regulate transcription (McDonel et al., 2009). As a key component of the complex, SIN3A is essential for the early development of mouse embryos 3–6 days after fertilization, with blastocyst inner cell mass cells unable to proliferate in its absence (Cowley et al., 2005; Dannenberg et al., 2005; McDonel et al., 2012). Surprisingly, SIN3A is not required for the development of trophectoderm (McDonel et al., 2012). Proliferation is also impaired in cultured embryonic stem (ES) cells, mouse embryonic fibroblasts (MEFs), and T-cells, but not in epidermal skin cells or Sertoli cells (Cowley et al., 2005; Dannenberg et al., 2005; Fazzio et al., 2008; Nascimento et al., 2011; Payne et al., 2010). Clearly, differences exist among various cell types with respect to the utilization of SIN3A through mechanisms that are not yet defined.
Within the muscle cell lineage of mice, cycling myoblasts lacking Sin3a result in perinatal death, while postmitotic myotubes ablated of Sin3a exhibit structural defects but allow pups to survive at least a week after birth (van Oevelen et al., 2010). Mitotic Sin3a-deleted Sertoli cells can support the early stages of spermatogenesis, yet following terminal differentiation such cells are unable to guide haploid round spermatids through the elongation process (Payne et al., 2010). These observations suggest that some, but not all mitotic cells require SIN3A for survival and proliferation, and that non-proliferating cells lacking Sin3a can exhibit cell cycle-independent defects that affect tissue development and differentiation.
Spermatogenesis is comprised of mitotic, meiotic, and postmeiotic phases with multiple stages of germ cell development within each phase. As such, it provides an ideal system in which to further define the roles of Sin3a within a single cell lineage. Upon the specification of indifferent fetal gonads to form testes in mice, the colonizing primordial germ cells become quiescent gonocytes and do not reenter the cell cycle until shortly after birth (De Felici and McLaren, 1983; McLaren, 1985). Once gonocytes resume proliferation, they become spermatogonia and follow one of two fates: to establish the germline stem cell reservoir or to begin immediate differentiation (Yoshida et al., 2006). Male germ cells exhibiting the potential for stem cell activity are referred to as undifferentiated spermatogonia, and are comprised by single, paired, and aligned Type A cells (Huckins, 1971; Nakagawa et al., 2010; Oakberg, 1971). Mitotic germ cells no longer exhibiting stem cell potential are referred to as differentiating spermatogonia, and consist of Type A1-A4, Intermediate, and B cells. Undifferentiated spermatogonia express the transcription factor promyelocytic leukemia zinc finger (PLZF; ZBTB16), a marker for germline stem cells, while differentiating spermatogonia, in response to retinoic acid, express the stimulated by retinoic acid gene 8 (Stra8) and upregulate the kit oncogene (Kit) (Buaas et al., 2004; Costoya et al., 2004; Zhou et al., 2008). Both undifferentiated and differentiated spermatogonia contain SIN3A, but until now its functions in these cell types had not been characterized (Payne et al., 2010).
Here we have generated two lines of conditional gene targeted mice: one in which Sin3a is ablated from perinatal quiescent male gonocytes, and one in which Sin3a is deleted from Stra8-expressing mitotic spermatogonia. We found strikingly distinct phenotypes between the two lines, reflecting differences in the cell types that are affected within the male germ cell lineage. Sin3a is required for the establishment of undifferentiated mitotic spermatogonia, but is not essential for differentiating germ cells. Our current results establish distinct roles for Sin3a during postnatal male germ cell development and highlight its importance for cell cycle progression in undifferentiated cells.
Materials and methods
Mice
All procedures and care of animals were carried out according to the Children’s Hospital of Chicago Research Center Animal Care and Use Committee. To generate SSKO animals, hemizygous FVB-Tg(Stra8-cre)1Reb/J (Stra8-cre) mice were mated with homozygous floxed 129-Sin3atmnRdp (Sin3afl/fl) mice to generate Stra8-cre;Sin3afl/+ offspring. These F1 animals were then mated with additional Sin3afl/fl mice, or in some cases interbred, to obtain Stra8-cre;Sin3aΔ/fl males. Control animals included Sin3a+/+ (wild type), Sin3aΔ/+ (F2 without transgene), Stra8-cre;Sin3a+/+ (transgenic only), and Stra8-cre;Sin3aΔ/+ (F2 with transgene). To generate VSKO animals, hemizygous FVB-Tg(Ddx4-cre)1Dcas/J (Vasa-cre) mice were mated with homozygous floxed 129-Sin3atmnRdp (Sin3afl/fl) mice to generate Vasa-cre;Sin3afl/+ offspring. These F1 animals were then mated with additional Sin3afl/fl mice, or in some cases interbred, to obtain Vasa-cre;Sin3aΔ/fl males. Control animals included Sin3a+/+ (wild type), Sin3aΔ/+ (F2 without transgene), Vasa-cre;Sin3a+/+ (transgenic only), and Vasa-cre;Sin3aΔ/+ (F2 with transgene). Mice were obtained from The Jackson Laboratory and genotyped by PCR analysis (primers and conditions re available upon request). To assess fertility of SSKO males and recombination efficiency of Stra8-cre on the floxed Sin3a allele, 3 SSKO males and 3 wild type males were mated individually with wild type females, and 3–5 litters were generated. To label cycling cells with 5-Bromo-2′-deoxyuridine (BrdU) at a dose of 100 mg/kg body weight, a single intraperitoneal injection of 10 mg/ml BrdU was administered to 6-wk-old male SSKO and control mice. 48 h later, injected mice were euthanized and testes were removed for analysis.
Histology and immunohistochemistry
Adult (6-wk-old), juvenile (4-, 2-, 1.5-, and 1-wk-old), and neonatal/perinatal (3-, 2-, and 0-days-old) testes were fixed for 8 h, 4–6 h, and 2 h, respectively, at 4°C in Bouin’s solution, rinsed in PBS, and dehydrated for paraffin embedding. Five-micron sections were cut, samples were deparaffinized and rehydrated, and some sections were stained with hematoxylin and eosin (H+E). Other sections were prepared for immunohistochemistry, initially subjecting them to antigen retrieval by boiling in 0.01 M sodium citrate, pH 6.0, for 10 min. Sections were blocked with 3% normal goat serum in PBS for 1 h at room temperature. Primary antibodies were diluted in PBS + 3% goat serum and added to the samples for overnight incubation at 4°C. Control reactions were performed by omitting primary antibodies from the incubations. Following PBS washes, samples were then incubated for 1 h in the dark at room temperature with appropriate fluorescence-conjugated secondary antibodies diluted in PBS + 3% goat serum. Vectashield anti-fade mounting medium (Vector Laboratories, Burlingame, CA) containing DAPI was applied to the samples, and sections were viewed using a Leica DMR-HC epifluorescence microscope. Images were captured by a QImaging Retiga 4000R camera. The following primary antibodies and dilutions were used in these studies: rabbit polyclonal anti-SIN3A (AK-11; sc-767, Santa Cruz Biotechnology, CA) at 1:50, mouse monoclonal anti-SIN3A (G-11; sc-5299, Santa Cruz) at 1:100, rat monoclonal anti-GCNA1 (gift of Dr. George Enders) at 1:100, mouse monoclonal anti-PLZF (D-9; sc-28319, Santa Cruz) at 1:50, rabbit anti-STRA8 (ab49405, Abcam, Cambridge, MA) at 1:200, mouse monoclonal anti-BrdU (clone G3G4, Developmental Studies Hybridoma Bank, Iowa City, IA) at 1:1000, mouse monoclonal anti-Serine139 phosphorylated histone H2A.X (clone JBW301; 05-636, Millipore, Temecula, CA) at 1:500, rabbit polyclonal anti-Ki67 (ab15580, Abcam) at 1:200, rabbit polyclonal anti-phosphorylated (Serine10) histone H3 (ab5176, Abcam) at 1:200, and rabbit polyclonal anti-caspase 3 (ab4051, Abcam) at 1:200. To detect apoptosis by TUNEL labeling, the In Situ Cell Death Detection Kit (fluorescein; 11684795910, Roche Applied Science, Mannheim, Germany) was used according to the manufacturer’s instructions.
Counting epididymal sperm and testicular germ cells
Whole epididymides from adult mice were individually placed into PBS, minced with scissors, and incubated for 60 min at 37°C. Samples were then centrifuged at 100 g for 5 min to remove debris and resuspended in 1 ml PBS. Sperm were counted using a hemocytometer. To assess motility, sperm were collected from excised cauda epididymides and capacitated for 90 min in T6 culture medium at 37°C. Sperm were considered motile if any movement was detected.
For counting GCNA1+ cells that also contained Ki67, pH3, or Casp3, whole testicular cross-sections representing the beginning (slide 1), middle (slide 2) and end (slide 3) of each testis were co-immunostained for 3 VSKO and 3 control replicate males. First, GCNA1+ cells were counted within all seminiferous tubules for slides 1–3 of each testis, and then double-positive (Ki67+GCNA1+, pH3+GCNA1+, or Casp3+GCNA1+) cells were counted in the same sections. Cell counts were performed on images captured using a QImaging Retiga 4000R camera with 2048×2048 pixel resolution, coupled to a Leica DMR-HC epifluorescence microscope. Data are expressed as the percent double-positive cells/total GCNA1+ cells. To reconstruct entire testis cross-sections, smaller image panels of immunostained cross-sections were assembled to line up interiors and edges using NIH ImageJ and Adobe Photoshop software. PLZF+ cells were counted as the number of cells per seminiferous tubule cross-section.
Preparation of single testicular cells and enrichment of spermatogonial subpopulations
The methods of Kofman et al. (2012b) were followed. Briefly, wild type testes were decapsulated and minced in ice-cold 1:1 Dulbecco’s Modified Eagle Medium–Ham’s F-12 Medium. Initial enzymatic digestions at 37°C for 30 min using collagenase IV (1mg/ml) and DNase I (2mg/ml) were administered to remove interstitial Leydig cells and peritubular myoid cells from seminiferous tubules. A second administration of enzymatic digestions at 37°C for 30 min using collagenase IV (1 mg/ml), DNase I (2mg/ml), hyaluronidase (1.5mg/ml), and trypsin (1mg/ml) facilitated the isolation of germ cells and Sertoli cells from the remaining tissue. A final suspension of single cells was prepared in ice-cold PBS containing 0.5% BSA and 2mM EDTA (MACS Buffer) for subsequent enrichment of spermatogonial stem cells and differentiating spermatogonia by magnetic-activated cell sorting (MACS).
Single testicular cells were resuspended with 80 μl MACS Buffer and 20 μl rabbit anti-GFRA1 antibodies (Santa Cruz) and incubated at 4°C for 20 min with rotation. After washes, second incubation of cells in 80 μl MACS Buffer with 10 μl go anti-rabbit antibody-conjugated MicroBeads and 10 μl anti-THY1 antibody-conjugated MicroBeads (Miltenyi Biotech, Auburn, CA) was administered at 4°C for 20 min. with rotation. The labeled cells were filtered through 30-μm pore size mesh to remove cell aggregates, and then sorted through a separation LS column attached to a MidiMACS separator (Miltenyi Biotec). THY1+ and GFRA1+ cells were retained inside the column within the magnetic field, while unlabeled cells passed through the column and were collected as the column-depleted THY1−/GFRA1− cell fraction (CD fraction). After washes with MACS Buffer, the LS column was removed from the magnetic field and the THY1+ and GFRA1+ cells representing the undifferentiated SSC fraction were flushed out. For the enrichment of differentiating spermatogonia, CD fraction cells were subsequently reconstituted in 90 μl MACS Buffer and incubated with 10 μl anti-CD117 (KIT) antibody-conjugated MicroBeads (Miltenyi Biotech) at 4°C for 20 min. with rotation. These samples were then sorted through a MidiMACS LS column to collect the KIT+ cells.
Spermatogonial stem cell cultures and treatment with retinoic acid
Approximately 200,000 THY+/GFRA1+ cells were seeded into 35-mm round dishes containing irradiated mouse embryonic fibroblast feeder layers (1.2 × 106 MEFs per dish). Colonies of spermatogonial stem cells (SSCs) formed within 1–2 wks, and were maintained in optimized culture medium (StemPro-34 supplemented with 1% FBS, 10 μg/ml GDNF, 10 ng/mL bFGF, 20 ng/mL EGF, 1,000 units/mL ESGRO/LIF) for >6 months, replenished with new medium every other day (Seandel et al., 2007). To induce differentiation, culture medium supplemented with 1μM all-trans retinoic acid in 0.1% EtOH (Sigma-Aldrich, St. Louis, MO) was added to SSCs and cells were harvested for analysis 24 h later.
Chromatin Immunoprecipitation
The methods of Dahl and Collas (2008) were followed, with minor changes. Briefly, approximately 100,000 THY1+/GFRA1+ cells were fixed for each chromatin shearing sample, using 1% (v/v) formaldehyde to cross-link at room temperature for 8 min. (Dahl and Collas, 2008). A Bioruptor™ Next Gen sonicator (Diagenode, Liège, Belgium) was used to shear the chromatin samples, with 12 cycles of 30 s. on, 30 s. off. Following immunoprecipitation with either anti-SIN3A antibodies or anti-IgG control antibodies, DNA was recovered in 10% Chelex-100 + Proteinase K (20mg/ml). Samples were incubated at 65°C for 15 min. and then boiled for 15 min. Genomic DNA primers were designed to amplify either the proximal (μ0.3 Kb) or distal (−1.6 Kb) promoter regions of Nr4a1, or the proximal promoter region (−0.3 Kb) of Actb as a negative control. SIN3A is not bound at the Actb promoter (Dannenberg et al., 2005). Primers available on request. Quantitative (q)PCR amplification of 40 cycles was performed, and protein enrichment was calculated as % input.
RNA isolation and quantitative RT-PCR analysis
Total RNA was extracted from samples using either the RNeasy Micro or the RNeasy Mini Kit from Qiagen (Valencia, CA) following the manufacturer’s protocol. RNA samples were treated with RNase-free DNase I (Qiagen) on-column to remove genomic DNA. Yield and quality of RNA samples were determined using the NanoDrop 2000 Spectrophotometer (ThermoScientific, Wilmington, DE). Total RNA was reverse transcribed into cDNA using random hexamer primers (New England Biolabs, Ipswich, MA). For qRT-PCR, cDNA was added to 2× Power SYBR® Green PCR Master Mix (Applied Biosystems, Foster City, CA) with specific oligonucleotide primer sets for the genes of interest. Samples from biological replicates were run in triplicate on an Applied Biosystems 7500 Real-Time PCR System using SYBR® Green dye for read-out and ROX™ dye as an internal reference. Each PCR reaction contained approximately 5–10 ng of cDNA, 1× Power SYBR® Green PCR Master Mix, and 500 nM of each forward and reverse primer for the desired gene. Nr4a1 primers were F (forward): 5′-ATCACTGATCGACACGGGCTCCA-3′ and R (reverse): 5′-TGTGGGTCTCCTGCCACGGT-3′. 18s rRNA was used as an endogenous control transcript. The threshold cycle (CT), indicating the relative abundance of a particular transcript, was calculated for each reaction by the system software. Quantification of the fold change in gene expression was determined by using the formula 2−ΔΔ CT, in which ΔΔCT = [(CT of gene of interest − CT of 18s)A − (CT of gene of interest − CT of 18s)B]. Fold change in transcript levels was plotted using Prism 5 software (GraphPad, La Jolla, CA). Values plotted are mean +/− SEM. Statistical analysis was performed using Prism 5, employing Student’s t-test; *p<0.05; **p<0.01; ***p<0.001.
Gene expression microarrays
For VSKO microarrays, total RNA samples were shipped to the laboratories of Miltenyi Biotec (Auburn, CA), where they were quality-checked prior to processing with the Agilent 2100 Bioanalyzer platform (Agilent Technologies, Santa Clara, CA). Linear T7-based amplification was performed using 100 ng of each RNA sample. Amplification and Cy3 labeling was performed using the Agilent Low Input Quick Amp Labeling Kit (Agilent Technologies) following the manufacturer’s protocol. Yields of cRNA and the dye-incorporation rate were measured using the NanoDrop 2000 Spectrophotometer (ThermoScientific). Hybridization was performed according to the Agilent 60-mer oligo microarray processing protocol using the Agilent Gene Expression Hybridization Kit (Agilent Technologies). The Cy3-labeled cRNAs were hybridized overnight (17 hours at 65°C) onto the Agilent Whole Mouse Genome oligo microarrays 8×60K using a hybridization chamber and oven. Fluorescence signals of the hybridized oligo microarrays were detected using Agilent’s DNA microarray scanner (Agilent Technologies).
Agilent Feature Extraction (AFE) software was used to process the VSKO microarray image files. AFE determines feature intensities and ratios (including background subtraction and normalization), rejects outliers and calculates statistical confidences (p-values). For determining differential gene expression, AFE-derived output data files were further analyzed using the Rosetta Resolver® gene expression data analysis system (Rosetta Biosoftware, Cambridge, MA). AFE-derived output data files contained the gene lists with complete raw data sets. Significantly changed genes were defined as having >2-fold differential expression and p-values <0.001.
For SSKO microarrays, total RNA samples were processed for the Affymetrix Mouse Gene 1.0 ST Array platform (Affymetrix, Santa Clara, CA). Linear T7-based amplification was performed using 200 ng of each RNA sample. Amplification and Cy3-/Cy5-labeling was performed using the WT-Ovation RNA Amplification System (NuGEN Inc., San Carlos, CA) following the manufacturer’s protocol. Yields of cRNA and the dye-incorporation rate were measured using the NanoDrop 2000 Spectrophotometer (ThermoScientific). Hybridization was performed according to the Affymetrix oligo microarray processing protocol (Affymetrix). The Cy3- and Cy5-labeled cRNAs were combined and hybridized overnight (17 hours at 65°C) onto the Affymetrix Mouse Gene 1.0 ST oligo microarrays using a hybridization chamber and oven. Fluorescence signals of the hybridized oligo microarrays were detected using Affymetrix’s DNA microarray scanner (Affymetrix). SSKO microarray image files were processed in a similar manner to the VSKO files, determining feature intensities, ratios, statistical confidences, and differential gene expression. Significantly changed genes were defined as having >1-fold differential expression (due to the smaller number of transcripts) and p-values <0.001.
Gene expression microarray data in this study are available in the public repository Gene Expression Omnibus, accession # GSE38096.
Transcriptome data analysis
Gene ontology (GO) analysis on differentially regulated genes was performed using findGO, a component of the HOMER (Hypergeometric Optimization of Motif EnRichment) software suite (http://biowhat.ucsd.edu/homer). The normalized intensities of VSKO microarray results were also hierarchically clustered using Multiple Experiment Viewer (MeV), a component of the TM4 Microarray software suite (http://www.tm4.org), generating heat maps of genes with similar expression patterns. To further analyze the VSKO and SSKO transcriptomes, we uploaded our data sets into Ingenuity Pathways Analysis (Ingenuity Systems, Redwood City, CA) to generate networks. Each gene identifier was mapped to its corresponding gene object in the Ingenuity Pathways Knowledge Base (IPKB). Genes whose expression was significantly differentially regulated were identified, overlaying them onto a global molecular network using information from the IPKB. Networks were algorithmically generated based on their connectivity, generating graphical representations of the molecular relationships among gene products (representing nodes) by depicting them as solid (direct) or dashed (indirect) lines. All lines are supported by at least one reference from the literature or from canonical information stored in the IPKB. Mouse, rat, and human orthologs of genes are stored as separate objects in the IPKB, but are represented as single nodes in the network. Nodes are displayed using various shapes that represent the functional class of the gene product (i.e. rectangle = nuclear receptor). Canonical pathway analysis utilizes well characterized metabolic and cell signaling pathways that are generated prior to data input and on which identified gene products are overlaid.
Results
Sin3a is not required for differentiating spermatogonia and subsequent spermatogenesis
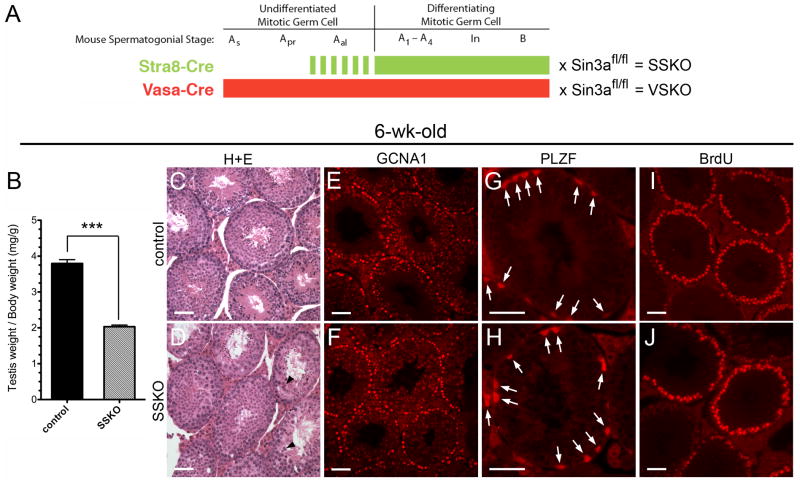
To begin examining the role of Sin3a within the postnatal male germ cell lineage, we generated conditional gene-targeted mice in which Sin3a was genetically ablated in differentiating spermatogonia. Floxed Sin3a mice were intercrossed with Stra8-cre mice that exhibit Cre recombinase activity in differentiating Type A, Intermediate, Type B, and a subset of undifferentiated Type A spermatogonia (Fig. 1A; (Sadate-Ngatchou et al., 2008). As the majority of spermatogonia with stem cell potential do not exhibit Stra8-cre activity, we reasoned that the resulting Sin3a ablation would predominately affect the differentiating mitotic germ cells and their progeny. These Stra8-cre;Sin3aΔ/fl mice will be referred to here as Stra8-cre mediated Sin3a Knock Out (SSKO) animals (Fig. 1A). At 6 weeks of age, SSKO males exhibited a 46.5% reduction in testis weight/body weight (Fig. 1B and Supplementary Fig. 1D), yet contained germ cells at all stages of development, including round and elongating spermatids (Fig. 1C, D). Approximately 20% of SSKO seminiferous tubule cross-sections displayed enlarged, multinucleated germ cells (Fig. 1D, arrowheads), though these abnormal cells were limited to fewer than 5–7 per cross-section. Antibodies raised against the Germ Cell Nuclear Antigen 1 (GCNA1; (Enders and May, 1994), which label germ cells through the diplotene/dictyate stage of the first meiotic division, revealed equivalent numbers of GCNA1+ cells between SSKO and control testes (169 ± 17 cells/cross-section and 187 ± 24 cells/cross-section, respectively; N=33 sections) (Fig. 1E, F). These findings suggest that post-meiotic germ cell loss accounts for the reduced weight of SSKO testes.
Fig. 1.
Testes of Stra8-cre; Sin3aΔ/fl male mice have reduced weights but contain equivalent numbers of undifferentiated spermatogonia and cycling, premeiotic germ cells. A. Schematic illustration denoting the expression patterns of Stra8-cre and Vasa-cre transgenes in mouse spermatogonia: the former is initially expressed in a subset of Type A aligned (Aal) undifferentiated cells (dashed green line) and continues its expression through late Type B differentiating cells (solid green line), while the latter is expressed throughout all spermatogonial stages – undifferentiated A single (As), A paired (Apr), Aal, and differentiating A1–A4, Intermediate (In), and B (solid red line). Matings of these cre driver mice with floxed Sin3a (Sin3afl/fl) mice generate SSKO (Stra8-cre; Sin3aΔ/fl) and VSKO (Vasa-cre; Sin3aΔ/fl) conditional gene-targeted mice, respectively. B. 6-wk-old SSKO male mice have testis weight/body weight ratios that are 46.5% reduced compared to littermate controls (N=7, ***p<0.001). C and D. Seminiferous tubule cross-sections of 6-wk-old testes stained with hematoxylin and eosin (H+E) reveal germ cells at all stages of development in SSKO and controls; arrowheads identify enlarged, multinucleated germ cells in SSKO tubules. E and F. Cross-sections of 6-wk-old seminiferous tubules immunostained for germ cell marker GCNA1. G and H. Cross-sections of 6-wk-old seminiferous tubules immunostained for germline stem cell marker PLZF (arrows). I and J. Seminiferous tubule cross-sections of 6-wk-old males injected with BrdU, examined 48 h later with anti-BrdU antibodies. Scale bars = 50μm.
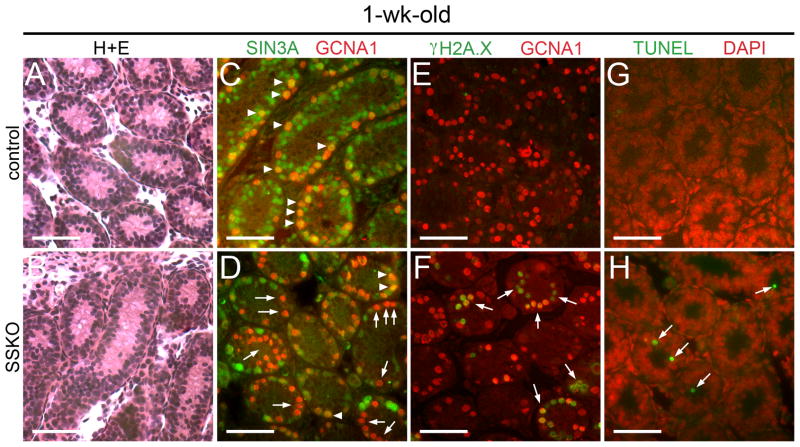
Given the significant reduction in testis weights, we next examined the epididymides of adult SSKO animals and found 54.2% fewer epididymal sperm when compared to controls (Table 1). No differences in the percentage of motile sperm were observed among the samples. To assess fertility, we outcrossed 3 SSKO males and 3 Sin3a+/+ control males to wild type females. Normal litter sizes, sex ratios of pups, and numbers of litters within a 4- or 6-month period were seen with breeding pairs involving the SSKO males (Table 2). Importantly, one of the SSKO males was over 10 months old when he produced the first of three litters, and was 1 year and 6 weeks of age when the third litter was born. We then examined the percentage of offspring carrying a Sin3aΔ allele among the pups born to SSKO fathers in order to determine the Stra8-cre recombination efficiency on the floxed Sin3a allele. All of the offspring born to the 3 SSKO males carried the deleted allele (65/65 pups), indicating that the floxed Sin3a allele underwent efficient recombination in the germ cells of SSKO males (Table 2). To confirm the specificity of recombination, we immunostained cross-sections of 1-week-old SSKO seminiferous tubules for SIN3A and GCNA1. The majority of GCNA1+ cells lacked SIN3A staining in the SSKO samples (84.5% ± 2.6%; N=55 cross-sections), compared with extensive co-localization between the two proteins in controls (97.4% ± 1.2%; N=53 cross-sections) (Fig. 2C, D). When 1-week-old SSKO samples were co-immunostained for germline stem cell marker PLZF and SIN3A (data not shown), co-expression of markers was observed in the majority of cells. However, when SSKO cross-sections were analyzed for the co-distribution of differentiation marker STRA8 and SIN3A, no co-expression could be discerned (Supplementary Fig. 1E). We conclude that the 15% of GCNA1+ that were positive for SIN3A were also positive for PLZF, but negative for STRA8. At 6 weeks of age, SSKO seminiferous tubules contained equivalent numbers of PLZF + spermatogonia (3.20 ± 0.35 cells/cross-section; N=60 sections) when compared to controls (3.23 ± 0.43 cells/cross-section; N=44 sections), suggesting that SSKO testes retain normal germline stem cell potential (Fig. 1G, H, Supplementary Fig. 1A; (Nakagawa et al., 2010). No phenotype was observed in SSKO ovaries, as the Stra8-cre transgene is not active in females (Sadate-Ngatchou et al., 2008).
Table 1.
Sperm analysis of SSKO mice
| Genotype | # of sperm per epididymis (× 106) | % motility |
|---|---|---|
| - ;Sin3a+/+ (n=5) | 3.74 ± 0.91a | 63.5 ± 8.9c |
| - ;Sin3aΔ/+ (n=3) | 3.41 ± 0.57a | 71.2 ± 10.7c |
| Stra8-cre;Sin3a+/+ (n=3) | 3.60 ± 0.83a | 66.7 ± 9.1c |
| Stra8-cre;Sin3aΔ/+ (n=5) | 3.86 ± 1.04a | 58.1 ± 13.5c |
| Stra8-cre;Sin3aΔ/Δ (n=3) | 1.67 ± 0.39b | 60.2 ± 7.8c |
Differences in values are not statistically significant when compared with each other (p > 0.05);
Difference in value compared with each of the a values is statistically significant (p < 0.05);
Differences in values are not statistically significant when compared with each other (p > 0.05)
Table 2.
Assessment of SSKO male fertility and Stra8-cre recombination efficiency on the floxed Sin3a allele
| Breeding pair ♂ × ♀ | % of offspring carrying Sin3aΔ allele | Ages of ♂ mice when litters born (wks) | |
|---|---|---|---|
| SSKO #7431 | WT #2256 | 100% (19/19; 3 litters) | 27 wks, 31 wks, 36 wks |
| SSKO #7463 | WT #2257 | 100% (17/17; 3 litters) | 46 wks, 53 wks, 59 wks |
| SSKO #7502 | WT #2258 | 100% (29/29; 5 litters) | 12 wks, 16 wks, 22 wks, 26 wks, 32 wks |
| Sin3a+/+ #7429 | WT #2263 | 0% (0/18; 3 litters) | 8 wks, 12 wks, 17 wks |
| Sin3a+/+ #7480 | WT #2265 | 0% (0/20; 3 litters) | 7 wks, 12 wks, 16 wks |
| Sin3a+/+ #7496 | WT #2271 | 0% (0/19; 3 litters) | 10 wks, 14 wks, 19 wks |
Fig. 2.
1-wk-old SSKO spermatogonia exhibit a modest increase in DNA double strand breaks and apoptosis. A and B. Seminiferous tubule cross-sections of 1-wk-old testes stained with H+E reveal equivalent numbers of cells in SSKO and controls. C and D. Cross-sections of 1-wk-old seminiferous tubules co-immunostained for GCNA1 (red) and SIN3A (green); arrowheads identify GCNA1+ cells containing SIN3A (C, D), while arrows denote GCNA1+ cells in SSKO sections lacking SIN3A (D). E and F. Cross-sections of 1-wk-old seminiferous tubules co-immunostained for GCNA1 (red) and γH2A.X (green); arrows identify GCNA1+ cells in SSKO sections that exhibit γH2A.X, signifying DNA double strand breaks. G and H. Cross-sections of 1-wk-old seminiferous tubules co-stained for DNA (DAPI, red) and apoptosis (TUNEL, green); arrows denote TUNEL+ cells in SSKO sections. Scale bars = 50μm.
SSKO spermatogonia progress through the cell cycle but exhibit increased DNA damage
Because SIN3A regulates cell cycle progression in other cell types, we next investigated the ability of SSKO spermatogonia to proceed through S-phase by assessing the incorporation of 5-bromo-2′-deoxyuridine (BrdU) 48 h following intraperitoneal administration in 6-week-old animals. No differences in the number of cycling spermatogonia were observed between SSKO and controls, confirming that SIN3A is not essential for the proliferation of differentiating spermatogonia (Fig. 1I, J). While our results suggested that the majority of SSKO germ cell loss occurred after the first meiotic division and throughout spermiogenesis, we wondered whether an early effect in differentiating spermatogonia might be an impaired DNA damage response (McDonel et al., 2012). To test this hypothesis, we immunostained cross-sections of 1-week-old SSKO seminiferous tubules for phosphorylated histone γH2A.X (S139), an indicator of DNA double strand breaks (Rogakou et al., 1998). SSKO testes contained numerous GCNA1+ cells that co-labeled with γH2A.X, while control testes exhibited no coincident staining for the two proteins (Fig. 2E, F). Furthermore, 1- and 6-week-old SSKO seminiferous tubules exhibited TUNEL-positive cells at a higher frequency than controls, providing evidence of increased apoptosis (Fig. 2G, H, Supplementary Fig. 1B, C). These data suggest that SIN3A promotes DNA damage repair in differentiating spermatogonia but that its absence likely involves compensation by other factors, as the majority of mitotic germ cells survive and enter meiosis.
Orphan nuclear receptor-encoding gene Nr4a1 is upregulated in SSKO testes
To enhance our understanding of SIN3A’s role in differentiating spermatogonia and to identify potential target genes, we performed gene expression microarray analysis on P9 SSKO testes, utilizing the Affymetrix Mouse Gene 1.0 ST Array platform. A total of 194 transcripts exhibited significantly changed expression levels between SSKO and control samples, with 57 upregulated and 137 downregulated transcripts (Supplementary Table 1). The top up- and downregulated genes contained members of the Eph receptor tyrosine kinase, Nr4a orphan nuclear receptor, and aldehyde dehydrogenase families (Table 3). One of the genes upregulated nearly 2-fold, Nr4a1 (Nur77), encodes an orphan nuclear receptor that has been shown to inhibit the proliferation of endothelial and smooth muscle cells when overexpressed in vitro (Arkenbout et al., 2003; de Waard et al., 2006). NR4A1 can also heterodimerize with retinoid X receptors to function in a 9-cis retinoic acid-dependent manner (Perlmann and Jansson, 1995). To integrate these microarray data into biological pathways, we used Ingenuity Pathway Analysis® (IPA), a knowledge-based database. IPA identified several potential interactions among the differentially regulated genes, with NR4A1 serving as a node to connect CREB binding protein/p300, E2F1 transcription factor, myocyte enhancer factor 2, Akt, platelet-derived growth factor beta, Jnk, interleukin 1 beta, calcium/calmodulin-dependent kinase 4, insulin, and SIN3A (Supplementary Fig. 2). These results identify a modest, yet potentially important set of genes in differentiating spermatogonia that are transcriptionally regulated by SIN3A.
Table 3.
Top differentially expressed genes from the SSKO microarray
| Upregulated (Sin3aΔ/Δ vs Sin3aΔ/+) | Fold Δ | Downregulated (Sin3aΔ/Δ vs Sin3aΔ/+) | Fold Δ |
|---|---|---|---|
| Ephb1 | +2.49 | H2-T22 | −3.23 |
| Nr4a1 | +1.86 | Rpl29 | −3.08 |
| Chrdl1 | +1.85 | Ccdc72 | −2.48 |
| Gmfg | +1.62 | Aldh1a2 | −2.35 |
| Mapkapk3 | +1.50 | Tdg | −2.14 |
| Oaz1 | +1.46 | Zfp345 | −1.83 |
| Steap4 | +1.46 | Ccdc73 | −1.73 |
| Klk1b11 | +1.41 | Fmod | −1.68 |
| Olfr1371 | +1.40 | Psma8 | −1.66 |
| Try10 | +1.38 | Dapk2 | −1.63 |
P values for all transcripts < 0.001
Sin3a is essential for male gonocytes to establish the postnatal germ cell lineage
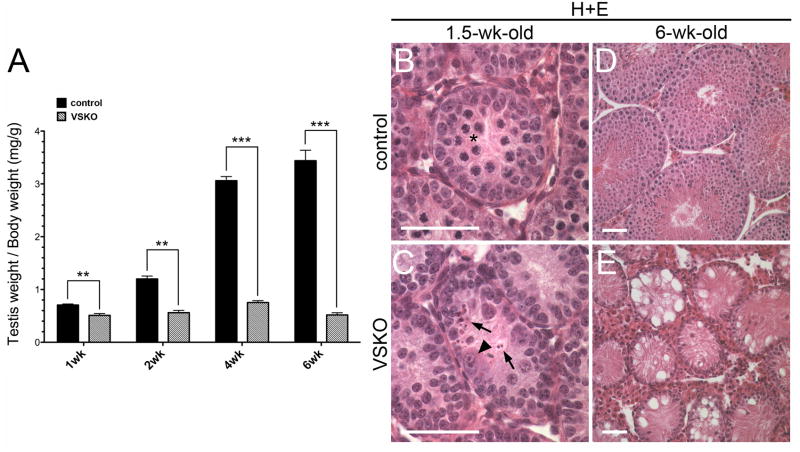
Because recombination of the Sin3a allele occurs in spermatogonia that are committing to differentiation in SSKO testes, we generated an alternative line of conditional gene-targeted mice in which recombination occurs at an earlier stage within the germ cell lineage to examine Sin3a loss-of-function effects in quiescent gonocytes and undifferentiated mitotic spermatogonia. Floxed Sin3a mice were intercrossed with Ddx4-cre (Mvh-cre; Vasa-cre) mice that exhibit Cre recombinase activity in both male and female germ cells beginning at embryonic day (E)15, a time point in which male germ cells are quiescent (Gallardo et al., 2007; McLaren, 1985). Vasa-cre mediated recombination is approximately 95% complete by birth (Gallardo et al., 2007). As a result, all Type A spermatogonia, including the spermatogonial stem cells, should exhibit deleted alleles (Fig. 1A). The Vasa-cre;Sin3aΔ/fl mice we generated are referred to here as Vasa-cre mediated Sin3a Knock Out (VSKO) animals (Fig. 1A). At 6 weeks of age, VSKO males exhibited an 84.9% reduction in testis weight/body weight, and their seminiferous tubules revealed a ‘Sertoli-cell only’ phenotype, completely devoid of germ cells (Fig. 3A, D, E). VSKO epididymides never contained sperm upon examination at 5, 6, and 8 weeks of age, indicating that spermatogenesis is never completed (data not shown). When testis weight/body weight ratios were assessed at 1, 2, and 4 weeks of age, they showed a significant, progressive reduction from 27.7% to 75.4% (Fig. 3A). Seminiferous tubule cross-sections from 1.5-week-old VSKO testes did not display any meiotic germ cells, but instead contained cells resembling spermatogonia that were either abnormally enlarged or fragmented into remnants (Fig. 3B, C). Meiotic spermatocytes were never detected in VSKO testes. Collectively, these results suggest that VSKO germ cell depletion occurs during the spermatogonial phase.
Fig. 3.
Testes of Vasa-cre; Sin3aΔ/fl male mice are completely depleted of germ cells between 1 and 6 wks of age. A. VSKO testis weight/body weight ratios undergo a significant, progressive reduction from 27.7% to 84.9% between 1 and 6 wks of age, relative to littermate controls (N=3 for each time point; **p<0.01, ***p<0.001). B and C. Seminiferous tubule cross-sections of 1.5-wk-old testes stained with H+E; asterisk denotes meiotic spermatocytes in control section (B). Arrowhead identifies enlarged, abnormal germ cell and arrows denote cell remnants in VSKO section (C). D and E. Cross-sections of 6-wk-old seminiferous tubules stained with H+E; while control sections contain germ cells at all stages and in expected numbers (D), VSKO sections are completely devoid of germ cells, exhibiting a ‘Sertoli cell only’ phenotype (E). Scale bars = 50 μm.
VSKO germ cell depletion due to DNA damage and apoptosis coincides with cell cycle reentry
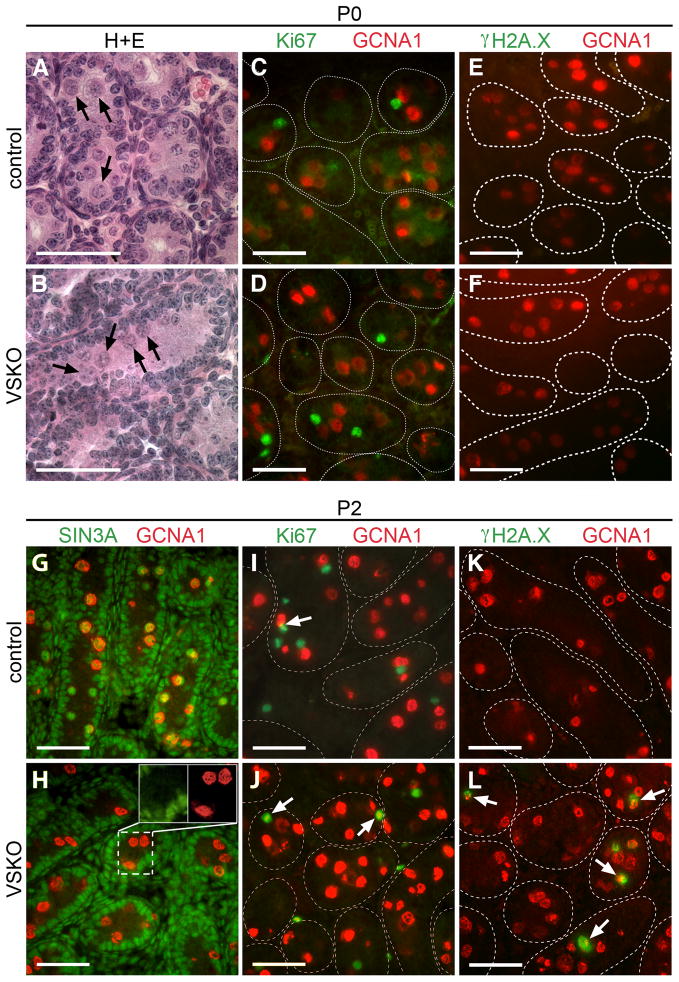
Given the significant reduction in testis weights and absence of meiotic cells in 1.5-week-old VSKO males, we next examined neonatal mutant testes beginning at P0 to determine precisely when germ cell loss occurs. Equivalent numbers of germ cells were detected in the lumen of seminiferous tubules in P0 VSKO and control testes (Fig. 4A, B, arrows). Quiescent male gonocytes reenter the cell cycle between birth and P3, depending on the mouse strain, and then migrate from the lumen to the basement membrane to occupy the appropriate niche for stem cell establishment or immediate differentiation (Bellve et al., 1977; Yoshida et al., 2006). Sertoli cells, on the other hand, are actively proliferating at birth, having been mitotically active during the period of gonocyte quiescence in the fetal testis (Vergouwen et al., 1991). A lack of co-immunostaining for Ki67 and GCNA1 in P0 VSKO and control seminiferous tubule cross-sections revealed that the male germ cells had not yet entered the cell cycle in either sample (Fig. 4C, D). To determine the efficiency and confirm the specificity of Vasa-cre recombination on the floxed Sin3a allele, we examined P0 and P2 VSKO testes and did not see any co-localization between SIN3A and GCNA1 (Fig. 4G, H, and data not shown). Nearly all control GCNA1+ cells exhibited SIN3A staining at these time points, while Sertoli cells, peritubular and interstitial cells in both VSKO and control testes contained equivalently abundant SIN3A (Fig. 4G, H; (Payne et al., 2010). From these data we conclude that the Vasa-cre recombination of Sin3a is highly efficient and specific to the germline.
Fig. 4.
VSKO germ cells begin to exhibit DNA double strand breaks upon cell cycle reentry. A and B. Seminiferous tubule cross-sections of postnatal day (P)0 testes stained with H+E; arrows identify quiescent gonocytes in the lumen of control (A) and VSKO (B) tubules. C and D. Cross-sections of P0 seminiferous tubules co-immunostained for GCNA1 (red) and Ki67 (green); note the absence of co-localization. E and F. Cross-sections of P0 seminiferous tubules co-immunostained for GCNA1 (red) and γH2A.X (green); no GCNA1+ cells exhibit γH2A.X in either control (E) or VSKO (F) sections. G and H. Cross-sections of P2 seminiferous tubules co-immunostained for GCNA1 (red) and SIN3A (green); control sections exhibit GCNA1+ cells containing SIN3A (G), while GCNA1+ cells in VSKO sections lack SIN3A (H). Inset features cells within the dashed lines in separate channels for SIN3A and GCNA1. I and J. Cross-sections of P2 seminiferous tubules co-immunostained for GCNA1 (red) and Ki67 (green); arrows identify GCNA1+ cells exhibiting Ki67, signifying reentry into the cell cycle. K and L. Cross-sections of P2 seminiferous tubules co-immunostained for GCNA1 (red) and γH2A.X (green); arrows identify GCNA1+ cells exhibiting γH2A.X in VSKO (L), but not in control (K) sections. The outlines of distinct seminiferous tubules in C-F and I-L are demarcated with dashed lines, representing basement membranes. Scale bars = 50μm.
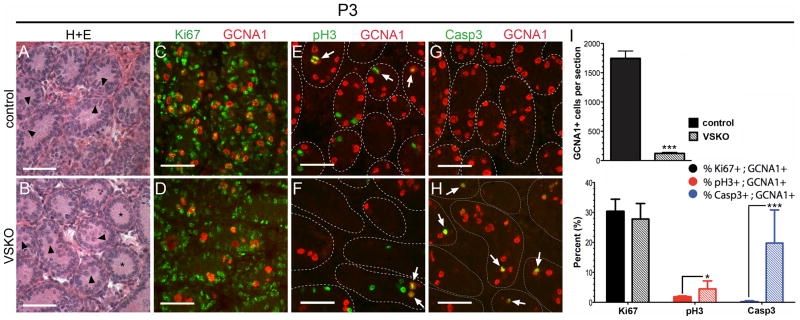
We next examined P2 VSKO and control seminiferous tubule cross-sections and detected a modest percentage of GCNA1+ cells co-staining for Ki67 in both samples, suggesting that germ cells were just beginning to enter the cell cycle at this time point (Fig. 4I, J, arrows). To determine whether these newly cycling VSKO germ cells exhibited an increase in DNA double strand breaks, as was seen with the 1-week-old SSKO spermatogonia, we co-immunostained P2 samples for γH2A.X and GCNA1 and frequently observed co-localization in the VSKO cells (Fig. 4L). No γH2A.X was ever detected in P2 control seminiferous tubule cross-sections (Fig. 4K). At P0, we never saw γH2A.X in either VSKO or control germ cells (Fig. 4E, F), indicating that DNA double strand breaks are not yet induced in Sin3a-deleted quiescent male gonocytes. In P3 VSKO testes, we noticed that the majority of germ cells remained in the lumen of seminiferous tubules, whereas most germ cells in control testes had already migrated to the basement membrane (Fig. 5A, B, arrowheads). We also observed that some VSKO seminiferous tubule cross-sections were completely devoid of germ cells (Fig. 5B, asterisks). Overall, the number of GCNA1+ cells in VSKO testes was significantly diminished (123 ± 18 GCNA1+ cells/complete testis section; N=27 sections) when compared to controls (1,744 ± 127 GCNA1+ cells/complete testis section; N=27 sections) (Fig. 5I top). Co-immunostaining for Ki67 and GCNA1 revealed that almost one-third of GCNA1+ cells were cycling in both VSKO and control seminiferous tubule cross-sections (27.8% ± 5.2% Ki67+GCNA1+ cells/complete testis section and 30.3% ± 4.1% Ki67+GCNA1+ cells/complete testis section, respectively; N=9 sections each) (Fig. 5C, D, I bottom). We next examined whether cycling P3 VSKO spermatogonia could enter M-phase, using phosphorylated histone H3 (pH3) as a marker (Perez-Cadahia et al., 2009). GCNA1+ cells in VSKO testes exhibited a 2.5-fold increase in co-immunostaining for pH3 compared with controls (4.5% ± 2.7% pH3+GCNA1+ cells/complete testis section versus 1.8% ± 0.4% pH3+GCNA1+ cells/complete testis section; N=9 sections each) (Fig. 5E, F, I bottom, Supplementary Fig. 3). These results suggested a possible G2/M block. To assess whether P3 VSKO spermatogonia were undergoing apoptosis, we used antibodies specific to activated caspase 3 (Casp3; (Porter and Janicke, 1999). GCNA1+ cells in VSKO testes exhibited a 50-fold increase in co-immunostaining for Casp3 compared with controls (19.8% ± 11.0% Casp3+GCNA1+ cells/complete testis section versus 0.4% ± 0.1% Casp3+GCNA1+ cells/complete testis section; N=9 sections each) (Fig. 5G, H, I bottom, Supplementary Fig. 4). Collectively, these findings reveal that perinatal gonocytes lacking Sin3a exhibit widespread DNA damage and undergo apoptosis coincident with cell cycle reentry in a manner distinct from that in differentiating spermatogonia: undifferentiated VSKO germ cells are completely lost from the testis while differentiating SSKO spermatogonia are largely retained in equivalent numbers to controls.
Fig. 5.
P3 VSKO germ cells exhibit a G2/M phase block and increased apoptosis that contribute to rapid cell depletion. A and B. Seminiferous tubule cross-sections of P3 testes stained with H+E; arrowheads identify germ cells that have migrated to the basement membranes in control (A) but not in VSKO (B) tubules. Asterisks denote VSKO cross-sections already devoid of germ cells (B). C and D. Cross-sections of P3 seminiferous tubules co-immunostained for GCNA1 (red) and Ki67 (green); almost one-third of GCNA1+ cells exhibit Ki67 in both control (C) and VSKO (D) sections. E and F. Cross-sections of P3 seminiferous tubules co-immunostained for GCNA1 (red) and phosphorylated histone H3 (pH3, green); arrows identify GCNA1+ cells containing pH3. G and H. Cross-sections of P3 seminiferous tubules co-immunostained for GCNA1 (red) and activated caspase 3 (Casp3; green); arrows identify GCNA1+ cells that exhibit Casp3 in VSKO (H), but not in control (G) sections. I. top, GCNA1+ cells per complete testis section in P3 VSKO males was significantly diminished when compared to controls (N=27 sections representing 3 animals for control, VSKO; ***p<0.001). I. bottom, Percentages of GCNA1+ cells that exhibited Ki67 (% Ki67+; GCNA1+, black), pH3 (% pH3+; GCNA1+, red), and Casp3 (% Casp3+; GCNA1+; blue) in P3 control (solid bars) and VSKO (hashed bars) testis sections (N=9 complete sections representing 3 animals for control, VSKO; *p<0.05, ***p<0.001). The outlines of distinct seminiferous tubules in E-H are demarcated with dashed lines, representing basement membranes. Scale bars = 50μm.
Nr4a1 is downregulated in VSKO testes and is a direct target of SIN3A in spermatogonia
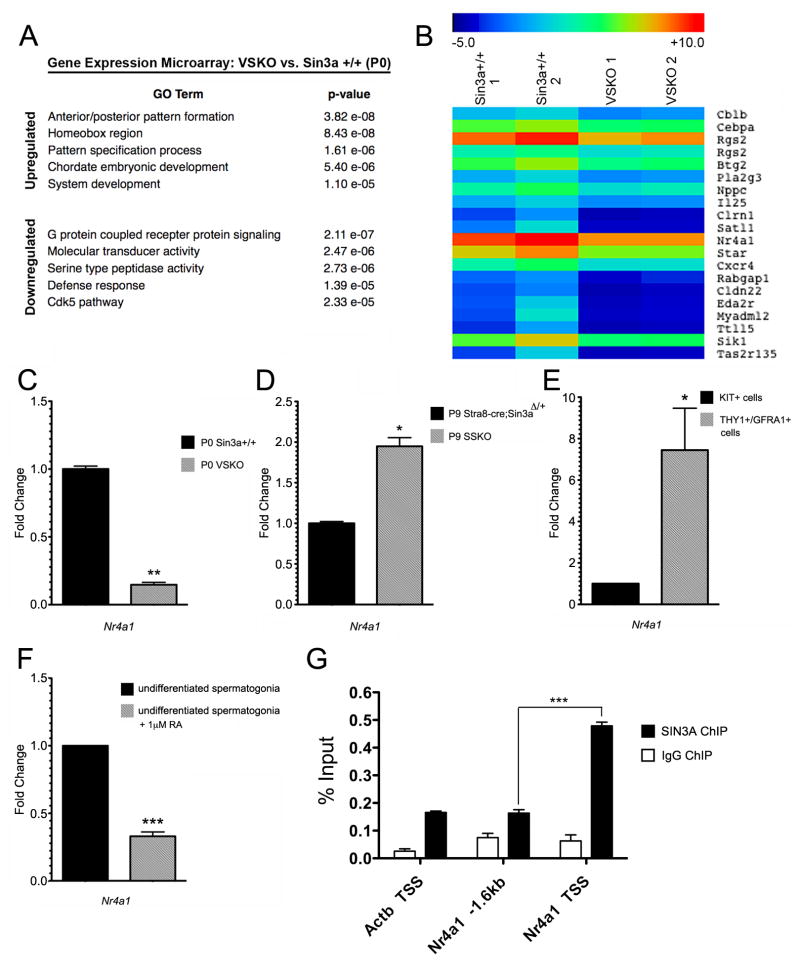
To determine what role SIN3A might play in undifferentiated male germ cells and to identify potential targets of SIN3A regulation in a cellular environment that is uncoupled from cell cycle activity, we performed gene expression microarray analysis on P0 VSKO and hemizygous Vasa-cre;Sin3aΔ/+ testes, utilizing the Agilent SurePrint G3 Mouse Gene Expression 8×60K Microarray platform. Comparison of VSKO with wild type Sin3a+/+ samples revealed that 1,426 transcripts were significantly upregulated while 1,325 transcripts were significantly downregulated (Supplementary Table 2). When VSKO was compared with hemizygous Vasa-cre;Sin3aΔ/+ samples, those numbers were 849 (up) and 708 (down) (Supplementary Table 3). We next performed gene ontology (GO) analysis on the differentially regulated genes between VSKO and wild type samples, using findGO, a component of the HOMER software suite (Heinz et al., 2010). Top categories for upregulated transcripts included anterior/posterior pattern formation, homeobox genes, and system development, while those for downregulated transcripts included G protein-coupled receptor signaling, molecular transducer activity, and cyclin-dependent kinase 5 (cdk5) pathway (Fig. 6A). The top up- and downregulated genes contained members of the ankyrin repeat domain, zinc finger and SCAN domain-containing, and serine/threonine kinase families (Table 4). For further data analysis, genes were hierarchically clustered using Multiple Experiment Viewer (MeV) software and a heat map was generated (Supplementary Fig. 5). From the testicular transcriptome heat map, individual clusters were examined in greater detail and a cluster containing the downregulated genes Cxcr4, Pla2g3, and Nr4a1 was identified (Fig. 6B). These genes were downregulated 3.05-fold, 2.51-fold, and 3.36-fold, respectively. Cxcr4 is essential for primordial germ cell migration and is expressed in postnatal male germ cells, while targeted mutations in Pla2g3 result in males exhibiting sperm defects, with reduced fertility and smaller litter sizes (Ara et al., 2003; Doitsidou et al., 2002; Gilbert et al., 2009; Molyneaux et al., 2003; Sato et al., 2010). The downregulation of Cxcr4 here is noteworthy, as germ cells in P3 VSKO testes failed to migrate to the basement membrane. Insufficient Cxcr4 levels could be a potential contributing factor to this migratory defect. IPA analysis identified several potential interactions among SIN3A, NR4A1, and CXCR4, with pathways connected through Jnk, histone H3, and C-terminal binding protein, among others (Supplementary Fig. 6).
Fig. 6.
VSKO testicular transcriptome at P0 exhibits an upregulation of genes important for development and patterning and a downregulation of Nr4a1, a gene conversely upregulated in P9 SSKO testes. A. Gene ontology (GO) categories and p-values for the top up- and downregulated genes expressed in P0 VSKO testes relative to wild type (Sin3a+/+) testes. B. Heat map of an individual cluster (hierarchically generated from 2 VSKO and 2 wild type testicular transcriptomes) of downregulated genes of interest. This specific cluster contains Cxcr4, Pla2g3, and Nr4a1. C and D. Quantitative (q)RT-PCR validation of microarray data, showing a 6.82-fold downregulation of Nr4a1 for P0 VSKO (C) and a 1.95-fold upregulation of Nr4a1 for P9 SSKO (D), relative to control samples (N=3, *p<0.05, **p<0.01). E. qRT-PCR revealing a 7.44-fold upregulation of Nr4a1 in THY1+/GFRA1+ cells (enriched from P7 wild type testes by magnetic-activated cell sorting, representing undifferentiated spermatogonia) relative to KIT+ cells (representing differentiating spermatogonia) (N=4, *p<0.05). F. qRT-PCR revealing a 3.03-fold downregulation of Nr4a1 in cultured wild type undifferentiated spermatogonia treated with 1 μM retinoic acid (RA) for 24 h (N=4, ***p<0.001). G. ChIP for SIN3A and IgG control on THY1+/GFRA1+ cells, showing SIN3A enrichment at the Nr4a1 transcription start site (TSS), but not at the Actb TSS or at a region −1.6kb upstream of Nr4a1 (N=3, ***p<0.001). 18s rRNA is the internal control transcript for C–F.
Table 4.
Top differentially expressed genes from the VSKO microarray
| Upregulated (Sin3aΔ/Δ vs Sin3a+/+) | Fold Δ | Downregulated (Sin3aΔ/Δ vs Sin3a+/+) | Fold Δ |
|---|---|---|---|
| Ankrd7 | +62.2 | Gm4736 | −35.3 |
| Masp1 | +59.8 | Fig2 | −35.0 |
| Dub1 | +50.0 | Gbp1 | −26.8 |
| Gm9125 | +43.4 | Mecom | −26.3 |
| Gm14347 | +41.1 | Cyp3a59 | −26.0 |
| Gm12794 | +40.0 | Kcna10 | −25.4 |
| Zscan4f | +36.6 | Olfr1123 | −25.3 |
| Gm13043 | +36.3 | Gm13125 | −24.8 |
| Krtap3-3 | +34.0 | Gabrg2 | −22.9 |
| Tcl1b5 | +33.5 | Stk4 | −22.9 |
P values for all transcripts < 0.001
The downregulation of Nr4a1 in P0 VSKO samples intrigued us, as this gene was upregulated nearly 2-fold in P9 SSKO samples. To validate the microarray data, we performed quantitative (q)RT-PCR on newly generated P0 VSKO and P9 SSKO samples for the Nr4a1 transcript. Our results showed a 6.82-fold reduction in Nr4a1 for VSKO and a 1.95-fold elevation in Nr4a1 for SSKO, confirming the microarray expression patterns (Fig. 6C, D). Nr4a1 expression has been described for both Leydig cells and Sertoli cells within the testis, but has not been characterized for germ cells beyond an observed enrichment of transcript levels in Type A and Type B spermatogonia relative to other cell types (Ding et al., 2011; Shima et al., 2004; Song et al., 2001). To determine whether Nr4a1 expression levels are higher in undifferentiated versus differentiating spermatogonia, as our microarray and qRT-PCR data implied, we utilized a magnetic-activated cell sorting (MACS) strategy to enrich for P7 wild type testicular cells expressing thymus cell antigen 1, theta (THY1) and glial cell line-derived neurotrophic factor family receptor alpha 1 (GFRA1), which represent undifferentiated spermatogonia. We also enriched for cells within the seminiferous tubules expressing surface marker kit oncogene (KIT), which represent differentiating spermatogonia (Gassei et al., 2009; Kofman et al., 2012; Kubota et al., 2003). Following MACS, the THY1+/GFRA1+ cells exhibited a 7.44-fold enrichment of Nr4a1 relative to KIT+ cells, indicating that indeed, Nr4a1 expression is higher in undifferentiated spermatogonia (Fig. 6E). As an additional test, we treated wild type undifferentiated spermatogonial cell cultures with 1 μM all-trans retinoic acid to induce the cells to differentiate, comparing Nr4a1 expression levels in RA-exposed cells with levels in EtOH vehicle-exposed cells. RA-treated cells exhibited a 3.03-fold reduction in Nr4a1 (Fig. 6F), revealing that Nr4a1 transcript levels diminish upon spermatogonial differentiation. To determine whether Nr4a1 is a direct target of SIN3A, we performed chromatin immunoprecipitation (ChIP) on THY1+/GFRA1+ cells. A 0.3Kb region of the Nr4a1 proximal promoter encompassing the transcription start site exhibited SIN3A enrichment, but not a more distal region 1.6Kb upstream (Fig. 6G). Collectively, these data identify Nr4a1 as a gene bound by SIN3A whose expression levels in gonocytes decrease when SIN3A is absent, and whose levels in differentiating spermatogonia increase when SIN3A is lost. Moreover, Nr4a1 expression diminishes in undifferentiated spermatogonia that commit to the differentiation process.
Discussion
Transcriptional regulator complexes carry out important functions during the development and differentiation of cell lineages within an organism, from the stem and progenitor cell stages to phases in which cells have fully matured or terminally differentiated. Increasing evidence suggests that such functions are distinct, depending on the state of cellular differentiation and the lineage itself. Here we have shown that quiescent male gonocytes require the SIN3A component of the SIN3 complex to maintain their cellular identity and to proliferate, whereas differentiation-committed, mitotic germ cells do not share such a requirement. Upon submission of this manuscript, we learned of a similar study by Pellegrino et al. (2012), who also used Vasa-cre to ablate Sin3a in mouse gonocytes, and observed the same cell cycle defects and apoptosis upon mitotic reentry (Pellegrino et al., 2012). We were surprised, then, that differentiating spermatogonia lacking Sin3a did not exhibit these problems. Differences between VSKO and SSKO outcomes were observed with respect to gene expression, DNA damage, and cell cycle progression.
Ablation of Sin3a from quiescent gonocytes allowed us to uncouple the regulatory activity of SIN3A from the cell cycle. Following Sin3a removal, genes important for development and patterning exhibited increased transcriptional activity, supporting the notion that the cells were inappropriately differentiating. An entire Hox cluster, Hoxb1-8, was upregulated between 2.15-fold and 9.32-fold (Supplementary Table 2). Pax1 and Pax6 levels were also elevated (2.46-fold and 4.07-fold, respectively), as were Six2 (2.33-fold), Six3 (2.42-fold), Nkx2-5 (2.14), and Cdx2 (2.59-fold). Interestingly, Cdx2 is a marker for trophectoderm, which is able to develop in the absence of Sin3a (McDonel et al., 2012). The increase in differentiation/patterning transcript levels was accompanied by a downregulation in genes normally expressed in perinatal testes. Dmrtc1b (Dmrt8.2), a gene closely related to the doublesex and mab-3 related transcription factor like family C2 (Dmrtc2; Dmrt7) that is expressed exclusively in the germ cells of testes, was downregulated 5.34-fold in P0 VSKO samples (Kim et al., 2003). Dmrt8.2 levels normally increase at birth in wild type testes (Veith et al., 2006). While DMRT7 functions to ensure that male germ cells can complete meiotic prophase, the role of DMRT8.2 is not currently known (Kim et al., 2007). Another transcript whose levels decreased was the ATP-binding cassette sub-family C (CFTR/MRP) member 6 (Abcc6; 2.13-fold). As an ABC transporter predominantly expressed in liver and kidney, the human ortholog of Abcc6 contains numerous mutations that associate with the autosomal recessive disease pseudoxanthoma elasticum (Le Saux et al., 2000). Recently, the transcripts of four ABC transporters were detected in wild type primordial germ cells and prenatal testes that could collectively promote the ability of SSCs to efflux dyes like Hoechst 33342, defining the “side-population” stem cell phenotype observed by flow cytometry (Scaldaferri et al., 2011). Abcc6 might be acting similarly. Thus, loss of Sin3a alters the transcriptome in quiescent gonocytes in a manner that shifts the cells towards differentiation.
In contrast, deletion of Sin3a from differentiating spermatogonia resulted in modest alterations to the transcriptome. While over 1,500 genes were significantly affected between P0 VSKO and Vasa-cre;Sin3aΔ/+ testes, only 194 genes reflected such changes between P9 SSKO and Stra8-cre;Sin3aΔ/+ testes (Supplementary Tables 1 and 3). One gene affected by the loss of Sin3a under both conditions, but misregulated in opposite directions, was Nr4a1. Highly homologous in genomic structure to its two family members, Nr4a2 (Nurr1) and Nr4a3 (Nor1), Nr4a1 functions as an immediate early response gene activated through numerous physiological signals (Hsu et al., 2004). It encodes a nuclear receptor that acts in a ligand-independent manner, with the activation-function 1 domain of the protein mediating transactivation and coactivator recruitment (Wansa et al., 2002). Notably, neither Nr4a2 nor Nr4a3 were differentially regulated in the VSKO or SSKO testicular transcriptomes, suggesting that SIN3A’s influence is specific to Nr4a1 within this family. Although implicated in processes involving cellular proliferation, differentiation, and apoptosis, the role of Nr4a1 in male germ cells is not currently known (Maxwell and Muscat, 2006). In Leydig cells, Nr4a1 is induced by luteinizing hormone, which results in NR4A1 protein binding to the promoters of steroidogenic enzyme encoding genes; this process is inhibited through androgens and the androgen receptor (Song et al., 2012; Song et al., 2001). In human Sertoli cells, follicle-stimulating hormone induces NR4A1, whereupon the nuclear receptor transcriptionally activates GDNF, whose gene product not only supports SSC self-renewal but also appears to mediate the commitment of aligned spermatogonia to differentiate (Ding et al., 2011; Grasso et al., 2012). Here we have shown that Nr4a1 is a direct target of SIN3A. Its expression values suggest that it is activated by SIN3A in quiescent gonocytes, but repressed by SIN3A in differentiating spermatogonia. The biological significance of this differential regulation is unclear, warranting further investigation.
For 1-week-old SSKO animals, approximately 15% of germ cells expressed both GCNA1 and SIN3A. We interpret this finding to represent the pre-Stra8-cre expressing spermatogonia, as SIN3A was not detected in spermatogonia positive for STRA8. Moreover, all of the offspring born to wild type females mated with SSKO males carried a deleted Sin3a allele, suggesting that all mature SSKO sperm capable of fertilization lack functional Sin3a. Interestingly, SSKO males remain fertile past 1 year of age. We conclude that spermatogonial self-renewal is not affected in the testes of SSKO mice.
Proliferating Sin3a-deleted, Stra8-expressing spermatogonia exhibited modest increases in DNA double strand breaks and apoptosis, but not to the extent that impeded fertility. Double strand breaks, repaired by non-homologous end-joining or homologous recombination mechanisms, are much more severe than single strand breaks, which are routinely corrected by base excision repair or nuclear excision repair events (Simonatto et al., 2007). Cells experiencing DNA double strand breaks display γH2A.X. Cycling Sin3a-deleted, undifferentiated spermatogonia not only exhibited increased γH2A.X, but also increased phosphorylated histone H3 and activated caspase 3. These observations suggest that these cells are unable to repair the extensive double strand breaks occurring in the absence of SIN3A and cannot proceed through the mitotic checkpoint, triggering apoptosis and, presumably, phagocytosis by Sertoli cells (Nakanishi and Shiratsuchi, 2004; Wang and Cho, 2004). As such, the VSKO phenotype resembles that of Sin3a-deleted blastocyst inner cell mass cells, ES cells, and MEFs (Dannenberg et al., 2005; Fazzio et al., 2008; McDonel et al., 2012). While VSKO spermatogonia could be prematurely entering meiosis, given that spermatocytes exhibit abundant levels of γH2A.X, we find this highly unlikely due to the rapid cell death occurring concomitantly with cell cycle resumption (Mahadevaiah et al., 2001). Apoptosis of spermatogonia and postmitotic germ cells occurs frequently throughout the lifespan of males, triggered through a variety of inputs (Meachem et al., 2005; Print and Loveland, 2000). From our data, we conclude that genome integrity promoted by SIN3A and the SIN3 complex inhibits apoptosis in undifferentiated mitotic spermatogonia, but that this process is dispensable for the maintenance of differentiating spermatogenic cells. These findings support the hypothesis that proliferating cells committed to differentiate respond to DNA damage differently than cycling, undifferentiated cells (Puri et al., 2002).
In conclusion, we have identified distinct requirements for Sin3a in postnatal male germ cells that reinforce the importance of the SIN3 complex in regulating cell cycle progression and DNA damage repair in undifferentiated cells, and in modulating the expression of genes involved in development and patterning. Collectively, our results provide important insights into how early postnatal male germ cells are regulated at the level of transcription, DNA replication, and cellular proliferation.
Supplementary Material
Highlights.
We generated two lines of germ cell-specific Sin3a gene-targeted mice (cKOs)
cKO undifferentiated spermatogonia (#1) rapidly die from DNA damage & G2/M arrest
cKO differentiating spermatogonia (#2) complete mitoses, meiosis & produce sperm
SIN3A influence on Nr4a1 expression is (+) in #1 and (−) in #2, switching roles
Nr4a1 is bound by SIN3A and decreases expression upon cell differentiation
Acknowledgments
The authors would like to thank Rachel Anderson for her technical assistance with these experiments, the Histology Core of the Center for Reproductive Research at Northwestern University for exemplary histology services, and the foundation of the Medical Research Institute Council at Children’s Hospital of Chicago Research Center for its generous financial support. Anti-GCNA1 antibodies were kindly provided by Dr. George Enders from the University of Kansas Medical Center. C.J.P. is the recipient of an NIH Pathway-to-Independence Award from the Eunice Kennedy Shriver National Institute of Child Health & Human Development. This work was supported by an NIH grant to C.J.P. (5R00 HD055330-05).
Footnotes
Competing interests statement: The authors declare no competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ara T, Nakamura Y, Egawa T, Sugiyama T, Abe K, Kishimoto T, Matsui Y, Nagasawa T. Impaired colonization of the gonads by primordial germ cells in mice lacking a chemokine, stromal cell-derived factor-1 (SDF-1) Proceedings of the National Academy of Sciences of the United States of America. 2003;100:5319–5323. doi: 10.1073/pnas.0730719100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arkenbout EK, van Bragt M, Eldering E, van Bree C, Grimbergen JM, Quax PH, Pannekoek H, de Vries CJ. TR3 orphan receptor is expressed in vascular endothelial cells and mediates cell cycle arrest. Arteriosclerosis, thrombosis, and vascular biology. 2003;23:1535–1540. doi: 10.1161/01.ATV.0000084639.16462.7A. [DOI] [PubMed] [Google Scholar]
- Bellve AR, Cavicchia JC, Millette CF, O’Brien DA, Bhatnagar YM, Dym M. Spermatogenic cells of the prepuberal mouse. Isolation and morphological characterization. The Journal of cell biology. 1977;74:68–85. doi: 10.1083/jcb.74.1.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buaas FW, Kirsh AL, Sharma M, McLean DJ, Morris JL, Griswold MD, de Rooij DG, Braun RE. Plzf is required in adult male germ cells for stem cell self-renewal. Nature genetics. 2004;36:647–652. doi: 10.1038/ng1366. [DOI] [PubMed] [Google Scholar]
- Costoya JA, Hobbs RM, Barna M, Cattoretti G, Manova K, Sukhwani M, Orwig KE, Wolgemuth DJ, Pandolfi PP. Essential role of Plzf in maintenance of spermatogonial stem cells. Nature genetics. 2004;36:653–659. doi: 10.1038/ng1367. [DOI] [PubMed] [Google Scholar]
- Cowley SM, Iritani BM, Mendrysa SM, Xu T, Cheng PF, Yada J, Liggitt HD, Eisenman RN. The mSin3A chromatin-modifying complex is essential for embryogenesis and T-cell development. Molecular and cellular biology. 2005;25:6990–7004. doi: 10.1128/MCB.25.16.6990-7004.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl JA, Collas P. A rapid micro chromatin immunoprecipitation assay (microChIP) Nature protocols. 2008;3:1032–1045. doi: 10.1038/nprot.2008.68. [DOI] [PubMed] [Google Scholar]
- Dannenberg JH, David G, Zhong S, van der Torre J, Wong WH, Depinho RA. mSin3A corepressor regulates diverse transcriptional networks governing normal and neoplastic growth and survival. Genes & development. 2005;19:1581–1595. doi: 10.1101/gad.1286905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Felici M, McLaren A. In vitro culture of mouse primordial germ cells. Experimental cell research. 1983;144:417–427. doi: 10.1016/0014-4827(83)90421-4. [DOI] [PubMed] [Google Scholar]
- de Waard V, Arkenbout EK, Vos M, Mocking AI, Niessen HW, Stooker W, de Mol BA, Quax PH, Bakker EN, VanBavel E, Pannekoek H, de Vries CJ. TR3 nuclear orphan receptor prevents cyclic stretch-induced proliferation of venous smooth muscle cells. The American journal of pathology. 2006;168:2027–2035. doi: 10.2353/ajpath.2006.050932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding LJ, Yan GJ, Ge QY, Yu F, Zhao X, Diao ZY, Wang ZQ, Yang ZZ, Sun HX, Hu YL. FSH acts on the proliferation of type A spermatogonia via Nur77 that increases GDNF expression in the Sertoli cells. FEBS letters. 2011;585:2437–2444. doi: 10.1016/j.febslet.2011.06.013. [DOI] [PubMed] [Google Scholar]
- Doitsidou M, Reichman-Fried M, Stebler J, Koprunner M, Dorries J, Meyer D, Esguerra CV, Leung T, Raz E. Guidance of primordial germ cell migration by the chemokine SDF-1. Cell. 2002;111:647–659. doi: 10.1016/s0092-8674(02)01135-2. [DOI] [PubMed] [Google Scholar]
- Enders GC, May JJ., 2nd Developmentally regulated expression of a mouse germ cell nuclear antigen examined from embryonic day 11 to adult in male and female mice. Developmental biology. 1994;163:331–340. doi: 10.1006/dbio.1994.1152. [DOI] [PubMed] [Google Scholar]
- Fazzio TG, Huff JT, Panning B. An RNAi screen of chromatin proteins identifies Tip60-p400 as a regulator of embryonic stem cell identity. Cell. 2008;134:162–174. doi: 10.1016/j.cell.2008.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallardo T, Shirley L, John GB, Castrillon DH. Generation of a germ cell-specific mouse transgenic Cre line, Vasa-Cre. Genesis. 2007;45:413–417. doi: 10.1002/dvg.20310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gassei K, Ehmcke J, Schlatt S. Efficient enrichment of undifferentiated GFR alpha 1+ spermatogonia from immature rat testis by magnetic activated cell sorting. Cell and tissue research. 2009;337:177–183. doi: 10.1007/s00441-009-0799-5. [DOI] [PubMed] [Google Scholar]
- Gilbert DC, Chandler I, McIntyre A, Goddard NC, Gabe R, Huddart RA, Shipley J. Clinical and biological significance of CXCL12 and CXCR4 expression in adult testes and germ cell tumours of adults and adolescents. The Journal of pathology. 2009;217:94–102. doi: 10.1002/path.2436. [DOI] [PubMed] [Google Scholar]
- Grasso M, Fuso A, Dovere L, de Rooij DG, Stefanini M, Boitani C, Vicini E. Distribution of GFRA1-expressing spermatogonia in adult mouse testis. Reproduction. 2012;143:325–332. doi: 10.1530/REP-11-0385. [DOI] [PubMed] [Google Scholar]
- Hsu HC, Zhou T, Mountz JD. Nur77 family of nuclear hormone receptors. Current drug targets. Inflammation and allergy. 2004;3:413–423. doi: 10.2174/1568010042634523. [DOI] [PubMed] [Google Scholar]
- Huckins C. The spermatogonial stem cell population in adult rats. I. Their morphology, proliferation and maturation. The Anatomical record. 1971;169:533–557. doi: 10.1002/ar.1091690306. [DOI] [PubMed] [Google Scholar]
- Kim S, Kettlewell JR, Anderson RC, Bardwell VJ, Zarkower D. Sexually dimorphic expression of multiple doublesex-related genes in the embryonic mouse gonad. Gene expression patterns : GEP. 2003;3:77–82. doi: 10.1016/s1567-133x(02)00071-6. [DOI] [PubMed] [Google Scholar]
- Kim S, Namekawa SH, Niswander LM, Ward JO, Lee JT, Bardwell VJ, Zarkower D. A mammal-specific Doublesex homolog associates with male sex chromatin and is required for male meiosis. PLoS genetics. 2007;3:e62. doi: 10.1371/journal.pgen.0030062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofman AE, Huszar JM, Payne CJ. Transcriptional Analysis of Histone Deacetylase Family Members Reveal Similarities Between Differentiating and Aging Spermatogonial Stem Cells. Stem cell reviews. 2012 doi: 10.1007/s12015-012-9392-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota H, Avarbock MR, Brinster RL. Spermatogonial stem cells share some, but not all, phenotypic and functional characteristics with other stem cells. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:6487–6492. doi: 10.1073/pnas.0631767100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Saux O, Urban Z, Tschuch C, Csiszar K, Bacchelli B, Quaglino D, Pasquali-Ronchetti I, Pope FM, Richards A, Terry S, Bercovitch L, de Paepe A, Boyd CD. Mutations in a gene encoding an ABC transporter cause pseudoxanthoma elasticum. Nature genetics. 2000;25:223–227. doi: 10.1038/76102. [DOI] [PubMed] [Google Scholar]
- Mahadevaiah SK, Turner JM, Baudat F, Rogakou EP, de Boer P, Blanco-Rodriguez J, Jasin M, Keeney S, Bonner WM, Burgoyne PS. Recombinational DNA double-strand breaks in mice precede synapsis. Nature genetics. 2001;27:271–276. doi: 10.1038/85830. [DOI] [PubMed] [Google Scholar]
- Maxwell MA, Muscat GE. The NR4A subgroup: immediate early response genes with pleiotropic physiological roles. Nuclear receptor signaling. 2006;4:e002. doi: 10.1621/nrs.04002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonel P, Costello I, Hendrich B. Keeping things quiet: roles of NuRD and Sin3 co-repressor complexes during mammalian development. The international journal of biochemistry & cell biology. 2009;41:108–116. doi: 10.1016/j.biocel.2008.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonel P, Demmers J, Tan DW, Watt F, Hendrich BD. Sin3a is essential for the genome integrity and viability of pluripotent cells. Developmental biology. 2012;363:62–73. doi: 10.1016/j.ydbio.2011.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaren A. Mouse germ cells: fertilization to birth. Archives d’anatomie microscopique et de morphologie experimentale. 1985;74:5–9. [PubMed] [Google Scholar]
- Meachem SJ, Ruwanpura SM, Ziolkowski J, Ague JM, Skinner MK, Loveland KL. Developmentally distinct in vivo effects of FSH on proliferation and apoptosis during testis maturation. The Journal of endocrinology. 2005;186:429–446. doi: 10.1677/joe.1.06121. [DOI] [PubMed] [Google Scholar]
- Molyneaux KA, Zinszner H, Kunwar PS, Schaible K, Stebler J, Sunshine MJ, O’Brien W, Raz E, Littman D, Wylie C, Lehmann R. The chemokine SDF1/CXCL12 and its receptor CXCR4 regulate mouse germ cell migration and survival. Development. 2003;130:4279–4286. doi: 10.1242/dev.00640. [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Sharma M, Nabeshima Y, Braun RE, Yoshida S. Functional hierarchy and reversibility within the murine spermatogenic stem cell compartment. Science. 2010;328:62–67. doi: 10.1126/science.1182868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi Y, Shiratsuchi A. Phagocytic removal of apoptotic spermatogenic cells by Sertoli cells: mechanisms and consequences. Biological & pharmaceutical bulletin. 2004;27:13–16. doi: 10.1248/bpb.27.13. [DOI] [PubMed] [Google Scholar]
- Nascimento EM, Cox CL, MacArthur S, Hussain S, Trotter M, Blanco S, Suraj M, Nichols J, Kubler B, Benitah SA, Hendrich B, Odom DT, Frye M. The opposing transcriptional functions of Sin3a and c-Myc are required to maintain tissue homeostasis. Nature cell biology. 2011;13:1395–1405. doi: 10.1038/ncb2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakberg EF. Spermatogonial stem-cell renewal in the mouse. The Anatomical record. 1971;169:515–531. doi: 10.1002/ar.1091690305. [DOI] [PubMed] [Google Scholar]
- Payne CJ, Gallagher SJ, Foreman O, Dannenberg JH, Depinho RA, Braun RE. Sin3a is required by sertoli cells to establish a niche for undifferentiated spermatogonia, germ cell tumors, and spermatid elongation. Stem Cells. 2010;28:1424–1434. doi: 10.1002/stem.464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrino J, Castrillon DH, David G. Chromatin associated Sin3A is essential for male germ cell lineage in the mouse. Developmental biology. 2012;369:349–355. doi: 10.1016/j.ydbio.2012.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Cadahia B, Drobic B, Davie JR. H3 phosphorylation: dual role in mitosis and interphase. Biochemistry and cell biology = Biochimie et biologie cellulaire. 2009;87:695–709. doi: 10.1139/O09-053. [DOI] [PubMed] [Google Scholar]
- Perlmann T, Jansson L. A novel pathway for vitamin A signaling mediated by RXR heterodimerization with NGFI-B and NURR1. Genes & development. 1995;9:769–782. doi: 10.1101/gad.9.7.769. [DOI] [PubMed] [Google Scholar]
- Porter AG, Janicke RU. Emerging roles of caspase-3 in apoptosis. Cell death and differentiation. 1999;6:99–104. doi: 10.1038/sj.cdd.4400476. [DOI] [PubMed] [Google Scholar]
- Print CG, Loveland KL. Germ cell suicide: new insights into apoptosis during spermatogenesis. BioEssays : news and reviews in molecular, cellular and developmental biology. 2000;22:423–430. doi: 10.1002/(SICI)1521-1878(200005)22:5<423::AID-BIES4>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- Puri PL, Bhakta K, Wood LD, Costanzo A, Zhu J, Wang JY. A myogenic differentiation checkpoint activated by genotoxic stress. Nature genetics. 2002;32:585–593. doi: 10.1038/ng1023. [DOI] [PubMed] [Google Scholar]
- Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. The Journal of biological chemistry. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- Sadate-Ngatchou PI, Payne CJ, Dearth AT, Braun RE. Cre recombinase activity specific to postnatal, premeiotic male germ cells in transgenic mice. Genesis. 2008;46:738–742. doi: 10.1002/dvg.20437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato H, Taketomi Y, Isogai Y, Miki Y, Yamamoto K, Masuda S, Hosono T, Arata S, Ishikawa Y, Ishii T, Kobayashi T, Nakanishi H, Ikeda K, Taguchi R, Hara S, Kudo I, Murakami M. Group III secreted phospholipase A2 regulates epididymal sperm maturation and fertility in mice. The Journal of clinical investigation. 2010;120:1400–1414. doi: 10.1172/JCI40493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaldaferri ML, Fera S, Grisanti L, Sanchez M, Stefanini M, De Felici M, Vicini E. Identification of side population cells in mouse primordial germ cells and prenatal testis. The International journal of developmental biology. 2011;55:209–214. doi: 10.1387/ijdb.092977ms. [DOI] [PubMed] [Google Scholar]
- Seandel M, James D, Shmelkov SV, Falciatori I, Kim J, Chavala S, Scherr DS, Zhang F, Torres R, Gale NW, Yancopoulos GD, Murphy A, Valenzuela DM, Hobbs RM, Pandolfi PP, Rafii S. Generation of functional multipotent adult stem cells from GPR125+ germline progenitors. Nature. 2007;449:346–350. doi: 10.1038/nature06129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shima JE, McLean DJ, McCarrey JR, Griswold MD. The murine testicular transcriptome: characterizing gene expression in the testis during the progression of spermatogenesis. Biology of reproduction. 2004;71:319–330. doi: 10.1095/biolreprod.103.026880. [DOI] [PubMed] [Google Scholar]
- Silverstein RA, Ekwall K. Sin3: a flexible regulator of global gene expression and genome stability. Current genetics. 2005;47:1–17. doi: 10.1007/s00294-004-0541-5. [DOI] [PubMed] [Google Scholar]
- Simonatto M, Latella L, Puri PL. DNA damage and cellular differentiation: more questions than responses. Journal of cellular physiology. 2007;213:642–648. doi: 10.1002/jcp.21275. [DOI] [PubMed] [Google Scholar]
- Song CH, Gong EY, Park JS, Lee K. Testicular steroidogenesis is locally regulated by androgen via suppression of Nur77. Biochemical and biophysical research communications. 2012;422:327–332. doi: 10.1016/j.bbrc.2012.04.161. [DOI] [PubMed] [Google Scholar]
- Song KH, Park JI, Lee MO, Soh J, Lee K, Choi HS. LH induces orphan nuclear receptor Nur77 gene expression in testicular Leydig cells. Endocrinology. 2001;142:5116–5123. doi: 10.1210/endo.142.12.8525. [DOI] [PubMed] [Google Scholar]
- van Oevelen C, Bowman C, Pellegrino J, Asp P, Cheng J, Parisi F, Micsinai M, Kluger Y, Chu A, Blais A, David G, Dynlacht BD. The mammalian Sin3 proteins are required for muscle development and sarcomere specification. Molecular and cellular biology. 2010;30:5686–5697. doi: 10.1128/MCB.00975-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veith AM, Klattig J, Dettai A, Schmidt C, Englert C, Volff JN. Male-biased expression of X-chromosomal DM domain-less Dmrt8 genes in the mouse. Genomics. 2006;88:185–195. doi: 10.1016/j.ygeno.2006.01.003. [DOI] [PubMed] [Google Scholar]
- Vergouwen RP, Jacobs SG, Huiskamp R, Davids JA, de Rooij DG. Proliferative activity of gonocytes, Sertoli cells and interstitial cells during testicular development in mice. Journal of reproduction and fertility. 1991;93:233–243. doi: 10.1530/jrf.0.0930233. [DOI] [PubMed] [Google Scholar]
- Wang JY, Cho SK. Coordination of repair, checkpoint, and cell death responses to DNA damage. Advances in protein chemistry. 2004;69:101–135. doi: 10.1016/S0065-3233(04)69004-8. [DOI] [PubMed] [Google Scholar]
- Wansa KD, Harris JM, Muscat GE. The activation function-1 domain of Nur77/NR4A1 mediates trans-activation, cell specificity, and coactivator recruitment. The Journal of biological chemistry. 2002;277:33001–33011. doi: 10.1074/jbc.M203572200. [DOI] [PubMed] [Google Scholar]
- Yoshida S, Sukeno M, Nakagawa T, Ohbo K, Nagamatsu G, Suda T, Nabeshima Y. The first round of mouse spermatogenesis is a distinctive program that lacks the self-renewing spermatogonia stage. Development. 2006;133:1495–1505. doi: 10.1242/dev.02316. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Li Y, Nie R, Friel P, Mitchell D, Evanoff RM, Pouchnik D, Banasik B, McCarrey JR, Small C, Griswold MD. Expression of stimulated by retinoic acid gene 8 (Stra8) and maturation of murine gonocytes and spermatogonia induced by retinoic acid in vitro. Biology of reproduction. 2008;78:537–545. doi: 10.1095/biolreprod.107.064337. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.