Abstract
Cytokines dimerize their receptors, with binding of the “second chain” triggering signaling. In the interleukin (IL)-4/13 system, different cell types express varying levels of alternative second receptor chains (γc or IL-13Rα1), forming functionally distinct Type-I or Type-II complexes. We manipulated the affinity and specificity of second chain recruitment by human IL-4. A Type-I receptor-selective IL-4 ‘superkine’ with 3700-fold higher affinity for γc was 3-10 fold more potent than wild-type IL-4. Conversely, a variant with high affinity for IL-13Rα1 more potently activated cells expressing the Type-II receptor, and induced differentiation of dendritic cells from monocytes, implicating the Type-II receptor in this process. Superkines exhibited signaling advantages on cells with lower second chain levels. Comparative transcriptional analysis reveals that the superkines induce largely redundant gene expression profiles. Variable second chain levels can be exploited to redirect cytokines towards distinct cell subsets and elicit novel actions, potentially improving the selectivity of cytokine therapy.
Cytokines regulate key cellular functions including differentiation, proliferation and apoptosis/anti-apoptosis 1, principally through dimerization of receptor subunits, which initiates intracellular JAK/STAT activation 2,3. Most cytokines mediate stimulation by first interacting with a high affinity cytokine-binding chain (usually designated “α”) followed by low affinity interaction with a receptor chain such as γc, gp130 or βc 4. The ultimate potency of the cytokine at inducing signaling is determined by the efficiency, i.e. affinity, of recruitment of the second chain 5,6. In many of these systems, different cell types express different amounts of the first and second chain 7. Thus, manipulation of the binding parameters for second chain recruitment could potentially skew the activity of a cytokine towards certain cell types8, potentially making these new engineered cytokines more specific and possibly less toxic, and therefore therapeutically advantageous.
IL-4 is a classical four α-helix bundle cytokine whose primary binding chain is IL-4Rα 9,10. The IL-4 /IL-4Rα complex serves as a ligand for the second component of the IL-4 receptor, which for the Type-I receptor is γc and for the Type-II receptor, IL-13Rα19. Formation of the IL-4/IL-4Rα/γc or IL-4/IL-4Rα/IL-13Rα1 complex on the cell surface activates intracellular signaling pathways including the Jak-STAT and the PI3K/Akt pathways 9,11. Recent resolution of the crystal structures of extracellular domains of the IL-4-bound Type-I and Type-II IL-4 receptors (Fig. 1a) showed that IL-4 sits between IL-4Rα and the second receptor chain and is in direct contact with the second receptor chain through binding surfaces on the D-helix of the cytokine 6. IL-4 binds to IL-4Rα with very high affinity (KD = ~10−10 M) through a highly charged interface 12, while the subsequent binding of the IL-4/IL-4Rα complex to either γc or IL-13Rα1 is of relatively low affinity 6,9,13,14. The very high affinity of IL-4 for IL-4Rα means that in most instances the formation of the signaling complex is largely determined by the expression level of the second chain(s)15. The alternative second chains have different patterns of cellular expression with γc being mainly expressed on hematopoietic cells and IL-13Rα1 mainly on non-hematopoietic cells. Much of IL-4’s regulatory activity is mediated by B cells and T cells that mainly express Type-I receptors whereas its effector functions, in which it mimics IL-13, are largely mediated by cells that uniquely express the Type-II receptor and that also respond to IL-13. Through its capacity to utilize both the Type-I and Type-II receptors, IL-4 is positioned to play a central role in regulatory functions (i.e. Th2 differentiation, immunoglobulin class switching, dendritic cell maturation, macrophage activation) as well as effector functions (i.e. airway hypersensitivity and goblet cell metaplasia). However, these latter activities are physiologically induced mainly by IL-13, which is made in far larger amounts than IL-4. Further, since IL-13 cannot bind to the Type-I receptor, which is dominantly expressed on hematopoietic cells, it has little or no “regulatory” activity.
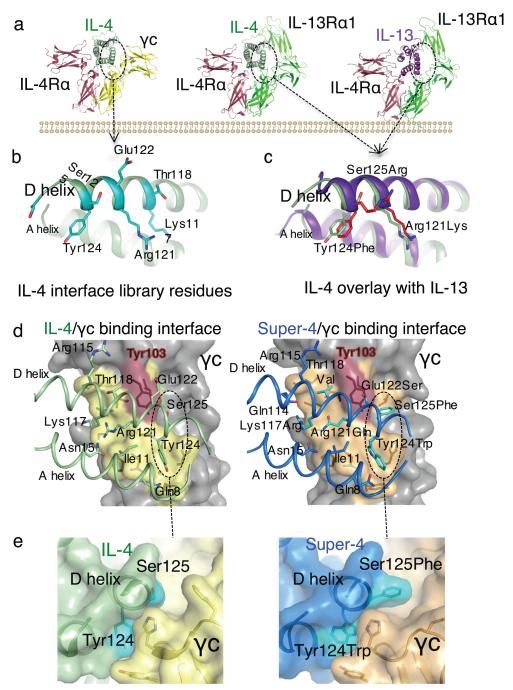
FIGURE 1. Structure-based engineering of IL-4 superkines.
(a) Crystal structures of the IL-4 and IL-13 Type-I and Type-II ternary ectodomain complexes 6. (b) and (c) The principal γc and IL-13Rα1 binding sites on the D-helices of IL-4 and IL-13, respectively. In (b) the positions randomized in the IL-4 site 2 library are shown, and in (c) a structural superposition of IL-4 and IL-13 in the receptor complexes shows that positions 121, 124, and 125 of IL-4 superimpose closely on the analogous positions of IL-13. In (c) IL-13 is in purple, and IL-4 is in light green, substituted residues are in red. (d) Isolated view of the site 2 interfaces in the WT (left) and super-4 (right) complexes with γc. The view shown is the ribbon representation of the A and D helices of the cytokines, with γc-interacting side chains shown, projected onto the semi-transparent molecular surface of γc. The interacting residues of γc underneath the surface are visible as dark outlines on the surface. The area contacted by the respective cytokines on γc is indicated in yellow on the surface, and the energetically critical Tyr103 of γc is colored red. A dashed oval encircles a region of the interface shown from the side in panel (e). In (e) a close-up is shown of interface packing and shape complementarity in super-4 (right) versus IL-4 (left).
Pharmacologically, utilization of IL-4 to regulate lymphocyte differentiation is complicated by its activity on non-hematopoietic cells through binding to the Type-II receptor and consequent effector function. There have been previous efforts to engineer IL-4 analogs 16, including the design of the antagonist Pitrakinra17. With the recent determination of the three dimensional structures of the complete liganded Type-I and Type-II receptor ternary complexes (Fig. 1a), we sought to engineer agonist IL-4 variants that would have altered relative binding activities for the second chains of the Type-I and Type-II receptors. In principle, these ‘superkines’ could have dose-dependent activities that allow optimal regulatory function while having reduced side effects.
Here we decouple the pleiotropy of IL-4 signaling through the engineering of Type I and Type II receptor-selective IL-4 superkines that exhibit cell-type specificity and novel activities, such as specific induction of dendritic cell maturation with a Type II receptor-specific superkine. Strikingly, the structure-activity relationships of these superkines do not reveal a linear correlation between superkine potency and receptor affinity, and the highest affinity superkines have a signaling advantage on cells with the lowest expression levels of second chain receptor chains. Thus, we demonstrate that cytokine affinity can be ‘tuned’ based on second receptor chain expression levels in order to selectively target desired cell types and potentially improve the selectivity of cytokine therapy.
RESULTS
Development of high affinity IL-4 variants
We used two different approaches to engineer IL-4 for higher affinity binding to γc (Fig. 1b) or IL-13Rα1 (Fig. 1c): directed mutagenesis and in vitro evolution. To increase the affinity of IL-4 for γc, we took a combinatorial library approach and used yeast surface display 18 (Supplementary Results, Supplementary Fig. 1a). We produced C-terminally biotinylated ectodomains of IL-4Rα, γc, and IL-13Rα1 for use as sorting reagents by coupling to Streptavidin-phycoerythrin (PE). We found that IL-4 displayed on yeast bound IL-4Rα with high affinity (Supplementary Fig. 1a) but did not bind to γc in the absence of IL-4Rα (Supplementary Fig. 1a). In the presence of IL-4Rα, IL-4 on yeast binds to the γc extracellular domain tetramer, indicating cooperative assembly of the heterodimeric receptor complex (Supplementary Fig. 1a). The use of high avidity tetramers of γc was essential for the detection of the initial weak γc binding in the early rounds of library sorting. To create a library of D helix variants of IL-4, which is the principal γc-interacting helix of the cytokine (Fig. 1b), we inspected the IL-4/γc interface in the crystal structure of the Type-I receptor ternary complex. We created a focused library in which eight residues on the face of helix D were randomized (Fig. 1b), resulting a yeast library with 2 × 108 variants. We carried out selections by decorating the yeast library with IL-4Rα to create the IL-4/IL-4Rα site 2 on the yeast and then sequentially enriched γc-binding yeast by decreasing the concentration of tetrameric, and finally monomeric γc (Supplementary Fig. 1b). Sequencing the IL-4-selected variants revealed two unique sequences, the ‘RQ’ and ‘RGA’ variants, in which one, RGA, was highly enriched (Supplementary Table 1).
To increase the affinity of IL-4 for IL-13Rα1, we took a rational, structure-based approach, rather than a combinatorial approach, based on inspection of the site 2 interfaces formed by IL-4 and by IL-13 with IL-13Rα1 (Fig. 1c). IL-13 binds with much higher affinity to IL-13Rα1 than does IL-4 (KD~30nM versus KD >1μM)6, so we aligned IL-4 with IL-13 from their structures in the two Type-II receptor ternary complexes (IL-4/IL-4Rα/IL-13Rα1 and IL-13/IL-4Rα/IL-13Rα1) to determine if we could “graft” important IL-13 receptor-interacting residues into the corresponding positions seen in IL-4 (Fig. 1c). We noted that three IL-4 D helix residues, Arg121, Tyr124, and Ser125, which form important contacts with γc in the IL-4 Type-II receptor ternary complex, are substituted in IL-13 6. We swapped these residues for their IL-13 positional equivalents (Fig. 1c) and made two IL-4 variants, a double mutant, Arg121Lys, Tyr124Phe, referred to as KF, and a triple mutant, KFR, in which all three residues are swapped.
Second receptor binding characteristics of the mutants
We expressed recombinant IL-4 and the variants KF, KFR, RQ and RGA using baculovirus and formed complexes with IL-4Rα in order to measure their binding affinities for IL-13Rα1 and γc by surface plasmon resonance (SPR) (see Supplementary Table 1 and Supplementary Fig. 2). The KD of WT IL-4/IL-4Rα for IL-13Rα1 and γc were 4200 nM and 3300 nM, respectively. KF/IL-4Rα had greater affinity for binding to both IL-13Rα1 (KD = 250 nM), and γc (KD = 330 nM). The addition of the Ser125Arg mutation in KFR resulted in a cytokine that had a 440-fold improvement over WT IL-4/IL-4Rα in affinity for IL-13Rα1 (KD = 9.6 nM) but a decreased affinity for γc (KD = 6400 nM). In this respect, the grafting was highly successful and resulted in a 3-log selectivity for IL-13Rα1 over γc.
The RQ and RGA variants complexed to IL-4Rα exhibited substantially higher affinity binding to γc (Supplementary Table 1, Supplementary Fig. 2). RQ/IL-4Rα showed a 36-fold higher affinity for γc (KD = 91 nM) and RGA/IL-4Rα exhibited a 3700-fold higher affinity (KD = 0.89 nM) than did IL-4/IL-4Rα. Both RQ and RGA superkines exhibited substantially decreased binding to IL-13Rα1 (KD = 29,000 nM and 21,000 nM, respectively), and would therefore be expected to exhibit negligible Type-II receptor binding. The structure-based and in vitro evolution approaches have therefore yielded higher affinity and receptor-selective IL-4 variants for functional testing. We refer to these cytokines as IL-4 “superkines” and specifically to the RGA variant as “super-4”.
Structural basis of IL-4 affinity enhancement for γc
We sought to understand whether the super-4 docking mode with the second chain, γc, was perturbed relative to WT IL-4 since this issue is important in interpreting signaling activity differences. We were able to crystallize the binary super-4/γc complex in the absence of IL-4Rα and obtain a structure with a resolution of 3.25Å (Fig. 1d and e, Supplementary Table 2, Supplementary Fig. 3). Superposition of the binary super-4/γc complex with the ternary Type-I signaling complex showed no major perturbations in cytokine-receptor orientation (Supplementary Fig. 3a-b). The position of γc bound to super-4 was essentially identical to the IL-4Rα/γc heterodimer geometry as observed in the complexes formed with WT IL-4. Therefore, any signaling changes we observe are very likely attributable to increased affinity and not structural differences.
In the super-4/γc interface, side chain density was clear for super-4 helix D residues 117-127 (Supplementary Fig. 3c); these engage the γc binding site in a topologically similar fashion to IL-4 with the γc hotspot residue Tyr124 occupying a central position (Fig. 1d). It seems clear that an important mechanism underlying super-4’s enhanced affinity was the replacement of Ser125 with Phe (Fig. 1e, left panel), which now inserts into a large hydrophobic pocket of γc that was previously unoccupied, contributing an additional 52.5 Å2 of buried surface area (BSA) (Fig. 1e, right panel). The hydrophobic groove in γc occupied by IL-4 Tyr124 gained a hydrogen bond from the Trp-N7 to a main chain carbonyl of γc. Based on the structure and SPR data, we propose that the major affinity gains in super-4 are derived from the Arg121Gln, Tyr124Trp, and Ser125Phe mutations. A detailed comparison of amino acid interactions of IL-4 and super-4 with γc is presented in Supplementary Table 3 and Supplementary Fig. 3d. We did not determine structure of the KFR/IL-13Rα1 complex as the mechanism for affinity enhancement seems obvious from the structure analysis and engineering strategy. The three side chains substituted on IL-4 D helix would endow IL-4 with “IL-13 like” contacts.
Cell activation in response to IL-4 superkines
To study responses to IL-4 and its superkines, we used Ramos, HH, A549, and U937 cells. We first measured the relative expression of mRNA (Supplementary Fig. 4a) and protein (Supplementary Fig. 4b) levels of the Type-I and Type-II receptor chains on these cells. Ramos cells have large amounts of IL-4Rα but their amounts of the Type-I receptor are limited by relatively low expression of γc. HH cells, although having less IL-4Rα than Ramos cells, have abundant γc. Both Ramos and HH cells have little or no IL-13Rα1. A549 cells have abundant Type-II receptor and little or no Type-I receptor. Finally, U937 cells have substantial amounts of both Type-I and Type-II receptor chains.
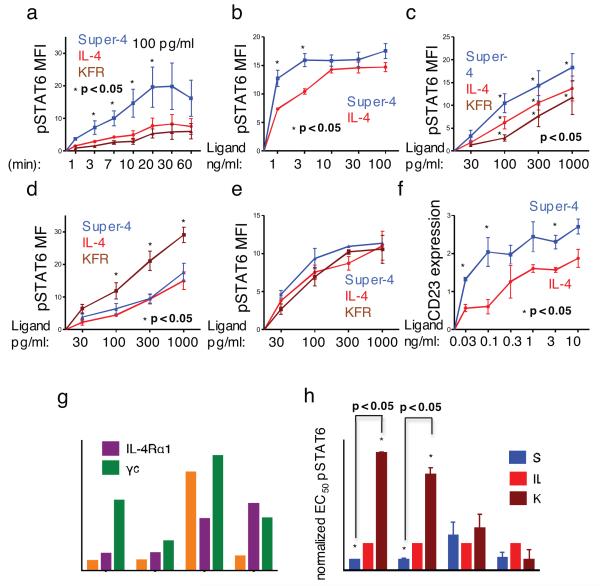
We initially tested the stimulatory activity of IL-4, super-4 and KFR. We used Ramos cells to study IL-4 responses dominated by the Type-I receptor complex (Supplementary Fig. 4). Stimulating Ramos cells with 100 pg/ml (~7 pM) of either IL-4, super-4 or KFR for various times, we found that the time course of stimulation of STAT6 phosphorylation by IL-4, super-4 and KFR is similar but super-4 induces substantially more phosphorylation than does IL-4 or KFR at all time points measured (Fig. 2a); after 20 minute stimulation, the mean MFI of STAT6 phosphorylation induced by super-4 is 19.6, by IL-4, 7.7 and by KFR, 5.4. In addition, dose/response experiments performed in Ramos cells with the three cytokines showed that super-4 was 10 fold more potent that KFR, although the three cytokines reach the same ‘plateau levels’ of STAT6 phosphorylation (Fig. 2b, Fig. 2c and Supplementary Fig. 5). However, the relative advantage of super-4 over IL-4 was relatively modest in comparison with the ~3700-fold difference in their solution equilibrium constants for γc when complexed to IL-4Rα (Supplementary Table 1).
FIGURE 2. Effect of IL-4 superkines on intracellular signaling.
(a) Overnight starved Ramos cells were unstimulated or stimulated for indicated times with 100 pg/ml of IL-4, super-4 or KFR. The cells were then fixed, permeabilized and stained with antibody against phosphorylated STAT6. (b-e) Ramos cells (b), Ramos cell starved overnight (c), A549 cells (d) and U937 cells (e) were stimulated for 15 minutes with increasing amounts of IL-4, super-4 and KFR, the analysis was then performed as in Figure 3a. (f) Ramos cells were stimulated for 8 hours either with IL-4 or super-4 as indicated, followed by surface staining of CD23. Means and SEMs from three independent experiments are shown for all experiments. (g) Expression of IL-4 Type-I and Type-II receptor chains on human PBLs from five donors. For the measurement of IL-4Rα, γc and IL-13Rα1 expression, B and T cells were gated by cell surface markers (CD19, CD4, CD8), while monocytes were identified as CD14+ cells. Appropriate isotype controls served as negative control. (h) Normalized pSTAT6 EC50 values obtained based on sigmoidal dose-response curves of IL-4 and the superkines (Supplementary Fig.6). pSTAT6 EC50 values from IL-4 wt were normalized to 1 and the EC50 values of the super-4 and KFR were calculated accordingly, means and SD are presented. Paired T-test was used to determine significant changes. In all the experiments, asterisk represent significant p values (p<0.05) obtained from the Paired T-test analysis.
A549 cells principally utilize IL-13Rα1 as their second chain (Supplementary Fig. 4). KFR was 3-10-fold more stimulatory than IL-4; super-4 was indistinguishable from IL-4 (Fig. 2d). Here again, while there was a qualitative agreement in that the highest affinity superkine caused a better response but the degree of signaling advantage by the variants did not mirror the absolute magnitudes of their solution affinity difference. In U937 monocytes, which express both γc and IL-13Rα1, super-4 slightly outperformed IL-4 but generally the differences between the superkines and IL-4 were modest (Fig. 2e).
To investigate whether the superior STAT6 activation by super-4 translates to the induction of STAT6-dependent gene products, we measured CD23 protein expression19 in Ramos cells that had been stimulated for 8 hours with either IL-4 or super-4. Super-4 was significantly more potent in inducing CD23 than was IL-4 (Fig. 2f), but again, super-4 showed less of an advantage over IL-4 than might have been expected from its far greater capacity, when complexed to IL-4Rα, to bind γc.
Primary human cell responses to IL-4 and superkines
We next studied STAT6 phosphorylation responses of human peripheral blood leukocytes (PBL) using Phospho-Flow cytometry coupled with fluorescent cell barcoding. We first measured IL-4Rα, γc and IL-13Rα1 expression in CD4 T cells, CD8 T cells, monocytes and B cells from 5 healthy donors by flow cytometry. IL-4Rα expression was highest on B cells while monocytes had intermediate expression and T cells had the least IL-4Rα (Fig. 2g). For γc, there was relatively little difference in expression between monocytes and CD4 T cells. B cells had slightly less γc and CD8 T cells had the lowest levels. As expected, IL-13Rα1 expression was highest on monocytes, whereas B and T cells showed very low expression of this chain. PBLs were either unstimulated or stimulated with IL-4 or the various superkines for 15 minutes; STAT6 Tyr641 phosphorylation was measured by flow cytometry. Super-4 induced stronger phosphorylation of STAT6 than IL-4 and much stronger phosphorylation than KFR in CD4 and CD8 T cells (Fig. 2h and Supplementary Fig. 6a-d). Monocytes showed little difference in their responses to IL-4, super-4 and KFR, in keeping with their expression of both γc and IL-13Rα1.
Modeling of receptor assemblage
The notion that super-4 was only ~3-10-fold more potent at activating STAT6 while its three dimensional equilibrium constant for γc was ~3700 higher than that of IL-4 left us wondering how signal-inducing receptor formation is dictated by the expression of the second chain. To address this question, we utilized a Matlab script slightly modified from that used in our previous publication7 (see Supplementary Methods) to calculate the assemblage of receptor complexes as a function of ligand concentration upon varying second chain numbers and varying second chain equilibrium constants. This matrix takes into account three parameters; the surface expression of IL-4Rα and γc receptor chains, the alteration in 2-dimensional binding affinities of ligand-bound IL-4Rα towards the γc chain and the ligand concentration. The calculation predicts the number of formed receptor complexes on cell surface and as such does not directly describe signaling particularly as it assumes that the availability of intracellular signaling molecules (Jak1, Jak3, Tyk2 or STAT6) does not limit the complex formation. Further, the calculation assumes that the physical interaction between cell membrane and all the receptor chains involved is similar and limits the free movement of the receptor chain equally on cell membrane.
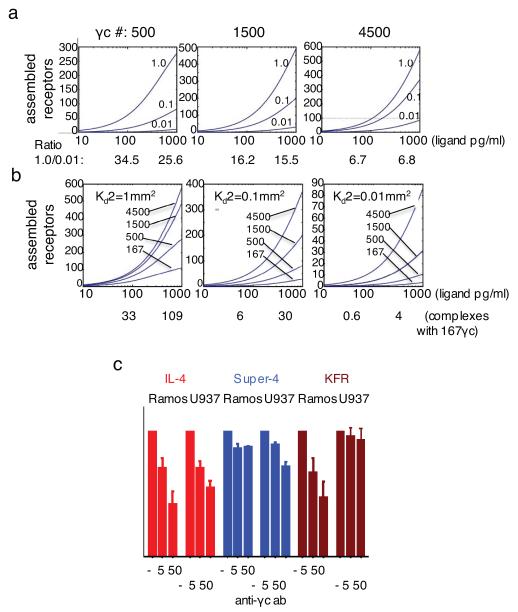
Since the number of IL-4Rα chains on Ramos cells has been reported to be ~1500 20, we determined the assemblage of receptors at this fixed IL-4Rα number. We used two equilibrium binding constants previously measured for IFN alpha receptor as “surrogate” values that roughly would be expected to correlate with Type-I and Type-II IL-4 receptors. 21. When γc number was set to 4500, there was a relatively modest effect of increasing second chain Ka2 from 0.01 to 1.0 μm2. However, when γc number was set to 500, the increase in second chain Ka2 had a strong impact on the number of receptor chains assembled (Fig. 3a). Thus, with a cytokine concentration of 100 pg/ml, the ratio of assembled complexes for Ka2=1μm2 compared to Ka2=0.01μm2 was 6.7 when the number of second chains was 4500 while that ratio was 34.5 when the γc number was set to 500. At 1000 pg/ml, the Ka2=1μm2 /Ka2=0.01μm2 ratio for 4500 γc molecules was 6.8 while it was 25.6 when the γc number was 500. Thus, increasing second chain Ka2 becomes more useful when the second chain number is relatively low. This would effectively mean that a cell that expresses low levels of γc or of IL-13Rα1 would most strongly benefit from enhanced affinity for the second chain. Indeed, when we calculated the number of formed receptor complexes at 100 pg/ml of ligand at only 167 γc receptor chains per cell, we found that the WT IL-4, with 2-dimensional Ka2 of 0.01μm2 assembled very few signaling complexes, as opposed to the 33 signaling complexes assembled by a superkine with a 100-fold higher 2-dimensional Ka2 (Fig. 3b).
FIGURE 3. Modeling of receptor assemblage in response to varying number of second chains.
A Matlab algorithm was used to calculate assemblage of IL-4 receptors on cell surfaces expressing only the Type-I IL-4 receptor. (a) IL-4Rα number was set to 1500. Second chain number was raised from 500 to 4500 and 2-dimensional equilibrium constant (Ka2) of IL-4Rα complexes for second chain were over a range from 0.01μm2 to 1μm2 as indicated. The ratio of assembled chains of highest (1.0 μm2) versus lowest (0.01 μm2) second chain Ka2 values was calculated for 100 and 1000 pg/ml at 500, 1500 and 4500 γc molecules per cell. (b) IL-4Rα number was set to 1500. 2-D equilibrium constant was varied from 1μm2 to 0.01μm2 and second chain number from 167 to 4500 per cell. Complexes assembled with 167 γc chains per cell at 100 and 1000 pg/ml of IL-4 or superkines at 2-D equilibrium constants of 1.0 μm2, 0.1 μm2 or 0.01 μm2 are shown. (c) Phosphorylation of STAT6 in Ramos and U937 cells in response to super-4, IL-4 and KFR in the presence of anti-γc antibody (0, 5, or 50 μg/ml). Response in the absence of anti-γc was normalized to 100% and responses in the presence of anti-γc expressed as in relation to the normalized value. Data (mean and SEM) are from three independent experiments.
As IL-4 and super-4 stimulate similar plateau values for STAT6 phosphorylation (Fig. 2b), we reasoned that assembling more signaling complexes than that induced by the lowest ligand concentration giving maximal stimulation would not result in any further signaling. As the plateau is achieved at 1,000 pg/ml of super-4 and 10,000 pg/ml of IL-4 in Ramos cells (Fig. 2b) we calculated the number of assembled complexes to be 65 for a ligand that had low affinity for the second chain (WT IL-4; 0.01 μm2) using an intermediate number of γc chains (1500) at 10,000 pg/ml.
Altering second receptor chain expression levels
Our modeling predicts that an increase in γc expression would be expected to decrease the advantage super-4 had over IL-4 and, conversely, limiting availability of γc would lead to clearer differences between IL-4 and super-4. We studied the sensitivity to IL-4 and super-4 of the HH cell line, which had much higher expression of γc than Ramos cells (Supplementary Fig. 4). Super-4 was not superior to IL-4 or KFR in inducing phosphorylation of STAT6 in HH cells at concentrations ranging from 10 to 10,000 pg/ml (Supplementary Fig. 6e).
An alternative test would be to diminish the accessibility of γc. For this purpose, we stimulated Ramos cells with 100 pg/ml of IL-4 or the super-4 and KFR superkines in the presence or absence of anti-γc, measured the phosphorylation of STAT6 by flow cytometry and calculated the percentage decrease in STAT6 phosphorylation caused by anti-γc. STAT6 phosphorylation induced by IL-4 was decreased 58% by 50 μg/ml of anti-γc whereas for super-4, the decrease was only 12% (Fig. 3c). For KFR, the inhibition was similar to that for IL-4. These results are consistent with the qualitative order of solution KD’s of IL-4 and the superkines for binding to γc (super-4>IL-4=KFR, Supplementary Table 1) and support the concept that increased affinity for the second chain results in greater stimulatory discrimination when the second chain expression is low.
In U937 cells, blocking γc would be predicted to diminish IL-4 responses whereas there should be little impact on the activity of the KFR superkine because of its principal utilization of the Type-II receptor. Indeed, blocking γc in U937 cells resulted in 44 % reduction in STAT6 phosphorylation in response to IL-4 but only a 7% reduction in response to KFR (Fig. 3c).
Immunomodulatory activities of IL-4 superkines
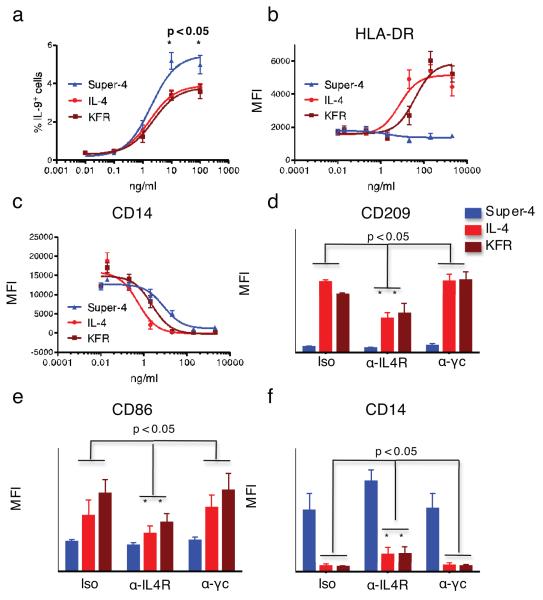
To study the functional specificity and immunomodulatory abilities of IL-4 and the superkines, we performed a series of experiments involving CD4 T cells and monocytes (Fig. 4). The combination of TGF-β and IL-4 promotes the differentiation of naïve human CD4 T cells into Th9 cells 22. To test whether super-4 more potently induces Th9 differentiation than WT IL-4, naïve CD4+ CD45RA+ CD45RO−CD25− T cells were isolated from human PBL and cultured with anti-CD3/anti-CD28-coated beads in the presence of TGF-β and varying concentrations of IL-4, super-4 or KFR for 4 days. Priming with 10 or 100 μg/ml of super-4 resulted in a significantly higher percentage of cells that produced IL-9 upon subsequent stimulation with PMA and ionomycin than did priming with the same concentrations of IL-4 or KFR (Fig. 4a).
FIGURE 4. Functional activities exhibited by IL-4 and superkines.
(a) Human naïve CD4+ CD45RA+ CD45RO− CD25− T cells were cultured with anti-CD3/anti-CD28-coated beads in the presence of TGF-β and the indicated concentrations of IL-4, super-4 or KFR. Cells were subsequently analyzed for intracellular expression of IL-9. Data (mean and SEM) are from 3 independent experiments with >4 donors. (b,c) CD14+ monocytes were isolated (>97% purity) from peripheral blood mononuclear cells obtained from healthy blood donors and cultured with 50 ng/mL GM-CSF alone or with the indicated concentrations of IL-4, KFR or super-4. Cells were subsequently stained with DAPI, fluorescently labeled isotype control mAbs, or mAbs against HLA-DR (b) and CD14 (c). Data (mean and SEM) are from 3 donors. (d,f) CD14+ monocytes were isolated (>97% purity) and cultured with 50 ng/mL GM-CSF and 2 μg/ml of IL-4, KFR or super-4 in the presence of the indicated antibodies. Cells were processed and subsequently stained with DAPI, fluorescently labeled isotype control mAbs, or mAbs against CD209 (d), CD86 (e) and CD14 (f). Data (mean and SEM) are from 3 donors. Paired T-test was used to determine significant changes.
IL-4, in combination with GM-CSF, induces the in vitro differentiation of dendritic cells (DCs) from human monocytes 23. Highly purified monocytes were cultured with GM-CSF alone or with varying concentrations of IL-4, super-4 or KFR. After 6 days, cells were analyzed for cell surface expression of the DC-associated molecules DC-SIGN (CD209), CD86 and HLA-DR. Strikingly, while IL-4 and KFR elicited monocyte differentiation into DCs that expressed CD209, CD86 and HLA-DR (Fig. 4b and Supplementary Fig.7), super-4 failed to do so suggesting that such differentiation is mainly driven by signaling through the Type-II IL-4 receptor complex, which is poorly engaged by super-4. Furthermore, super-4 was somewhat less effective than KFR or IL-4 in down-regulating CD14, a process also associated with the differentiation of monocytes into DCs (Fig. 4c). Additionally, analysis of further markers used to distinguish different DC subsets show that cells induced by GM-CSF with or without super-4 are phenotypically identical (Supplementary Fig. 8), implying that super-4-induced cells were incompletely differentiated rather than differentiated into a distinct DC subset.
To confirm the relative roles of Type-I and Type-II IL-4 receptor complexes in DC differentiation, we showed that anti-IL-4Rα, which would block both the Type-I and Type-II receptors, diminished the expression of CD86 and CD209 in response to IL-4 and KFR while anti-γc, which would only block the Type-I receptor failed to do so (Fig. 4d-f). Super-4 caused very modest induction of these markers. Super-4-induced CD14 down-regulation was partially inhibited by anti-IL-4Rα but not anti-γc. Thus, when γc was blocked, IL-4 and KFR still induced the same level of DC maturation as in the control condition (Fig. 4d-f), confirming that the Type-II IL-4 receptor complex plays an important role in GM-CSF/IL-4-mediated DC differentiation.
Signaling profile of IL-4 and superkines in monocytes
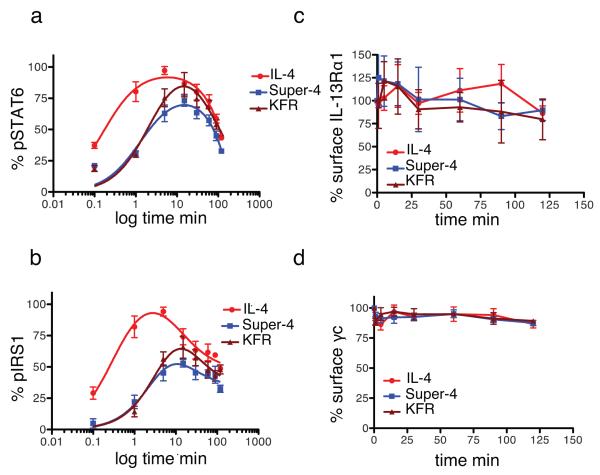
Since IL-4 and the two superkines activated STAT6 to the same extent in monocytes (Supplementary Fig. 6c), we sought to understand why super-4 was unable to induce DC differentiation. Purified monocytes were treated with two doses of cytokines, one dose corresponding to the pSTAT6 EC50 value (30pM) (Figure 5) and another dose corresponding to saturation (50nM) (Supplementary Fig. 9). The levels of STAT6 and IRS1 phosphorylation as well as the downregulation of the γc and IL-13Rα1 receptors were analyzed at the indicated times. At low doses, super-4 and KFR exhibited delayed activation of STAT6 and IRS1 (Fig. 5a-b) when compared to IL-4. No significant internalization of either γc or IL-13Rα1 was observed (Figure 5c-d). At high doses, the three cytokines induced the same kinetics profile of STAT6 and IRS1 activation (Supplementary Fig. 9a-b). KFR exhibited stronger internalization of IL-13Rα1 at later times of stimulation (Supplementary Figure 9c-d). Overall, these results show a lack of correlation between surface receptor internalization and signaling activation. Moreover, the delayed kinetics of signaling activation alone cannot explain the inefficiency of super-4 to induce DC differentiation, suggesting that Type-II receptor specific signaling is required for DC differentiation.
FIGURE 5. Signaling and internalization kinetics of IL-4 and the two superkines in monocytes.
CD14+ monocytes (>97% pure) were stimulated with 30pM of IL-4, super-4 or KFR for the indicated times. (a,b) Kinetics of STAT6 (a) and IRS1 phosphorylation (b) were measured by flow cytometry using phospho-specific antibodies coupled to fluorescence dyes. (c,d) Surface IL-13Rα1 (c) and γc internalization (d) was assayed by flow cytometry using receptor chain specific antibodies fluorescently labeled. In both cases data (mean and SEM) are from 4 healthy donors.
Gene expression profiling of IL-4 and superkines
To gain qualitative insights into the extent of redundancy of genetic programs induced by IL-4 and superkines in differentiating DCs, we performed genome-wide analysis of gene expression in response to WT IL-4 and the two superkines in monocytes treated simultaneously with GM-CSF. Monocytes from 5 healthy donors were stimulated for six hours with GM-CSF with or without IL-4, KFR or super-4 and RNA expression was analyzed as described in Materials and Methods. As shown by scatter plot correlation, the three cytokines induce the vast majority of genes to the same extent (Supplementary Figure 10a). However, interestingly, minor pockets of gene expression specificity can also be observed between IL-4 and the two superkines. A considerable number of genes were significantly induced by only one or two of the cytokines used. IL-4 regulated specifically 16 genes while super-4 and KFR regulated 72 and 45 respectively (Supplementary Figure 10b). The heatmap in Supplementary Figure 10c shows a representative set of cytokine-selective genes where clear differences in the expression patterns induced by IL-4 and the two superkines were observed. A more complete list of genes regulated differentially by superkines and IL-4 in monocytes is presented in Supplementary Table 4. DC-specific genes such as TPA1, HLA-DPA and CISH were clearly induced to a higher level by IL-4 and KFR than by super-4, consistent with specific signals coming from the Type-II IL-4 receptor that could bias the dendritic cell differentiation process induced by IL-4.
Cytokine secretion profiling of IL-4 and superkines
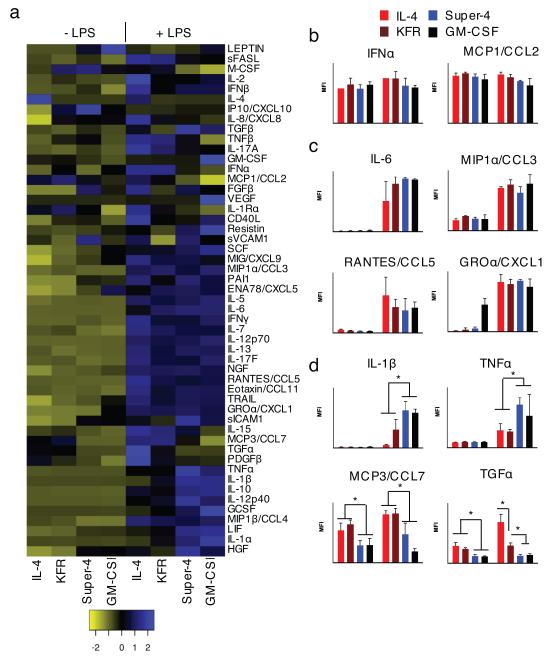
To further assess the functionality of the DCs induced by the engineered cytokines, we compared the secretion patterns of cytokines, chemokines and growth factors by performing a Luminex assay on supernatant of cells cultured for 8 days with or without LPS stimulation during the last 24 hours (Fig. 6a). Among the 51 analytes, 20 showed no difference in expression between treatments (superkines and LPS) (Fig. 6b) and 19 were up-regulated by LPS (most notably IL-6, CCL3, CCL5, and CXCL1) without difference between IL-4 and the superkines (Fig. 6c). The expression of the remaining 12 products discriminated the cells induced by GM-CSF only or GM-CSF + super-4 from the DCs induced by GM-CSF + IL-4 or KFR (Fig. 6d and Supplementary Fig. 11). The former two subsets were very similar and produced more G-CSF, HGF, IL-1α, IL-1β, IL-10, IL-12p40, LIF, TNFα, and less MCP3, MIP1β, PDGF, TGFα than the latter two, also very similar, subsets. Most of the differences were seen after LPS stimulation but some also existed in non-activated cells. Altogether, these data demonstrate that super-4 had no effect over that of GM-CSF alone on monocytes, while the addition of IL-4 or KFR led to phenotypically and functionally different DCs. Thus, the engineered cytokines appear to possess new and distinct functional activities.
FIGURE 6. Distinct patterns of cytokine secretion induced by IL-4 and the two superkines in immature and LPS-matured DCs.
Purified monocytes from 3 healthy donors were cultured for 7 days with GM-CSF (50 ng/ml) alone or combined with IL-4, KFR or super-4 (20 ng/ml), then stimulated (or not) with LPS (2 μg/ml) for another 24 hours. Culture supernatant was assessed by Luminex for relative amounts of 51 cytokines, chemokines and growth factors (listed by the heatmap (a)). (b-d) The panels on the right of the heatmap are representative examples of products whose secretion was either (b) unchanged (n=19), (c) increased by LPS stimulation only (n=20), or (d) modulated by superkines in the presence or absence of LPS (n=12). Data represent mean and SD from 3 healthy donors (normalized to GM-CSF alone group). Paired T-test was used to determine significant changes, *p<0.05).
DISCUSSION
Many cytokines being developed in the pharmaceutical sector are associated with dose-limiting toxicities or inadequate efficacy. One possibility for improving cytokines as pharmacologic agents is to bias them for preferential activity on certain desired cell types. Indeed, in a recent report by our lab, we succeeded in biasing the action of IL-2 to different leukocyte subsets by enhancing IL-2 affinity for IL-2Rβ 24. Cytokines that act through heterodimeric receptor complexes, such as those in the γc, gp130 and βc families, are particularly amenable to this approach given that the relative expression levels of the specific α chains of their receptors and the shared “second” chains often vary on different cell types.
Here, guided by structures of IL-4 receptor complexes, we have altered the agonistic properties of IL-4 in a way to redirect cell subset selectivity through engineering based on the metric of differential second receptor chain expression level. While the signaling experiments qualitatively confirmed the superiority of super-4 over IL-4 for a cell line predominantly using the Type-I receptor (Ramos) and of KFR over IL-4 for a cell line predominantly using the Type-II receptor (A549), the differences between IL-4 and super-4 on Ramos cells or IL-4 and KFR on A549 cells were much less dramatic than might have been anticipated. These results could be accounted for in several ways. The measurement of the equilibrium constant of the binding of soluble IL-4 (or superkine) complexed to IL-4Rα to immobilized γc or IL-13Rα1 may overestimate the differences in the 2-dimensional equilibrium constants among these proteins for second chain recruitment on the cell surface when both ligand and receptor are membrane bound and have greater diffusion limits. Another possibility is that the receptor heterodimers could exist, in a pre-associated form or alternatively, localized in membrane compartments in close proximity, as seen for IL-2 25.
IL-4 is not currently in use as a therapeutic agent but it had been considered for such use in the past and, if free of toxicity, might be considered for purposes such as directing CD4 T cell differentiation during vaccination or altering an established pattern of differentiation in view of the recent recognition of the plasticity of differentiated CD4 T cells 26. In the early 1990’s, clinical trials were performed in which IL-4 was administrated to cancer patients with the hope of boosting T cell responses or of engaging the innate immune system. However, intravenous administration of high dose (600 μg/m2/day) IL-4 resulted in a vascular leak syndrome in two out of three patients in the study group 27. Other toxicities were encountered in these studies and in preclinical analysis. The production of IL-4 superkines that cannot activate the Type-II receptor or in which activation of the Type-I receptor can be achieved at substantially lower concentrations than activation of the Type-II receptor might mitigate these problems, since most non-hematopoietic cells use only the Type-II receptor and cells of the monocyte/ macrophage lineage tend to express similar numbers of both receptors. Indeed, our observation that super-4 was relatively inefficient in inducing DC differentiation favor this hypothesis and agrees with previous work describing the requirement of Type-II IL-4 receptor for the surface expression of DC co-stimulatory molecules in mouse bone-marrow precursor cells 28.
The use of IL-4 to redirect T cell differentiation from more inflammatory phenotypes (i.e. Th1 or Th17) could be contemplated since CD4 T cells utilize the Type-I receptor virtually exclusively. Our data strongly suggest that super-4 would have greater efficacy for this purpose than IL-4 by combining a stronger activation of Type-I responses, which are required for T cell effects, and by reducing the activation of Type-II responses including DC differentiation. Indeed, super-4 more potently enhances TH9 differentiation, and may provide greater clinical benefit than IL-4 in boosting Th9 immunity.
While delivery of IL-4 by various means generally has a beneficial outcome in several preclinical models of autoimmunity such as the non-obese diabetic or the collagen-induced arthritis mouse models, the interpretation of the mechanisms of action has been made difficult by the pleiotropic nature of IL-4 binding. Thus, the use of receptor-selective superkines in mouse models will help to both better delineate the mode of action of therapies involving IL-4 and improve their efficacy 29,30.
The development of superkines can be considered as proof of the feasibility of this approach to achieve cell subset-specific cytokine effects. In principle, this approach can be attempted with many different cytokines whose signaling is dependent on the biophysical parameters of second chain recruitment.
METHODS
For Protein expression, yeast surface display, biophysical analysis and microarray analysis see Supplemental Information.
Cell lines and stimulations with IL-4 and superkines
Ramos, U937, A549 and HH cells were grown in RPMI containing 10% FBS, penicillin/streptomycin and l-glutamine (2mM) and maintained at 37°C with 5% CO2. Prior to stimulation, cells were cultured overnight in growth medium containing 2% FBS (“starved”). For γc-blocking experiments, overnight starved Ramos or U937 cells were incubated for 1 hour at 37°C with blocking antibody (R&D). Various concentrations and stimulation times of superkines are indicated in figures.
Flow cytometric stainings and antibodies
Cell surface expression of IL-4 receptor chains was performed after blocking Fc receptors I, II and III. Antibodies to CD23 (ref. 555711), IL-4Rα (ref. 552178) and γc (ref. 555900) antibodies were purchased from BD and for IL-13Rα1 (ref. FAB1462F) from R&D. Intracellular pSTAT6 and pIRS-1 staining was performed after ice-cold methanol (90%) permeabilization. Anti-pSTAT6 Ax488 (ref. 612600) and anti-pIRS-1 (ref. 558440) antibodies were purchased from BD. The induction of STAT6 phosphorylation was calculated by subtracting the Mean Fluorescence Intensity (MFI) of the stimulated sample from unstimulated sample. For primary human cells, analysis of STAT6 activation was performed as previously described 31. Briefly, PBMC samples from five donors were purified and stimulated with increasing concentrations of the appropriate cytokine for 15 min. Samples were then fixed in PFA for 15 min at 37°C. Cells were pelleted, washed with PBS, permeabilized with cold (4°C) methanol. Samples were then diluted with PBS to a final concentration of 50% and fluorescently barcoded with DyLight 800 and Pacific Orange dyes as previously described 31. After barcoding and combining, samples were stained for one hour with CD19 PE (ref. 302209), CD4 Brilliant Violet (ref. 300531), CD14 PerCP-Cy5.5 (ref. 325621), CD8 PE-Cy5 (ref. 301009) purchased from Biolegend and pSTAT6 Ax488. Analysis was performed on a BD Aria. Data analysis was performed in Cytobank software. Log median fluorescence intensity values were plotted against cytokine concentration to yield dose-response curves in cell subsets against pSTAT6.
RT-PCR
RNA was isolated from starved cells with RNeasy Kit (Qiagen). RNA was reverse-transcribed to cDNA using SuperScript II First-Strand Synthesis System for RT-PCR (Invitrogen). Quantitative PCR reactions were performed using a 7900HT sequence detection system (Applied Biosystems). The primer/probe sets to detect IL-4Rα, IL-13Rα1 and γc (FAM-MGB probe), and TaqMan Ribosomal RNA Control Reagents for detecting the 18S ribosomal RNA (VIC-MGB probe) were from Applied Biosystems. The mRNA levels were normalized to 18S ribosomal RNA.
Th9 differentiation assay
Enriched CD4 T cells were prepared from buffy coats obtained from healthy donors (Stanford Blood Center) using RosetteSep™ Human CD4+ T Cell Enrichment (Stem Cell Technologies) prior to density gradient centrifugation with Ficoll-Paque PLUS (GE Healthcare). Naïve CD4+ CD45RA+ CD45RO− CD25− T cells were magnetically sorted with Naïve CD4+ T Cell Isolation Kit II (Miltenyi Biotec). Cells were cultured at 37°C in 48-well flat-bottomed plates (Falcon) in X-VIVO 15 media (Lonza) supplemented with 10% Human Serum Type AB (Lonza), 100 units/ml penicillin/streptomycin, L-glutamine (Invitrogen) and 50 μM β-mercaptoethanol (Sigma-Aldrich). Cells were cultured at 2.5×105 cells/mL with anti-CD3/CD28 coated beads (Invitrogen) at a 1:1 bead-to-cell ratio in the presence of 5 ng/mL TGF-β (eBioscience) and the indicated concentrations of IL-4, super-4 or KFR. After 4 days in culture, beads were magnetically removed and cells were re-stimulated with 25 ng/mL PMA and 750 ng/mL Ionomycin (Invitrogen) in the presence of Brefeldin A (eBioscience) for 4 hours. Cells were then stained with a LIVE/DEAD Fixable Aqua Dead Cell Stain Kit (Invitrogen), then fixed and permeablized (eBioscience) according to manufacturer’s protocols. Subsequently, cells were stained with fluorescently labeled Abs against IL-9 and Foxp3 (eBioscience). Labeled cells were acquired on a BD LSRII (BD Bioscience), and data were analyzed on gated live single cells by FlowJo software (Treestar).
Dendritic cells differentiation: phenotyping and cytokine profiling
CD14+ monocytes were isolated (>97% purity) from peripheral blood mononuclear cells obtained from healthy blood donors (Stanford Blood Center) by density centrifugation using a RosetteSep Human Monocyte Enrichment Cocktail (Stem Cell Technologies) followed by magnetic separation with anti-CD14 conjugated microbeads (Miltenyi Biotec). 0.5-1×106 CD14+ monocytes were subsequently cultured with 50 ng/mL GM-CSF alone or with the indicated concentrations of IL-4, KFR or super-4 in 12-well plates (Corning) containing IMDM medium (Gibco) supplemented with 10% human AB serum, 100 U/mL penicillin, 100 μg/mL streptomycin, 2 mM L-glutamine, sodium pyruvate, non-essential amino acids and 50 μM 2-ME. Fresh cytokines were added on days 2 and 4. Cells were processed on day 6-7 with 5 mM EDTA and subsequently stained with DAPI (Invitrogen), fluorescently labeled antibodies against CD11c (ref. 561356), CD16 (ref. 560195), CD80 (ref. 555683), CD86 (ref. 555660), CD209 (ref. 551265), HLA-DR (ref. 560944) (BD Biosciences), CD1a (ref. 8017-0017-025), CD123 (ref. 48-1239-42) (eBioscience), CD1c (ref. 331514), CD40 (ref. 334312), CD14 (ref. 325619) (Biolegend), CD141 (ref. 130-090-513) and CD304 (ref. 130-090-9533) (Miltenyi Biotec), or appropriate isotype controls. Dendritic cell differentiation was assessed by flow cytometry with a BD LSRII flow cytometer and the mean fluorescent intensity (MFI) was determined on the FlowJo software (Treestar).
For Luminex cytokine profiling, culture supernatant was collected on day 8 (with or without 24h stimulation with 2 μg/ml LPS). Human 51-plex kits were purchased from Affymetrix and used according to the manufacturer’s recommendations with modifications as described below. Briefly, samples were mixed with antibody-linked polystyrene beads on 96-well filter-bottom plates and incubated at room temperature for 2 h followed by overnight incubation at 4°C. Plates were vacuum-filtered and washed twice with wash buffer, then incubated with biotinylated detection antibody for 2 h at room temperature. Samples were then filtered and washed twice as above and resuspended in streptavidin-PE. After incubation for 40 minutes at room temperature, two additional vacuum washes were performed, and the samples resuspended in Reading Buffer. Plates were read using a Luminex 200 instrument with a lower bound of 100 beads per sample per cytokine. MFIs were normalized to values from unstimulated cells cultured with GM-CSF only.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Dr. Josh Gregorio and Dr. Kipp Weiskopf for additional help and reagents, and the Stanford Human Immune Monitoring Center. Supported by National Institute of Allergy and Infectious Diseases Division of Intramural Research, (ISJ, WEP) and Finnish Medical Foundation (ISJ), the Howard Hughes Medical Institute (KCG), NIH RO1-AI51321(KCG) and NIH UO1-DK078123 (CGF), and the Stanford Immunology Program Training Grant (DLB).
Footnotes
AUTHOR CONTRIBUTION STATEMENT:
KCG conceived project, designed approaches for engineering of IL-4 and initiated subsequent cellular and functional experiments. DLB and PL performed protein engineering and biophysical experiments. ISJ, RJC, IM, and WEP designed and performed signaling experiments. RJC, MMS, and IM performed transcriptional analysis and Luminex experiments. WEP, IJS and MMS performed mathematical modeling using matlab. MTW, MNA, MMS and IM performed DC experiments. ISJ, RJC, IM, DLB, CGF, PJU, WEP and KCG analyzed the data. PJU and EGE provided reagents and guidance for human primary cell experiments. ISJ, RJC, IM, WEP and KCG wrote the manuscript.
COMPETING FINANCIAL INTERESTS STATEMENT
The authors have no competing financial interests.
REFERENCES
- 1.Leonard WJ. Type I Cytokines and Interferons and their Receptors. In: Paul W, editor. Fundamental Immunology. Lippinscott-Raven; Philadelphia: 1999. pp. 741–774. [Google Scholar]
- 2.Ihle JN. Cytokine receptor signalling. Nature. 1995;377:591–594. doi: 10.1038/377591a0. [DOI] [PubMed] [Google Scholar]
- 3.Stroud RM, Wells JA. Mechanistic diversity of cytokine receptor signaling across cell membranes. Sci. STKE. 2004;2004:re7. doi: 10.1126/stke.2312004re7. [DOI] [PubMed] [Google Scholar]
- 4.Wang X, Lupardus P, Laporte SL, Garcia KC. Structural biology of shared cytokine receptors. Annu. Rev. Immunol. 2009;27:29–60. doi: 10.1146/annurev.immunol.24.021605.090616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cunningham BC, et al. Dimerization of the extracellular domain of the human growth hormone receptor by a single hormone molecule. Science. 1991;254:821–825. doi: 10.1126/science.1948064. [DOI] [PubMed] [Google Scholar]
- 6.LaPorte SL, et al. Molecular and structural basis of cytokine receptor pleiotropy in the interleukin-4/13 system. Cell. 2008;132:259–272. doi: 10.1016/j.cell.2007.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Junttila IS, et al. Tuning sensitivity to IL-4 and IL-13: differential expression of IL-4Ralpha, IL-13Ralpha1, and gammac regulates relative cytokine sensitivity. J. Exp. Med. 2008;205:2595–2608. doi: 10.1084/jem.20080452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ito T, et al. Distinct structural requirements for interleukin-4 (IL-4) and IL-13 binding to the shared IL-13 receptor facilitate cellular tuning of cytokine responsiveness. J. Biol. Chem. 2009;284:24289–24296. doi: 10.1074/jbc.M109.007286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mueller TD, Zhang JL, Sebald W, Duschl A. Structure, binding, and antagonists in the IL-4/IL-13 receptor system. Biochim. Biophys. Acta. 2002;1592:237–250. doi: 10.1016/s0167-4889(02)00318-x. [DOI] [PubMed] [Google Scholar]
- 10.Nelms K, Keegan AD, Zamorano J, Ryan JJ, Paul WE. The IL-4 receptor: signaling mechanisms and biologic functions. Annu. Rev. Immunol. 1999;17:701–738. doi: 10.1146/annurev.immunol.17.1.701. [DOI] [PubMed] [Google Scholar]
- 11.Kelly-Welch AE, Hanson EM, Boothby MR, Keegan AD. Interleukin-4 and interleukin-13 signaling connections maps. Science. 2003;300:1527–1528. doi: 10.1126/science.1085458. [DOI] [PubMed] [Google Scholar]
- 12.Hage T, Sebald W, Reinemer P. Crystal structure of the interleukin-4/receptor alpha chain complex reveals a mosaic binding interface. Cell. 1999;97:271–281. doi: 10.1016/s0092-8674(00)80736-9. [DOI] [PubMed] [Google Scholar]
- 13.Andrews AL, Holloway JW, Holgate ST, Davies DE. IL-4 receptor alpha is an important modulator of IL-4 and IL-13 receptor binding: implications for the development of therapeutic targets. J. Immunol. 2006;176:7456–7461. doi: 10.4049/jimmunol.176.12.7456. [DOI] [PubMed] [Google Scholar]
- 14.Andrews AL, Holloway JW, Puddicombe SM, Holgate ST, Davies DE. Kinetic analysis of the interleukin-13 receptor complex. J. Biol. Chem. 2002;277:46073–46078. doi: 10.1074/jbc.M209560200. [DOI] [PubMed] [Google Scholar]
- 15.Heller NM, et al. Type I IL-4Rs selectively activate IRS-2 to induce target gene expression in macrophages. Sci. Signal. 2008;1:ra17. doi: 10.1126/scisignal.1164795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kraich M, et al. A modular interface of IL-4 allows for scalable affinity without affecting specificity for the IL-4 receptor. B.M.C. Biol. 2006;4:13. doi: 10.1186/1741-7007-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wenzel S, Wilbraham D, Fuller R, Getz EB, Longphre M. Effect of an interleukin-4 variant on late phase asthmatic response to allergen challenge in asthmatic patients: results of two phase 2a studies. Lancet. 2007;370:1422–1431. doi: 10.1016/S0140-6736(07)61600-6. [DOI] [PubMed] [Google Scholar]
- 18.Boder ET, Wittrup KD. Yeast surface display for screening combinatorial polypeptide libraries. Nat. Biotechnol. 1997;15:553–557. doi: 10.1038/nbt0697-553. [DOI] [PubMed] [Google Scholar]
- 19.Conrad DH, et al. Effect of B cell stimulatory factor-1 (interleukin 4) on Fc epsilon and Fc gamma receptor expression on murine B lymphocytes and B cell lines. J. Immunol. 1987;139:2290–2296. [PubMed] [Google Scholar]
- 20.Siegel JP, Mostowski HS. A bioassay for the measurement of human interleukin-4. J. Immunol. Methods. 1990;132:287–295. doi: 10.1016/0022-1759(90)90040-3. [DOI] [PubMed] [Google Scholar]
- 21.Gavutis M, Jaks E, Lamken P, Piehler J. Determination of the two-dimensional interaction rate constants of a cytokine receptor complex. Biophys. J. 2006;90:3345–3355. doi: 10.1529/biophysj.105.072546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wong MT, et al. Regulation of human Th9 differentiation by type I interferons and IL-21. Immunol. Cell Biol. 2010;88:624–631. doi: 10.1038/icb.2010.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dauer M, et al. Mature dendritic cells derived from human monocytes within 48 hours: a novel strategy for dendritic cell differentiation from blood precursors. J. Immunol. 2003;170:4069–4076. doi: 10.4049/jimmunol.170.8.4069. [DOI] [PubMed] [Google Scholar]
- 24.Levin AM, et al. Exploiting a natural conformational switch to engineer an interleukin-2 ‘superkine’. Nature. 2012;484:529–533. doi: 10.1038/nature10975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pillet AH, et al. IL-2 induces conformational changes in its preassembled receptor core, which then migrates in lipid raft and binds to the cytoskeleton meshwork. J. Mol. Biol. 2010;403:671–692. doi: 10.1016/j.jmb.2010.08.056. [DOI] [PubMed] [Google Scholar]
- 26.O’Shea JJ, Paul WE. Mechanisms underlying lineage commitment and plasticity of helper CD4+ T cells. Science. 2010;327:1098–1102. doi: 10.1126/science.1178334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sosman JA, Fisher SG, Kefer C, Fisher RI, Ellis TM. A phase I trial of continuous infusion interleukin-4 (IL-4) alone and following interleukin-2 (IL-2) in cancer patients. Ann. Oncol. 1994;5:447–452. doi: 10.1093/oxfordjournals.annonc.a058878. [DOI] [PubMed] [Google Scholar]
- 28.Lutz MB, et al. Differential functions of IL-4 receptor types I and II for dendritic cell maturation and IL-12 production and their dependency on GM-CSF. J. Immunol. 2002;169:3574–3580. doi: 10.4049/jimmunol.169.7.3574. [DOI] [PubMed] [Google Scholar]
- 29.Creusot RJ, et al. A short pulse of IL-4 delivered by DCs electroporated with modified mRNA can both prevent and treat autoimmune diabetes in NOD mice. Mol. Ther. 2010;18:2112–2120. doi: 10.1038/mt.2010.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Creusot RJ, et al. Tissue-targeted therapy of autoimmune diabetes using dendritic cells transduced to express IL-4 in NOD mice. Clin. Immunol. 2008;127:176–187. doi: 10.1016/j.clim.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krutzik PO, Nolan GP. Fluorescent cell barcoding in flow cytometry allows high-throughput drug screening and signaling profiling. Nat. Methods. 2006;3:361–368. doi: 10.1038/nmeth872. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.