Abstract
Gene-expression microarrays allow researchers to characterize biological phenomena in a high-throughput fashion but are subject to technological biases and inevitable variabilities that arise during sample collection and processing. Normalization techniques aim to correct such biases. Most existing methods require multiple samples to be processed in aggregate; consequently, each sample's output is influenced by other samples processed jointly. However, in personalized-medicine workflows, samples may arrive serially, so renormalizing all samples upon each new arrival would be impractical. We have developed Single Channel Array Normalization (SCAN), a single-sample technique that models the effects of probe-nucleotide composition on fluorescence intensity and corrects for such effects, dramatically increasing the signal-to-noise ratio within individual samples while decreasing variation across samples. In various benchmark comparisons, we show that SCAN performs as well as or better than competing methods yet has no dependence on external reference samples and can be applied to any single-channel microarray platform.
Keywords: Method, normalization, microarray, linear model, mixture model, single-sample technique
1 Introduction
Gene-expression microarrays enable high-throughput characterizations of biological phenomena. They are used widely in the investigation of biological and clinical phenotypes and have begun to be disseminated in clinical settings [1, 2, 3] via “personalized-medicine” workflows. For example, expression profiles representing prognostic subclasses of breast cancer are being used to guide treatment decisions and have been shown to influence medical-oncologist treatment recommendations [4] . Developing such a workflow involves two distinct steps: 1) identification of expression profiles that reliably characterize subclasses and 2) comparison of new breast-cancer samples against those profiles to predict each sample's subclass. However, a critical preliminary step is to normalize each microarray sample to adjust for technological biases and non-biological variabilities that arise during sample collection and processing and that obfuscate true signal. Traditional normalization methods require groups of samples to be processed jointly—thus each time a new sample arrived, all samples would need to be renormalized. Clearly, this type of approach presents serious logistical and reproducibility challenges and obviates the need for methods that process samples individually. To address this need, we developed Single Channel Array Normalization (SCAN), a normalization technique that uses data observed within a given microarray sample to correct for probe-nucleotide biases and to produce standardized output values that remain constant irrespective of any other sample. Furthermore, unlike competing methods, SCAN requires no ancillary reference samples to perform these corrections and can be applied to any one-color microarray.
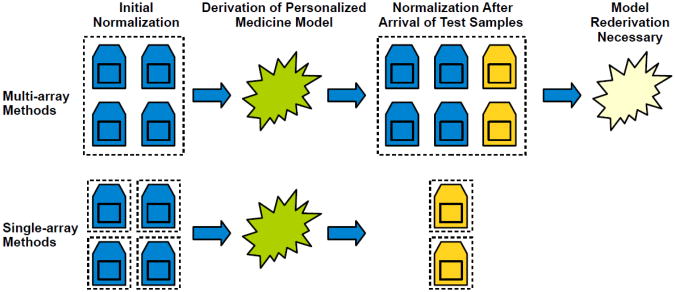
Many normalization methods—including the popular Robust Multichip Average (RMA) [5] and its probe-sequence-adjusting counterpart, GeneChip RMA (GC-RMA) [6]—borrow information from other samples to estimate probe-level effects and to standardize variances across arrays. However, with such approaches, output values for any given array depend on the quantity and quality of other arrays that are processed jointly (see Figure 1A). In an attempt to address such limitations, two normalization techniques capable of processing individual samples have gained popularity. The MAS5 algorithm [7], developed by Affymetrix, uses mismatch probes with single-base differences to correct for background variation. As such, this method attempts to minimize probe-level effects by observing such effects in probes with nearly identical sequences. Despite the relative simplicity of its algorithmic approach, MAS5 has performed well against multi-array methods [8]; however, this algorithm can only be applied to platforms containing mismatch probes, which excludes the newer Affymetrix Gene ST, Exon ST, and GG-H arrays (see Figure 1B). Recently, McCall, et al. introduced another single-sample approach, frozen RMA (fRMA) [9], which borrows information from publicly available arrays of the same platform. A key prerequisite to applying fRMA is that a reference database be derived from a large set of biologically diverse samples from a wide variety of studies. For the array platforms currently supported by fRMA, approximately 6,000 “well-annotated” samples (from which 850 were randomly selected) were used to derive each reference database. However, obtaining such a large and diverse sample set is currently impossible for many platforms, and this approach requires arbitrary decisions of which samples to consider and a deliberate effort to construct a reference set for each unique platform.
Figure 1.
Multi-array versus single-array normalization. A) With multi-array methods, such as RMA, samples are processed in groups. Thus when new samples have been hybridized—for example, in personalized-medicine settings—all samples, old and new, may need to be renormalized as a group, which may require reanalysis of the data or recalibration of biomarkers. Contrarily, with single-array methods, including SCAN, each sample is normalized individually. Thus newly arrived samples remain separate during normalization, and data values for existing samples do not change. B) Affymetrix offers many different array versions to quantify human gene expression. SCAN can normalize any version. However, fRMA does not currently support most array versions because an inadequate number and diversity of previously hybridized samples have been made available publicly. And because MAS5 relies on mismatch probes, it is unable to normalize samples from newer array versions.
To overcome these limitations, SCAN estimates the effect of probe-nucleotide composition on fluorescence intensity through a linear statistical model and distinguishes between background noise and biological signal with a mixture-modeling approach. Probe-level values are standardized against the variance observed for other probes on the same array that have similar binding-affinity-adjusted intensities. In this light, our approach is more appropriately deemed a standardization—rather than the more commonly termed normalization—approach; however, to be consistent with current terminology in the field, we use both terms interchangeably here. Because SCAN processes a given array only using information intrinsic to that array, normalized values are independent of any other sample that may have been hybridized in the same batch. Also importantly, to process a large number of arrays, computer memory must only be adequate to process a single array at a time, making SCAN a versatile approach that can be used on low-memory desktop computers or compute nodes, even when existing methods fail to process the set of arrays simultaneously. However, in the case where a large number of processors and memory are available, SCAN can be used to process multiple arrays in parallel.
In this study, we demonstrate that SCAN corrects for probe-level effects associated with GC bias and drastically increases the signal-to-noise ratio within each array. We show that SCAN compares favorably against RMA, fRMA, and MAS5 in its ability to detect differences in expression across a wide spectrum of RNA spike-in concentrations. Using data from the Connectivity Map (CMAP) [10], we demonstrate SCAN's methodological advantages in dealing with batch effects [11] and differences between array versions. Strikingly, we show that SCAN enables effective concurrent use of independent gene-expression datasets. In particular, in an analysis of two independent microarray compendia that profile identical cell lines—Broad-Novartis Cancer Cell Line Encyclopedia (CCLE) and GSK Cancer Cell Line Genomic Profiling Data—we show that SCAN produces highly consistent values for samples grown, processed, and hybridized by independent groups at separate times. Finally, in a pathway-based analysis, we show that SCAN-based predictions are highly concordant across the data sets which are known to contain batch effects.
2 Results
2.1 General description of SCAN standardization approach
The most common approaches to normalizing and summarizing microarray data simultaneously process a series of samples, causing measurements for each sample to be dependent on other samples in the group. This approach can lead to significant batch effects and can create discrepant results between datasets that are normalized separately, thus creating problems for applications in personalized medicine. Furthermore, processing large datasets may require excessive amounts of physical memory or splitting large datasets in order to successfully complete the normalization process. SCAN has been developed to resolve these limitations, and is highly effective at standardizing array data to control for background and individual array signal.
SCAN utilizes a modification of the MAT standardization approach developed for Affymetrix tiling arrays [12]. The MAT method models expected probe behavior based on the individual probe sequence and other probes on the array with similar nucleotide composition and removes effects due to base-pair content and array bias. Kapur, et al. successfully applied the MAT model to Affymetrix Exon ST arrays [13], but in their approach parameters were estimated using all data from each array. This assumption is reasonable for tiling arrays where the number of probes measuring signal is small with respect to the number of probes measuring background signal, but is not appropriate for RNA-based expression array experiments due to the high percentage of probes that measure true signal. Therefore, our novel modeling procedure is based on a mixture of two Gaussian distributions with MAT-like models for the means of both distributions—one mixture component measuring background noise and the other measuring biological signal (plus background). The component of interest is the background distribution, which is used by SCAN to standardize the probe-level data by subtracting out the background mean and then standardizing the variance based on the estimated variance of other probes with similar expected background. As illustrated below, this approach is highly effective at removing GC effects and other probe and array-specific variation from each sample individually while leaving the biological signal intact, thus increasing the signal-to-noise ratio within the array while reducing technical variation between arrays.
2.2 SCAN corrects for GC bias inherent to one-color arrays
SCAN accounts for GC bias using a refined linear statistical model that includes effects for the counts of each nucleotide type separately, the nucleotide count squared, and also the location of each nucleotide type on the probe. Using publicly available CEL files downloaded from Gene Expression Omnibus (GEO) [14], we have tested SCAN on a wide variety of Affymetrix platforms, including those that are widely used— including Human Genome U133A, Human Genome U133 Plus 2.0, and Human Exon 1.0 ST—as well as those that have been released recently or that have been less utilized. Upon analyzing these samples, we observed a strong GC bias prior to normalization—probes associated with higher GC content tended to have higher raw-intensity values (see Figure 2A). We assessed for GC bias after normalization and observed that SCAN abrogated this bias (see Figure 2B). This result illustrates that SCAN can correct for binding-affinity biases using only data from within a given sample.
Figure 2.
Proportion GC content versus probe intensity. A) For an Affymetrix Human Exon ST 1.0 array (GSE25219), this figure illustrates the relationship between probe-level GC content and expression intensities. B) As GC content increases, raw intensity also tends to increase. However, after SCAN processing, this bias is removed.
2.3 Overall microarray signal-to-noise explained by SCAN model
A unique advantage of SCAN is that it directly identifies systematic within-array variation that can be explained as probe effects and removes this variation from the data. For probes measuring both signal and noise, this method only removes noise and leaves the signal intact. When applied to a set of 100 Affymetrix Human Exon ST 1.0 arrays (GSE25219), SCAN estimated that an average of 79% of probes are only measuring background noise and exhibit no signal above background. In these arrays, we estimate that an average of 35% of the total squared variation in the array data is attributable to background signal. In all, the probe-mixture model accounts for an average of 79% of the variation in the data, and 44% of this explained variation is attributable to background noise (probe effects) and thus removed. Therefore, we estimate the average overall signal-to-noise ratio to be 1.4 in the raw data; this is improved to 6.5 after removing probe-specific noise, resulting in a relative increase of 482% in the overall signal-to-noise ratio. Table 1 summarizes these results and gives ranges for these values across all arrays in the dataset we examined. Therefore these results show that SCAN is highly effective at removing background variation and magnifying signal within each array.
Table 1.
Summary of probe-level metrics for SCAN normalization. SCAN was applied to 100 Affymetrix Human Exon 1.0 ST arrays from publicly available data representing spatio-temporal transcription patterns in the human brain (GSE25219) [26]. This table lists various probe-level metrics that characterize SCAN's ability to account for variation and to magnify signal in the data.
| Metric | Average | Minimum | Maximum |
|---|---|---|---|
| Proportion of active probes in sample | 0.208 | 0.154 | 0.263 |
| Proportion of total variation explained by model | 0.792 | 0.757 | 0.814 |
| Proportion of variation attributed to background noise | 0.439 | 0.280 | 0.634 |
| Estimated signal-to-noise ratio before application of SCAN | 1.357 | 0.578 | 2.574 |
| Estimated signal-to-noise ratio after SCAN | 6.554 | 2.632 | 13.13 |
| Percent increase in signal-to-noise ratio after SCAN | 482% | 412% | 538% |
2.4 SCAN detects transcription levels with high sensitivity and specificity
Next we assessed SCAN's ability to detect known transcription levels in the Affymetrix Human Genome U133 Latin Square data (http://www.affymetrix.com/support/technical/sample_data/datasets.affx), which is designed to support comparisons of analysis methods. This data set contains 14 distinct concentrations of spiked transcripts, ranging from 0.125 picomolar (pM) to 512 pM, which were hybridized to Affymetrix U133A microarrays. After normalization and summarization, we sorted expression levels for the human cell-line probeset groups by pM concentration and then averaged the values across all samples. For each concentration, we generated receiver-operating-characteristic (ROC) curves to evaluate how well “expressed” transcripts could be differentiated from “non-expressed” transcripts, using that concentration as a threshold. We then calculated the mean area under the ROC (AUC) across all thresholds and used this value as a comparison metric. Identical processing steps were followed for the SCAN, fRMA, RMA, and MAS5 normalization methods. As shown in Supplementary Figures 1 and 2, MAS5 and SCAN performed best, attaining mean AUCs of 0.977 and 0.976, respectively. Results for fRMA and RMA were slightly lower (0.960 and 0.959, respectively). Note that although these differences are only small in these spike-in data, the differences will be magnified in more complex analysis scenarios, as illustrated in the examples below. These results suggest that SCAN is highly effective at detecting relatively minor differences in abundance, a crucial capability for “real-world” studies where differences in transcription between conditions may be subtle.
2.5 SCAN adjusts for batch and platform effects
In processing large data sets, differences in personnel, equipment, and laboratory conditions may lead to non-biological variability in expression, known as batch effects [11]. When multiple array versions are used for profiling, such effects may be amplified further. CMAP contains thousands of microarrays that were used to profile cancer cell line responses to dosage levels of many drugs. We examined the subset that was used to profile MCF7 cell lines treated with valproic acid and observed strong batch effects in the raw data (see Figure 3A). However, after SCAN normalization, values fell within a similar range for each array, irrespective of batch or array type (see Figure 3B). RMA also standardized the value ranges within each array type; however, because RMA can only be applied to samples from a single platform at a time, substantial differences persisted across samples measured on different platforms; a similar pattern was observed for MAS5 (see Supplementary Figure 4).
Figure 3.
SCAN adjusts for sample-level variations in expression intensity arising from platform and batch effects. In A), log2 intensity values are shown for CMAP samples treated with valproic acid. In B), SCAN normalized values are shown for the same samples. Before normalization, value ranges varied largely by batch and/or platform (each color represents a distinct batch of samples profiled on a specific Affymetrix platform). After normalization, values fell within a similar range, irrespective of batch or platform.
For an additional comparison, we tested how well valproic acid response could be estimated across the various concentration levels. For each normalization method, we generated a gene signature using cell-line data from Cohen, et al. [15] and projected this signature onto the CMAP samples using BinReg 2.0 [16] with no quantile or shift-scale normalization that might bias the comparison of methods. As illustrated in Supplementary Figure 5, each method produced response predictions that increase by concentration. However, SCAN produced predictions that ranged nearly from zero to one, whereas RMA and MAS5 predictions ranged between 0.4 and 0.8. Accordingly, SCAN appeared to be more effective at characterizing cellular activity associated with valproic acid treatment, thus enabling a clearer distinction between samples that had been treated at lower versus higher concentrations.
It is important to note that the fRMA software currently provides external reference vectors for only one (HG-U133A) of the three array versions used in CMAP. The other two versions (HT_HG-U133A and HT_HG-U133_EA), which constitute 89.0% of perturbed samples in CMAP, are unsupported because an inadequate number and diversity of reference samples have been deposited in GEO. Thus we did not include fRMA in the CMAP comparisons. This scenario illustrates an important methodological advantage of SCAN over fRMA; SCAN uses only data from a given array for normalization and thus can be applied to any one-color array version, whereas fRMA requires platform-specific ancillary samples that may or may not be available. Additionally, because fRMA reference vectors are derived from different sets of samples for each platform, platform-specific noise may be introduced.
2.6 Concordance between two cancer cell-line compendia
Next we assessed whether the various normalization methods would produce consistent values for identical cell lines that were prepared and hybridized by different personnel, in different facilities, and at different times. Such differences may cause non-biological noise that could confound downstream conclusions. We posited that an effective normalization procedure would reduce the effects of such noise. To perform this comparison, we obtained raw files from the Cancer Cell Line Encyclopedia (CCLE) (http://www.broadinstitute.org/ccle) and a corresponding data set generated by GlaxoSmithKline (GSK) [17]. For both data sets, the Affymetrix Human Genome U133 Plus 2.0 platform was used for profiling. We identified samples that had been profiled in both CCLE and GSK (n = 222). For the GSK data set, which contained triplicate hybridizations of each cell line, we selected the first sample that had a GAPDH 3′/5′ ratio greater than 3.0. Next we calculated the mean Pearson correlation coefficient between corresponding samples in CCLE and GSK. SCAN produced highly consistent values across the data sets and had overall higher correlation coefficients than the other methods (see Figure 4 and 3).
Figure 4.
Correlation of sample-wise expression levels between CCLE and GSK cell lines. CCLE and GSK data were processed using each normalization method, and sample-wise Pearson correlation coefficents were calculated. Across the samples, SCAN values were more highly correlated than for the other normalization methods, suggesting its ability to produce consistent output values for analogous samples processed in independent facilities by different personnel at different times. (Correlation coefficients were sorted for each normalization method independently before plotting.)
In a follow-on analysis, we estimated RAS pathway activation within the treated cell lines using a methodology developed by Bild, et al. [18] In this approach, a pathway signature is constructed by comparing expression levels of human primary mammary epithelial cell cultures infected with recombinant adenoviruses and control cells infected with green fluorescent protein. After renormalizing RAS samples from the Bild, et al. study, we derived expression signatures representing pathway activation and projected those signatures onto the CCLE and GSK samples using the BinReg 2.0 algorithm. Under the expectation that normalization methods better at filtering non-biological artifacts would result in greater consistency between the two data sets, we calculated Pearson correlation coefficients based on the predicted probabilities of pathway activation. As illustrated in Figure 5, SCAN and fRMA performed substantially better (r = 0.870, 0.869) than MAS5 and RMA (r = 0.757, 0.323).
Figure 5.
Comparison of RAS pathway activation probabilities for CCLE and GSK cell line data. The probability of RAS pathway activation was estimated for individual samples in CCLE and GSK cell lines. For each normalization method, a pathway signature was derived through a comparison of expression levels in RAS-activated cell cultures and controls. The signatures were then projected onto the cell line data, and a probability of pathway activation was calculated. For SCAN and fRMA (A and B), probabilities were highly concordant across the data sets, whereas for RAS and MAS5 (C) and D) the probabilities were less concordant and fell within narrower ranges. Lack of replication between data sets may lead to markedly different biological conclusions, depending on which data set is examined.
Taken together, these findings suggest that SCAN produces standardized data that account for unavoidable variations that occur in microarray studies.
3 Discussion
Prior capital investments in microarray equipment and enormous troves of institutional and publicly available data will surely lead to microarrays' continuance as a prominent research tool for years to come. In the Gene Expression Omnibus alone, hundreds of thousands of raw data files are available for secondary analyses [14]. Optimization of microarray normalization procedures remains imperative to genomic research by enabling more accurate characterizations of biomedical phenomena and providing insights into the design of RNA-sequencing projects. In this study, we have described SCAN, a novel normalization technique capable of processing one microarray sample at a time. Through analyses of spike-in data and cell-line experiments, we have demonstrated that SCAN performs similarly or better than several existing methods, including other single-sample techniques and the most popular multi-sample method.
Various normalization techniques have been proposed over the years, but most depend on groups of samples for processing. Although in some simple cases such an approach may be adequate for standardizing values within a given data set, sample aggregation introduces logistical challenges that may limit their usefulness in settings where samples accrue over time—for example, in personalized-medicine workflows or meta-analyses that span multiple studies. SCAN takes advantage of the large amount of raw data produced by a given array and corrects for binding-affinity biases on a single-sample basis. This approach provides various methodological advantages, including that no information is borrowed from external samples. Additionally, SCAN is robust to differences among microarray platforms because the same linear model is applied for every platform. Indeed, we observed that SCAN produced highly concordant (see Supplementary Figure 6) gene-summarized values for Human Exon ST 1.0 and HG-U133A arrays from the same ovarian-cancer sample (TCGA-04-1335) [19], even though the array platforms differ considerably in design and in the number of probes they contain.
Across our analyses, SCAN performed as well as or better than all other techniques, which suggests its superior utility and consistency as a gene-expression normalization method. In our analyses, the most popular [9] multi-array method, RMA, underperformed in relation to the single-array methods. In particular, RMA faltered in its attempts to produce congruous expression values for the cancer cell-line data that had been processed and hybridized at different facilities. Conceivably, RMA's attempt to standardize across arrays introduces subtle biases that currently hamper cross-study replicability. In fact, Giorgi, et al. demonstrated that RMA's median-polish summarization step can introduce between-array correlation artifacts [20]. fRMA attempts to overcome the limitations of multi-array processing by constructing an external reference database of probe-level effects. Indeed, across the various analyses we performed, fRMA performed better than its multi-array counterpart. However, fRMA comes with practical limitations that hamper its broad utility. Researchers who desire to apply this method to samples from array versions that are currently unsupported by fRMA must go through an extensive process of constructing and validating a new reference database for each platform. Importantly, variation in the reference data can itself affect data quality. The SCAN algorithm, on the other hand, can be applied to any single-channel array (such as those manufactured by Affymetrix and Nimblegen) that provides a support file matching probe sequences with probe coordinates. In some of our analyses, MAS5 performed similarly to SCAN; however, it also lacked consistency when applied to the cell-line data. And perhaps most importantly, MAS5 can only be applied to older generations of Affymetrix arrays that contain mismatch probes. Affymetrix has also provided the probe logarithmic intensity error (PLIER) method (http://media.affymetrix.com/support/technical/technotes/plier_technote.pdf), which corrects for GC content and can be applied to older- and newer-generation arrays. However, PLIER relies upon external samples for its summarization approach and thus was not evaluated in this study.
By accurately distinguishing between true biological signal and background noise, SCAN helps elucidate the cellular mechanisms driven by complex variations in transcriptional activity. Effectively correcting for technological biases on a single-sample basis is particularly important for microarrays to gain acceptance in the clinic.
4 Materials and methods
4.1 Model-based adjustment for probe-nucleotide composition
To correct for binding-affinity biases, we developed a linear model that is a variation on the Model-based Analysis of Tiling arrays (MAT) method [12], which was designed for Affymetrix tiling arrays. MAT has been shown to account for as much as 50% of the variation that results from binding-affinity and cross-hybridization effects in tiling-array samples [12]. As with MAT, our model contains a nucleotide indicator for each position on a given 25-mer probe sequence, along with quadratic terms representing nucleotide counts. However, the novel contribution of SCAN, which makes it appropriate for expression-array experiments, is the use of a mixture of two Gaussian distributions, one mixture component measuring background noise and the other measuring signal (plus background). The main component of interest is the background distribution, as the other component will likely result in a confounded distribution of true biological signal of interest, cross-hybridization, etc. More specifically, letting Pi represent the probe-intensity value for probe i=1,…,N, we assume that log(Pi) = (1 - Δi)Y 1i + Δi Y2i where Y1i ∼ N(Xi θ 1, σ12) and Y2i ∼ N(Xiθ2, σ22), where Xiθm is the mth mixture-component MAT model for probe i presented in the following equation for a given m:
| (1) |
where αm is the baseline value based on the number of T's in the probe sequence, nil is the nucleotide l count, βmjk is the effect of each nucleotide k (except T which is already modeled in αm) at each position j, Iijk is an indicator function such that Iijk=1 if the nucleotide at position j is k in probe i, γml is the effect of nucleotide count squared, and εmi is the probe-specific error term, assumed to follow a normal distribution. The probe mixture component indicators, Δi, take on the values either 0 or 1, indicating from which mixture component each probe was drawn. However, these indicators are not observable and will be estimated in the mixture-modeling procedure. We formulate the above model based on a ‘missing data’ approach (where the Δis are the missing data), and apply the Expectation-Maximization (EM) algorithm to estimate the model parameters [21]. Specifically, assuming the previously described Gaussian mixture for log(Pi) and assuming Δi∼ Bernoulli(π) where π is the proportion of non-background probes on the array, we obtain the complete data likelihood of
| (2) |
where f1(yi| θ1) and f2(yi| θ2) are Gaussian densities representing the distributions of the ‘background’ and ‘background + signal’ distributions of probe respectively. Because the complete data log-likelihood is linear in the missing data (Δis), applying the EM algorithm is straight-forward in this case. The Expectation Step (E-Step) consists of imputing the missing data in the complete data log-likelihood (2) with their expected values using the most recent estimates of the model parameters from the previous Maximization Step (M-Step). The M-Step then consists of maximizing the model parameters given using (2) and the most recent imputed missing data from the E-step. The E-Step and M-Step are then iterated until the model parameters converge to the Maximum Likelihood Estimates (MLEs). These MLEs are then used to standardize the data as described below.
The model (for each mixture component) contains 80 parameters, 1 for α, 3 × 25 for β, and 4 for γ. SCAN estimates these parameters by ordinary least squares using a subset of probes (n = 50,000) on the array. After the parameters have been estimated, a baseline intensity is predicted for each probe. After fitting the model, we use the background mixture component to obtain an estimate for the background expected intensity for probe i, mi, to remove the probe-specific variation. We bin probes into groups of 5,000 with similar mi and normalize the data as follows:
| (3) |
where mi is the baseline intensity predicted by the model based on the sequence of probe i, and si bin is the standard deviation of the bin to which probe i belongs.
The MAT statistical model was previously applied to Affymetrix exon arrays [13]. However, we have incorporated modifications that were not included in their approach. Most importantly, their approach did not take into account that most of the variation in an expression experiment is likely due to the true biological signal of interest. The original MAT approach estimates parameters based on all the variation in the data (background and signal), which will remove some biological signal from the data and result in reduction of the signal-to-noise ratio. SCAN's novel introduction of a mixture modeling approach results in the removal of mostly background while preserving the biological signal in the data.
4.2 Software
SCAN has been implemented as an open-source software package that can be downloaded freely from http://www.bu.edu/jlab/software/scan. It can be applied to any single-channel platform for any species, so long as metadata describing the coordinates and sequence of each probe is available. SCAN outputs probe-level expression values, which by default are summarized to gene-level values using a trimmed mean.
In this study, the R statistical software [22] and its associated ROCR package [23] were used for analyzing results and producing graphics. The implementations of fRMA, RMA, and MAS included in the affy [24] and frma [9] R/Bioconductor packages were applied with default parameters.
4.3 Data summarization
Affymetrix offers a variety of microarray platforms that target various applications. For each platform, Affymetrix provides a tab-delimited file that indicates the position and sequence of each probe. We downloaded these files from http://www.affymetrix.com and parsed the relevant information into consistently formatted “probe-tab” files. (The probe-sequence information for the recently released Affymetrix GG-H arrays was downloaded from http://gluegrant1.stanford.edu/∼DIC/GGHarray.) By design, Affymetrix places a variety of background and control probes on the Affymetrix ST and GG-H arrays. To avoid overfitting the linear model, SCAN ignores these probes when estimating model parameters but does output normalized expression values for these probes. Unlike previous generations of Affymetrix microarrays, the ST and GG-H arrays also include a small proportion of probes that have sequences shorter than 25 base pairs; because the linear model requires all probes to have the same length, SCAN ignores these probes.
Upon normalizing a CEL file, SCAN outputs a tab-delimited text file containing three columns. The first column lists a unique identifier for each probe (a concatenation of the probe's X and Y position on the array). The second column lists the normalized expression value for each probe. And the third column contains values representing the probability that each probe is expressed above background noise. (A companion publication describing the latter is pending.)
To support gene-level summaries, probe values are mapped to genes using either the mapping files provided by Affymetrix or custom mapping files. By default, we use custom mapping files provided by the BrainArray resource [25]; if no such mapping file is available, we default to mappings provided by Affymetrix. However, users also have the option to specify alternative mapping files that adhere to a simple tab-delimited format. Currently, each human GeneChip platform listed on http://www.affymetrix.com is supported.
After probes have been mapped to genes, gene-level values are calculated using a 10% trimmed mean. If fewer than 3 probes are associated with a gene, no gene-level value is calculated.
Supplementary Material
Highlights.
Designed for personalized medicine workflows where samples must be processed serially
Outperforms existing methods on spike-in experiments and ‘real-world’ data sets
Amplifies signal-to-noise ratio by correcting for probe-level biases
Supports combining data across different Affymetrix microarray platforms
Acknowledgments
We thank Adam Cohen for help with microarray quality control. We gratefully acknowledge an allocation of computer time from the Fulton Supercomputing Lab at Brigham Young University. We thank Affymetrix, the Broad Institute, the Novartis Institutes for Biomedical Research, and GlaxoSmithKline for making raw data available publicly. This work was partially supported by funds from the United States National Institutes of Health (1R01HG00569 and 5T32CA093247).
Footnotes
Author contributions: SRP, AHB, and WEJ conceived and designed the study. SRP performed all data analyses. SRP and YS developed software. MEL and JDC provided feedback on the manuscript. WEJ developed the statistical approach. AHB designed experiments testing SCAN on external datasets. SRP, AHB, and WEJ wrote the manuscript. All authors read and approved the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.van 't Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AAM, Mao M, Peterse HL, van der Kooy K, Marton MJ, Witteveen AT, Schreiber GJ, Kerkhoven RM, Roberts C, Linsley PS, Bernards R, Friend SH. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–6. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 2.Paik S, Shak S, Tang G, Kim C, Baker J, Cronin M, Baehner FL, Walker MG, Watson D, Park T, Hiller W, Fisher ER, Wickerham DL, Bryant J, Wolmark N. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351:2817–26. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 3.McDermott U, Downing JR, Stratton MR. Genomics and the continuum of cancer care. N Engl J Med. 2011;364:340–50. doi: 10.1056/NEJMra0907178. [DOI] [PubMed] [Google Scholar]
- 4.Lo SS, Mumby PB, Norton J, Rychlik K, Smerage J, Kash J, Chew HK, Gaynor ER, Hayes DF, Epstein A, Albain KS. Prospective multicenter study of the impact of the 21-gene recurrence score assay on medical oncologist and patient adjuvant breast cancer treatment selection. J Clin Oncol. 2010;28:1671–6. doi: 10.1200/JCO.2008.20.2119. [DOI] [PubMed] [Google Scholar]
- 5.Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu Z, Irizarry RA, Gentleman R, Martinez-Murillo F, Spencer F. A model-based background adjustment for oligonucleotide expression arrays. J Am Stat Assoc. 2004;99:909–917. [Google Scholar]
- 7.Hubbell E, Liu W, Mei R. Robust estimators for expression analysis. Bioinformatics. 2002;18:1585–92. doi: 10.1093/bioinformatics/18.12.1585. [DOI] [PubMed] [Google Scholar]
- 8.Lim WK, Wang K, Lefebvre C, Califano A. Comparative analysis of microarray normalization procedures: Effects on reverse engineering gene networks. Bioinformatics. 2007;23:i282–8. doi: 10.1093/bioinformatics/btm201. [DOI] [PubMed] [Google Scholar]
- 9.McCall MN, Bolstad BM, Irizarry RA. Frozen robust multiarray analysis (fRMA) Biostatistics. 2010;11:242–53. doi: 10.1093/biostatistics/kxp059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, Wrobel MJ, Lerner J, Brunet JP, Subramanian A, Ross KN, Reich M, Hieronymus H, Wei G, Armstrong SA, Haggarty SJ, Clemons PA, Wei R, Carr SA, Lander ES, Golub TR. The connectivity map: Using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 11.Leek JT, Scharpf RB, Bravo HC, Simcha D, Langmead B, Johnson WE, Geman D, Baggerly K, Irizarry RA. Tackling the widespread and critical impact of batch effects in high-throughput data. Nature Reviews Genetics. 2010;11:733–739. doi: 10.1038/nrg2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson WE, Li W, Meyer CA, Gottardo R, Carroll JS, Brown M, Liu XS. Model-based analysis of tiling-arrays for ChIP-chip. Proc Natl Acad Sci U S A. 2006;103:12457–62. doi: 10.1073/pnas.0601180103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kapur K, Xing Y, Ouyang Z, Wong WH. Exon arrays provide accurate assessments of gene expression. Genome Biol. 2007;8:R82. doi: 10.1186/gb-2007-8-5-r82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barrett T, Troup DB, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Muertter RN, Holko M, Ayanbule O, Yefanov A, Soboleva A. NCBI GEO: Archive for functional genomics data sets–10 years on. Nucleic Acids Res. 2011;39:D1005–10. doi: 10.1093/nar/gkq1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cohen AL, Soldi R, Zhang H, Gustafson AM, Wilcox R, Welm BE, Chang JT, Johnson E, Spira A, Jeffrey SS, Bild AH. A pharmacogenomic method for individualized prediction of drug sensitivity. Mol Syst Biol. 2011;7:513. doi: 10.1038/msb.2011.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.West M, Blanchette C, Dressman H, Huang E, Ishida S, Spang R, Zuzan H, Olson JA, Marks JR, Nevins JR. Predicting the clinical status of human breast cancer by using gene expression profiles. Proc Natl Acad Sci U S A. 2001;98:11462–7. doi: 10.1073/pnas.201162998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Greshock J, Bachman KE, Degenhardt YY, Jing J, Wen YH, Eastman S, McNeil E, Moy C, Wegrzyn R, Auger K, Hardwicke MA, Wooster R. Molecular target class is predictive of in vitro response profile. Cancer Res. 2010;70:3677–86. doi: 10.1158/0008-5472.CAN-09-3788. [DOI] [PubMed] [Google Scholar]
- 18.Bild AH, Potti A, Nevins JR. Linking oncogenic pathways with therapeutic opportunities. Nat Rev Cancer. 2006;6:735–741. doi: 10.1038/nrc1976. [DOI] [PubMed] [Google Scholar]
- 19.The Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Giorgi FM, Bolger AM, Lohse M, Usadel B. Algorithm-driven artifacts in median polish summarization of microarray data. BMC Bioinformatics. 2010;11:553. doi: 10.1186/1471-2105-11-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dempster AP, Laird NM, Rubin DB. (Series B (Methodological)).Maximum likelihood from incomplete data via the EM algorithm, Journal of the Royal Statistical Society. 1977;39:1–38. [Google Scholar]
- 22.R Development Core Team. R: A language and environment for statistical computing [computer program] 2011 Available from http://www.r-project.org/
- 23.Sing T, Sander O, Beerenwinkel N, Lengauer T. ROCR: Visualizing the performance of scoring classifiers [computer program] 2009 Available from http://cran.r-project.org/package=ROCR.
- 24.Gautier L, Cope L, Bolstad BM, Irizarry RA. Affy–analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–15. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 25.Dai M, Wang P, Boyd AD, Kostov G, Athey B, Jones EG, Bunney WE, Myers RM, Speed TP, Akil H, Watson SJ, Meng F. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res. 2005;33:e175. doi: 10.1093/nar/gni179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kang HJ, Kawasawa YI, Cheng F, Zhu Y, Xu X, Li M, Sousa AMM, Pletikos M, Meyer KA, Sedmak G, Guennel T, Shin Y, Johnson MB, Krsnik Z, Mayer S, Fertuzinhos S, Umlauf S, Lisgo SN, Vortmeyer A, Weinberger DR, Mane S, Hyde TM, Huttner A, Reimers M, Kleinman JE, Sestan N. Spatio-temporal transcriptome of the human brain. Nature. 2011;478:483–9. doi: 10.1038/nature10523. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.