Abstract
Cancer arises from normal cells that acquire a series of molecular changes; however, the founding events that create the clonogens from which a tumor will arise and progress have been the subject of speculation. Through the efforts of several generations of cancer biologists it has been established that the malignant phenotype is an amalgamation of genetic and metabolic alterations. Numerous theories have suggested their either, or both of these elements might serve as the impetus for cancer formation. Recently, the epigenetic origins of cancer have been suggested as an additional mechanism to give rise to the malignant phenotype. When coupled with the discovery that the enzymes responsible for initiating and perpetuating epigenetic events are linked to metabolism by their cofactors a new paradigm for the origins of cancer can be created. Here, we summarize the foundation for such a paradigm on the origins of cancer in which metabolic alterations create an epigenetic progenitor which clonally expands to become cancer. We suggest that the metabolic alterations disrupt the production and availability of cofactors like S-adenosylmethionine, α-ketoglutarate, NAD+, and acetyl-CoA to modify the epigenotype of cells. We further speculate that redox biology can change epigenetic events through oxidation of enzymes and alterations of metabolic cofactors that affect epigenetic events like DNA methylation. Combined, these metabolic and redox changes serve as the foundation for altering the epigenotype of normal cells and creating the epigenetic progenitor of cancer.
Keywords: Warburg effect, anaplerotic metabolite, glycolysis, demethylation, epigenetic progenitor
Introduction
Cancer is a constellation of individual diseases arising from different tissues and cell types of origin. In addition to their fundamental irreversible genetic bases of origin (initiation), diverse types of cancer share certain other fundamental underlying similarities including aberrant gene expression, an atypical redox state, and fundamental defect in metabolism often called the Warburg effect. Characterizing malignant phenotypes has proven to be a relatively straight forward process; however, elucidating factors causal in its manifestation have proven more difficult. Such as gap in knowledge left us to ponder and speculate about mechanisms responsible for forming and sculpting malignancies, and about how a single or finite number of genotypes, through adaptation and selection by the tumor microenvironment, can give rise to numerous different phenotypes in a relatively short period of time. The discovery by Bishop and Varmus that human genomic DNA contains sequences homologous to transforming retroviral oncogenes led to the formation of the genetic theory of cancer in the 1980’s. Since then, numerous studies have examined genetic alterations of cancer and genomic instability thus firmly cementing the concepts of how activating driver mutations in genes that normally stimulate proliferation (oncogenes) and/or inactivating mutations in genes that inhibit proliferation (tumor suppressor genes). In spite of these analyses, and while much insight has been gained from these studies, the mutation frequency of normal cells is paradoxically low, suggesting cancer should rarely develop in the first place [1]. Consistent with these views are the numerous inheritable disorders that predispose individuals to developing certain malignancies [2].
Prior to the genetic theory of cancer that has predominated over the last four decades; a prevailing theory for the development and progression of cancer was the Metabolic theory of cancer. Over 70 years ago Otto Warburg observed increased glucose metabolism in tumor cells, suggesting a metabolic defect may be causal in the development of cancer [3]. This increased need for glucose by cancer cells did not appear to be necessary for them to make ATP because tumor cells could make sufficient ATP by respiration but still displayed increased glucose demand. In contrast, glucose-derived carbon is likely used for the increased biosynthetic demand of rapidly growing cell populations as well for providing reducing equivalents in the form of NADH generated during glycolysis. The NADH and pyruvate resulting from glycolysis are both potent protectors against the increased oxidative stress associated with tumor cells. An extension of these concepts is the Free Radical theory of cancer rendered by Oberley and Buettner in the late 1970’s [4]. They suggested that free radicals, produced by aberrant reduction of oxygen, initiate cancer (mutationally as by DNA oxidation to 8-OHdG) and additionally provide the momentum fro proliferation (promotion) to drive disease progression to malignancy. More recently the epigenetic origins of cancer initiation have been suggested as a further mechanism to fashion the malignant phenotype [5]. In addition, the mounting discoveries of mutations in TCA cycle genes that act in interesting ways to perturb epigenetics through central metabolism portend a paradigm shift in the thinking regarding the role of how metabolism drives oncogenic alterations in gene expression during cancer progression.
Epigenetic processes are involved in many facets of molecular and cell biology including regulation of transcription, the cell cycle, DNA repair and DNA replication. Involvement in these processes puts epigenetics in direct control of Hanahan and Weignberg’s phenotypes of cancer: limitless replicative potential, self-sufficiency in growth signals, insensitivity to anti-growth signals, ability to evade apoptosis, increased tissue invasion, and sustained angiogenesis [6]. The dossier of cancer epigenetics includes the detailed information on its ability to create each of these characteristics by influencing transcription, the cell cycle, DNA repair and DNA replication (For reviews on these topics see [7–9]. Cancer can be conceptualized as a disease originating via the clonal expansion of a single transformed cell (clonogen). As the clonogenic progenitor divides, something intrinsic to the progenitor’s biology allows it to incur additional epigenetic and genetic alterations. The accumulation of these genetic and epigenetic flaws over several generations creates the malignant phenotype. This epigenetic basis for cancer is summed up by the epigenetic progenitor origin of human cancer suggested by Feinberg, Ohlsson, and Henikoff [5]. Their model aptly suggests that the stochastic epigenetic inactivation of key genes in clonogens gives rise to many malignancies; however, it does not suggest what flaw in the progenitor’s biology is responsible for creating its founding epigenetic alterations. Extending this model to provide a means to generate founding epigenetic alterations in a clonogenic progenitor cell would lend further credence this model. Recently the discovery and characterization of the enzymes involved in creating, maintaining, and removing epigenetic marks has become an area of interest. This is led to a wealth of information regarding these enzymes’ biochemistry. We propose that founding epigenetic events in cancer are forged by changes in redox biology and metabolism. We have previously explored the link between epigenetic processes, cancer, development, and disease [10–12]. Here, we seek to couple the recent burst of knowledge regarding the biochemistry of epigenetic enzymes with cancer’s altered gene expression, atypical redox state, and defective metabolism.
II. Epigenetic cofactors
Overview
The basic sources from which cells derive energy include glucose, fatty acids, ketone bodies, amino acids, and lactate. Tissues do exhibit specificity with respect to the fuel source(s) they burn and create. Eukaryotes can interchange one energy source to the other in various anabolic and catabolic pathways. This is dictated by the enzymatic pathways present within a cell, or via regulatory mechanisms such as substrate availably/inhibition, post-translational modification, and the ability to transport fuel sources across the plasma membrane. When a normal cell undergoes oncogenic transformation it exhibits marked changes in its intermediary metabolism [13, 14]. This phenomenon presents as the Warburg and Crabtree effects along with an atypical redox state compared to the normal cells of origin. How can these perturbations in cellular biochemistry and redox alter manifest epigenetic derangements in the nucleus?
Epigenetic marks are initiated, perpetuated, and removed via the activity of enzymes. Chromatin is a pliable structure, responding to the biochemistry within a cell created by its surroundings. Creating the ever-changing epigenetic landscape enables cellular adaptation to microenvironmental cues and requires the collaboration of numerous enzymes that continually sculpt chromatin to further refine its function. Because they create chromatin structure by altering epigenetic marks, these enzymes constitute a connection between gene expression and biochemistry. Epigenetic enzymes perceive metabolic changes by requiring cofactors from numerous biochemical pathways to power their effort. A paradigm illustrating this linkage is functional dyad of HIF1-α and proyl hydroxlases (PHDs). Proyl hydroxylases require the cofactors molecular oxygen, iron (II), α-ketoglutarate, and ascorbate to hydroxylate proline residues on HIF1-α [15]. This marks them for ubititinlyation by VHL, and degradation by the 26S proteosome. When one or more of these cofactors becomes limiting PHDs’ function is lost, HIF1-α is stabilized, and the transcription of hypoxia inducible genes is initiated. Histone and DNA modifying enzymes follow a similar pattern, requiring basic metabolic substrates from numerous biochemical pathways.
Histone acetylation
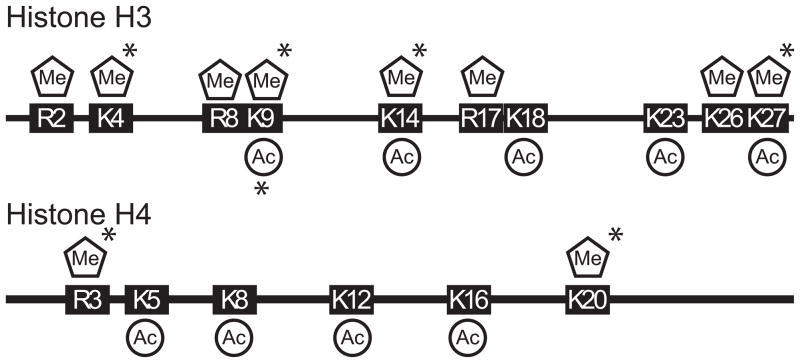
DNA is organized into higher ordered structures by the nucleosome and its associated proteins. Since the first report of histone acetylation has generally been associated with open chromatin and active transcription [16]. Histone acetyltransferases (HATs) catalyze the acetylation of lysines using acetyl-CoA as a cofactor to create ε-N-acetyl lysine [17]. This reaction is driven by the energy derived from the cleavage of acety-CoA’s thioester linkage, making it excellent donor of the acetate moiety. Mammalian cells have several HATs targeting lysines in histones, and non-histone proteins alike (for review see [18]). Countering histone acetylation and repressing gene expression are four major classes of histone deacetylases (HDACs). Classes I, II, and IV rely on a coordinated metal ion to remove acetyl groups from histones by hydrolysis [19]. Class III HDACS encompass the Sirtuin family of proteins that use NAD+ to break bond between lysine and acetyl group, resulting in the production of the unique metabolite O-acetyl-ADP-ribose [20]. The requirement for NAD+ by sirtuins reveals an obvious link between metabolism and gene expression and vividly exemplifies how metabolism can be a driver of gene expression patterns. HATs and HDACs target specific lysines within histone tails with their respective activities (Fig. 2). These opposing activities actuate chromatin structure to alter DNA’s function.
Figure 2. Sites of post-translation acetylation and methylation in mammalian histone tails.
Histone tails consist of basic amino acids that are amendable to several types of post-translational modifications. Several of these basic amino acids (Lysine, Arginine) are targeted for acetylation and methylation by numerous histone modifying enzymes (* denotes lysine can be mono, di, or tri methylated).
Metabolic cofactors
The metabolic connection to histone acetylation
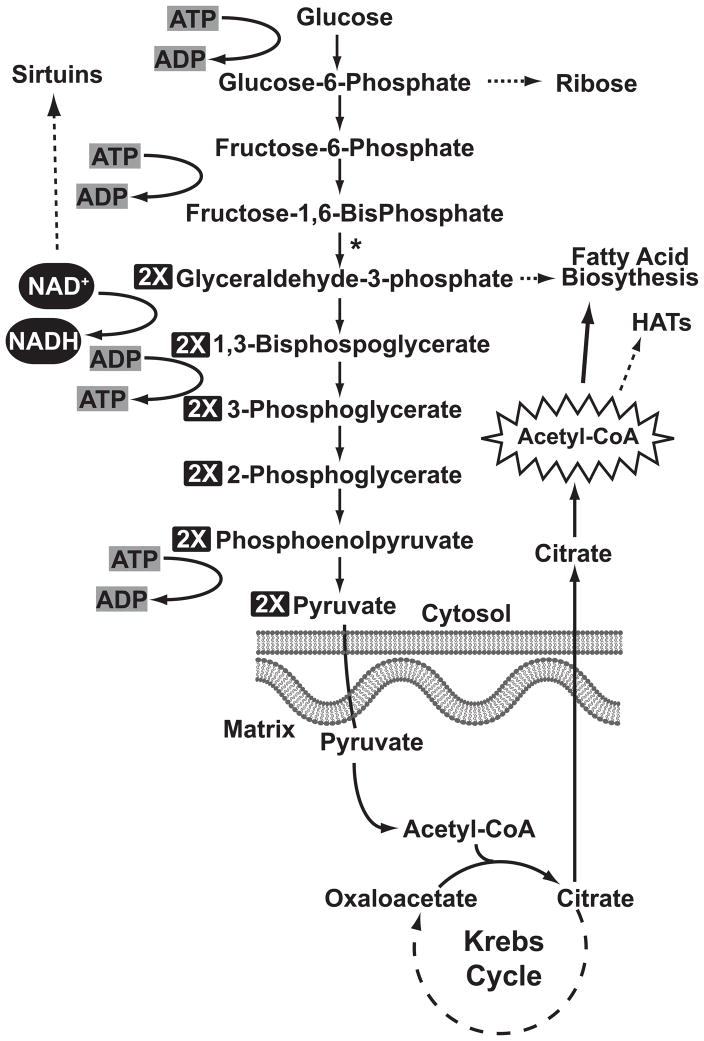
Numerous studies have demonstrated that histone acetylation is altered in cancer. Changes in the level of acetylation at numerous histone lysines are altered in cancer and my correlate with disease progression [21–23]. Indeed, while the expression of many HATS and HDACs are altered in cancer, their activity is still dependent upon the availability of their cofactors [24–27]. A stunning association exists between protein acetylation and glycolysis. The abnormally high levels of aerobic glycolysis (termed the Warburg Effect) in cancer has often been described as a way tumor cells produce energy. Here, we will conceptualize the Warburg effect not only as manner in which tumor cells produce energy, but a process by which glucose can be smelted into raw materials for anabolic processes need to support cancers rapid proliferation such as lipid and protein biosynthesis (Fig. 3). Histone acetylation’s epigenetic link to glycolysis is acetyl-CoA. The cytosolic pool of acetyl-CoA derived from glucose requires numerous steps. First is the conversion of glucose to pyruvate during glycolysis in the cytosol. Pyruvate is then imported into the mitochondria where it is decarboxylated to form acetyl-CoA by the holoenzyme pyruvate dehydrogenase [28]. Since acetyl-CoA is not mobilized through the membranes of the mitochondria, it must be assimilated into the TCA cycle by condensing with oxaloacetate, forming citrate, which can be exported from the mitochondria [29]. Tumor cells favor the export of Citrate from the mitochondria rather than its continued oxidation in the TCA cycle due to enzyme dysfunction in the conversion of citrate to isocitrate by aconitase [30]. Aconitase activity is altered in cancer by several mechanisms. The Fe-S cluster within the aconitase active site is amenable to oxidation, and attack by heavy metals such as Zinc, creates an inactive enzyme in the mitochondrial matrix [31]. Furthermore, tumor cells also often exhibit lower expression of aconitase compared to their normal counterparts [32]. These mechanisms make it favorable for citrate to accumulate and be exported to the cytosol where it is broken down by ATP Citrate Lyase (ACL) into acetyl-CoA. This process creates a cytosolic acetyl-CoA pool shared by fatty acid production, and histone acetylation [33]. Given the high glycolytic activity of the Warburg Effect, it seems assured that high levels of acetyl-CoA are produced to power fully operate both processes. However, transformation is accompanied by an elevation in de novo fatty acid synthesis, which is mediated in part by increasing the increased expression of Fatty Acid Synthase (FAS) [34–36]. FAS, which condenses malony-CoA and acetyl-CoA in the presence of NADPH to produce palmitate, depletes acetyl-CoA from other biochemicals processes to support its own activity. Increased de novo fatty acid synthesis in tumors is also supported through the aberrant expression of enzymes involved such as Acetyl-CoA carboxylase, monoacylglycerol lipase and cytosolic aconitase [37]. Inhibiting fatty acid synthesis has been shown some promise as a treatment in cancer [38]. Overall, it seems likely that epigenetic processes mediated by HATs would become disrupted in cancer.
Figure 3. Glucose metabolism is intimately linked to histone acetylation and deacetylation through cofactors.
The oxidation of glucose to pyruvate influences NAD+/NADH ratio in cells. Pyruvate from glycolysis is converted into acetyl-CoA via the mitochondria and TCA cycle. The Warburg effect, and the increased proliferation of cancer, are anticipated to change decrease the activities of sirtuins, and histone acetyltransferases. (* denotes that two molecules of each metabolite are being processed from this point on)
HDAC activity is often altered in cancer and associated with the silencing of tumor suppressor genes by creating a closed chromatin structure [18]. This has instigated the development and usage in clinical trials of several HDAC inhibitors [39]. A majority of these compounds modulate the activity of class I and II HDACs by binding to their metal ion containing catalytic domains. These inhibitors are effective at inhibiting the activity of class I, II HDACs since these enzymes require no additional cofactors. The NAD+ dependent mechanisms of sirtuins make them unresponsive to many of these inhibitors. Current strategies aimed at decreasing sirtutin HDAC activity in cancer is aimed at severing their link to metabolism by inhibiting NAD+ [40]. NAD+ creates a metabolic link between Sirutins and the Warburg effect of cancer. The activity of these enzymes is sensitive to the cellular NAD+/NADH ratio as a means to perceive metabolism so an appropriate transcriptional response can be mounted. In cancer, the increased flux of glucose through glycolysis decreases the cellular NAD+/NADH ratio. Defects in electron transport further decrease the ratio by limiting the conversion of NADH back into NAD+. To regenerate NAD+ tumor cells convert pyruvate into lactate, which is secreted into the microenvironment and has recently been identified as a putative HDAC inhibitor itself [41]. Sirtuin activity could also be altered in the early stages of cancer development. Immortalization of normal cells increases glycolytic activity, and decreases their NAD/NADH ratio [42]. Many germ-line mutations may indirectly influence the availability of NAD+ sirutins. Loss of von Hippel-Lindau leads to increased transcriptional activation of pyruvate dehydrogenase kinase 1 (PDK1) by HIF-1α [43]. PDK1 inactivates Pyruvate dehydrogenase by phosphorylating it. This effectively stops the entry of carbon derived from glycolysis into the mitochondria and the TCA cycle. The NAD+/NADH ratio forms an effective linkage between metabolism and epigenetic regulation of gene expression by histone acetylation.
Methylation as an epigenetic process
S-adenosylmethionine (SAM)
Epigenetic regulation of gene expression by methylation is initiated and perpetuated by DNA and histone methyltransferases. DNA is methylated almost exclusively at CpG (cytosine followed by guanine) dinucleotides. This nucleotide doublet is recognized by DNA methyltransferases that add a methyl group to the 5 position of the pyrimidine ring to create 5-methylcytosine (5-MeC). Mammalian cells contain three DNMTs (DNMT1, 3a, and 3B) each of which exist in multiple isoforms to catalyze this reaction (For review see [44–47]). We categorize these enzymes based on their operational activity as either de novo or maintenance DNA methyltransferases. DNMT3a, and b initiate new epigenetic events by targeting CpGs for methylation that are previously ummethylated. These events are perpetuated during DNA replication by DNMT1, which targets hemimethylated CpGs for methylation. Typically only a fraction of the CpGs within an organisms’ genome are methylated and coincide with condensed transcriptionally inert heterochromatin. Histones are also targeted for DNA methylation by several histone metyltransferasese (HMTs). HMT substrate specificity and activity is centralized in SET domains and surrounding motifs [48, 49]. Histone lysines can mono, di, or tri methylated. The same HMT can be responsible for all three reactions, while progressive methylation from mono to tri can require several enzymes. Progressive methylation endows this epigenic process with a complexity that allows for dynamic changes in chromatin structure. DNMTs and HMTs are reliant upon the co-factor S-adenosylmethionine (SAM) as a methyl group donor while producing S-adenosylhomocysteine (Fig. 4)[50]. As a component of the methionine cycle, SAM is primarily used as a cofactor in for enzymes that catalyze numerous transmethylation reactions, or in the synthesis of polyamines [51]. One of the central purposes of the methionine cycle is to preserve sulfur so it can be recycled and used by various biochemical pathways including transsulfuration (glutathione), polyamine biosynthesis, generation of the amino acid methionine and as a methyl group donor (transmethylation) [52]. Redox can alter the flow of metabolites through the methionine cycle to convey sulfur to the biochemical pathway requiring it most. We suggest that the atypical redox state of cancer modifies the flow of metabolites through the methionine cycle, and thus limits SAM availability for use in epigenetic transmethylation reactions. Numerous studies have established SAM’s role in carcinogenesis. During tumor promotion studies in rodent models, the level of SAM, and the SAM/SAH ratio become decreased in preneoplastic lesions [53]. Conversely, exogenous SAM decreases the transformation frequency and inhibits the development of some forms of cancer [54, 55]. SAM’s affect on carcinogenesis can be directly related to its ability to influence DNA methylation and gene expression. Studies by Franceso Feo’s group have shown that SAM can inhibit the expression of proto-oncogenes by maintaining DNA methylation at their promoters [56]. Exogenous SAM also attenuates the malignant phenotype of human prostate cancer cell lines grown in mouse xenograft by altering the pattern of DNA methylation in tumors [57]. Likewise we speculate that concomitant changes in histone methylation would also be observed following oxidative stress and the addition of exogenous SAM. In what manner can redox influence the availability of SAM, and promote the creation of the malignant phenotype?
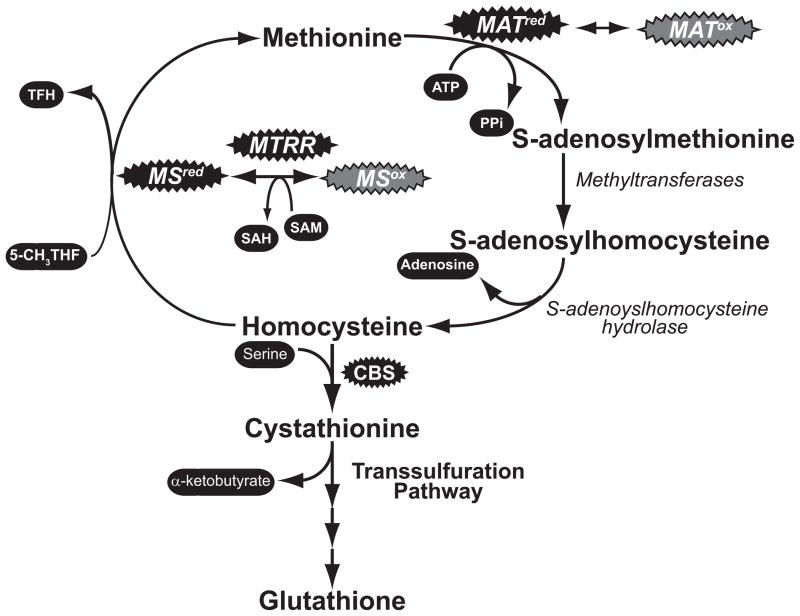
Figure 4. The flow of metabolites through one-carbon metabolism is influenced by redox.
The oxidation of key enzymes (shown in grey bubbles) limits their activity, and stops the flow of metabolites through the methionine cycle. SAM synthetase (MAT) can become inactivated through reversible oxidation. Oxidation of the Cbl core of Methionine synthase (MS) leads to its inactivation, however Methionine synthase reductase (MTRR) can use SAM as a cofactor to restart the enzyme. At the same time, an oxidizing environment can increase the activity of enzymes such Cystathionine β-synthase (CBS) to siphon sulfur into the production of glutathione.
Redox influences the production of SAM
The altered redox state of cancer influences SAM levels through aberrant regulation of the methionine cycle. SAM is generated by the enzyme SAM synthetase (also known methionine adenosyltransferase) by combining methionine, with ATP [50]. Humans have three forms of this enzyme: MATI, MATII and MATIII. Both MATI and MATIII are encoded by the mat1α gene, which expressed specifically in the liver [52]. While the primary amino acid sequence is identical between MATI and MATIII, the two enzymes exist as tetramers and dimers respectively [50]. Other tissues utilize MATII, which is a heterodimer of MAT2α and MAT2β subunits [58]. All forms of the enzyme are critical in controlling the level of SAM, and the progression of metabolites thought the methionine cycle. Furthermore, all three forms of the enzyme contain a conserved cysteine that might function as a redox “switch”. Decreasing the redox buffering capacity switches MAT activity off, while a more reducing environment increases its production of SAM [59–61]. Oxidation of this cysteine has been suggested to be a contributing factor in the development of liver cirrhosis in some models [62, 63]. Since cancer cells contain a more oxidized environment, it would suggest that MAT activity, and subsequently SAM levels, would be decreased in cancer.
A second redox sensitive point in the methionine cycle is the conversion of homocysteine into methionine. This critical step that converts homocysteine into methionine facilitated by Methionine Synthase (MS) Methionine synthase reductase (MTRR) couple. Methionine Synthase is the part of this duo responsible for producing methionine from homocysteine using a folate/cobalamin (Cbl or Vitamin B12) dependent mechanism [64]. MS essentially “loads” the sulfur of homocysteine with another methyl group which eventually gets donated by SAM during subsequent transmethylation reactions in the methionine cycle. Oxidation of the Cbl in MS renders the enzyme inactive, halting the methionine cycle. MS can be recycled by MTRR, however it requires SAM and produces SAH [65] [66]. Combined, free radical mediated inactivation of MS influences epigenetics in two ways: by halting the production of SAM, and requiring SAM during MTRR mediated reactivation of MS. This suggests that this pathway should significantly influence DNA methylation. Numerous mutations have been identified in MS, and MTRR that increase homocysteine levels and the potential risk for developing cancer [67]. Several groups have suggested that these alterations may have a role in altering DNA methylation in tumor cells [68].
The mechanisms we describe allow normal tissues to counter acute oxidative stress. However, during the persistent oxidative stress of cancer, these compensatory mechanisms are exploited to shunt sulfur containing metabolites into the production of glutathione. In some tissues homocysteine derived from the methionine cycle accounts for up to fifty percent of the glutathione produced [52, 69]. Oxidative stress also enhances the exit of homocysteine from the methionine cycle and into glutathione production by increasing the activity of cystathione β-synthase [70]. It has been proposed that the source of oxidative stress in most cancers can is related to a fundamental defect in glucose metabolism, and that this change is causal in carcinogenesis. We suggest that this defect in metabolism exerts an influence on epigenetics by drawing metabolites away from SAM production, and into synthesis of glutathione. The chemical depletion of glutathione decreases SAM levels, increase glutathione production, and leads to genome wide DNA hypomethylation [71–73]. From these studies we can draw a parallel to what is observed in cancer. Aberrant reduction of molecular oxygen to superoxide by the defective electron transport chains of cancer cells creates and maintains an altered redox state. To counter, tumor cells increase their production of glutathione, and therefore SAM becomes depleted. The metabolic defect of cancer may also affect the ability of tumor cells to recycle GSSG back into GSH. Tumor cells have low GSH/GSSG ratios compared to most normal tissues, suggesting they are less efficient at recycling glutathione. Transformed cells would compensate for this by producing more GSH de novo, further sapping metabolites that contain sulfur from other biochemical pathways [74] . Because glutathione is so intimately linked with SAM availability, and therefore epigenetics, we perceive oxidative stress as both an initiator, and promoter of epigenetic changes in cancer. Chronic oxidative stress would alter the initial epigenotype of cells (initiate), and then serve as the momentum to create additional epigenetic changes as the initiated oxidatively stressed clone expands. This presents one mechanisms by which oxidative stress can facilitate the creation of epigenetic changes in cancer. As we have discussed above free radical production could decrease the level of SAM in tumor cells. Therefore it seems likely that decreasing SAM levels is one mechanism by which free radicals could lead to the activation of oncogenes in cancer.
Demethylation of 5-Methylcytosine and Histones
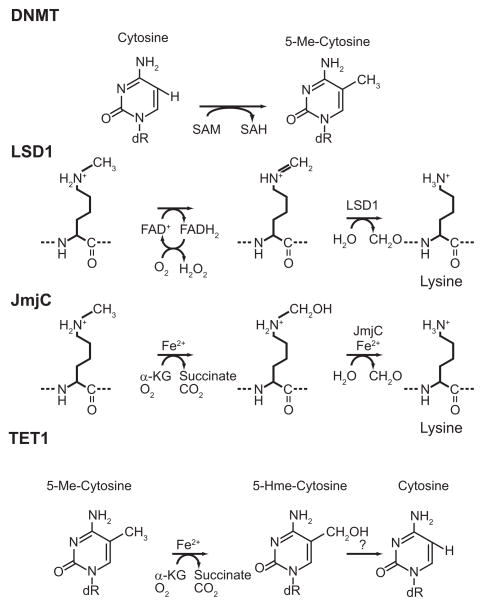
Until the recent discovery of enzymes that remove methyl groups from DNA and histones, methylation was believed a static epigenetic mark. Two families of histone demethylases are known to exist: lysine specific demethylase 1 (LSD1) and the JmjC family of histone demethylases. LSD1 uses its histone demethylase activity to function both as a transcriptional repressor and activator, depending on the context in which it is functioning [75]. The active site of LSD1 has flavin dependent amine oxidase activity, which uses oxygen to progressively oxidize methyl groups to formaldehyde, and generates hydrogen peroxide (Fig. 1) [76]. The amine oxidase mechanisms can efficiently demethylate mono, and di-methylated lysines, however its mechanism prohibits it from having activity on tri-methylated lysines. In mammalian cells demethylation of trimethylated lysines is catalyzed by the JmjC superfamily of lysine demethylases [77, 78]. Each member of this superfamily contains a common histone demethylating jumonji domain [77]. This activity, housed within each protein’s jumonji domain, requires Fe(II), α-ketoglutarate, and oxygen to progressively oxidize methyl groups to formaldehyde. Ascorbate is also important for the function of the enzyme to keep the iron reduced as the Fe(II) is loosely coordinated and subject to auto-oxidation. Demethylation can have significant effects on the epigenetic landscape. Histone lysines can be mono, di, or tri methylated. Shifting a lysine’s methylation status between different degrees of methylation can quickly alter chromatin structure and affect the function of DNA that nucleosome is associated with. Likewise, demethylation of DNA can quickly remodel the epigenetic landscape and influence gene expression. In the first reported enzymatic conversion of 5-MeC to 5-hydroxymethylcytosine (5-HmeC) by TET1 was reported [79]. These 5-MeC oxidases use the same cofactors as jumonji histone demethylases. While the conversion of 5-MeC to 5-HmeC is not true demethylation per se, oxidation may mark methylated cytosine for removal by base excision repair (BER) enzymes, or by an as of yet unidentified mechanism [79]. While acetylation appears to be linked to glycolysis, methylation is related to metabolites within the TCA cycle.
Figure 1. The mechanisms of various epigenetic enzymes are reliant upon metabolic cofactors.
The activities of epigenetic enzymes draw upon cofactors from various biochemical pathways to form distinct linkages between metabolism and gene expression. Because metabolic reprogramming alters these pathways in cancer, the activities of these enzymes during would be expected to change during carcinogenesis. The result is epigenetic reprogramming including widespread disruption of chromatin structure and aberrant gene expression.
Anaplerotic sources of carbon
From our discussion above it is clear that the channel by which carbon flows through metabolism is dramatically altered in cancer. These cancer associated metabolic changes have the potential to influence DNA and histone methylation in a manner similar to acetylation. Warburg’s initial explanation for the effect bearing his name was the mitochondrial of tumor cells must be inactive. Today, several fundamental defects in mitochondria function have been described in cancer that influences the flux of metabolites through the TCA cycle [80]. In normal cells pyruvate derived from glucose is the primary source of carbon for the TCA cycle. This allows anaplerotic sources of carbon, such as glutamine, to be used in other biochemical processes. In normal cells glutamine is readily converted into glutamate, and then processed into α-ketoglutarate. This generates a pool of α-ketoglutarate for and enzymes regulating gene expression such as prolyl hydroxylases (HIF1α), jumonji, and TET1 [30]. Sharing α-ketoglutarate links metabolism and epigenetics and suggests that limited α-ketoglutarate availability may alter gene expression in cancer. How this occurs is best illustrated in normal cells during hypoxia.
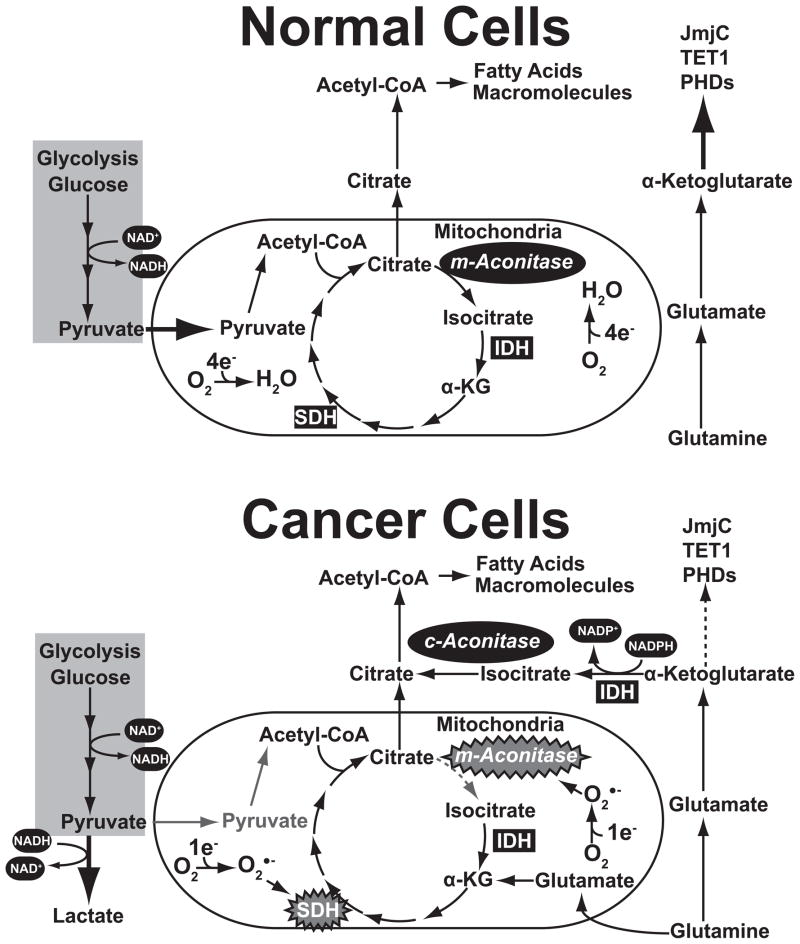
Increased anaerobic glycolysis during hypoxia diverts glycolytic products into lactate as a means to regenerate NAD+, instead of entering the mitochondria for assimilation into the TCA cycle. Under hypoxic conditions the carbon source of choice to produce raw materials for the biosynthesis of macromolecules becomes α-ketoglutarate derived from glutamine [30, 81]. This occurs because the molecular mechanisms of hypoxia limit pyruvate’s entry into the TCA cycle. With limited citrate being created in the mitochondria, cells draw upon anaplerotic sources of carbon to fill the gap. Glutamine is a classical example of this, as it can be readily converted into α-ketoglutarate, then into acetyl-CoA utilizing a pathway running in reverse from the canonical TCA cycle (Fig. 5) [81]. The pathway for generating acetyl-CoA from α-ketoglutarate begins with isocitrate dehydrogenase (IDH) [82]. Isocitrate dehydrogenase exists as three isoforms in mammalian cells, IDH1, IDH2, and IDH3. It is generally depicted as catalyzing the third step of the TCA cycle by converting isocitrate into α-ketoglutarate while generating reducing equivalents in the process. IDH3 is the canonical NAD+ dependent form of the enzyme responsible for moving the TCA cycle in the forward direction by producing α-ketoglutarate from isocitrate in the mitochondria. The other isoforms (IDH1 and IDH2) use NADP+ as a cofactor, and is expressed in mitochondrial and cytosolic forms [83]. Like IDH3, these enzymes generate reducing equivalents while converting isocitrate into α-ketoglutarate. However, both IDH1 and IDH2 can also catalyze the NADPH dependent reductive carboxylation of α-ketoglutarate into isocitrate [84]. The production of acetyl-CoA from anaplerotic sources hinges upon the reductive carboxylation activity of both IDH1 and IDH2 isoforms. Reductive carboxylation occurs in both the cytosol and mitochondria (Fig. 5). Once complete, isocitrate can be converted into citrate by aconitase, and then processed into acetyl-CoA by ACL as described above. Disrupting the electron transport chain by ETC blockers or hypoxia momentary increases the reductive carboxylation activity of IDH in order to keep acetyl-CoA production going. Thus, reductive carboxylation might serve as a means to activate HIF-1α during times of hypoxia. During oxidative stress in cancer the production of acetyl-CoA most likely occurs in the cytosol because c-aconitase is much less amenable to oxidative inactivation than m-aconitase [31]. This suggests that reductive carboxylation is likely occurring at a high rate which conveys α-ketoglutarate produced from anaplerotic glutamine into citrate to fatty acid synthesis. Can perturbations created by reductive carboxylation truly alter α-ketoglutarate levels and modify a cell’s epigenome? If the activity of these enzymes parallels HIF modifying prolyl hydroxylases the answer is likely yes. Hypoxia is a state in which both oxygen and α-ketoglutarate availably is altered. Changes in one or both have the same net effect on cells, the stabilization of HIF and the activation of hypoxia related genes. Hypoxia suggests that α-ketoglutarate levels are a fulcrum by which metabolism can alter gene activity. We surmise that oxidative stress leverages epigenetic changes via alterations in α-ketoglutarate levels. Oxidative stress inhibits the activity of many TCA cycle enzymes, thus altering the flow of metabolites like α-ketoglutarate through the mitochondria. Furthermore, many cancers contain dysfunctional mitochondria which harbor defects in the electron transport chain. This occurs by inheriting germline mutations in critical electron transport chain components like Fumarase, and Succinate Dehydrogenase [80]. Tumors with these germline mutations must manufacture sufficient amounts of acetyl-CoA to support lipogenesis and other biochemical processes. Unlike normal cells, the abnormal amounts of pyruvate they produce from glucose via the Warburg effect is utilized in lactic acid fermentation as a means to regenerate NAD+[85]. The confluence of these events requires that these tumors derive a bulk of their carbon units of anaplerotic sources. This manifests itself as the glutamine dependency exhibited by many tumors, and tumor derived cell lines. Likewise, anaplerotic glutamine becomes the source of carbons entering the TCA cycle. In tumors without inherited defects in electron transport the dependence upon anaplerotic glutamine is likely driven by a combination of lactate fermentation and the inactivation of mitochondrial aconitase by free radical attack. Dubbed the “Truncated TCA cycle” by Loris Baggeto, it involves the almost exclusive utilization of α-ketoglutarate derived from glutamine as a carbon source to keep metabolites flowing through the TCA cycle[86]. Collectively, these metabolic alterations unbalance the use and availability of α-ketoglutarate in cells.
Figure 5. Cancer cells utilize glutamine as an anaplerotic carbon source to fuel the TCA cycle and fatty acid biosynthesis.
In normal cells glucose is used as the principle source of carbon for the TCA cycle and fatty acid biosynthesis, allowing glutamine to be used to create a pool of α-ketoglutarate to fully support the activities of epigenetic enzymes. Cancer cells have an altered flow of carbon into the TCA cycle the flow of carbon into the TCA cycle
We suggest that using glutamine as an anaplerotic carbon source for the TCA cycle, or to produce acetyl-CoA, limit α-ketoglutarate availability to histone and DNA demethylases. Expression a catalytically dead, dominant negative version, of cytosolic IDH1 decreases the availability of α-ketoglutarate in the cytosol, decreases PHD activity, and HIF-1α becomes stabilized [87]. At this time it is unknown if expression of these mutant IDH1 forms can have a concomitant effect is exhibited by histone and DNA demethylases, but similar experiments would directly answer this question. Mutations in cytosolic IDH1 and IDH2 have also been identified and characterized. Theses mutations (IDH1 R132, IDH2 R172 and IDH2 R140) alter the active site of the enzyme and results in the production of the oncometabolite 2-hydroxyglutarate [88–95]. The oncogenic function of 2-hydroxyglutarate can be attributed to its substrate inhibition of enzymes that require α-ketoglutarate. Conferring 2-hydroxyglutarate production upon cells decreases their production of α-ketoglutarate, and activates HIF mediated transcription. Recently, it has been reported that 2-hydroxyglutarate has a similar affect on histone demethylases and histone methylation [96, 97]. Increased utilization of glutamine by tumor cells as a means to produce α-ketoglutarate for fatty acid synthesis could have another unintended consequence, creating oxidative stress. Since reductive carboxylation via IDH1 and IDH2 requires NADPH as an electron donor, it is no longer available in sufficient quantities to reduce glutathione disulfide back into glutathione.
Influence of oxidation on Epigenetics
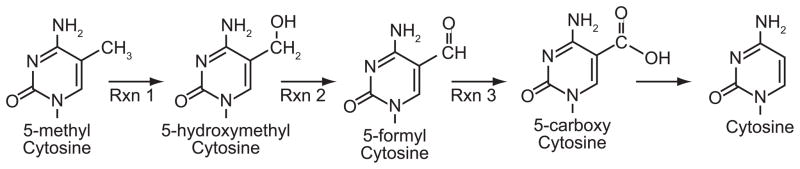
The prooxidant state of cancer can directly influence epigenetic processes. This can be accomplished through the oxidation of DNA, oxidizing histones. The formation of 8-oxoguanine (8-OG) is a paradigm of oxidative damage to DNA in free radical biology. 8-OG is highly mutagenic because it can be perceived as adenine or guanine by DNA repair machinery. However its presence is equally likely to induced epigenetic changes if the oxidation event occurs at a CpG dinucleotide. The present of 8-OG within a CpG inhibits DNMT binding, and can likely induced passive demethylathion of that CpG [98, 99]. Given the high frequency at which the enzymes that remove 8-OG from DNA are mutated in cancer it seems likely that increasing the steady state level of 8-OG in DNA can contribute to the hypomethylation of DNA observed in many cancers [100, 101]. The direct oxidation of 5-MeC can also lead to epigenetic changes. Oxidizing the methyl groups of 5-MeC can change how the CpG is recognized by proteins that bind 5-MeC, and possibly lead to its demethylation. Free radicals can directly attack the methyl group of 5-MeC, to create 5-HmC in a manner similar to the TET1 demethylases [79, 102, 103]. The production of free radicals has been shown to induce DNA demethylation in several model systems [104]. More recently 5-formyl cytosine and 5-carboxy cytosine have been identified in genomic DNA. These bases are likely produced by the progressive oxidation of 5-HmC, and constitute a means to demethylate cytosine (Fig. 6) [105–109]. 5-HmC is possibly removed from DNA before it can be completely oxidized by HMC glycosylase in a similar manner that occurs following TET1 oxidation [79]. If 5-HmC is allowed to persist in DNA it can affect epigenetic regulation of gene expression by other means. Methylated CpGs are recognized and bound by several methyl binding proteins. These proteins “read” the methylated CpG and interpret its function into the condensed chromatin often associated with hypermethylated DNA. An oxidation event at a methylated CpG, occurring at either a guanine or 5-MeC, inhibits the binding of these MBPs, and alters chromatin structure [107]. Oxidation of a methylated CpG can also create a novel epigenetic signature. Oxidizing a methylated cytosine in a CpG doublet can dramatically increase, or decrease the affinity by which different MBPs bind it [110].
Figure 6. Demethylation of 5-methylcytosine to cytosine by progressive oxidation.
The addition of one electron by free radical attack, or TET1 demethylases to the methyl group generates the stable modified base 5-hydroxymethylcytosine (5-Hm-C) in genomic DNA (Rxn 1). Additional oxidation events create 5-formyl cytosine and 5-carboxy cytosine (Rxns 2 and 3). Each derivative of 5-Me-C is unique, and can potentially alter the function of the CpG dinucleotide at which it occurs.
Summary
Characterization of the malignant phenotype has yielded an abundance of information about cancer. However, even with this wealth of information we are still left speculating as to what mechanisms manifest its existence. Cancer has extreme defects in cellular metabolism required to produce its energy and support its increased rate of replication. Could these universal defects in metabolism be the impetus for cancer’s development? Phenotype can be defined as the expression of a cell’s genotype in response to environment. A cell’s intracellular environment is a product of its metabolism interacting with gene expression. Warburg hypothesized over 70 years ago that a tumor cell’s increased glycolytic activity may play an important role in carcinogenesis. Likewise, Oberley and Buettner suggested in the 1970’s that unchecked superoxide production in mitochondria, forged from a metabolic defect, is the basis for cancer to development. The genetics of cancer is often interpreted as what is seen after the malignant phenotype as developed. This differs markedly to the genotype of the normal cell from which the disease arose. Others have aptly suggested that the metabolic defects of cancer such as increased glycolysis, dysfunctional mitochondrial electron transport, aberrant production of oxidants, and the formation of an atypical redox state are causal in the initiation, promotion, and progression of the malignant phenotype [4, 111–113]. These hypothesizes have centered on the ability of these metabolic changes to create genetic alterations to create cancer. While the discussion here focuses on the connection between epigenetic alterations and redox, our model by no means precludes the possibility that the metabolic defects of cancer can create oxidative modifications to DNA that lead to mutations. In multi-cellular organisms, epigenetic regulation of gene expression allows for different radically different interpretations of an organism’s genome. This is a central tenet of the epigenetic progenitor model, in which the genome of an organism can be interpreted differently to create a novel phenotype (i.e. cancer). Epigenetics is also the means by which a cell’s phenotype can be extracted from its genome during a response to its environment. We suggest that the founding epigenetic events in cancer’s clonogenic progenitor are fashioned via the aberrant cues within a cell.
A biological triad of environment, metabolism and genome intersect at epigenetics to create phenotype. We surmise that abiotic and biotic cues disrupt the triad, resulting in aberrant epigenetic changes that are bequeathed by a progenitor to its decendents. Exogenous stimuli can create oxidative stress and initiate carcinogenesis in numerous model systems. This is exemplified by the numerous carcinogens that create oxidative stress as part of their biological activity. From our discussions above it is likely that during these scenarios of oxidative stress a cell’s metabolism is affected, altering the flow of metabolites through a multitude of biochemical pathways. These biochemical changes would cue epigenetic alterations that manipulate the genome and altered gene expression. Because we are not yet fully aware of the scope in which epigenetics dictates usage of the genome, we can only speculate on primary targets for epigenetic manipulation. If a cell’s response to these cues is interpreted incorrectly, persists, or targets the wrong genes; aberrant epigenetic events could be created. Thus, our triad of environment, metabolism, and genome conspire via epigenetics to create the progenitor of cancer. Overall it seems likely that redox changes might serve as the impetus for the creating of the founding epigenetic events in the progenitor. Redox could also be the momentum that drives malignant progression. Once a progenitor of cancer has been created, redox can create new epigenetic alterations that manifest the malignant phenotype.
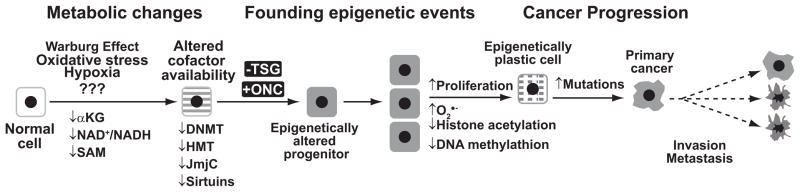
Our current model for the how each of these metabolic defects influences the epigenotype control of gene expression is shown in Fig. 7. In our model, founding epigenetic events are created in a stochastic manner when aberrant metabolism limits cofactor availability. Additional epigenetic alterations are incurred when this progenitor starts to divide and exhibits persistent changes in metabolism such as the use of anaplerotic glutamine as a carbon source. Epigenetics can be conceptualized as how the genome is interpreted in response to metabolism, which makes sense given that chromatin evolved in the foreground of the existing roots of glycolysis and the TCA cycle. Indeed, central metabolism and redox biology, the deliberate and controlled movement of electrons to oxygen, preceded the evolution of chromatin and thus it seems clear that chromatin would have mechanisms to respond to cues from central metabolism. When the sensors and regulators of these pathways are mutated or perturbed as in cancer, they contribute to epigenomic instability and phenotypic diversity during cancer progression.
Figure 7. Integrating metabolic defects into the epigenetic progenitor model of the origins of cancer.
We can operationally divide the origins of cancer into three discrete steps: creation of metabolic changes, formation of founding epigenetic events, and cancer progression. Metabolic changes are created through alterations in the cellular microenvironment. These in turn limit the availability of cofactors to epigenetic enzymes. Aberrant epigenetic events can be created that silence tumor suppressor genes (TSG) and activate oncogenes (ONC) and thus creating founding epigenetic events in an epigenetic progenitor. As the epigenetic progenitor clonally expands at an accelerated rate additional changes occur in cellular metabolism to form epigenetic plasticity. The clonogen can expand further and acquire additional mutations to cause cancer progression and manifest the malignant phenotype.
Highlights.
Cancer is a disease associated with changes in metabolism, redox and gene expression
Mechanisms leading to epigenetic alterations during carcinogenesis are unknown
Epigenetic disruption of cancer genes is linked to aberrant mitochondrial metabolism
Metabolic redox changes alter cofactor availability to enzymes that alter chromatin
Aberrant metabolic and redox changes cause epigenetic instability in cancer
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Loeb LA. Mutator phenotype may be required for multistage carcinogenesis. Cancer Res. 1991;51:3075–3079. [PubMed] [Google Scholar]
- 2.Eng C. Mendelian genetics of rare--and not so rare--cancers. Ann N Y Acad Sci. 1214:70–82. doi: 10.1111/j.1749-6632.2010.05789.x. [DOI] [PubMed] [Google Scholar]
- 3.Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 4.Oberley LW, Buettner GR. Role of superoxide dismutase in cancer: a review. Cancer research. 1979;39:1141–1149. [PubMed] [Google Scholar]
- 5.Feinberg AP, Ohlsson R, Henikoff S. The epigenetic progenitor origin of human cancer. Nat Rev Genet. 2006;7:21–33. doi: 10.1038/nrg1748. [DOI] [PubMed] [Google Scholar]
- 6.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 7.Alabert C, Groth A. Chromatin replication and epigenome maintenance. Nat Rev Mol Cell Biol. 13:153–167. doi: 10.1038/nrm3288. [DOI] [PubMed] [Google Scholar]
- 8.Bao Y. Chromatin response to DNA double-strand break damage. Epigenomics. 3:307–321. doi: 10.2217/epi.11.14. [DOI] [PubMed] [Google Scholar]
- 9.Mendez-Acuna L, Di Tomaso MV, Palitti F, Martinez-Lopez W. Histone post-translational modifications in DNA damage response. Cytogenet Genome Res. 128:28–36. doi: 10.1159/000296275. [DOI] [PubMed] [Google Scholar]
- 10.Hitchler MJ, Domann FE. Metabolic defects provide a spark for the epigenetic switch in cancer. Free Radic Biol Med. 2009;47:115–127. doi: 10.1016/j.freeradbiomed.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hitchler MJ, Domann FE. An epigenetic perspective on the free radical theory of development. Free Radic Biol Med. 2007;43:1023–1036. doi: 10.1016/j.freeradbiomed.2007.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cyr AR, Domann FE. The redox basis of epigenetic modifications: from mechanisms to functional consequences. Antioxid Redox Signal. 15:551–589. doi: 10.1089/ars.2010.3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Biswas S, Lunec J, Bartlett K. Non-glucose metabolism in cancer cells-is it all in the fat? Cancer Metastasis Rev. doi: 10.1007/s10555-012-9384-6. [DOI] [PubMed] [Google Scholar]
- 14.Bayley JP, Devilee P. The Warburg effect in 2012. Curr Opin Oncol. 24:62–67. doi: 10.1097/CCO.0b013e32834deb9e. [DOI] [PubMed] [Google Scholar]
- 15.Jokilehto T, Jaakkola PM. The role of HIF prolyl hydroxylases in tumour growth. J Cell Mol Med. 14:758–770. doi: 10.1111/j.1582-4934.2010.01030.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Allfrey VG, Faulkner R, Mirsky AE. Acetylation and Methylation of Histones and Their Possible Role in the Regulation of Rna Synthesis. Proc Natl Acad Sci U S A. 1964;51:786–794. doi: 10.1073/pnas.51.5.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Phillips DM. The presence of acetyl groups of histones. Biochem J. 1963;87:258–263. doi: 10.1042/bj0870258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang K, Dent SY. Histone modifying enzymes and cancer: going beyond histones. J Cell Biochem. 2005;96:1137–1148. doi: 10.1002/jcb.20615. [DOI] [PubMed] [Google Scholar]
- 19.Witt O, Deubzer HE, Milde T, Oehme I. HDAC family: What are the cancer relevant targets? Cancer Lett. 2009;277:8–21. doi: 10.1016/j.canlet.2008.08.016. [DOI] [PubMed] [Google Scholar]
- 20.Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 21.Song JS, Kim YS, Kim DK, Park SI, Jang SJ. Global histone modification pattern associated with recurrence and disease-free survival in non-small cell lung cancer patients. Pathol Int. 62:182–190. doi: 10.1111/j.1440-1827.2011.02776.x. [DOI] [PubMed] [Google Scholar]
- 22.Seligson DB, Horvath S, McBrian MA, Mah V, Yu H, Tze S, Wang Q, Chia D, Goodglick L, Kurdistani SK. Global levels of histone modifications predict prognosis in different cancers. Am J Pathol. 2009;174:1619–1628. doi: 10.2353/ajpath.2009.080874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seligson DB, Horvath S, Shi T, Yu H, Tze S, Grunstein M, Kurdistani SK. Global histone modification patterns predict risk of prostate cancer recurrence. Nature. 2005;435:1262–1266. doi: 10.1038/nature03672. [DOI] [PubMed] [Google Scholar]
- 24.Zhang Z, Yamashita H, Toyama T, Sugiura H, Ando Y, Mita K, Hamaguchi M, Hara Y, Kobayashi S, Iwase H. Quantitation of HDAC1 mRNA expression in invasive carcinoma of the breast*. Breast Cancer Res Treat. 2005;94:11–16. doi: 10.1007/s10549-005-6001-1. [DOI] [PubMed] [Google Scholar]
- 25.Mariadason JM. HDACs and HDAC inhibitors in colon cancer. Epigenetics. 2008;3:28–37. doi: 10.4161/epi.3.1.5736. [DOI] [PubMed] [Google Scholar]
- 26.Weichert W. HDAC expression and clinical prognosis in human malignancies. Cancer Lett. 2009;280:168–176. doi: 10.1016/j.canlet.2008.10.047. [DOI] [PubMed] [Google Scholar]
- 27.Glozak MA, Seto E. Histone deacetylases and cancer. Oncogene. 2007;26:5420–5432. doi: 10.1038/sj.onc.1210610. [DOI] [PubMed] [Google Scholar]
- 28.Israel M, Schwartz L. The metabolic advantage of tumor cells. Mol Cancer. 10:70. doi: 10.1186/1476-4598-10-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Santos CR, Schulze A. Lipid metabolism in cancer. FEBS J. doi: 10.1111/j.1742-4658.2012.08644.x. [DOI] [PubMed] [Google Scholar]
- 30.Anastasiou D, Cantley LC. Breathless cancer cells get fat on glutamine. Cell Res. 22:443–446. doi: 10.1038/cr.2012.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gardner PR, Raineri I, Epstein LB, White CW. Superoxide radical and iron modulate aconitase activity in mammalian cells. J Biol Chem. 1995;270:13399–13405. doi: 10.1074/jbc.270.22.13399. [DOI] [PubMed] [Google Scholar]
- 32.Tsui KH, Feng TH, Lin YF, Chang PL, Juang HH. p53 downregulates the gene expression of mitochondrial aconitase in human prostate carcinoma cells. Prostate. 71:62–70. doi: 10.1002/pros.21222. [DOI] [PubMed] [Google Scholar]
- 33.Wellen KE, Hatzivassiliou G, Sachdeva UM, Bui TV, Cross JR, Thompson CB. ATP-citrate lyase links cellular metabolism to histone acetylation. Science. 2009;324:1076–1080. doi: 10.1126/science.1164097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou Y, Niu C, Li Y, Gao B, Zheng J, Guo X, Ma W. Fatty acid synthase expression and esophageal cancer. Mol Biol Rep. doi: 10.1007/s11033-012-1838-y. [DOI] [PubMed] [Google Scholar]
- 35.Makino K, Nakamura H, Hide TI, Yano S, Kuroda JI, Iyama KI, Kuratsu JI. Fatty acid synthase is a predictive marker for aggressiveness in meningiomas. J Neurooncol. doi: 10.1007/s11060-012-0907-3. [DOI] [PubMed] [Google Scholar]
- 36.Notarnicola M, Altomare DF, Calvani M, Orlando A, Bifulco M, D’Attoma B, Caruso MG. Fatty acid synthase hyperactivation in human colorectal cancer: relationship with tumor side and sex. Oncology. 2006;71:327–332. doi: 10.1159/000107106. [DOI] [PubMed] [Google Scholar]
- 37.Calvisi DF, Wang C, Ho C, Ladu S, Lee SA, Mattu S, Destefanis G, Delogu S, Zimmermann A, Ericsson J, Brozzetti S, Staniscia T, Chen X, Dombrowski F, Evert M. Increased lipogenesis, induced by AKT-mTORC1-RPS6 signaling, promotes development of human hepatocellular carcinoma. Gastroenterology. 140:1071–1083. doi: 10.1053/j.gastro.2010.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pandey PR, Liu W, Xing F, Fukuda K, Watabe K. Anti-cancer drugs targeting fatty acid synthase (FAS) Recent Pat Anticancer Drug Discov. 7:185–197. doi: 10.2174/157489212799972891. [DOI] [PubMed] [Google Scholar]
- 39.Venugopal B, Evans TR. Developing histone deacetylase inhibitors as anti-cancer therapeutics. Curr Med Chem. 18:1658–1671. doi: 10.2174/092986711795471284. [DOI] [PubMed] [Google Scholar]
- 40.Sivaraman P, Mattegunta S, Subbaraju GV, Satyanarayana C, Padmanabhan B. Design of a novel nucleoside analog as potent inhibitor of the NAD dependent deacetylase, SIRT2. Syst Synth Biol. 4:257–263. doi: 10.1007/s11693-011-9069-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Latham T, Mackay L, Sproul D, Karim M, Culley J, Harrison DJ, Hayward L, Langridge-Smith P, Gilbert N, Ramsahoye BH. Lactate, a product of glycolytic metabolism, inhibits histone deacetylase activity and promotes changes in gene expression. Nucleic Acids Res. 40:4794–4803. doi: 10.1093/nar/gks066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schwartz JP, Passonneau JV, Johnson GS, Pastan I. The effect of growth conditions on NAD+ and NADH concentrations and the NAD+:NADH ratio in normal and transformed fibroblasts. J Biol Chem. 1974;249:4138–4143. [PubMed] [Google Scholar]
- 43.Boulahbel H, Duran RV, Gottlieb E. Prolyl hydroxylases as regulators of cell metabolism. Biochem Soc Trans. 2009;37:291–294. doi: 10.1042/BST0370291. [DOI] [PubMed] [Google Scholar]
- 44.Bestor TH. The DNA methyltransferases of mammals. Hum Mol Genet. 2000;9:2395–2402. doi: 10.1093/hmg/9.16.2395. [DOI] [PubMed] [Google Scholar]
- 45.Jeltsch A. Circular permutations in the molecular evolution of DNA methyltransferases. J Mol Evol. 1999;49:161–164. doi: 10.1007/pl00006529. [DOI] [PubMed] [Google Scholar]
- 46.Jeltsch A. Beyond Watson and Crick: DNA methylation and molecular enzymology of DNA methyltransferases. Chembiochem. 2002;3:274–293. doi: 10.1002/1439-7633(20020402)3:4<274::AID-CBIC274>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 47.Jeltsch A. Molecular enzymology of mammalian DNA methyltransferases. Curr Top Microbiol Immunol. 2006;301:203–225. doi: 10.1007/3-540-31390-7_7. [DOI] [PubMed] [Google Scholar]
- 48.Wilson JR, Jing C, Walker PA, Martin SR, Howell SA, Blackburn GM, Gamblin SJ, Xiao B. Crystal structure and functional analysis of the histone methyltransferase SET7/9. Cell. 2002;111:105–115. doi: 10.1016/s0092-8674(02)00964-9. [DOI] [PubMed] [Google Scholar]
- 49.Zhang X, Yang Z, Khan SI, Horton JR, Tamaru H, Selker EU, Cheng X. Structural basis for the product specificity of histone lysine methyltransferases. Mol Cell. 2003;12:177–185. doi: 10.1016/s1097-2765(03)00224-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cai J, Sun WM, Hwang JJ, Stain SC, Lu SC. Changes in S-adenosylmethionine synthetase in human liver cancer: molecular characterization and significance. Hepatology (Baltimore, Md. 1996;24:1090–1097. doi: 10.1002/hep.510240519. [DOI] [PubMed] [Google Scholar]
- 51.Wang YC, Tang FY, Chen SY, Chen YM, Chiang EP. Glycine-N methyltransferase expression in HepG2 cells is involved in methyl group homeostasis by regulating transmethylation kinetics and DNA methylation. J Nutr. 141:777–782. doi: 10.3945/jn.110.135954. [DOI] [PubMed] [Google Scholar]
- 52.Stipanuk MH. Sulfur amino acid metabolism: pathways for production and removal of homocysteine and cysteine. Annual review of nutrition. 2004;24:539–577. doi: 10.1146/annurev.nutr.24.012003.132418. [DOI] [PubMed] [Google Scholar]
- 53.Garcea R, Daino L, Pascale R, Simile MM, Puddu M, Ruggiu ME, Seddaiu MA, Satta G, Sequenza MJ, Feo F. Protooncogene methylation and expression in regenerating liver and preneoplastic liver nodules induced in the rat by diethylnitrosamine: effect of variations of S-adenosylmethionine:S-adenosylhomocysteine ratio. Carcinogenesis. 1989;10:1183–1192. doi: 10.1093/carcin/10.7.1183. [DOI] [PubMed] [Google Scholar]
- 54.Pascale RM, Simile MM, Satta G, Seddaiu MA, Daino L, Pinna G, Vinci MA, Gaspa L, Feo F. Comparative effects of L-methionine, S-adenosyl-L-methionine and 5′-methylthioadenosine on the growth of preneoplastic lesions and DNA methylation in rat liver during the early stages of hepatocarcinogenesis. Anticancer Res. 1991;11:1617–1624. [PubMed] [Google Scholar]
- 55.Pascale RM, Marras V, Simile MM, Daino L, Pinna G, Bennati S, Carta M, Seddaiu MA, Massarelli G, Feo F. Chemoprevention of rat liver carcinogenesis by S-adenosyl-L-methionine: a long-term study. Cancer Res. 1992;52:4979–4986. [PubMed] [Google Scholar]
- 56.Simile MM, Pascale R, De Miglio MR, Nufris A, Daino L, Seddaiu MA, Gaspa L, Feo F. Correlation between S-adenosyl-L-methionine content and production of c-myc, c-Ha-ras, and c-Ki-ras mRNA transcripts in the early stages of rat liver carcinogenesis. Cancer Lett. 1994;79:9–16. doi: 10.1016/0304-3835(94)90056-6. [DOI] [PubMed] [Google Scholar]
- 57.Shukeir N, Pakneshan P, Chen G, Szyf M, Rabbani SA. Alteration of the methylation status of tumor-promoting genes decreases prostate cancer cell invasiveness and tumorigenesis in vitro and in vivo. Cancer Res. 2006;66:9202–9210. doi: 10.1158/0008-5472.CAN-06-1954. [DOI] [PubMed] [Google Scholar]
- 58.Halim AB, LeGros L, Geller A, Kotb M. Expression and functional interaction of the catalytic and regulatory subunits of human methionine adenosyltransferase in mammalian cells. The Journal of biological chemistry. 1999;274:29720–29725. doi: 10.1074/jbc.274.42.29720. [DOI] [PubMed] [Google Scholar]
- 59.Corrales F, Ochoa P, Rivas C, Martin-Lomas M, Mato JM, Pajares MA. Inhibition of glutathione synthesis in the liver leads to S-adenosyl-L-methionine synthetase reduction. Hepatology (Baltimore, Md. 1991;14:528–533. [PubMed] [Google Scholar]
- 60.Corrales F, Gimenez A, Alvarez L, Caballeria J, Pajares MA, Andreu H, Pares A, Mato JM, Rodes J. S-adenosylmethionine treatment prevents carbon tetrachloride-induced S-adenosylmethionine synthetase inactivation and attenuates liver injury. Hepatology (Baltimore, Md. 1992;16:1022–1027. doi: 10.1002/hep.1840160427. [DOI] [PubMed] [Google Scholar]
- 61.Pajares MA, Duran C, Corrales F, Pliego MM, Mato JM. Modulation of rat liver S-adenosylmethionine synthetase activity by glutathione. The Journal of biological chemistry. 1992;267:17598–17605. [PubMed] [Google Scholar]
- 62.Pajares MA, Corrales FJ, Ochoa P, Mato JM. The role of cysteine-150 in the structure and activity of rat liver S-adenosyl-L-methionine synthetase. The Biochemical journal. 1991;274(Pt 1):225–229. doi: 10.1042/bj2740225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mato JM, Alvarez L, Ortiz P, Mingorance J, Duran C, Pajares MA. S-adenosyl-L-methionine synthetase and methionine metabolism deficiencies in cirrhosis. Adv Exp Med Biol. 1994;368:113–117. doi: 10.1007/978-1-4615-1989-8_11. [DOI] [PubMed] [Google Scholar]
- 64.Yamada K, Gravel RA, Toraya T, Matthews RG. Human methionine synthase reductase is a molecular chaperone for human methionine synthase. Proc Natl Acad Sci U S A. 2006;103:9476–9481. doi: 10.1073/pnas.0603694103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Banerjee RV, Harder SR, Ragsdale SW, Matthews RG. Mechanism of reductive activation of cobalamin-dependent methionine synthase: an electron paramagnetic resonance spectroelectrochemical study. Biochemistry. 1990;29:1129–1135. doi: 10.1021/bi00457a005. [DOI] [PubMed] [Google Scholar]
- 66.Elmore CL, Wu X, Leclerc D, Watson ED, Bottiglieri T, Krupenko NI, Krupenko SA, Cross JC, Rozen R, Gravel RA, Matthews RG. Metabolic derangement of methionine and folate metabolism in mice deficient in methionine synthase reductase. Mol Genet Metab. 2007;91:85–97. doi: 10.1016/j.ymgme.2007.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhou D, Mei Q, Luo H, Tang B, Yu P. The polymorphisms in methylenetetrahydrofolate reductase, methionine synthase, methionine synthase reductase, and the risk of colorectal cancer. Int J Biol Sci. 8:819–830. doi: 10.7150/ijbs.4462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wettergren Y, Odin E, Carlsson G, Gustavsson B. MTHFR, MTR, and MTRR polymorphisms in relation to p16INK4A hypermethylation in mucosa of patients with colorectal cancer. Mol Med. 16:425–432. doi: 10.2119/molmed.2009.00156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mosharov E, Cranford MR, Banerjee R. The quantitatively important relationship between homocysteine metabolism and glutathione synthesis by the transsulfuration pathway and its regulation by redox changes. Biochemistry. 2000;39:13005–13011. doi: 10.1021/bi001088w. [DOI] [PubMed] [Google Scholar]
- 70.Prudova A, Bauman Z, Braun A, Vitvitsky V, Lu SC, Banerjee R. S-adenosylmethionine stabilizes cystathionine beta-synthase and modulates redox capacity. Proc Natl Acad Sci U S A. 2006;103:6489–6494. doi: 10.1073/pnas.0509531103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lertratanangkoon K, Orkiszewski RS, Scimeca JM. Methyl-donor deficiency due to chemically induced glutathione depletion. Cancer Res. 1996;56:995–1005. [PubMed] [Google Scholar]
- 72.Lertratanangkoon K, Savaraj N, Scimeca JM, Thomas ML. Glutathione depletion-induced thymidylate insufficiency for DNA repair synthesis. Biochem Biophys Res Commun. 1997;234:470–475. doi: 10.1006/bbrc.1997.6623. [DOI] [PubMed] [Google Scholar]
- 73.Lertratanangkoon K, Wu CJ, Savaraj N, Thomas ML. Alterations of DNA methylation by glutathione depletion. Cancer Lett. 1997;120:149–156. doi: 10.1016/s0304-3835(97)00300-5. [DOI] [PubMed] [Google Scholar]
- 74.Huang ZZ, Chen C, Zeng Z, Yang H, Oh J, Chen L, Lu SC. Mechanism and significance of increased glutathione level in human hepatocellular carcinoma and liver regeneration. Faseb J. 2001;15:19–21. doi: 10.1096/fj.00-0445fje. [DOI] [PubMed] [Google Scholar]
- 75.You A, Tong JK, Grozinger CM, Schreiber SL. CoREST is an integral component of the CoREST-human histone deacetylase complex. Proc Natl Acad Sci U S A. 2001;98:1454–1458. doi: 10.1073/pnas.98.4.1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 77.Klose RJ, Kallin EM, Zhang Y. JmjC-domain-containing proteins and histone demethylation. Nat Rev Genet. 2006;7:715–727. doi: 10.1038/nrg1945. [DOI] [PubMed] [Google Scholar]
- 78.Klose RJ, Yamane K, Bae Y, Zhang D, Erdjument-Bromage H, Tempst P, Wong J, Zhang Y. The transcriptional repressor JHDM3A demethylates trimethyl histone H3 lysine 9 and lysine 36. Nature. 2006;442:312–316. doi: 10.1038/nature04853. [DOI] [PubMed] [Google Scholar]
- 79.Wu H, Zhang Y. Mechanisms and functions of Tet protein-mediated 5-methylcytosine oxidation. Genes Dev. 25:2436–2452. doi: 10.1101/gad.179184.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gottlieb E, Tomlinson IP. Mitochondrial tumour suppressors: a genetic and biochemical update. Nat Rev Cancer. 2005;5:857–866. doi: 10.1038/nrc1737. [DOI] [PubMed] [Google Scholar]
- 81.Filipp FV, Scott DA, Ronai ZA, Osterman AL, Smith JW. Reverse TCA cycle flux through isocitrate dehydrogenases 1 and 2 is required for lipogenesis in hypoxic melanoma cells. Pigment Cell Melanoma Res. 25:375–383. doi: 10.1111/j.1755-148X.2012.00989.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Metallo CM, Gameiro PA, Bell EL, Mattaini KR, Yang J, Hiller K, Jewell CM, Johnson ZR, Irvine DJ, Guarente L, Kelleher JK, Vander Heiden MG, Iliopoulos O, Stephanopoulos G. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature. 481:380–384. doi: 10.1038/nature10602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Smolkova K, Jezek P. The Role of Mitochondrial NADPH-Dependent Isocitrate Dehydrogenase in Cancer Cells. Int J Cell Biol. 2012:273947. doi: 10.1155/2012/273947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Leonardi R, Subramanian C, Jackowski S, Rock CO. Cancer-associated isocitrate dehydrogenase mutations inactivate NADPH-dependent reductive carboxylation. J Biol Chem. 287:14615–14620. doi: 10.1074/jbc.C112.353946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schwartz JP, Lust WD, Lauderdale VR, Passonneau JV. Glycolytic metabolism in cultured cells of the nervous system. II. Regulation of pyruvate and lactate metabolism in the C-6 glioma cell line. Mol Cell Biochem. 1975;9:67–72. doi: 10.1007/BF01732197. [DOI] [PubMed] [Google Scholar]
- 86.Baggetto LG. Deviant energetic metabolism of glycolytic cancer cells. Biochimie. 1992;74:959–974. doi: 10.1016/0300-9084(92)90016-8. [DOI] [PubMed] [Google Scholar]
- 87.Zhao S, Lin Y, Xu W, Jiang W, Zha Z, Wang P, Yu W, Li Z, Gong L, Peng Y, Ding J, Lei Q, Guan KL, Xiong Y. Glioma-derived mutations in IDH1 dominantly inhibit IDH1 catalytic activity and induce HIF-1alpha. Science. 2009;324:261–265. doi: 10.1126/science.1170944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ward PS, Cross JR, Lu C, Weigert O, Abel-Wahab O, Levine RL, Weinstock DM, Sharp KA, Thompson CB. Identification of additional IDH mutations associated with oncometabolite R(-)-2-hydroxyglutarate production. Oncogene. 31:2491–2498. doi: 10.1038/onc.2011.416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Andersson AK, Miller DW, Lynch JA, Lemoff AS, Cai Z, Pounds SB, Radtke I, Yan B, Schuetz JD, Rubnitz JE, Ribeiro RC, Raimondi SC, Zhang J, Mullighan CG, Shurtleff SA, Schulman BA, Downing JR. IDH1 and IDH2 mutations in pediatric acute leukemia. Leukemia. 25:1570–1577. doi: 10.1038/leu.2011.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bleeker FE, Atai NA, Lamba S, Jonker A, Rijkeboer D, Bosch KS, Tigchelaar W, Troost D, Vandertop WP, Bardelli A, Van Noorden CJ. The prognostic IDH1( R132 ) mutation is associated with reduced NADP+-dependent IDH activity in glioblastoma. Acta Neuropathol. 119:487–494. doi: 10.1007/s00401-010-0645-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, Fantin VR, Jang HG, Jin S, Keenan MC, Marks KM, Prins RM, Ward PS, Yen KE, Liau LM, Rabinowitz JD, Cantley LC, Thompson CB, Vander Heiden MG, Su SM. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 465:966. doi: 10.1038/nature09132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lin J, Yao DM, Qian J, Chen Q, Qian W, Li Y, Yang J, Wang CZ, Chai HY, Qian Z, Xiao GF, Xu WR. IDH1 and IDH2 mutation analysis in Chinese patients with acute myeloid leukemia and myelodysplastic syndrome. Ann Hematol. 91:519–525. doi: 10.1007/s00277-011-1352-7. [DOI] [PubMed] [Google Scholar]
- 93.Paugh BS, Qu C, Jones C, Liu Z, Adamowicz-Brice M, Zhang J, Bax DA, Coyle B, Barrow J, Hargrave D, Lowe J, Gajjar A, Zhao W, Broniscer A, Ellison DW, Grundy RG, Baker SJ. Integrated molecular genetic profiling of pediatric high-grade gliomas reveals key differences with the adult disease. J Clin Oncol. 28:3061–3068. doi: 10.1200/JCO.2009.26.7252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ward PS, Patel J, Wise DR, Abdel-Wahab O, Bennett BD, Coller HA, Cross JR, Fantin VR, Hedvat CV, Perl AE, Rabinowitz JD, Carroll M, Su SM, Sharp KA, Levine RL, Thompson CB. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell. 17:225–234. doi: 10.1016/j.ccr.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, Kos I, Batinic-Haberle I, Jones S, Riggins GJ, Friedman H, Friedman A, Reardon D, Herndon J, Kinzler KW, Velculescu VE, Vogelstein B, Bigner DD. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim SH, Ito S, Yang C, Xiao MT, Liu LX, Jiang WQ, Liu J, Zhang JY, Wang B, Frye S, Zhang Y, Xu YH, Lei QY, Guan KL, Zhao SM, Xiong Y. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases. Cancer Cell. 19:17–30. doi: 10.1016/j.ccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chowdhury R, Yeoh KK, Tian YM, Hillringhaus L, Bagg EA, Rose NR, Leung IK, Li XS, Woon EC, Yang M, McDonough MA, King ON, Clifton IJ, Klose RJ, Claridge TD, Ratcliffe PJ, Schofield CJ, Kawamura A. The oncometabolite 2-hydroxyglutarate inhibits histone lysine demethylases. EMBO Rep. 12:463–469. doi: 10.1038/embor.2011.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Turk PW, Laayoun A, Smith SS, Weitzman SA. DNA adduct 8-hydroxyl-2′-deoxyguanosine (8-hydroxyguanine) affects function of human DNA methyltransferase. Carcinogenesis. 1995;16:1253–1255. doi: 10.1093/carcin/16.5.1253. [DOI] [PubMed] [Google Scholar]
- 99.Weitzman SA, Turk PW, Milkowski DH, Kozlowski K. Free radical adducts induce alterations in DNA cytosine methylation. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:1261–1264. doi: 10.1073/pnas.91.4.1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Tsuzuki T, Nakatsu Y, Nakabeppu Y. Significance of error-avoiding mechanisms for oxidative DNA damage in carcinogenesis. Cancer Sci. 2007;98:465–470. doi: 10.1111/j.1349-7006.2007.00409.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Nohmi T, Kim SR, Yamada M. Modulation of oxidative mutagenesis and carcinogenesis by polymorphic forms of human DNA repair enzymes. Mutat Res. 2005;591:60–73. doi: 10.1016/j.mrfmmm.2005.03.033. [DOI] [PubMed] [Google Scholar]
- 102.Ehrlich M, Gama-Sosa MA, Huang LH, Midgett RM, Kuo KC, McCune RA, Gehrke C. Amount and distribution of 5-methylcytosine in human DNA from different types of tissues of cells. Nucleic Acids Res. 1982;10:2709–2721. doi: 10.1093/nar/10.8.2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gong Z, Zhu JK. Active DNA demethylation by oxidation and repair. Cell Res. 21:1649–1651. doi: 10.1038/cr.2011.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lim SO, Gu JM, Kim MS, Kim HS, Park YN, Park CK, Cho JW, Park YM, Jung G. Epigenetic changes induced by reactive oxygen species in hepatocellular carcinoma: methylation of the E-cadherin promoter. Gastroenterology. 2008;135:2128–2140. doi: 10.1053/j.gastro.2008.07.027. [DOI] [PubMed] [Google Scholar]
- 105.Penn NW, Suwalski R, O’Riley C, Bojanowski K, Yura R. The presence of 5-hydroxymethylcytosine in animal deoxyribonucleic acid. The Biochemical journal. 1972;126:781–790. doi: 10.1042/bj1260781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Castro GD, Diaz Gomez MI, Castro JA. 5-Methylcytosine attack by hydroxyl free radicals and during carbon tetrachloride promoted liver microsomal lipid peroxidation: structure of reaction products. Chem Biol Interact. 1996;99:289–299. doi: 10.1016/0009-2797(95)03680-6. [DOI] [PubMed] [Google Scholar]
- 107.Valinluck V, Tsai HH, Rogstad DK, Burdzy A, Bird A, Sowers LC. Oxidative damage to methyl-CpG sequences inhibits the binding of the methyl-CpG binding domain (MBD) of methyl-CpG binding protein 2 (MeCP2) Nucleic Acids Res. 2004;32:4100–4108. doi: 10.1093/nar/gkh739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Inoue A, Shen L, Dai Q, He C, Zhang Y. Generation and replication-dependent dilution of 5fC and 5caC during mouse preimplantation development. Cell Res. 21:1670–1676. doi: 10.1038/cr.2011.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Valinluck V, Sowers LC. Endogenous cytosine damage products alter the site selectivity of human DNA maintenance methyltransferase DNMT1. Cancer research. 2007;67:946–950. doi: 10.1158/0008-5472.CAN-06-3123. [DOI] [PubMed] [Google Scholar]
- 110.Hashimoto H, Liu Y, Upadhyay AK, Chang Y, Howerton SB, Vertino PM, Zhang X, Cheng X. Recognition and potential mechanisms for replication and erasure of cytosine hydroxymethylation. Nucleic Acids Res. 40:4841–4849. doi: 10.1093/nar/gks155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Oberley LW, Oberley TD, Buettner GR. Cell division in normal and transformed cells: the possible role of superoxide and hydrogen peroxide. Med Hypotheses. 1981;7:21–42. doi: 10.1016/0306-9877(81)90018-9. [DOI] [PubMed] [Google Scholar]
- 112.Gius D, Spitz DR. Redox signaling in cancer biology. Antioxidants & redox signaling. 2006;8:1249–1252. doi: 10.1089/ars.2006.8.1249. [DOI] [PubMed] [Google Scholar]
- 113.Spitz DR, Sim JE, Ridnour LA, Galoforo SS, Lee YJ. Glucose deprivation-induced oxidative stress in human tumor cells. A fundamental defect in metabolism? Annals of the New York Academy of Sciences. 2000;899:349–362. doi: 10.1111/j.1749-6632.2000.tb06199.x. [DOI] [PubMed] [Google Scholar]