Abstract
Within the central nervous system, microRNAs have emerged as important effectors of an array of developmental, physiological, and cognitive processes. Along these lines, the CREB-regulated microRNA miR-132 has been shown to influence neuronal maturation via its effects on dendritic arborization and spinogenesis. In the mature nervous system, dysregulation of miR-132 has been suggested to play a role in a number of neurocognitive disorders characterized by aberrant synaptogenesis. However, little is known about the inducible expression and function of miR-132 under normal physiological conditions in vivo. Here, we begin to explore this question within the context of learning and memory. Using in situ hybridization, we show that the presentation of a spatial memory task induced a significant ~1.5-fold increase in miR-132 expression within the CA1, CA3, and GCL excitatory cell layers of the hippocampus. To examine the role of miR-132 in hippocampal-dependent learning and memory, we employ a doxycycline-regulated miR-132 transgenic mouse strain to drive varying levels of transgenic miR-132 expression. These studies revealed that relatively low levels of transgenic miR-132 expression, paralleling the level of expression in the hippocampus following a spatial memory task, significantly enhanced cognitive capacity. In contrast, higher (supra-physiological) levels of miR-132 (>3-fold) inhibited learning. Interestingly, both the impaired cognition and elevated levels of dendritic spines resulting from supra-physiological levels of transgenic miR-132 were reversed by doxycycline suppression of transgene expression. Together, these data indicate that miR-132 functions as a key activity-dependent regulator of cognition, and that miR-132 expression must be maintained within a limited range to ensure normal learning and memory formation.
Keywords: miRNA, CREB, Hippocampus, Neuronal plasticity, Memory, Learning
Introduction
MicroRNAs (miRNAs) are small (~22 nt) evolutionarily conserved single-stranded RNA species that function as potent negative regulators of gene expression (Ambros 2004; Bartel 2004; Lim e t al. 2005). miRNAs repress gene expression via their ability to hybridize with the 3′ UTR of target mRNAs, and, in turn, trigger mRNA degradation and/or translational repression (reviewed in Bartel 2009; Eulalio et al. 2008; Carthew and Sontheimer 2009). miR-NA expression is often cell-type and tissue-specific (Lagos-Quintana et al. 2002; Kim et al. 2004b; Kosik 2006; Cao et al. 2006), and within the central nervous system (CNS), recent work has indicated that they play key roles in a wide range of physiological processes. Along these lines, during development, the disruption of miRNA expression through genetic deletion of Dicer results in aberrant neuronal development and differentiation, as well as altered morphogenesis and neuronal signaling (Cuellar et al. 2008; Giraldez et al. 2005). Furthermore, recent work reveals specific roles for miRNAs (including miR-124a, miR-134, and miR-375) in dendritogenesis and axonal path finding (Schratt et al. 2006; Abdelmohsen et al. 2010; Fiore et al. 2009; Sanuki et al. 2011).
As more neuronal-enriched miRNA have been identified, the possibility of a prominent role for miRNA in the regulation of neuronal activity and cognition has emerged (Konopka et al. 2011; Im et al. 2010). For example, brain-specific miR-128b was found to regulate fear-extinction memory formation (Lin et al. 2011), and SIRT1 regulation of miR-134 was shown to mediate hippocampal plasticity and fear conditioning (Gao et al. 2010). Interestingly, Konopka et al. (2010) demonstrated that conditional deletion of Dicer1 enhanced learning and memory capacity in mice (Konopka et al. 2010).
Within the CNS, several miRNAs are known to be expressed in an activity-dependent manner, suggesting that miRNAs could sculpt the synaptically evoked transcriptome (Khudayberdiev et al. 2009; Wibrand et al. 2010; Hansen et al. 2011). Among these is the CREB-regulated miRNA-132 (miR-132) (Cheng et al. 2007; Remenyi et al. 2010; Wibrand et al. 2010; Nudelman et al. 2010). miR-132 is transcribed from the intron of a non-coding RNA and has been shown to alter both neuronal morphology and synaptic physiology (Hansen et al. 2010b; Vo et al. 2005). Along these lines, the effects of miR-132 on dendritic morphogenesis appears, in part, to be mediated by its suppression of p250GAP, thus leading to a de-repression of Rac1-PAK-mediated spinogenesis (Impey et al. 2010; Wayman et al. 2008). In cultured hippocampal neurons, overexpressing miR-132 alters short-term synaptic plasticity, characterized by decreased synaptic depression and an increased paired-pulse ratio, whereas the repression of miR-132 led to a decreased mEPSC frequency (a result likely related to the effects of miR-132 on synapse formation) (Lambert et al. 2010). Furthermore, selective deletion of the miR-132/212 locus led to significant alterations in neuronal morphology in newborn adult hippocampal neurons, including decreased dendritic complexity and spine density (Magill et al. 2010). It was also recently demonstrated that the inhibition of miR-132 alters dendritic spine maturation and prevents ocular dominance plasticity in neurons within the visual cortex (Tognini et al. 2011; Mellios et al. 2011).
Given its complex and context-specific effects on neuronal structure and function, an examination of the potential role of miR-132 as a regulator of cognition is highly merited. In line with this, we recently reported that robust transgenic overexpression of miR-132 in the forebrain increased spine density and caused deficits in novel object recognition memory (Hansen et al. 2010b). Interestingly, in this study, transgenic miR-132 expression levels were quite high (>5-fold over basal expression), approximating the expression level of endogenous miR-132 following seizure activity (Jimenez-Mateos et al. 2011). This high level of transgenic miR-132 raised the possibility that our reported effects on novel object recognition memory may not serve as a useful guide to understand the role of miR-132 under normal physiological conditions such as following learning/ memory tasks. Consistent with this idea, transgenic overexpression of a number of genes implicated in learning and memory formation has been shown to negatively affect cognitive capacity (Croll et al. 1999; Pietropaolo et al. 2007; Viosca et al. 2009). Hence, given that (1) miR-132 is CREB-regulated, (2) CREB has been shown to play a role in cognition (Sakamoto et al. 2011), and (3) miR-132 modulates neuronal morphology, we hypothesized that under normal physiological conditions, miR-132 could couple neuronal activity to enhanced cognition.
To begin to address this question, we employed a combination of in vivo detection methods and transgenic mouse models to assess the functional effects of miR-132 on hippocampal-dependent learning and memory. Here, we show that endogenous miR-132 is induced in response to a spatial memory task, and that this ‘induced’ level of miR-132 enhances cognition. Moreover, overexpression (~3-fold) of miR-132 leads to profound cognitive deficits. Together, these data indicate that miR-132 is part of a normal cellular signaling pathway that couples associative learning paradigms to memory formation, and that dysregulation of its expression leads to impaired cognition.
Materials and methods
Ethics statement
All animal breeding and experimental procedures were approved by the Ohio State University Animal Care and Use Committee (protocol number: 2008A0227).
tTA::miR-132 mice
Generation of the tetracycline-regulated bidirectional miR-132/cyan fluorescent protein (CFP) transgenic mouse line (here referred to as: miR-132 transgenic mice) was recently described (Hansen et al. 2010b). To drive transgene expression, the TRE-miR-132 transgenic mice were crossed with the CaMKII tTA driver line (Mayford et al. 1996), which allowed for robust transgene expression within excitatory forebrain neurons (Hansen et al. 2010b). Temporal regulation of tTA (i.e., ‘Tet-off’) inducible miR-132 transgene expression was accomplished by administration of doxycycline (0.40–200 µg/mL) to the drinking water.
Tissue processing and fluorescent in situ hybridization (FISH)
Mature (6–8 weeks), sex-matched C57/Bl6 mice were used. All tissue preparation was carried out under RNAse-free conditions and with the use of DEPC-treated solutions. Brain tissue was removed after cervical dislocation and post-fixed in 4 % paraformaldehyde for 4 h at 4 °C and cryoprotected with 30 % sucrose in PBS. Hippocampus-containing sections were then thin cut (40 µm) on a freezing microtome. Dorsal hippocampal sections were incubated (5 min) with the nuclear stain DRAQ5 (1:10,000; BioStatus Limited, UK) and washed 3× in PBS.
Fluorescent in situ hybridization against miRNA was carried out according to procedures outlined by Nuovo (2010). In brief, tissue was mounted onto slides, followed by a short digestion with pepsin. Tissue was then hybridized overnight with fluorescein-conjugated locked nucleic acid (LNA) probes (Obernosterer et al. 2007) to mouse miR-132, snRNA U6, or an Exiqon-designed ‘scrambled’ negative control probe (Exiqon). The signal from the fluorescein-conjugated probes was amplified with an anti-fluorescein Alexa 488 signal detection kit (Invitrogen).
Images were acquired at 40× using a Zeiss 510 confocal microscope and LSM Software. Images of the relevant cellular layers of FISH-labeled sections were traced digitally, and the intensity levels were analyzed with MetaMorph software (Molecular Devices). Intensity levels were normalized by background subtraction and presented relative to naïve CA1 staining.
RT-PCR miRNA quantitation
The NCode™ VILO™ miRNA cDNA Synthesis Kit (Invitrogen) was used to create a cDNA library of total hippocampal RNA that was extracted by TRIzol isolation (Invitrogen) from monotransgenic and tTA::miR-132 animals. miRNAs were polyadenylated with Poly(A)poly-merase and reverse transcribed with universally tagged 5′ oligo-dT primer (Invitrogen). cDNA amplification was performed using SYBR green reporter-based qPCR (Applied Biosystems) and the miScript Primers (Qiagen). Data were normalized to RNU6B_2 cDNA levels and presented relative to the tTA (No dox) condition, which was set equal to a value of 1.
Barnes maze analysis
Mature (6–8 weeks), age-and sex-matched animals were used for all behavioral paradigms. Because of the limited availability of transgenic animals, both male and female mice were used for these paradigms, but were balanced in equal numbers between each group. Animals were housed five per cage, and maintained under a standard 12 h light/dark cycle. Barnes maze exposure was initiated at the end of the light phase (1 h before light off) each day. Barnes maze testing followed procedures outlined by Sunyer et al. (2007) and Barnes (1979). In summary, a circular and elevated maze platform (92 cm diameter, 76 cm above the floor) with 20 equally spaced holes was positioned in a brightly lit room. An escape box was placed under one of the holes, and visual cues (geometric shapes) were placed on the walls that surround the maze. Bright light (585 lux) and an electronic metronome [Boss DB-66, 74 dB (measured at the center of the maze), 120 beats per minute (bpm)] were used as aversive stimuli. Of note, increasing the decibel level beyond 74 dB led to excessive freezing behavior, and thus was not conducive to the assessment of learning in these animals.
Examination of learning-induced miR-132 expression in wild-type animals was conducted using a modified Barnes maze paradigm in order to capture changes in early miRNA expression. Animals were given 2 days of Barnes maze training, by which point spatial memory appeared to be largely established, as demonstrated by improved performance across measures of both latency and errors (Fig. 3). Each day of training, mice experienced three 5-min trials of free exploration with a 10-min inter-trial interval. A trial ended once the mouse entered the escape box or after 5 min of exploration had elapsed (at which point it was gently guided to the target box). To limit the possibility of olfactory cues, all surfaces of the maze and the target box were cleaned with 70 % ethanol after each trial. Control animals were also handled and experienced the same auditory and visual cues as treated animals, but were not trained to escape to the target box. Tissue was isolated for in situ hybridization 12 h after the last trial on day 2, reflecting both the time course of miRNA processing (Schmittgen et al. 2008), as well as similar paradigms used to detect early signaling mechanisms of memory formation (Vázquez et al. 2000; Kim et al. 2004a).
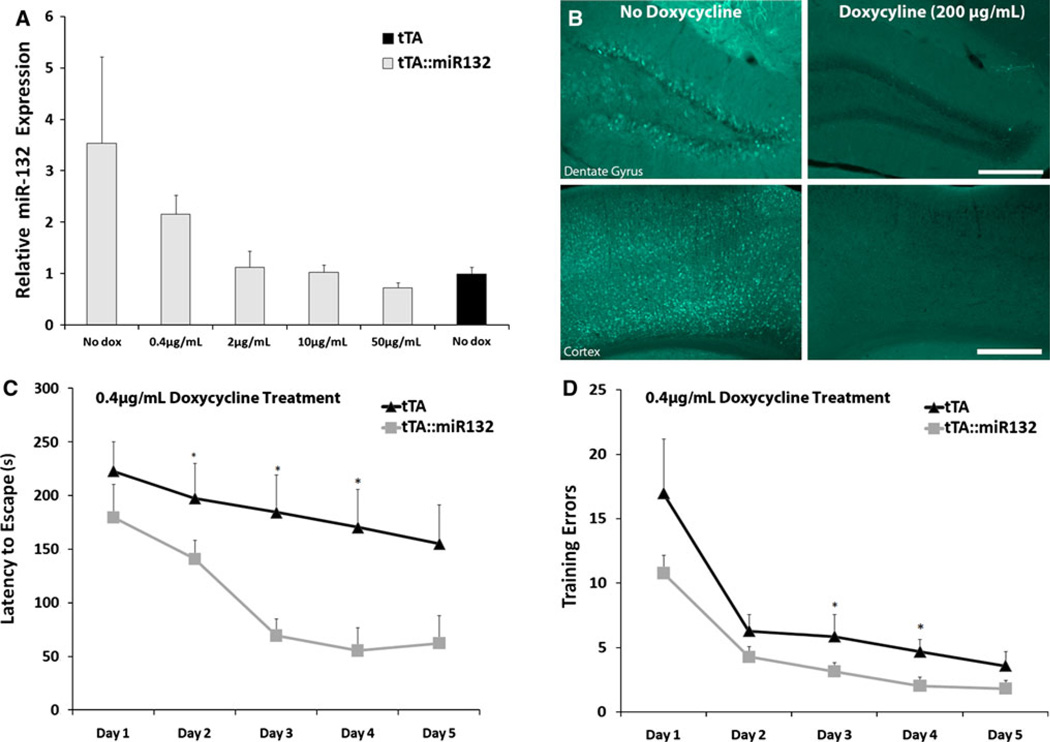
Fig. 3.
Limited expression of transgenic miR-132 enhances Barnes maze performance. a tTA::miR-132 animals exhibited robust expression of miR-132 in the hippocampus by qPCR in the absence of doxycycline, while the addition of doxycycline administration to the drinking water for 2 weeks led to a dose-dependent reduction in miR-132 expression. From 2–10 µg/mL of doxycycline, miR-132 levels in tTA::miR-132 mice were not significantly different from levels in monotransgenic animals (tTA). Data are presented as mean ± SEM (n = 3 per group). b Immunofluorescent analysis of cyan fluorescent protein (CFP) expression in tTA::miR-132 mice confirms robust transgene expression in the absence of doxycycline administration, and potent suppression of transgene expression following 2 weeks of doxycycline administration (200 µg/mL). Scale bars 200 µm. c, d Animals (tTA and tTA::miR-132) were administered doxycycline (0.4 µg/mL) for 3 weeks prior to Barnes maze testing. Under this condition, tTA::miR-132 animals exhibited faster escape latency (c) and fewer training errors (d) than monotransgenic littermates (*P <0.05, n = 11 animals per group). All groups improved significantly across 5 days of acquisition
When used as a measure of spatial learning capacity, monotransgenic and tTA::miR-132 mice were given 5 days of training, following the same procedure outlined above. Nose pokes into incorrect holes and latency to enter the target hole were recorded as measures of maze performance over all trials (scored by an experimenter blind to experimental conditions). To distinguish the potentially subtle phenotypic effects of graded transgene expression, Barnes maze testing on animals with titered doxycycline administration (Fig. 3) was conducted under slightly less aversive conditions (500 lux, 62 dB, 100 bpm) than those used to test the effects of robust transgenic miR-132 (Fig. 4; as in WT learning-induction trials: 585 lux, 74 dB, 120 bpm). By decreasing the motivation to escape, we allow for greater resolution of the performance of enhanced-cognition animals, and minimize the likelihood of a ‘ceiling effect’ related to minimum training errors and latency to escape. Thus, the combination of these paradigms allows for a detailed and subtle examination of both increases and decreases in cognition.
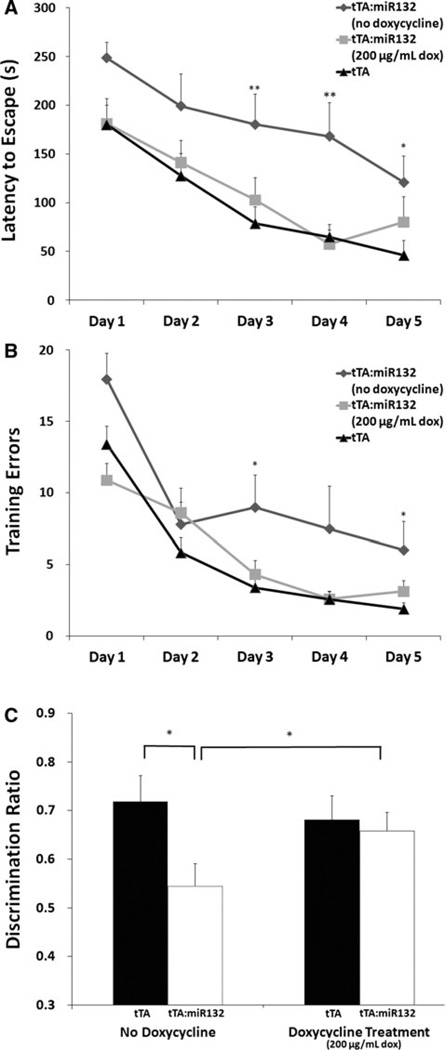
Fig. 4.
tTA::miR-132 animals under conditions of robust transgenic miR-132 show cognitive impairment. a, b In the absence of doxycycline treatment, tTA::miR-132 animals undergoing Barnes maze testing showed increased latency to escape (a), as well as increased training errors (b) relative to monotransgenic littermates (tTA) (*P <0.05, **P < 0.01; n = 12 animals per group). In a separate group of tTA::miR-132 mice, doxycycline treatment (200 mg/mL: 3 weeks) suppressed the cognitive impairment observed in the tTA::miR-132 animals that were not treated with doxycycline. tTA animals were also treated with doxycycline, but showed no differences from untreated controls (Online Resource 1). All experimental groups improved significantly across 5 days of acquisition. c In the absence of doxycycline, tTA::miR-132 animals showed significant impairment in novel object recognition memory. Hence, tTA::miR-132 mice did not exhibit greater exploratory bias toward the novel versus the familiar object. In contrast, blocking transgenic miR-132 expression via doxycycline administration (200 µg/mL: 3 weeks) suppressed the novel object recognition impairment of tTA::miR-132 animals. Thus, doxycycline-treated tTA::miR-132 mice exhibited significantly greater exploration time with the novel object than with the familiar one, paralleling the behavioral phenotype of monotransgenic littermates (tTA). Data are presented as mean discrimination ratio ± SEM, n = 5 animals per group, *P <0.05
Successful acquisition of the Barnes maze task was demonstrated by a probe trial that occurred 24 h after the final acquisition day. The probe trial consisted of a single 90-s exploration of the maze with the escape box removed.
Of note, the visual ability of each mouse was established by suspending the animal by the tail and slowly lowering it toward a solid dark surface (a table) for three successive trials. Visual acuity was demonstrated by animals reaching for the surface before vibrissae made contact with it. Visual acuity testing was performed under both ambient white light and red light conditions. Auditory capacity for each mouse was established by freezing behavior in response to three successive, sudden claps. A positive freeze response was taken as an indication of hearing capability. All mice demonstrated similar visual and auditory acuity. Animals exhibiting inappropriate freezing or lack of exploration for more than 200 s per trial (suggestive of high levels of anxiety) were excluded from the analysis (a total of three monotransgenic animals and one tTA::miR-132 animal across all experiments).
Novel object recognition
Novel object recognition was adapted from procedure described by Bevins and Besheer (Bevins and Besheer 2006). Briefly at CT 12, each animal was allowed a 10-min training session with exposure to two identical, non-toxic objects (glass or hard plastic items). After training, animals were returned to the home cage for a 30-min delay period. After the retention interval, each animal was returned to the testing arena in which one familiar object was replaced with a novel object. Animals were given 5 min to explore while being video recorded for later scoring by an experimenter blind to experimental conditions. Subsequently, exploration time for each object was timed, as defined by the animal’s nose being within 2 cm of, and pointed toward, the object. After each exposure, the surfaces of the objects and the exploration environment were cleaned with 70 % ethanol to prevent recognition by olfactory cues. Objects were counterbalanced between novel and familiar to account for preference bias, and the placement of the objects were also alternated to avoid direction bias. The discrimination ratio was calculated as [‘exploration time with novel object’/(‘time with novel object’ + ‘time with familiar object’)]. Of note, all animals demonstrated similar ambulatory capacity, as demonstrated by midline crossings and total exploration time. Animals were tested at the beginning of the dark phase of the 24 h cycle. So as not to disrupt the circadian rhythm of the mice, animals were tested under red light (200 lux) conditions.
GFP and CFP immunolabeling and morphometric analysis
For the examination of neuronal spine density, tTA::miR-132 mice were crossed with a transgenic line expressing green fluorescent protein (GFP) in a subpopulation of hippocampal neurons under the control of the thy-1 promoter (Morris 1985). Thy-1 GFP mice (Feng et al. 2000) were generously provided by Gouping Feng (Duke University). Of note, the Thy-1-driven morphological marker does not affect the electrophysiological or the morphological properties (i.e., dendrite length and number, spine number and density, soma size) of hippocampal neurons (Vuksic et al. 2008).
To immunolabel for GFP expression, tissue was fixed and prepared as described above. Sections were then washed and permeabilized in PBS with 1 % Triton X-100 PBST (3×, 10 min each). Sections were blocked for 1 h in 10 % normal goat serum in PBS. Sections were incubated overnight at 4 °C in rabbit polyclonal anti-GFP antibody (acquired from Dr. Luc G. Berthiaume, University of Alberta, Canada). The GFP antibody was used at 1:2,500 to detect the TRE-regulated CFP transgene, and 1:20,000 to detect the Thy-1-regulated GFP transgene. Of note, the expression of the tet-responsive CFP transgene is markedly lower than the Thy-1-driven GFP, and thus, by using this relatively low concentration of the primary antibody, we are able to distinguish the Thy-1 GFP transgene from the CFP transgene. A series of control data sets which confirm successful discrimination of the two signals is presented in Hansen et al. (2010b). Sections were then incubated in Alexa Fluor-488-conjugated goat anti-mouse IgG antibody (1:1,000; Invitrogen, Carlsbad, CA) for 3 h at room temperature. Tissue was washed in PBST for a total of 30 min between each labeling step. Sections were mounted on slides with Fluoromount-G (SouthernBiotech, Birmingham, Alabama, AL). For clarity, CFP transgene immunolabeling (Fig. 3b) is presented as a blue/cyan fluorescent signal.
CA1 basal dendrites (ten per animal) were examined for morphological changes in 40-µm-thick coronal sections. Images of Thy-1 GFP immunofluorescence of 20 µm dendritic segments extending 90–120 µm from the cell soma were captured using confocal microscopy. Processes extending >0.5 µm from the dendrite were counted as dendritic spines and included both mushroom-shaped and filopodia-like protrusions. Images were acquired at 63× with 4.3× optical zoom, using a Zeiss 510 confocal microscope and LSM Software.
Western blotting
Isolated hippocampal tissue was lysed in 100 µL radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris– HCl, 150 mM NaCl, 1 mM EDTA, 1 % Triton X-100, 0.1 % sodium dodecyl sulfate, 1 % sodium deoxycholate, 1 mM sodium vanadate, 1 mM NaF, and 1× protease inhibitor cocktail [Roche]). Total protein (25 µg/lane) was loaded into a 10 % SDS-PAGE gel, electrophoresed, and transblotted onto polyvinylidene difluoride membranes (Immobilon-P; Millipore, Billerica, MA). After blocking in 5 % milk in PBST for 1 h, membranes were incubated (overnight, 4 °C) in 5 % BSA in PBST with rabbit anti-phospho-Akt substrate, rabbit anti-phospho-PKA substrate, or mouse anti-phospho-tyrosine antibodies (all: 1:2,000; Cell Signaling, Danvers, MA). Membranes were also probed for total protein using anti-Tau and anti-Neun antibodies (both: 1:2,000; Millipore). The following day, membranes were incubated in 5 % milk (in PBST) with an anti-goat or anti-mouse IgG horseradish peroxidase-conjugated antibody (1:2,000; PerkinElmer Life Sciences, Waltham, MA). Membranes were washed a minimum of four times (10 min per wash) in PBST between each antibody treatment. The Western Lighting Chemiluminescence light-emitting system (PerkinElmer) and HyBlot CL autoradiography film (Denville Scientific, Metuchen, NJ) were used to visualize and document antibody signals.
Bioinformatic profiling
Asurvey of algorithmically derived predicted targets of miR-132 was performed using the August 2010 release of microRNA. org (http://www.microrna.org/microrna/home.do).
Statistics
All values presented here are given as mean ± SEM and were calculated using SPSS 19.0. Unless otherwise noted, a value of P < 0.05 was accepted as statistically significant. Comparisons between two groups were made by Student’s t test. Significance for qPCR, spine density, and within-day Barnes maze analysis were assessed using one-way ANOVA analysis, followed by Fisher’s least significant difference (LSD) test. Repeated measures mixed-model ANOVA was used for multi-day Barnes maze training comparison.
Results
Expression of miR-132 in the hippocampus
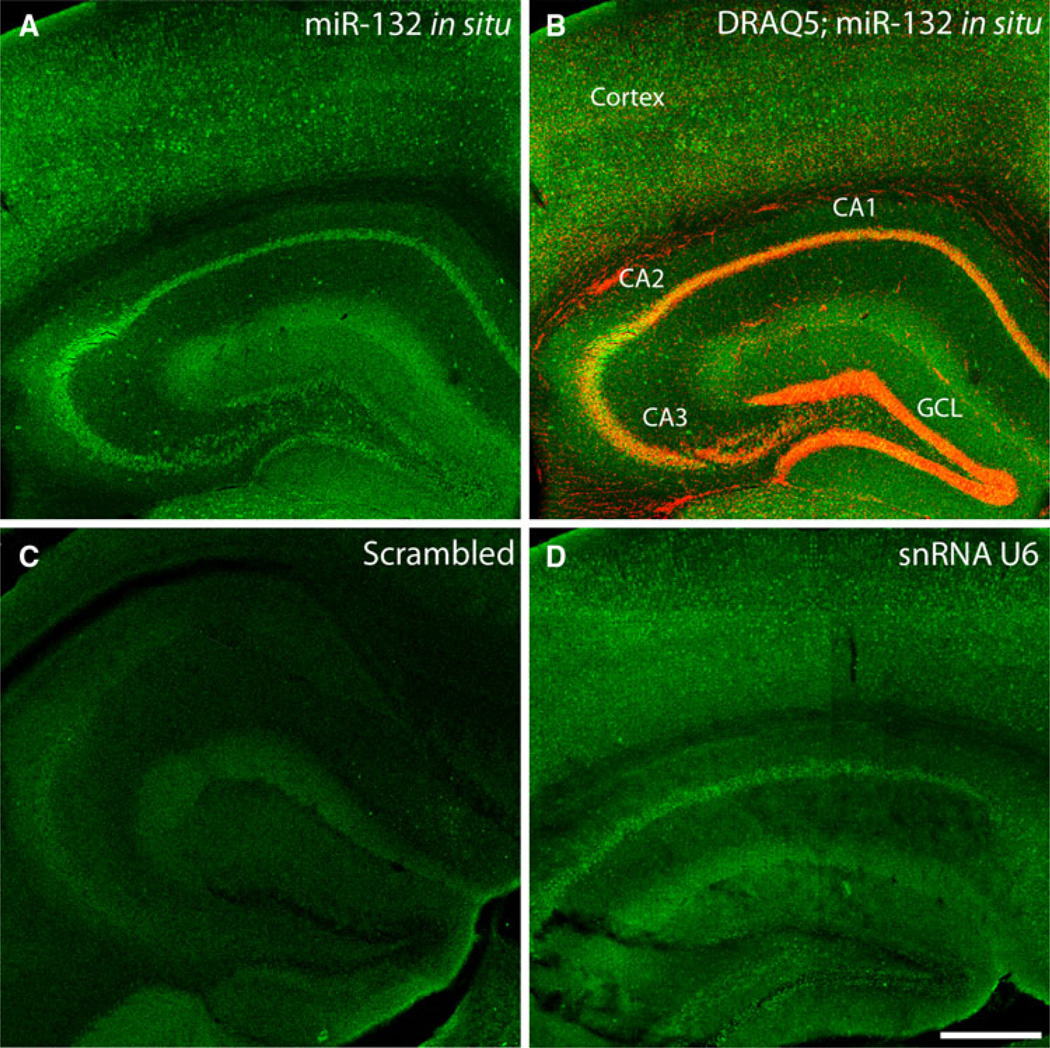
To begin our analysis, we profiled miR-132 expression in the mature hippocampus using FISH (Fig. 1). Representative data reveal marked expression of miR-132 in all excitatory sublayers of the hippocampus (i.e., CA1, CA2, CA3, and GCL), as well as in the cortex (Fig. 1a, b). Higher magnification imaging of tissue labeling confirmed that miR-132 expression was largely restricted to neuronal cell populations (Fig. 2). Hence, in contrast to the robust expression detected in the CA1 and GCL sublayers, minimal labeling was detected within cells (largely non-neuronal) located in the molecular layer of the dentate gyrus (Fig. 2a–c). A control set of experiments in which tissue was labeled with a ‘scrambled’ fluorescein-conjugated LNA probe that does not detect known microRNA species revealed little labeling (Fig. 1c). As a positive control, tissue was labeled with an LNA probe against the ubiquitously expressed snRNA U6. As expected, U6 labeling was detected throughout the hippocampus. Together, these indicate neuronal enrichment of miR-132 in the adult hippocampus.
Fig. 1.
miR-132 expression in the hippocampus. a Fluorescent in situ hybridization (FISH) using an LNA anti-sense probe directed against miR-132 reveals robust miR-132 expression throughout the cortex and hippocampus of wild-type animals. b Within the hippocampus, marked excitatory layer-specific expression of miR-132 was confirmed by merging the miR-132 fluorescence micrograph in a with a fluorescence micrograph image of DRAQ5 labeling (red). c Representative FISH labeling using a ‘scrambled’ miRNA probe: minimal labeling was detected. d As a positive control for the FISH labeling approach, hippocampal sections were incubated with an anti-sense probe against the snRNA U6. CA1–3 hippocampal subfields, GCL granule cell layer. Scale bar 400 µm
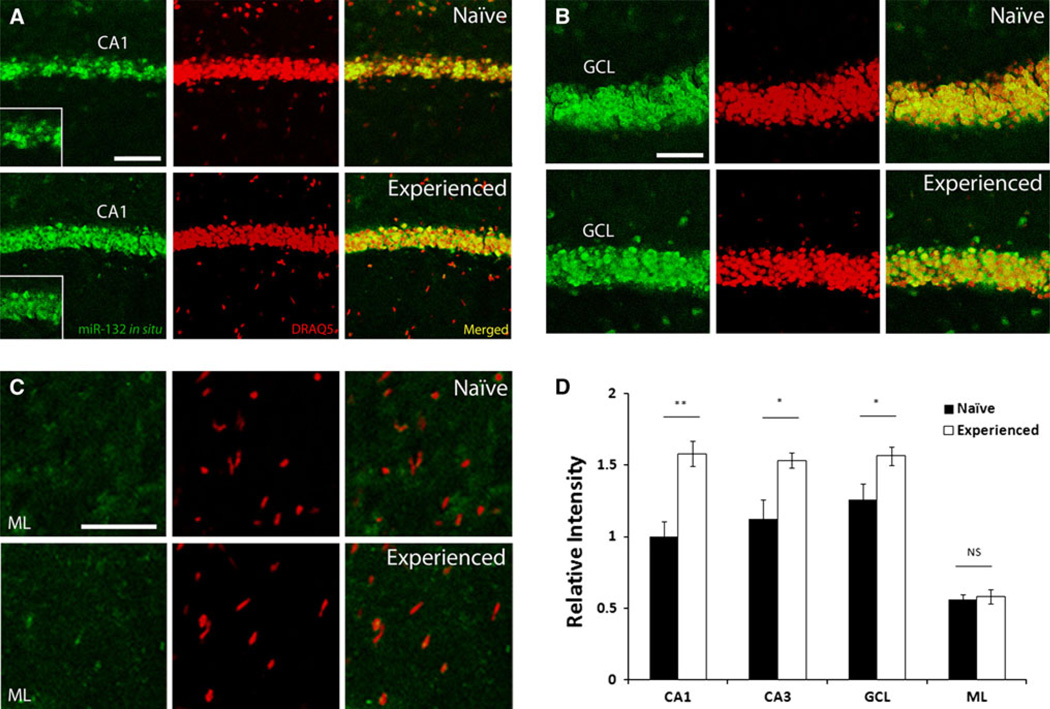
Fig. 2.
miR-132 expression is induced in response to the Barnes maze learning paradigm. Hippocampal miR-132 expression was examined by FISH in naïve animals, and in animals that experienced 2 days of Barnes maze learning. a–c In animals exposed to the learning paradigm, expression of miR-132 (green) was increased in the CA1 (a), CA3 (not shown), and GCL (b) excitatory cell layers relative to naïve animals, but not in the molecular layer (ML, c). Tissue was also labeled with the DNA stain DRAQ5 (red). Scale bar 100 µm. d Quantification of fluorescent intensity in CA1, CA3, GCL, and ML relative to naïve CA1, ± SEM; NS not significant. *P <0.05, **P <0.01, two-tailed t test, n = 6 animals for each group
Induction of miR-132 after spatial learning
A number of studies have shown that miR-132 expression is tightly regulated by CREB (Jimenez-Mateos et al. 2011; Cheng et al. 2007). Given that CREB plays a central role in transcriptionally-dependent forms of learning and memory, it was reasonable to posit that miR-132 may be inducibly expressed during memory formation. To begin to test this idea, we profiled hippocampal miR-132 expression in wild-type animals undergoing Barnes maze spatial memory training (described in detail below). Because the greatest period of memory formation appears to occur during the first 2 days of training (as demonstrated by both reduced latency and errors, and successful escape to the target box: Fig. 3d), animals were given 2 days of training (three trials per day). Tissue was then collected 12 h later, and miR-132 expression was measured via FISH (Fig. 2). Quantitative analysis of the FISH signal revealed that miR-132 expression was significantly higher (~1.5-fold) in the CA1 [t(10) = 3.82, P <0.01], CA3 [t(10) = 2.87, P <0.05], and GCL [t test, t(10) = 2.38, P <0.05] cell layers of Barnes maze-trained animals, relative to control animals that were naïve to the learning paradigm (Fig. 2a, b, d). In naïve mice, miR-132 expression in the CA1 layer was heterogeneous, with limited expression in some cells, while other cells expressed relatively high levels. Interestingly, training resulted in an elevated and more uniform miR-132 signal across the CA1 sublayer (Fig. 2a). Of note, learning did not increase miR-132 expression in non-neuronal cells, including those occupying the molecular layer of the dentate gyrus [t(10) = 0.361, P >0.05; Fig. 2c, d]. Together, these data show that miR-132 expression is significantly increased in response to the learning challenge.
Cognitive enhancement in animals with moderate increases in transgenic miR-132
Our previous work using a transgenic miR-132 mouse model revealed that robust overexpression (~5-fold, relative to endogenous levels) of miR-132 impaired learning and memory (Hansen et al. 2010b). Given our work here showing a relatively low level of miR-132 induction following a learning paradigm, coupled with a number of studies showing that miR-132 affects neuronal plasticity (Impey et al. 2010; Wayman et al. 2008), we hypothesized that robust transgenic overexpression may not be a useful model to dissect the contribution of endogenous miRNA-132 to cognition. Hence, in an attempt to provide a better model, we returned to the miR-132 transgenic mouse, and tested the effects of low-level transgene expression on Barnes maze learning. As outlined in our previous paper (Hansen et al. 2010b), to drive transgene expression in forebrain excitatory neurons, the tetracycline-regulated miR-132/CFP bidirectional transgenic mouse line was crossed with a CaMKII tTA driver line (Mayford et al. 1996) (denoted as ‘tTA::miR-132’ mice). To negatively regulate transgenic miR-132 expression, varying concentrations of doxycycline were added to the drinking water, and miR-132 expression was profiled 2 weeks later. As expected, increasing the concentration of doxycycline in the drinking water (0.4–50 µg/mL) resulted in an incremental repression of transgene expression in the hippocampus, as measured by qPCR (Fig. 3a). Consistent with this, administration of 200 µg/mL of doxycycline led to a suppression of CFP expression, as detected by immunofluorescent staining (Fig. 3b). To approximate the wildtype expression level of miR-132 after 2 days of Barnes maze learning, we chose an intermediate doxycycline concentration (0.4 µg/mL) that yielded an ~2-fold increase in transgene expression over control tTA mice (Fig. 3a). For the behavioral assay, tTA::miR-132 animals were treated with this dose of doxycycline for 3 weeks and then tested on the Barnes maze paradigm. Animals were given five consecutive days of training (three trials per day), during which they were allowed to explore the circular Barnes maze platform. To avoid potential ceiling effects of performance with cognitive enhancement, this assay was performed under less aversive conditions (see ‘‘Materials and methods’’). Remarkably, doxycycline-treated tTA::miR-132 animals showed excellent Barnes maze performance, exhibiting both reduced latencies and fewer errors relative to monotransgenic (tTA) littermates that also received doxycycline (Fig. 3c, d). Doxycycline administration to monotransgenic controls had no significant effect relative to untreated animals (Online Resource 1). Hence, moderate increases in transgenic miR-132 significantly enhanced cognitive capacity, suggesting that the learning-evoked increase in endogenous miR-132 also enhances cognition. Of note, all groups successfully demonstrated a capacity to locate the target hole by the final training day, showing significant improvement across all 5 days of acquisition [latency: F(4,80) = 6.685, P <0.001; training errors: F(4,80) = 17.723, P <0.001]. There were no significant differences between groups on the probe day (data not shown).
Cognitive impairment in mice overexpressing miR-132
As noted, our previous work demonstrated that robust transgenic overexpression of miR-132 (i.e., no doxycycline treatment) impaired novel object recognition memory (Hansen et al. 2010b). Here, we furthered this line of inquiry by testing the effects of transgenic miR-132 over-expression on spatial memory capacity, and by testing the plastic nature of the learning phenotype.
In the Barnes maze assay, mice that expressed high levels of transgenic miR-132 (no doxycycline) showed significant impairment in spatial memory capacity, as demonstrated by poor performance in maze acquisition. Of note, the assay for cognitive impairment was performed using levels of aversion stimuli more motivating than those used in Fig. 3. While all groups improved across 5 days of acquisition, the transgenic group without doxycycline exhibited both a longer latency to escape to the target hole, as well as an increase in errors made before escape, as compared to monotransgenic littermates (Fig. 4a, b). These data are consistent with our prior work using the novel object recognition test, and thus provide further support for the idea that miR-132 shapes cognitive capacity.
To examine whether the cognitive deficits observed in tTA::miR-132 were reversible, a separate group of miR-132:tTA animals received doxycycline through the drinking water at a dose sufficient to fully suppress trans-gene expression (200 µg/mL). After 3 weeks of treatment, these animals exhibited Barnes maze performance that was indistinguishable from tTA monotransgenic mice (Fig. 4a, b). All three groups were capable of learning the task, and showed significant improvement across 5 days of acquisition [latency: F(4,160) = 33.429, P <0.001; training errors: F(4,156) = 45.65, P <0.001]. Similarly, the poor performance of transgenic miR-132:tTA animals assayed via the novel object recognition test (Hansen et al. 2010b) was reversed by maintaining animals on doxycycline treatment for 3 weeks (Fig. 4c). A two-way ANOVA revealed a significant treatment by genotype interaction effect, F(3,16) = 4.7. Post hoc tests revealed a significant difference between untreated monotransgenic and tTA::-miR-132 animals, as well as between treated and untreated tTA::miR-132 animals. Doxycycline administration to monotransgenic controls yielded no significant change relative to untreated animals (Fig. 4c). Thus, these data indicate that cognitive capacity is tightly and dynamically linked to the expression level of miR-132.
Reversibility of increased spine density in tTA::miR-132 animals
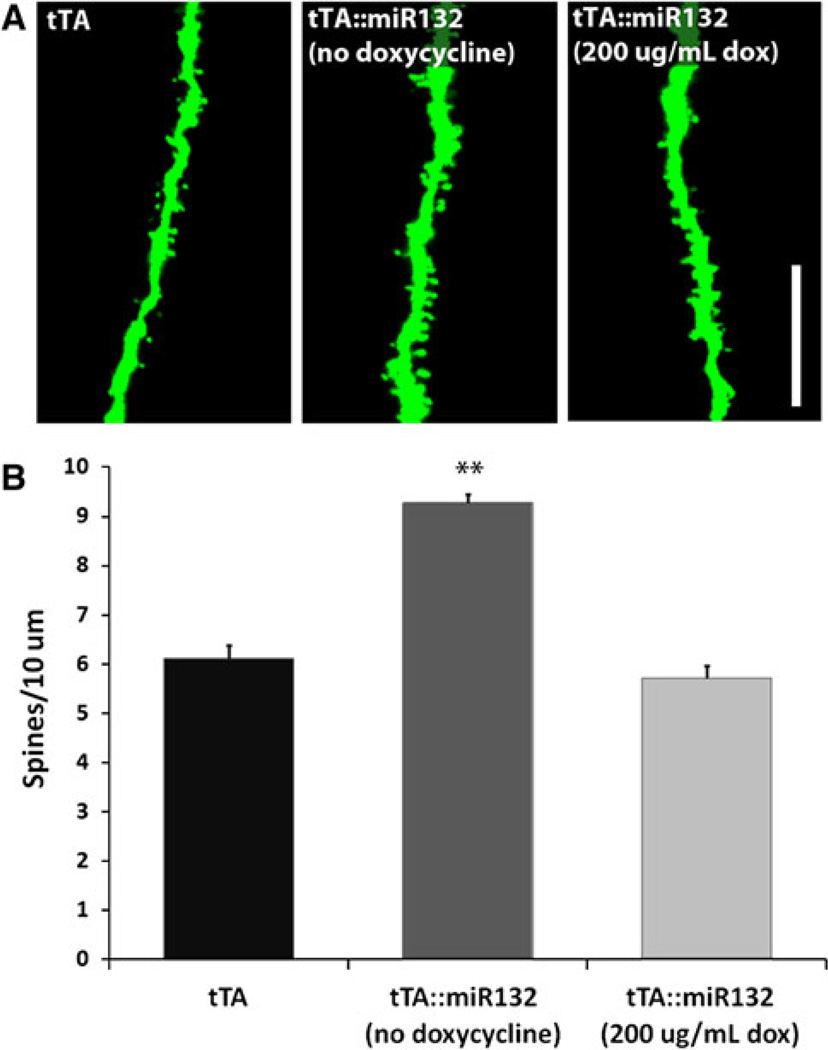
Our previous work demonstrated that, in the absence of doxycycline treatment, tTA::miR-132 mice exhibit a marked increase in dendritic spine density relative to monotransgenic mice (Hansen et al. 2010b). Although increased spine density is often associated with enhanced cognitive capacity, excessive spine formation has been linked to cognitive deficits (Hutsler and Zhang 2010). Here, we examined spine density in mature tTA::miR-132 animals after receiving 3 weeks of doxycycline treatment (200 µg/mL). For these experiments, the miR-132 transgenic mice were crossed with a Thy-1 GFP mouse line that has been used extensively to examine neuronal morphology and plasticity (Vuksic et al. 2008; Feng et al. 2000; Hansen et al. 2010b). Morphometric analysis revealed that spine density in CA1 basal dendrites of tTA::miR-132 mice treated with doxycycline was indistinguishable from tTA monotransgenic mice (Fig. 5). Further, consistent with our prior work, spine density was significantly enhanced in tTA::miR-132 mice that were not treated with doxycycline [F(2,12) = 83.563]. Doxycycline administration to monotransgenic animals showed no effect on neuronal morphology (data not shown). This ostensible doxycyclinemediated reversibility of the miR-132 spine density phenotype parallels the reversibly of the miR-132 cognitive phenotype, and thus suggests that miR-132 is a dynamic and transient regulator of neuronal plasticity.
Fig. 5.
Transgenic miR-132-induced increases in spine density are suppressed by the administration of doxycycline. A Thy-1-driven GFP transgenic marker was used to examine spine density in tTA::miR-132 mice. a Confocal images reveal an increase in spine density under conditions of robust miR-132 expression [tTA::miR-132 (no doxycycline)] that is absent upon suppression of the transgene with doxycycline [tTA::miR-132 (200 µg/mL dox)]. Scale bar 10 µm. b Graphical representation of the mean ± SEM spine density, **P < 0.01, n = 5 animals for each group
miR-132::tTA transgenic mice exhibit marked alterations in kinase activity
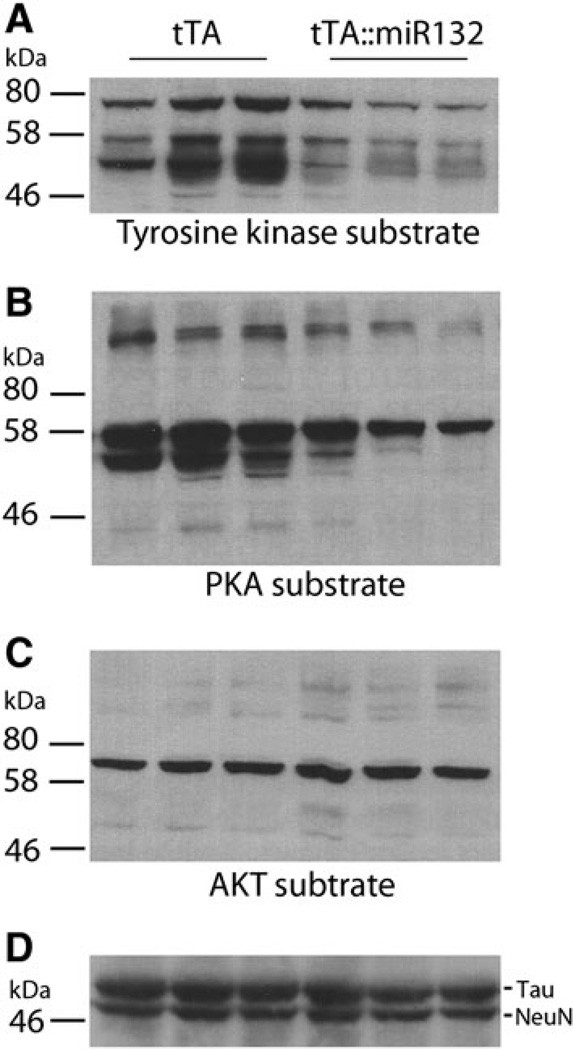
miR-132 has been shown to affect the expression of a functionally diverse set of mRNA targets, including MeCP2, p250GAP, and p300 (Klein et al. 2007; Wayman et al. 2008; Lagos et al. 2010). However, this may only be a small subset of the physiological effectors of miR-132. For example, a bioinformatic analysis using microRNA.org identifies over 7,000 putative targets. Here, we attempted to examine the question of miR-132 and cellular function by employing an alternative strategy that profiles the functional impact of miR-132 on kinase signaling pathways. Although these kinases may not be direct targets of miR-132, insight into how miR-132 affects intracellular communication, possibly via both direct and indirect actions, may provide new clues as to how this miRNA affects neuronal plasticity. To this end, hippocampal lysates from tTA monotransgenic and tTA::miR-132 animals (no doxycycline treatment) were probed via Western analysis with phosphorylation motif-specific antibodies against broad classes of kinase families. Interestingly, the tyrosine phosphorylation motif pattern (a marker of tyrosine kinase activity) was reduced in tTA::miR-132 animals compared to tTA littermates (Fig. 6a). Likewise, phosphorylation of the protein kinase A (PKA) substrate motif (RRXS*/T*: * denotes phosphorylated amino acid residue) was also reduced in tTA::miR-132 tissue. However, phosphorylation of the AKT target motif (RXXS*/T*) was not markedly different between the tTA monotransgenic and tTA::miR-132 animals (Fig. 6). These data suggest that broad, but not universal, kinomic dysregulation accompanies miR-132 overexpression. Together, these data, along with the noted work on miR-132 targets, indicate a key role for this noncoding RNA in activity-inducible CNS physiology and function.
Fig. 6.
tTA::miR-132 mice exhibit altered kinomic activity. To begin to identify molecular mechanisms by which miR-132 influences cognitive capacity, hippocampal lysates from tTA and tTA::miR-132 animals were probed with phospho motif-specific antibodies that infer the activation state of protein tyrosine kinases (a, tyrosine kinase substrate), protein kinase A (b PKA substrate), and AKT (c AKT substrate). a Transgenic animals showed decreased phosphorylation of the tyrosine kinase substrate motif and b the PKA substrate motif (RRXS*/T*: * denotes phosphorylated amino acid residue) relative to tTA mice. Phosphorylation of the AKT motif (RXXS*/T*) remained relatively unaffected in tTA::miR-132 mice. As a protein loading control, membranes were also probed for Tau and NeuN; for clarity, only a subset of the banding pattern is presented. Lysates from three animals of each genotype were profiled (i.e., biological triplicate determinations)
Discussion
The central goal of this project was to further our understanding of the role of miR-132 in hippocampal-dependent learning and memory formation. To this end, we demonstrated that endogenous miR-132 is expressed in excitatory cell layers throughout the hippocampus, and that miR-132 is induced in response to a spatial learning task. Using these data as a guide, we employed a tetracycline-regulated operon to drive the expression of transgenic miR-132 and assess its effects on cognition. These data reveal that cognitive capacity is tightly regulated by miR-132. Hence, physiological levels of miR-132 enhance cognition, whereas supra-physiological levels of the miRNA lead to cognitive deficits. Together, these data indicate that miR-132 plays a key role in shaping cognitive capacity.
As noted, our in situ hybridization approach revealed cytoplasmic miR-132 expression in all excitatory cell layers of the wild-type hippocampus, and relatively modest expression in other hippocampal regions. This expression pattern of miR-132 in the CNS is consistent with several recent reports. Within the visual cortex, miR-132 is mainly localized to the neuronal cytoplasm (Tognini et al. 2011; Mellios et al. 2011), and Wibrand et al. (2010) also noted cytoplasmic miR-132 expression within the hippocampus, although they also detected limited miR-132 in proximal dendrites (Wibrand et al. 2010). Interestingly, in situ hybridization of cultured hippocampal neurons not only detected miR-132 expression within the cell body, but also within the processes (Wayman et al. 2008). Given the recognized role of miR-132 in spinogenesis, it may not be unreasonable to suspect that miR-132 could also be localized to dendritic terminals in vivo. Further, other miRNAs have been detected in neuronal processes, where they have been reported to affect cell morphology and function (Schratt et al. 2006; Cohen et al. 2011). Clearly, a detailed examination of the subcellular expression pattern of miR-132 in vivo is merited and could be achieved via cell fractionation approaches, or more sensitive hybridization methods than those employed here.
Next, we turned to the question of miR-132 inducible expression following the presentation of a spatial learning task. One key motivation for choosing a relatively short time point following initiation of the learning paradigm was the desire to capture ostensible early-stage gene expression, which would be expected to play a central role in the induction of learning. At this time point, significant upregulation of miR-132 was detected in the excitatory cell layers of the hippocampus, suggesting that miR-132 induction occurs during a period of memory consolidation. Interestingly, these data are consistent with recent PCR-based profiling data, which showed that contextual fear conditioning triggers a rapid increase in the expression of the primary transcript of miR-132 (Nudelman et al. 2010), although a learning-evoked increase in the mature form of miR-132 was not detected in the noted study. Clearly, to gain deeper insights into the role of miR-132 in learning and memory, a much more detailed characterization of the time dependence and duration of learning-evoked miR-132 expression will be required.
In many respects, the learning-evoked induction of miR-132 that we report parallels the expression pattern of other activity-responsive, CREB-regulated genes. Along these lines, a number of learning paradigms have been shown to induce the expression of immediate early genes, including Fos, Zif268, and EGR1 (Hall et al. 2001; Porte et al. 2008; Pollak et al. 2005). Likewise, numerous studies have shown that other spatial learning tasks, analogous to the Barnes maze paradigm used here, trigger both an increase in CREB phosphorylation at Serine 133 (Porte et al. 2008; Colombo et al. 2003; Mizuno et al. 2002), and an increase in CRE-mediated transcription (Impey et al. 1998). Together, these data suggest that miR-132 is part of the CREB-regulated, learning-evoked transcriptional response, and, given the time course of induction, raises the prospect that miR-132 plays a role in CREB-dependent memory consolidation.
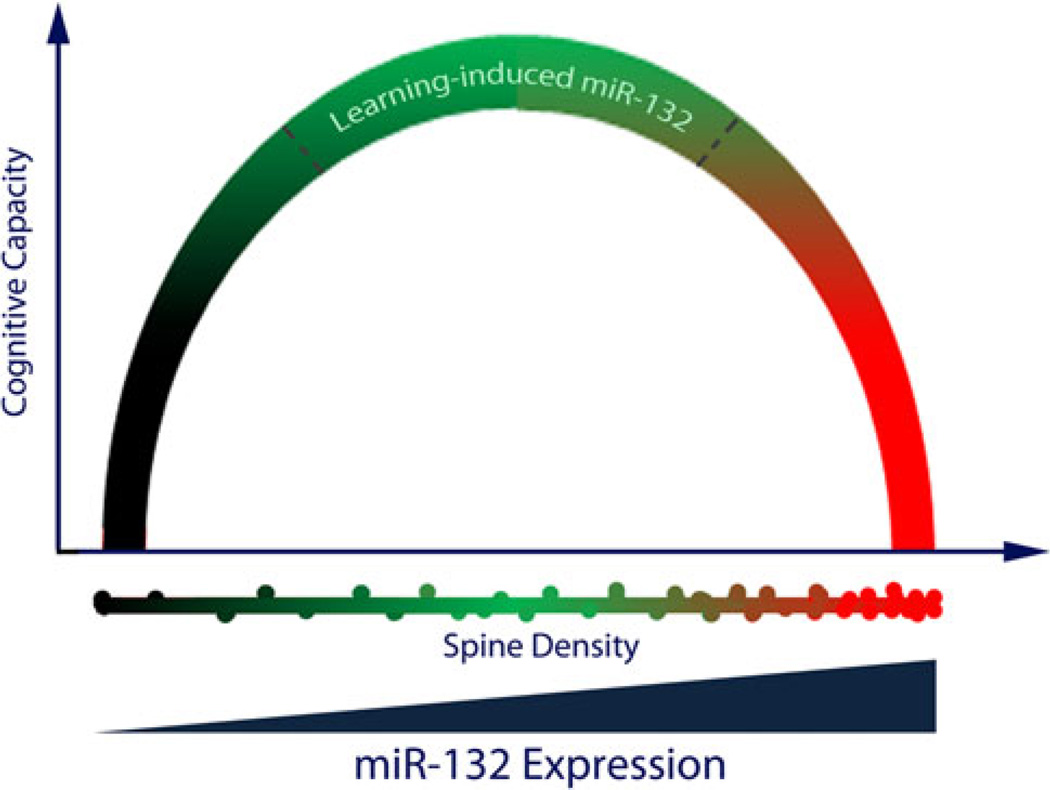
In our prior report, we found that transgenic miR-132 expression led to an increase in overall dendritic spine density in the hippocampus, and a deficit in novel object recognition memory (Hansen et al. 2010b). Here, the examination of transgenic miR-132’s action on learning and memory was extended to the Barnes maze, where we found that overexpression of miR-132 (no doxycycline treatment) led to a profound deficit in spatial learning capacity. On its face, pairing our current data showing that learning induces an increase in miR-132 expression with our prior data showing that transgenic overexpression of miR-132 triggers cognitive deficits would suggest that endogenous miR-132 functions as a negative regulator of learning and memory. However, a more circumspect examination of the data raises issues with this interpretation. Central to this concern is the marked difference between the miR-132 induction level evoked by learning and the level driven by the tet-operon. Notably, wild-type animals showed learning-evoked increases in expression of ~1.5-fold, whereas in transgenic mice miR-132 expression was elevated >3-fold over control levels (in the absence of doxycycline). This transgenic, supra-physiological level of miR-132 could profoundly dysregulate neuronal physiology and, in turn, cognition. To address this possibility, we employed the doxycycline regulatory system to modulate transgenic miR-132 levels, and assess the effects of moderate miR-132 induction that approximates the learningevoked expression level. Under these conditions, tTA::-miR-132 animals showed significantly enhanced spatial memory capacity relative to monotransgenic littermates. The expression level-specific effects of miR-132 led us to envision miRNA functionality as an inverted U-shaped curve, where low or supra-physiological levels of the miRNA result in cognitive impairment, and where miR-132 enhances cognitive capacity only within a fairly limited expression range (Fig. 7). Here, it is worth noting that the effects of the transgene and the endogenous gene cannot be conclusively disambiguated with the experimental approaches used here. Hence, within the context of the Barnes maze paradigm, the inducible expression of endogenous miR-132 would likely contribute to both the enhanced cognitive capacity resulting from relatively low levels of transgenic miR-132, and the cognitive impairment resulting from relatively high levels of transgenic miR-132. In this light, our inverted U-shaped model would suggest that a range of miR-132 (1.5-to 3.5-fold over basal levels) facilitates learning and memory. Interestingly, Tognini et al. (2011) posit a model for miR-132 within the visual cortex in which miR-132 mediation of spine stability allows for appropriate levels of plasticity. Furthermore, they suggest that both excessive and minimized miR-132 expression leads to inappropriately low plasticity, due to excessive stability or instability, respectively, of spine formation. Clearly, a combination of transgenic and knockout miR-132 mouse models would need to be used to gain a clearer insight into how both the level and duration of miR-132 expression modulates cognition.
Fig. 7.
Model depicting the proposed relationship among miR-132, neuronal morphology, and cognition. In this model, we propose that there is an optimal, and limited, range of miR-132 expression which enhances cognitive capacity. Supra-physiological expression of miR-132 leads to excessive spine density and cognitive deficits that compromise learning and memory formation. Further, limited basal and/or damped inducible expression of miR-132 is hypothesized to inhibit cognition, possibly via a paucity of spine formation. Hence, deviation from an optimized miR-132 expression level (either excessive induction or too little expression) may lead to the severe learning deficits that are characteristic of a variety of neurocognitive disorders
The compromised cognitive capacity associated with relatively high levels of transgenic miR-132 expression is reminiscent of several lines of work in which robust expression of genes shown to enhance neuronal plasticity and memory formation surprisingly led to a disruption of cognition (Pineda et al. 2004; Bourtchouladze et al. 2006; Migaud et al. 1998). Similarly, there appears to be an upper limit to the facilitatory effects that CREB/CRE pathway signaling has on learning and memory. Along these lines, transgenic approaches that drive relatively low levels of CRE-mediated gene expression enhances cognition, whereas robust overexpression of constitutively active CREB interferes with the retrieval of spatial information (Viosca et al. 2009; Suzuki et al. 2011; Kaitsuka et al. 2011; Lopez de Armentia et al. 2007). Hence, similar to what we propose for miR-132, there appears to be an optimal range of CREB/CRE signaling, within which cognitive capacity is enhanced. In fact, Viosca et al. (2009) also proposed an inverted U-shaped function to describe the effects of CREB on cognition. Given the connection between CREB and miR-132, it is interesting to posit that the deleterious cognitive effects of excessive CRE-mediated transcription may result in part from high levels of miR-132.
There are numerous mechanisms by which high levels of miR-132 would result in cognitive deficits. Within this context, the noted increase in spine density reported here bares further discussion. Though spine density is often positively correlated with cognitive capacity (Leuner et al. 2003; Kleim et al. 1998; Trommald et al. 1996), hyperconnectivity and heighted spinogenesis are also associated with cognitive deficits, as observed in the autism spectrum disorders and in fragile X syndrome (Hutsler and Zhang 2010; Irwin et al. 2001). Our data raise the prospect that a limited range of miR-132 expression leads to morphological plasticity conducive to normal cognitive capacity, and that relatively high levels disrupt cognition via aberrant spinogenesis/synaptic connections (Fig. 7). Consistent with this idea, our data reveal that doxycycline suppression of transgenic miR-132 reversed both the cognitive deficits and the spinogenesis phenotype. Additional studies that utilize varied levels of doxycycline to examine the relationship between cognitive capacity and spine density could provide important new insights into how even ostensibly modest changes in spine morphology/density trigger marked cognitive effects.
Of the miR-132 targets identified to date, p250GAP provides the best mechanistic connection between miR-132 and enhanced spinogenesis. p250GAP appears to function as a potent negative regulator of spinogenesis via its inhibitory effects on Rho family GTPase activity (Moon et al. 2003; Nakayama et al. 2000; Van Aelst and Cline 2004). Of note, in cultured hippocampal neurons, the onset of spine formation corresponds with an increase in miR-132 expression, a decrease in p250GAP expression, and an increase in Rac activation (Wayman et al. 2008). Hence, inducible regulation of p250GAP expression via miR-132 could be a key mechanism by which synaptic activity regulates Rho family-dependent structural plasticity (i.e., spinogenesis). Clearly, one could develop a set of experiments that tests whether miR-132 functions via p250GAP to give rise to the spinogenesis phenotype. However, with respect to the cognitive phenotype described here, the sheer complexity of miRNA signaling raises some doubt that these effects of transgenic miR-132 could be ascribed to a single mRNA target. In support of this idea, in addition to p250GAP, miR-132 has also been shown to affect the expression of MECP2 and p300 (Hansen et al. 2010b; Lagos et al. 2010; Klein et al. 2007; Vo et al. 2005; Vo and Goodman 2001), two proteins involved in gene transcription and chromatin structure (Goodman and Smolik 2000; Sterner and Berger 2000; Hansen et al. 2010a). Clearly, dysregulation of either MECP2 or p300 target would have profound effects on complex gene networks that underlie neuronal plasticity. Consistent with this idea of gene network modulation, the data presented here reveal that transgenic miR-132 expression leads to marked changes in the functionality of a number of kinase families.
Expression of miR-132 is known to be dysregulated in a number of disorders such as Rett Syndrome, Huntington’s disease, Schizophrenia, and Alzheimer’s disease (Klein et al. 2007; Packer et al. 2008; Kim et al. 2010; Cogswell et al. 2008). Thus, insights into miR-132 regulation and dysfunction may provide meaningful lines of inquiry with regard to these disorders. Given the complexity of miRNA expression and the plethora of miR-132 targets that have yet to be validated, newly developed approaches, including array and deep sequencing assays designed to quantify gene expression levels and identify gene networks, may be required to begin to unravel the mechanisms by which miR-132 sculpts cognition.
Supplementary Material
Acknowledgments
We thank Heather Dziema, and Andrea M. Hesse for technical support. National Institutes of Health Grant numbers: F31-MH096460-01, NS066345, MH062335.
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s00429-012-0431-4) contains supplementary material, which is available to authorized users.
Conflict of interest The authors have declared that no competing interests exist.
Contributor Information
Katelin F. Hansen, Department of Neuroscience, Ohio State University, Graves Hall, Rm 4118, 333 W. 10th Ave, Columbus, OH 43210, USA
Kate Karelina, Department of Neuroscience, Ohio State University, Graves Hall, Rm 4118, 333 W. 10th Ave, Columbus, OH 43210, USA.
Kensuke Sakamoto, Department of Neuroscience, Ohio State University, Graves Hall, Rm 4118, 333 W. 10th Ave, Columbus, OH 43210, USA.
Gary A. Wayman, Department of Veterinary and Comparative Anatomy, Pharmacology and Physiology, Washington State University, Pullman, WA, USA
Soren Impey, Oregon Stem Cell Center, Oregon Health and Science University, Portland, OR, USA.
Karl Obrietan, Email: obrietan.1@osu.edu, Department of Neuroscience, Ohio State University, Graves Hall, Rm 4118, 333 W. 10th Ave, Columbus, OH 43210, USA.
References
- Abdelmohsen K, Hutchison ER, Lee EK, Kuwano Y, Kim MM, Masuda K, Srikantan S, Subaran SS, Marasa BS, Mattson MP, Gorospe M. miR-375 inhibits differentiation of neurites by lowering HuD levels. Mol Cell Biol. 2010;30(17):4197–4210. doi: 10.1128/MCB.00316-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambros V. The functions of animal microRNAs. Nature. 2004;431(7006):350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- Barnes CA. Memory deficits associated with senescence: a neurophysiological and behavioral study in the rat. J Comp Physiol Psychol. 1979;93(1):74–104. doi: 10.1037/h0077579. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. (pii: S0092867404000455) [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevins RA, Besheer J. Object recognition in rats and mice: a one-trial non-matching-to-sample learning task to study ‘recognition memory’. Nat Protoc. 2006;1(3):1306–1311. doi: 10.1038/nprot.2006.205. [DOI] [PubMed] [Google Scholar]
- Bourtchouladze R, Patterson SL, Kelly MP, Kreibich A, Kandel ER, Abel T. Chronically increased Gsalpha signaling disrupts associative and spatial learning. Learn Mem. 2006;13(6):745–752. doi: 10.1101/lm.354106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X, Yeo G, Muotri AR, Kuwabara T, Gage FH. Noncoding RNAs in the mammalian central nervous system. Annu Rev Neurosci. 2006;29:77–103. doi: 10.1146/annurev.neuro.29.051605.112839. [DOI] [PubMed] [Google Scholar]
- Carthew RW, Sontheimer EJ. Origins and mechanisms of miRNAs and siRNAs. Cell. 2009;136(4):642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng HY, Papp JW, Varlamova O, Dziema H, Russell B, Curfman JP, Nakazawa T, Shimizu K, Okamura H, Impey S, Obrietan K. microRNA modulation of circadian-clock period and entrainment. Neuron. 2007;54(5):813–829. doi: 10.1016/j.neuron.2007.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogswell JP, Ward J, Taylor IA, Waters M, Shi Y, Cannon B, Kelnar K, Kemppainen J, Brown D, Chen C, Prinjha RK, Richardson JC, Saunders AM, Roses AD, Richards CA. Identification of miRNA changes in Alzheimer’s disease brain and CSF yields putative biomarkers and insights into disease pathways. J Alzheimers Dis. 2008;14(1):27–41. doi: 10.3233/jad-2008-14103. [DOI] [PubMed] [Google Scholar]
- Cohen JE, Lee PR, Chen S, Li W, Fields RD. MicroRNA regulation of homeostatic synaptic plasticity. Proc Natl Acad Sci USA. 2011;108(28):11650–11655. doi: 10.1073/pnas.1017576108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo PJ, Brightwell JJ, Countryman RA. Cognitive strategy-specific increases in phosphorylated cAMP response element-binding protein and c-Fos in the hippocampus and dorsal striatum. J Neurosci. 2003;23(8):3547–3554. doi: 10.1523/JNEUROSCI.23-08-03547.2003. (pii: 23/8/3547) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croll SD, Suri C, Compton DL, Simmons MV, Yancopoulos GD, Lindsay RM, Wiegand SJ, Rudge JS, Scharfman HE. Brain-derived neurotrophic factor transgenic mice exhibit passive avoidance deficits, increased seizure severity and in vitro hyperexcitability in the hippocampus and entorhinal cortex. Neuroscience. 1999;93(4):1491–1506. doi: 10.1016/s0306-4522(99)00296-1. (pii: S0306-4522(99)00296-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuellar TL, Davis TH, Nelson PT, Loeb GB, Harfe BD, Ullian E, McManus MT. Dicer loss in striatal neurons produces behavioral and neuroanatomical phenotypes in the absence of neurodegeneration. Proc Natl Acad Sci USA. 2008;105(14):5614–5619. doi: 10.1073/pnas.0801689105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A, Huntzinger E, Izaurralde E. Getting to the root of miRNA-mediated gene silencing. Cell. 2008;132(1):9–14. doi: 10.1016/j.cell.2007.12.024. [DOI] [PubMed] [Google Scholar]
- Feng G, Mellor RH, Bernstein M, Keller-Peck C, Nguyen QT, Wallace M, Nerbonne JM, Lichtman JW, Sanes JR. Imaging neuronal subsets in transgenic mice expressing multiple spectral variants of GFP. Neuron. 2000;28(1):41–51. doi: 10.1016/s0896-6273(00)00084-2. (pii: S0896-6273(00)00084-2) [DOI] [PubMed] [Google Scholar]
- Fiore R, Khudayberdiev S, Christensen M, Siegel G, Flavell SW, Kim TK, Greenberg ME, Schratt G. Mef2-mediated transcription of the miR379-410 cluster regulates activity-dependent dendritogenesis by fine-tuning Pumilio2 protein levels. EMBO J. 2009;28(6):697–710. doi: 10.1038/emboj.2009.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Wang WY, Mao YW, Gräff J, Guan JS, Pan L, Mak G, Kim D, Su SC, Tsai LH. A novel pathway regulates memory and plasticity via SIRT1 and miR-134. Nature. 2010;466(7310):1105–1109. doi: 10.1038/nature09271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldez AJ, Cinalli RM, Glasner ME, Enright AJ, Thomson JM, Baskerville S, Hammond SM, Bartel DP, Schier AF. MicroRNAs regulate brain morphogenesis in zebrafish. Science. 2005;308(5723):833–838. doi: 10.1126/science.1109020. [DOI] [PubMed] [Google Scholar]
- Goodman RH, Smolik S. CBP/p300 in cell growth, transformation, and development. Genes Dev. 2000;14(13):1553–1577. [PubMed] [Google Scholar]
- Hall J, Thomas KL, Everitt BJ. Fear memory retrieval induces CREB phosphorylation and Fos expression within the amygdala. Eur J Neurosci. 2001;13(7):1453–1458. doi: 10.1046/j.0953-816x.2001.01531.x. (pii: ejn1531) [DOI] [PubMed] [Google Scholar]
- Hansen JC, Ghosh RP, Woodcock CL. Binding of the Rett syndrome protein, MeCP2, to methylated and unmethylated DNA and chromatin. IUBMB Life. 2010a;62(10):732–738. doi: 10.1002/iub.386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen KF, Sakamoto K, Wayman GA, Impey S, Obrietan K. Transgenic miR-132 alters neuronal spine density and impairs novel object recognition memory. PLoS One. 2010b;5(11):e15497. doi: 10.1371/journal.pone.0015497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen KF, Sakamoto K, Obrietan K. MicroRNAs: a potential interface between the circadian clock and human health. Genome Med. 2011;3(2):10. doi: 10.1186/gm224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutsler JJ, Zhang H. Increased dendritic spine densities on cortical projection neurons in autism spectrum disorders. Brain Res. 2010;1309:83–94. doi: 10.1016/j.brainres.2009.09.120. [DOI] [PubMed] [Google Scholar]
- Im HI, Hollander JA, Bali P, Kenny PJ. MeCP2 controls BDNF expression and cocaine intake through homeostatic interactions with microRNA-212. Nat Neurosci. 2010;13(9):1120–1127. doi: 10.1038/nn.2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Impey S, Davare M, Lasiek A, Fortin D, Ando H, Varlamova O, Obrietan K, Soderling TR, Goodman RH, Wayman GA. An activity-induced microRNA controls dendritic spine formation by regulating Rac1-PAK signaling. Mol Cell Neurosci. 2010;43(1):146–156. doi: 10.1016/j.mcn.2009.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Impey S, Smith DM, Obrietan K, Donahue R, Wade C, Storm DR. Stimulation of cAMP response element (CRE)-mediated transcription during contextual learning. Nat Neurosci. 1998;1(7):595–601. doi: 10.1038/2830. [DOI] [PubMed] [Google Scholar]
- Irwin SA, Patel B, Idupulapati M, Harris JB, Crisostomo RA, Larsen BP, Kooy F, Willems PJ, Cras P, Kozlowski PB, Swain RA, Weiler IJ, Greenough WT. Abnormal dendritic spine characteristics in the temporal and visual cortices of patients with fragile-X syndrome: a quantitative examination. Am J Med Genet. 2001;98(2):161–167. doi: 10.1002/1096-8628(20010115)98:2<161::aid-ajmg1025>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- Jimenez-Mateos EM, Bray I, Sanz-Rodriguez A, Engel T, McKiernan RC, Mouri G, Tanaka K, Sano T, Saugstad JA, Simon RP, Stallings RL, Henshall DC. miRNA expression profile after status epilepticus and hippocampal neuroprotection by targeting miR-132. Am J Pathol. 2011 doi: 10.1016/j.ajpath.2011.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaitsuka T, Li ST, Nakamura K, Takao K, Miyakawa T, Matsushita M. Forebrain-specific constitutively active CaMKKα transgenic mice show deficits in hippocampus-dependent long-term memory. Neurobiol Learn Mem. 2011;96(2):238–247. doi: 10.1016/j.nlm.2011.04.017. [DOI] [PubMed] [Google Scholar]
- Khudayberdiev S, Fiore R, Schratt G. MicroRNA as modulators of neuronal responses. Commun Integr Biol. 2009;2(5):411–413. doi: 10.4161/cib.2.5.8834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Krichevsky A, Grad Y, Hayes GD, Kosik KS, Church GM, Ruvkun G. Identification of many microRNAs that copurify with polyribosomes in mammalian neurons. Proc Natl Acad Sci USA. 2004b;101(1):360–365. doi: 10.1073/pnas.2333854100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim IH, Park SK, Sun W, Kang Y, Kim HT, Kim H. Spatial learning enhances the expression of inositol 1,4,5-trisphosphate 3-kinase A in the hippocampal formation of rat. Brain Res Mol Brain Res. 2004a;124(1):12–19. doi: 10.1016/j.molbrainres.2003.12.016. [DOI] [PubMed] [Google Scholar]
- Kim AH, Reimers M, Maher B, Williamson V, McMichael O, McClay JL, van den Oord EJ, Riley BP, Kendler KS, Vladimirov VI. MicroRNA expression profiling in the prefrontal cortex of individuals affected with schizophrenia and bipolar disorders. Schizophr Res. 2010;124(1–3):183–191. doi: 10.1016/j.schres.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleim JA, Swain RA, Armstrong KA, Napper RM, Jones TA, Greenough WT. Selective synaptic plasticity within the cerebellar cortex following complex motor skill learning. Neurobiol Learn Mem. 1998;69(3):274–289. doi: 10.1006/nlme.1998.3827. [DOI] [PubMed] [Google Scholar]
- Klein ME, Lioy DT, Ma L, Impey S, Mandel G, Goodman RH. Homeostatic regulation of MeCP2 expression by a CREB-induced microRNA. Nat Neurosci. 2007;10(12):1513–1514. doi: 10.1038/nn2010. [DOI] [PubMed] [Google Scholar]
- Konopka W, Kiryk A, Novak M, Herwerth M, Parkitna JR, Wawrzyniak M, Kowarsch A, Michaluk P, Dzwonek J, Arnsperger T, Wilczynski G, Merkenschlager M, Theis FJ, Köhr G, Kaczmarek L, Schütz G. MicroRNA loss enhances learning and memory in mice. J Neurosci. 2010;30(44):14835–14842. doi: 10.1523/JNEUROSCI.3030-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopka W, Schütz G, Kaczmarek L. The microRNA contribution to learning and memory. Neuroscientist. 2011 doi: 10.1177/1073858411411721. [DOI] [PubMed] [Google Scholar]
- Kosik KS. The neuronal microRNA system. Nat Rev Neurosci. 2006;7(12):911–920. doi: 10.1038/nrn2037. [DOI] [PubMed] [Google Scholar]
- Lagos D, Pollara G, Henderson S, Gratrix F, Fabani M, Milne RS, Gotch F, Boshoff C. miR-132 regulates antiviral innate immunity through suppression of the p300 transcriptional coactivator. Nat Cell Biol. 2010;12(5):513–519. doi: 10.1038/ncb2054. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12(9):735–739. doi: 10.1016/s0960-9822(02)00809-6. (pii: S0960982202008096) [DOI] [PubMed] [Google Scholar]
- Lambert TJ, Storm DR, Sullivan JM. MicroRNA132 modulates short-term synaptic plasticity but not basal release probability in hippocampal neurons. PLoS One. 2010;5(12):e15182. doi: 10.1371/journal.pone.0015182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leuner B, Falduto J, Shors TJ. Associative memory formation increases the observation of dendritic spines in the hippocampus. J Neurosci. 2003;23(2):659–665. doi: 10.1523/JNEUROSCI.23-02-00659.2003. (pii: 23/2/659) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433(7027):769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- Lin Q, Wei W, Coelho CM, Li X, Baker-Andresen D, Dudley K, Ratnu VS, Boskovic Z, Kobor MS, Sun YE, Bredy TW. The brain-specific microRNA miR-128b regulates the formation of fear-extinction memory. Nat Neurosci. 2011;14(9):1115–1117. doi: 10.1038/nn.2891. [DOI] [PubMed] [Google Scholar]
- Lopez de Armentia M, Jancic D, Olivares R, Alarcon JM, Kandel ER, Barco A. cAMP response element-binding protein-mediated gene expression increases the intrinsic excitability of CA1 pyramidal neurons. J Neurosci. 2007;27(50):13909–13918. doi: 10.1523/JNEUROSCI.3850-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magill ST, Cambronne XA, Luikart BW, Lioy DT, Leighton BH, Westbrook GL, Mandel G, Goodman RH. microRNA-132 regulates dendritic growth and arborization of newborn neurons in the adult hippocampus. Proc Natl Acad Sci USA. 2010;107(47):20382–20387. doi: 10.1073/pnas.1015691107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayford M, Bach ME, Huang YY, Wang L, Hawkins RD, Kandel ER. Control of memory formation through regulated expression of a CaMKII transgene. Science. 1996;274(5293):1678–1683. doi: 10.1126/science.274.5293.1678. [DOI] [PubMed] [Google Scholar]
- Mellios N, Sugihara H, Castro J, Banerjee A, Le C, Kumar A, Crawford B, Strathmann J, Tropea D, Levine SS, Edbauer D, Sur M. miR-132, an experience-dependent microRNA, is essential for visual cortex plasticity. Nat Neurosci. 2011;14(10):1240–1242. doi: 10.1038/nn.2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migaud M, Charlesworth P, Dempster M, Webster LC, Watabe AM, Makhinson M, He Y, Ramsay MF, Morris RG, Morrison JH, O’Dell TJ, Grant SG. Enhanced long-term potentiation and impaired learning in mice with mutant postsynaptic density-95 protein. Nature. 1998;396(6710):433–439. doi: 10.1038/24790. [DOI] [PubMed] [Google Scholar]
- Mizuno M, Yamada K, Maekawa N, Saito K, Seishima M, Nabeshima T. CREB phosphorylation as a molecular marker of memory processing in the hippocampus for spatial learning. Behav Brain Res. 2002;133(2):135–141. doi: 10.1016/s0166-4328(01)00470-3. (pii: S0166432801004703) [DOI] [PubMed] [Google Scholar]
- Moon SY, Zang H, Zheng Y. Characterization of a brain-specific Rho GTPase-activating protein, p200RhoGAP. J Biol Chem. 2003;278(6):4151–4159. doi: 10.1074/jbc.M207789200. [DOI] [PubMed] [Google Scholar]
- Morris R. Thy-1 in developing nervous tissue. Dev Neurosci. 1985;7(3):133–160. doi: 10.1159/000112283. [DOI] [PubMed] [Google Scholar]
- Nakayama AY, Harms MB, Luo L. Small GTPases Rac and Rho in the maintenance of dendritic spines and branches in hippocampal pyramidal neurons. J Neurosci. 2000;20(14):5329–5338. doi: 10.1523/JNEUROSCI.20-14-05329.2000. (pii: 20/14/5329) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nudelman AS, DiRocco DP, Lambert TJ, Garelick MG, Le J, Nathanson NM, Storm DR. Neuronal activity rapidly induces transcription of the CREB-regulated microRNA-132, in vivo. Hippocampus. 2010;20(4):492–498. doi: 10.1002/hipo.20646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuovo GJ. In situ detection of microRNAs in paraffin embedded, formalin fixed tissues and the co-localization of their putative targets. Methods. 2010;52(4):307–315. doi: 10.1016/j.ymeth.2010.08.009. [DOI] [PubMed] [Google Scholar]
- Obernosterer G, Martinez J, Alenius M. Locked nucleic acid-based in situ detection of microRNAs in mouse tissue sections. Nat Protoc. 2007;2(6):1508–1514. doi: 10.1038/nprot.2007.153. [DOI] [PubMed] [Google Scholar]
- Packer AN, Xing Y, Harper SQ, Jones L, Davidson BL. The bifunctional microRNA miR-9/miR-9* regulates REST and CoREST and is downregulated in Huntington’s disease. J Neurosci. 2008;28(53):14341–14346. doi: 10.1523/JNEUROSCI.2390-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietropaolo S, Paterna JC, Büeler H, Feldon J, Yee BK. Bidirectional changes in water-maze learning following recombinant adenovirus-associated viral vector (rAAV)-mediated brain-derived neurotrophic factor expression in the rat hippocampus. Behav Pharmacol. 2007;18(5–6):533–547. doi: 10.1097/FBP.0b013e3282da0bf6. [DOI] [PubMed] [Google Scholar]
- Pineda VV, Athos JI, Wang H, Celver J, Ippolito D, Boulay G, Birnbaumer L, Storm DR. Removal of G(ialpha1) constraints on adenylyl cyclase in the hippocampus enhances LTP and impairs memory formation. Neuron. 2004;41(1):153–163. doi: 10.1016/s0896-6273(03)00813-4. (pii: S0896627303008134) [DOI] [PubMed] [Google Scholar]
- Pollak DD, Herkner K, Hoeger H, Lubec G. Behavioral testing upregulates pCaMKII, BDNF, PSD-95 and egr-1 in hippocampus of FVB/N mice. Behav Brain Res. 2005;163(1):128–135. doi: 10.1016/j.bbr.2005.04.010. [DOI] [PubMed] [Google Scholar]
- Porte Y, Buhot MC, Mons NE. Spatial memory in the Morris water maze and activation of cyclic AMP response element-binding (CREB) protein within the mouse hippocampus. Learn Mem. 2008;15(12):885–894. doi: 10.1101/lm.1094208. [DOI] [PubMed] [Google Scholar]
- Remenyi J, Hunter CJ, Cole C, Ando H, Impey S, Monk CE, Martin KJ, Barton GJ, Hutvagner G, Arthur JS. Regulation of the miR-212/132 locus by MSK1 and CREB in response to neurotrophins. Biochem J. 2010;428(2):281–291. doi: 10.1042/BJ20100024. [DOI] [PubMed] [Google Scholar]
- Sakamoto K, Karelina K, Obrietan K. CREB: a multifaceted regulator of neuronal plasticity and protection. J Neurochem. 2011;116(1):1–9. doi: 10.1111/j.1471-4159.2010.07080.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanuki R, Onishi A, Koike C, Muramatsu R, Watanabe S, Muranishi Y, Irie S, Uneo S, Koyasu T, Matsui R, Chérasse Y, Urade Y, Watanabe D, Kondo M, Yamashita T, Furukawa T. miR-124a is required for hippocampal axogenesis and retinal cone survival through Lhx2 suppression. Nat Neurosci. 2011;14(9):1125–1134. doi: 10.1038/nn.2897. [DOI] [PubMed] [Google Scholar]
- Schmittgen TD, Lee EJ, Jiang J, Sarkar A, Yang L, Elton TS, Chen C. Real-time PCR quantification of precursor and mature microRNA. Methods. 2008;44(1):31–38. doi: 10.1016/j.ymeth.2007.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schratt GM, Tuebing F, Nigh EA, Kane CG, Sabatini ME, Kiebler M, Greenberg ME. A brain-specific microRNA regulates dendritic spine development. Nature. 2006;439(7074):283–289. doi: 10.1038/nature04367. [DOI] [PubMed] [Google Scholar]
- Sterner DE, Berger SL. Acetylation of histones and transcription-related factors. Microbiol Mol Biol Rev. 2000;64(2):435–459. doi: 10.1128/mmbr.64.2.435-459.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunyer B, Lubec G, Höger H, Patil S. Barnes maze, a useful task to assess spatial reference memory in the mice. 2007 [Google Scholar]
- Suzuki A, Fukushima H, Mukawa T, Toyoda H, Wu LJ, Zhao MG, Xu H, Shang Y, Endoh K, Iwamoto T, Mamiya N, Okano E, Hasegawa S, Mercaldo V, Zhang Y, Maeda R, Ohta M, Josselyn SA, Zhuo M, Kida S. Upregulation of CREB-mediated transcription enhances both short- and long-term memory. J Neurosci. 2011;31(24):8786–8802. doi: 10.1523/JNEUROSCI.3257-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tognini P, Putignano E, Coatti A, Pizzorusso T. Experience-dependent expression of miR-132 regulates ocular dominance plasticity. Nat Neurosci. 2011;14(10):1237–1239. doi: 10.1038/nn.2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trommald M, Hulleberg G, Andersen P. Long-term potentiation is associated with new excitatory spine synapses on rat dentate granule cells. Learn Mem. 1996;3(2–3):218–228. doi: 10.1101/lm.3.2-3.218. [DOI] [PubMed] [Google Scholar]
- Van Aelst L, Cline HT. Rho GTPases and activity-dependent dendrite development. Curr Opin Neurobiol. 2004;14(3):297–304. doi: 10.1016/j.conb.2004.05.012. [DOI] [PubMed] [Google Scholar]
- Vázquez SI, Vázquez A, Peña de Ortiz S. Different hippocampal activity profiles for PKA and PKC in spatial discrimination learning. Behav Neurosci. 2000;114(6):1109–1118. doi: 10.1037//0735-7044.114.6.1109. [DOI] [PubMed] [Google Scholar]
- Viosca J, Malleret G, Bourtchouladze R, Benito E, Vronskava S, Kandel ER, Barco A. Chronic enhancement of CREB activity in the hippocampus interferes with the retrieval of spatial information. Learn Mem. 2009;16(3):198–209. doi: 10.1101/lm.1220309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vo N, Goodman RH. CREB-binding protein and p300 in transcriptional regulation. J Biol Chem. 2001;276(17):13505–13508. doi: 10.1074/jbc.R000025200. [DOI] [PubMed] [Google Scholar]
- Vo N, Klein ME, Varlamova O, Keller DM, Yamamoto T, Goodman RH, Impey S. A cAMP-response element binding protein-induced microRNA regulates neuronal morphogenesis. Proc Natl Acad Sci USA. 2005;102(45):16426–16431. doi: 10.1073/pnas.0508448102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuksic M, Del Turco D, Bas Orth C, Burbach GJ, Feng G, Müller CM, Schwarzacher SW, Deller T. 3D-reconstruction and functional properties of GFP-positive and GFP-negative granule cells in the fascia dentata of the Thy1-GFP mouse. Hippocampus. 2008;18(4):364–375. doi: 10.1002/hipo.20398. [DOI] [PubMed] [Google Scholar]
- Wayman GA, Davare M, Ando H, Fortin D, Varlamova O, Cheng HY, Marks D, Obrietan K, Soderling TR, Goodman RH, Impey S. An activity-regulated microRNA controls dendritic plasticity by down-regulating p250GAP. Proc Natl Acad Sci USA. 2008;105(26):9093–9098. doi: 10.1073/pnas.0803072105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wibrand K, Panja D, Tiron A, Ofte ML, Skaftnesmo KO, Lee CS, Pena JT, Tuschl T, Bramham CR. Differential regulation of mature and precursor microRNA expression by NMDA and metabotropic glutamate receptor activation during LTP in the adult dentate gyrus in vivo. Eur J Neurosci. 2010;31(4):636–645. doi: 10.1111/j.1460-9568.2010.07112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.