Abstract
Understanding the signals that guide neuronal development and direct formation of axons, dendrites, and synapses during wiring of the brain is a fundamental challenge of developmental neuroscience. Discovering how local signals shape developing neurons has been impeded by the inability of conventional culture methods to interrogate micro-environments of complex neuronal cytoarchitectures, where different sub-domains encounter distinct chemical, physical, and fluidic features. Micro-fabrication techniques are enabling the creation of micro-environments tailored to neuronal structures and sub-domains, with unprecedented access and control. The design, fabrication, and properties of microfluidic devices offer significant advantages for addressing unresolved issues of neuronal development. These high-resolution approaches are poised to contribute new insights into mechanisms for restoring neuronal function and connectivity compromised by injury, stress, and neurodegeneration.
Keywords: Microfluidic devices, neuronal polarity, developmental neuroscience, axon guidance, dendrite, synapse
Introduction
The brain forms through a remarkable process of self-organization. Neuroblasts migrate and terminally differentiate into the neurons and glia of each brain region. They extend multiple protrusions, each of which encounters distinct, complex, and dynamic micro-environments. Emergence of neuronal polarity and synaptic specializations generates nascent networks along which flow the earliest signals of communication. Simple networks elaborate so that they sense, coordinate, and regulate the range of metabolic, endocrine, physiologic, and cognitive functions that enable complex behaviors and thoughts. Deciphering the mosaic of signals in the chemical and physical landscape that guide the development of each neuron and direct formation of its axon, dendrites, their synapses, and functional domains during this wiring process is a formidable task.
The mature neuron is elongate, highly branched, and forms thousands of synaptic connections with other neurons at extensions distant from the soma. Within the complex cytoarchitecture of the brain, neurons and their extensions are densely packed, filling all available space (Figure 1a, b)[1,2]; this impedes sub-cellular analysis in vivo. In dish cultures, neurons develop similar complex morphologies, albeit at lower densities and with relatively less oriented growth (Figure 1c). Consequently, neurites often reside in different chemical, physical, and fluidic micro-environments. Whereas in vivo and conventional cell culture approaches have achieved remarkable insights on axonal, dendritic, and synaptic development, discovering how local, sub-cellular signals of individual neurons influence differentiation and function has been elusive. Nevertheless, it is at the sub-cellular level that dynamic changes during development and throughout life determine information flow and processing (e.g., whether or not specific experiences form or recall memories). Restoring brain function impaired by damage, stress, or degeneration is especially difficult. Local cues and micro-environments that guide brain wiring during development are often ephemeral and may have been expressed distant from adult termini. Solutions to these issues require new approaches that enable local high-resolution analyses of the interplay between myriad extracellular signals and the intracellular responses that shape the developing brain.
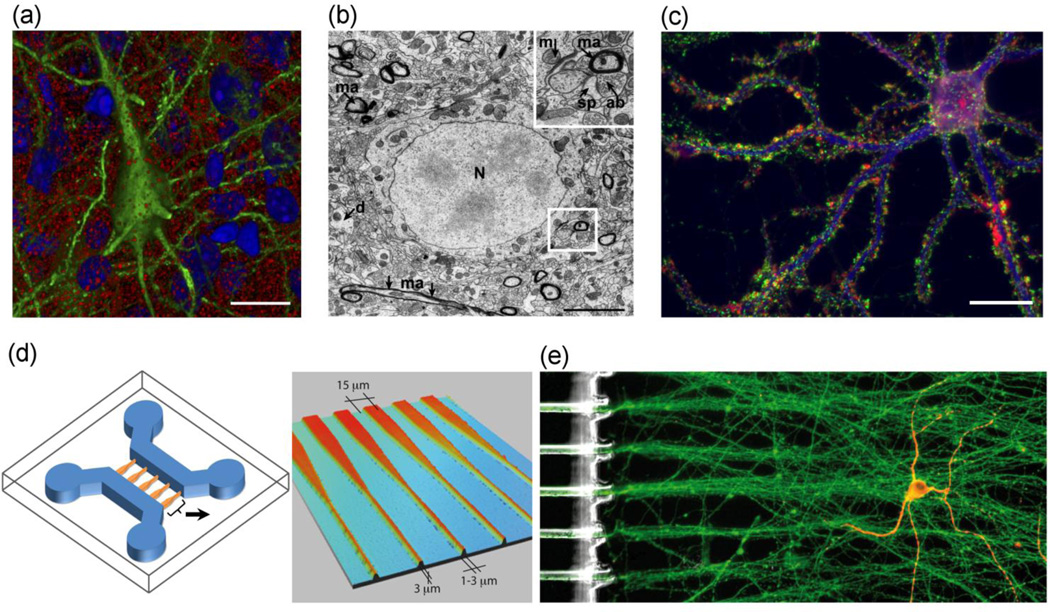
Figure 1. Cytoarchitectures of neurons in vivo compared with dish or microfluidic-device (µFD) cultures.
(a) The cytoarchitecture of brain tissue is remarkably complex, as seen in cell packing and interwoven processes of the cerebral cortex in situ imaged by array tomography. The three-dimensional space of a mouse cortical neuron (green) is surrounded by other cells (marked by DAPI-labeled nuclei, blue) and myriad presynaptic boutons (synapsin-labeled, red). Figure modified and used with permission.[2] Scale bar = 10 µm. (b) Electron micrograph of layer IV of the rat somatosensory cortex reveals densely packed structures with essentially no free space around the cell body (nucleus, N), myelinated axons (ma), axon bundles (ab), dendritic spines (sp), and dendrites (d); mitochrondria also are shown (m). Scale bar = 5 µm. Original image contributed by Graham Knott. (c) At low densities in a conventional dish culture, neurons develop complex cytoarchitectures and connections despite open space between neurites and cell bodies. Pre-synaptic terminals (synapsin, green) stud the dendritic arbor (MAP 2, blue) of a primary hippocampal neuron (33 days in vitro); rhodamine-phalloidin labels filamentous actin (red), scale bar = 20 µm. Modified and used with permission.[80] (d) A µFD fabricated to organize cellular connections and control the local fluidic micro-environment. Depicted is a two-channel µFD (left) with tapering interconnects (enlarged at right), which facilitate directional orientation of neurons for analyzing interactions. Neurons can be cultured in each channel; interactions can be restricted to and guided by the interconnecting tunnels (i.e., interconnects). Cortical neurons were seeded in one channel (left, blue) and striatal neurons in another channel (right, blue). Interactions between channels are restricted to the interconnects (orange, enlarged on right). Tapered interconnects funnel cortical axons toward the channel on the right, while restricting the reverse migration of afferent axons back through the interconnects. Reproduced and modified by permission of The Royal Society of Chemistry (RSC).[52] (e) Complex and elaborate architectures of neurons can be guided and organized using a µFD. A single striatal neuron (yellow, right channel) is innervated by organized axon bundles from cortical neurons (green) cultured in the adjacent channel and directed via the interconnecting tunnels. Striatal neurons immunolabeled for MAP2 (red), cortical neurons labeled for α-tubulin (green). Reproduced and modified by permission of the RSC.[52]
Microfluidic devices (µFDs) - cell-culture environments with channels of micrometer-scale dimensions containing nano-liter volumes - are addressing these needs. Interfacing engineering technologies with biological methodologies enables fabrication and application of microfluidic-based systems with new capabilities for maintaining and studying brain cells and circuits in stable micro-culture (Figure 1d, e). By using replica molding (Figure 2), environments approximating not only single cells, but even single neuronal processes, can be fabricated in the laboratory setting (Box 1). Advances in chemistry and materials science over the last decade have transformed and propelled the ability to control spatial and temporal signals within channels of the µFD and to enable co-cultures and compartmentalization for studying neuronal-glial interactions,[3,4] disease progression,[5,6] and repair of injury.[7–10] Exquisite spatial and temporal control of the cellular micro-environments offers significant advantages for studying neurons and their processes. In this review, we summarize and evaluate new perspectives on neuronal development at sub-cellular, cellular, and tissue levels resulting from the dissemination and adoption of µFDs.
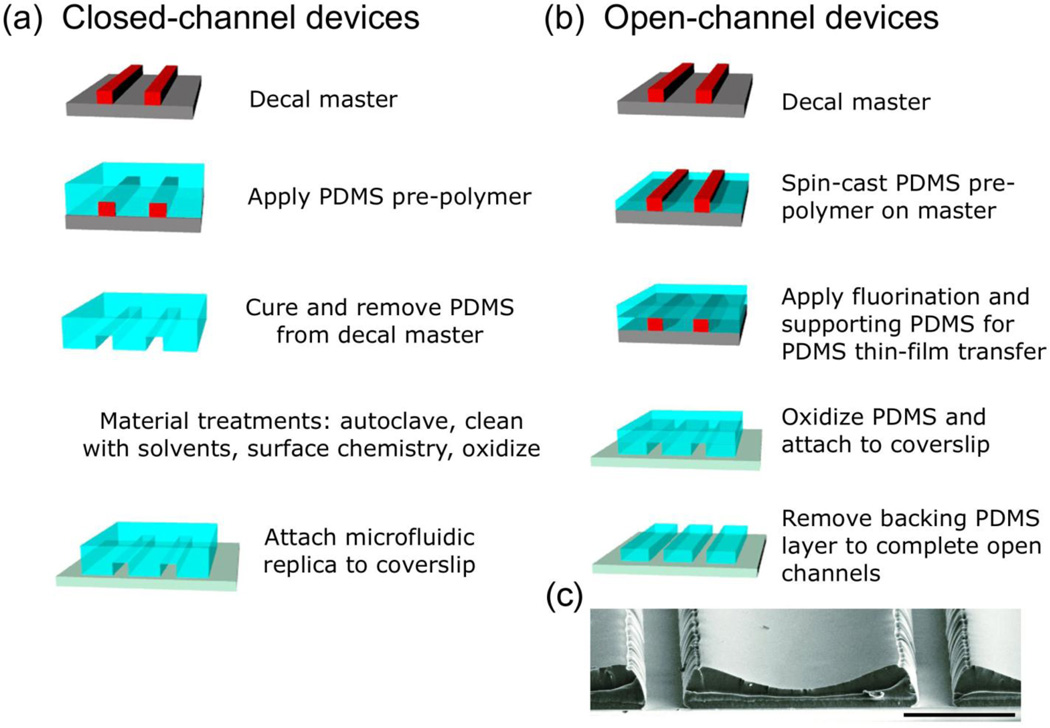
Figure 2. Replica molding enables fabrication of open- and close-channel µFDs.
Masters are made by patterning SU-8 photoresist on silicone, generating the decal, a three-dimensional negative of the final structure, and treating with a “no stick” layer to allow PDMS release.[81] (a) For closed-channel devices, bulk PDMS is poured onto the master, cured, cleaned, treated (if appropriate), and assembled onto clean coverslips. Channels or coverslips may be coated with substrate guidance cue(s), e.g., poly-lysine, and rinsed with media prior to cell culture. (b) For one type of open-channel device, PDMS is spin-cast onto the master and cured, fluorinated, and covered with bulk PDMS to support the PDMS thin-film decal during transfer. The multilayer replica is removed, oxidized, and covalently attached to a clean coverslip, after which bulk PDMS is removed. (c) Scanning electron micrograph of open-channel µFD. Scale bar = 100 µm. Reproduced and modified by permission of The Royal Society of Chemistry (RSC).[64]
BOX 1. Advantages of microfluidic devices for studying neuronal development.
µFDs offer unparalleled spatial, chemical, and temporal control of the micro-environments that shape differentiating neurons, enabling neuronal development to be investigated in new ways. From simple to complex, µFDs can be fabricated in the lab, are available through commercial sources for “off the shelf” use, or they can be designed and manufactured for specific research applications. Some resources for µFDs include: KNI Foundry at CalTech, Micralyne, Microfluidic ChipShop, Microliquid, Micronit Microfluidics, Stanford Microfluidics Foundry, and Xona Microfluidics.
The main advantages of microfluidic devices include:
High reproducibility: Small variations on the micro-environmental scale could cause significant variables at the sub-cellular level; therefore, highly reproducibility is essential. Replica molding enables the fabrication of highly reproducible, disposable µFD of moldable gels and polymers in a hood or on a benchtop (Figure 2).[84] PDMS, an advanced silicone material, is the most widely used fabrication material; it can be molded easily to form simple, complex, or multilayered channel systems. A large number of PDMS devices can be made from a single master, reducing the time and cost of fabrication.
Easy assembly: PDMS-based µFD are easily sterilized through conventional means and can be reversibly, or irreversibly, sealed to glass, polystyrene, or silicone wafers to produce a 3D microenvironment.[64]
Fluidic control: Controlling the fluidic environment is critical to establishing the local chemical and fluidic features of sub-cellular domains. µFDs enable chemical substrate features and the fluidic surround to be controlled with spatial and temporal dynamics and precision.[32,49]
Material versatility: Probing specific developmental issues may benefit from fabricating µFDs from different moldable polymers that possess distinct physical and/or chemical features. A variety of moldable polymers can be used.[84,85] Choice of material is influenced by fabrication requirements and experimental design.
Design flexibility: Channel design is established using photolithography, which uses light to selectively transfer the design pattern via a photomask to a light-sensitive chemical photoresist on a substrate. This process either engraves the pattern or enables deposition of material in the pattern upon the substrate beneath the photoresist. This permits micro-fabrication of a range of designs from simple channels to highly complex fluid networks, and with resolution down to ~500 nm. This design flexibility renders µFDs broadly applicable to specific problems in neuroscience, enabling the study of subcellular regions, individual cells, neuron pairs, and networks of neurons in highly controllable ways.[35,64,86]
Experimental feasibility: Intrinsic properties of µFDs make them amenable for cell culture, imaging, and biochemical analysis:[60,63,64,87] local fluid exchange, optical transparency and imaging compatibility, high gas permeation, low water permeability, thermal stability, and the ability to physically confine neurons to control connectivity.[88–90] Substrate properties that can be altered by design include hydrophobicity, stiffness, topography, functionalization with bound biomolecules, and substrate patterning.
Neuronal differentiation and polarity
Neurons developing in vitro undergo sequential differentiation, polarization, and specification of neurites as axon or dendrites, recapitulating developmental stages in vivo.[11] In the absence of patterned instructive cues in dish cultures, processes tend to be stellate (Figure 1c) and overlapping. µFDs enable significantly greater control over neuronal development in vitro. The chemical and topographic disposition of developmental cues with micropatterning techniques can induce pre-determined morphologies.[12–18] Attractive and/or repulsive cues can be patterned with highly controlled geometries. Cues can be patterned singly, within close proximity, in combination with other substrates, or in direct apposition to them (Figure 3a–b). Physical cues on the scale of individual neurites can be fabricated to control the navigation of neurites.[19] Presenting spatially defined cues permits the influence of local extracellular signals on neuronal differentiation to be resolved.
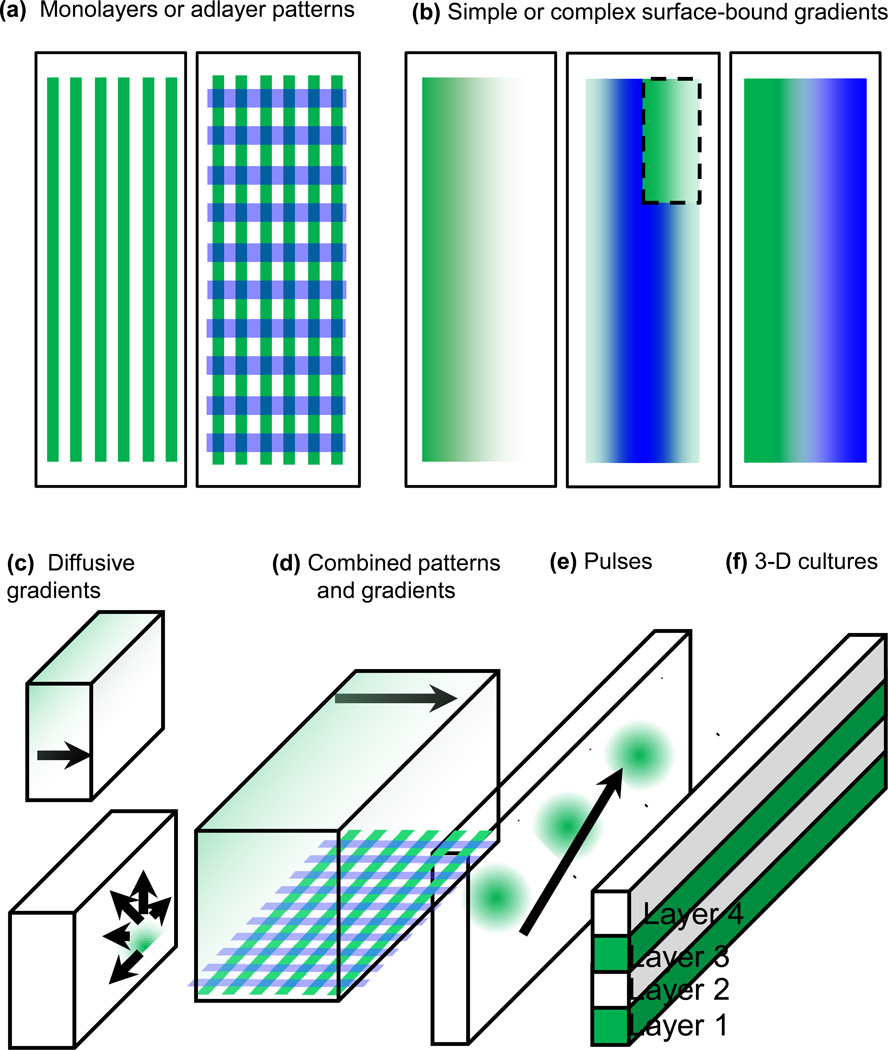
Figure 3. µFDs enable a high degree of spatial and temporal control of neuronal micro-environments.
(a) Binary patterns of substrate molecules can be defined with a high degree of spatial resolution to generate monolayer stripes of one or two cues patterned in apposition (left). Adlayers of binary protein patterns can also be formed as culture substrates (e.g., laminin, polylysine, neurotrophins) (right). (b) Through laminar flow, surface-bound gradients of guidance cues can be patterned onto planar culture substrates. Gradients can be of single (left) or multiple cues aligned onto each other (middle), or the gradients can be laid down with opposing slopes of increasing concentrations (right).[43] Inset is a cut-away of the top layer of the two-layer pattern. (c) Diffusive gradients established via different three-dimensional (3D) fluidic channel domains. Upper left: The arrow signifies the direction of diffusion from high to low concentration; the source can be from adjacent laminar-flow streams or gels.[43,48,82] Lower right: A focal source of a chemical factor can be introduced via a small inlet channel or ‘jetting’ stream.[83] (d) Fluid-phase and surface-bound cues can be used simultaneously. (e) Chemical factors can be introduced into channels with pulsatile temporal control to deliver dynamic patterns of chemical agents to cells and sub-regions within.[49] (f) 3D gel structures can be fabricated via µFDs. Layers can alternately incorporate neurons, then diffusive growth factors, or various combinations for axonal growth toward a lateral or vertical target region.[67]
The stripe assay, also known as the Bonhoeffer stripe assay, has long been used in dish cultures to interrogate the behavior of the axons of tissue explants in response to neutral vs. known or potentially novel guidance cues. Application of the stripe assay demonstrated that the first hippocampal neurite to encounter a locally presented growth-promoting molecule [laminin (LN) or neuron-glia cell adhesion molecule (NgCAM)], is specified immediately to become the axon.[14] A recent modification using sole-source silicone matrices in a complex fabrication protocol extended the stripe resolution to 100 µm.[20] µFDs have been used to refine stripe patterns for subcellular analyses. Stripes can be laid down more easily, with greater line resolution (< 10 µm) and sharper edges, and are more versatile for multiple chemical depositions compared to the conventional stripe assay protocol. The µFD-based approach identified signals that induce axon initiation during the polarization process.[21] When one neurite of a hippocampal neuron is exposed selectively to a high resolution stripe of substrate bearing brain-derived neurotrophic factor (BDNF), the local encounter initiates a cascade of events, activating cAMP signaling, directional elongation, and axonal differentiation. Asymmetric, reciprocal regulation of cAMP vs. cGMP evokes axon or dendritic formation, respectively.[22] Thus, upstream effectors that regulate cAMP/cGMP commit naive neurites to their developmental path. Presenting the developing axon with spatially and chemically distinct substrates in a µFD-based stripe assay revealed that the phosphorylation state of the E3 ubiquitin ligase, Smurf1, determines substrate preferences.[23] This approach identified transforming growth factor β (TGF-β) as an extrinsic cue that induces neuronal polarity during patterning of neural circuits;[24] how TGF-β signal transduction engages intrinsic regulators, such as cAMP, is yet to be determined.
Advances in fabrication methods, such as vacuum soft lithography, enable the patterning of multiple geometrically-defined instructive cues in a single procedure. They are refining the stripe assay by enhancing chemical and spatial complexity. [25] Such precise patterning methods offer a high degree of control in guiding formation of neuronal networks with pre-determined connectivity.[25,26] Intricate large-scale patterns, such as substrate chemicals in interconnected or juxtaposed shapes, letters, or designs, can be printed so as to design the arrangement of neurons and their connections.[27] We envision that advancements in patterning techniques will permit real-time study of neuronal signaling pathways, including exquisite resolution of sub-cellular processes, neuronal network communications, neural repair mechanisms, and the construction of functionally integrated neuronal systems.
Axon guidance and differentiation
After polarity has been established, the axon begins a phase of rapid growth. While traversing its developmental course, the axon passes many cells, extra-cellular matrices, fluid microdomains, and structures, such as capillaries, before arriving at its target. What cues guide the course of each axon? Studies in culture dishes effectively used the stripe assay to determine axon preference for single substrate molecules; ephrin/Eph receptor-signaling identified in this way was instrumental in elucidating axon guidance in vivo.[28] µFDs enable great flexibility in designing microenvironments to evaluate responsiveness, sensitivity, and dynamics of population or individual axonal growth cones toward single or multiple guidance cues.[26,29–31] Furthermore, spatial control of neuronal morphology by µFDs can be exploited to selectively isolate axons, and then assess their behavior optically, as well as determine their molecular content (e.g., proteomic profiles or axonal mRNAs).[32–34]
Patterned biochemical substrates
Similar to the stripe assay, patterned lines of binary substrates can align axons within a population. In µFDs, this substrate pattern forms parallel fibers of axonal bundles without axonal intersections. This avoids potential confounds in conventional, un-patterned dish cultures where many axons overlap. These populations of axons can be used to study organized axotomy, axonal transport, and analysis of recovery.[7,8,35] Such patterned neuronal cultures also are useful for titrating substrate concentrations that guide the growth and behavior of single growth cones, and for dissecting axonal segments for molecular analysis.[36,37]
To elucidate the mechanism of a putative axon guidance cue, a compartmentalized microfluidic chamber can be positioned onto micro-patterned stripes designed to expose subcellular regions of neurons to defined substrates. For example, micro-patterned stripes were used to resolve mechanisms of axonal avoidance of semaphorins (Sema3f).[38] Materials can be applied throughout the µFD or only in sub-compartments enabling parsing of global vs. local signaling mechanisms. Somata can be restricted to a channel used for seeding cultures (Fig. 1d). Axons grow down interconnecting tunnels to an adjacent, fluidically isolated channel.[10,39] By using stripes of N-cadherin or laminin (LN) within the channels, patterned growth substrates are restricted to axons. Selectively inhibiting fibroblast growth factor receptors (FGFR) of fluidically isolated axons reduced the rate, but not the direction, of axonal growth.[39] FGFR inhibition restricted to the soma had no effect on axonal growth rate or navigation. Thus, behavior of the growth cone and morphology of the axon is mediated locally, by FGFRs on the axon or growth cone, rather than soma.
Factors accelerating axon elongation have been probed selectively using µFDs with complex substrates in an effort to identify common targets that mediate axon growth inhibition and thus develop interventions that enhance the axon regenerative capacity of mature neurons. Dorsal root ganglia neurons were cultured so that elongating axons isolated in channels were exposed to permissive–inhibitory substrate borders, including chondroitin sulfate proteoglycans (CSPG), an inhibitor of axon elongation via non-muscle myosin II (NMII). Local application of blebbistatin, which is thought to increase microtubule extension toward the growth cone leading edge, markedly accelerated axon growth over the inhibitory CSPG substrate.[40] Because mature central neurons fail to regenerate axons after injury due in part to the diminished intrinsic axonal growth capacity, non-muscle myosin II may be a target for modulating regenerative capacity. These findings demonstrate the power of spatial control offered by µFDs to reveal local regulatory mechanisms for guidance, growth, and inhibition of axons.[41] They enable roles of specific substrates and regulatory mechanisms to be resolved locally, within sub-cellular domains of axons.
Biochemical gradients
In addition to patterned lines of substrate, developing neurons respond to biochemical gradients. External gradients, which may be both long-range diffusive cues and short-range contact cues,[42] guide axonal growth and trajectory. Patterning of neural circuits is determined by cues that are non-uniform, transient, and changing during critical developmental periods. Compared to binary patterns, which provide sharp contrasts in surface cues, gradients offer subtle, yet meaningful contrasts of environmental signals that promote and/or repel axon growth.[29,43] µFDs permit patterning of micro-scale substrate and diffusive gradients far more controlled and refined than is possible in dishes and, thus, achieve a scale and complexity that more closely approximates nature.[44]
Gradients of potentially instructive molecules can be controlled through µFD-based laminar flow and presented either tethered to the substrate or as diffuse cues.[29,43–46] Substrate-tethered gradient cues can be deposited in single or multiple layers using laminar flow (Figure 3a, b). When early post-natal rat hippocampal neurons were cultured on laminin substrate gradients, behaviors of individual axons within the population could be tracked, and outcomes scored. On average, 68% of axons showed preference for increasing laminin concentrations, whereas 10% migrate away from laminin, 5% were at right angles, and 17% were indifferent toward it.[43]
Fluid-phase gradient cues within µFDs can be tuned on-the-fly through micro-channel mixers, jets, or valves to expose cells to transient or stable diffusive gradients (Figure 3c–f). Such approaches are fully compatible with real-time, high-resolution microscopic imaging. Microfluidic jets can be used to create chemical gradients within culture wells for testing the influence of an array of candidate agents on axonal outgrowth, and even the dynamics of vanguard filopodia.[47] Large-scale diffusive, fluid-phase gradients of putative axon guidance factors (e.g., netrin) were generated microfluidically in cultures of cortical neurons from embryonic mouse. While the majority of neurons (~73%) extended axons toward increasing netrin concentrations, axons of the minor population showed degrees of aversive growth trajectories, suggesting heterogeneity in the responding population.[47] Indeed, this study found a distributed response of axons to fluid-phase netrin gradients. How this distribution of axon directionalities in populations where cells are in contact may differ from isolated neurons remains to be resolved.
A hybrid microfluidic-collagen gel gradient generator was developed to screen neurite responses to growth-factor gradients.[48] This high-throughput analysis of putative neuronal guidance cues confirmed netrin-1 (attractive) and slit-2 (repellant) as regulators of axonal guidance, and also revealed that homogenized mouse brain pulp contains significant guidance cues for directing axonal growth of embryonic hippocampal neurons. Thus, µFDs minimize experimental confounds inherent in vivo and, compared with dish cultures, facilitate analyzing signaling cues that govern axonal growth and guidance.
Dendritic signaling and synapse formation
Dendrites are conduits of information influx and integration, from synapse to nucleus and back. Interconnected compartments within µFDs (Figure 1d, Figure 4) also facilitate controlled probing of dendrites and sub-regions. Ultra-thin fluid streams of chemo-attractants can be positioned or moved across neurons in µFDs to administer pulses of neurotransmitter,[49,50] mimicking synaptic release. The ability to control micro-environments of dendrites has enabled studies addressing fundamental questions pertaining to mechanisms of synapse formation and regulation.[46,49]
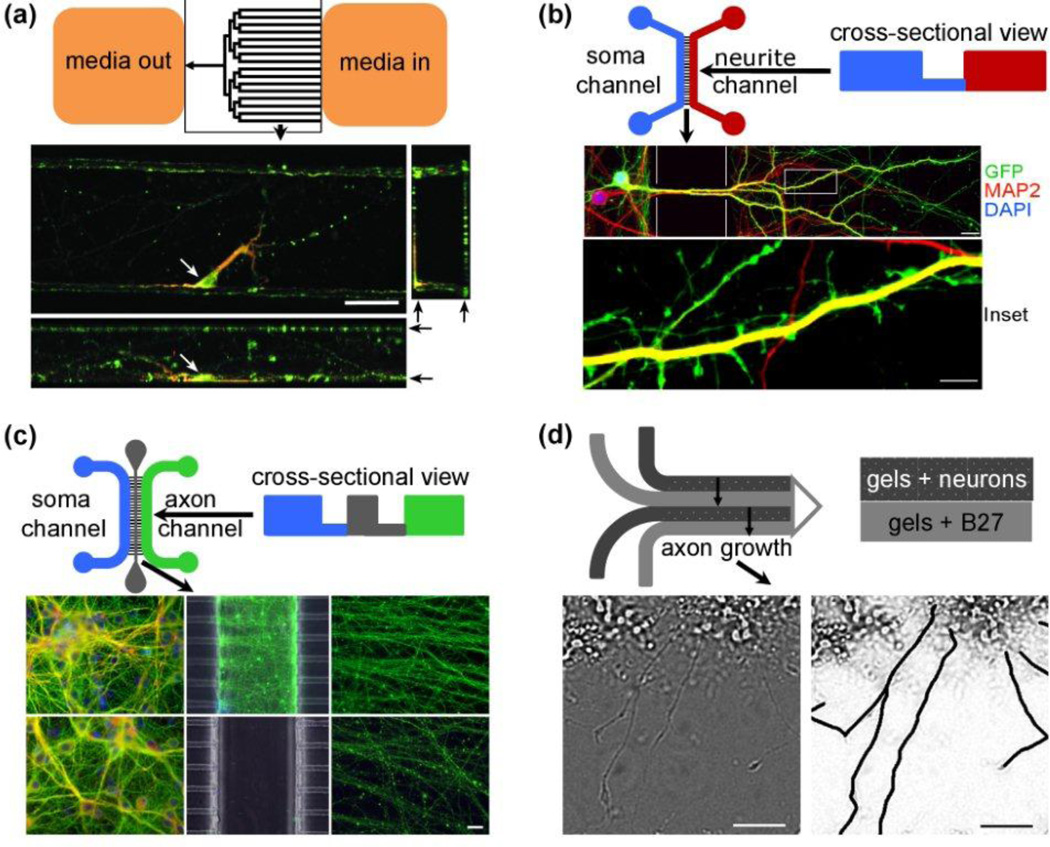
Figure 4. µFDs offer design flexibility for probing developing neurons.
Top-down, and some cross-sectional, schematics of four types of µFDs appear above images of the neurons within them. (a) Ultra-low densities of primary post-natal rat hippocampal neurons differentiate and extend complex processes in µFDs fabricated with extracted PDMS. An individual neuron is identified (cell body, white arrow). Dendrites (labeled for MAP2, red) and axons (labeled for tau, green) navigate all surfaces of the microfluidic channels, but especially corners, where they contact ≥ 2 surfaces. Black arrows (right) mark the top and bottom of the channel for compressed side-/end-view images. Image orientations: top left: XY, top-down view; bottom: XZ, side-view; top right, YZ, end-view. 7 DIV, scale bar = 50 µm. Reproduced and modified by permission of The Royal Society of Chemistry (RSC).[64] (b) Regulation of communication via dendrite-to-soma signaling can be studied using a multi-domain µFD. This µFD has two channels that communicate fluidically via a series of parallel interconnecting tunnels. One channel (left, blue) is where the neurons are seeded. As they develop, the neurons extend their processes through the interconnects into the second channel (right, red). The interconnects restrict somal migration while allowing the processes to extend down the interconnects into neurite channel, where they differentiate into dendrites with spines. Fluorescence microscopy reveals cellular components: dendritic cytoskeleton (microtubule-associated protein 2 MAP2, red) of green fluorescent protein GFP-expressing cortical neurons (GFP, diffuses throughout the cytoplasm, green), and nucleus (DNA marker, 4',6-diamidino-2-phenylindole, DAPIblue). Because dendritic shafts co-localize MAP2 and GFP, they fluoresce yellow. Spines (bulbous, GFP-filled protrusions from the dendritic shaft) develop in the right channel, where processes are fluidically isolated from the soma. This precise subcellular fluidic exposure and cellular manipulation is not achievable in dish cultures. Scale bars = 20 µm (top), 2 µm (bottom). Modified and used with permission.[51] (c) Spatially controlled axotomy and degeneration of central neurons in a compartmentalized µFD. This µFD has 3 channels, one in which primary mouse cortical neurons are seeded (left middle and bottom, blue), and two where axons extend (center middle and bottom, gray, and right middle and bottom, green). Axons extend through interconnects traverse the central channel (middle, control, and bottom experimental) and a second set of interconnects before emerging in parallel to invade the right channel (middle and bottom). The central channel is the ‘surgical suite’ (bottom center), where detergent treatment transects axons synchronously and in geometric register. Fluidic isolation protects somata (bottom left) from damage. Axonal channels of control cultures contain healthy axons (middle right) vs. axons undergoing Wallerian-like degeneration on the distal side of the transection (bottom right). This design provides a model system for studying axonal degeneration and death mechanisms vs. regeneration processes, with high spatial and temporal control of axonal states and molecular pathways. Neurons are immunostained for β-III-tubulin (green) and F-actin (red), scale bar = 20 µm. Modified and used with permission.[54] (d) Creating cortical lamina in vitro. Multi-layered µFD (top left) were used to study alternating layers of embryonic rat cortical neurons (dark grey layer) and trophic factors (i.e., B27, light grey layer), as illustrated in the schematic on the top right. Axons, the longest neurites, traverse from one layer into an adjoining layer (left), as shown in axonal traces (bottom right), generating a laminar structure of cortical neurons. Scale bars = 50 µm. Modified and used with permission.[67]
Noteworthy examples illustrate the merits of this approach for resolving questions of subcellular regulation of dendritic function. How signals at dendrites change synaptic state is a long-standing question. Toward resolving mechanisms of BDNF induction of transcription-mediated synapse strengthening, µFDs were used to compartmentalize and fluidically isolate dendrites from the somata of rat embryonic cortical neurons.[51] Restricted dendritic or somatic exposure to BDNF and pharmacological agents revealed a novel, dendrite-localized neurotrophin signaling pathway not discernible in dish cultures where dendrites and axons cannot be isolated. This study discovered that dendrite-to-nucleus induction of c-Fos expression by BDNF is: (1) Ca2+-independent, (2) not dependent on Trk activity in dendrites, (3) mediated by mitogen-activated protein kinase kinase (MAPKK or MEK1/2), wherein dentritic signal processing differs from MEK5-mediated axonal retrograde signaling, and (4) dependent on intra-dendritic mRNA translation for immediate early-gene expression.[51] These elements distinguish dendritic pathways from retrograde neurotrophin signaling in axons.
Discriminating mediators of axo-dendritic synaptic differentiation and degeneration in neurons of the cortico-striatal pathway is challenging in vivo. Exogenous cues are difficult to control precisely in conventional dishes due to plume dynamics, diffusion from the source micropipette, and static-fluidic conditions. Using µFDs that control neurite directionality enables analysis of en passant axo-dendritic connections and cortico-striatal synapses. Tapered interconnecting channels funnel cortical axons toward an adjacent chamber containing striatal neurons (Figure 1d, e).[52] This microfluidic approach circumvents many of the confounds of conventional methods, by enabling identification of specific types of neurons within co-cultured populations and segregation of two different populations[53] while permitting ample en passant axo-dendritic interactions. Further, the environment of the µFD promotes striatal neuron differentiation that results in longer dendrites, greater spine density, increased phosphorylated extracellular signal-regulated kinase (p-ERK) activation, and spontaneous Ca2+ oscillations synchronous with cortical neuron activity.
Analyzing neuronal stimulus-response
µFDs enable precise, controlled delivery of neuromodulators, imaging reagents, or inhibitors to cultured neurons compared with conventional approaches. They permit focal stimulation of dendrites, visualization and manipulation of synapses and single-molecule axonal transport, monitoring real-time responses to specific axon guidance cues, analysis of molecular changes, and investigation of traumatic and neurodegenerative conditions.[4,47,49,54–56] Large-order arrays of neurons can be probed to determine the proportion of the population responsive to specific neuromodulators. This design facilitated molecular profiling and resolved responses of over 2,900 olfactory sensory neurons to four different odorant molecules.[57] Absolute spatial and fluidic control permitted discrimination of fifteen classes of possible combinatorial responses. This type of population analysis of stimulus-response characteristics should be applied to developing neuronal networks, where it will facilitate understanding components and circuits.
Neurochemical identification of substances released by neurons, including discriminating those released locally at specific sub-structures, can be accomplished by coupling sampling via µFDs with analytical chemistry.[58] With the ability to perform spatially targeted sampling of low-density primary neurons,[59] media containing local cellular releasates can be collected and processed by mass spectrometry.[60] Competitive signals and confounding variables of surrounding tissues and cells are diminished. Thus, neurochemical analysis of secreted molecules in volume- and analyte-limited samples can be achieved. Due to these advantages, an increasing number of studies are utilizing µFDs as tools for analyses of neuropeptides, mRNA, and subcellular pH.[60–63]
Advancing developmental neurobiology at cell population and tissue levels
To more fully understand the signals that shape the forming brain, cultures will need to incorporate multiple cell types in three dimensions (3D). The central nervous system comprises heterogeneous populations of neuronal, glial, microglial, and endothelial cells that mutually interact. Recapitulating the microenvironment of the brain, in the simplest form, should achieve topographical neural structures. This requires detailed characterization of critical environmental cues. To construct organized 3D cultures, cell-compatible features must be incorporated into the material properties, fabrication design, and imaging processes. For example, in microchannels of the highly biocompatible silicone gel, solvent-extracted polydimethylsiloxane (PDMS), rat hippocampal neurons prefer extracted PDMS over glass substrates, and neurites show a strong affinity for topographic cues where walls meet at right angles (Figure 4a).[64] Furthermore, when presented with competitive substrate cues, hippocampal neurons unexpectedly prefer physical contact guidance cues over chemical ligands.[65,66] Thus, cellular tension versus chemical substrate cues must access different, competitive signaling pathways.
Microfluidic channels can be combined with layered hydrogel scaffolds to more closely approximate the physical, three-dimensional structure of the developing brain (Figure 4d). An agarose scaffold can be fabricated with four hydrogel layers on the scale of layers within the cerebral cortex. Layers with embedded neurons intercalate adjacent layers that encapsulate growth factors (serum, B27 supplement, and nerve growth factor, NGF), which are released diffusively and guide developing neurons.[46,67] As a result, embedded neurons, axonal growth, and synapse formation within the 3D matrixes self-organize into laminar brain tissue, which is useful for studying corticogenesis in vitro.
Large, interconnecting populations of dissociated cells (e.g., neurons, glia, microglia)[3,68] and organotypic tissue slices can be achieved through multi-channel µFDs. Organotypic slices from different brain regions having different culture requirements can be grown so that they interconnect through microchannels, enabling synchronized neural activity between them.[69] These complex tissue-level constructs can used to precisely manipulate neural circuits to advance understanding of the development of neural plasticity, circadian-clock coupling, and neuropathologies.
The application of µFDs to studying developmental interactions of neurons, glia, and cells of interacting brain regions is in its infancy. This approach has the power to interrogate cell-to-cell contact and communication, while achieving differential fluidic exposure, sample manipulation, and collection.[26,70] For example, Alzheimers disease-like pathological states occur in only a subset of neuronal networks grown within µFDs;[5] similar approaches could be applied to probe developing neuronal networks. Thus, the flexibility of µFD design makes it adaptable to dispersed neurons in compartmentalized microdomains and 3D tissue constructs for addressing a multitude of developmental questions.
Technical considerations
Increased experimental complexity and micro-environmental manipulation in µFDs require stringent evaluation and controls. Greater scrutiny is required by both reviewers and readers in assessing the implications of µFD-based studies. Diligence and care are required to prove that observations and conclusions are free of unintended variables or experimental confounds. Even culture media can bias experimental results, demonstrating influences of common reagents on experimental outcomes. Defined formulations of culture media are widely accepted for sustaining primary neurons without the need for complex additives, such as fetal serum. However, defined media formulations are under renewed scrutiny for the influences that albumin, selenium, and antioxidants impose on developing neurons.[71–73]
µFD materials, themselves, may contribute to media biases through absorption, adsorption, and leaching (Box 2).[74–76] As a result, materials used to fabricate µFDs and the physical influences they exert may affect biological processes of the cells within them. Because µFDs can be fabricated to a wide range of dimensions and architectures with different materials, potential confounds may depend on process, scale, and material.[77] These subtle variables may explain why some laboratories experience confounds while others do not. For example, some researchers have found that mild washing of PDMS with alcohol is less effective at removing unpolymerized oligomers,[74] while rigorous solvent extraction of PDMS is effective at removing oligomers and improving neuronal viability at low cell densities.[64]
BOX 2. Potential factors limiting neuronal development in microfluidic devices.
Whereas µFDs generally are considered highly biocompatible, some reports suggest adverse effects of materials or the environment on cell growth or viability.[64,74] Influences that potentially could limit or bias viability, development, or function, and thus need controls, are:
Shear stress: Flowing fluid through microchannels can present cells with a range of shear forces due to flow velocities and channel architectures, stresses that could bias neuronal development and function.[91,92]
Gas solubility, permeability, and diffusibility: Defined cell-culture media formulations were developed with levels of antioxidants and buffers for specific concentrations of gases, e.g., O2 and CO2. Materials, channel dimensions, and flow regimes potentially could alter gas concentrations available to cells and chemical properties of the culture media.[77]
Absorption: Porous polymers have the ability to absorb chemicals from the fluid, thus altering fluidic composition within the µFD or its channels.[74,75]
Adsorption: Fluids or media components may adhere differentially onto the surface(s) of the µFD, and thus become depleted.[76]
Desorption: Chemicals or elements of the material may have the capacity to dissolve in the media, bind to, and/or accumulate within cells, and thereby influence cell function.[74]
Evaporation: Water can evaporate through porous materials, increasing media osmolarity.[93]
Innovative design improvements undoubtedly will minimize or eliminate undesirable effects of the µFD environment. Because fluid flow can influence neuronal growth and function, methods have been developed to minimize neuron exposure to the shear stresses of fluidic perfusion, while providing the desired instructive gradient guidance cues for axons.[47] Specific requirements of different biological models also may explain differences in outcomes between groups using µFDs. For example, locust neurons in µFDs require different modifications to culture conditions than rat neurons.[78] Whereas species-based differences are not surprising, significant culture-induced differences have been found. Conditions have been developed that enhance mRNA profiles of cultured astrocytes from 10% to nearly 80% of mRNAs expressed in vivo.[79] This remarkable advance in refining glial cell culture conditions predicts that we may yet discover different, more optimal, requirements for specific neuronal types or brain regions.
Exposing fragile and sensitive neurons to new materials or physical conditions requires careful characterization and validation of the material and device for experimental compatibility before adopting new techniques with confidence. Physical, chemical, and flow properties of µFDs may influence experimental design and applications to developmental neurobiology (Box 1). Adhering to these principles will drive studies to improve fabrication processes and the material composition, and enable µFDs to further advance developmental neuroscience.
Concluding remarks
The emergence of µFDs that enable the manipulation of microenvironments surrounding neuronal substructures endows neuroscientists with a new toolset that elevates the sophistication of neuroscience in vitro to a fundamentally new level of refinement. No single method or device can yet fully recreate the natural environment of the brain to permit studying neurons under the range of contexts and dynamics in which they develop and function. Nevertheless, remarkable insights have been gained from these reduced neuronal preparations. From decoding neuronal polarity to probing axonal guidance and dendritic function, µFDs are advancing neuroscience on multiple fronts. Thus, as µFDs are refined to address unanswered questions (Box 3), we anticipate that they will reveal new insights into how emergent developmental processes shape the nervous system.
Box 3. Outstanding questions.
Outstanding questions, accessible through µFD technologies, include the following:
How are the self-organizing events of early neuronal morphogenesis coordinated through extrinsic cues?
What mechanisms and extracellular factors govern asymmetric modulation of selective protein translation and protein degradation during neuronal polarization?
How does establishing neuronal polarity regulate dendrite development?
What molecular mechanisms regulate intracellular signaling and trafficking between dendrites and the nucleus to modulate gene expression and then alter dendritic functions selectively?
What cellular processes regulate dendritic growth and branching?
How do nascent dentritic filopodia form synapses?
How is synaptogenesis between different types of neurons orchestrated?
Manipulating the microenvironment of the developing neuron and monitoring changes in cell structure and function has never been approachable at the resolution enabled by µFDs. Like other new technologies, exploiting the full potential of µFDs requires care, skill, insight, and creativity. The unparalleled ability to control local chemical, physical, and fluidic environments while accessing and probing microdomains of the developing neuron offers extraordinary opportunities.
Acknowledgements
The authors thank Jonathan Sweedler, Ralph Nuzzo, and the ‘Neuro-Nano Group’ for insightful discussions, as well as Stephanie Ceman, Anika Jain, and Lori Raetzman for review and Maureen Holtz for preparation of the manuscript. We are grateful to Graham Knott for the gift of his original image. The authors are not affiliated with, nor endorse, µFDs suppliers listed in this work. Development of this review was supported by the National Institute of Mental Health (R21MH085220), National Heart, Lung and Blood Institute (RO1HL092571 Z ARRA, RO1HL086870), and National Science Foundation (NSF; IOS 0818555 and STC CBET 0939511) to M.U.G.. L.J.M. was supported by the National Institute of Child Health and Human Development Developmental Psychobiology and Neurobiology Training Grant (T32HD007333). Content is solely the responsibility of the authors and does not necessarily represent official views of the National Institutes of Health or NSF.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Knott G, et al. Serial section scanning electron microscopy of adult brain tissue using focused ion beam milling. J Neurosci. 2008;28(12):2959–2964. doi: 10.1523/JNEUROSCI.3189-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Micheva KD, Smith SJ. Array tomography: a new tool for imaging the molecular architecture and ultrastructure of neural circuits. Neuron. 2007;55(1):25–36. doi: 10.1016/j.neuron.2007.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Majumdar D, et al. Co-culture of neurons and glia in a novel microfluidic platform. J Neurosci Methods. 2011;196(1):38–44. doi: 10.1016/j.jneumeth.2010.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hosmane S, et al. Toll/Interleukin-1 receptor domain-containing adapter inducing interferon-β mediates microglial phagocytosis of degenerating axons. J Neurosci. 2012;32(22):7745–7757. doi: 10.1523/JNEUROSCI.0203-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kunze A, et al. Co-pathological connected primary neurons in a microfluidic device for Alzheimer studies. Biotechnol Bioeng. 2011;108(9):2241–2245. doi: 10.1002/bit.23128. [DOI] [PubMed] [Google Scholar]
- 6.Poon WW, et al. beta-Amyloid impairs axonal BDNF retrograde trafficking. Neurobiol Aging. 2011;32(5):821–833. doi: 10.1016/j.neurobiolaging.2009.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hellman AN, et al. Examination of axonal injury and regeneration in micropatterned neuronal culture using pulsed laser microbeam dissection. Lab Chip. 2010;10(16):2083–2092. doi: 10.1039/b927153h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim YT, et al. Neuro-optical microfluidic platform to study injury and regeneration of single axons. Lab Chip. 2009;9(17):2576–2581. doi: 10.1039/b903720a. [DOI] [PubMed] [Google Scholar]
- 9.Kim HJ, et al. Integrated microfluidics platforms for investigating injury and regeneration of CNS axons. Ann Biomed Eng. 2012;40(6):1268–1276. doi: 10.1007/s10439-012-0515-6. [DOI] [PubMed] [Google Scholar]
- 10.Yang IH, et al. Compartmentalized microfluidic culture platform to study mechanism of paclitaxel-induced axonal degeneration. Exp Neurol. 2009;218(1):124–128. doi: 10.1016/j.expneurol.2009.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barnes AP, Polleux F. Establishment of axon-dendrite polarity in developing neurons. Annu Rev Neurosci. 2009;32:347–381. doi: 10.1146/annurev.neuro.31.060407.125536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shi P, et al. Local presentation of L1 and N-cadherin in multicomponent, microscale patterns differentially direct neuron function in vitro. Dev Neurobiol. 2007;67(13):1765–1776. doi: 10.1002/dneu.20553. [DOI] [PubMed] [Google Scholar]
- 13.Chang JC, et al. A modified microstamping technique enhances polylysine transfer and neuronal cell patterning. Biomaterials. 2003;24(17):2863–2870. doi: 10.1016/s0142-9612(03)00116-9. [DOI] [PubMed] [Google Scholar]
- 14.Esch T, et al. Local presentation of substrate molecules directs axon specification by cultured hippocampal neurons. J Neurosci. 1999;19(15):6417–6426. doi: 10.1523/JNEUROSCI.19-15-06417.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oliva AA, Jr, et al. Patterning axonal guidance molecules using a novel strategy for microcontact printing. Neurochem Res. 2003;28(11):1639–1648. doi: 10.1023/a:1026052820129. [DOI] [PubMed] [Google Scholar]
- 16.Withers GS, et al. Effects of substrate geometry on growth cone behavior and axon branching. J Neurobiol. 2006;66(11):1183–1194. doi: 10.1002/neu.20298. [DOI] [PubMed] [Google Scholar]
- 17.Steedman MR, et al. Enhanced differentiation of retinal progenitor cells using microfabricated topographical cues. Biomed Microdevices. 2010;12(3):363–369. doi: 10.1007/s10544-009-9392-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang J, et al. Microfluidics: a new cosset for neurobiology. Lab Chip. 2009;9(5):644–652. doi: 10.1039/b813495b. [DOI] [PubMed] [Google Scholar]
- 19.Baranes K, et al. Topographic cues of nano-scale height direct neuronal growth pattern. Biotechnol Bioeng. 2012;109(7):1791–1797. doi: 10.1002/bit.24444. [DOI] [PubMed] [Google Scholar]
- 20.Knöll B, et al. Stripe assay to examine axonal guidance and cell migration. Nat Protoc. 2007;2(5):1216–1224. doi: 10.1038/nprot.2007.157. [DOI] [PubMed] [Google Scholar]
- 21.Shelly M, et al. LKB1/STRAD promotes axon initiation during neuronal polarization. Cell. 2007;129(3):565–577. doi: 10.1016/j.cell.2007.04.012. [DOI] [PubMed] [Google Scholar]
- 22.Shelly M, et al. Local and long-range reciprocal regulation of cAMP and cGMP in axon/dendrite formation. Science. 2010;327(5965):547–552. doi: 10.1126/science.1179735. [DOI] [PubMed] [Google Scholar]
- 23.Cheng PL, et al. Phosphorylation of E3 ligase Smurf1 switches its substrate preference in support of axon development. Neuron. 2011;69(2):231–243. doi: 10.1016/j.neuron.2010.12.021. [DOI] [PubMed] [Google Scholar]
- 24.Yi JJ, et al. TGF-beta signaling specifies axons during brain development. Cell. 2010;142(1):144–157. doi: 10.1016/j.cell.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nevill JT, et al. Vacuum soft lithography to direct neuronal polarization. Soft Matter. 2011;7(2):343–347. [Google Scholar]
- 26.Pirlo RK, et al. Biochip/laser cell deposition system to assess polarized axonal growth from single neurons and neuron/glia pairs in microchannels with novel asymmetrical geometries. Biomicrofluidics. 2011;5(1):13408. doi: 10.1063/1.3552998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Millet LJ, et al. Pattern analysis and spatial distribution of neurons in culture. Integr Biol (Camb) 2011;3(12):1167–1178. doi: 10.1039/c1ib00054c. [DOI] [PubMed] [Google Scholar]
- 28.Murai KK, Pasquale EB. ‘Eph’ective signaling: forward, reverse and crosstalk. J Cell Sci. 2003;116(pt 14):2823–2832. doi: 10.1242/jcs.00625. [DOI] [PubMed] [Google Scholar]
- 29.Lang S, et al. Growth cone response to ephrin gradients produced by microfluidic networks. Anal Bioanal Chem. 2008;390(3):809–816. doi: 10.1007/s00216-007-1363-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fozdar DY, et al. Selective axonal growth of embryonic hippocampal neurons according to topographic features of various sizes and shapes. Int J Nanomedicine. 2011;6:45–57. doi: 10.2147/IJN.S12376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Francisco H, et al. Regulation of axon guidance and extension by three-dimensional constraints. Biomaterials. 2007;28(23):3398–3407. doi: 10.1016/j.biomaterials.2007.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Taylor AM, et al. A microfluidic culture platform for CNS axonal injury, regeneration and transport. Nat Methods. 2005;2(8):599–605. doi: 10.1038/nmeth777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hosie KA, et al. Chronic excitotoxin-induced axon degeneration in a compartmented neuronal culture model. ASN neuro. 2012;4(1):47–57. doi: 10.1042/AN20110031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu HI, et al. A lab-on-a-chip platform for studying the subcellular functional proteome of neuronal axons. Lab Chip. 2010;10(5):647–653. doi: 10.1039/b918217a. [DOI] [PubMed] [Google Scholar]
- 35.Hoffman-Kim D, et al. Topography, cell response, and nerve regeneration. Annu Rev Biomed Eng. 2010;12:203–231. doi: 10.1146/annurev-bioeng-070909-105351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.von Philipsborn AC, et al. Substrate-bound protein gradients for cell culture fabricated by microfluidic networks and microcontact printing. Sci STKE. 2007;2007(414):16. doi: 10.1126/stke.4142007pl6. [DOI] [PubMed] [Google Scholar]
- 37.Jing G, et al. Precise cell patterning using cytophobic self-assembled monolayer deposited on top of semi-transparent gold. Biomed Microdevices. 2010;12(5):935–948. doi: 10.1007/s10544-010-9448-8. [DOI] [PubMed] [Google Scholar]
- 38.Nédelec S, et al. Concentration-dependent requirement for local protein synthesis in motor neuron subtype-specific response to axon guidance cues. J Neurosci. 2012;32(4):1496–1506. doi: 10.1523/JNEUROSCI.4176-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shi P, et al. Combined microfluidics/protein patterning platform for pharmacological interrogation of axon pathfinding. Lab Chip. 2010;10(8):1005–1010. doi: 10.1039/b922143c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hur EM, et al. Engineering neuronal growth cones to promote axon regeneration over inhibitory molecules. Proc Natl Acad Sci U S A. 2011;108(12):5057–5062. doi: 10.1073/pnas.1011258108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dajas-Bailador F, et al. microRNA-9 regulates axon extension and branching by targeting Map1b in mouse cortical neurons. Nat. Neurosci. 2012;15(5):697–701. doi: 10.1038/nn.3082. [DOI] [PubMed] [Google Scholar]
- 42.Tessier-Lavigne M, Goodman CS. The molecular biology of axon guidance. Science. 1996;274(5290):1123–1133. doi: 10.1126/science.274.5290.1123. [DOI] [PubMed] [Google Scholar]
- 43.Millet LJ, et al. Guiding neuron development with planar surface gradients of substrate cues deposited using microfluidic devices. Lab Chip. 2010;10(12):1525–1535. doi: 10.1039/c001552k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Joanne Wang C, et al. A microfluidics-based turning assay reveals complex growth cone responses to integrated gradients of substrate-bound ECM molecules and diffusible guidance cues. Lab Chip. 2008;8(2):227–237. doi: 10.1039/b713945d. [DOI] [PubMed] [Google Scholar]
- 45.Dertinger SK, et al. Gradients of substrate-bound laminin orient axonal specification of neurons. Proc Natl Acad Sci U S A. 2002;99(20):12542–12547. doi: 10.1073/pnas.192457199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kunze A, et al. Synergistic NGF/B27 gradients position synapses heterogeneously in 3D micropatterned neural cultures. PLoS One. 2011;6(10):e26187. doi: 10.1371/journal.pone.0026187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bhattacharjee N, et al. A neuron-benign microfluidic gradient generator for studying the response of mammalian neurons towards axon guidance factors. Integr Biol (Camb) 2010;2(11–12):669–679. doi: 10.1039/c0ib00038h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kothapalli CR, et al. A high-throughput microfluidic assay to study neurite response to growth factor gradients. Lab Chip. 2011;11(3):497–507. doi: 10.1039/c0lc00240b. [DOI] [PubMed] [Google Scholar]
- 49.Taylor AM, et al. Microfluidic local perfusion chambers for the visualization and manipulation of synapses. Neuron. 2010;66(1):57–68. doi: 10.1016/j.neuron.2010.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Botzolakis EJ, et al. Achieving synaptically relevant pulses of neurotransmitter using PDMS microfluidics. J Neurosci Methods. 2009;177(2):294–302. doi: 10.1016/j.jneumeth.2008.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cohen MS, et al. Neurotrophin-mediated dendrite-to-nucleus signaling revealed by microfluidic compartmentalization of dendrites. Proc Natl Acad Sci U S A. 2011;108(27):11246–11251. doi: 10.1073/pnas.1012401108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Peyrin JM, et al. Axon diodes for the reconstruction of oriented neuronal networks in microfluidic chambers. Lab Chip. 2011;11(21):3663–3673. doi: 10.1039/c1lc20014c. [DOI] [PubMed] [Google Scholar]
- 53.Kaufman AM, et al. Opposing roles of synaptic and extrasynaptic NMDA receptor signaling in cocultured striatal and cortical neurons. J Neurosci. 2012;32(12):3992–4003. doi: 10.1523/JNEUROSCI.4129-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kilinc D, et al. Wallerian-like degeneration of central neurons after synchronized and geometrically registered mass axotomy in a three-compartmental microfluidic chip. Neurotox Res. 2011;19(1):149–161. doi: 10.1007/s12640-010-9152-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Park J, et al. Microfluidic compartmentalized co-culture platform for CNS axon myelination research. Biomed Microdevices. 2009;11(6):1145–1153. doi: 10.1007/s10544-009-9331-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang K, et al. Single-molecule imaging of NGF axonal transport in microfluidic devices. Lab Chip. 2010;10(19):2566–2573. doi: 10.1039/c003385e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Figueroa XA, et al. Large-scale investigation of the olfactory receptor space using a microfluidic microwell array. Lab Chip. 2010;10(9):1120–1127. doi: 10.1039/b920585c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mitchell JW, et al. Direct cellular peptidomics of hypothalamic neurons. Front Neuroendocrinol. 2011;32(4):377–386. doi: 10.1016/j.yfrne.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Millet LJ, et al. Direct cellular peptidomics of supraoptic magnocellular and hippocampal neurons in low-density co-cultures. ACS Chem Neurosci. 2010;1(1):36–48. doi: 10.1021/cn9000022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jo K, et al. Mass spectrometric imaging of peptide release from neuronal cells within microfluidic devices. Lab Chip. 2007;7(11):1454–1460. doi: 10.1039/b706940e. [DOI] [PubMed] [Google Scholar]
- 61.Taylor AM, et al. Axonal mRNA in uninjured and regenerating cortical mammalian axons. J Neurosci. 2009;29(15):4697–4707. doi: 10.1523/JNEUROSCI.6130-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vitzthum L, et al. Study of Na+/H+ exchange-mediated pHi regulations in neuronal soma and neurites in compartmentalized microfluidic devices. Integr Biol (Camb) 2010;2(1):58–64. doi: 10.1039/b918440f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhong M, et al. Label-free quantitation of peptide release from neurons in a microfluidic device with mass spectrometry imaging. Lab Chip. 2012;12(11):2037–2045. doi: 10.1039/c2lc21085a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Millet LJ, et al. Microfluidic devices for culturing primary mammalian neurons at low densities. Lab Chip. 2007;7(8):987–994. doi: 10.1039/b705266a. [DOI] [PubMed] [Google Scholar]
- 65.Gomez N, et al. Polarization of hippocampal neurons with competitive surface stimuli: contact guidance cues are preferred over chemical ligands. J R Soc Interface. 2007;4(13):223–233. doi: 10.1098/rsif.2006.0171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gomez N, et al. Immobilized nerve growth factor and microtopography have distinct effects on polarization versus axon elongation in hippocampal cells in culture. Biomaterials. 2007;28(2):271–284. doi: 10.1016/j.biomaterials.2006.07.043. [DOI] [PubMed] [Google Scholar]
- 67.Kunze A, et al. Micropatterning neural cell cultures in 3D with a multi-layered scaffold. Biomaterials. 2011;32(8):2088–2098. doi: 10.1016/j.biomaterials.2010.11.047. [DOI] [PubMed] [Google Scholar]
- 68.Hosmane S, et al. Circular compartmentalized microfluidic platform: Study of axon-glia interactions. Lab Chip. 2010;10(6):741–747. doi: 10.1039/b918640a. [DOI] [PubMed] [Google Scholar]
- 69.Berdichevsky Y, et al. Building and manipulating neural pathways with microfluidics. Lab Chip. 2010;10(8):999–1004. doi: 10.1039/b922365g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lovchik RD, et al. A microfluidic device for depositing and addressing two cell populations with intercellular population communication capability. Biomed Microdevices. 2010;12(2):275–282. doi: 10.1007/s10544-009-9382-9. [DOI] [PubMed] [Google Scholar]
- 71.Chen Y, et al. NS21: re-defined and modified supplement B27 for neuronal cultures. J Neurosci Methods. 2008;171(2):239–247. doi: 10.1016/j.jneumeth.2008.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Roth S, et al. Development of a serum-free supplement for primary neuron culture reveals the interplay of selenium and vitamin E in neuronal survival. J Trace Elem Med Biol. 2010;24(2):130–137. doi: 10.1016/j.jtemb.2010.01.007. [DOI] [PubMed] [Google Scholar]
- 73.van der Valk J, et al. Optimization of chemically defined cell culture media--replacing fetal bovine serum in mammalian in vitro methods. Toxicol In Vitro. 2010;24(4):1053–1063. doi: 10.1016/j.tiv.2010.03.016. [DOI] [PubMed] [Google Scholar]
- 74.Regehr KJ, et al. Biological implications of polydimethylsiloxane-based microfluidic cell culture. Lab Chip. 2009;9(15):2132–2139. doi: 10.1039/b903043c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Toepke MW, Beebe DJ. PDMS absorption of small molecules and consequences in microfluidic applications. Lab Chip. 2006;6(12):1484–1486. doi: 10.1039/b612140c. [DOI] [PubMed] [Google Scholar]
- 76.Li N, et al. PDMS compound adsorption in context. J Biomol Screen. 2009;14(2):194–202. doi: 10.1177/1087057108327326. [DOI] [PubMed] [Google Scholar]
- 77.Zahorodny-Burke M, et al. Finite element analysis of oxygen transport in microfluidic cell culture devices with varying channel architectures, perfusion rates, and materials. Chem Eng Sci. 2011;66(23):6244–6253. [Google Scholar]
- 78.Gobbels K, et al. Low density cell culture of locust neurons in closed-channel microfluidic devices. J Insect Physiol. 2010;56(8):1003–1009. doi: 10.1016/j.jinsphys.2010.05.017. [DOI] [PubMed] [Google Scholar]
- 79.Foo LC, et al. Development of a method for the purification and culture of rodent astrocytes. Neuron. 2011;71(5):799–811. doi: 10.1016/j.neuron.2011.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang Z, et al. Spatial light interference microscopy (SLIM) Opt Express. 2011;19(2):1016–1026. doi: 10.1364/OE.19.001016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Childs WR, Nuzzo RG. Decal transfer microlithography: a new soft-lithographic patterning method. J Am Chem Soc. 2002;124(45):13583–13596. doi: 10.1021/ja020942z. [DOI] [PubMed] [Google Scholar]
- 82.Ambravaneswaran V, et al. Directional decisions during neutrophil chemotaxis inside bifurcating channels. Integr Biol (Camb) 2010;2(11–12):639–647. doi: 10.1039/c0ib00011f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Keenan TM, et al. Microfluidic “jets” for generating steady-state gradients of soluble molecules on open surfaces. Appl Phys Lett. 2006;89(11):114103. [Google Scholar]
- 84.Velve-Casquillas G, et al. Microfluidic tools for cell biological research. Nano Today. 2010;5(1):28–47. doi: 10.1016/j.nantod.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Paguirigan A, Beebe DJ. Gelatin based microfluidic devices for cell culture. Lab Chip. 2006;6(3):407–413. doi: 10.1039/b517524k. [DOI] [PubMed] [Google Scholar]
- 86.Previtera ML, et al. Effects of substrate stiffness and cell density on primary hippocampal cultures. J Biosci Bioeng. 2010;110(4):459–470. doi: 10.1016/j.jbiosc.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chronis N, et al. Microfluidics for in vivo imaging of neuronal and behavioral activity in Caenorhabditis elegans. Nat Methods. 2007;4(9):727–731. doi: 10.1038/nmeth1075. [DOI] [PubMed] [Google Scholar]
- 88.Kartalov EP, et al. The analytical approach to polydimethylsiloxane microfluidic technology and its biological applications. J Nanosci Nanotechnol. 2006;6(8):2265–2277. doi: 10.1166/jnn.2006.504. [DOI] [PubMed] [Google Scholar]
- 89.Mehta G, et al. Quantitative measurement and control of oxygen levels in microfluidic poly(dimethylsiloxane) bioreactors during cell culture. Biomed Microdevices. 2007;9(2):123–134. doi: 10.1007/s10544-006-9005-7. [DOI] [PubMed] [Google Scholar]
- 90.Merkel TC, et al. Gas sorption, diffusion, and permeation in poly(dimethylsiloxane) J Polym Sci, Part B: Polymer Physics. 2000;38(3):415–434. [Google Scholar]
- 91.Shin HS, et al. Shear stress effect on transfection of neurons cultured in microfluidic devices. J Nanosci Nanotechnol. 2009;9(12):7330–7335. doi: 10.1166/jnn.2009.1769. [DOI] [PubMed] [Google Scholar]
- 92.van der Meer AD, et al. Analyzing shear stress-induced alignment of actin filaments in endothelial cells with a microfluidic assay. Biomicrofluidics. 2010;4(1):11103. doi: 10.1063/1.3366720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Heo YS, et al. Characterization and resolution of evaporation-mediated osmolality shifts that constrain microfluidic cell culture in poly(dimethylsiloxane) devices. Anal Chem. 2007;79(3):1126–1134. doi: 10.1021/ac061990v. [DOI] [PMC free article] [PubMed] [Google Scholar]