Abstract
We have produced and analyzed transgenic birdsfoot trefoil (Lotus corniculatus L.) plants harboring antisense dihydroflavonol reductase (AS-DFR) sequences. In initial experiments the effect of introducing three different antisense Antirrhinum majus L. DFR constructs into a single recipient genotype (S50) was assessed. There were no obvious effects on plant biomass, but levels of condensed tannins showed a statistical reduction in leaf, stem, and root tissues of some of the antisense lines. Transformation events were also found, which resulted in increased levels of condensed tannins. In subsequent experiments a detailed study of AS-DFR phenotypes was carried out in genotype S33 using pMAJ2 (an antisense construct comprising the 5′ half of the A. majus cDNA). In this case, reduced tannin levels were found in leaf and stem tissues and in juvenile shoot tissues. Analysis of soluble flavonoids and isoflavonoids in tannin down-regulated shoot tissues indicated few obvious default products. When two S33 AS-DFR lines were outcrossed, there was an underrepresentation of transgene sequences in progeny plants and no examples of inheritance of an antisense phenotype were observed. To our knowledge, this is the first report of the genetic manipulation of condensed tannin biosynthesis in higher plants.
Condensed tannins are polymeric flavonoid molecules that are found in a range of higher plant species. These secondary compounds are of particular value in forage crops because their presence in the foliage of herbage legumes at levels of up to 3 to 4% dry weight is regarded as being a beneficial agronomic trait. The accumulation of tannins in shoot tissues is beneficial for grazing livestock from the perspective of the prevention of pasture bloat (Waghorn et al., 1990); as these highly hydroxylated polymers collapse, protein foams within the rumen in a dose-dependent manner (Tanner et al., 1995). Another advantage of condensed tannins is that they increase the levels of bypass protein, passing through the rumen of grazing animals (Barry and Duncan, 1984), and therefore the accumulation of these end products can increase livestock productivity in an environmentally sensitive manner. Because the major European forage legumes white clover and lucerne do not contain tannins in leaf tissues, strategic work aims at transferring the foliar tannin trait from birdsfoot trefoil (Lotus corniculatus L.) to these major pasture crop species (Morris and Robbins, 1997).
The focus of this paper, however, is to explore the possibility of decreasing the levels of condensed tannins in crop species using antisense technology. Lowering the amounts of condensed tannins in animal feedstuff is important both for ruminant and nonruminant (monogastric) livestock. At high levels (>3–4% dry weight) tannins in forage and fodder are deleterious for use with ruminants, and such levels reduce both palatability and nutritive value (for review, see Mueller-Harvey and McAllan, 1992). This phenomenon mitigates against the effective use of a number of potential forage crop species that have good levels of primary productivity, often in nonoptimal environments, but are of limited nutritive value because of accumulation of high levels of antinutritive secondary compounds. For monogastric species such as pigs and chickens, tannins in feedseeds are generally detrimental at both high and low levels (Griffiths, 1989). Methods of reducing levels of condensed tannins in tanniferous forages, fodders, and feedseeds are therefore of considerable interest (Bavage et al., 1997a; Morris and Robbins, 1997).
The genetic control of tannin biosynthesis has been elucidated in recent years in the studies of tannin mutants in barley (Jende-Strid, 1991) and Lotus japonicus (Regel) Larsen (M.P. Robbins and T.E. Evans, unpublished data). One particular feature of interest is that the initial enzymatic steps of the anthocyanin pathway are also common with condensed tannin biosynthesis. Therefore, the cloning of genes involved in anthocyanin and flavonoid biosynthesis (for review, see Forkmann, 1993) has direct significance for workers studying the agronomically important pathways that lead to tannins and related polyphenolics.
DFR is a particularly attractive target with reference to tannin biosynthesis. The gene corresponding to this enzyme activity has been cloned from a number of plant species, including maize, barley, Gerbera sp., and Antirrhinum majus L. DFR is the most distal gene in the tannin biosynthetic pathway that has been cloned and identified to date, and it shows high levels of sequence homology when comparisons are made between clones isolated from a range of higher plant species (Charrier et al., 1995). Therefore, if one wishes to make specific modifications to the tannin biosynthetic pathway, manipulative strategies aimed at DFR appear to be of potential value. The position of DFR with respect to other enzymes and related secondary pathways is shown in Figure 1.
Figure 1.
Interrelationship of different classes of flavonoids in higher plants. Letters beside arrows indicate enzymes catalyzing conversions between classes. CHI, Chalcone isomerase; F3OH, flavanone 3-hydroxylase; FDR, flavan 3,4-diol reductase; FS, flavone synthase; FlS, flavonol synthase; AS, anthocyanin synthase; 3GT, 3-glycosyl transferase; CE, condensing enzyme; and PE, polymerizing enzyme. Additional enzymatic steps exist in this pathway that alter B-ring hydroxylation and the stereospecificity of monomer units, but these have been omitted for the sake of clarity.
In previous work (Carron et al., 1994) we reported the introduction of three different antisense A. majus DFR constructs into three recipient Lotus corniculatus genotypes. In genotypes that accumulated low (S33) or medium (S50) levels of tannin, antisense root culture lines were produced with decreased levels of condensed tannin accumulation. In genotype S33 three lines were found, RFD8, RFD19, and RFD38, in which 80% reductions of tannin accumulation were noted relative to controls. In genotype S50 low tannin lines were also produced with reductions of up to 70%, RFD40 and RFD28. Although root cultures have been subjected to study, no results have been previously presented regarding a detailed analysis of AS-DFR plants. In this paper we report the analysis of cotransformed S50 and S33 plants harboring these AS-DFR constructs. Condensed tannin levels in leaf, stem, and root tissues under a variety of environmental growth conditions have been determined, and in addition to PCR and RT-PCR analysis we report, to our knowledge, the first preliminary analysis of default products in tissues of AS-DFR plants. Progeny were also produced by outcrossing control and antisense lines, and we report data regarding inheritance of transgene sequences.
MATERIALS AND METHODS
Source Material
Transgenic lines subjected to study in this paper are outlined in Tables I and II. Transformed and cotransformed plants were regenerated from “hairy root” lines grown at 25°C on Gamborg's B5 basal medium (Imperial Laboratories, Ltd., Andover, UK) supplemented with 1% agar and 3% Suc (Carron et al., 1994). After 4 to 6 weeks of growth in the dark, plates were transferred to light (80 μmol m−2 s−1) and shoots started to appear after 1 to 2 weeks. After 1 month in the light, shoots, together with basal nodes, were excised and transferred to 50-mL Sterilin tubes (Bibby Sterilin Ltd., Stones, Staffordshire, UK) containing solid Murashige and Skoog medium (Imperial Laboratories, Ltd.) supplemented with 1% agar and 3% Suc and then allowed to root. In genotype S33 control transformed lines (C26, C28, and C32) and vector controls (C33/3, C33/5, and C33/10) regenerated shoots poorly when grown on solid medium. These lines were therefore cultured in liquid B5 basal medium plus 3% Suc in the light, and the resultant shoots were then detached and transferred to solid Murashige and Skoog medium for rooting.
Table I.
Control and RFD lines in the S50 genotype analyzed in this study and a summary of PCR analysis
| Description of Line | Designation | nptII | CaMV-nos |
|---|---|---|---|
| Recipient (nontransformed control) | S50 | − | − |
| A. rhizogenes transformed | C22 | − | − |
| Cotransformed with pROK2 (vector control) | C50/1, C50/2 | + | + |
| Cotransformed with pMAJ1 | RFD60, RFD61, RFD62, RFD63, RFD64, RFD65 | + | +a |
| Cotransformed with pMAJ2 | RFD28, RFD31, RFD40 | + | + |
| Cotransformed with pMAJ3 | RFD9, RFD66 | + | +b |
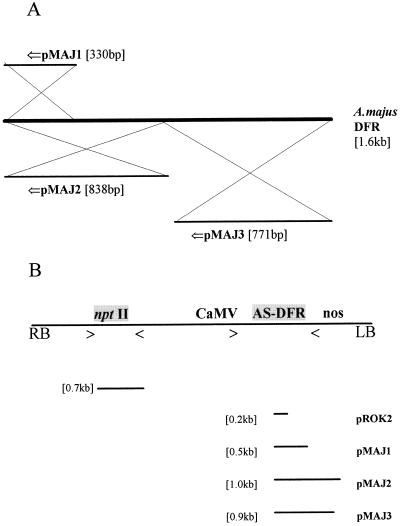
Details of control and antisense constructs (pROK2, pMAJ1, pMAJ2, and pMAJ3) and derived amplification products are shown in Figure 2. Amplification products with nptII primers confirm the transfer of the selectable marker from binary vector T-DNA to L. corniculatus genomic DNA. CaMV-nos amplification products confirm the presence of promoter and terminator sequences and produce characteristically sized products from each of the four constructs in this study. +, Product noted; −, no product after amplification.
No amplification product from RFD65.
Truncated amplification product from RFD66 (750 bp only).
Table II.
Control and RFD lines in the S33 genotype analyzed in this study and a summary of PCR analysis
| Description of Line | Designation | nptII | CaMV-nos |
|---|---|---|---|
| Recipient (nontransformed control) | S33 | − | − |
| A. rhizogenes transformed | C26, C28, C32 | − | − |
| Cotransformed with pROK2 (vector control) | C33/3, C33/5, C33/10 | + | + |
| Cotransformed with pMAJ2 | RFD4, RFD7, RFD8, RFD19, RFD38, RFD49, RFD50 | + | + |
Details of control and antisense constructs (pROK2, pMAJ1, pMAJ2, and pMAJ3) and derived amplification products are shown in Figure 2. Amplification products with nptII primers confirm the transfer of the selectable marker from binary vector T-DNA to L. corniculatus genomic DNA. CaMV-nos amplification products confirm the presence of promoter and terminator sequences and produce characteristically sized products from each of the four constructs in this study. +, Product noted; −, no product after amplification.
Growth of Plants
Stock plants were established by transferring 4-month-old rooted shoots to John Innes Number 1/Levington's 50:50 medium in 7.5-cm pots. Plants were grown in a growth room at 18°C with an 18-h day under 530 μmol m−2 s−1 light from “white” fluorescent tubes (General Electric). Plants were gradually hardened off in plastic bags and then nodulated with Rhizobium loti Lc 3011 (Institute of Grassland and Environmental Research nomenclature) over a 3-week period. Replicate, whole-shoot cuttings were taken from 10-week-old plants and rooted in sand as described above. Established plants were grown under containment-B greenhouse conditions (June–July, 1995) at an average day temperature of 25 ± 4°C and a minimum night temperature of 15°C, and leaf and stem tissues were harvested and analyzed for condensed tannins after 10 weeks of growth. Rooted and nodulated cuttings were also transferred and grown in vermiculite medium in 7.5-cm pots in a growth room (20°C, 18-h day, 530 μmol m−2 s−1 fluorescent light). Replicated plants were harvested after 6 weeks (harvest 1) or cut back to 5 cm, grown for another 6 weeks, and harvested (harvest 2).
Transgenic and control plants were therefore grown under a number of specified growth conditions for comparative purposes. S50 lines were grown in a growth cabinet in 3-inch pots containing vermiculite and watered with one-half-strength Hoagland nutrient solution without nitrogen, which permitted analysis of both shoot and root tissues. S50 lines were also grown in 12.5-cm pots containing John Innes Number 1/Levington's 50:50 medium under greenhouse conditions The S33 data reported here are from lines grown in 12.5-cm pots containing John Innes Number 1/Levington's 50:50 medium in a growth cabinet watered with 50% nutrient solution with no added nitrogen.
Chemical Analysis of Plant Tissues
Condensed Tannins
Plant tissues were harvested, freeze dried, and powdered before the measurement of condensed tannins. Juvenile shoot tissues were harvested directly, whereas stem and leaf tissues from older plants were subjected to botanical separation after freeze drying. Roots from vermiculite-grown plants were washed vigorously and then used for tannin analysis; however, it was not possible to make accurate determinations of tannin levels in roots from soil-grown plants. All tannin determinations were carried out in duplicate on 30 to 50 mg dry weight of freeze-dried powdered tissues using the butanol/HCl-hydrolysis method of Terrill et al. (1992), and levels were determined by summing values derived from residue and aqueous hydrolysates. Statistical analysis of tannin levels in transgenic plants was made by comparing levels of condensed tannins in individual AS-DFR lines with those in corresponding controls derived from the parental genotype (S33 or S50), wild-type Agrobacterium rhizogenes transformants, and vector transformants of these genotypes (Tables I and II).
Leaf Flavonoids
Freeze-dried powder samples of leaves (100 mg) were extracted three times with 25 mL of 80% MeOH for 2 h with shaking at 20°C. After filtration and MeOH removal under a vacuum at 50°C, the remaining aqueous extracts were concentrated on an activated C18 Sep-Pak cartridge (500 mg) (Waters) and bound phenolic compounds were eluted with 4 mL of 100% MeOH. Flavonoid profiles were obtained by gradient HPLC onto a μNovapak C18 RCM cartridge (Waters) with a linear MeOH:acetic acid (5%) gradient from 0 to 100% MeOH in 50 min at a flow rate of 2 mL min−1. Eluting peaks were monitored with a diode array detector (model 990, Waters) at 350 nm, and spectra were recorded between 240 and 400 nm to obtain their UV/visible absorption spectra. Acid, alkali, and enzymic hydrolysis was carried out on 1-mL extracts with 1 m HCl at 100°C for 1 h, with 0.5 m NaOH at 50°C for 30 min, or with 100 units of β-glucosidase (sweet almond), pH 5.0, at 37°C for 2 h. Hydrolyzed samples were diluted to 5 mL, adjusted to pH 6.0 with NaOH or HCl, and concentrated on an activated C18 Sep-Pak cartridge, and the flavonoids were eluted with 4 mL of MeOH. Flavonoid glycosides and aglycones were identified by their retention times and UV/visible spectra compared with authentic standards obtained from Sigma-Aldrich and Apin Chemicals, Ltd. (Oxford, UK).
PCR Analysis of Transgenic Lines
In this study plants were produced that harbored a range of transgene constructs (Table I). PCR from genomic DNA derived from leaf tissues was employed using well-characterized PCR primers (see Fig. 2). Genomic DNA was extracted from leaf tissues of control and antisense plants using the method of Dellaporta et al. (1983), as modified by Robbins et al. (1991). Amplification of nptII sequences used a primer pair (nptIIa: 5′ GAG GCT ATT CGG CTA TGA CGT; and nptIIb: 3′ ATC GGG AGC GGC GAT ACC GTA), which amplified a 700-bp sequence from the nptII gene in the T-DNA of constructs derived from pROK2. Antisense sequences were amplified using CaMV-specific (5′ CTG ATA TCT CCA CTG AC) and nos-specific (3′ TCA TCG CAA GAC CGG C) primers, which amplified characteristic fragments from different antisense constructs. Amplified fragment sizes were 200 bp for pROK2, 500 bp for pMAJ1, 1 kb for pMAJ2, and 900 bp for pMAJ3. Similar methods were used when analyzing for the presence of the transgene in T1 progeny derived from original transgenic lines.
Figure 2.
A, Origin of RFD constructs used in this study. Three constructs (pMAJ1, pMAJ2, and pMAJ3) were prepared in the binary vector pROK2 from a cDNA corresponding to an A. majus DFR gene. The 5′→3′ of the original DFR sequence is indicated by ⇐. B, Schematic representation of T-DNA of control and antisense gene constructs. RB, Right border of T-DNA; LB, left border of T-DNA. Positions of nptII gene and AS-DFR constructions within T-DNA are indicated. nptII, CaMV, and nos primer positions are marked with > and <. Sizes of predicted PCR products from the control construct and from the three AS-DFR constructs are indicated.
Transcript Analysis
RNA was extracted from Lotus corniculatus shoot and root tissues using the method of Ougham and Davies (1990). Plant material was harvested, frozen in liquid nitrogen, and stored at −70°C before extraction. Nucleic acid was quantified on a UV spectrophotometer (model PU 8720, Pye Unicam Ltd., Cambridge, UK) and 10-μg samples were run on 1% formaldehyde gels. RNA was blotted onto Hybond N membranes (Amersham) and probed using a labeled L. corniculatus fragment, pLCDFRA (accession no. X97576), which contains 683 bp of an endogenous DFR gene amplified from genomic DNA (Bavage and Robbins, 1994; Bavage et al., 1997b).
However, it proved impossible to detect transgene or endogenous DFR gene expression in RNA extracted from leaf or root tissues using this protocol, so we adopted a RT-PCR approach to assess transcript abundance. The basic method was as described by Bavage et al. (1997b). Total RNA was extracted from leaf tissues using the cetyl-trimethyl-ammonium bromide method of Chang et al. (1993), and mRNA was isolated using Dynal oligo(dT) Dynabeads (Dynal AS, Oslo, Norway). Then, first-strand cDNA was synthesized from 0.5 μg of mRNA using Moloney murine leukemia virus RT and oligo(dT) primer (Stratagene) in a total reaction volume of 50 μL. Five-microliter aliquots of resultant cDNA were diluted one-tenth with sterile, distilled water and used as a target for PCR analysis.
To determine the expression of endogenous DFR and transgene antisense-DFR genes, sequence-specific primers were designed. For A. majus DFR, primers were 5′ CAA GCA AAA ACC GTC AAG and 3′ GCG ATA TCA TCA TAA ATT GTT GC; predicted product size was 450 bp. For L. corniculatus DFR, primers were 5′ ATA AAC GGG GTG CTA GAC and 3′ ACA ACA AGG GAT GGA ATG; predicted product size was 300 bp. Cycling conditions for RT-PCR were 94°C for 3 min; 10 cycles of 94°C for 1 min, 53°C for 1 min, and 72°C for 1.5 min; 20 cycles of 94°C for 1 min, 55°C for 1 min, and 72°C for 1.5 min; followed by a final extension of 72°C for 3 min. Products of PCR amplification were analyzed by electrophoresis of 20-μL aliquots of reactions on 0.7% agarose gels in Tris-borate-EDTA buffer and visualized with ethidium bromide. This protocol has been used successfully to detect and discriminate between A. majus and L. corniculatus DFR transcripts in cDNA from “hairy root” cultures (Bavage et al., 1997b).
Southern-Blot Analysis of Parental Lines
For lines selected for progeny analysis transgene copy numbers were estimated using the following method. Genomic DNA was isolated as for PCR analysis and after fluorimetric quantification using bisbenzimide (Brunk et al., 1979), 11 μg of L. corniculatus genomic DNA was cut with appropriate restriction enzymes: HindIII and XbaI. After ethanol precipitation, digests of genomic DNA were run on a 0.8% gel and transferred to Zetaprobe membrane by alkaline blotting. The membrane was probed with the 1.6-kb A. majus DFR fragment from pJAM212 (Beld et al., 1989) and insert copy numbers were estimated by counting numbers of hybridizing border fragments noted in XbaI digests.
Production of Progeny and Analysis of T1 Generation
For production of progeny, S33, C26, RFD8, and RFD19 plants were grown to flowering in a containment-B greenhouse. Lines were then manually outcrossed to genotype S50. This protocol was necessary because L. corniculatus is a typical outbreeding forage species and because Agrobacterium rhizogenes-transformed plants exhibit reduced female fertility (Webb et al., 1990).
Statistical Analysis of Condensed Tannin Levels in Transgenic Plants
The data were assessed for normality using the Shapiro-Wilk test (Shapiro and Wilk, 1965). In most of the experiments described in this paper, the Shapiro-Wilk test indicated that data were from a nonnormal population, which could not be corrected using data transformations. Therefore, we used the sign test, which is thought to be robust to nonnormality and asymmetric data (Sprent, 1993). The sign test was used to construct 85% (maximum possible from the available tables) confidence limits for the control lines. Any antisense line that had all of its values outside of the confidence interval of the control lines was defined as having a statistically different condensed tannin content in comparison with the control lines. Such lines are marked with asterisks in Figures 3–6.
Figure 3.
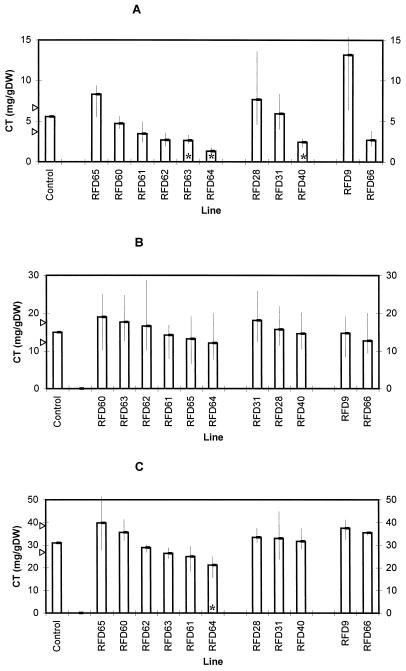
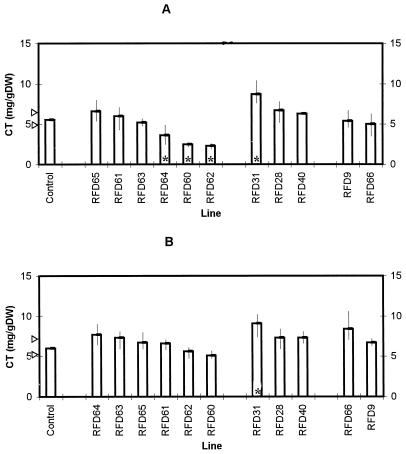
Levels of condensed tannin (CT) in S50 plants harboring control and AS-DFR constructs at harvest 1. Bars represent mean values, with the control bars corresponding to the mean of lines S50, C22, C50/1, and C50/2. For each control and antisense line four replicate plants were analyzed. Arrows indicate 85% confidence limits for the median of the control condensed tannin values using the sign test. Vertical lines indicate the range of values noted for each antisense line. Asterisks indicate antisense ranges outside of the control confidence intervals. A, Leaves, growth room (harvest 1); B, stems, growth room (harvest 1); and C, roots, growth room (harvest 1). DW, Dry weight.
Figure 6.
Levels of condensed tannin (CT) in S33 control plants and lines harboring the pMAJ2 construct. Bars represent mean values, and the control bars correspond to the mean of lines S33, C26, C28, and C32. For each control and antisense line three replicate plants were analyzed. Arrows indicate 85% confidence limits for the median of the control condensed tannin values using the sign test. Vertical lines indicate the range of values noted for each antisense line. Asterisks indicate antisense ranges outside of the control confidence intervals. A, Juvenile growth; B, leaves; and C, stems. DW, Dry weight.
RESULTS
PCR Analysis and Initial Characterization of Control and AS-DFR Lines
The lines in this study are outlined in Tables I and II. There were six classes of lines in the S50-line background and these corresponded to the original recipient genotype, an A. rhizogenes control line, vector control lines, and transgenic lines harboring three types of the AS-DFR gene construct. In the S33-line background, the recipient genotype was analyzed together with A. rhizogenes control lines, vector control lines, and lines harboring pMAJ2, which was the construct that produced optimal antisense phenotypes in cotransformed root cultures (Carron et al., 1994).
A schematic representation of the control binary vector and the three classes of AS-DFR construct is given in Figure 2 (Carron et al., 1994). The binary vector used in these experiments was pROK2, which includes the CaMV and nos sequences within its T-DNA. A. majus DFR sequences resided between these sequences in an antisense orientation relative to the CaMV. To verify the cotransformed nature of the lines in this study, PCR analysis was carried out on genomic DNA extracted from control and antisense plants. We used both nptII and CaMV plus nos primer pairs to detect transgene sequences inserted into genomic DNA in these lines. The generation of predictable amplification fragments is shown schematically in Figure 2B.
A summary of the PCR analysis of primary transformant lines is shown in Tables I and II. All vector and AS-DFR lines were scored positive using nptII primers. Using CaMV and nos primers, all amplification products were as predicted, with two exceptions. RFD66 had a truncated transgene, whereas no transgene was detectable by PCR in RFD65. This latter result may be attributable to poor amplification using this primer pair or alternatively to a partial T-DNA deletion from the left-hand border of the T-DNA of pMAJ1. We did not perform Southern-blot analysis to clarify the status of RFD65.
Analysis of S50 AS-DFR Lines Grown in Controlled Environments
Our initial observations concentrated on the effects of AS-DFR constructs in an S50-line background. Replicate plants were set up and grown in vermiculite under growth-room conditions. Leaf, stem, and root tissues were harvested and subjected to analysis. No obvious effects were noted on biomass (data not shown) or plant morphologic characteristics, although some of the antisense lines appeared to be paler green and had a more erect phenotype.
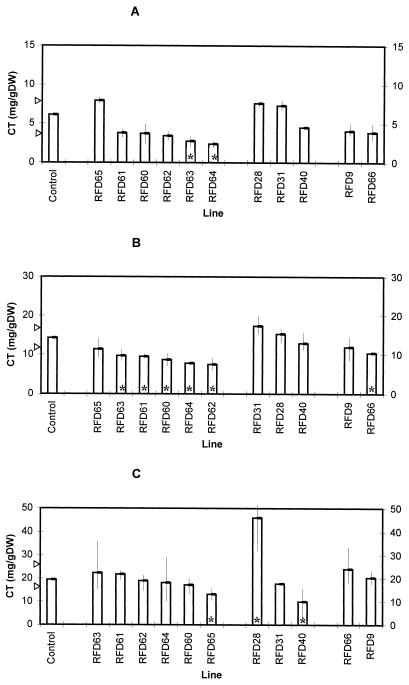
We analyzed levels of condensed tannins after 6 weeks of initial growth (harvest 1) and 6 weeks after cutting and regrowth (harvest 2). This protocol was used because forages and fodder crops are regularly grazed or cut throughout the growing year, so phenotypes noted after defoliation are of agronomic significance. In initial growth (Fig. 3) and regrowth (Fig. 4), levels of condensed tannins in leaf and stem tissues of S50 L. corniculatus plants were similar to those previously reported by Carter et al. (1997). However, statistically significant differences were apparent between control and AS-DFR lines. Levels of tannins in leaf tissues were reduced in RFD63 and RFD64 (in both harvests), whereas levels in RFD40 were lower than levels in controls at harvest 1 only. Phenotypes were also noted in stem tissues at the second harvest, with reductions in condensed tannins being detected in RFD60, RFD61, RFD62, RFD63, RFD64, and RFD66. Because these plants had been grown in vermiculite, it was also possible to estimate the tannin levels in root tissues of S50 control and AS-DFR lines (Figs. 3C and 4C). Root results were difficult to fully interpret, but statistically significant reductions in RFD64, RFD65, and RFD40, and an increase in condensed tannin levels in the roots of RFD28 were apparent.
Figure 4.
Levels of condensed tannin (CT) in S50 plants harboring control and AS-DFR construct at harvest 2. Bars represent mean values, and control bars correspond to the mean of lines S50, C22, C50/1, and C50/2. For each control and antisense line three replicate plants were analyzed. Arrows indicate 85% confidence limits for the median of the control condensed tannin values using the sign test. Vertical lines indicate the range of values noted for each antisense line. Asterisks indicate antisense ranges outside of the control confidence intervals. A, Leaves, growth room (harvest 2); B, stems, growth room (harvest 2); and C, roots, growth room (harvest 2). DW, Dry weight.
S50 AS-DFR Phenotypes in Greenhouse-Grown Plants
We were also interested to know whether tannin-down-regulated phenotypes were also maintained in plants grown under other environmental conditions, because growth temperature in particular has been found to affect tannin accumulation in L. corniculatus and Lotus uliginosus L. (bigfoot trefoil) (Lees et al., 1994; Carter et al., 1995). S50 AS-DFR lines were therefore grown in soil as described in Methods under summer greenhouse conditions. In this case low-tannin phenotypes were also noted in leaf tissues, with levels below those found in control tissues being observed in RFD60, RFD62, and RFD64 (Fig. 5A). No lines were detected that showed statistically reduced tannin levels in stem tissues (Fig. 5B), but RFD31 had significantly elevated tannin levels in both leaves and stems.
Figure 5.
Levels of condensed tannin (CT) in S50 control and AS-DFR plants grown in a greenhouse. Bars represent mean values, and control bars correspond to the mean of lines S50, C22, C50/1, and C50/2. For each control and antisense line three replicate plants were analyzed. Arrows indicate 85% confidence limits for the median of the control condensed tannin values using the sign test. Vertical lines indicate the range of values noted for each antisense line. Asterisks indicate antisense ranges outside of the control confidence intervals. A, Leaves; B, stems. DW, Dry weight.
Results from growth-room- and greenhouse-grown plants are summarized in Table III. It is interesting to note that some lines (RFD61, RFD63, RFD65, RFD28, RFD40, and RFD66) showed a phenotype only under growth-room conditions. RFD31 expressed a phenotype only under greenhouse conditions, whereas RFD60, RFD62, and RFD64 showed effects in both environments. When different constructs are compared, it is clear that cotransforming with pMAJ1, pMAJ2, or pMAJ3 resulted in lines that accumulated reduced levels of condensed tannins relative to controls. However, two pMAJ2 lines, RFD28 and RFD31, surprisingly showed increases in condensed tannin levels; these effects were not found in any of the S50 pMAJ1 and pMAJ3 lines analyzed in this study.
Table III.
Summary of RFD phenotypes in the S50 background
| Organ/Environment | pMAJ1
|
pMAJ2
|
pMAJ3
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Line | Line | Line | |||||||||
| 60 | 61 | 62 | 63 | 64 | 65 | 28 | 31 | 40 | 9 | 66 | |
| Leaf (harvest 1) | – | – | – | ↓ | ↓ | – | – | – | ↓ | – | – |
| Leaf (harvest 2) | – | – | – | ↓ | ↓ | – | – | – | – | – | – |
| Leaf (greenhouse) | ↓ | – | ↓ | – | ↓ | – | – | ↑ | – | – | – |
| Stem (harvest 1) | – | – | – | – | – | – | – | – | – | – | – |
| Stem (harvest 2) | ↓ | ↓ | ↓ | ↓ | ↓ | – | – | – | – | – | ↓ |
| Stem (greenhouse) | – | – | – | – | – | – | – | ↑ | – | – | – |
| Root (harvest 1) | – | – | – | – | ↓ | – | – | – | – | – | – |
| Root (harvest 2) | – | – | – | – | – | ↓ | ↑ | – | ↓ | – | – |
Results summarize changes in condensed tannin levels in leaf, stem, and root tissues and under different growth conditions. Transgenic lines analyzed harbored the pMAJ1 construct (RFD60, 61, 62, 63, 64, 65), pMAJ2 (RFD28, 31, and 40), or pMAJ3 (RFD9 and 66). ↓, Statistically significant reduction in condensed tannin levels relative to four independent control lines using the sign test under a given environment; ↑, a statistically significant increase relative to control lines; –, not statistically different from control lines.
Analysis of S33 AS-DFR Lines
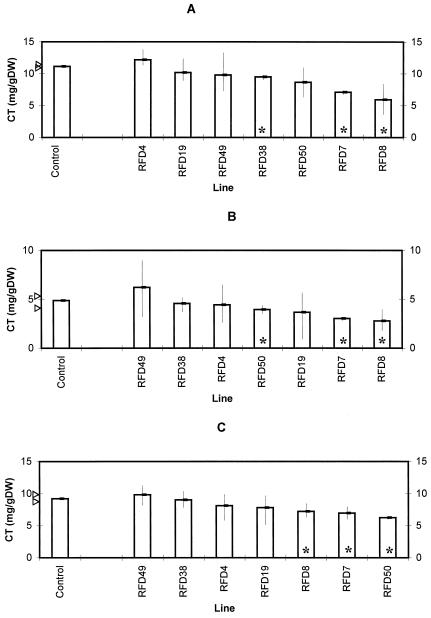
In view of the complexity of interpreting results when analyzing three constructs in an S50 background, we decided to perform a detailed analysis of pMAJ2 cotransformants in the S33 genetic background. These lines had previously been well characterized at the root-culture level (Carron et al., 1994), and there were a number of examples (RFD8, RFD19, and RFD38) that showed large reductions in condensed tannin levels relative to S33 control lines when analyzed as root cultures. Therefore, we analyzed tannin levels in juvenile shoots (Fig. 6A), leaves (Fig. 6B), and stems (Fig. 6C) in control and AS-DFR plants. In juvenile tissues, RFD7, RFD8, and RFD38 had tannin levels lower than the control lines. In leaves and stems of 6-week-old plants of RFD7, RFD8, and RFD50, tannin levels were reduced relative to controls. These results are summarized in Table IV. Although none of the reductions in the quantity of condensed tannins was on the order of 80%, as found in root cultures, reductions in levels of condensed tannins were similar. In juvenile shoot tissues reductions were in the order of 4 mg/g dry weight, and in leaves and stems they were in the order of 2 mg/g dry weight. Corresponding tannin reductions in root cultures for effective antisense lines were on the order of 0.25 mg/g fresh weight, which translates to approximately 2.5 mg/g dry weight.
Table IV.
Summary of RFD phenotypes in the S33 background
| Organ | pMAJ2
|
||||||
|---|---|---|---|---|---|---|---|
| Line | |||||||
| 4 | 7 | 8 | 19 | 38 | 49 | 50 | |
| Juvenile shoot | – | ↓ | ↓ | – | ↓ | – | – |
| Mature leaf | – | ↓ | ↓ | – | – | – | ↓ |
| Mature stem | – | ↓ | ↓ | – | – | – | ↓ |
Results summarize changes in condensed tannin levels in leaf, stem, and juvenile shoot tissues. All transgenic lines harbor the pMAJ2 construct (RFD4, 7, 8, 19, 38, 49, and 50). ↓, Statistically significant reduction in condensed tannin levels relative to four independent control lines using the sign test. –, Not statistically different from control lines.
Another point of interest is that when previously analyzed as root cultures, only RFD8, RFD19, and RFD38 showed a reduction relative to controls; RFD4 and RFD7 were ineffective antisense lines and RFD49 and RFD50 were not analyzed. Therefore, of the effective lines at root culture level, only RFD8 gave a corresponding phenotype in shoot tissues. RFD7, which was ineffective at the root culture level, had reduced tannin levels in leaves, stems, and juvenile shoot tissues. A conclusion from this data set would appear to be that antisense phenotypes noted at the root culture level do not necessarily correspond to phenotypes in regenerated plant tissues.
Molecular Analysis of S33 AS-DFR Lines
Measurement of condensed tannin levels in S33 AS-DFR lines identified a number of interesting effective and ineffective antisense lines. To further our understanding of these antisense phenotypes, transcript, RT-PCR, and HPLC analyses were carried out on selected AS-DFR lines to characterize underlying and associated molecular parameters.
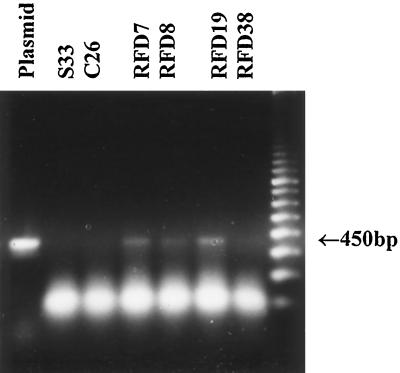
Initial experiments focused on attempting to measure AS-DFR and endogenous DFR transcripts in root, leaf, and stem tissues of transgenic L. corniculatus lines. In previous work (Carron et al., 1994), we found it straightforward to detect antisense transcript in S33 and S50 root cultures. However, when using either the A. majus DFR sequence or a L. corniculatus DFR probe (Bavage et al., 1997b) on RNA blots from these plant tissues, no transcript was detectable in either case. One possible explanation might be that flavonoid gene and antisense transcripts are at very low levels in regenerated plant tissues compared with corresponding root cultures. Alternatively, the high levels of soluble and insoluble condensed tannins in regenerated plants may compromise conventional northern blotting. Therefore, we tried to detect transcripts using RT-PCR from cDNA derived from control and antisense lines. We have found this method to be convenient for the detection of endogenous and introduced A. majus DFR genes in L. corniculatus root cultures (Bavage et al., 1997b). Using this method we obtained the results shown in Figure 7. No antisense amplification product was detectable in S33 (recipient genotype) or C26 (control line); however, a 450-bp fragment was noted in RFD7, RFD8, and RFD19, but not in RFD38. The faintness of these amplification products was consistent with low levels of expression of the antisense transgene, and it is interesting to note the detectable product in both effective RFD7, RFD8, and ineffective RFD19 lines.
Figure 7.
RT-PCR analysis of cDNA prepared from selected S33 control and pMAJ2 lines. Amplifications from selected lines were performed as outlined in Methods. The Plasmid lane is the amplification product from 20 pg of pJAM212 (A. majus DFR cDNA). The arrow indicates the predicted size of the amplification product derived from pMAJ2. DNA-marker lane is 2.5 μg of a 100-bp ladder (Pharmacia-Biotech).
Chemical Analysis of S33 AS-DFR Lines
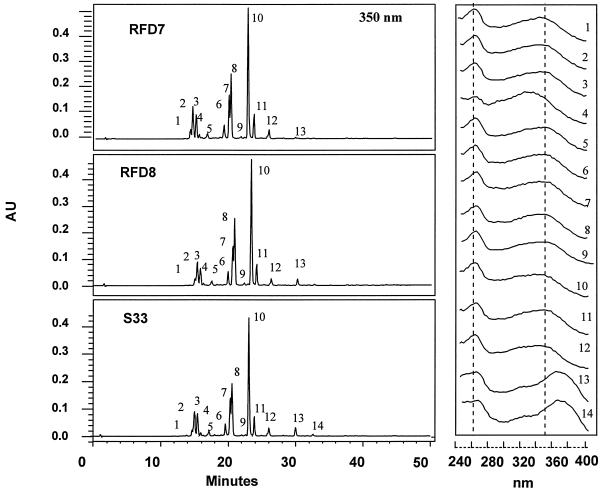
Analysis of the MeOH-soluble flavonoids of leaves of AS-DFR lines showed no major qualitative or quantitative changes in the levels of flavonols and flavonol glycosides compared with extracts from control lines (Fig. 8). Previous studies on the leaf flavonoid glycosides of L. corniculatus (Raynaud et al., 1982) indicated the presence of 13 kaempferol and quercetin glycosides, with kaempferol- and quercetin-3,7-dirhamnoside, and the corresponding 3-rhamnoside-7-glucosides, as the major components. However, we were able to detect only kaempferol glycosides (Fig. 8, λ max band I, 260 nm) in extracts of genotype S33. This was confirmed by acid hydrolysis of extracts that yielded only kaempferol (peak 13). A combination of alkali and enzymatic hydrolysis indicated other peaks as kaempferol-3,7-dirhamnoside (peak 10), kaempferol-3-rhamnoside-7-glucoside (peak 7), kaempferol-3-glucoside (peak 8), kaempferol-7-rhamnoside (peak 11), and kaempferol-3-rhamnoside (peak 9). Both kaempferol and quercetin glycosides, however, were found in leaves and stems of genotype S50.
Figure 8.
HPLC profile and UV/visible spectra of MeOH extracts of 6-week-old shoot tissues from control (S33) and AS-DFR lines (RFD7 and RFD 8) of L. corniculatus. Condensed tannin contents of these lines were: S33, 10.7 mg/g dry weight; RFD7, 7.1 mg/g dry weight (66% of control level); and RFD8, 5.9 mg/g dry weight (55% of control level). Extracts from equivalent amounts of tissue were subjected to HPLC with diode-array detection. HPLC profiles were monitored at 350 nm and spectra were recorded between 240 and 400 nm. Chemical analysis indicated that the majority of peaks were either kaempferol or kaempferol glycosides (peaks 1–3 and 5–13). Identified peaks are: kaempferol (peak 13), kaempferol-3-rhamnoside-7-glucoside (peak 7), kaempferol-3-glucoside (peak 8), kaempferol-3-rhamnoside (peak 9), kaempferol-3,7-dirhamnoside (peak 10), and kaempferol-7-rhamnoside (peak 11).
Genetic Analysis of Parental Lines and Inheritance of Transgene in Progeny
Following on from the characterization of AS-DFR phenotypes in primary cotransformed lines, we chose to study transgene inheritance in four crosses. These were S33×S50 (recipient genotype), C26×S50 (control transformant), RFD8×S50 (effective antisense line), and RFD19×S50 (ineffective antisense line). As a prelude to this experiment, we performed an analysis of transgene sequences in RFD8 and RFD19. Southern blotting indicated that after digestion with XbaI, RFD8 had five border fragments and RFD19 had three border fragments. HindIII digests were also consistent with three transgene copies in RFD19 and three or four transgene copies in RFD8 (data not shown). These copy numbers are similar to those determined by genomic slot blotting (Carron et al., 1994) and are typical for binary T-DNA sequences introduced into L. corniculatus using A. rhizogenes.
Because some of the antisense plants showed decreased tannin phenotypes in shoot tissues, we studied the inheritance of the antisense T-DNA and analyzed the progeny for the corresponding chemical phenotype. The crosses were made using S33, C26, RFD8, and RFD19 as pollen donors against S50, and seed from resulting pods was collected. There were no obvious effects on seed number and on numbers of hard seeds. When seeds were planted, resulting plants were grown in a growth room and analyzed for the presence of transgenes and for levels of condensed tannin in leaves and stems. Tables V and VI show the results of PCR analysis for 24 progeny from RFD8 and 21 progeny from RFD19. nptII primers gave highly reproducible results, and 6 of 24 (25%) of the progeny from the RFD8 cross were scored as positive. With regard to the RFD19 cross, 10 of 21 (48%) of the progeny gave PCR products with nptII primers. The results from RFD8 are clearly indicative of gross underrepresentation of transgenes in the progeny of this cross. Even if all of the transgene copies in RFD8 noted on Southern-blot analysis shared linked integration sites on a single chromosome, one would still expect 50% of the progeny to be transgene positive, and in fact this was the result noted in the progeny of RFD19.
Table V.
PCR analysis of progeny from an effective RFD (RFD8)
| Line | nptII | CaMV-nos |
|---|---|---|
| RFD8 (five copies of transgene) | + | + |
| S50 | − | − |
| 7 | + | + |
| 13 | + | − |
| 15 | + | + |
| 18 | + | − |
| 22 | + | + |
| 23 | + | + |
| Remainder | − | − |
| Transgene inheritance | 25% | 17% |
Parental lines were analyzed together with 24 progeny lines (1–24). Amplification products with nptII primers confirm the presence of the selectable marker from binary vector T-DNA in genomic DNA of parental and progeny lines. CaMV-nos amplification products confirm the presence of promoter and terminator sequences and the antisense DFR fragment from pMAJ2. +, Product noted; −, no product noted after amplification.
Table VI.
PCR analysis of progeny from an ineffective RFD (RFD19)
| Line | nptII | CaMV-nos |
|---|---|---|
| RFD19 (three copies of transgene) | + | − |
| S50 | − | − |
| 1 | + | + |
| 2 | + | + |
| 4 | + | + |
| 9 | + | + (−) |
| 11 | + | − |
| 12 | + | + |
| 13 | + | + |
| 14 | + | + |
| 20 | + | + (−) |
| 22 | + | − |
| Remainder | − | − |
| Transgene inheritance | 48% | 38 (29)% |
Parental lines were analyzed together with 21 progeny lines (1–22). Amplification products with nptII primers confirm the presence of the selectable marker from binary vector T-DNA in genomic DNA of parental and progeny lines. CaMV-nos amplification products confirm the presence of promoter and terminator sequences and the antisense DFR fragment from pMAJ2. +, Product noted; −, no product noted; +(−), product not consistently observed after amplification.
PCR analysis of progeny using CaMV and nos primers to amplify the AS-DFR sequence gave results that are a little more difficult to explain. Two of the RFD8 progeny (13 and 18) and four of the RFD19 progeny (9, 11, 20, and 22) were nptII positive but did not reproducibly yield PCR products using CaMV and nos. There are several possible explanations for these observations. One possibility is that this primer pair was not fully optimized for amplification and that these are false-negative results (Luo et al., 1992). Another possibility is that in at least some of these lines there has been truncation of transgene sequences at the left-hand border of binary vector T-DNA.
To study the inheritance of the antisense phenotype in progeny, six plants were selected from all four crosses. Six seed-grown plants from S33 and C26 crosses were selected, together with 6 nptII-positive progeny from the RFD8 and RFD19 crosses. Mean condensed tannin contents for progeny were as follows: S33 (stems, 20.0 ± 1.4 mg/g dry weight; leaves, 8.0 ± 1.9 mg/g dry weight); C26 (stems, 17.0 ± 1.1 mg/g dry weight; leaves, 10.3 ± 2.9 mg/g dry weight); RFD8 (stems, 22.8 ± 2.4 mg/g dry weight; leaves, 12.3 ± 3.8 mg/g dry weight); and RFD19 (stems, 15.9 ± 2.1 mg/g dry weight; leaves, 10.4 ± 2.3 mg/g dry weight). Therefore, transgene-positive progeny from RFD8 and RFD19 showed no significant reduction in mean tannin levels relative to control progeny.
DISCUSSION
Antisense is a powerful technology that may be useful for the analysis of plant metabolic pathways. Secondary pathways are an area of metabolism that is particularly attractive for plant genetic manipulation (Bavage et al., 1997a; Robbins et al., 1997). The results presented in this paper can be compared with other examples of heterologous antisense in higher plants. Notable examples include the expression of antisense sequences of petunia (Petunia hybrida) CHS genes in tobacco (van der Krol et al., 1988) and the antisense expression of a lignin-specific O-methyltransferase from Populus tremuloides in tobacco (Dwivedi et al., 1994). In the experiments reported here, an A. majus DFR gene has been expressed in antisense orientation in L. corniculatus and phenotypes have been noted in leaf, root, and stem tissues.
One noteworthy aspect of this study is the observation that all three constructs (pMAJ1, pMAJ2, and pMAJ3) derived from different sections of the A. majus cDNA gave rise to transgenic plants with reduced levels of condensed tannins. However, individual lines exhibited decreases in tannin levels only in some tissues and under some environmental growth conditions (Tables III and IV). For example, RFD61 and RFD66 showed reduced tannin levels only in stems; RFD60, RFD62, and RFD63 showed reduced tannin levels in leaves and stems; RFD40 showed reduced tannin levels in leaves and roots; RFD65 showed reduced tannin levels in roots only; and RFD64 showed reduced tannin levels in leaves, stems, and roots. This phenomenon of organ-specific antisense phenotypes found with a construct driven by a “constitutive” promoter is curious. Previous evidence from a number of sources indicates that the 35S CaMV promoter expresses consistently in different tissues and at different growth stages when introduced into L. corniculatus (Forde et al., 1991; Webb et al., 1994). The data presented here are at variance with these observations. One possible reason is that the CaMV promoter is positioned next to different tissue-specific or development-specific enhancers in different transformation events, and that each transgenic plant therefore may show differences in tissue-specific expression even though all plants in the study share a common promoter sequence. Another possibility is that a number of different DFR genes are expressed in L. corniculatus, and that different antisense transformation events suppress DFR genes that are expressed in individual tissues.
Another surprising observation is the production of lines (e.g. RFD28 and RFD31) with enhanced levels of condensed tannins. It is interesting that statistically significant increases in levels were noted only with one construct, pMAJ2, in one recipient genotype, S50. In this context, we also note that in previous work on AS-DFR root cultures, one pMAJ2 line in an S50 background RFD3 was also identified with increased tannin levels. There appears to be no obvious explanation for this antisense up-regulation, but it may result from genetic compensation, i.e. there may be expression of more than one L. corniculatus DFR gene in line S50 leaf, stem, and root tissues, and in these events suppression of one of these genes results in overcompensation by another member of the gene family. Some supporting evidence for this has been found recently in the experiments involving the expression of an antisense β-1,3 glucanase in tobacco (Beffa et al., 1993) and in antisense expression of CHS in L. corniculatus root cultures (Colliver et al., 1997). However, to fully test this hypothesis one would need to clone all of the expressed members of the DFR gene family in S50 and analyze their expression in control and antisense up-regulated lines.
In addition to analysis of lines in the S50 genetic background, this study also focused on pMAJ2 cotransformants in genotype S33. As discussed above, only reductions in tannin accumulation were noted in this genotype. Lines RFD7, RFD8, and RFD50 were identified with low tannin levels in leaf and stem tissues, whereas phenotypes were found in juvenile shoot tissues in lines RFD7, RFD8, and RFD38. Unfortunately, because these plants were grown in soil, it was not possible to accurately determine condensed tannin levels in root tissues. However, it is clear that when lines showed antisense phenotypes in root cultures, these lines did not necessarily show corresponding phenotypes in regenerated plants. Therefore, analysis of root cultures may give a description of the effect of a particular construct (such as increasing or decreasing levels of condensed tannins) but lines showing such phenotypes in root cultures will not necessarily show a phenotype when regenerated to transgenic plants. Therefore, it is not possible to predict the phenotype of a plant from an individual transgenic line by screening at the root-culture level.
Analysis of low-molecular-weight flavonoid and isoflavonoid end products in AS-DFR lines proved to be of interest. One prediction was that in effective antisense lines there would be a decreased flux into condensed tannins and an increase in the levels of proximal end products (such as flavonols and flavones) or alternative end products (isoflavonoids and coumestans). In previous work on AS-DFR root cultures, no obvious default products were noted (P. Morris, unpublished data). Similarly in the work reported here, HPLC analysis indicated few obvious changes in MeOH-soluble flavonoids between RFD7, RFD8, and a corresponding control line; this observation is supported by data from AS-DFR lines in the S50 background. A conclusion from these studies may be that feedback mechanisms exist that limit flux into flavonoid pathways when the synthesis of condensed tannins is reduced. It would be of great interest to measure steady-state transcript levels of earlier steps in the flavonoid pathway. In this context it is notable that we have found a decrease in CHS transcript levels in some AS-DFR root cultures (M.P. Robbins, unpublished data) and this may be mediated by the low-level accumulation of pathway intermediates between CHS and DFR. Similarly, measurement of CHS enzyme activities would be of value in assessing the significance of feedback mediated by posttranscriptional mechanisms in these antisense lines.
Finally, inheritance studies have yielded some unexpected results. In the crosses from one of the effective primary transformants, RFD8, there was clear evidence of gross underrepresentation of transgenes in the T1 generation. One possibility is that functional AS-DFR sequences are detrimental to the viability of pollen or alternatively underrepresentation of sequences may relate to problems with genetic loading in pollen from transgenic parental lines. We note that there are few examples of studies relating to transgene inheritance in outbreeding forage species. Webb et al. (1994) reported underrepresentation of Agrobacterium-derived TR-DNA sequences and of GUS expression in the progeny of L. corniculatus, and further studies of the inheritance of transgenes in forage species may be of considerable interest.
ACKNOWLEDGMENTS
Work on genetically manipulated plants was carried out under Ministry of Agriculture, Fisheries, and Food license 162A/61/80. We thank a number of people for assistance with this study: Wendy Thornley, Ian Davies, Eunice Carter, and Jenny Phillips (Terrill tannin determinations) and Teri Evans (molecular analysis of parental and progeny lines). We also thank Dan Dhanoa for critical comments on the manuscript.
Abbreviations:
- AS-DFR
antisense dihydroflavonol reductase
- CaMV
cauliflower mosaic virus promoter
- CHS
chalcone synthase
- DFR
dihydroflavonol reductase
- MAJ
antisense DFR construct
- MeOH
methanol
- nos
nopaline synthase terminator
- RFD
antisense DFR line
- RT
reverse transcriptase
Footnotes
The Institute of Grassland and Environmental Research is grant funded by the Biotechnology and Biological Sciences Research Council (BBSRC). This work was supported by the BBSRC Plant Molecular Biology Initiative (parts 1 and 2).
LITERATURE CITED
- Barry TN, Duncan SJ. The role of condensed tannins in the nutritional value of Lotus pedunculatus for sheep. 1. Voluntary intake. Br J Nutr. 1984;51:485–491. doi: 10.1079/bjn19840054. [DOI] [PubMed] [Google Scholar]
- Bavage AD, Davies IG, Robbins MP, Morris P (1997a) Progress and potential for the genetic manipulation of plant quality. In G Cadisch, KE Giller, eds, Driven by Nature: Plant Litter Quality and Decomposition. CAB International, Wallingford, CT, pp 201–211
- Bavage AD, Davies IG, Robbins MP, Morris P. Expression of an Antirrhinum dihydroflavonol reductase gene results in changes in condensed tannin structure and accumulation in root cultures of Lotus corniculatus (bird's foot trefoil) Plant Mol Biol. 1997b;35:443–458. doi: 10.1023/a:1005841214470. [DOI] [PubMed] [Google Scholar]
- Bavage AD, Robbins MP. Dihydroflavonol reductase a Lotus corniculatus L. tannin biosynthesis gene: isolation of a partial gene clone by PCR. Lotus Newsletter. 1994;25:37–40. [Google Scholar]
- Beffa RS, Neuhaus J-M, Meins F. Physiological compensation in antisense transformants: specific induction of an “ersatz” glucan endo-1,3-glucosidase in plants infected with necrotizing viruses. Proc Natl Acad Sci USA. 1993;90:8792–8796. doi: 10.1073/pnas.90.19.8792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beld M, Martin C, Huits H, Stuitje AR, Gerats AGM. Flavonoid synthesis in Petunia hybrida: partial characterisation of dihydroflavonol 4-reductase genes. Plant Mol Biol. 1989;13:491–502. doi: 10.1007/BF00027309. [DOI] [PubMed] [Google Scholar]
- Brunk CF, Jones KC, James TW. Assay for nanogram quantities of DNA in cellular homogenates. Anal Biochem. 1979;92:497–500. doi: 10.1016/0003-2697(79)90690-0. [DOI] [PubMed] [Google Scholar]
- Carron TR, Robbins MP, Morris P. Genetic modification of condensed tannin biosynthesis in Lotus corniculatus. 1. Heterologous antisense dihydroflavonol reductase down-regulates tannin accumulation in “hairy root” cultures. Theor Appl Genet. 1994;87:1006–1015. doi: 10.1007/BF00225796. [DOI] [PubMed] [Google Scholar]
- Carter EB, Morris P, Theodorou MK (1995) Environmental effects on the chemistry and nutritive value of Lotus corniculatus (birdsfoot-trefoil). In GE Pollott, ed, Grassland into the 21st Century: Challenges and Opportunities. British Grassland Society Symposium No. 29. British Grassland Society, Harrogate, UK, pp 166–168
- Carter EB, Theodorou MK, Morris P. Responses of Lotus corniculatus to environmental change. 1. Effects of elevated CO2, temperature and drought on growth and plant development. New Phytol. 1997;136:245–253. [Google Scholar]
- Chang S, Puryear J, Cairney J. A simple and efficient method for isolating RNA from pine trees. Plant Mol Biol Rep. 1993;11:113–116. [Google Scholar]
- Charrier B, Coronado C, Kondorosi A, Ratet P. Molecular characterisation and expression of alfalfa (Medicago sativa L.) flavanone 3-hydroxylase and dihydroflavonol 4-reductase encoding genes. Plant Mol Biol. 1995;29:773–786. doi: 10.1007/BF00041167. [DOI] [PubMed] [Google Scholar]
- Colliver SP, Morris P, Robbins MP. Differential modification of flavonoid and isoflavonoid biosynthesis with an antisense chalcone synthase construct in transgenic Lotus corniculatus. Plant Mol Biol. 1997;35:509–522. doi: 10.1023/a:1005821801228. [DOI] [PubMed] [Google Scholar]
- Dellaporta SL, Wood J, Hicks JB. A plant DNA minipreparation: version II. Plant Mol Biol Rep. 1983;1:19–21. [Google Scholar]
- Dwivedi UN, Campbell WH, Yu J, Datla RSS, Bugos RC, Chiang VL, Podila GP. Modification of lignin biosynthesis in transgenic Nicotiana through expression of an antisense O-methyltransferase gene from Populus. Plant Mol Biol. 1994;26:61–71. doi: 10.1007/BF00039520. [DOI] [PubMed] [Google Scholar]
- Forde BG, Day HM, Turton JF, Wen-Jun S, Cullimore JV, Oliver JE. Two glutamine synthetase genes from Phaseolus vulgaris L. display contrasting developmental and spatial patterns of expression in transgenic Lotus corniculatus plants. Plant Cell. 1991;1:391–401. doi: 10.1105/tpc.1.4.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forkmann G. Genetics of flavonoids. In: Harborne JB, editor. The Flavonoids: Advances in Research since 1986. London: Chapman & Hall; 1993. pp. 538–564. [Google Scholar]
- Griffiths DW. Polyphenols and their possible effect on nutritive value. Aspects of Applied Biology. 1989;19:93–103. [Google Scholar]
- Jende-Strid B. Gene-enzyme relations in the pathway of flavonoid biosynthesis in barley. Theor Appl Genet. 1991;81:668–674. doi: 10.1007/BF00226735. [DOI] [PubMed] [Google Scholar]
- Lees GL, Hinks CF, Suttill NH. Effect of high temperature on condensed tannin accumulation in leaf tissue of bigfoot trefoil (Lotus uliginosus) J Sci Food Agric. 1994;65:415–421. [Google Scholar]
- Luo G, Hepburn AG, Widholm JD. Preparation of plant DNA for PCR analysis: a fast, good and reliable procedure. Plant Mol Biol Rep. 1992;10:319–323. [Google Scholar]
- Morris P, Robbins MP (1997) Manipulating condensed tannins in forage legumes. In BD McKersie, DCW Brown, eds, Biotechnology and the Improvement of Forage Legumes. CAB International, Wallingford, CT, pp 147–173
- Mueller-Harvey I, McAllan AB. Tannins: their biochemistry and anti-nutritional properties. Adv Plant Cell Biochem Biotechnol. 1992;1:149–213. [Google Scholar]
- Ougham HJ, Davies TGE. Leaf development in Lolium temulentum: gradients of RNA complement plastid and non-plastid transcripts. Physiol Plant. 1990;79:331–338. [Google Scholar]
- Raynaud J, Jay M, Raynaud J. Flavonoid glycosides of Lotus corniculatus (Leguminosae) Phytochemistry. 1982;21:2604–2605. [Google Scholar]
- Robbins MP, Bavage AD, Morris P. Options for the genetic manipulation of astringent and antinutritional metabolites in fruit and vegetables. In: Tomas-Barberan FA, Robins RJ, editors. Phytochemistry of Fruit and Vegetables. Oxford, UK: Clarendon Press; 1997. pp. 251–261. [Google Scholar]
- Robbins MP, Evans TE, Morris P, Carron TR. Lotus Newsletter. 1991;22:18–21. [Google Scholar]
- Shapiro SS, Wilk MB. An analysis of variance test for normality (complete samples) Biometrika. 1965;52:591–611. [Google Scholar]
- Sprent P. Applied Nonparametric Statistical Methods, Ed 2. London: Chapman & Hall; 1993. [Google Scholar]
- Tanner GJ, Moate PJ, Davis LH, Laby RH, Yuguang L, Larkin PA. Proanthocyanidins (condensed tannin) destabilise plant protein foams in a dose dependent manner. Aust J Agric Res. 1995;46:1101–1109. [Google Scholar]
- Terrill TH, Douglas GB, Foote AG, Purchas RW, Wilson GF, Barry TN. Determination of extractable and bound condensed tannin concentrations in forage plants, protein concentrate meals and cereal grains. J Sci Food Agric. 1992;58:321–329. [Google Scholar]
- van der Krol AR, Lenting PE, Veenstra J, van der Meer IM, Koes RE, Gerats AGM, Mol JNM, Stuitje AR. An anti-sense chalcone synthase gene in transgenic plants inhibits flower pigmentation. Nature. 1988;333:866–869. [Google Scholar]
- Waghorn GC, Jones WT, Shelton ID, McNabb WC. Condensed tannins and the nutritive value of herbage. Proceedings of the New Zealand Grassland Association. 1990;51:171–176. [Google Scholar]
- Webb KJ, Jones S, Robbins MP, Minchin FR. Characterisation of transgenic root cultures of Trifolium repens, Trifolium pratense and Lotus corniculatus and transgenic plants of Lotus corniculatus. Plant Sci. 1990;70:243–254. [Google Scholar]
- Webb KJ, Robbins MP, Mizen S. Expression of GUS in primary transformants and segregation of GUS, TL and TR-DNA in the T1 generation of hairy root transformants of Lotus corniculatus. Transgenic Research. 1994;3:232–240. [Google Scholar]