Abstract
The derivation of stably cultured cell lines has been critical to the advance of molecular biology. We profiled gene expression in the first two generally available cell lines derived from zebra finch. Using Illumina RNA-seq, we generated ~93 million reads and mapped the majority to the recently assembled zebra finch genome. Expression of most Ensembl-annotated genes was detected, but over half of the mapped reads aligned outside annotated genes. The male-derived G266 line expressed Z-linked genes at a higher level than did the female-derived ZFTMA line, indicating persistence in culture of the distinctive lack of avian sex chromosome dosage compensation. Although these cell lines were not derived from neural tissue, many neurobiologically relevant genes were expressed, although typically at lower levels than in a reference sample from auditory forebrain. These cell lines recapitulate fundamental songbird biology and will be useful for future studies of songbird gene regulation and function.
Keywords: zebra finch, RNA-seq, song learning, gene expression, Illumina, dosage compensation, bird, sex chromosome
1. Introduction
Songbirds are intensively studied across a diversity of fields ranging from ecology to neuroscience. Songbirds are arguably the best animal models for the study of learned vocal communication [1] and have yielded important insights into the mechanisms of evolutionary adaptation [2,3] and sexual differentiation of brain and behavior [4-7]. Genomic tools for songbirds have now come of age with publication of the complete genome sequence [8] and development of high throughput gene expression assays [9-11] for the zebra finch, the most common songbird in laboratory research. These tools have now been used to identify genes associated with perception of song [12-14], singing behavior [8,11], seasonally regulated courtship and territorial behavior [15,16] and sex-specific brain development [17].
A critical task now is to develop functional tests of specific genes identified through songbird genomics. To this end, recent studies in vivo have used pharmacological manipulations [18] and RNA interference [19] to affect zebra finch behavior, and transgenic zebra finches have also now been produced [20]. However, whole-animal manipulations are laborious and expensive and many basic aspects of functional characterization could be carried out more efficiently in cell lines (e.g., assaying consequences of specific gene knockdown on gene expression networks, probing gene dosage compensation mechanisms, or testing microRNA-mRNA interactions). Recently, cultured cell lines from zebra finches been established [21]. Although some experimental objectives may be accomplished using cell lines from other organisms, it remains possible and even likely that transcriptional control networks (e.g., for dosage compensation) and specific molecular interactions are sufficiently different to warrant specific study in cells and tissues from the zebra finch.
In this report we contribute to the characterization of the two tumor-derived cell lines of Itoh and Arnold [21]. One of the lines was derived from a male, which in birds are the homogametic sex (ZZ), and the other from a female bird (ZW). Both tumors were removed from non-neural tissues (although the exact cellular origin of neither line is known [21]). We used Illumina mRNA sequencing (RNA-seq) to generate gene expression profiles of these two cell lines, and analyzed the data specifically to evaluate potential utility of the cell lines for study of sex differences and for genes of neurobiological interest. RNA-seq expression profiling is still a relatively new and evolving methodology [22-25], and our study is one of the first applications of this method in songbird research [see also 8,26,27], or for de novo characterization of cell lines from any species.
2. Results
2.1. RNA-seq and read mapping
One lane of sequencing of the zebra finch cell lines on the Illumina HiSeq2000 platform yielded 92,609,701 reads. These were distributed nearly evenly between the two cell lines, male G266 (45,268,521 reads) and tetraploid female ZFTMA (47,340,550 reads). Using Tophat [28] we were able to map over half (G266: 58.8%; ZFTMA: 53.7%) of the reads unambiguously to a single location in the genome. Allowing for multireads (reads mapping up to 20 places in the genome), we were able to map 69.8% and 71.2% of G266 and ZFTMA reads, respectively. Of the uniquely mapped reads, only approximately 40 percent mapped to genome regions covered by current Ensembl coding regions (Figure 1, Supplementary Table1). The remaining reads mapped to regions currently annotated as introns, intergenic regions or UTRs.
Figure 1.
Distribution of uniquely mapping RNA-seq reads among genomic compartments. Flanking regions are defined as being within 1kb up or downstream of the Ensembl cDNA. Reads outside of annotated gene models are termed “intergenic” although it is possible likely of these are actually novel genes. The “other” category includes reads of ambiguous mapping location (reads spanning two compartments) as well as reads mapping to annotated telomeres.
To provide a point of reference for cell line gene expression profiles we also sequenced RNA from the auditory forebrain (“auditory lobule” or “AL” [29]) of female zebra finches. The auditory lobule is composed of three forebrain subregions, Field L, the caudomedial nidopallium (NCM) and the caudomedial mesopallium (CMM). We focused on the auditory lobule as it is a focal point for recent research on gene expression in the zebra finch brain [8,12,18,30]. Three lanes of sequencing of zebra finch auditory forebrain samples on an Illumina Genome Analyzer produced 69,836,901 reads of which 68.1% were mapped uniquely (these results were generated using a different library preparation and sequencing platform, see Methods).
Across all samples (cell lines and auditory lobule), less than six percent of reads mapped to known UTR regions, likely reflecting the incomplete state of zebra finch gene annotations. The large proportion of reads mapped to regions flanking Ensembl gene models (~11-15%) suggests that these areas are in fact transcribed regions that have yet to be formally annotated. In particular, a relatively large proportion of reads mapped to the region within 1kb of the 3’ end of Ensembl models (Figure 1).
Read mapping from cell lines versus auditory lobule also showed some distinctive differences. Despite a higher overall mapping rate (68.1% versus < 60% for cell lines), a lower proportion of the auditory forebrain reads (only 30.0%) mapped to Ensembl coding regions (Figure 1). We also note that a much higher proportion of auditory lobule reads mapped to the mitochondrial genome (19.69%) than the samples from the cell lines (G266 = 3.42%, ZFTMA = 7.03%; Figure 1).
2.2. Functional annotation of genes present in cell lines
We detected 13,333 Ensembl-annotated genes with at least one read in each cell line (Figure 2), and we carried out further functional analyses of this set of “cell line expressed” genes. Statistical over and under-representation of Gene Ontology (GO) terms and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway representation was assessed using Fisher's Exact Test and p-values were adjusted for multiple testing. Canonical signaling pathways are well represented among this set (Table 1). Only one KEGG pathway was significantly underrepresented among the 132 detected pathways: Calcium signaling pathway (gga04020, p = 0.00018). One pathway, Ribosome (gga03010) was significantly enriched (p = 0.043). Genes associated with 44 Gene Ontology (GO) terms were underrepresented (p < 0.05) and another 45 terms were over-represented (p < 0.05) in the cell lines (Supplementary Table 2). Categories that were enriched often involved cellular components (e.g., cytoplasm, mitochondrion, endoplasmic reticulum) whereas categories that were underrepresented included a number of signaling processes (olfactory receptor activity, G-protein coupled receptor activity) and immune components (immune response, MHC Class II protein complex, MHC Class I protein complex). We also specifically examined the list of expressed genes for GO terms related to neurobiology (121 GO categories containing “neuro” or “synap”). Of these, only three terms were significantly underrepresented among expressed genes, indicating a relative lack of neuronal post-synaptic proteins and receptors (Table 2).
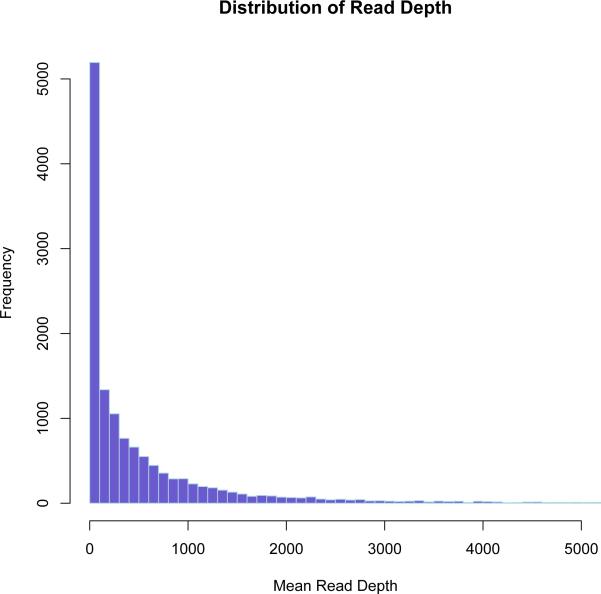
Figure 2.
Distribution of normalized read count across 13,333 genes that were represented by an average of one read across both cell lines.
Table 1.
Canonical signaling pathway (KEGG) representation in the two cell lines.
| KEGG id | description | total | expected | observed | adjusted-p |
|---|---|---|---|---|---|
| gga04340 | Hedgehog signaling pathway | 39 | 34 | 28 | 0.22 |
| gga04630 | Jak-STAT signaling pathway | 63 | 54 | 49 | 0.35 |
| gga04010 | MAPK signaling pathway | 145 | 125 | 117 | 0.3 |
| gga04150 | mTOR signaling pathway | 32 | 28 | 29 | 1 |
| gga04330 | Notch signaling pathway | 27 | 23 | 23 | 1 |
| gga04350 | TGF-beta signaling pathway | 47 | 41 | 43 | 1 |
| gga04370 | VEGF signaling pathway | 37 | 32 | 35 | 1 |
| gga04310 | Wnt signaling pathway | 95 | 82 | 79 | 0.93 |
Table 2.
Fifteen largest Gene Ontology terms containing “neuro” or “synap”. Of 121 such GO categories, only three (all shown here) are significantly under-represented in the cell lines.
| GO id | description | total | expected | observed | adjusted-p |
|---|---|---|---|---|---|
| 0004983 | neuropeptide Y receptor activity | 91 | 72 | 47 | 2.00E-08 |
| 0045211 | postsynaptic membrane | 59 | 46 | 24 | 7.10E-08 |
| 0008021 | synaptic vesicle | 49 | 39 | 36 | 1 |
| 0030594 | neurotransmitter receptor activity | 42 | 33 | 14 | 8.40E-08 |
| 0001764 | neuron migration | 38 | 30 | 26 | 1 |
| 0007218 | neuropeptide signaling pathway | 35 | 28 | 29 | 1 |
| 0043524 | negative regulation of neuron apoptosis | 32 | 25 | 28 | 1 |
| 0045202 | synapse | 31 | 24 | 27 | 1 |
| 0006836 | neurotransmitter transport | 26 | 20 | 17 | 1 |
| 0007268 | synaptic transmission | 26 | 20 | 19 | 1 |
| 0005328 | neurotransmitter:sodium symporter activity | 24 | 19 | 15 | 1 |
| 0030182 | neuron differentiation | 23 | 18 | 13 | 0.50 |
| 0050885 | neuromuscular process controlling balance | 23 | 18 | 19 | 1 |
| 0008188 | neuropeptide receptor activity | 21 | 17 | 15 | 1 |
| 0045665 | negative regulation of neuron differentiation | 18 | 14 | 12 | 1 |
2.3. Gene expression differences between cell lines
We identified 98 genes that were differentially expressed between the two cell lines (FDR p < 0.01 Fig. 3). This gene list was significantly enriched for a number of GO terms (Table 3). The most obvious differences between cell lines indicate variation in the gene regulatory landscape (GO terms “transcription factor activity” and “regulation of transcription - DNA dependent”) and homophilic cell adhesion. The transcription factor activity category was composed of a set of 13 genes with striking similarity in structure/function and genomic location. Eight of the 13 are homeodomain proteins (Hox A3, Hox A7, Hox A10, HoxD10, IRX1 (Iroquois homeobox 1), IRX2, SIX2). Five of these (Hox A3, Hox A7, Hox A10, IRX1, IRX2) as well as RARB (Retinoic Acid Receptor B) and TBX20 are located on chromosome 2. Another TBX gene, TBX4, located on chromosome 19, is also differentially expressed as is another HOX protein HoxD10 on chromosome 7. All of the HOX genes above are highly expressed (> 200 normalized reads) in the ZFTMA line whereas they are barely expressed (< 2 normalized reads) at all in the G266 line. Six other HOX genes (HOXA6, HOXA4, HOXA5, HOXA11, HOXA9 and HOXD11) show a similar pattern, but fall short of statistical significance. Three HOX genes (HOXD3, HOXD4-2, HOXA2) show the opposite pattern (> 500 reads in G266 and < 100 reads in ZFTMA) but these differences are also non-significant (p > 0.01). Both IRX genes are highly expressed in G266 and absent (read count = 0) in ZFTMA.
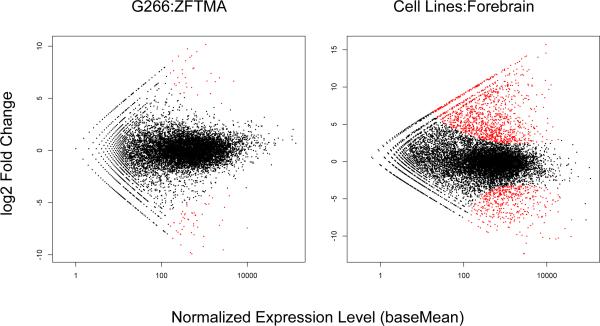
Figure 3.
Scatterplot of normalized expression level versus log2 fold change generated by DE-Seq analysis. Points colored in red are those show a significant difference in expression at FDR < 0.01. 98 genes are differentially expressed between cell lines and 2120 are differentially expression between the two cell lines and the auditory forebrain sample.
Table 3.
Over-represented GO terms among genes differentially expressed between cell lines. Only GO categories represented by at least five genes among the expressed genes are shown.
| GO id | description | total | expected | observed | adjusted-p |
|---|---|---|---|---|---|
| 0007156 | homophilic cell adhesion transmembrane receptor protein tyrosine | 57 | 0 | 6 | 0.0026 |
| 0007169 | kinase signaling pathway | 42 | 0 | 5 | 0.0029 |
| 0003700 | transcription factor activity | 457 | 4 | 13 | 0.0069 |
| 0043565 | sequence-specific DNA binding | 309 | 3 | 10 | 0.011 |
| 0005509 | calcium ion binding | 439 | 4 | 12 | 0.011 |
| 0006355 | regulation of transcription, DNA-dependent | 566 | 5 | 13 | 0.023 |
| 0005578 | proteinaceous extracellular matrix | 96 | 1 | 5 | 0.031 |
| 0016020 | membrane | 1249 | 10 | 20 | 0.048 |
Comparison of RNA-seq data from the two cell lines reveals higher expression of Z linked genes in male (ZZ) G266 than female (ZW) ZFTMA cells (Kolmogorov-Smirnov Test p = 0.02; Figure 4). We validated this specific finding, and the results of our analysis more broadly, with quantitative PCR (qPCR). We used mRNA and genomic DNA to assay six genes that had higher RNA-Seq expression values in G266 than ZFTMA cells. qRT-PCR confirmed higher mRNA levels in the G266 relative to the ZFTMA cells. (Table 4). Three of these genes (FST, UHRF2-2 and RIOK2) have previously been shown to be expressed at higher levels in male compared to female zebra finches across tissue types (brain, kidney and liver) [7]. We also used qPCR to confirm that all six genes showed the expected higher genomic DNA concentration in the male G266 cells compared to the ZFTMA. As expected, Z-linked gene concentration was consistently higher (1.31- to 1.88-fold; Table 4) in males (ZZ) than females (ZW).
Figure 4.
Distribution of fold change estimates for Z-linked (Z) versus autosomal (A) genes. The distribution of fold changes for autosomal genes is centered around zero whereas the distribution of fold changes for Z-linked genes is shifted to right. Z linked genes tend to be more highly expressed in the male G266 cell line than the female ZFTMA cell line.
Table 4.
qPCR analysis of Z-linked genes in the two cell lines. The p-value for each gene is the result of t-test. Inf stands for infinite.
| RNA-seq G266:ZFTMA | qPCR (RNA) G266:ZFTMA | qPCR (DNA) G266:ZFTMA | |||||
|---|---|---|---|---|---|---|---|
| Ensembl transcript | gene symbol | ratio | p-value | ratio | p-value | ratio | p-value |
| ENSTGUT00000002529 | FST | 104.99 | 1.89E-07 | 249.60 | 2.83E-03 | 1.48 | 1.59E-03 |
| ENSTGUT00000005161 | UHRF2-2 | 255.71 | 9.33E-05 | 51.30 | 2.26E-03 | 1.33 | 1.98E-02 |
| ENSTGUT00000004232 | CRHBP | Inf | 3.14E-07 | 1030.89 | 6.84E-05 | 1.31 | 3.92E-02 |
| ENSTGUT00000004793 | FREM1 | 131.20 | 1.73E-07 | 30.62 | 3.71 E-03 | 1.42 | 4.64E-03 |
| ENSTGUT00000005664 | VLDLR | 34.87 | 4.24E-06 | 50.17 | 1.24E-03 | 1.60 | 1.54E-02 |
| ENSTGUT00000001276 | RIOK2 | 2.25 | 0.225 | 3.22 | 2.11E-02 | 1.84 | 5.52E-05 |
2.4. Differences between cell lines and auditory lobule
Distance based clustering of the three expression profiles shows that the two tumor-derived cell lines were more similar to each other than they were to the auditory lobule expression profile (Figure 4). Despite broad representation of neural genes in the cell lines, mRNA levels were often very different between the cell lines and the auditory forebrain. 2120 genes were differentially expressed at FDR < 0.01 (Figure 2). This gene set is described by 67 over-represented Gene Ontology terms (p < 0.01). Among the most strongly over represented terms are those related to ion channels (e.g., GO:0005216, Ion channel activity; GO:0006811 ion transport; GO:0005509, calcium ion binding; GO:0006813, potassium ion transport) and a diversity of neural components and functions, (e.g., GO:0045211, post-synaptic membrane; GO:0006813, synaptic vesicle, GO:0045202, synapse; GO:0030424, axon; GO:0030594 neurotransmitter receptor activity; Supplementary Table 3).
3. Discussion
We have broadly characterized the transcriptional landscape of two recently created zebra finch cell lines. Using Illumina HiSeq2000 sequencing on a single lane of a flow cell, we were able to detect the expression of the large majority (82%) of known genes (14,253 of 17,475 of genes annotated by Ensembl). Both cell lines were derived from spontaneous tumors and were not derived from brain tissue. Nevertheless, we find strong representation of genes associated with neural structure and function. We suggest, therefore, that these two cell lines will have broad utility for studies of avian biology in general, and for specific studies of neurobiologically relevant genes.
Our study is among the first to use Illumina RNA-seq to profile gene expression in the zebra finch; an initial analysis by RNA-seq of gene expression in juvenile and adult zebra finch forebrain was included in the primary publication of the zebra finch genome assembly and annotation [8]. RNA-seq of small RNAs has also been used to describe the microRNA landscape of zebra finches [8,13]. RNA-seq offers a well-known advantage over microarrays in that it is not restricted to previously known transcripts.
As in many genome profiling studies, we found evidence of extensive transcription outside of currently annotated genes. Some of our putative intergenic reads may arise from bona fide novel genes, whereas others may represent unannotated exonic regions of known genes. In particular, we find strong evidence of transcription within 1kb up and down-stream of Ensembl gene models, suggesting that many exons, likely UTRs, extend well beyond their currently defined boundaries. De novo transcript prediction algorithms like Cufflinks [31-33] offer the promise of improving upon the current understanding of avian transcript structures. Our current read coverage depth here is not sufficient to confidently assess alternative splicing. In particular, splice site prediction appeared to be confounded by reads that map to introns. These reads may represent incomplete splicing of transcribed RNAs or genomic DNA contamination of RNA preparations despite the DNase treatment step in our protocol. Deeper sequence coverage and paired-end (as opposed to single-end) sequencing will doubtless enhance our ability to revise existing annotations. Distinguishing whether the observed widespread transcription across the genome is functional or merely biological noise remains an ongoing challenge for all gene expression studies, including those utilizing RNA-seq technology [34,35].
We identified a small number of statistically significant differences between the two tumor cell lines. For example, we discovered a set of eight differentially expressed homeodomain proteins. Both Hox genes and IRX genes have been implicated in cancer [36,37]. We speculate that different levels of RNAs for these genes may be related to differences between the cell lines in their mechanism of tumorigenesis. Further, the observed differences in gene expression plausibly contribute to one phenotypic difference between the lines: ZFTMA cells easily detached from the surface of culture dish whereas G266 cells tended to grow on top of each other in stable multi-layers. Thus in the difference in cell adhesion and contact inhibition properties, may be mediated through differential expression of homophilic adhesion genes (Table 3). The genes we identified as differentially expressed had large fold changes (average log2 fold change = 6.99) and high read depth (mean normalized read counts = 784, median= 222). There are almost certainly also smaller quantitative differences in expression of additional genes that would be revealed by more intensive sampling of replicate cell line populations.
Birds are of special interest for the study of sex chromosome evolution and function because genes on the homogametic (ZZ) male sex chromosomes do not undergo complete dosage compensation. As a consequence, Z genes tend to be more highly expressed in males than females (ZW) [6-8]. Here we found that the male and female zebra finch cell lines maintained sex differences in both Z:autosome ratio (of DNA) and Z-linked gene expression ratio. These ratios were maintained even though the female cell line (ZFTMA) is tetraploid. This stability supports the hypothesis that the lack of sex chromosome dosage compensation in birds has some functional benefit that is retained even through the selective forces of cell transformation and adaptation to culture, though more observations would be needed to establish the generality of this conclusion.
Sex differences in RNA were observed consistently for six different Z-linked genes, using both RNA-seq and qRT-PCR (Table 4), supporting the overall statistical integrity of our analyses. We used qPCR of genomic DNA to validate the copy number difference for these genes and confirmed that the homogametic (ZZ) male cell line has higher Z-linked DNA concentration than the ZW female line. Across all six genes, however, our estimate of the ratio of DNA content was less than the expected two-fold difference. This may reflect the loss of some autosomal DNA following tetraploidy, or it may be an artifact of the different DNA preparations [38]. We also note that expression differences for some Z-linked genes measured by qRT-PCR (Table 4) were much higher than the average difference measured for all Z genes (Figure 3). This presumably reflects the contribution of other regulatory influences beyond simple gene dosage, e.g., the operation of distinct “male” and “female” transcriptional control networks, or differences in the tissue origins of the two lines.
Although a large proportion of all known genes are expressed in both cell lines, we find that many RNAs identified in the auditory lobule are absent or present only at low levels in the cell lines. Our results therefore suggest differences in the transcriptional landscapes between cell lines derived from spontaneous non-brain tumors and the heterogeneous pool of cells that comprise the auditory forebrain. This is not surprising given the different source tissue and the genomic distinctions recently reported even across brain regions [39]. We also note that auditory forebrain and cell line libraries were generated using different library preparation pipelines and sequencing technology. Differences in sequencing pipelines may contribute to the variation in the distribution of derived reads among genomic compartments (Figure 1). Even with these technical and biological considerations, we found neurobiologically relevant gene ontology categories to be well-represented in the cell lines, indicating that they are appropriate addition to the post-genomic resources available to songbird neurobiologists.
The zebra finch, and birds in general, are important model systems in a number of disciplines of biological study. One of current limitations of bird research has been the lack of tools for experimental genetic manipulation in vitro. The development of immortalized cell lines [21] is an important step towards such functional manipulations in the songbird model system. The utility of these cell lines for functional investigation increases with this description of their gene expression profiles.
4. Material and Methods
4.1. Cell culture
Two zebra finch cell lines, ZFTMA and G266, derived from spontaneous tumors, were obtained from the laboratory of Dr. Arthur Arnold at the University of California Los Angeles. ZFTMA cells were derived from a bird with an abnormally enlarged thigh, and G266 cells were cultured from tissue beneath the skin on the head of a male bird. ZFTMA is a tetraploid female cell line and G266 is a diploid male cell line [21]. Conditions for cell culture are described by Itoh and Arnold [21] and were followed accordingly.
Total RNA was extracted using TRI Reagent (Ambion) and treated with TURBO DNase (Ambion) according to the manufacturer's instructions. After DNase treatment, the RNA samples were purified using an RNeasy Mini Kit (Qiagen). RNA samples were analyzed on Bioanalyzer (Agilent) to ensure adequate quality and quantity of RNA. Genomic DNA was extracted by DNeasy Blood & Tissue Kit (Qiagen). The concentration of each DNA sample was determined by ND-1000 spectrophotometer (NanoDrop).
4.2. RNA-seq of zebra finch cell lines
Library preparation and sequencing were done at the University of Illinois Roy J. Carver Biotechnology Center. RNA libraries for ZFTMA and G266 RNA samples were prepared using Illumina's TruSeq RNA-Seq Sample Prep Kit following manufacturer's instructions. Libraries were pooled and sequenced for 100 cycles on one lane of an Illumina HiSeq2000. Sequencing was done using a TruSeq SBS sequencing kit version 2 and analyzed with Illumina RNA-Seq pipeline version 1.8. These Illumina kits include library-specific tagging steps and their standard post-sequencing pipeline includes steps for separating reads based on tags.
4.3. RNA-seq of zebra finch auditory forebrain
Three libraries were generated and sequenced on an Illumina Genome Analyzer using cluster kits V4, sequencing V4 and pipeline 1.6. Each library was derived from a pool of ten female zebra finches to control for individual variation and variability in dissection. The details of the experimental manipulation of these birds will be presented elsewhere (London et al., in prep). Here, we treated the transcriptomes of three experimental groups as replicates, as the aim was to have broad representation of auditory forebrain RNAs for comparison to the unmanipulated cell lines. Individual samples within each library were not tagged prior to sequencing so information on individual animal expression profiles is not available. All Illumina read data will be deposited to the NCBI Short Read Archive.
4.4. Read mapping
Reads were mapped using Tophat version 1.5.0 as implemented on the Galaxy public server (build: $Rev 6056:338ead4737ba$) and on a local installation. Our final set of analyses used options requiring 8 base pairs to flank a putative splice site (-a 8), allowed one mismatch in the seed regions (-m 1), and allowed for introns between 70 and 500,000 bp in length. We only included reads that mapped to unambiguously to a single location in the genome (-g 1). We set segment length at 25 and allowed for two segment-mismatches. For the coverage search we set the minimum introns length at 50, and the maximum at 20,000. The BAM file generated by Tophat [28] was converted to SAM format using SAMtools [40]. We then used htseq-count (part of the HT-Seq package of python scripts [41]) to convert the mapped reads to read counts per transcript. Transcripts were defined using Ensembl Build 56 gene models. Reads within 1kb up and downstream of Ensembl models were terms “flanking”, and genes were termed “intergenic” if they were outside of Ensembl annotated genes.
DE-seq does not incorporate information on splicing so information on alternative isoforms is ignored in our analysis. This is appropriate because we do not have the sequencing depth to confidently assess alternative splicing frequency. We used Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis implemented using CORNA [42] to describe gene representation among the sequenced and mapped reads. We tested for over- and under-representation by comparing genes expressed in both cell lines relative to the full list of annotated Ensembl Genes and associated GO and KEGG terms. All GO and KEGG statistical tests were corrected for multiple comparisons (Benjamini Hochberg method in R).
4.5. Differential expression analysis
We used DE-Seq version 1.5.22 [43] implemented in R to test for differential gene expression between the two cell lines and between the cell lines and the sample of reads from the zebra finch auditory forebrain. DE-Seq treats gene expression data as count data modeled under a negative binomial distribution. For the comparison of the two cell lines, we used the option [method = ‘blind’] that allows comparisons between unreplicated experimental groups. This method is based on the negative relationship between expression level and variance. In lieu of experimental replication, information on variance in expression of transcripts of similar expression level is used to derive a null expectation for variance and significance threshold. Approaches that do not require biological replication were also used in benchmark studies describing other methods for differential expression using RNA-seq (e.g., Cufflinks [32]). Under this approach in DE-Seq, experimental groups were treated as technical replicates and a threshold for statistical significance was derived under the assumption that most genes are not differentially expressed. We also used the options [sharingMode= ‘fit-local’] and [fitType = ‘local’]. These settings match those described in the published version of DE-seq [43].
For the comparison of the cell lines with the auditory forebrain data we did not use [method=“blind”] but rather treated the two cell lines as replicates. This approach is reasonable because relatively few genes are differentially expressed between the two cell lines and clustering analysis reveals them to be much more similar to each other than either is to the auditory forebrain sample. We also treated the three auditory forebrain samples as replicates. Clustering analysis was done using the dist function in R and Euclidean distances after data were transformed using the variance stabilizing transformation implemented in DE-Seq (Figure 4).
In both sets of comparisons (G266 vs. ZFTMA and Cell Lines vs. auditory forebrain) we identified differentially expressed genes as those that were significant at FDR < 0.01 (Benjamini Hochberg method as implemented in DE-Seq). Gene lists of differentially expressed genes were described using Gene Ontology analysis relative the total pool of genes expressed both groups (mean expression ≥ 0). Statistical tests for GO terms were done using Fisher's Exact tests and were corrected for multiple comparisons.
4.6. Quantitative RT-PCR (qRT-PCR) validation
We used qPCR to validate the RNA-Seq findings with a specific focus on genes of the Z chromosome. Gene-specific primers (Table 5) were designed by Primer3 [44]. Relative PCR efficiencies between each gene of interest and the internal control gene were assayed by amplification of five log2 serial dilutions of the template DNA (9.375 ng to 150 ng). The absolute value of the slope of log input amount vs. ΔCt was < 0.1 demonstrating that relative efficiencies of the genes of interest and the internal control gene are approximately equal (data not shown).
Table 5.
Primer sequences used for RT-PCR.
| Ensembl transcript | gene symbol | forward | reverse |
|---|---|---|---|
| ENSTGUT00000002529 | FST | AAGCCAAATCCTGTGAAGAC | CGCTTGGGTAAGTTGTGTTA |
| ENSTGUT00000005161 | UHRF2-2 | ATGAATTTGTCCTGTTGTGC | TTATAGTAAGAAAGAGAACCCACA |
| ENSTGUT00000004232 | CRHBP | GGAAGCAGAAACAAGAAAGG | TTATAGAAAGGCCCGACATC |
| ENSTGUT00000004793 | FREM1 | GGAATGGAGGAGAACCTGTA | TTTGACCTTCTTGCAGTCAG |
| ENSTGUT00000005664 | VLDLR | GTTGTCAGCACAGATGATG | TTCCAAGAATGGAGGAAG |
| ENSTGUT00000001276 | RIOK2 | CAAATGGAAGATCCTGCTG | CATCATCTGAGGGAAGTCAA |
For analysis of gene expression between two cell lines, two micrograms of total RNA was reverse transcribed by RETROscript Kit (Ambion). Twenty-five nanograms of cDNA were then used as the template in the qRT-PCR reaction; a total of 6 RNA samples from 3 passages of G266 and 3 passages of ZFTMA were analyzed. For analysis of genomic DNA content between the two cell lines, 50 nanograms of genomic DNA was used in the qPCR reaction; a total of 6 DNA samples from 3 passages of G266 and 3 passages of ZFTMA were analyzed. The qPCR reactions were run in triplicates using FastStart Universal SYBR Green chemistry (Roche) on the ABI 7900 HT machine (Applied Biosystems). The dissociation curve for each gene of interest was checked to ensure that a single PCR product was amplified. The data analysis and statistics were performed in R.
Supplementary Material
Highlights.
Illumina RNA-seq was used to profile gene expression in two new zebra finch cell lines
The two cell lines show a broad representation of known genes, but also extensive transcription outside of annotated regions
Genes of known neural function are also well-represented
The two cell lines, derived from birds of different sex, maintain the lack of dosage compensation shown previously in birds
The two cell lines will have broad utility for the studies of avian molecular biology and neuroscience.
Figure 5.
Heatmap showing clustering by Euclidean distances between two cell line libraries and three auditory lobule (AL) libraries. The two tumor cell lines, although derived from birds of different sexes and tissue from different parts of the birds, were more similar to each other than they were to profiles of the auditory lobule.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Doupe AJ, Kuhl PK. Birdsong and human speech: common themes and mechanisms. Annu Rev Neurosci. 1999;22:567–631. doi: 10.1146/annurev.neuro.22.1.567. [DOI] [PubMed] [Google Scholar]
- 2.Abzhanov A, Kuo WP, Hartmann C, Grant BR, Grant PR, Tabin CJ. The calmodulin pathway and evolution of elongated beak morphology in Darwin's finches. Nature. 2006;442:563–567. doi: 10.1038/nature04843. [DOI] [PubMed] [Google Scholar]
- 3.Cheviron ZA, Whitehead A, Brumfield RT. Transcriptomic variation and plasticity in rufous-collared sparrows (Zonotrichia capensis) along an altitudinal gradient. Mol Ecol. 2008;17:4556–4569. doi: 10.1111/j.1365-294X.2008.03942.x. [DOI] [PubMed] [Google Scholar]
- 4.Arnold AP. Sex chromosomes and brain gender. Nat Rev Neurosci. 2004;5:701–708. doi: 10.1038/nrn1494. [DOI] [PubMed] [Google Scholar]
- 5.Itoh Y, Kampf K, Arnold AP. Comparison of the chicken and zebra finch Z chromosomes shows evolutionary rearrangements. Chromosome Research. 2006;14:805–815. doi: 10.1007/s10577-006-1082-1. [DOI] [PubMed] [Google Scholar]
- 6.Itoh Y, Melamed E, Yang X, Kampf K, Wang S, Yehya N, Van Nas A, Replogle K, Band MR, Clayton DF, Schadt EE, Lusis AJ, Arnold AP. Dosage compensation is less effective in birds than in mammals. J Biol. 2007;6:2. doi: 10.1186/jbiol53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Itoh Y, Replogle K, Kim YH, Wade J, Clayton DF, Arnold AP. Sex bias and dosage compensation in the zebra finch versus chicken genomes: general and specialized patterns among birds. Genome Res. 2010;20:512–518. doi: 10.1101/gr.102343.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Warren WC, Clayton DF, Ellegren H, Arnold AP, Hillier LW, Kunstner A, Searle S, White S, Vilella AJ, Fairley S, Heger A, Kong L, Ponting CP, Jarvis ED, Mello CV, Minx P, Lovell P, Velho TA, Ferris M, Balakrishnan CN, Sinha S, Blatti C, London SE, Li Y, Lin YC, George J, Sweedler J, Southey B, Gunaratne P, Watson M, Nam K, Backstrom N, Smeds L, Nabholz B, Itoh Y, Whitney O, Pfenning AR, Howard J, Volker M, Skinner BM, Griffin DK, Ye L, McLaren WM, Flicek P, Quesada V, Velasco G, Lopez-Otin C, Puente XS, Olender T, Lancet D, Smit AF, Hubley R, Konkel MK, Walker JA, Batzer MA, Gu W, Pollock DD, Chen L, Cheng Z, Eichler EE, Stapley J, Slate J, Ekblom R, Birkhead T, Burke T, Burt D, Scharff C, Adam I, Richard H, Sultan M, Soldatov A, Lehrach H, Edwards SV, Yang SP, Li X, Graves T, Fulton L, Nelson J, Chinwalla A, Hou S, Mardis ER, Wilson RK. The genome of a songbird. Nature. 2010;464:757–762. doi: 10.1038/nature08819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Naurin S, Bensch S, Hansson B, Johansson T, Clayton DF, Albrekt AS, V.O.N.S. T, Hasselquist D. TECHNICAL ADVANCES: A microarray for large-scale genomic and transcriptional analyses of the zebra finch (Taeniopygia guttata) and other passerines. Mol Ecol Resour. 2008;8:275–281. doi: 10.1111/j.1471-8286.2007.01979.x. [DOI] [PubMed] [Google Scholar]
- 10.Replogle K, Arnold AP, Ball GF, Band M, Bensch S, Brenowitz EA, Dong S, Drnevich J, Ferris M, George JM, Gong G, Hasselquist D, Hernandez AG, Kim R, Lewin HA, Liu L, Lovell PV, Mello CV, Naurin S, Rodriguez-Zas S, Thimmapuram J, Wade J, Clayton DF. The Songbird Neurogenomics (SoNG) Initiative: community-based tools and strategies for study of brain gene function and evolution. BMC Genomics. 2008;9:131. doi: 10.1186/1471-2164-9-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wada K, Howard JT, McConnell P, Whitney O, Lints T, Rivas MV, Horita H, Patterson MA, White SA, Scharff C, Haesler S, Zhao S, Sakaguchi H, Hagiwara M, Shiraki T, Hirozane-Kishikawa T, Skene P, Hayashizaki Y, Carninci P, Jarvis ED. A molecular neuroethological approach for identifying and characterizing a cascade of behaviorally regulated genes. Proc Natl Acad Sci U S A. 2006;103:15212–15217. doi: 10.1073/pnas.0607098103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dong S, Replogle KL, Hasadsri L, Imai BS, Yau PM, Rodriguez-Zas S, Southey BR, Sweedler JV, Clayton DF. Discrete molecular states in the brain accompany changing responses to a vocal signal. Proc Natl Acad Sci U S A. 2009;106:11364–11369. doi: 10.1073/pnas.0812998106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gunaratne PH, Lin YC, Benham AL, Drnevich J, Coarfa C, Tennakoon JB, Creighton CJ, Kim JH, Milosavljevic A, Watson M, Griffiths-Jones S, Clayton DF. Song exposure regulates known and novel microRNAs in the zebra finch auditory forebrain. BMC Genomics. 2011;12:277. doi: 10.1186/1471-2164-12-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.London SE, Dong S, Replogle K, Clayton DF. Developmental shifts in gene expression in the auditory forebrain during the sensitive period for song learning. Dev Neurobiol. 2009;69:437–450. doi: 10.1002/dneu.20719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mukai M, Replogle K, Drnevich J, Wang G, Wacker D, Band M, Clayton DF, Wingfield JC. Seasonal differences of gene expression profiles in song sparrow (Melospiza melodia) hypothalamus in relation to territorial aggression. PLoS One. 2009;4:e8182. doi: 10.1371/journal.pone.0008182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thompson CK, Meitzen J, Replogle K, Drnevich J, Lent KL, Wissman AM, Farin FM, Bammler TK, Beyer RP, Clayton DF, Perkel DJ, Brenowitz EA. Seasonal changes in patterns of gene expression in avian song control brain regions. PLoS One. 2012;7:e35119. doi: 10.1371/journal.pone.0035119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tomaszycki ML, Peabody C, Replogle K, Clayton DF, Tempelman RJ, Wade J. Sexual differentiation of the zebra finch song system: potential roles for sex chromosome genes. BMC Neurosci. 2009;10:24. doi: 10.1186/1471-2202-10-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.London SE, Clayton DF. Functional identification of sensory mechanisms required for developmental song learning. Nat Neurosci. 2008;11:579–586. doi: 10.1038/nn.2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haesler S, Rochefort C, Georgi B, Licznerski P, Osten P, Scharff C. Incomplete and inaccurate vocal imitation after knockdown of FoxP2 in songbird basal ganglia nucleus Area X. PLoS Biol. 2007;5:e321. doi: 10.1371/journal.pbio.0050321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Agate RJ, Scott BB, Haripal B, Lois C, Nottebohm F. Transgenic songbirds offer an opportunity to develop a genetic model for vocal learning. Proc Natl Acad Sci U S A. 2009;106:17963–17967. doi: 10.1073/pnas.0909139106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Itoh Y, Arnold AP. Zebra finch cell lines from naturally occurring tumors. In Vitro Cell Dev Biol Anim. 2011;47:280–282. doi: 10.1007/s11626-011-9392-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 23.Singh D, Orellana CF, Hu Y, Jones CD, Liu YF, Chiang DY, Liu JZ, Prins JF. FDM: a graph-based statistical method to detect differential transcription using RNA-seq data. Bioinformatics. 2011;27:2633–2640. doi: 10.1093/bioinformatics/btr458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tarazona S, Garcia-Alcalde F, Dopazo J, Ferrer A, Conesa A. Differential expression in RNA-seq: a matter of depth. Genome Res. 2011;21:2213–2223. doi: 10.1101/gr.124321.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou YH, Xia K, Wright FA. A powerful and flexible approach to the analysis of RNA sequence count data. Bioinformatics. 2011;27:2672–2678. doi: 10.1093/bioinformatics/btr449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ekblom R, Balakrishnan CN, Burke T, Slate J. Digital gene expression analysis of the zebra finch genome. BMC Genomics. 2010;11:219. doi: 10.1186/1471-2164-11-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ekblom R, Slate J, Horsburgh GJ, Birkhead T, Burke T. Comparison between Normalised and Unnormalised 454-Sequencing Libraries for Small-Scale RNA-Seq Studies. Comp Funct Genomics. 2012;2012:281693. doi: 10.1155/2012/281693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheng HY, Clayton DF. Activation and habituation of extracellular signal-regulated kinase phosphorylation in zebra finch auditory forebrain during song presentation. J Neurosci. 2004;24:7503–7513. doi: 10.1523/JNEUROSCI.1405-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mello CV. Mapping vocal communication pathways in birds with inducible gene expression. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 2002;188:943–959. doi: 10.1007/s00359-002-0347-1. [DOI] [PubMed] [Google Scholar]
- 31.Roberts A, Pimentel H, Trapnell C, Pachter L. Identification of novel transcripts in annotated genomes using RNA-Seq. Bioinformatics. 2011;27:2325–2329. doi: 10.1093/bioinformatics/btr355. [DOI] [PubMed] [Google Scholar]
- 32.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Clark MB, Amaral PP, Schlesinger FJ, Dinger ME, Taft RJ, Rinn JL, Ponting CP, Stadler PF, Morris KV, Morillon A, Rozowsky JS, Gerstein MB, Wahlestedt C, Hayashizaki Y, Carninci P, Gingeras TR, Mattick JS. The reality of pervasive transcription. PLoS Biol. 2011;9:e1000625. doi: 10.1371/journal.pbio.1000625. discussion e1001102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Bakel H, Nislow C, Blencowe BJ, Hughes TR. Most “Dark Matter” Transcripts Are Associated With Known Genes. Plos Biology. 2010;8 doi: 10.1371/journal.pbio.1000371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guo X, Liu W, Pan Y, Ni P, Ji J, Guo L, Zhang J, Wu J, Jiang J, Chen X, Cai Q, Li J, Zhang J, Gu Q, Liu B, Zhu Z, Yu Y. Homeobox gene IRX1 is a tumor suppressor gene in gastric carcinoma. Oncogene. 2010;29:3908–3920. doi: 10.1038/onc.2010.143. [DOI] [PubMed] [Google Scholar]
- 37.Shah N, Sukumar S. The Hox genes and their roles in oncogenesis. Nature Reviews Cancer. 2010;10:361–371. doi: 10.1038/nrc2826. [DOI] [PubMed] [Google Scholar]
- 38.Fernandez-Jimenez N, Castellanos-Rubio A, Plaza-Izurieta L, Gutierrez G, Irastorza I, Castano L, Vitoria JC, Bilbao JR. Accuracy in copy number calling by qPCR and PRT: a matter of DNA. PLoS One. 2011;6:e28910. doi: 10.1371/journal.pone.0028910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Drnevich J, Replogle K, Lovell P, Hahn TP, Johnson F, Mast TG, Strand C, London SE, Mukai M, Wingfield JC, Arnold AP, Ball GF, Brenowitz E, Wade J, Mello C, Clayton DF. The impact of experience-dependent and -independent factors on gene expression in songbird brain. (submitted) [DOI] [PMC free article] [PubMed]
- 40.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.HTSeq: Analysing high-throughput sequencing data with Python. doi: 10.1093/bioinformatics/btac166. [ http://www-huber.embl.de/users/anders/HTSeq/] [DOI] [PMC free article] [PubMed]
- 42.Wu X, Watson M. CORNA: testing gene lists for regulation by microRNAs. Bioinformatics. 2009;25:832–833. doi: 10.1093/bioinformatics/btp059. [ http://www.ark-genomics.org/tools/GOfinch, http://www.arkgenomics.org/tools/KEGGfinch] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.