Abstract
Membranes are flexible barriers that surround the cell and its compartments. To execute vital functions such as locomotion or receptor turnover, cells need to control the shapes of their membranes. In part, this control is achieved through membrane-bending proteins, such as the bin/amphiphysin/rvs domain (BAR) proteins. Many open questions remain about the mechanisms by which membrane-bending proteins function. Addressing this shortfall, recent structures of BAR protein:membrane complexes support existing mechanistic models, but also produced novel insights into how BAR-domain proteins sense, stabilize and generate curvature. Here we review these recent findings, focusing on how BAR proteins interact with the membrane, and how the resulting scaffold structures might aid the recruitment of other proteins to the sites where membranes are bent.
Keywords: BAR-domain, membrane curvature, scaffold, cryoEM, membrane remodeling, amphipathic wedge
Membrane curvature: a vital property of cells
Separation of ‘inside’ from ‘outside’ was a pivotal event in the creation of life; it happened when organic amphiphiles, known as phospholipids, became abundant enough to form selfsealing, curved and semipermeable barriers that established chemically defined compartments. The same breakthrough, however, created many challenges because the newly formed flexible envelopes, known as biological membranes, needed to allow the steady influx of nutrients and the secretion of wastes. Moreover, cells needed to sense changes in the environment and communicate signals across membranes. To meet these constraints, cells had to be able to deliberately change the shape of their membranes and with high spatial and temporal accuracy, because many cellular processes such as cell migration, cell division or endocytosis crucially depend on membrane remodeling reactions. Although tremendous progress has been made in the understanding of membrane transport and signaling, understanding how cells change the shape of their membranes is still at an early stage. To date, studies of how cells change shape have largely focused on a geometric measure known as membrane curvature, which describes the degree to which a membrane bilayer is bent (Figure 1). The sign (and thus direction) of curvature is an arbitrary measure: when viewed from the cytosol, plasma membrane invaginations predominantly contain segments with positive curvature, whereas protrusions consist of membranes with negative curvature (Figure 1A). Membrane curvature is not a passive property of the membrane but rather has emerged as a highly regulated state, likely because it actively influences diverse processes including membrane fusion and fission, the activity of membrane proteins, and the recruitment of proteins to the membrane-cytosol interface.
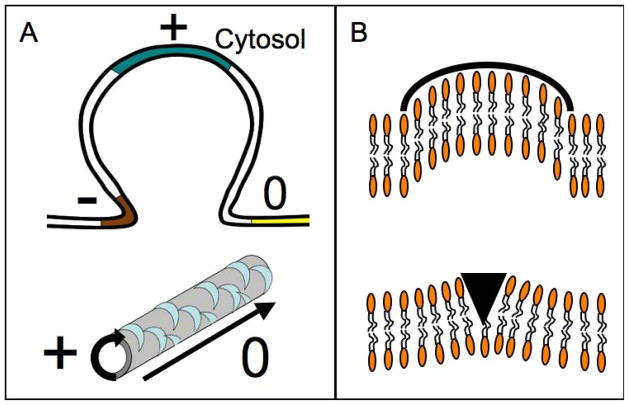
Figure 1. Curvature and membrane structures.
(A) Membrane structures, like invaginations, are composed of areas with different curvatures. Note that the sign of the curvature depends on the vantage point; hereafter curvature will be referred to as seen from the cytosol. On the top is a schematic view of a plasma membrane invagination displaying various curvatures. The flat membrane has zero (0) curvature (yellow). At the base of the invagination (brown) the curvature is negative (−). The part of the membrane that reaches further into the cytosol possesses positive (+) curvature. The bottom shows a membrane tubule, which is a ubiquitous structure in cells (e.g. T-tubules in muscle cells). These structures are created and maintained by proteins (blue crescents). The curvature is positive along the circumference of the tubule and zero along the tubular axis. (C) Schematic view of the mechanisms by which BAR domains can generate curvature. Upper panel: the crescent shaped BAR domain interacts with the bilayer through electrostatic interactions, imposing its intrinsic shape onto the membrane. This mechanism is known as the ‘scaffolding mechanism’. Lower panel: alternatively, BAR domains can introduce an amphipathic structure, such as a helix, into one leaflet of the membrane. This ‘wedge’ displaces lipids, which will cause the membrane to bend towards the BAR-domain. This mechanism is referred to as the ‘wedging mechanism’.
The number of publications containing ‘membrane curvature’ in their title has doubled over the past 8 years compared to the two decades before 2005 (180 vs. 394, PubMed), marking membrane remodeling as a rapidly emerging new field. Why has this accelerating progress been made only so recently? In part, the answer is that the direct visualization and investigation of the dynamics of curvature generation is difficult. Put into perspective, the publication of electron micrographs by Roth and Porters in 1964 of tissues undergoing endocytosis presented the first visual proof for curvature-generating proteins [1]. Yet almost 20 years went by until dynamin, the protein that fissions the membrane, was identified, and another 8 years passed until this protein was cloned [2–4]. Extending this initial discovery, membrane-bending proteins such as the Bin/amphiphysin/Rvs161 domain (BAR) proteins or the epsin N-terminal homology (ENTH) domain proteins [5, 6] have been described since the mid 1990s, but their structure and mechanisms of action remained elusive. This knowledge gap began to close with the emergence of high-resolution structures [7–11], which in the case of BAR-domains immediately suggested possible mechanisms of action because of the characteristic banana-shaped structure of the molecules (discussed later). Moreover, the increasing number of protein structures capable of bending membranes allowed testing and improvement of the theoretical framework of curvature induction that had existed since the 1970s [12, 13].
Despite this tremendous progress, however, the molecular mechanisms by which membrane-bending domains sense, generate and stabilize membrane curvature and how these events enable the coordinated progression of curvature-dependent cellular processes remains unclear. One impediment to work in this area is the chemical and physical complexity of the bilayer, which profoundly influences its susceptibility to membrane remodeling and the recruitment of specific membrane remodelers. Adding to this challenge, the field lacks standardized methods to study membrane remodeling and the tools to computationally describe the complex lipid mixtures that are naturally found in cellular membranes. As a consequence, current studies are largely focused on answering questions such as how membrane-remodeling proteins organize on the membrane surface, how their scaffold structure is controlled, and what aspects of the surface lattices are likely to contribute to the management of membrane curvature. Although these questions are among the simplest questions one can ask, the emerging structural biology of membrane-associated scaffolds is an exciting new chapter in a rapidly growing field that is focused on understanding how cells adapt the shape of bilayers to meet their individual needs.
Architecture and classification of BAR-domain proteins
Among the plethora of membrane bending proteins, the superfamily of BAR proteins has become a main focus of attention over recent years. Numerous reports reaffirm the notion that these proteins belong to an essential arsenal that cells deploy to shape membranes from yeast to mammals (see [14] for a review). Lacking characteristic signature sequence motifs in their primary structure, membership in the BAR-domain superfamily is not readily recognizable. However, at the structural level BAR domains are remarkably conserved, featuring a three helix coiled coil core that, in most cases, forms curved homo- or heterodimers, which results in the characteristic ’banana-shape’ (Figure 2) [14]. The factors that determine the formation of homodimers and exclude the existence of heterodimers are largely unknown. A notable exception is sorting nexin 33, for which the ability to homodimerize has been attributed to specific amino acids [15]. Regardless, disruption of the dimerization interface renders BAR proteins non-functional and is one way to regulate their membrane remodeling activity [15, 16]. This functional impairment occurs because proper interaction of the BAR-domains with negatively charged lipid headgroups such as in phosphoinositides or phosphatidylserine requires the appropriate positioning of positively charged residues in the context of the banana-shaped dimer [17–19].
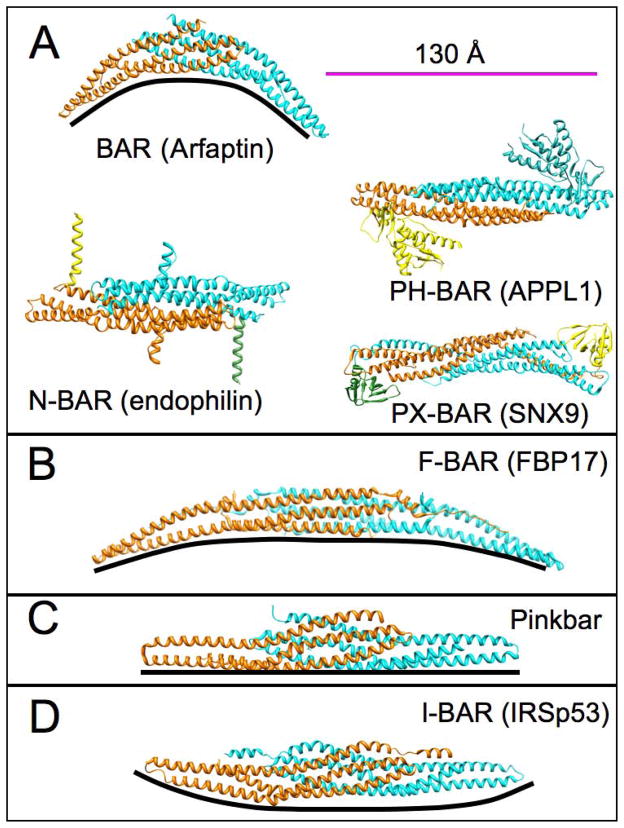
Figure 2. Structures of BAR domains from different subfamilies.
All structures are depicted as dimers (cyan and orange) (scale bar 130Å). The black line underneath each structure was introduced to show the intrinsic curvature of each domain. (A) Left, above: side view of the BAR domain of arfaptin 1 (pdb ID 1I49). BAR domains exhibit the highest positive curvature within the BAR superfamily. Classic BAR domains are further subdivided based on additional membrane binding domains. For clarity, different BAR domains are shown as top view. The N-BAR domain (endophilin, pdb courtesy of G. Voth from [76], left, below) contains a N-terminal helix (yellow and green) and a second set of amphipathic helices known as ‘insert helices’ (orange and cyan). APPL1 (right, above) is an example of a PH-BAR domain that contains an additional pleckstrin-homology domain (PH) (pdb ID 2Z0O). Lastly, PX-BAR proteins (right, below), such as sorting nexin 9 (SNX9, pdb ID 2RAI), are comprised of a phox-homology domain (PX) and a BAR domain. (B) Sideview of the F-BAR domain from FBP (pdb ID 2EFL) exemplifies a shallow, positive curvature that is typical for this subfamily. (C) The Pinkbar is the only instance of a BAR protein that promotes zero curvature; its BAR domain shows no intrinsic curvature (pdb ID 3OK8). (D) I-BAR domain proteins are the only subfamily that can bend membranes to generate structures with negative curvature (IRSp53, pdb ID 1Y2O).
Extending past the BAR-domain, most BAR-proteins have at least one additional domain, such as a src-homology 3 (SH3) domain, which enables BAR proteins to interact with proline-rich domain (PRD) containing proteins. Thus, BAR-domain proteins are scaffolding proteins that organize a variety of other proteins in a curvature-dependent manner. BAR proteins are categorized based on the general type of their BAR domain: a classical BAR domain, extended FCH domain (F-BAR), or IMD/Inverse BAR domain (I-BAR) (Figure 2).
Classical BAR domains, as found in arfaptin, were the first to be discovered and have the highest degree of intrinsic curvature (Figure 2). This branch of BAR-domains is further subdivided based on additional membrane binding domains such as an amphipathic N-terminal helix (N-BAR, e.g. amphiphysins and endophilins), the pleckstrin homology domain (PH-BAR, e.g. in APPL1), or the phox domain (PX-BAR, e.g. sorting nexin 9)[20]. Moreover, a recent structure of Arfaptin-2 complexed to the GTPase Alr 1 suggests that BAR proteins can integrate domains into the scaffold that are not part of the BAR protein itself [21]. Regardless of the complement of additional membrane binding domains, all classical BAR-domains support and promote positive membrane curvature.
F-BAR domain proteins, such as Cdc42-interacting protein 4 (CIP4), formin binding protein 17 (FBP17), or FCHO (Fes-CIP4 homology domain) represent the largest and most diverse family and is further subdivided into six distinct subfamilies [22]. Notably, the intrinsic curvature of the F-BAR domain ranges from high (syndapins) to almost planar (FCHO) [23], which allows these proteins to support a large spread of membrane curvatures. Just as classical BAR-domains, F-BAR domains are associated with positive membrane curvature.
Lastly, I-BAR domains are another variation of the BAR domain theme. These domains have a negative curvature, and represent a tool for the cell to generate extrusions [24]. A special variation of I-BAR proteins is the recently discovered PinkBAR domain [25], which in contrast to the other BAR-subfamilies, has no intrinsic curvature. This lack of intrinsic curvature allows PinkBAR domains to create scaffolds on flat membrane surfaces.
BAR-dependent membrane remodeling has largely been associated with the cellular process of endocytosis. However, there is rapidly emerging evidence that BAR proteins play a more general role in membrane scaffolding, organelle creation, organismal patterning [26], and disease (Box 1). Regardless of their physiological role, high-resolution crystal structures of BAR-domains have solidly established that, with the exception of PinkBar-domains, all BAR-domain-dimers are intrinsically curved. That the physical shape of BAR-domain dimers corresponds with the type of curvature they promote is consistent with the idea that the intrinsic shape of the dimer contributes to the bending of membranes and the recognition and support of curved membrane states. As will be described in more detail later, however, some BAR-domains exploit additional features, such as amphipathic wedges, to accomplish their tasks in membrane remodeling.
Box 1. BAR proteins in health and disease: case studies.
Bin1 and the T-tubule system in myocytes
The T-tubule system in skeletal and cardiac muscle is a specialized structure that relays current formed at the plasma membrane. Bin1 (or amphisphysin 2) is highly expressed in muscle cells and constitutes the major component of T-tubules [27]. Genetic studies in Drosophila showed that the Drosophila Bin1 homologue is dispensable for endocytosis but crucial for the formation of T-tubules [28]. Interestingly, myocytes from patients with a failing heart lose T-tubules [29], which is correlated with significantly reduced expression of Bin1 and Ca2+-influx. This reduction in Ca2+-influx is because Bin1 is also the main scaffolding protein for calcium channels in the T-tubules [30, 31]. Similarly, skeletal myopathies have also been linked to dysfunctional Bin1 protein or reduced expression levels of Bin1. For instance, alternative splicing of Bin1 mRNA has been identified as the cause of several types of myotonic dystrophy, which is a common muscular dystrophy in adults [32]. In addition, some forms of centronuclear myopathies, a rare group of diseases that manifest in abnormal localization of nuclear material within myocytes and muscle weakness, have also been attributed to mutation in Bin1 [33]. The histology of both myopathies identifies disrupted T-tubules as the major cause for the loss of muscle strength. These diseases confirm the importance of a BAR domain as a membrane scaffold that maintains organelles.
BAR proteins in cell migration and cancer
Filopodia and lamellopodia are specialized structures of the plasma membrane that are necessary for cell motility and the maturation of neurons [34]. Although actin polymerization was originally thought to create these structures, it is becoming increasingly clear that I-BAR proteins play a major role in scaffolding the plasma membrane during these processes [35–37]. However, the precise mechanism by which I-BAR proteins control the formation of filopodia remains to be fully elucidated. I-BAR proteins might drive the formation of protrusions by bending membranes or activating actin polymerization by sensing curvature. The latter mechanism was reported for F-BAR proteins [38], which recently have also been shown to play roles in cancer cell invasion and the formation of invadopodia [39, 40]. Interestingly, besides their role in enhancing actin polymerization, several I-BAR proteins have been shown to bind to actin directly, and to steer cell migration by inhibiting endocytosis of chemotaxis receptors [41]
Molecular mechanisms of membrane bending
In the past, crystal structures and molecular dynamics simulations have strongly influenced how the field thinks about membrane bending by BAR domains [20]. Among the most intuitive implications arising from the structures was the hypothesis that the intrinsic curvature of BAR-domain dimers aids membrane remodeling by simply imposing its shape on the membrane substrate (Figure 1B, upper panel). This mode of action has become known as the ‘scaffolding’ mechanism [13, 42] and is well supported by molecular dynamics simulations [43, 44]. Contrasting with scaffolding is a second mechanism that poses that the insertion of amphipathic wedges into the bilayer can sense membrane curvature and promote its formation through the concerted displacement of lipids in the leaflet proximal to the site of insertion [45] (Figure 1B, lower panel). Just as for scaffolding, there is solid experimental and computational evidence to support the membrane insertion of amphipathic wedges during membrane remodeling processes [19, 45]. However, the relative contribution of each mechanism remains an area of active investigation in cases such as endophilin and amphiphysin, which contain at least one amphipathic wedge in addition to their BAR-domain. For instance, studies that investigated whether an N-BAR amphipathic wedge can generate curvature by itself have yielded seemingly conflicting data, which in part might be rooted in the use of different lipid substrates for these studies [19, 46–48]. Supporting the idea that substrate choice might be a possible cause for experimental bias, there is growing evidence that the composition of the bilayer and pre-existing curvature are major factors determining membrane susceptibility for BAR-domain recruitment and remodeling [49, 50]. Further complicating the picture, the N-terminal helix in N-BAR proteins as well as amphipathic wedges in other membrane remodeling proteins have recently been described to also play a role in membrane fission. Notably, this study reported that membrane bending and fission are reciprocally related and that the length and composition of the amphipathic wedges are major factors determining their potential to effect fission [51]. These recent discoveries vividly illustrate the complexity of membrane remodeling processes and emphasize that much remains to be learned from studying these systems.
Visualizing membrane-bound scaffolds: the new frontier
Complementing high-resolution structural and spectroscopic studies, electron microscopic reconstructions of membrane-bound F-BAR and N-BAR proteins have emerged over the past four years [17, 52]. These reconstructions confirmed the scaffolding and wedging mechanisms, and for the first time revealed design details of a few select lattices that BAR-domains are capable of forming on the surface of membranes. Although the helical arrays used for these studies were more extensive, and likely more ordered, than those found inside cells, their structures nevertheless appear relevant. In support of this notion, T-tubules in muscle cells are N-BAR protein-coated membrane tubules (Box 1) that are several μm in size, illustrating that BAR-scaffolds can grow to large sizes even under physiological conditions [28]. Moreover, mutational analysis of sites predicted to be important for scaffold formation impaired the formation of invaginations both in vitro and in cells [8, 17]. It therefore seems likely that the electron microscopic reconstructions reflect major aspects of lattices formed in biological structures such as the endocytotic neck.
Comparison of the available N-BAR and F-BAR scaffold structures reveals that their general design principles are remarkably different and that membrane-bound BAR-domains display previously unknown, yet functionally highly significant properties (Figure 3). For instance, extensive lateral interactions between the cores of CIP4 F-BAR domain dimers result in oligomerization and tight packing of the molecules on curved membrane surfaces. These interactions were shown to be necessary for curvature generation both in vitro and in vivo [17], but were not observed in high-resolution crystal structures [8], which emphasizes the importance of electron microscopic imaging in the exploration of BAR-dependent membrane remodeling. Supporting this idea further, there is evidence that the scaffolds of other F-BAR subfamilies might follow different design rules [17, 53]; therefore, achieving a mechanistic understanding of these proteins will ultimately require direct visualization of their membrane bound states.
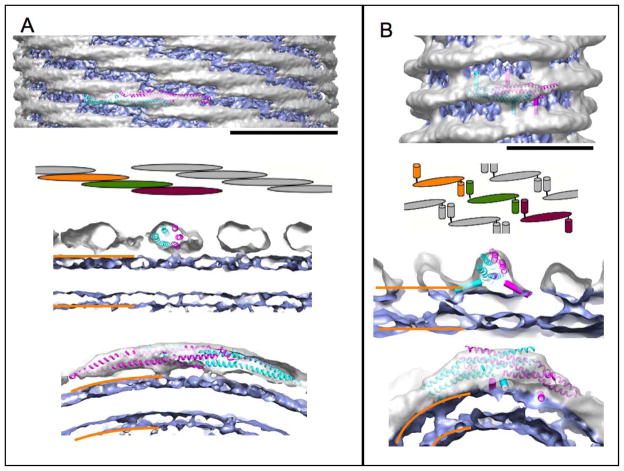
Figure 3. Structures of membrane-bound BAR domains.
All structures are of protein-coated membrane tubules that have been reconstructed from electron microscopic images. The density of the protein is colored in silver, whereas the membrane is blue. (A) Structure the CIP4 F-BAR domain scaffold (pdb ID 2EFL) (EMDB accession code 1471). Top: side view of the membrane tubule. One copy of the F-BAR domain of the closely related FBP17 was fitted to highlight organization of the protein on the membrane (scale bar 220Å). F-BAR dimers form extensive lateral interactions that give rise to a stiff protein coat, which imposes its shape on the membrane. The cartoon highlights the high degree of overlap between adjacent F-BAR dimers. Middle: cross-section along the membrane tubule axis. Bottom: cross-section perpendicular to the membrane tubule axis. The orange bars demarcate the approximate boundaries of the membrane bilayer. (B) Structure of the endophilin N-BAR scaffold (EMDB accession code 5365). Top: side view of the membrane tubule. One dimer of the N-BAR domain (pdb ID 1ZWW) was fitted to show the density occupied by the protein (scale bar 130Å). Additionally, the amphipathic helices present in endophilin are depicted as cylinders for better visibility. N-BAR scaffolds are held together by interactions between the N-terminal H0 helices (see cartoon). Middle: cross-section along the membrane tubule axis, illustrating the positioning of the BAR-domain and the slope of the insert helices. Bottom: cross-section perpendicular to the membrane tubule axis. The leaflet closest to the N-BAR domain is continuous with the density of the protein, indicating a loss of order of the lipid head groups.
Adding to the variety of potential lattices, the study by Frost et al. demonstrated that the F-BAR domain dimer of FBP17 harbors an alternate lipid-binding interface that allows the dimer to form two-dimensional arrays on the surface of flat membranes [17]. Mutagenesis studies confirmed that this novel and previously unknown lipid-binding mode is essential for curvature induction in vivo, raising the intriguing possibility that the two-dimensional arrays are a curvature-neutral storage form of FBP17. Such a storage form would have two advantages over dispersed FBP17 dimers: a cooperative transition of several dimers from a ‘flat’ to ‘curved’ membrane binding mode would facilitate the generation of macroscopic curvature, and it would overcome the resistance of the bilayer towards deformation by forming the observed extensive interactions within the curved lattice [54]. How this hypothetical process would be triggered and regulated remains to be shown. More excitingly, however, the existence of a novel membrane-binding mode in one F-BAR subfamily raises the possibility that alternate binding modes also exist in other BAR-domain proteins, opening a potentially new chapter in the mechanistic and cell biological understanding of BAR-dependent membrane remodeling.
Sharply contrasting with the design of CIP4/FBP17 scaffolds, electron microscopic reconstructions by Mim et al. revealed that lattices of the N-BAR protein endophilin lack direct lateral interactions between the BAR-domain cores and expose large areas of membrane surface (Figure 3) that might allow other proteins to access the membrane [52]. Lattice coherence is achieved through antiparallel interactions between the N-terminal amphipathic wedge helices that are known as H0. These interactions are independently supported by experimental data [46] and have been extensively used in modeling of N-BAR lattices [55]. Moreover, the experimental scaffold structures of endophilin revealed that its second amphipathic wedge slopes towards the bilayer interior. This was unexpected based on crystallographic (pdb ID 2Z0V) and spectroscopic data [56], and suggests that curvature induction by endophilin is accompanied by defined conformational changes. Also unexpected was the finding that H0:H0 interactions are very degenerate, non-specific, and of low affinity. An important functional implication of this discovery is that, in principle, endophilin might be able to form mixed lattices with another N-BAR protein such as amphiphysin, allowing the close spatial clustering of several functionalities, while also excluding other types of BAR-domain proteins such as F-BAR proteins, whose lattice packing arrangement is fundamentally different [17]. Whether such co-polymers exist and how their assembly is regulated will need to be addressed by future studies. A different study by Mizuno et al. also visualized helical arrays of endophilin N-BAR domains bound to very narrow lipid tubules consisting of pure palmitoyl-oleoyl-phosphatidylglycerol [57]. Although at a coarse scale these reconstructions share some similarity, the study by Mizuno et al. did not provide detailed insights into the packing interactions between N-BAR dimers or the disposition of the H0-helices. Interestingly, though, some of the observed structures were too narrow to contain a complete bilayer at the core, suggesting that endophilin N-BAR domains assembled on the surface of lipid micelles. Such structures were not observed in the study by Mim et al. [52], who focused their analysis on lattices assembled on tubes with diameters close to those reported to be relevant for endocytotic membrane fission by the GTPase dynamin. Nevertheless, the reconstructions by Mim et al. revealed significant bilayer stress that was caused by the high local concentration of amphipathic helices in the bilayer leaflet proximal to the BAR-domain. This direct observation of bilayer stress ties in with the finding that amphipathic wedges can drive membrane fission [51], and emphasizes how direct visualization of membrane-associated scaffolds by electron microscopy complements other approaches to understand membrane bending and remodeling.
Just as different as the general design of their surface-associated lattices, BAR-domains seem to employ various strategies to accommodate a distribution of curvature states. For instance, the strong lateral interactions between CIP4 and FBP17 F-BAR domain cores allow the entire lattice to tilt in the plane of the membrane. Such reorientation of the lattice was identified as the main mechanism that allows CIP4/FBP17 lattices to accommodate a broad and smooth distribution of curvature states [17]. By contrast, the absence of direct lateral interactions between endophilin BAR-domain cores precludes a significant role for lattice reorientation in curvature accommodation by this protein. Instead, endophilin accommodates different curvature states primarily by the removal or addition of entire endophilin dimers from the helical array. It will be interesting to see to what extent these two extremes contribute to curvature accommodation in the various BAR-domain subfamilies. However, even at this early stage, it is becoming increasingly clear that direct visualization of membrane-bound scaffolds by EM will be important for uncovering the mechanistic details of BAR-dependent membrane remodeling.
Control of membrane binding
The ability of BAR domains to engage bilayers and recruit specific interaction partners creates a need to control when and where a given BAR-domain protein can gain access to its targets. A crucial parameter for controlling membrane access is membrane curvature. Numerous studies have shown that BAR proteins have a preference for membranes with specific curvatures in vitro, and that the curvature preferences correlate with the intrinsic curvature of the BAR-domain [19, 45, 50]. Adding to this intuitive mechanism, sizeable evidence has emerged in support of a second, less intuitive mechanism that utilizes the ability of amphipathic sequences, such as the N-terminal helix in N-BAR proteins, to detect and bind to areas where curvature stress causes packing defects in the lipid headgroup region of the bilayer [45, 58, 59]. The number of defects correlates with the degree of curvature; therefore, the ability of some BAR-domain proteins to recognize these defects is an efficient way to control differential binding to membranes of different curvatures. Notably, it is the dual role of amphipathic wedges in curvature sensing and generation that fuels much of the current discussion about the mechanistic contributions of wedges in membrane remodeling.
Independent of membrane curvature and adding to the repertoire of mechanisms that regulate BAR-membrane interactions, intramolecular inhibition (often referred to as autoinhibition) can also restrict membrane binding by BAR proteins. For instance, PICK1 (protein interacting with protein kinase C alpha) and syndapin 1 (also known as PACSIN) are proteins for which their PDZ (PSD95-DLG1-Zo1 homology) or SH3 domains, respectively, obstruct membrane binding [60–62]. In the case of PICK1, membrane recruitment and release of the autoinhibited ‘closed conformation‘ requires a membrane-bound ligand. [63]. By contrast, syndapin 1 autoinhibition can be released by soluble ligands.
Finally, phosphorylation of specific residues of BAR proteins has been demonstrated to block membrane binding and bending. For instance, phosphorylation of the F-BAR domain of syndapin 1 was shown to impair oligomerization in vivo and in vitro [64]. Similarly, reversible hyperphoshorylation of the F-BAR protein cdc15 causes it to adopt an autoinhibited ‘closed’ conformation that can no longer engage the membrane [65]. No doubt, these initial studies are only the beginning in a journey that will reveal how frequently the different mechanisms of membrane recruitment are found throughout various BAR-protein families. Regardless, the existence of diverse mechanisms to regulate membrane access and partner recruitment, as described in the next section, suggest that regulatory networks are likely to emerge in the foreseeable future, adding yet another layer of complexity to BAR-dependent membrane remodeling.
Recruitment of BAR-protein-interacting proteins
As the structures of BAR-domain scaffolds begin to emerge, new and puzzling observations take center stage. For instance, the experimentally determined lattice structures allow estimation of the actual concentrations of SH3- or other interactor domains that are tethered to the membrane surface through the BAR-lattice. The results of a ‘back of the envelope’ calculation yields a surprising result: assuming a 100nm annulus above the membrane surface, the concentrations of interactor domains are ~3–5 mM irrespective of type of BAR-protein (F-BAR vs N-BAR) and the actual membrane curvature (e.g. ~28,000 SH3-domains/μm2 in case of a CIP4 F-BAR tube with diameter of 56nm, or ~21,000 SH3-domains/μm2 in case of an endophilin N-BAR tube with diameter of 28nm) [17, 52]. This estimate is in good agreement with a report by Arasada and Pollard who found the density of SH3-domains to be ~16,500/μm2 above the F-BAR-scaffold that is present in the neck region of fission yeast endocytotic invaginations [66]. An immediate implication of these numbers is that membrane-bound BAR protein scaffolds seem to act as kinetic traps. More specifically, BAR-scaffolds will non-specifically engage any potential binding partner whose binding affinity is in the low micromolar to nanomolar range, which is typically observed for protein:protein interactions [67–69]. This observation raises several questions. For instance, how is this result compatible with the spatial and temporal accuracy of processes such as endocytosis, which involves over 30 different proteins, including several BAR-domain proteins, that act at different stages along the time axis [70]? Similarly, how can the apparent non-specificity of interactions with potential interaction partners explain why, for instance, the most notable defect in endophilin knockout mice is the failure to recruit only one of the PRD-containing ligands, synaptojanin [71]? In the latter case, a potential answer might lie in the properties of synaptojanin itself, because its lipid-binding domains and curvature-dependent enzymatic phosphatase activity [38, 72, 73] allow for a coincidence detection mechanism that assures proper recruitment to sites where synaptojanin function is needed. In other cases, ‘binary’ switches such as phosphorylation could bias retention of interaction partners above the BAR-lattice [74]. Going beyond these known and familiar mechanistic solutions, the reconstructions of endophilin lattices suggest two additional mechanisms that might govern specificity in the kinetically controlled recruitment of interaction partners. First, lattices formed by endophilin and the F-BAR domain of CIP4 expose different amounts of membrane surface. Therefore, simple steric exclusion or permissivity might bias recruitment of components to the membrane surface. Second, reconstructions of endophilin lattices on the surface of membranes with different curvatures revealed that the SH3 domains of endophilin can dimerize above some, but not all scaffolds [52]. This suggests that specific spatial presentation patterns above the scaffolds might guide recruitment and stable retention of binding partners in a curvature-dependent manner. For example, close spatial apposition of the SH3 domains in the context of an SH3 domain dimer could guide the preferential recruitment of partners such as dynamin that have spatially close proline-rich sequences. That said, experimentally determined scaffold structures challenge longstanding ideas about the recruitment of biological machinery to membrane interfaces and simultaneously suggest how these macromolecular arrays might achieve specificity by several putative mechanisms. Although some of the emerging ideas need to be validated experimentally, there is no doubt that recent progress in the field is opening up many new directions for further inquiry.
Concluding remarks
Curvature and its generation is a rapidly expanding field in membrane biochemistry and biophysics. The past few years have seen dramatic improvements in the structure determination of membrane-bound scaffolds, information technology, and the development of suitable models that allow the systematic study of membrane remodeling in living organisms. The original, simplistic view that differences in intrinsic curvatures define the spatial and temporal patterns of BAR-domain protein deployment [75] has given way to the realization that the recruitment and action of BAR-domains are subject to complex and poorly understood regulatory mechanisms [70]. Moreover, direct visualization of membrane-bound BAR-scaffolds has revealed novel and unexpected aspects of BAR biology such as the ability of some F-BAR domains to engage both flat and curved bilayers [17, 53]. In addition, rapidly emerging evidence of the involvement of BAR-domain proteins and other membrane remodeling proteins in a large number of cellular processes, ranging from organelle biogenesis to malignant cell invasion, brings membrane remodeling to the forefront of research in the life sciences. With methodology improving continuously, future efforts will likely focus on visualizing the structure of membrane-bound multiprotein scaffolds and cracking the code of regulatory mechanisms that enable the recruitment of process-specific components to generic membrane remodeling scaffolds. Understanding these processes at the molecular level will provide unprecedented insight into one of the most dazzling yet elusive biological problems: membrane dynamics.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Roth TF, Porter KR. Yolk Protein Uptake in the Oocyte of the Mosquito Aedes Aegypti. L. The Journal of cell biology. 1964;20:313–332. doi: 10.1083/jcb.20.2.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen MS, et al. Multiple forms of dynamin are encoded by shibire, a Drosophila gene involved in endocytosis. Nature. 1991;351:583–586. doi: 10.1038/351583a0. [DOI] [PubMed] [Google Scholar]
- 3.van der Bliek AM, Meyerowitz EM. Dynamin-like protein encoded by the Drosophila shibire gene associated with vesicular traffic. Nature. 1991;351:411–414. doi: 10.1038/351411a0. [DOI] [PubMed] [Google Scholar]
- 4.Kosaka T, Ikeda K. Possible temperature-dependent blockage of synaptic vesicle recycling induced by a single gene mutation in Drosophila. Journal of neurobiology. 1983;14:207–225. doi: 10.1002/neu.480140305. [DOI] [PubMed] [Google Scholar]
- 5.Takei K, et al. Functional partnership between amphiphysin and dynamin in clathrin-mediated endocytosis. Nature cell biology. 1999:33–39. doi: 10.1038/9004. [DOI] [PubMed] [Google Scholar]
- 6.Wendland B, et al. Yeast epsins contain an essential N-terminal ENTH domain, bind clathrin and are required for endocytosis. The EMBO journal. 1999;18:4383–4393. doi: 10.1093/emboj/18.16.4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ford MGJ, et al. Curvature of clathrin-coated pits driven by epsin. Nature. 2002:361–366. doi: 10.1038/nature01020. [DOI] [PubMed] [Google Scholar]
- 8.Shimada A, et al. Curved EFC/F-BAR-domain dimers are joined end to end into a filament for membrane invagination in endocytosis. Cell. 2007:761–772. doi: 10.1016/j.cell.2007.03.040. [DOI] [PubMed] [Google Scholar]
- 9.Peter B, et al. BAR domains as sensors of membrane curvature: the amphiphysin BAR structure. Science (New York, NY. 2004:495. doi: 10.1126/science.1092586. [DOI] [PubMed] [Google Scholar]
- 10.Tarricone C, et al. The structural basis of Arfaptin-mediated cross-talk between Rac and Arf signalling pathways. Nature. 2001;411:215–219. doi: 10.1038/35075620. [DOI] [PubMed] [Google Scholar]
- 11.Mills IG, et al. EpsinR: an AP1/clathrin interacting protein involved in vesicle trafficking. The Journal of cell biology. 2003;160:213–222. doi: 10.1083/jcb.200208023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Helfrich W. Elastic properties of lipid bilayers: theory and possible experiments. Zeitschrift fur Naturforschung. 1973;28:693–703. doi: 10.1515/znc-1973-11-1209. [DOI] [PubMed] [Google Scholar]
- 13.Zimmerberg J, Kozlov MM. How proteins produce cellular membrane curvature. Nature reviews. 2006;7:9–19. doi: 10.1038/nrm1784. [DOI] [PubMed] [Google Scholar]
- 14.Ren G, et al. The BAR domain proteins: molding membranes in fission, fusion, and phagy. Microbiology and molecular biology reviews. 2006:37. doi: 10.1128/MMBR.70.1.37-120.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dislich B, et al. Specific amino acids in the BAR domain allow homodimerization and prevent heterodimerization of sorting nexin 33. The Biochemical journal. 2011;433:75–83. doi: 10.1042/BJ20100709. [DOI] [PubMed] [Google Scholar]
- 16.Gortat A, et al. Single point mutation in Bin/Amphiphysin/Rvs (BAR) sequence of endophilin impairs dimerization, membrane shaping, and Src homology 3 domain-mediated partnership. The Journal of biological chemistry. 2012;287:4232–4247. doi: 10.1074/jbc.M111.325837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frost A, et al. Structural basis of membrane invagination by F-BAR domains. Cell. 2008:807–817. doi: 10.1016/j.cell.2007.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Farsad K, et al. Generation of high curvature membranes mediated by direct endophilin bilayer interactions. The Journal of cell biology. 2001;155:193–200. doi: 10.1083/jcb.200107075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gallop J, et al. Mechanism of endophilin N-BAR domain-mediated membrane curvature. The EMBO journal. 2006:2898–2910. doi: 10.1038/sj.emboj.7601174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qualmann B, et al. Let’s go bananas: revisiting the endocytic BAR code. The EMBO journal. 2011:3501–3515. doi: 10.1038/emboj.2011.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakamura K, et al. Structural Basis for Membrane Binding Specificity of the Bin/Amphiphysin/Rvs (BAR) Domain of Arfaptin-2 Determined by Arl1 GTPase. The Journal of biological chemistry. 2012;287:25478–25489. doi: 10.1074/jbc.M112.365783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heath RJ, Insall RH. F-BAR domains: multifunctional regulators of membrane curvature. Journal of cell science. 2008;121:1951–1954. doi: 10.1242/jcs.023895. [DOI] [PubMed] [Google Scholar]
- 23.Roberts-Galbraith RH, Gould KL. Setting the F-BAR: functions and regulation of the F-BAR protein family. Cell cycle (Georgetown, Tex. 2010;9:4091–4097. doi: 10.4161/cc.9.20.13587. [DOI] [PubMed] [Google Scholar]
- 24.Zhao H, et al. I-BAR domain proteins: linking actin and plasma membrane dynamics. Current opinion in cell biology. 2011;23:14–21. doi: 10.1016/j.ceb.2010.10.005. [DOI] [PubMed] [Google Scholar]
- 25.Pykalainen A, et al. Pinkbar is an epithelial-specific BAR domain protein that generates planar membrane structures. Nature structural & molecular biology. 2011;18:902–907. doi: 10.1038/nsmb.2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Umasankar PK, et al. Distinct and separable activities of the endocytic clathrin-coat components Fcho1/2 and AP-2 in developmental patterning. Nature cell biology. 2012;14:488–501. doi: 10.1038/ncb2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee E, et al. Amphiphysin 2 (Bin1) and T-tubule biogenesis in muscle. Science (New York, NY. 2002:1193–1196. doi: 10.1126/science.1071362. [DOI] [PubMed] [Google Scholar]
- 28.Razzaq A, et al. Amphiphysin is necessary for organization of the excitation-contraction coupling machinery of muscles, but not for synaptic vesicle endocytosis in Drosophila. Genes & development. 2001;15:2967–2979. doi: 10.1101/gad.207801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lyon AR, et al. Loss of T-tubules and other changes to surface topography in ventricular myocytes from failing human and rat heart. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:6854–6859. doi: 10.1073/pnas.0809777106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hong TT, et al. BIN1 is reduced and Cav1.2 trafficking is impaired in human failing cardiomyocytes. Heart Rhythm. 2010 doi: 10.1016/j.hrthm.2011.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hong TT, et al. BIN1 is reduced and Cav1.2 trafficking is impaired in human failing cardiomyocytes. Heart Rhythm. 2011 doi: 10.1016/j.hrthm.2011.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fugier C, et al. Misregulated alternative splicing of BIN1 is associated with T tubule alterations and muscle weakness in myotonic dystrophy. Nature medicine. 2011;17:720–725. doi: 10.1038/nm.2374. [DOI] [PubMed] [Google Scholar]
- 33.Toussaint A, et al. Defects in amphiphysin 2 (BIN1) and triads in several forms of centronuclear myopathies. Acta neuropathologica. 2011;121:253–266. doi: 10.1007/s00401-010-0754-2. [DOI] [PubMed] [Google Scholar]
- 34.Dent EW, et al. Filopodia are required for cortical neurite initiation. Nature cell biology. 2007;9:1347–1359. doi: 10.1038/ncb1654. [DOI] [PubMed] [Google Scholar]
- 35.Mattila PK, et al. Missing-in-metastasis and IRSp53 deform PI(4,5)P2-rich membranes by an inverse BAR domain-like mechanism. The Journal of cell biology. 2007;176:953–964. doi: 10.1083/jcb.200609176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Suetsugu S, et al. The RAC binding domain/IRSp53-MIM homology domain of IRSp53 induces RAC-dependent membrane deformation. The Journal of biological chemistry. 2006;281:35347–35358. doi: 10.1074/jbc.M606814200. [DOI] [PubMed] [Google Scholar]
- 37.Millard TH, et al. Structural basis of filopodia formation induced by the IRSp53/MIM homology domain of human IRSp53. The EMBO journal. 2005;24:240–250. doi: 10.1038/sj.emboj.7600535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takano K, et al. EFC/F-BAR proteins and the N-WASP-WIP complex induce membrane curvature-dependent actin polymerization. The EMBO journal. 2008;27:2817–2828. doi: 10.1038/emboj.2008.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hu J, et al. Transducer of Cdc42-dependent actin assembly promotes epidermal growth factor-induced cell motility and invasiveness. The Journal of biological chemistry. 2011;286:2261–2272. doi: 10.1074/jbc.M110.157974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pichot CS, et al. Cdc42-interacting protein 4 promotes breast cancer cell invasion and formation of invadopodia through activation of N-WASp. Cancer research. 2010;70:8347–8356. doi: 10.1158/0008-5472.CAN-09-4149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Quinones GA, et al. I-BAR protein antagonism of endocytosis mediates directional sensing during guided cell migration. The Journal of cell biology. 2010;189:353–367. doi: 10.1083/jcb.200910136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McMahon HT, Gallop JL. Membrane curvature and mechanisms of dynamic cell membrane remodelling. Nature. 2005;438:590–596. doi: 10.1038/nature04396. [DOI] [PubMed] [Google Scholar]
- 43.Ayton GS, et al. Membrane remodeling from N-BAR domain interactions: insights from multi-scale simulation. Biophysical journal. 2007;92:3595–3602. doi: 10.1529/biophysj.106.101709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Blood PD, et al. Factors influencing local membrane curvature induction by N-BAR domains as revealed by molecular dynamics simulations. Biophysical journal. 2008;95:1866–1876. doi: 10.1529/biophysj.107.121160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bhatia V, et al. Amphipathic motifs in BAR domains are essential for membrane curvature sensing. The EMBO journal. 2009:3303–3314. doi: 10.1038/emboj.2009.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fernandes F, et al. Role of helix 0 of the N-BAR domain in membrane curvature generation. Biophysical journal. 2008;94:3065–3073. doi: 10.1529/biophysj.107.113118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Domanov YA, Kinnunen PK. Antimicrobial peptides temporins B and L induce formation of tubular lipid protrusions from supported phospholipid bilayers. Biophysical journal. 2006;91:4427–4439. doi: 10.1529/biophysj.106.091702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Varkey J, et al. Membrane curvature induction and tubulation are common features of synucleins and apolipoproteins. The Journal of biological chemistry. 2010;285:32486–32493. doi: 10.1074/jbc.M110.139576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hatzakis NS, et al. How curved membranes recruit amphipathic helices and protein anchoring motifs. Nature chemical biology. 2009;5:835–841. doi: 10.1038/nchembio.213. [DOI] [PubMed] [Google Scholar]
- 50.Sorre B, et al. Nature of curvature coupling of amphiphysin with membranes depends on its bound density. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:173–178. doi: 10.1073/pnas.1103594108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Boucrot E, et al. Membrane fission is promoted by insertion of amphipathic helices and is restricted by crescent BAR domains. Cell. 2012;149:124–136. doi: 10.1016/j.cell.2012.01.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mim C, et al. Structural basis of membrane bending by the N-BAR protein endophilin. Cell. 2012;149:137–145. doi: 10.1016/j.cell.2012.01.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Henne WM, et al. FCHo proteins are nucleators of clathrin-mediated endocytosis. Science (New York, N Y. 2010;328:1281–1284. doi: 10.1126/science.1188462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lyman E, et al. Water under the BAR. Biophysical journal. 2010;99:1783–1790. doi: 10.1016/j.bpj.2010.06.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yin Y, et al. Simulations of membrane tubulation by lattices of amphiphysin N-BAR domains. Structure. 2009;17:882–892. doi: 10.1016/j.str.2009.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jao CC, et al. Roles of amphipathic helices and the bin/amphiphysin/rvs (BAR) domain of endophilin in membrane curvature generation. The Journal of biological chemistry. 2010;285:20164–20170. doi: 10.1074/jbc.M110.127811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mizuno N, et al. Multiple modes of endophilin-mediated conversion of lipid vesicles into coated tubes: implications for synaptic endocytosis. The Journal of biological chemistry. 2010;285:23351–23358. doi: 10.1074/jbc.M110.143776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Suresh S, Edwardson JM. The endophilin N-BAR domain perturbs the structure of lipid bilayers. Biochemistry. 2010:5766–5771. doi: 10.1021/bi100760e. [DOI] [PubMed] [Google Scholar]
- 59.Cui H, et al. Mechanism of membrane curvature sensing by amphipathic helix containing proteins. Biophysical journal. 2011:1271–1279. doi: 10.1016/j.bpj.2011.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rao Y, et al. Molecular basis for SH3 domain regulation of F-BAR–mediated membrane deformation. Proceedings of the National Academy of Sciences. 2010:8213. doi: 10.1073/pnas.1003478107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rocca DL, et al. Inhibition of Arp2/3-mediated actin polymerization by PICK1 regulates neuronal morphology and AMPA receptor endocytosis. Nature cell biology. 2008;10:259–271. doi: 10.1038/ncb1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lu W, Ziff EB. PICK1 interacts with ABP/GRIP to regulate AMPA receptor trafficking. Neuron. 2005;47:407–421. doi: 10.1016/j.neuron.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 63.Madsen K, et al. Membrane localization is critical for activation of the PICK1 BAR domain. Traffic. 2008:1327–1343. doi: 10.1111/j.1600-0854.2008.00761.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Quan A, et al. Phosphorylation of syndapin I F-BAR domain at two helix-capping motifs regulates membrane tubulation. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:3760–3765. doi: 10.1073/pnas.1108294109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Roberts-Galbraith RH, et al. Dephosphorylation of F-BAR protein Cdc15 modulates its conformation and stimulates its scaffolding activity at the cell division site. Molecular cell. 2010;39:86–99. doi: 10.1016/j.molcel.2010.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Arasada R, Pollard TD. Distinct roles for F-BAR proteins Cdc15p and Bzz1p in actin polymerization at sites of endocytosis in fission yeast. Curr Biol. 2011;21:1450–1459. doi: 10.1016/j.cub.2011.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Demers JP, Mittermaier A. Binding mechanism of an SH3 domain studied by NMR and ITC. Journal of the American Chemical Society. 2009;131:4355–4367. doi: 10.1021/ja808255d. [DOI] [PubMed] [Google Scholar]
- 68.Harkiolaki M, et al. Structural basis for SH3 domain-mediated high-affinity binding between Mona/Gads and SLP-76. The EMBO journal. 2003;22:2571–2582. doi: 10.1093/emboj/cdg258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhou P, et al. Modeling and prediction of binding affinities between the human amphiphysin SH3 domain and its peptide ligands using genetic algorithm-Gaussian processes. Biopolymers. 2008;90:792–802. doi: 10.1002/bip.21091. [DOI] [PubMed] [Google Scholar]
- 70.Taylor MJ, et al. A high precision survey of the molecular dynamics of mammalian clathrin-mediated endocytosis. PLoS biology. 2011;9:e1000604. doi: 10.1371/journal.pbio.1000604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Milosevic I, et al. Recruitment of Endophilin to Clathrin-Coated Pit Necks Is Required for Efficient Vesicle Uncoating after Fission. Neuron. 2011 doi: 10.1016/j.neuron.2011.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Roux A, et al. Membrane curvature controls dynamin polymerization. Proceedings of the National Academy of Sciences. 2010;107:4141. doi: 10.1073/pnas.0913734107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chang-Ileto B, et al. Synaptojanin 1-mediated PI(4,5)P2 hydrolysis is modulated by membrane curvature and facilitates membrane fission. Developmental cell. 2011;20:206–218. doi: 10.1016/j.devcel.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Solomaha E, et al. Kinetics of Src homology 3 domain association with the proline-rich domain of dynamins: specificity, occlusion, and the effects of phosphorylation. The Journal of biological chemistry. 2005;280:23147–23156. doi: 10.1074/jbc.M501745200. [DOI] [PubMed] [Google Scholar]
- 75.Itoh T, Takenawa T. Mechanisms of membrane deformation by lipid-binding domains. Progress in lipid research. 2009;48:298–305. doi: 10.1016/j.plipres.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 76.Cui H, et al. Membrane binding by the endophilin N-BAR domain. Biophysical journal. 2009;97:2746–2753. doi: 10.1016/j.bpj.2009.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]