Abstract
An important step in PET brain kinetic analysis is the registration of functional data to an anatomical MR image. Typically, PET-MR registrations in nonhuman primate neuroreceptor studies used PET images acquired early post-injection, (e.g., 0–10 min) to closely resemble the subject’s MR image. However, a substantial fraction of these registrations (~25%) fail due to the differences in kinetics and distribution for various radiotracer studies and conditions (e.g., blocking studies). The Multi-Transform Method (MTM) was developed to improve the success of registrations between PET and MR images. Two algorithms were evaluated, MTM-I and MTM-II. The approach involves creating multiple transformations by registering PET images of different time intervals, from a dynamic study, to a single reference (i.e., MR image) (MTM-I) or to multiple reference images (i.e., MR and PET images pre-registered to the MR) (MTM-II). Normalized mutual information was used to compute similarity between the transformed PET images and the reference image(s) to choose the optimal transformation. This final transformation is used to map the dynamic dataset into the animal’s anatomical MR space, required for kinetic analysis. The chosen transformed from MTM-I and MTM-II were evaluated using visual rating scores to assess the quality of spatial alignment between the resliced PET and reference. One hundred twenty PET datasets involving eleven different tracers from 3 different scanners were used to evaluate the MTM algorithms. Studies were performed with baboons and rhesus monkeys on the HR+, HRRT, and Focus-220. Successful transformations increased from 77.5%, 85.8%, to 96.7% using the 0–10 min method, MTM-I, and MTM-II, respectively, based on visual rating scores. The Multi-Transform Methods proved to be a robust technique for PET-MR registrations for a wide range of PET studies.
Keywords: registration, transformation, normalized mutual information, intermodality, nonhuman primate
Introduction
Analysis of 4D PET brain data usually involves co-registration with MR data, where the MR image serves as a reference for anatomical localization (Gholipour et al., 2007; Myers, 2002). In order to attain the optimal transformation to map PET image data into MR image space, optimization functions are employed based on similarities between the target and reference images (Jenkinson et al., 2002; Maintz and Viergever, 1998). One conventional approach for PET-MR processing of human brain images is to use an early summed PET image (e.g., 0–10 min post-injection, during initial tracer uptake). If tracer delivery is limited primarily by blood flow, brain uptake is typically much higher than activity outside the brain, thus providing good correspondence between PET and MR brain tissue classes. This “0–10 min method” for PET-MR registration was used in many studies in the nonhuman primate (NHP), however, registration failures were a frequent occurrence, with a diversity of neuroreceptor studies. This suggested that using the first ten minutes of the PET image was not always optimal for registration to MR.
PET-MR registrations can be challenging in the NHP because the brain occupies approximately 25% of the head volume in the field-of-view (FOV), with the rest composed of muscle around the skull, jaw, and snout, where tracer can accumulate. On the other hand, the human brain occupies the majority of the head voxels in the scanner FOV and is much larger in volume than that of the NHP (~1250 mL in humans vs. ~80 mL in NHPs) (Baare et al., 2001; Cheverud et al., 1990). The smaller size of the NHP also allows more of the body to appear within the scanner FOV, where the lungs, heart, and spinal cord can be visible. Intensity-based registration algorithms (i.e., AIR (Woods et al., 1993)) are likely to succeed when there is good correspondence between PET and MR brain voxels belonging to gray matter, white matter, and cerebrospinal fluid. Additionally, high contrast between the brain and non-brain structures is required in order to achieve a successful registration to a skull- and muscle-stripped MR brain.
For PET-MR intermodality registrations, mutual information is a robust, intensity-based optimization method (Studholme et al., 1997). Studholme and others developed normalized mutual information (NMI) to decrease sensitivity to image overlap, where large misalignments can occur with respect to the FOV between images from different modalities (Studholme et al., 1999). For within-subject registrations, the NMI cost function (C) is maximized, by finding a linear (rigid) 6-parameter (3 translations + 3 rotations), transformation matrix that maximizes the shared information between two images, for instance a reference (R) MR image and a target (P) PET image. C is defined in terms of entropy (H), specifically the joint entropy of the two images, H(R, P), normalized by the marginal entropy of each (H(R) and H(P)), i.e.,
| (Eq. 1) |
H(X) is defined by
| (Eq. 2) |
where p denotes the probability of a given pixel value, x, from a possible set of binned values. Each pixel value x in the image is histogrammed into bins based on the minimum and maximum value in the image, and the probability of x is computed.
Eq. 1 is used to compute a similarity metric between the reference and transformed PET images, where 0≤C≤1. For instance, the maximum C between two identical images will equal 1. When the two image volumes are distinctly dissimilar, for instance, a PET image with both brain and muscle uptake and a reference MR image that was stripped of skull and muscle, C will be a value closer to 0. For iterative registration algorithms in these cases, depending on the quality of the initial guess, it is more likely that the algorithm will fail to converge or converge to an incorrect registration at a local maximum of the cost function.
The robustness of a registration algorithm is nontrivial due to the fact that functional PET and anatomical MR have very different image content (Skerl et al., 2006). Heterogeneity exists across different tracers and experimental conditions, thus affecting tracer distribution. Distribution within the first ten minutes is driven by delivery kinetics, typically referred to as the uptake image. Some tracers are known to have high early cortical uptake, so it may be sufficient to always use an early image (e.g., 0–10 min) for registration. If the initial kinetics of the tracer are relatively slow, registration algorithms tend to be less robust and may not converge to a reasonable result. The distribution in subsequent images relies both on delivery and binding kinetics of the tracer. Pharmacological intervention can also affect the kinetics and distribution, e.g., blocking the receptor site to study the binding specificity of an experimental tracer or increasing endogenous neurotransmitter to study changes in tracer binding with respect to a baseline study.
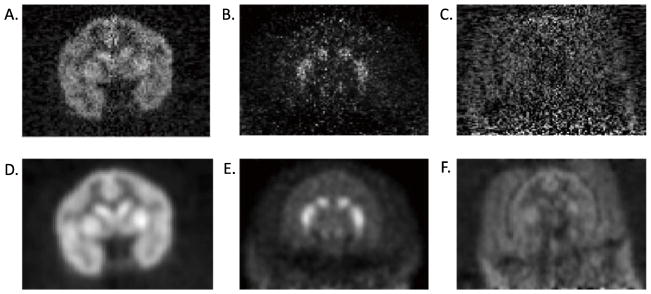
Successful registrations are most common when there is uniform distribution of radiotracer in the gray matter with high gray/white matter contrast and low extra-brain uptake (Fig. 1A). A challenging distribution to register is a tracer that binds non-uniformly, e.g., only to subcortical structures even at early times (Fig. 1B). Difficulty in PET-MR registrations can also occur when tracer activity outside of the brain is comparable to brain radioactivity levels, making it difficult to distinguish the brain from muscle (low contrast between brain and extra-brain) (Fig. 1C). These characteristics are examples of PET image distributions that pose challenges for NHP PET-MR registrations.
Fig. 1.
Examples of PET tracer distribution for the Focus-220 scanner (10–20 min post-injection). A. [18F]FPEB images have good gray/white matter contrast and low extra-brain uptake B. [11C]PHNO images are noisy with high uptake in the striatum compared to the rest of the brain. C. [11C]OMAR images have high noise and high extra-brain uptake. D, E, and F are post-smoothed (3×3×3 voxel FWHM Gaussian filter) versions of A, B, and C, respectively.
Thus, the aim of this study was to develop an automated algorithm that optimizes PET-MR NHP registrations and is broadly applicable to a wide variety of PET studies. We refer to this approach as the Multi-Transform Method (MTM). The main concept of the MTM algorithm entails registering multiple static PET images from a dynamic study (e.g., from different time intervals or using different levels of smoothing) to one or more reference images, creating a group of possible transformations. Then, a figure of merit is used to choose the optimal transformation. Note that this method is applied to data from anesthetized NHPs, so it is assumed that there is no motion between scan frames.
Two algorithms, MTM-I and MTM-II, were evaluated. In MTM-I, multiple transforms were created by registering the PET images to one reference image, the same NHPs anatomical MR image. In an extension to the method, MTM-II, PET images were registered to multiple reference images including the NHPs anatomical MR plus additional PET reference images. PET reference images are PET images that were pre-registered to the MR image of the same NHP. These methods were tested against our conventional method of registering an average image from the first 10 min of the PET acquisition to the MR. The algorithm was tested on control and blocking studies from various tracers performed in PET scanners having different resolutions.
Materials and Methods
Animals
The PET datasets evaluated were from eight rhesus monkeys and four baboons from various radiotracer studies performed under protocols approved by the Institutional Animal Care and Use Committee. Animals were initially anesthetized with an intramuscular injection of ketamine hydrochloride then transported to the PET facility. Once intubated, the animals were maintained on oxygen and isoflurane (1.75–2.50%) throughout the study. Rhesus monkeys were scanned with the HRRT and Focus-220 and baboons were scanned with the HR+ (Lehnert et al., 2006; van Velden et al., 2009) (Siemens/CTI, Knoxville, TN, USA). For head positioning, animals in the HRRT and HR+ were placed prone, with their head in a stereotaxic head holder. Monkeys scanned on the Focus-220 were placed head first on their back, left, or right side (depending on catheter placement) on the scanner bed. Once positioned in the scanner, a transmission acquisition was performed followed by an emission acquisition lasting for at least 120 min.
Datasets
One hundred twenty PET datasets using eleven tracers were evaluated. Studies were performed with three scanners having different resolutions, defined in terms of full-width-at-half-maximum (FWHM). PET images from the Focus-220, HRRT, and HR+ are shown in Fig. 2. Focus-220 (FWHM=1–2 mm) studies included radiotracers [11C]GR103545 (kappa opioid agonist, n=13) (Nabulsi et al., 2011), [11C]OMAR (cannabinoid receptor subtype 1, n=14) (Wong et al., 2010), [11C]PHNO (dopamine D2/D3 receptor agonist, n=12) (Graff-Guerrero et al., 2008), and [18F]FPEB (metabotropic glutamate receptor 5, n=8) (Wang et al., 2007). HRRT (FWHM=2–3 mm) studies included radioligands [11C]AFM (serotonin transporter, n=9) (Huang et al., 2004), [11C]MRB (norephinephrine transporter (NET), n=10) (Gallezot et al., 2011), and [11C]P943 (5-HT1B receptor antagonist, n=12) (Nabulsi et al., 2010). HR+ (FWHM=5–6 mm) studies included radioligands [11C]carfentanil (mu opioid receptor agonist, n=10) (Wand et al., 2011), [11C]raclopride (dopamine D2/D3 receptor antagonist, n=4) (Graff-Guerrero et al., 2008), [11C]GR205171 (NK1 receptor antagonist, n=24) (Zamuner et al., 2012), and [18F]FEPPA (peripheral benzodiazepine receptor, n=4) (Wilson et al., 2008). In addition to the variety in range of studies, control and blocking scans were included. For instance, data was used from the [11C]MRB occupancy study with atomoxetine, a NET reuptake inhibitor, where Gallezot and others reported a dose-dependent decrease in [11C]MRB binding at NET. In addition, baseline radioligand binding may be altered by self-blocking with cold compound prior to tracer administration or via displacement of tracer with a drug (e.g., with the bolus infusion paradigm, after equilibrium is achieved) as exhibited in [11C]P943 studies (Cosgrove et al., 2011; Nabulsi et al., 2010).
Fig. 2.
Sagittal PET images from 3 scanners. A. HR+ image of [11C]GR205171 (FWHM=5–6 mm, FOV=128×128×63, voxel size=2.06×2.06×2.4 mm) B. HRRT image of [11C]AFM (FWHM=2–3 mm, FOV=256×256×207, voxel size=1.2×1.2×1.2 mm) C. Focus-220 image of [18F]FPEB (FWHM=1–2mm, FOV=256×256×95, voxel size=0.95×0.95×0.80 mm). The larger axial FOV of the HRRT (25.5 cm) and HR+ (15.2 cm) included more non-brain structures compared with the Focus-220 image (7.6 cm).
MRI scanning and processing
MR images were acquired on a Siemens Magnetom 3.0T Trio scanner, using an extremity coil. T1-weighted images were acquired in the transverse plane with spin echo sequence (TE=3.34, TR=2530, flip angle=7°, section thickness=0.50 mm, FOV=140 mm, image matrix=256×256×176 pixels, matrix size=0.55×55×0.50 mm). The MR image volume was then cropped to 176×176×176 pixels and re-oriented into coronal slices using MEDx software (Medical Numerics Inc, Germantown, MD, USA). For processing, the MR images were stripped of skull and muscle so that only the brain remained in the image (FMRIB’s Brain Extraction Tool, http://www.fmrib.ox.ac.uk/fsl/bet2/index.html). This skull and muscle stripping procedure was performed once for each monkey MR image prior to co-registration with the PET images.
PET Images
PET data were collected on three different scanners: HR+, High Resolution Research Tomograph (HRRT), and Focus-220. A sequence of 33 frames was reconstructed with the following timing: 6 × 30 sec; 3 × 1 min; 2 × 2 min; 22 × 5 min. HR+ data were collected in sinograms and reconstructed using FBP with all corrections (attenuation, normalization, scatter, randoms, and deadtime). HRRT dynamic listmode data were reconstructed with all corrections using the MOLAR algorithm (Carson et al., 2004). Focus-220 dynamic listmode data were reconstructed with all corrections using the OSEM algorithm via ASIPro 2.3 microPET software. Final image dimensions and voxel sizes from each scanner were: HR+ (image matrix=128×128×63, matrix size=2.06×2.06×2.43 mm), HRRT (image matrix=256×256×207, matrix size =1.22×1.22×1.23 mm), and Focus-220 (image matrix=256×256×95, matrix size=0.95×0.95×0.80 mm).
PET-MR registrations
For HR+ and HRRT scanners, the axial FOV is 15.2 and 25.5 cm, respectively. The animal’s brain only occupies about 7.4 cm for both scanners, or 49% and 30% of the respective axial FOV. The remainder of the FOV in HR+ and HRRT PET images contained non-brain including the snout, salivary glands, heart, and lungs (Figs. 2A and 2B). In some cases, the radioactivity concentration in the lungs was much higher than the brain. To deal with this issue, HR+ and HRRT images were first registered to the MR image with the intermodality 6-parameter rigid automated image registration (AIR) algorithm as an initialization step, providing a reasonable starting point (using AIR 3.08 in MEDx). The AIR algorithm seeks a transformation that minimizes the overall standard deviation of PET values, grouped by the corresponding MR values (Hill et al., 2001; Woods et al., 1993). Thus, to initialize PET-MR registrations for HR+ and HRRT images, a rigid AIR registration was performed prior to implementing the FLIRT-NMI registration. This initialization step was necessary because FLIRT-NMI alone produced inadequate registrations. The PET image was then resliced with the AIR transform and registered to the MR via a 6-parameter rigid registration using the NMI cost function with FSL FLIRT (FMRIB’s Linear Image Registration Tool, www.fmrib.ox.ac.uk/fsl/flirt/index.html). The final transformation was the product of both the AIR and FLIRT-NMI transformations. FLIRT-NMI alone always produced visually inadequate transformations. AIR alone achieved better results, however, still misregistered (See Discussion). The Focus-220 axial FOV is only 7.6 cm, with the brain present in 70% of the axial FOV. Since the brain occupies much of the FOV in the Focus-220 (Fig. 2C), the preliminary AIR registration was unnecessary.
MTM Algorithm
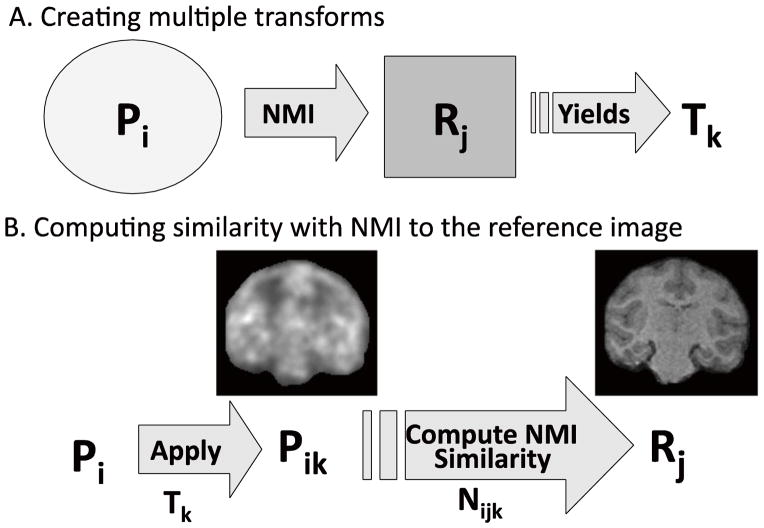
The MTM algorithm is defined as follows:
PET summed images, Pi, i=1,…,nP were created from specific time intervals from a 120-min acquisition: 0–10, 10–20, 20–40, 40–60, 60–90, and 90–120 min (n=6). For Focus-220 data, in addition to using different time intervals, both non-smoothed (Figs. 1A–C) and post-smoothed (3×3×3 voxel FWHM Gaussian filter, Figs. 1D–F) PET summed images were created (n=12); the use of additional PET images was found to be necessary due to the higher noise in the Focus-220 images.
The MTM algorithm is shown in Fig. 3. Each of nP PET images (indexed by i) was registered to one MR reference image, R1, and any number of additional PET reference images, R2,…,Rj, where i=1,..,nP and j=1, …,nR. The resulting number of transformations was n=nP·nR, the total number of PET images (nP) times the number of reference images (nR) (Fig. 3A). MTM-I uses only one MR as a reference image, R1 (nR=1), whereas MTM-II uses multiple reference images, specifically, PET images pre-registered to the MR (R2, …,Rj), in addition to R1 (nR> 1). Then, to choose the best of the n transforms, all transforms Tk, k=1, …,n, are applied to all PET images Pi, i=1, …,nP to yield i·k resliced PET images in MR image space, Pik, where
Fig. 3.
Multi-Transform Method (MTM) algorithm. A. Registration of a PET image (Pi) to a reference image (Rj) to yield transforms (Tk) where i=1,..,nP, j=1,…,nR, and k=1,…,nPxnR. B. Apply each Tk transform to each Pi and compute NMI similarity (Nijk) between resliced PET (Pik) and each reference, Rj. See Table 1 for example of selection of the optimal transform.
| (Eq. 3) |
Thus, Pik is PET image i mapped by transform k. Note that each reference image produced a set of transforms (i.e., 6 PET images × 3 references=18 transforms), which were applied to each Pi. If the transform is accurate, this image will be well registered to the NHPs MR space (Fig. 3B). The similarity, Nijk, is then computed between the resliced PET image, Pik, and each reference image, Rj, where
| (Eq. 4) |
The calculation of Nijk, the similarity between Pik and Rj, was limited to pixels masked within the MR image volume. Using Eq. 1, Nijk was computed based on the minimum and maximum value in each image where pixel values were histogrammed into 256 bins to compute marginal and joint entropy. In MTM-I, Nijk is computed between Pik and R1 (Table 1, Eq. 4). Nijk is computed between Pik and all reference images, Rj, in MTM-II (see Inline Supplementary Table 1, Eq. 4).
Table 1.

|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n=1 | T1 (0–10) | T2 (10–20) | T3 (20–40) | T4 (40–60) | T5 (60–90) | T6 (90–120) | T7 (0–10s) | T8 (10–20s) | T9 (20–40s) | T10 (40–60s) | T11 (60–90s) | T12 (90–120s) | |
|
|
P1 | 0.65 | 0.88 | 0.90 | 0.99 | 0.61 | 0.90 | 0.83 | 0.95 | 0.86 | 0.99 | 1.00 | 0.57 |
| P2 | 0.42 | 0.90 | 0.90 | 0.98 | 0.40 | 0.77 | 0.76 | 1.00 | 0.85 | 0.97 | 0.87 | 0.45 | |
| P3 | 0.37 | 0.93 | 0.93 | 1.00 | 0.37 | 0.77 | 0.77 | 1.00 | 0.89 | 0.99 | 0.90 | 0.38 | |
| P4 | 0.54 | 0.80 | 0.82 | 1.00 | 0.55 | 0.77 | 0.69 | 0.87 | 0.80 | 0.99 | 0.89 | 0.56 | |
| P5 | 0.69 | 0.78 | 0.81 | 0.96 | 0.72 | 0.83 | 0.72 | 0.87 | 0.76 | 1.00 | 0.92 | 0.65 | |
| P6 | 0.82 | 0.84 | 0.85 | 0.95 | 0.84 | 0.88 | 0.77 | 0.86 | 0.82 | 1.00 | 0.95 | 0.81 | |
| P7 | 0.53 | 0.86 | 0.87 | 0.95 | 0.52 | 0.95 | 0.83 | 0.93 | 0.84 | 1.00 | 1.00 | 0.40 | |
| P8 | 0.33 | 0.90 | 0.91 | 0.97 | 0.33 | 0.82 | 0.80 | 1.00 | 0.86 | 1.00 | 0.93 | 0.32 | |
| P9 | 0.35 | 0.93 | 0.94 | 0.99 | 0.35 | 0.81 | 0.81 | 0.99 | 0.90 | 1.00 | 0.93 | 0.31 | |
| P10 | 0.45 | 0.76 | 0.79 | 0.96 | 0.48 | 0.80 | 0.67 | 0.83 | 0.75 | 1.00 | 0.93 | 0.39 | |
| P11 | 0.63 | 0.76 | 0.78 | 0.95 | 0.70 | 0.89 | 0.69 | 0.83 | 0.75 | 1.00 | 0.99 | 0.52 | |
| P12 | 0.80 | 0.81 | 0.82 | 0.94 | 0.89 | 0.96 | 0.74 | 0.86 | 0.79 | 0.99 | 1.00 | 0.73 | |
| Sk | 0.55 | 0.85 | 0.86 | 0.97 | 0.56 | 0.85 | 0.76 | 0.92 | 0.82 | 0.99 | 0.94 | 0.51 | |
| VRS for T | 3 | 1 | 1 | 1 | 3 | 2 | 2 | 1 | 2 | 1 | 1 | 3 | |
MTM-I algorithm for choosing the transform with the best score (S)
Normalized NMI similarity (Nijk) values between resliced PET (Pik) and MR (R1) (Eq. 4), Sk scores (Eq.6), and visual rating scores, where Mijk values are shown (Eq.5). This is an example from a Focus-220 study where non-smoothed and smoothed (s) summed PET images (i.e., T1–T6 and T7–T12, respectively) were used to create the transforms (k=12). For this study, the MTM-I algorithm chose the transform T10(40–60s) with the highest Sk score (highlighted grey), created by registering the 40–60 min post-smoothed summed PET image to the MR. An example of the MTM-II algorithm for choosing the transform using multiple reference images is shown in Inline Suppl. Table 1.
For MTM-I and MTM-II, Nijk scores were first normalized to the highest value across n transforms (Table 1), i.e., for a given PET image and reference image, the C values for each transform were divided by the maximum C attained by any transform. The normalized value, Mijk, was computed where the highest value in each row was equal to 1 (Eq. 5). Row normalization across the k transforms was especially important for MTM-II that uses multiple reference images, because Nijk values with each PET image i and reference image j have a different range of values due to the nature of the types of images being compared (i.e., resliced PET and MR vs. resliced PET and PET reference).
| (Eq. 5) |
A final score for each transform (Sk) was computed by taking the mean of Mijk for each transform (k), i.e., a column-mean in Table 1, yielding
| (Eq. 6) |
The final transform was chosen for MTM-I and MTM-II as the one with the maximum Sk score. See Table 1 and Inline Supplementary Table 1, for MTM-I and MTM-II, respectively. The MTM algorithms were coded with IDL Version 8.0.
In summary, the MTM algorithm is as follows:
Create multiple PET images using different time intervals and post-smoothing (for Focus-220 images only). For each PET image, create transformation matrices by registering to the MR reference image (MTM-I) or to multiple reference images (MTM-II) (Fig 3A).
Reslice every PET image with each transform (Eq. 3, Fig. 3B) and compute a similarity metric between each resliced PET images and the reference image(s) (Eq. 4, Fig. 3B).
Row normalize Nijk scores to the maximum value for each transform yielding Mijk, to account for variability in similarity measures between resliced PET and different reference images (Eq. 5).
Choose the best transform based on the overall NMI measures for each transform (Eq. 6, Table 1).
Evaluation
The two new algorithms MTM-I and MTM-II were compared to our original method of registering a single summed image (0–10 min post-injection) to the NHPs MR image. Thus, the original method is a special case of MTM-I with nP=1. Of the total number of studies for each tracer, the percentage of PET time intervals that generated the chosen transform was computed.
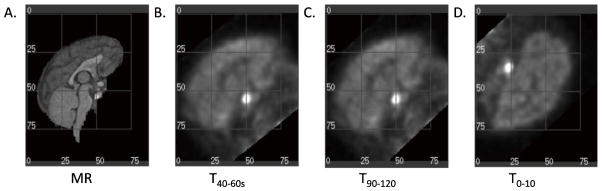
The quality of all the transforms was evaluated visually. For MTM-I and MTM-II, each summed PET image was resliced with the transform generated from registering the same summed PET to the MR image. The resulting registration was given a visual rating score (VRS) of 1=good, 2=slightly misregistered, or 3=failed. MEDx software (Medical Numerics Inc, Germantown, MD, USA) was used to assess the quality of registration from each transform in sagittal, coronal, and transverse views, between resliced PET images and the MR and reference PET images. An example of registrations and their visual rating scores is shown in Fig. 4. Transforms that were designated a VRS of 1 had good spatial alignment of both images (Figs. 4A and 4B). Transforms that were assigned a VRS of 2 produced slightly tilted resliced PET images in either sagittal, coronal, or transverse views compared with the MR, where the magnitude of error was 2–3 mm or 5°–10° degree rotation in either x, y, or z (Fig. 4C). Transforms that were assigned a VRS of 3 produced completely misaligned resliced PET images, compared with the MR (Fig. 4D). To obtain a qualitative VRS for each transformation, the studies were split amongst two individuals (CS and DW) experienced at assessing NHP PET-MR registration quality. The percentage of registrations with VRS=1, 2, and 3 for the chosen transforms were computed for the 0–10 transform method, MTM-I, and MTM-II for each study.
Fig. 4.
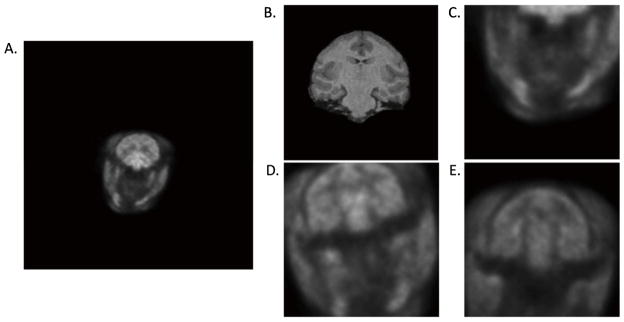
Visual rating scores from the study example in Table 1. Transforms were used to reslice the 10–20 min post-smoothed PET image (3×3×3 voxel FWHM Gaussian filter) into MR space and were given a visual rating score (VRS) for one monkey study with the tracer [11C]GR103545 on the Focus-220. The grid with increments of 25 pixels is shown for visualization. A. MR reference image, R1. B. T40–60s was designated a VRS=1 where there is good alignment between the PET and MR images. C. T90–120 was designated a VRS=2 where the PET and MR images are slightly misaligned with a 5º rotation about the frontal tip of the brain. D. T0–10 was designated a VRS=3, where the transform produced a failed registration.
Results
MTM-I
Early summed PET images (e.g., 0–10 min) were used for PET-MR registrations with the assumption that brain contrast is high in these images, providing similar brain tissue content as in the MR images, thus improving the accuracy of registration. The time period 0–10 min was one option available to the MTM-I algorithm, however, the transforms were selected from all time intervals across scanners as optimum for registration. The percentages of chosen transforms from each time period across 120 studies evaluated were: T0–10 (33%), T10–20 (33%), T20–40 (13%), T40–60 (10%), T60–90 (8%), and T90–120 (3%). The percentage of chosen transforms from each PET time interval was calculated for each study, with those exceeding 20% highlighted in gray (Table 2). Preferred transforms for the Focus-220 and HRRT images were taken 34% and 35%, respectively, from the 10–20 min summed PET image registered to the MR, whereas the favored transform for the HR+ was taken 60% from the 0–10 min summed PET-MR registration. Note that the choice of optimal time period may have more to do with the tracer and the study (control or blocking), rather than the scanner.
Table 2.
| Tracer | n | T(0–10) | T(10–20) | T(20–40) | T(40–60) | T(60–90) | T(90–120) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Focus-220 | ns | s | ns | s | ns | s | ns | s | ns | s | ns | s | |
| [11C]GR103545 | 13 | 0 | 0 | 38 | 8 | 8 | 8 | 23 | 8 | 8 | 0 | 0 | 0 |
| [11C]OMAR | 14 | 29 | 7 | 29 | 0 | 7 | 14 | 0 | 7 | 0 | 0 | 0 | 7 |
| [11C]PHNO | 12 | 8 | 8 | 42 | 17 | 0 | 0 | 8 | 17 | 0 | 0 | 0 | 0 |
| [18F]FPEB | 8 | 0 | 0 | 25 | 0 | 38 | 0 | 13 | 0 | 0 | 0 | 25 | 0 |
| HRRT | |||||||||||||
| [11C]AFM | 9 | 22 | 14 | 11 | 11 | 11 | 0 | ||||||
| [11C]MRB | 10 | 10 | 40 | 10 | 10 | 30 | 0 | ||||||
| [11C]P943 | 12 | 33 | 25 | 25 | 8 | 8 | 0 | ||||||
| HR+ | |||||||||||||
| [11C] carfentanil | 10 | 100 | 0 | 0 | 0 | 0 | 0 | ||||||
| [11C] raclopride | 4 | 100 | 0 | 0 | 0 | 0 | 0 | ||||||
| [11C]GR205171 | 24 | 38 | 29 | 13 | 0 | 17 | 4 | ||||||
| [18F]FEPPA | 4 | 50 | 50 | 0 | 0 | 0 | 0 | ||||||
Percentage of chosen transforms from each PET time interval (min) for MTM-I.
Focus-220 images were smoothed (s) and non-smoothed (ns) prior to registration to create transforms. HRRT and HR+ images were not smoothed prior to registration. Grayed cells highlight the time interval/smoothing combinations that exceeded 20% of the test.
For Focus-220 images, smoothing the image prior to registration was helpful for 13 of the 47 (28%) of studies analyzed: [11C]GR103545 (3 of 13 studies), [11C]OMAR (4 of 14 studies), [11C]PHNO (5 of 12 studies). No transforms from registrations of smoothed PET images were chosen for [18F]FPEB, presumably due to its higher statistical quality.
A higher percentage of chosen transforms with MTM-I (86%) were visually better (VRS=1) than those from the 0–10 min transform method (78%) (Table 3). For MTM-I, 3% of the cases had failed registrations (VRS=3) from chosen transformations. In these cases, registrations from all time intervals failed. For MTM-I transforms given a VRS score of 2 (11%), there were 8 cases with transforms that provided visually better registrations, yet had lower overall S values and were not chosen for [11C]PHNO (4 out of 4 cases), [11C]GR103545 (1 out of 2 cases), and [11C]GR205171 (3 out of 5 cases) (Table 3). For the remaining 5 studies with chosen transform given a VRS=2, no better transform choices were observed. Focus-220 studies produced 47% of MTM-I algorithm failures (chosen transforms visually designated a 2 or 3), HRRT studies had no failures, and HR+ studies contributed 53% of failures.
Table 3.
Comparison of Visual Rating Scores (VRS) for 0–10 min transforms, MTM-I, and MTM-II
| 0–10 min VRS | MTM-I VRS | MTM-II VRS | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Tracer | n | 1 | 2 | 3 | 1 | 2 | 3 | 1 | 2 | 3 |
| Focus-220 | ||||||||||
| [11C]GR103545 | 13 | 2 | 2 | 9 | 11 | 2 | 0 | 13 | 0 | 0 |
| [11C]OMAR | 14 | 8 | 0 | 6 | 12 | 0 | 2 | 14 | 0 | 0 |
| [11C]PHNO | 12 | 10 | 2 | 0 | 8 | 4 | 0 | 11 | 1 | 0 |
| [18F]FPEB | 8 | 7 | 1 | 0 | 8 | 0 | 0 | 8 | 0 | 0 |
| HRRT | ||||||||||
| [11C]AFM | 9 | 9 | 0 | 0 | 9 | 0 | 0 | 9 | 0 | 0 |
| [11C]MRB | 10 | 10 | 0 | 0 | 10 | 0 | 0 | 10 | 0 | 0 |
| [11C]P943 | 12 | 12 | 0 | 0 | 12 | 0 | 0 | 12 | 0 | 0 |
| HR+ | ||||||||||
| [11C] carfentanil | 10 | 8 | 2 | 0 | 8 | 2 | 0 | 8 | 2 | 0 |
| [11C] raclopride | 4 | 4 | 0 | 0 | 4 | 0 | 0 | 4 | 0 | 0 |
| [11C]GR205171 | 24 | 19 | 2 | 3 | 17 | 5 | 2 | 23 | 1 | 0 |
| [18F]FEPPA | 4 | 4 | 0 | 0 | 4 | 0 | 0 | 4 | 0 | 0 |
| %VRS of total n | 120 | 77.5 | 7.5 | 15.0 | 85.8 | 10.8 | 3.3 | 96.7 | 3.3 | 0.0 |
Transforms from the 0–10 min method, MTM-I, and MTM-II were given a visual rating score (VRS) of 1=good, 2=slightly misregistered, or 3=failed for each tracer. The total percentage of each VRS was calculated for the 3 methods.
MTM-II
MTM-II is an extension of MTM-I using multiple reference images to generate additional transforms. By adding more reference possibilities, the percentage of failed cases from MTM-I (14%: VRS of 2 or 3) was reduced to 3% with MTM-II; successful registrations (VRS=1) increased from 86% to 97% (Table 3). Across studies, a PET reference was selected over the MR reference to generate the best transforms in many cases: Focus-220 (70%), HRRT (71%), and HR+ (74%).
The MR reference was used to generate the chosen transform for 28% of the studies. For 30 out of 34 of these cases, MTM-I and MTM-II provided consistent results, where the same summed PET image was registered to the MR to yield the chosen transformation.
MTM-II eliminated all transforms with VRS=3 (Table 3). However, in the 4 cases where the chosen transform was given a VRS of 2, visual comparison suggested that there were better transform options available.
Discussion
Summary
The Multi-Transform Method (MTM) was developed to optimize PET-MR NHP registrations across various PET studies including: radiotracers with varying kinetics and distributions, conditions (e.g. baseline vs. blocking studies), and scanners of different resolutions. The goal of the method is to provide a robust registration over the wide range of studies performed. The previous method used, registering the 0–10 min summed PET image to the MR, would often result in failed transformations: ~23% of the time. Thus, MTM-I was developed to produce multiple transformations by registering different time intervals (or smoothing) of the PET dynamic dataset to the MR. Then, the NMI similarity metric was computed between the resliced PET images and the MR to choose the best transformation. Chosen transforms were given a visual rating score (VRS), where successful registrations (good alignment between the resliced PET and MR) were given a score of ‘1-good’ and poor registrations were given a ‘2-slightly misregistered’ or ‘3-failed’ (Fig. 4). Implementing MTM-I improved the success of registrations by 8% (from 78% to 86%) when compared with the 0–10 min registration method.
An extension of the algorithm, MTM-II, was developed to improve upon MTM-I, providing more possible transformations by registering the summed/smoothed PET images to the MR plus additional references, i.e., PET images pre-registered to MR space (PET references). NMI similarity was computed between the resliced PET images and all of the reference images. MTM-II further improved success in the chosen transforms by 11% compared with MTM-I to a 97% overall success rate. In addition, 72% of ‘selected’ transforms were registered to PET references. A PET reference may have been chosen in cases where the tracer had a more similar distribution to the PET reference than the MR reference. Interestingly, good transformations were generated for all registration methods for tracer studies performed in the HRRT. In summary, successful transformations increased from 77.5%, 85.8%, to 96.7% using the 0–10 min method, MTM-I, and MTM-II, respectively, based on visual rating scores.
Necessity for MTM algorithm
Analysis of dynamic PET data often requires PET-MR image co-registration, where the MR serves as an anatomical reference for functional PET images. The resulting transformation must be sufficient to map the entire PET dynamic dataset into the space of the MR, assuming that the NHP does not move throughout the scan while under anesthesia. Brain regions of interest (ROIs) are delineated on the MRI or via a template (Black et al., 2004; McLaren et al., 2009), to generate time-activity curves (TACs). Analysis of TACs, with different modeling methods, provides estimates of physiological parameters (e.g., flow, receptor binding, metabolism, etc.). The accuracy of brain ROI TACs are dependent on accurate spatial alignment between the PET and the MR.
MTM was developed to automate PET-MR registrations, where the choice of transformation is based on mutual information measures. Prior to MTM, the transformation was determined by registering the 0–10 min summed image to the MR to generate a transformation. If the registration failed based on visual inspection, multiple other summed PET images were created from the dynamic study and registered individually to the MR until a ‘good’ transformation was found. When comparing PET-MR registrations of consecutive time intervals, it was often difficult to determine visually whether one registration was better than another. This process was time consuming and tedious. When a transformation is chosen via the MTM algorithm, the user can perform a visual inspection to check the alignment between the PET and the MR before ROI TACs are generated.
MTM-I algorithm performance
MTM-I was successful for 83%, 100%, and 79% of Focus-220, HRRT, and HR+ studies, respectively. MTM-I algorithm failures typically occurred when the distribution of the tracer was dissimilar to that of the MR image having either low brain uptake or poor brain contrast. [11C]OMAR contributed 12% of failures (2/17), where all registrations from all time intervals failed. [11C]GR103545 also contributed 12% (2/17) of algorithm failures where, in one of two cases, there was a better transformation that was not selected. Both tracers exhibited low contrast between brain and muscle (Figs. 1C, 1F, and 4). [11C]PHNO contributed 24% (4/17) of failures, where uptake was high in striatum, but low in the rest of the brain (Figs. 1B and 1E). All 4 failures for [11C]PHNO had better transform options, where typically the 0–10 min registration to the MR was visually better. It may be that NMI may not provide the optimal metric for this particular type of brain distribution and that registering 0–10 min summed image may be more appropriate for [11C]PHNO studies than MTM-I.
Forty-one percent (7/17) of MTM-I algorithm failures were attributed to [11C]GR205171, where 3 of the 7 failures had better transform options. [11C]GR205171 studies were performed on baboons in the HR+, where images show extra-brain uptake in the salivary glands and snout (Fig. 2A). Two studies with [11C]carfentanil had similar registration failures. For HR+ studies, a contributing factor to the misregistrations may be the scanner resolution (FWHM=5–6mm), giving rise to partial volume effects.
It is worth noting that there were 9 cases in MTM-I where a suboptimal transformation was chosen because none of the possible registrations were good (VRS>1). This was due to the FLIRT-NMI registration algorithm that either failed to converge or converged to a local maximum. Thus, true MTM-I algorithm failures included the 8 cases where a better transform option was available upon visual inspection, but were not chosen. For these cases, it is likely that the algorithm converged to an incorrect registration at a local maximum of the cost function
MTM-II algorithm performance
MTM-II increased successful registrations to 98% for the Focus-220, 100% for the HRRT, and 93% for the HR+. Increasing the number of transform options optimized the chances of generating better transformations, especially when registering to a reference PET image that may have a more similar distribution to the tracer study of interest. Seventy-two percent of the selected transforms were from registrations of a summed PET image to a PET reference image. Between MTM-I and MTM-II, when the MR reference was chosen to generate the ‘best’ transform, there were only 4 of 34 instances when a different summed PET image was used. When a different summed PET image was chosen for registration to the MR, the next adjacent time interval was used from MTM-I to MTM-II (i.e., if the MTM-I chosen transform was 10–20 min PET to MR registration, then the MTM-II chosen transform was 20–40 min PET to MR registration). This means that the quality of registration and the total NMI scores were very similar between the two transforms. For MTM-I and MTM-II, 12 of 120 studies chose the 0–10 min transform to the MR. The four cases with MTM-I where all PET registrations to the MR failed (VRS of 3) were eliminated with MTM-II. However, all 4 MTM-II cases with a VRS of 2 had visually better transform options; two had better transform options from either the ‘0–10 min’ registration method or from MTM-I, and two had better transform options chosen by neither of the two methods.
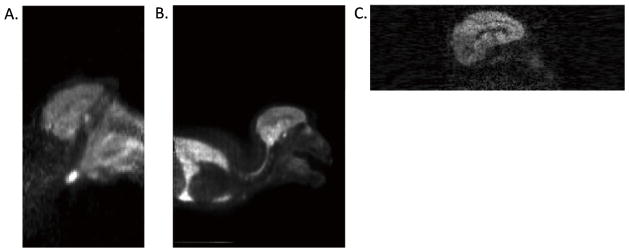
AIR pre-registration step
Uptake in the snout, salivary glands, and lungs appeared in the PET FOV, where the brain comprised only 49% and 30% of the image in the HR+ and HRRT, respectively (Figs. 2A and 2B). FLIRT-NMI alone with HR+ and HRRT images consistently produced misregistered results (Fig. 5C). This could either mean that FLIRT converged to a local maximum that was not registered to the brain, or that the initial search parameters of FLIRT were not optimized. The AIR algorithm was not sensitive to uptake in non-brain organs and achieved better registrations than FLIRT-NMI alone (Fig. 5D); however, the FLIRT-NMI registration was needed to improve the AIR-registered PET image (Fig. 5E). The AIR registration step was not needed for the Focus-220 images because the head comprised 70% of the axial FOV.
Fig. 5.
The AIR initialization registration step was helpful for HR+ and HRRT images. The following are registration examples of A. [11C]P943 PET HRRT image and the B. MR image. A. and B. were registered using C. FLIRT-NMI algorithm D. AIR algorithm, and E. AIR+FLIRT-NMI algorithms. Notice that FLIRT-NMI produced a failed registration, and AIR produced a misregistered, but a slightly better registration. The AIR-registered image, D, was then re-registered to the MR with FLIRT-NMI. The product of the two transformations produced a better registration, as shown in E. AIR alone was not sufficient to produce a quality registration.
Choice of registration software
Another widely used registration tool that implements NMI for intermodality registrations is included in the SPM software (http://www.fil.ion.ucl.ac.uk/spm/doc/, (Collignon et al., 1995). For human studies, SPM has been used for PET-MR registrations without requiring skull/muscle editing to achieve a reasonable result. This may not be the case for NHP registrations, especially with more difficult tracer distributions (Figs. 1B and 1C). Pre-processing in SPM typically includes smoothing the PET images and segmenting the PET and MR images to create partitioned images of gray matter, white matter, and cerebrospinal fluid prior to registration. The segmentation step may be nontrivial for PET images with activity in non-brain organs in the FOV or nonuniform distribution within brain tissue classes (Figs. 1B, 1C, 2A, 2B). It remains to be tested whether the SPM registration implementation of NMI can handle the various tracer distributions for NHP PET studies, but there is potential for integrating SPM registrations within the MTM algorithm.
Caveats of MTM algorithm design
To design the MTM algorithm, a number of decisions were made. For instance, the time intervals for the PET image were arbitrarily chosen to cover the entire dynamic PET scan, avoid overlap in time intervals, and maintain similar number of counts as the tracer decays over time. The intervals increased from ten, twenty, and thirty between 0–20, 20–60, 60–120 min, respectively.
Next, the choice of smoothing (3×3×3 voxel FWHM Gaussian filter) was made for the Focus-220 images, but not the HR+ or HRRT images that had poorer resolution (Fig. 2). Because the Focus-220 images are of higher resolution (FWHM=1–2 mm), reconstructed images tended to be noisy, especially for tracers that are poorly distributed in the brain (Figs. 1B and 1C). Without smoothing, for initial evaluations with MTM-I, it was difficult to achieve a good registration at any time interval. The idea was that creating additional smoothed versions of the PET images would helpful for the FLIRT-NMI algorithm (Figs. 1D–F). Smoothing was helpful for 23% [11C]GR103545, 29% of [11C]OMAR, and 42% of [11C]PHNO studies. No transforms from smoothed PET-MR registrations were chosen for [18F]FPEB because the images were less noisy and generally had high brain to extra-brain contrast and high gray/white matter contrast. [18F]FPEB binds to metabotropic glutamate-5 receptor, abundant in cortical gray matter. In addition, radiotracers labeled with the 18F isotope tend to have less noise due to a longer half-life than those labeled with 11C (110 min vs. 20 min, respectively). Although smoothing was not required for [18F]FPEB registrations, 26% of all Focus-220 studies with MTM-I chose transformations created by the registration of smoothed PET images to the MR. Another approach would be to try smoothed versions of the MR image to closer match the resolution of the PET images.
A visual rating score (VRS) was given for each transform, by applying it to the PET image that was used to create that transform after registration to the MR. Two authors, CS and DW, adept at assessing NHP PET-MR registrations, defined the rating system and acted as raters. To test the reliability of the rating system, the individuals independently scored five of the same Focus-220 studies. When VRS were compared between the two, there was 100% agreement.
For MTM-II, PET references were created from registered studies of the same animal. Appropriate PET references were chosen for each NHP for MTM-II. The criteria used for this decision was based on availability and range of tracer studies for the animal. MTM-I was utilized to choose the appropriate transform and reslice the PET image that was used to generate it into MR space (pre-registered to the MR) with a VRS of 1. The range in types of PET references varied across scanners (e.g., an individual NHP was scanned in the Focus-220 with [11C]PHNO and the HRRT with [11C]P943), and across studies (e.g., an individual NHP participated in both an [18F]FPEB and an [11C]OMAR study). Some of the rhesus monkeys were scanned in both the HRRT and Focus-220 scanners, and some were scanned in the Focus-220 only. Therefore, a set of PET reference images was chosen for each animal, independent of scanner type. The baboons were primarily scanned on the HR+, and were limited to the studies performed on that scanner in terms of number PET reference images.
Extensions of MTM algorithm
The MTM algorithm is not limited to the choice of PET time intervals, references, or level of smoothing used in this study. MTM can obviously be extended to create more transformation possibilities. For instance, overlapping times (e.g., 0–30 min and 10–30 min) could be used to register to the MR image to generate another transform. In addition, using a summed imaged from the full 0–120 min scan period was tested with MTM-I to create an additional transform. This approach was applied for a subset of scans that included all cases where 1) MTM-I failed (n=17), and 2) the 0–10 min PET-MR transform was chosen for both MTM-I and MTM-II (n=12). The 0–120 min PET-MR transform was only chosen as best once of the 29 re-processed studies. The additional transform did not reduce the number of failures based on visual inspection (subset 1), and the 0–10 min PET-MR transform was still chosen as optimal (subset 2). For these data, the 0–120 min summed PET image was not the best choice for registration to the MR. However, given the flexibility of the MTM algorithm, there is no disadvantage to include this additional PET image in producing the possible transforms.
T1-weighted MR images were used for this study, but if available, other MR sequences, such as T2-weighted MR images, may be useful as an additional reference. Registering the PET images to the full MR image, including the skull and muscle outside of the brain, was also considered as an additional reference image. The ability to register directly to the entire MR image would avoid the tedious step of skull/muscle stripping (although this must only be performed once per animal). The utility of unstripped MR images was investigated for one NHP with MTM-II using 2 reference images: the skull-and muscle-stripped MR brain (R1) and the whole head MR (R2) for [11C]AFM (n=3) and [18F]FPEB (n=3) studies performed in the HRRT and Focus-220 scanners, respectively. Preliminary MTM-II results in 6 studies still chose transforms from PET images registered to the skull/muscle stripped MR. Upon visual inspection, registrations with all summed PET images to the whole head MR were failures (VRS of 3). Non-brain features in the MR image can increase the probability that intensity-based registration algorithms would converge to incorrect local maxima. For example, the muscles around the head and the gray matter have very similar MR voxel intensities, but typically have dramatically different PET activities, depending on the tracer.
To strengthen MTM-II, for animals that have participated in several studies, it may be possible to generate PET reference templates in MR space. For example, if a monkey participated in a few control and blocking studies for a given tracer, MTM-I would be used to choose the best transform and image to reslice the images into the NHPs MR space for each case. Then an averaged PET reference can be created for each of the control and blocking conditions. In addition, an averaged PET reference can be created for many different tracers with various distributions.
In this study, a set of PET references from each animal was used in MTM-II. However, this is not possible for NHPs with no previous studies. PET references from other NHPs may be helpful for the case of having a new animal participating in a new tracer study. In the event were MTM-I fails to produce a good transform, it may be possible to use MTM-II to perform an affine (12-parameter) registration to a different monkey’s set of PET reference images to acquire a useful transformation.
Limitations of MTM algorithm
The MTM algorithm works under the assumption that the NHP does not move for the entire duration of the scan while under anesthesia. For example, if the MTM algorithm chooses a transform from registering an early PET image to the MR (i.e., T10–20), and the monkey’s head moves as little as a few millimeters half-way through the scan, the concentration values in the TACs can be as much as 15% wrong in a larger region and as much as 25% in a smaller region in an infusion study. This example was based on an infusion study with [18F]FPEB where the animal’s head moved during the equilibrium period. In order for the MTM algorithm to work, the image must be realigned or motion-corrected. Otherwise, separate registrations may need to be performed to correct the post-movement portion of the images.
Another limitation of the MTM algorithms is computation time. Using a single processor, MTM-I requires 20–30 min, whereas, MTM-II requires one hour or more, depending on the number of PET images and references images being registered. Further, although the MTM algorithm has been designed to be applied across a variety of PET studies, one must not assume that the chosen transform to the MR is optimal in all cases (i.e., [11C]PHNO studies). The final transform should be visually inspected for registration quality.
The absolute accuracy of the final registration cannot be determined for real data. In principle, a careful simulation of dynamic PET data can be used to assess the absolute accuracy of a registration. Studies have been published to test the accuracy of different registration algorithms by generating human PET brain data via a segmented MR and applying known transformations, applying the registration algorithms, and measuring the error between the known and estimated transforms. (Davatzikos et al., 2001; Kiebel et al., 1997). To simulate the human PET data, relative concentration values were assigned to gray matter, white matter, and cerebrospinal fluid, smoothed to the resolution of the scanner, and noise was added. However, simulating PET images for NHP brain data to test the MTM algorithms would additionally require knowledge of tracer kinetics and distribution for various tracers throughout the scan, i.e., in brain and non-brain tissues. This would be particularly challenging for tracers that distribute in the muscle that surrounds the brain and in other organs visible in the image FOV (Figs. 1B, 1C, 2A, 2B).
Conclusions
The MTM algorithm has been developed with the goal of producing a robust PET-MR registration method for nonhuman primate PET studies. Normalized mutual information was used to compute a similarity metric to choose the optimal transformation, where a group of transformations were created by registering PET images of different time intervals to the MR or PET reference images. MTM-I chosen transforms increased the success of registrations over the conventional method (registration to a 0–10 min image), however, MTM-II proved to be more robust for a wide range of PET studies. The MTM algorithm can be extended by using alternate time intervals, degrees of smoothing, or PET references, to work across many different types of PET studies.
Supplementary Material
Highlights.
A PET-MR registration algorithm is proposed for nonhuman primate brain studies.
Transforms from our original method often resulted in misaligned PET-MR images.
The algorithm was tested in 120 datasets, and transforms were assessed visually.
Successful transforms improved from 78% to 97% with the new registration method.
Acknowledgments
The authors acknowledge the staff at the Yale PET Center, especially Krista Fowles and the NHP team, for conducting the numerous NHP PET experiments used in this study. Special thanks to Jean-Dominique Gallezot, Beata Planeta, Edward Fung, and Xiao Jin for scientific discussions and technical assistance. Research support was provided by the training grant 1-T90-DK070068. This publication was also made possible by CTSA Grant Number UL1 RR024139 from the National Center for Research Resources (NCRR) and the National Center for Advancing Translational Science (NCATS), components of the National Institutes of Health (NIH), and NIH roadmap for Medical Research. Its contents are solely the responsibility of the authors and do not necessarily represent the official view of NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Baare WF, Hulshoff Pol HE, Boomsma DI, Posthuma D, de Geus EJ, Schnack HG, van Haren NE, van Oel CJ, Kahn RS. Quantitative genetic modeling of variation in human brain morphology. Cereb Cortex. 2001;11:816–824. doi: 10.1093/cercor/11.9.816. [DOI] [PubMed] [Google Scholar]
- Black KJ, Koller JM, Snyder AZ, Perlmutter JS. Atlas template images for nonhuman primate neuroimaging: baboon and macaque. Methods Enzymol. 2004;385:91–102. doi: 10.1016/S0076-6879(04)85006-7. [DOI] [PubMed] [Google Scholar]
- Carson RE, Barker WC, Liow JS, Johnson CA. Design of a motion-compensation OSEM list-mode algorithm for resolution-recovery reconstruction for the HRRT. 2003 Ieee Nuclear Science Symposium, Conference Record; 2004. pp. 3281–3285. [Google Scholar]
- Cheverud JM, Falk D, Vannier M, Konigsberg L, Helmkamp RC, Hildebolt C. Heritability of brain size and surface features in rhesus macaques (Macaca mulatta) J Hered. 1990;81:51–57. doi: 10.1093/oxfordjournals.jhered.a110924. [DOI] [PubMed] [Google Scholar]
- Collignon A, Maes F, Delaere D, Vandermeulen D, Suetens P, Marchal G. Automated multi-modality image registration based on information theory. Information Processing in Medical Imaging. 1995;3:263–274. [Google Scholar]
- Cosgrove KP, Kloczynski T, Nabulsi N, Weinzimmer D, Lin SF, Staley JK, Bhagwagar Z, Carson RE. Assessing the sensitivity of [(1)(1)C]p943, a novel 5-HT(1B) radioligand, to endogenous serotonin release. Synapse. 2011;65:1113–1117. doi: 10.1002/syn.20942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davatzikos C, Li HH, Herskovits E, Resnick SM. Accuracy and sensitivity of detection of activation foci in the brain via statistical parametric mapping: a study using a PET simulator. Neuroimage. 2001;13:176–184. doi: 10.1006/nimg.2000.0655. [DOI] [PubMed] [Google Scholar]
- Gallezot JD, Weinzimmer D, Nabulsi N, Lin SF, Fowles K, Sandiego C, McCarthy TJ, Maguire RP, Carson RE, Ding YS. Evaluation of [(11)C]MRB for assessment of occupancy of norepinephrine transporters: Studies with atomoxetine in non-human primates. Neuroimage. 2011;56:268–279. doi: 10.1016/j.neuroimage.2010.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gholipour A, Kehtarnavaz N, Briggs R, Devous M, Gopinath K. Brain functional localization: A survey of image registration techniques. Ieee Transactions on Medical Imaging. 2007;26:427–451. doi: 10.1109/TMI.2007.892508. [DOI] [PubMed] [Google Scholar]
- Graff-Guerrero A, Willeit M, Ginovart N, Mamo D, Mizrahi R, Rusjan P, Vitcu I, Seeman P, Wilson AA, Kapur S. Brain region binding of the D2/3 agonist [11C]-(+)-PHNO and the D2/3 antagonist [11C]raclopride in healthy humans. Hum Brain Mapp. 2008;29:400–410. doi: 10.1002/hbm.20392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill DL, Batchelor PG, Holden M, Hawkes DJ. Medical image registration. Physics in Medicine and Biology. 2001;46:R1–45. doi: 10.1088/0031-9155/46/3/201. [DOI] [PubMed] [Google Scholar]
- Huang Y, Hwang DR, Bae SA, Sudo Y, Guo N, Zhu Z, Narendran R, Laruelle M. A new positron emission tomography imaging agent for the serotonin transporter: synthesis, pharmacological characterization, and kinetic analysis of [11C]2-[2-(dimethylaminomethyl)phenylthio]-5-fluoromethylphenylamine ([11C]AFM) Nucl Med Biol. 2004;31:543–556. doi: 10.1016/j.nucmedbio.2003.11.008. [DOI] [PubMed] [Google Scholar]
- Jenkinson M, Bannister P, Brady M, Smith S. Improved optimization for the robust and accurate linear registration and motion correction of brain images. Neuroimage. 2002;17:825–841. doi: 10.1016/s1053-8119(02)91132-8. [DOI] [PubMed] [Google Scholar]
- Kiebel SJ, Ashburner J, Poline JB, Friston KJ. MRI and PET coregistration - A cross validation of statistical parametric mapping and automated image registration. Neuroimage. 1997;5:271–279. doi: 10.1006/nimg.1997.0265. [DOI] [PubMed] [Google Scholar]
- Lehnert W, Meikle SR, Siegel S, Newport D, Banati RB, Rosenfeld AB. Evaluation of transmission methodology and attenuation correction for the microPET Focus 220 animal scanner. Physics in Medicine and Biology. 2006;51:4003–4016. doi: 10.1088/0031-9155/51/16/008. [DOI] [PubMed] [Google Scholar]
- Maintz JB, Viergever MA. A survey of medical image registration. Med Image Anal. 1998;2:1–36. doi: 10.1016/s1361-8415(01)80026-8. [DOI] [PubMed] [Google Scholar]
- McLaren DG, Kosmatka KJ, Oakes TR, Kroenke CD, Kohama SG, Matochik JA, Ingram DK, Johnson SC. A population-average MRI-based atlas collection of the rhesus macaque. Neuroimage. 2009;45:52–59. doi: 10.1016/j.neuroimage.2008.10.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers R. The application of PET-MR image registration in the brain. British Journal of Radiology. 2002;75:S31–S35. doi: 10.1259/bjr.75.suppl_9.750031. [DOI] [PubMed] [Google Scholar]
- Nabulsi N, Huang Y, Weinzimmer D, Ropchan J, Frost JJ, McCarthy T, Carson RE, Ding YS. High-resolution imaging of brain 5-HT 1B receptors in the rhesus monkey using [11C]P943. Nucl Med Biol. 2010;37:205–214. doi: 10.1016/j.nucmedbio.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabulsi NB, Zheng MQ, Ropchan J, Labaree D, Ding YS, Blumberg L, Huang YY. [(11)C]GR103545: novel one-pot radiosynthesis with high specific activity. Nuclear Medicine and Biology. 2011;38:215–221. doi: 10.1016/j.nucmedbio.2010.08.014. [DOI] [PubMed] [Google Scholar]
- Skerl D, Likar B, Pernus F. A protocol for evaluation of similarity measures for rigid registration. Ieee Transactions on Medical Imaging. 2006;25:779–791. doi: 10.1109/tmi.2006.874963. [DOI] [PubMed] [Google Scholar]
- Studholme C, Hill DL, Hawkes DJ. Automated three-dimensional registration of magnetic resonance and positron emission tomography brain images by multiresolution optimization of voxel similarity measures. Med Phys. 1997;24:25–35. doi: 10.1118/1.598130. [DOI] [PubMed] [Google Scholar]
- Studholme C, Hill DLG, Hawkes DJ. An overlap invariant entropy measure of 3D medical image alignment. Pattern Recognition. 1999;32:71–86. [Google Scholar]
- van Velden FH, Kloet RW, van Berckel BN, Buijs FL, Luurtsema G, Lammertsma AA, Boellaard R. HRRT versus HR+ human brain PET studies: an interscanner test-retest study. Journal of Nuclear Medicine. 2009;50:693–702. doi: 10.2967/jnumed.108.058628. [DOI] [PubMed] [Google Scholar]
- Wand GS, Weerts EM, Kuwabara H, Frost JJ, Xu XQ, McCaul ME. Naloxone-induced cortisol predicts mu opioid receptor binding potential in specific brain regions of healthy subjects. Psychoneuroendocrinology. 2011;36:1453–1459. doi: 10.1016/j.psyneuen.2011.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JQ, Tueckmantel W, Zhu A, Pellegrino D, Brownell AL. Synthesis and preliminary biological evaluation of 3-[(18)F]fluoro-5-(2-pyridinylethynyl)benzonitrile as a PET radiotracer for imaging metabotropic glutamate receptor subtype 5. Synapse. 2007;61:951–961. doi: 10.1002/syn.20445. [DOI] [PubMed] [Google Scholar]
- Wilson AA, Garcia A, Parkes J, McCormick P, Stephenson KA, Houle S, Vasdev N. Radiosynthesis and initial evaluation of [18F]-FEPPA for PET imaging of peripheral benzodiazepine receptors. Nucl Med Biol. 2008;35:305–314. doi: 10.1016/j.nucmedbio.2007.12.009. [DOI] [PubMed] [Google Scholar]
- Wong DF, Kuwabara H, Horti AG, Raymont V, Brasic J, Guevara M, Ye W, Dannals RF, Ravert HT, Nandi A, Rahmim A, Ming JE, Grachev I, Roy C, Cascella N. Quantification of cerebral cannabinoid receptors subtype 1 (CB1) in healthy subjects and schizophrenia by the novel PET radioligand [11C]OMAR. Neuroimage. 2010;52:1505–1513. doi: 10.1016/j.neuroimage.2010.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods RP, Mazziotta JC, Cherry SR. Mri-Pet Registration with Automated Algorithm. Journal of Computer Assisted Tomography. 1993;17:536–546. doi: 10.1097/00004728-199307000-00004. [DOI] [PubMed] [Google Scholar]
- Zamuner S, Rabiner EA, Fernandes SA, Bani M, Gunn RN, Gomeni R, Ratti E, Cunningham VJ. A pharmacokinetic PET study of NK receptor occupancy. Eur J Nucl Med Mol Imaging. 2012;39:226–235. doi: 10.1007/s00259-011-1954-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.