Abstract
Visualization of the reaction coordinate undertaken by glycosyltransferases has remained elusive, but is critical for understanding this important class of enzyme. Using substrates and substrate mimics, we describe structural snapshots of all species along the kinetic pathway for human O-GlcNAc transferase, an intracellular enzyme that catalyzes installation of a dynamic post-translational modification. The structures reveal key features of the mechanism and show that substrate participation is important during catalysis.
Glycosyltransferases are ubiquitous enzymes that play vital cellular roles in organisms found in all kingdoms of life. These enzymes catalyze transfer of a sugar from a donor, most commonly a nucleotide sugar, to a variety of acceptors including proteins1. One glycosyltransferase that has received considerable recent attention is human O-GlcNAc transferase (OGT)2,3. OGT transfers N-acetylglucosamine from the sugar donor UDP-GlcNAc onto specific serine or threonine residues of nucleocytoplasmic proteins with inversion of configuration at the anomeric center (Fig. 1a)3. This post-translational modification has been implicated in gene transcription4,5, stress response6, and nutrient sensing7. Given recent advances in OGT structural characterization8, the development of a substrate-mimicking inhibitor9, and current interest in the O-GlcNAc modification, we felt that OGT would be an ideal candidate to dissect glycosyltransferase catalysis. Here we report crystal structures of human OGT complexed to substrates and products, including ternary complexes that provide a comprehensive set of snapshots of stable species that define the trajectory of the reaction.
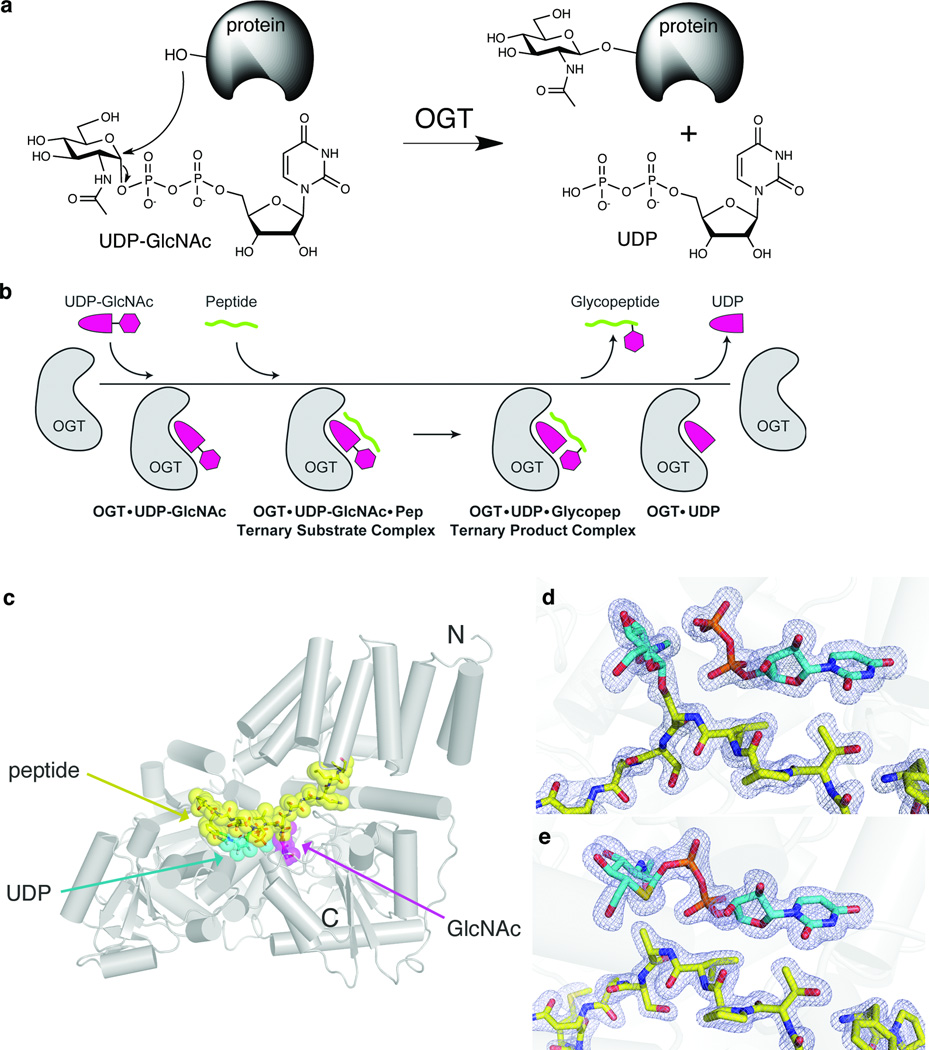
Figure 1. Structural snapshots define the kinetic mechanism of OGT.
(a) OGT catalyzes transfer of GlcNAc from UDP-GlcNAc to serine or threonine residues of nucleocytoplasmic proteins. (b) Schematic of the ordered bi-bi kinetic mechanism of OGT and the complexes obtained along the trajectory. The final binary complex, OGT-UDP, was previously obtained8. (c) Crystal structure of the ternary product complex of human OGT4.5, shown as a cartoon representation. The CKII peptide is shown in yellow, the UDP in cyan, and the GlcNAc moiety of the glycopeptide is shown in magenta. The N and C termini of the protein are indicated. (d) Electron density of the ternary product complex with the fo-fc map, shown at 3σ. The GlcNAc and the UDP are shown in cyan, and the peptide is shown in yellow. (e) Electron density of the ternary substrate complex. The fo-fc map, shown at 3σ, clearly indicates that the density connects the sugar to the UDP. The UDP-5SGlcNAc is shown in cyan, with the sulfur atom in the sugar shown in yellow.
OGT uses an ordered bi-bi mechanism with UDP-GlcNAc binding first, followed by the polypeptide acceptor (Fig. 1b)8. We wanted to visualize the complete set of stable species along the kinetic pathway to gain insight into how glycosyltransferases, and OGT in particular, catalyze sugar transfer. We obtained a structure of OGT containing UDP-GlcNAc but no acceptor substrate (Supplementary Results, Supplementary Fig. 1), which represents the first complex in the ordered bi-bi mechanism (Fig. 1b). To observe the next species along the reaction pathway, we attempted to crystallize OGT with UDP-GlcNAc and a previously characterized peptide substrate from CKII10,11 (sequence YPGGSTPVS*SANMM, where S* is the only O-GlcNAc modification site) and obtained a 1.7 Å ternary complex structure (Fig. 1c). Unambiguous electron density in the active site revealed the presence of UDP and the O-GlcNAc modified peptide, indicating that O-GlcNAc transfer had taken place during crystallization to yield the product complex (Fig 1d).
Because glycosyltransfer is fast compared with the time scale of crystallization, an alternative approach was needed to capture a ternary substrate complex. Efforts to crystallize UDP-GlcNAc with an incompetent peptide acceptor, the CKII peptide containing a Ser* to Ala mutation (CKIIA), yielded structures without density for the sugar (data not shown), presumably due to hydrolysis of UDP-GlcNAc. We therefore turned to a chemically modified analogue of UDP-GlcNAc, UDP-5SGlcNAc, wherein the endocyclic oxygen atom is replaced by sulfur (Supplementary Methods). This molecule is an effective inhibitor of OGT9, but we find it to be a very poor (3200 times slower) donor substrate compared to UDP-GlcNAc (Supplementary Fig. 2). This reduced reactivity allowed us to obtain a 1.85 Å ternary substrate complex with the CKIIA peptide (Fig. 1e).
Since the use of non-natural substrates could alter the binding conformation we obtained two additional complexes: a binary complex with UDP-5SGlcNAc and a ternary 5SGlcNAc-CKII product complex. The relative position of the GlcNAc and the 5SGlcNAc are indistinguishable in both the binary and ternary product complexes (Supplementary Figs. 1 and 3). Moreover, the conformation and position of the CKII and CKIIA peptides are the same in all the ternary complexes. Therefore, the use of catalytically incompetent substrates does not alter the binding mode, enabling meaningful insight into the reaction coordinate from these structures.
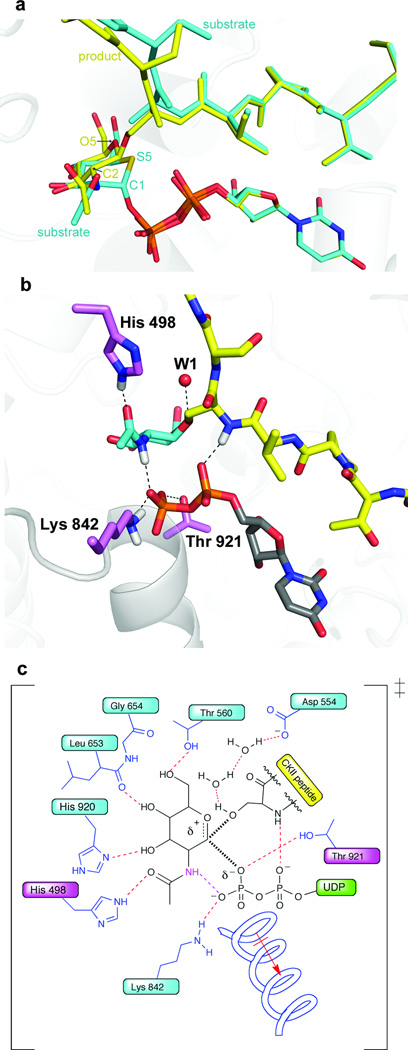
Structural alignment of these complexes reveals several features of glycosyltransferase catalysis. OGT catalyzes transfer with inversion of anomeric stereochemistry, with concomitant departure of the leaving group and nucleophilic attack by the acceptor. The ternary substrate and product complexes flank the transition state for glycosyltransfer, thus offering insight into how OGT facilitates catalysis. Superposition of the ternary substrate and product complexes (Fig. 2a) reveals that both the UDP moiety and the peptide acceptor retain their positions. Motion along the reaction coordinate is limited primarily to GlcNAc atoms C2, C1, and O5 such that the anomeric carbon moves from bonded contact with the pyrophosphate group to bonded contact with the nucleophilic serine hydroxyl. The structures are thus consistent with an electrophilic migration mechanism in which an oxocarbenium ion-like species moves away from the leaving group toward the acceptor12,13. While this type of mechanism is clearly operative for glycosidases14–17, structural evidence that glycosyltransferases may also use such a mechanism has been lacking until now.
Figure 2. Glycosylation reaction trajectory and interactions facilitate glycosyltransfer.
(a) Overlay of the ternary substrate complex (cyan) and product complex (yellow). (b) Selected interactions in the ternary product complex. Proposed hydrogen bonds are illustrated with a dashed line. The positions of putative hydrogen atoms are shown in white for the His, acetamido group, Lys, and the peptide backbone. New interactions to the phosphate observed only in the product complex are the acetamide interaction and the Thr921 interaction18. An important water molecule that may shuttle away the proton after reaction is shown as W1. (c) Putative transition state schematic. Important interactions to the transition state are indicated, based on an analysis of the substrate and product structures. The partial bonds in the transition state are shown as thick, hashed lines. The new interactions in the product complex that may reflect directly on the late transition state are shown in magenta.
Analysis of the ternary complexes reveals several contacts to the β-phosphate oxygen atoms of the leaving group that may stabilize the dissociative transition state implied by an electrophilic migration mechanism. For example, a previously described8 contact from Lys842 to one of the β-phosphate oxygens helps mitigate the increasing negative charge on the leaving group during the transition from substrates to products (Fig. 2b and Supplementary Fig. 4). Additional stabilization is provided by contacts from the side chain of Thr921 and the amide backbone of the N-terminus of an active site helix. Notably, the acetamido group of GlcNAc undergoes a substantial rotation upon conversion to product, enabling it to contact the β-phosphate oxygen. This structural change suggests that the sugar acetamide plays a critical role in catalysis (Figs. 2a and 2b). The acetamide group may also be important for ground state binding since a different set of contacts is observed in the substrate complexes (Supplementary Fig. 4). Consistent with the importance of the acetamide, we have found that OGT is able to transfer UDP-GalNAc, but not UDP-Glucose or UDP-2-ketoGlc to peptide and protein substrates (Supplementary Figs. 5 and 6).
General base catalysis is generally thought to play a role in glycosyltransferase mechanism, but we found no obvious candidate general base in OGT. His558 and His498, which were previously proposed as candidate bases8,18, are too far from the acceptor hydroxyl to fulfill this role, although both play important roles in catalysis. Tyr841 has also been proposed as the general base19 but is likewise too far from the serine. In retaining glycosyltransferases, it has been suggested the β-phosphate acts as a general base20,21 but this group is inappropriately positioned for such a role in OGT. One of the α-phosphate oxygens is within 3.5 Å of the serine hydroxyl, raising the possibility that this phosphate could serve as a base; however, the basicity of the candidate oxygen is attenuated by a hydrogen bond to the peptide backbone. Furthermore, the serine hydroxyl rotamer that would be suitably positioned to engage in a hydrogen bond with the α-phosphate would not be well aligned for attack on the anomeric carbon (Supplementary Fig. 7). Hence, the structures do not support a role for the α-phosphate acting as a general base. Accordingly, we note that deprotonation of the incoming nucleophile during bond formation may not be required in an electrophilic migration mechanism involving an oxocarbenium ion-like species, although a process for removing the proton from the active site may still be important. In this regard, we observed a chain of bound waters in the structures that may link this proton to the carboxylate side chain of conserved residue Asp554 (Fig. 2c), possibly enabling proton relay by a Grotthus-type mechanism. Further studies will be needed to clarify these alternative mechanistic scenarios.
While these structural snapshots have demonstrated features that OGT uses to facilitate catalysis, it is less obvious how OGT favors glycosylation over the energetically wasteful hydrolysis. Since water is not excluded from the active site, the likely explanation is that interactions between UDP-GlcNAc and the protein substrate facilitate catalysis. Indeed, the contact interface between the donor sugar and peptide is extensive (Supplementary Fig. 8), and includes the previously mentioned hydrogen bond between the Ser* amide and the α-phosphate. In addition to enforcing the productive orientation of the incoming nucleophile in a manner not possible for water, this contact may further activate the leaving group. Supporting the importance of this extensive interaction for activating the donor, substantial differences were recently observed in the Km values for UDP-GlcNAc when studying different proteins as substrates for OGT22.
Considerable effort has been dedicated to improved understanding of glycosyltransferase-catalyzed reactions, yet structural studies of glycosyltransferases have only yielded incomplete snapshots along the reaction pathway20,23–25. Here we have described a set of OGT complexes that provide an unprecedented, complete structural view of enzyme-catalyzed glycosyltransfer. These structures provide compelling structural support that OGT promotes catalysis via an electrophilic migration mechanism. They further imply that crucial contacts between the two substrates limit unproductive hydrolysis while contributing to both UDP-GlcNAc donor and peptide acceptor specificity. These structures therefore pave the way to an improved understanding of this ubiquitous class of enzyme and will facilitate engineering substrate specificity, and development of inhibitors, for OGT and possibly other glycosyltransferases.
Supplementary Material
Acknowledgements
This work was supported by the National Institutes of Health (NIH) grant R01GM094263 to S.W. and the Natural Sciences and Engineering Research Council of Canada (NSERC) and Simon Fraser University. T.M.G. is a Sir Henry Wellcome postdoctoral fellow and a Michael Smith for Health Research (MSFHR) trainee award holder. D.J.V. is a scholar of MSFHR and holds a Canada Research Chair in Chemical Glycobiology. We thank L. Deng for assistance with the chemical synthesis. We thank L. Cai (University of South Carolina, Salkehatchie) for the kind gift of UDP-2-ketoGlc.
Footnotes
Accession codes. Protein Data Bank: Coordinates and structure factors have been deposited under accession codes 4GYW, 4GYY, 4GZ3, 4GZ5, and 4GZ6.
Author contributions
M.B.L. carried out all structural experiments. J.J. helped with X-ray data collection. J.J. and M.B.L. performed kinetics assays. W.F.Z. and T.M.G. performed thiosugar kinetics assays and peptide assays. G.E.W. performed the synthesis of peptide and glycopeptides substrates. M.B.L., J.J., T.M.G., D.J.V., and S.W. designed experiments, analyzed data and wrote the manuscript.
Competing Financial Interests
The authors declare no competing financial interests.
Additional information
Supplementary information is available in the online version of the paper. Reprints and permissions information is available online at http://www.nature.com/reprints/index.html
References
- 1.Lairson LL, Henrissat B, Davies GJ, Withers SG. Annu Rev Biochem. 2008;77:521–555. doi: 10.1146/annurev.biochem.76.061005.092322. [DOI] [PubMed] [Google Scholar]
- 2.Torres CR, Hart GW. J Biol Chem. 1984;259:3308–3317. [PubMed] [Google Scholar]
- 3.Kreppel LK, Blomberg MA, Hart GW. J Biol Chem. 1997;272:9308–9315. doi: 10.1074/jbc.272.14.9308. [DOI] [PubMed] [Google Scholar]
- 4.Sinclair DA, et al. Proc Natl Acad Sci U S A. 2009;106:13427–1332. doi: 10.1073/pnas.0904638106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Love DC, et al. Proc Natl Acad Sci U S A. 2010;107:7413–7418. doi: 10.1073/pnas.0911857107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kazemi Z, Chang H, Haserodt S, McKen C, Zachara NE. J Biol Chem. 2010;285:39096–39107. doi: 10.1074/jbc.M110.131102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dentin R, Hedrick S, Xie J, Yates J, 3rd, Montminy M. Science. 2008;319:1402–1405. doi: 10.1126/science.1151363. [DOI] [PubMed] [Google Scholar]
- 8.Lazarus MB, Nam Y, Jiang J, Sliz P, Walker S. Nature. 2011;469:564–567. doi: 10.1038/nature09638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gloster TM, et al. Nat Chem Biol. 2011;7:174–181. doi: 10.1038/nchembio.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iyer SP, Hart GW. J Biol Chem. 2003;278:24608–24616. doi: 10.1074/jbc.M300036200. [DOI] [PubMed] [Google Scholar]
- 11.Gross BJ, Kraybill BC, Walker S. J Am Chem Soc. 2005;127:14588–14589. doi: 10.1021/ja0555217. [DOI] [PubMed] [Google Scholar]
- 12.Lee SS, et al. Nat Chem Biol. 2011;7:631–638. doi: 10.1038/nchembio.628. [DOI] [PubMed] [Google Scholar]
- 13.Burkart MD, et al. Bioorg Med Chem. 2000;8:1937–1946. doi: 10.1016/s0968-0896(00)00139-5. [DOI] [PubMed] [Google Scholar]
- 14.Vocadlo DJ, Davies GJ. Curr Opin Chem Biol. 2008;12:539–555. doi: 10.1016/j.cbpa.2008.05.010. [DOI] [PubMed] [Google Scholar]
- 15.Schramm VL, Shi W. Curr Opin Struct Biol. 2001;11:657–665. doi: 10.1016/s0959-440x(01)00269-x. [DOI] [PubMed] [Google Scholar]
- 16.Vocadlo DJ, Davies GJ, Laine R, Withers SG. Nature. 2001;412:835–838. doi: 10.1038/35090602. [DOI] [PubMed] [Google Scholar]
- 17.Davies GJ, Planas A, Rovira C. Acc Chem Res. 2012;45:308–316. doi: 10.1021/ar2001765. [DOI] [PubMed] [Google Scholar]
- 18.Martinez-Fleites C, et al. Nat Struct Mol Biol. 2008;15:764–765. doi: 10.1038/nsmb.1443. [DOI] [PubMed] [Google Scholar]
- 19.Dorfmueller HC, et al. Amino Acids. 2011;40:781–792. doi: 10.1007/s00726-010-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Errey JC, et al. Angew Chem Int Ed Engl. 2010;49:1234–1237. doi: 10.1002/anie.200905096. [DOI] [PubMed] [Google Scholar]
- 21.Reinert DJ, Jank T, Aktories K, Schulz GE. J Mol Biol. 2005;351:973–981. doi: 10.1016/j.jmb.2005.06.071. [DOI] [PubMed] [Google Scholar]
- 22.Shen DL, Gloster TM, Yuzwa SA, Vocadlo DJ. J Biol Chem. 2012;287:15395–15408. doi: 10.1074/jbc.M111.310664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ramakrishnan B, Ramasamy V, Qasba PK. J Mol Biol. 2006;357:1619–1633. doi: 10.1016/j.jmb.2006.01.088. [DOI] [PubMed] [Google Scholar]
- 24.Persson K, et al. Nat Struct Biol. 2001;8:166–175. doi: 10.1038/84168. [DOI] [PubMed] [Google Scholar]
- 25.Ni L, et al. Biochemistry. 2007;46:6288–6298. doi: 10.1021/bi700346w. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.