Abstract
Background
Negative-staining (NS), a rapid, simple and conventional technique of electron microscopy (EM), has been commonly used to initially study the morphology and structure of proteins for half a century. Certain NS protocols however can cause artifacts, especially for structurally flexible or lipid-related proteins, such as lipoproteins. Lipoproteins were often observed in the form of rouleau as lipoprotein particles appeared to be stacked together by conventional NS protocols. The flexible components of lipoproteins, i.e. lipids and amphipathic apolipoproteins, resulted in the lipoprotein structure being sensitive to the NS sample preparation parameters, such as operational procedures, salt concentrations, and the staining reagents.
Scope of review
The most popular NS protocols that have been used to examine lipoprotein morphology and structure were reviewed.
Major conclusions
The comparisons show that an optimized NS (OpNS) protocol can eliminate the rouleau artifacts of lipoproteins, and that the lipoproteins are similar in size and shape as statistically measured from two EM methods, OpNS and cryo-electron microscopy (cryo-EM). OpNS is a high-throughput, high-contrast and high-resolution (near 1 nm, but rarely better than 1 nm) method which has been used to discover the mechanics of a small protein, 53 kDa cholesterol ester transfer protein (CETP), and the structure of an individual particle of a single protein by individual-particle electron tomography (IPET), i.e. a 14 Å-resolution IgG antibody three-dimensional map.
General significance
It is suggested that OpNS can be used as a general protocol to study the structure of proteins, especially highly dynamic proteins with equilibrium-fluctuating structures.
Keywords: protein structure, lipoprotein structure, electron microscopy, negative-staining, optimized negative-staining protocol, individual-particle electron tomography
1. Introduction
Most transmission electron microscopy (TEM) has the capability to display the atomic structure of hard materials [1]. However, when examining the structure of soft (especially biological) materials such as proteins it is challenging to obtain structure at even near one nanometer resolution [2], due to radiation damage, low contrast of images, structural collapse or flattening, and dehydration [3, 4].
Cryo-electron microscopy (cryo-EM) is an advanced approach to determining protein structure at an atomic resolution under near physiological conditions [5–7]. The cryo-EM technique involves preparing the sample in vitreous ice by flash freezing the specimen, and then examining the specimen under cryogenic conditions, such as liquid nitrogen or helium temperatures [6–10]. Advantages of cryo-EM include absence of artifacts, and the ability to examine the protein with near-native state images [4]. Despite its advantages cryo-EM has many complications. The primary disadvantages involve low contrast and necessary expensive equipment that is not readily accessible to many laboratories, including specimen cryo-holders and a cryo-plunger that is needed for sample preparation apparatuses with liquid nitrogen. In addition, preparing a cyro-EM sample with perfect ice thickness and imaging under low-dose conditions requires highly intensive specialized training [4, 11, 12]. Namely knowledge with image processing software to enhance the poor signal-to-noise ratio (SNR) of cryo-EM [3] images which has a steep learning curve and is time consuming. In addition, the one-time usage of the cryo-specimens limits the chance for catching unexpected discoveries, especially when the sample is difficult to purify or is unstable after isolation [13].
Historically, the study of protein structure via negative-staining (NS) was initially developed with viruses by Brenner and Horne a half century ago [14]. The concept of NS began with light microscopy by submerging bacteria into a dense stain to provide darkness around the specimen, thus illuminating the sample utilizing a negative contrast [15]. NS-EM involves coating the specimen with a thin stain layer of cationic or anionic heavy-metal salts. NS-EM can produce high-contrast images with these coated heavy-metal stains [4], since the heavy-metal stains more strongly scatter electrons than do the lighter atoms within the proteins themselves [16–19].
Preparation of the sample for NS-EM can be easily adapted in any laboratory. Heavy metal stains allow for higher electron dose tolerance, improved contrast, and also function as a fixative for the specimen. NS-EM also delivers images that contain high SNR, and an intermediate (near a nanometer) resolution [3] for reconstruction of three-dimensional (3D) models [20]. However, certain effects of the heavy-metal stain produce undesirable outcomes due to interactions with the specimen, such as aggregation, molecular dissociation, and artifacts of stacking [4, 21, 22].
Since the early 1970s, the NS method has been used to examine the structure and morphology of lipoproteins [15, 23–30]. Lipoproteins are composed of lipids and apolipoproteins (apo), and are known to be structurally dynamic and flexible in nature [8, 29–34]. The structures of lipoproteins are also sensitive to the buffer pH, salt concentration and chemical reagents. An obvious artifact of lipoprotein structure observed for decades is how the lipoprotein particles regularly present stacking rouleau in NS images [35–40]. This is significant in that rouleau formation is absent from serum analysis, native-gel, small-angle scattering, and cryo-EM studies [12, 22, 29, 30, 36, 39, 41–44]. Considering that rouleau formation displayed in NS images could lead to inaccurate interpretations of lipoprotein structure and function, a thorough investigation or comparison of the NS protocols used to examine the lipoprotein structure is necessary.
Here, we have reviewed the literature by focusing on the structure and morphology of lipoprotein-related NS sample preparation parameters. The comparisons were performed from three parameters: operational procedures, salt concentrations, and the staining reagents themselves. These comparisons show that the optimized NS (OpNS) protocol eliminates the artifact of rouleau in lipoproteins and delivers structural features that are most similar in size and shape to that from cryo-EM images. OpNS features make it a suitable protocol to study lipoprotein structure and morphology. Moreover, OpNS displays itself as a reliable and powerful protocol revealing the mechanics and structures of small and asymmetric proteins, such as 53 kDa cholesteryl ester transfer protein (CETP) [45], and an individual 160 kDa IgG antibody (with ~1 nm high-resolution 3D reconstruction) [11].
2. Rouleau artifact in lipoproteins under negative-staining
With certain popular and conventional NS protocols [15, 46], lipoprotein particles regularly present stacking into rouleau formation [35–40]. For example, apoE4-contained high-density lipoprotein (HDL) (Fig. 1A), apoA-I-contained discoidal nascent HDL (Fig. 1B), and even apoB-contained human plasma LDL (Fig. 1C) form rouleau by the conventional NS method [29, 30]. The rouleau formation seems unrelated to comprising apolipoprotein types or particle concentrations, although it may relate to lipid component type (cationic or ionic). However, rouleau formation is absent in the results from nondenaturing polyacrylamide gradient gel electrophoresis, mass spectrometry [47, 48], X-ray crystallography [49, 50], small-angle X-ray diffraction data [43], or cryo-EM studies [29, 47, 51–53]. Thus, rouleau formation is generally believed to be an artifact of the conventional NS-EM protocol [29, 30]. Unfortunately, the observation of rouleau formation by conventional NS protocol (because of the challenges in imaging the lipoproteins in their near native-state by cryo-EM) has for decades been an indicator of successful synthesis of HDL particles.
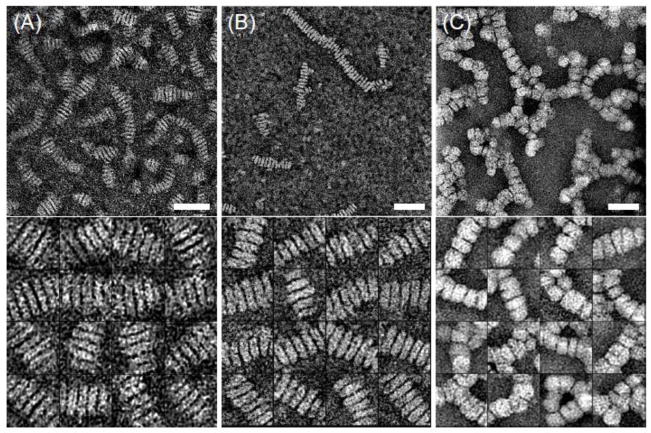
Figure 1.
Electron micrographs presented with rouleau of different lipoproteins by NS with parameters of PTA, and the mix procedure. (A) apoE4-contained rHDL. (B) 9.6-nm nascent rHDL. (C) LDL. Micrographs (up panel) and selected particles (down panel) are shown. Bars: 50 nm. Particle window size: A and B, 30 nm; C, 60 nm. This research was originally published in the Journal of Lipid Research as reference [29, 30]© the American Society for Biochemistry and Molecular Biology.
Recently, Ren et al. have faced these same challenges in imaging lipoproteins under near physiological buffer conditions by cryo-EM [8, 11, 12, 29, 44, 54]. We investigated the NS protocols by using the images obtained from cryo-electron microscopy (cryo-EM) of apoE4 HDL as a standard control for comparison, and reported an optimized negative-staining (OpNS) protocol (Fig. 2) [29]. OpNS eliminates the rouleau artifact of apoE4 HDL, and provides images that are highly similar (difference < 5%) in size and shape to that from cryo-EM (Fig. 2A), but with higher contrast [29]. A validation of OpNS protocol has been conducted by examining the elimination of rouleau artifact in various lipoprotein samples [30].
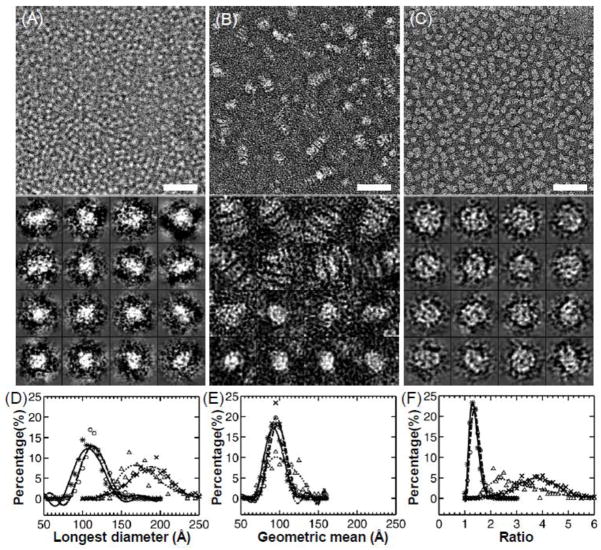
Figure 2.
Electron micrographs presented with and without rouleau formation of apoE4•POPC rHDL by (A) cryo-EM, (B) wash protocol with PTA as stain, and (C) OpNS. Bars: 50 nm. Particle window size: A and C, 20 nm; B, 30 nm. Micrographs (up panel) and selected particles (down panel) are shown. Histograms of particle size and shape are also shown by (D) histogram of longest particle diameter, (E) histogram of particle diameter geometric mean, and (F) histogram of particle diameter ratio. Legend: cryo-EM, asterisks and solid line; PTA, crosses and dash-dot line; PTA with washing, triangles and dotted line; and UF, circles and dashed line. These images and histograms suggested that the OpNS protocol could produce near native images of the apoE4•POPC particles similar to those produced by cryo-EM with much higher contrast. This research was originally published in the Journal of Lipid Research as reference [29]© the American Society for Biochemistry and Molecular Biology.
To further illuminate the different NS protocol effects in the structure and conformation of lipoproteins, we cataloged the effects of NS protocols from the three NS parameters, i.e. NS sample preparation procedures, salt concentrations, and staining reagents.
3. NS procedure effect on the rouleau formation of lipoprotein
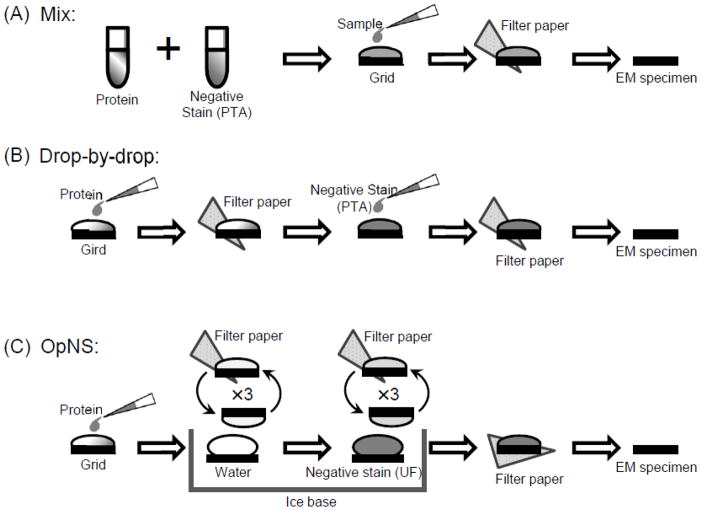
NS protocols used in examining lipoproteins [30, 35–40] can be cataloged into three major groups based on their NS sample preparation procedures: mix [46], drop-by-drop [15], and washing procedure [29] (Fig. 3). The mix procedure (Fig. 3A) requires mixing the stain and the lipoprotein sample using a certain ratio, applying the mixture to a carbon-coated grid, blotting with filter paper to remove excess solution, and then finally drying the grid completely for analyzing the sample with an electron microscope [46]. This procedure has been demonstrated with electron micrographs showing rouleau with micellar complexes of DPPC-apoA-I, egg-PC-apoA-I [55], very low-density lipoproteins (VLDL) [56], and Hep G2 fractions of lipoproteins [57].
Figure 3.
A schematic diagram illustrating the procedure parameter commonly used with NS of lipoproteins as reported previously. (A) Mix; (B) Drop-by-Drop; (C) OpNS.
The drop-by-drop procedure (Fig. 3B) entails direct application of the sample onto a carbon-coated EM grid, and blotting filter paper to remove excess solution. A drop of stain is then directly adhered to the EM grid, blotted again to remove excess solution, and finally dried before imaging [15, 58–60]. Many electron micrographs employing this procedure show rouleau formation with various size discoidal high-density lipoprotein (HDL) [34], plasma low-density lipoprotein (LDL), and plasma VLDL [61].
The washing procedure (Fig. 3C) requires that the lipoprotein sample be affixed to the carbon-coated EM grid with excess solution blotted with filter paper immediately before washing with deionized water and blotting with filter paper three times, and then three series of touching the stain solution and blotting before drying [3, 29, 30]. Washing the sample before staining reduces protein buffer-salt concentration and reduces potential reactions with the buffer, therefore reducing the probability of artifacts that may develop between the stain, protein, and salt. Using a uranyl formate (UF) stain, no rouleau is formed after washing the sample (Fig. 2C). However, with phosphotungstic acid (PTA) as the stain, a small amount of rouleau has been shown with an apoE4•POPC (1-palmitoyl-2-oleoyl phosphatidylcholine) complex (Fig. 2B) [3, 29]. The washing procedure seems to reduce rouleau phenomena, but may not be a direct factor in eliminating the rouleau artifact.
4. NS stain reagent effect on the rouleau formation of lipoprotein
Heave metal stains provide stronger electron scattering than the protein [16–19, 62], allowing the sample to tolerate higher radiation doses, and generating a higher negative contrast [4–6]. Heavy metal stains are cataloged as anionic, which include stains such as PTA [15], methylamine tungstate [13] and silicon tungstate [63], and cationic which include stains such as uranyl acetate (UA) [64], UF [3, 29, 30], uranyl nitrate (UN) [65]. Both anionic and cationic stains have been used with various NS protocols of biological macromolecules.
PTA is one of the most commonly used anionic NS stains to examine lipoproteinstructure [37, 66–69]. PTA is a heteropoly acid containing several negative charges, used at a pH range of 7.0 to 7.5, and is also used for positive staining [3, 15, 29, 30, 70]. Since the negative charges of PTA can interact electrostatically with the positively charged head groups of phospholipids on the surface of lipoproteins and even liposomes, PTA can serve as a mediator between lipoproteins causing rouleau formation [29, 30] under regular buffer salt conditions [29, 30] (Fig. 1). For example, apoE4 HDL stained by PTA displayed short rouleau formation even with the inclusion of a washing procedure (Fig. 2B) [29]. To test the rouleau formation related to either surface lipids or apolipoproteins, Ren et. al. examined POPC liposome vesicles using the conventional NS-EM protocol (PTA as stains). EM micrographs revealed particles stacked into rouleau, suggesting that rouleau formation is due to the interaction between the POPC molecules of neighboring liposome vesicles [29, 30]. Similar results were also reported in earlier experiments with POPC and dimyristoyl phosphatidylcholine (DMPC) liposome vesicles, which formed stacks when PTA was used for NS-EM [22, 71].
UA is a popular choice of heavy metal cationic stains and is frequently used in the NS of various biological specimens [64, 72, 73]. The structure of UA is a preferred stain parameter for NS because it is positively charged, and will not interact with any positively charged lipid groups of the lipoprotein, and it also serves as a fixative [3]. Less frequently used cationic stains for NS of lipoproteins are UF and UN, which are also positively charged, again avoiding interactions with the positively charged groups of the lipoprotein [3, 29, 30, 65]. UA/UF/UN can be used at a lower pH with a pH range of 3.5 to 4.6. Precipitation of the salt can occur at certain pH values and a low pH may not be suitable for some physiological specimen states [13, 74]. Interestingly, UA/UF fixes protein structure on a millisecond timescale [75] so that the mildly acidic pH, 4.2 – 4.6 should not be lipolytic. The choline group contains a positive charge whereas the phosphoryl moiety is negatively charged, giving its well-known zwitterionic structure. The mechanism of PTA adsorption to surfaces is electrostatic, rather than via hydrogen bonding because adsorption was not affected by pH [76]. In contrast to PTA, UA/UF is a salt, not an acid. As far as we know, using UA/UF, various lipoprotein particles were all displayed in isolated form, including a high-density lipoprotein-C (HDL-c) [64], and lipoprotein LP-X [77].
UF has been reported that it can provide better results than UA [78]. An interesting feature of UF and UN stains are their smaller than UA grain sizes (UF: 0.3 nm, UN: ~0.5 nm) [3, 79] that permits for EM analysis of small proteins (<100 kDa) [3, 29, 80–82]. For example, we used UF as a stain to image the high-resolution structure of cholesteryl ester transfer protein (CETP, ~53 kDa), in which, some secondary structural information could be revealed from raw electron micrographs [45].
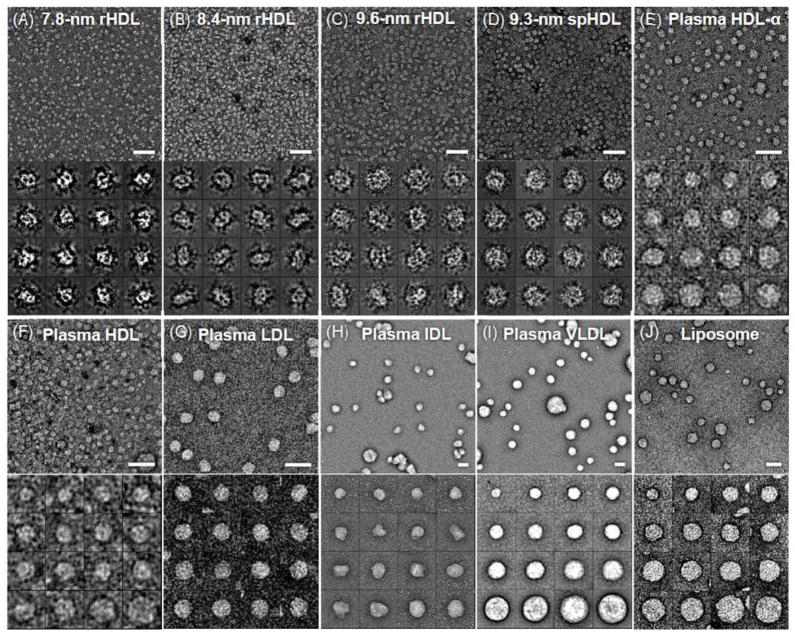
The drawbacks of UF are: i) UF is sensitive to light, ii) has a short half-life, and iii) requires higher standard of safety to handle because UF has radioactivity (though it is weak). UF has been utilized at a lower pH in which rouleau has not been observed, thus UF serves as a model stain parameter to eliminate artifacts of lipoproteins including: reconstituted HDL (Fig. 4A–D), human plasma HDL (Fig. 4E and F), LDL (Fig. 4G), IDL (Fig. 4H), VLDL (Fig. 4I), and liposomes (Fig. 4J) [30]. UF has been proven to be a worthy and viable NS stain for examining lipoproteins.
Figure 4.
Electron micrographs presented without rouleau of different lipoproteins and liposome by OpNS. (A) 7.8-nm rHDL. (B) 8.4-nm rHDL. (C) 9.6-nm rHDL. (D) 9.3-nm spherical rHDL. (E) Human plasma HDL. (F) Human plasma HDL. (G) Human plasma LDL. (H) Human plasma IDL. (I) Human plasma VLDL. (J) Liposome. Micrographs (up panel) and selected particles (down panel) are shown. Bars: 50 nm. Particle window size: A–D, 20 nm; E and F, 25 nm; G, 50 nm; H and I, 100 nm; I, 80 nm. This research was originally published in the Journal of Lipid Research as reference [30]© the American Society for Biochemistry and Molecular Biology.
5. Buffer salt concentration effect on the rouleau formation of lipoprotein
To lower these unfavorable salt effects, washing the sample with deionized water after adherence to the EM grid has been shown to reduce the level of rouleau formation, but does not entirely eliminate it with the PTA stain parameter. A few drops of water can be enough to wash the sample to reduce salt effects. However, in one particular extreme case, ten drops of water were used to wash the sample prior to staining with PTA, and some rouleaux were still observed in electron micrographs [83]. A noteworthy study was conducted by Ren et. al. to reveal rouleau formation in liposome vesicles via PTA after interactions with variable salt concentrations, and with the mix procedure. Liposome vesicles with high salt (0.5 M NaCl), regular salt (0.25 M NaCl), low salt (0.1M NaCl), and absence of salt all showed rouleau formation in electron micrographs [30] (Fig. 5). Notably, the higher salt concentrations in liposomes showed tighter rouleau (Fig. 5A and B). In comparison, with decreased levels of salt concentration, shorter rouleau was observed (Fig. 5C and D). Interestingly, using the parameters of UF and the optimized procedure presented no rouleau formation and isolated particles were observed of the same liposome vesicle [30] (Fig. 4J). Thus, salt concentrations provide for a level of vesicle-vesicle interaction that can cause rouleau. Similar results were also observed with apoE4 HDL [29]. These results suggest that a procedure to reduce the buffer salt concentration is a necessary step, but not an independently sufficient step for elimination of rouleau in POPC-containing biological samples.
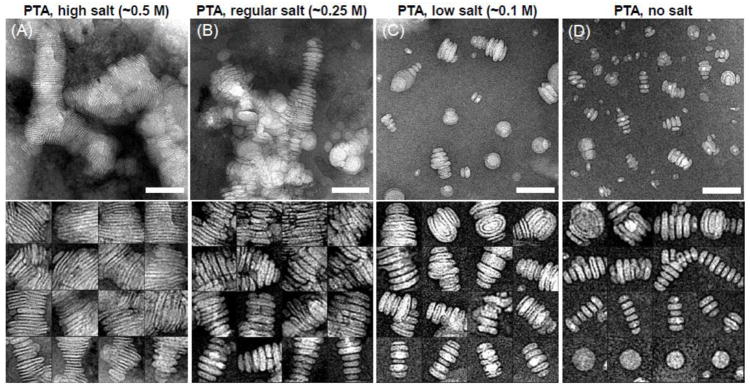
Figure 5.
Electron micrographs presented rouleau formation of liposomes by mix NS protocol (PTA as stain) with different salt concentrations. (A) High salt concentration (~0.5 M NaCl). (B) Regular salt concentration (~0.25 M NaCl). (C) Low salt concentration (~0.1 M NaCl). (D) No salt. Micrographs (up panel) and selected particles (down panel) are shown. Bars: 100 nm. Particle window size: 80 nm. This research was originally published in the Journal of Lipid Research as reference [30]© the American Society for Biochemistry and Molecular Biology.
6. High-resolution images of individual particles of protein by OpNS
Ren et. al. reported that OpNS as a general NS protocol can be used to examine a series of lipoprotein species and complexes without introducing the rouleau artifact [29, 30]. The lipoproteins that have been validated [30] include nascent HDL (Fig. 4A–C), spherical HDL (Fig. 4D), plasma HDL (Fig. 4E and F), LDL, IDL and VLDL (Fig. 4G–I). Moreover, the complexes formed by the interactions between lipoproteins and proteins can also be imaged by the OpNS, such as LDL/CETP, LDL/CETP/HDL, LDL/antibody, and HDL/antibody [45]. Moreover, Ren et. al have used the OpNS for high resolution (~1 nm) imaging of one of smallest proteins in the EM field, 53 kDa CETP (Fig. 6) [45].
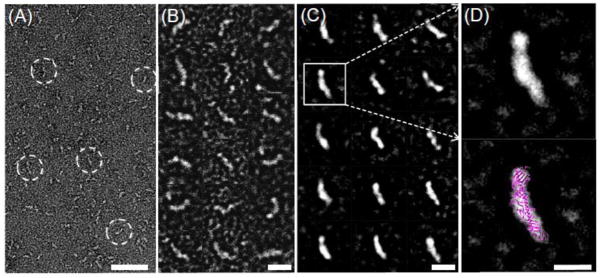
Figure 6.
Electron micrographs of CETP by the OpNS. (A) Survey view of the micrograph shows the banana-shaped CETP (dashed circles). (B) Selected particles of CETP. (C) In these randomly oriented particles selected, reference-free, and class-averaged images usually show one distal end of CETP larger and denser than the other. (D) A reference-free, class average image demonstrating a larger, dense and more globular end and a smaller, less dense and more tapered end (top panel). Overlaying the CETP crystal structure (PDB entry: 2OBD) onto this class average image shows a near-perfect match in structural shape and size (bottom panel). Bars: A, 50 nm; B–C, 10 nm; D, 5nm. This research was originally published in the Nature Chemical Biology as reference [45].
CETP mediates the transfer of neutral lipids, including cholesteryl esters (CEs) and triglycerides (TGs), between HDL, LDL and very low-density lipoproteins (VLDL) [84]. An elevated level of LDL-cholesterol (LDL-C) and/or a low level of HDL-cholesterol (HDL-C) in human plasma are major risk factors for cardiovascular disease (CVD) [85, 86]. Since increased CETP can reduce HDL-C concentration [87] and CETP deficiency is associated with elevated HDL-C levels [88, 89], CETP inhibitors, including torcetrapib, anacetrapib and dalcetrapib, have been investigated in clinical trials for treating CVD [90–92]. Despite the intense clinical interest in CETP inhibition, little is known concerning the molecular mechanisms of CETP-mediated lipid transfer among lipoproteins, or even how CETP interacts with lipoproteins.
Ren et. al. used OpNS protocol as a high-throughput and high-resolution method and prepared more than 300 EM specimens to reveal how CETP interacts with various lipoproteins [45]. They also used single-particle image processing for the 3D reconstruction of CETP and CETP bound to HDL. And, they used molecular dynamics simulation to study and better understand the CETP mechanism in CE transfer among lipoproteins. They discovered that CETP bridges a ternary complex with its N-terminal β-barrel domain penetrating into HDL and its C-terminal domain interacting with LDL or VLDL, and discovered a tunnel mechanism. In this mechanistic model, the CETP lipoprotein-interacting regions, which are highly mobile, form pores that connect to a hydrophobic central cavity, thereby forming a tunnel for transfer of neutral lipids from donor to acceptor lipoproteins [45]. These new insights into CETP transfer provide a molecular basis for analyzing mechanisms for CETP inhibition.
Another exciting application of OpNS is the 3D reconstruction of the first individual protein structure, an individual IgG antibody (Fig. 7) [11, 93, 94]. The OpNS EM images of individual antibody particles from a series of tilt-angle images provide high-resolution and high-contrast structural information that allow the successful reconstruction of an individual particle of protein by individual particle electron tomography (IPET) (Fig. 7D–F) [11]. In IPET 3D reconstruction [11], the specimen movements between different exposures for different tilted-view images were precisely computed, and then the serials of those targeted IgG antibody images were precisely aligned to their global center for 3D reconstruction calculated via a focus-electron tomography reconstruction (FETR) algorithm [11]. Notably, the successfully reconstructed individual IgG antibody particle has opened the gate to study the dynamics and fluctuations of small proteins [11, 44, 54, 94–96].
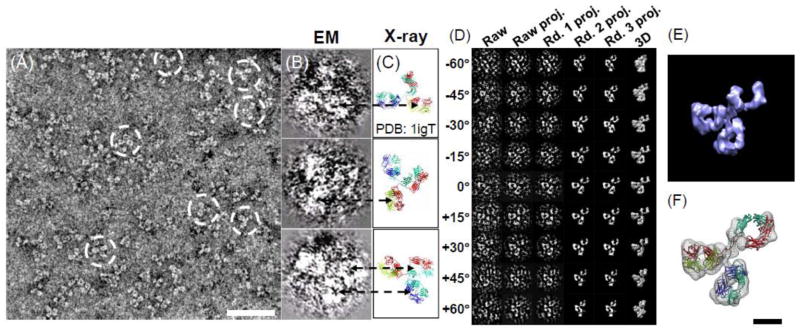
Figure 7.
Human IgG antibody particles imaged and reconstructed by OpNS. (A) Survey view of human IgG antibody imaged. The white-circled particles clearly displayed three domains within each particle. (B) Selected three particles display low-density regions (holes indicated by dash arrows) within domains. (C) Their corresponding orientations of the crystal structure (PDB entry: 1IGT) displayed in their corresponding holes within the corresponding domains can also be visualized, suggesting the holes are the intrinsic structure features instead of the artifact from neither negative-staining nor defocus-related contrast transfer function (CTF). (D–E) 3D reconstruction of one single-instance of IgG antibody by IPET is shown. (D) The process of IPET. (E) The final 3D reconstruction of a targeted individual antibody particle was displayed. The reconstruction displayed three ring-shaped domains that corresponding to three domain of IgG antibody. (F) Docking the crystal structure (PDB entry: 1IGT) of each domain of the IgG antibody into each ring-shaped density of IgG showed a good fit. Bars: A, 50 nm; F, 5 nm. This research was originally published in the PloS ONE as reference [11].
7. Optimized NS (OpNS) protocol
Considering that the OpNS is a reliable protocol to be used to examine the structure of various lipoproteins, that OpNS is a high-throughput approach for studying protein mechanism, and that it provides a high-contrast and high-resolution imaging method for studying the individual particle 3D structure of protein, it is necessary to give a complete description of OpNS protocol below [29, 30].
Prepare 100 ml of a 1% (w/v) UF solution. Put UF powder in deionized water and stir it overnight in a dark room at room temperature. Cover the bottle of solution with aluminum foil. Filter 5 ml of the 1% solution with the NORM-JECT syringe and the Anotop filter of 0.02 μm, and aliquot it into 2 ml vials, wrapped in aluminum foil to keep the solution in the dark. Immediately after aliquoting the 1% UF solution, place the vials into liquid nitrogen by using long handle forceps.
Store the 2 ml vials of the 1 % solution in an −80 °C freezer until use.
Before use, thaw a vial in a 4 °C water bath, and make sure it remains wrapped (cover it) in aluminum foil to keep the vial in the dark.
Once the UF is thawed and in liquid form, filter the UF again, using a 1 ml NORM-JECT syringe, and using the Anotop filter of 0.02 μm pore size (Anotop 10). Cover it with aluminum foil and store it on ice or at 4 °C.
Place ice in a uniformly level manner into the flat ice chamber, and cover it.
Designate 3 rows of 6 small circular regions in Parafilm. Place the Parafilm in the flat ice chamber and then place ~35 μl drops of deionized water in the first three circle regions in each row. Subsequently place ~35 μl drops of the filtered UF in the next three small circle regions in each row.
Fill the icebox with ice, cover it, and let it stand for ~5 minutes.
Obtain thin carbon film-coated copper EM grids (Cu-300CN, Pacific Grid-tech, San Francisco, CA) with Dumont #5 medical tweezers with clamping ring, put the carbon film side up on a clean glass microscope slide, perform glow-discharge for 15 seconds with an EMS 100, and place the slide on a clean filter paper in a petri dish and cover it.
Open the icebox and hold the grid with tweezers at a 45° angle; place ~3 μl of the lipoprotein sample (~0.005 mg/ml, protein, diluted by Dulbecco’s Phosphate Buffered Saline) on the EM grid carbon film side and incubate for ~1 minute.
After ~1 minute, remove excess solution by gently touching the edge of the grid with filter paper (#1, Whatman). Wash the grid by briefly placing the surface of the grid with a drop (~35 μl) of deionized water on Parafilm and then blot with filter paper to remove the excess solution. The touching and blotting steps are to be performed quickly three times, each with a clean drop of deionized water. Perform the same touching and blotting steps with three successive drops (~ 35 μl) of 1% UF solution applied on Parafilm, and remove the excess solution by blotting similarly with water. Contact the grid with the last UF drop with the sample side down for 1–3 min in the dark (close the lid of the flat ice chamber) before removing excess stain by blotting again in the entire backside parallel to the grid (non-carbon side) with filter paper. Subsequently, air-dry the sample by a low-flow of nitrogen gas at room temperature.
Store the grid on filter paper in a petri dish, and partially cover it for ~30 minutes.
Send the grid to EM or store it in a grid storage box.
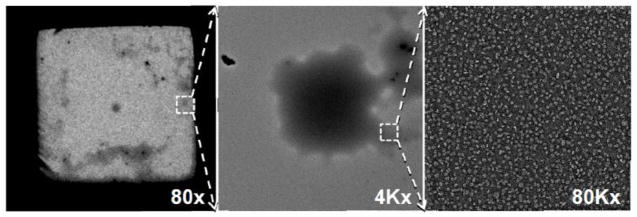
To be noted, the thickness of the stain of the carbon-coated grid is not even (Fig. 8). Areas of thicker stain look like “cloud” on the grid when using low magnification (< 400x). These cloudy areas generally can provide better images.
Figure 8.
EM imaging area. Micrographs show thicker stain area as cloudy areas that are usually ideal imaging areas for EM imaging. Cloud (highlighted by box) of lipoprotein to designate lipoprotein location at 80x magnification (left), same designated area of cloud (highlighted by box) of lipoprotein further magnified at 4Kx (middle), and same designated cloud area further magnified at 80Kx (right).
8. Conclusions
OpNS protocol is a reliable protocol that can eliminate the rouleau artifact, and provide high-resolution structural details. The OpNS protocol has some disadvantages, such as more complicated steps of specimen preparation; the need to deal with a radioactive substance, UF; the shorter half-life of the UF stain requires keeping the stain fresh; necessary storage of the UF solution in −80°C requires thawing before use; and it involves the handling of hazardous waste. However, after investigating several other techniques with their benefits, drawbacks, and key results illustrated via electron micrographs, we have determined that the OpNS protocol is the best approach to investigate structure and morphology of lipoproteins. This NS protocol that eliminates rouleau while maintaining near native conformations can be quickly adapted into any laboratory, and then exploration of lipoprotein function related to structure can be more thoroughly investigated.
Highlights.
Negative-staining (NS) EM is a rapid, common method to examine protein structure.
We reviewed the most popular NS protocols used to examine lipoproteins.
Rouleau artifact of lipoprotein was commonly observed using conventional NS protocol.
The optimized NS (OpNS) protocol can eliminate the rouleau artifacts.
OpNS can be used as a general protocol to study the structure of proteins.
Acknowledgments
We thank Mr. Matthew J. Rames for editing comments. This work was supported by the Office of Science, Office of Basic Energy Sciences of the United States Department of Energy (contract no. DE-AC02-05CH11231) and the National Heart, Lung, And Blood Institute of the National Institutes of Health (no. R01HL115153).
Abbreviations
- EM
electron microscopy
- NS
negative staining
- OpNS
optimized negative staining
- cryo-EM
cryo-electron microscopy
- IPET
individual-particle electron tomography
- HDL
high-density lipoprotein
- LDL
low-density lipoprotein
- VLDL
very low-density lipoprotein
- IDL
intermediate-density lipoprotein
- UF
uranyl formate
- PTA
phosphotungstic acid
- SNR
signal-to-noise ratio
- CVD
cardiovascular disease
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wang ZL. Transmission electron microscopy of shape-controlled nanocrystals and their assemblies. J Phys Chem B. 2000;104:1153–1175. [Google Scholar]
- 2.Henderson R. Realizing the potential of electron cryo-microscopy. Quarterly reviews of biophysics. 2004;37:3–13. doi: 10.1017/s0033583504003920. [DOI] [PubMed] [Google Scholar]
- 3.Ohi M, Li Y, Cheng Y, Walz T. Negative Staining and Image Classification - Powerful Tools in Modern Electron Microscopy. Biological procedures online. 2004;6:23–34. doi: 10.1251/bpo70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sander B, Golas MM. Visualization of bionanostructures using transmission electron microscopical techniques. Microsc Res Tech. 2011;74:642–663. doi: 10.1002/jemt.20963. [DOI] [PubMed] [Google Scholar]
- 5.Liu H, Jin L, Koh SB, Atanasov I, Schein S, Wu L, Zhou ZH. Atomic structure of human adenovirus by cryo-EM reveals interactions among protein networks. Science. 2010;329:1038–1043. doi: 10.1126/science.1187433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ren G, Reddy VS, Cheng A, Melnyk P, Mitra AK. Visualization of a water-selective pore by electron crystallography in vitreous ice. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:1398–1403. doi: 10.1073/pnas.041489198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ren G, Cheng A, Reddy V, Melnyk P, Mitra AK. Three-dimensional fold of the human AQP1 water channel determined at 4 A resolution by electron crystallography of two-dimensional crystals embedded in ice. Journal of molecular biology. 2000;301:369–387. doi: 10.1006/jmbi.2000.3949. [DOI] [PubMed] [Google Scholar]
- 8.Ren G, Rudenko G, Ludtke SJ, Deisenhofer J, Chiu W, Pownall HJ. Model of human low-density lipoprotein and bound receptor based on cryoEM. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:1059–1064. doi: 10.1073/pnas.0908004107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ren G, Quispe J, Leppla SH, Mitra AK. Large-scale structural changes accompany binding of lethal factor to anthrax protective antigen: a cryo-electron microscopic study. Structure. 2004;12:2059–2066. doi: 10.1016/j.str.2004.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ren G, Cheng A, Melnyk P, Mitra AK. Polymorphism in the packing of aquaporin-1 tetramers in 2-D crystals. Journal of structural biology. 2000;130:45–53. doi: 10.1006/jsbi.2000.4211. [DOI] [PubMed] [Google Scholar]
- 11.Zhang L, Ren G. IPET and FETR: experimental approach for studying molecular structure dynamics by cryo-electron tomography of a single-molecule structure. PloS one. 2012;7:e30249. doi: 10.1371/journal.pone.0030249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jones MK, Zhang L, Catte A, Li L, Oda MN, Ren G, Segrest JP. Assessment of the validity of the double superhelix model for reconstituted high density lipoproteins: a combined computational-experimental approach. The Journal of biological chemistry. 2010;285:41161–41171. doi: 10.1074/jbc.M110.187799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bremer A, Henn C, Engel A, Baumeister W, Aebi U. Has negative staining still a place in biomacromolecular electron microscopy? Ultramicroscopy. 1992;46:85–111. doi: 10.1016/0304-3991(92)90008-8. [DOI] [PubMed] [Google Scholar]
- 14.Brenner S, Horne RW. A negative staining method for high resolution electron microscopy of viruses. Biochim Biophys Acta. 1959;34:103–110. doi: 10.1016/0006-3002(59)90237-9. [DOI] [PubMed] [Google Scholar]
- 15.Forte TM, Nordhausen RW. Electron microscopy of negatively stained lipoproteins. Methods Enzymol. 1986;128:442–457. doi: 10.1016/0076-6879(86)28086-6. [DOI] [PubMed] [Google Scholar]
- 16.Colliex C, Cowley JM, Dudarev SL, Fink M, Gjonnes J, Hilderbrandt R, Howie A, Lynch DF, Peng LM, Ren G, Ross AW, Smith VH, Jr, Spence JCH, Steeds JW, Wang J, Whelan MJ, Zvyagin BB. Electron Diffraction. In: Prince E, editor. International Tables For Crystallography. C. Kluwer Academic Publishers; 2006. pp. 259–429. [Google Scholar]
- 17.Ren G, Zuo JM, PL-M Accurate Measurements of Crystal Structure Factors Using a FEG Electron Microscope Using Digital Micrographs. Micron. 1997;28:459–467. [Google Scholar]
- 18.Peng LM, Ren G, Dudarev SL, Whelan MJ. Robust parameterization of elastic and absorptive electron atomic scattering factors. Acta Crystallogr A. 1996;52:257–276. [Google Scholar]
- 19.Peng LM, Ren G, Dudarev SL, Whelan MJ. Debye-Waller factors and absorptive scattering factors of elemental crystals. Acta Crystallogr A. 1996;52:456–470. [Google Scholar]
- 20.Ren G, Gao K, Bushman FD, Yeager M. Single-particle image reconstruction of a tetramer of HIV integrase bound to DNA. Journal of molecular biology. 2007;366:286–294. doi: 10.1016/j.jmb.2006.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.De Carlo S, Harris JR. Negative staining and cryo-negative staining of macromolecules and viruses for TEM. Micron. 2011;42:117–131. doi: 10.1016/j.micron.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Melchior V, Hollingshead CJ, Cahoon ME. Stacking in Lipid Vesicle Tubulin Mixtures Is an Artifact of Negative Staining. J Cell Biol. 1980;86:881–884. doi: 10.1083/jcb.86.3.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Allan V, Vale R. Movement of Membrane Tubules Along Microtubules in-Vitro - Evidence for Specialized Sites of Motor Attachment. Journal of Cell Science. 1994;107:1885–1897. doi: 10.1242/jcs.107.7.1885. [DOI] [PubMed] [Google Scholar]
- 24.Catte A, Patterson JC, Jones MK, Jerome WG, Bashtovyy D, Su Z, Gu F, Chen J, Aliste MP, Harvey SC, Li L, Weinstein G, Segrest JP. Novel changes in discoidal high density lipoprotein morphology: a molecular dynamics study. Biophys J. 2006;90:4345–4360. doi: 10.1529/biophysj.105.071456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Forester GP, Tall AR, Bisgaier CL, Glickman RM. Rat intestine secretes spherical high density lipoproteins. The Journal of biological chemistry. 1983;258:5938–5943. [PubMed] [Google Scholar]
- 26.Gantz DL, Walsh MT, Small DM. Morphology of sodium deoxycholate-solubilized apolipoprotein B-100 using negative stain and vitreous ice electron microscopy. Journal of Lipid Research. 2000;41:1464–1472. [PubMed] [Google Scholar]
- 27.Pentikainen MO, Lehtonen EM, Kovanen PT. Aggregation and fusion of modified low density lipoprotein. J Lipid Res. 1996;37:2638–2649. [PubMed] [Google Scholar]
- 28.Tall AR, Green PH, Glickman RM, Riley JW. Metabolic fate of chylomicron phospholipids and apoproteins in the rat. The Journal of clinical investigation. 1979;64:977–989. doi: 10.1172/JCI109564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang L, Song J, Newhouse Y, Zhang S, Weisgraber KH, Ren G. An optimized negative-staining protocol of electron microscopy for apoE4 POPC lipoprotein. Journal of lipid research. 2010;51:1228–1236. doi: 10.1194/jlr.D002493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang L, Song J, Cavigiolio G, Ishida BY, Zhang S, Kane JP, Weisgraber KH, Oda MN, Rye KA, Pownall HJ, Ren G. Morphology and structure of lipoproteins revealed by an optimized negative-staining protocol of electron microscopy. Journal of lipid research. 2011;52:175–184. doi: 10.1194/jlr.D010959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carnemolla R, Ren X, Biswas TK, Meredith SC, Reardon CA, Wang J, Getz GS. The specific amino acid sequence between helices 7 and 8 influences the binding specificity of human apolipoprotein A-I for high density lipoprotein (HDL) subclasses: a potential for HDL preferential generation. The Journal of biological chemistry. 2008;283:15779–15788. doi: 10.1074/jbc.M710244200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sivashanmugam A, Yang Y, Murray V, McCullough C, Chen B, Ren X, Li Q, Wang J. Structural basis of human high-density lipoprotein formation and assembly at sub nanometer resolution. Methods in cell biology. 2008;90:327–364. doi: 10.1016/S0091-679X(08)00815-7. [DOI] [PubMed] [Google Scholar]
- 33.Cavigiolio G, Shao B, Geier EG, Ren G, Heinecke JW, Oda MN. The interplay between size, morphology, stability, and functionality of high-density lipoprotein subclasses. Biochemistry. 2008;47:4770–4779. doi: 10.1021/bi7023354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen B, Ren X, Neville T, Jerome WG, Hoyt DW, Sparks D, Ren G, Wang J. Apolipoprotein AI tertiary structures determine stability and phospholipid-binding activity of discoidal high-density lipoprotein particles of different sizes. Protein science: a publication of the Protein Society. 2009;18:921–935. doi: 10.1002/pro.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gong EL, Nichols AV, Weisgraber KH, Forte TM, Shore VG, Blanche PJ. Discoidal complexes containing apolipoprotein E and their transformation by lecithin-cholesterol acyltransferase. Biochim Biophys Acta. 1989;1006:317–328. doi: 10.1016/0005-2760(89)90019-2. [DOI] [PubMed] [Google Scholar]
- 36.Schneeweis LA, Koppaka V, Lund-Katz S, Phillips MC, Axelsen PH. Structural analysis of lipoprotein E particles. Biochemistry. 2005;44:12525–12534. doi: 10.1021/bi050872j. [DOI] [PubMed] [Google Scholar]
- 37.Raussens V, Drury J, Forte TM, Choy N, Goormaghtigh E, Ruysschaert JM, Narayanaswami V. Orientation and mode of lipid-binding interaction of human apolipoprotein E C-terminal domain. The Biochemical journal. 2005;387:747–754. doi: 10.1042/BJ20041536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li X, Kan HY, Lavrentiadou S, Krieger M, Zannis V. Reconstituted discoidal ApoE-phospholipid particles are ligands for the scavenger receptor BI. The amino-terminal 1–165 domain of ApoE suffices for receptor binding. The Journal of biological chemistry. 2002;277:21149–21157. doi: 10.1074/jbc.M200658200. [DOI] [PubMed] [Google Scholar]
- 39.Innerarity TL, Pitas RE, Mahley RW. Binding of arginine-rich (E) apoprotein after recombination with phospholipid vesicles to the low density lipoprotein receptors of fibroblasts. The Journal of biological chemistry. 1979;254:4186–4190. [PubMed] [Google Scholar]
- 40.Lu B, Morrow JA, Weisgraber KH. Conformational reorganization of the four-helix bundle of human apolipoprotein E in binding to phospholipid. The Journal of biological chemistry. 2000;275:20775–20781. doi: 10.1074/jbc.M003508200. [DOI] [PubMed] [Google Scholar]
- 41.Peng DC, Song C, Reardon CA, Liao SS, Getz GS. Lipoproteins produced by ApoE−/− astrocytes infected with adenovirus expressing human ApoE. J Neurochem. 2003;86:1391–1402. doi: 10.1046/j.1471-4159.2003.01950.x. [DOI] [PubMed] [Google Scholar]
- 42.Peters-Libeu CA, Newhouse Y, Hall SC, Witkowska HE, Weisgraber KH. Apolipoprotein E*dipalmitoylphosphatidylcholine particles are ellipsoidal in solution. J Lipid Res. 2007;48:1035–1044. doi: 10.1194/jlr.M600545-JLR200. [DOI] [PubMed] [Google Scholar]
- 43.Peters-Libeu CA, Newhouse Y, Hatters DM, Weisgraber KH. Model of biologically active apolipoprotein E bound to dipalmitoylphosphatidylcholine. The Journal of biological chemistry. 2006;281:1073–1079. doi: 10.1074/jbc.M510851200. [DOI] [PubMed] [Google Scholar]
- 44.Zhang L, Cavigiolio G, Oda M, Rye K-A, Ren G. A Method to Study the Time-Dependent Remodeling of High Density Lipoproteins by Individual-Particle Electron Cryo-Tomography. Biophysical Journal. 2010;98:440a. [Google Scholar]
- 45.Zhang L, Yan F, Zhang S, Lei D, Charles MA, Cavigiolio G, Oda M, Krauss RM, Weisgraber KH, Rye KA, Pownall HJ, Qiu X, Ren G. Structural basis of transfer between lipoproteins by cholesteryl ester transfer protein. Nat Chem Biol. 2012;8:342–349. doi: 10.1038/nchembio.796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Forte T, Norum KR, Glomset JA, Nichols AV. Plasma lipoproteins in familial lecithin: cholesterol acyltransferase deficiency: structure of low and high density lipoproteins as revealed by elctron microscopy. The Journal of clinical investigation. 1971;50:1141–1148. doi: 10.1172/JCI106586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Silva RA, Huang R, Morris J, Fang J, Gracheva EO, Ren G, Kontush A, Jerome WG, Rye KA, Davidson WS. Structure of apolipoprotein A-I in spherical high density lipoproteins of different sizes. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:12176–12181. doi: 10.1073/pnas.0803626105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Davidson WS, Silva RA. Apolipoprotein structural organization in high density lipoproteins: belts, bundles, hinges and hairpins. Current opinion in lipidology. 2005;16:295–300. doi: 10.1097/01.mol.0000169349.38321.ad. [DOI] [PubMed] [Google Scholar]
- 49.Newhouse Y, Peters-Libeu C, Weisgraber KH. Crystallization and preliminary X-ray diffraction analysis of apolipoprotein E-containing lipoprotein particles. Acta crystallographica. Section F, Structural biology and crystallization communications. 2005;61:981–984. doi: 10.1107/S1744309105032410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ajees AA, Anantharamaiah GM, Mishra VK, Hussain MM, Murthy HM. Crystal structure of human apolipoprotein A-I: insights into its protective effect against cardiovascular diseases. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:2126–2131. doi: 10.1073/pnas.0506877103. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 51.van Antwerpen R, La Belle M, Navratilova E, Krauss RM. Structural heterogeneity of apoB-containing serum lipoproteins visualized using cryo-electron microscopy. J Lipid Res. 1999;40:1827–1836. [PubMed] [Google Scholar]
- 52.van Antwerpen R, Chen GC, Pullinger CR, Kane JP, LaBelle M, Krauss RM, Luna-Chavez C, Forte TM, Gilkey JC. Cryo-electron microscopy of low density lipoprotein and reconstituted discoidal high density lipoprotein: imaging of the apolipoprotein moiety. J Lipid Res. 1997;38:659–669. [PubMed] [Google Scholar]
- 53.van Antwerpen R. Preferred orientations of LDL in vitreous ice indicate a discoid shape of the lipoprotein particle. Archives of biochemistry and biophysics. 2004;432:122–127. doi: 10.1016/j.abb.2004.08.031. [DOI] [PubMed] [Google Scholar]
- 54.Zhang L, Cavigiolio G, Wang J, Rye KA, Oda M, Ren G. Structure of 9.6nm Discoidal High-Density Lipoprotein Revealed by Individual-Particle Electron Tomography. Biophysical Journal. 2010;98:440a. [Google Scholar]
- 55.Matz CE, Jonas A. Micellar complexes of human apolipoprotein A-I with phosphatidylcholines and cholesterol prepared from cholate-lipid dispersions. The Journal of biological chemistry. 1982;257:4535–4540. [PubMed] [Google Scholar]
- 56.Glomset JA, Applegate K, Forte T, King WC, Mitchell CD, Norum KR, Gjone E. Abnormalities in lipoproteins of d < 1.006 g/ml in familial lecithin:cholesterol acyltransferase deficiency. J Lipid Res. 1980;21:1116–1127. [PubMed] [Google Scholar]
- 57.Thrift RN, Forte TM, Cahoon BE, Shore VG. Characterization of lipoproteins produced by the human liver cell line, Hep G2, under defined conditions. J Lipid Res. 1986;27:236–250. [PubMed] [Google Scholar]
- 58.Fang Y, Gursky O, Atkinson D. Lipid-binding studies of human apolipoprotein A-I and its terminally truncated mutants. Biochemistry. 2003;42:13260–13268. doi: 10.1021/bi0354031. [DOI] [PubMed] [Google Scholar]
- 59.Gursky O, Ranjana, Gantz DL. Complex of human apolipoprotein C-1 with phospholipid: thermodynamic or kinetic stability? Biochemistry. 2002;41:7373–7384. doi: 10.1021/bi025588w. [DOI] [PubMed] [Google Scholar]
- 60.Jayaraman S, Gantz DL, Gursky O. Effects of protein oxidation on the structure and stability of model discoidal high-density lipoproteins. Biochemistry. 2008;47:3875–3882. doi: 10.1021/bi7023783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li CM, Chung BH, Presley JB, Malek G, Zhang X, Dashti N, Li L, Chen J, Bradley K, Kruth HS, Curcio CA. Lipoprotein-like particles and cholesteryl esters in human Bruch’s membrane: initial characterization. Invest Ophthalmol Vis Sci. 2005;46:2576–2586. doi: 10.1167/iovs.05-0034. [DOI] [PubMed] [Google Scholar]
- 62.Ren G, Peng LM. The Analytic Doyle-Turner Representation of High Energy Electron Absorptive Structure Factors. Acta Physica Sinica. 1996;45:1344–1349. [Google Scholar]
- 63.Camejo G, Suarez ZM, Munoz V. The apo-lipoproteins of human plasma high density lipoprotein: a study of their lipid binding capacity and interaction with lipid monolayers. Biochim Biophys Acta. 1970;218:155–166. doi: 10.1016/0005-2760(70)90103-7. [DOI] [PubMed] [Google Scholar]
- 64.Kondo A, Muranaka Y, Ohta I, Kanno T. Dynamic reaction in a homogeneous HDL-cholesterol assay visualized by electron microscopy. Clin Chem. 1999;45:1974–1980. [PubMed] [Google Scholar]
- 65.Tufteland M, Ren G, Ryan RO. Nanodisks derived from amphotericin B lipid complex. J Pharm Sci. 2008;97:4425–4432. doi: 10.1002/jps.21325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Clay MA, Pyle DH, Rye KA, Barter PJ. Formation of spherical, reconstituted high density lipoproteins containing both apolipoproteins A-I and A-II is mediated by lecithin: cholesterol acyltransferase. Journal of Biological Chemistry. 2000;275:9019–9025. doi: 10.1074/jbc.275.12.9019. [DOI] [PubMed] [Google Scholar]
- 67.Desilva HV, Masoliva J, Taylor JM, Mahley RW. Identification of Apolipoprotein B-100 Low-Density Lipoproteins, Apolipoprotein B-48 Remnants, and Apolipoprotein E-Rich High-Density-Lipoproteins in the Mouse. Journal of Lipid Research. 1994;35:1297–1310. [PubMed] [Google Scholar]
- 68.Fagan AM, Holtzman DM, Munson G, Mathur T, Schneider D, Chang LK, Getz GS, Reardon CA, Lukens J, Shah JA, LaDu MJ. Unique lipoproteins secreted by primary astrocytes from wild type, apoE (−/−), and human apoE transgenic mice. Journal of Biological Chemistry. 1999;274:30001–30007. doi: 10.1074/jbc.274.42.30001. [DOI] [PubMed] [Google Scholar]
- 69.Musliner TA, Long MD, Forte TM, Nichols AV, Gong EL, Blanche PJ, Krauss RM. Dissociation of high density lipoprotein precursors from apolipoprotein B-containing lipoproteins in the presence of unesterified fatty acids and a source of apolipoprotein A-I. J Lipid Res. 1991;32:917–933. [PubMed] [Google Scholar]
- 70.Silverman L, Glick D. The reactivity and staining of tissue proteins with phosphotungstic acid. J Cell Biol. 1969;40:761–767. doi: 10.1083/jcb.40.3.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Guo LS, Hamilton RL, Goerke J, Weinstein JN, Havel RJ. Interaction of unilamellar liposomes with serum lipoproteins and apolipoproteins. J Lipid Res. 1980;21:993–1003. [PubMed] [Google Scholar]
- 72.Hamilton RL, Williams MC, Fielding CJ, Havel RJ. Discoidal bilayer structure of nascent high density lipoproteins from perfused rat liver. The Journal of clinical investigation. 1976;58:667–680. doi: 10.1172/JCI108513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pollard H, Scanu AM, Taylor EW. On the geometrical arrangement of the protein subunits of human serum low-density lipoprotein: evidence for a dodecahedral model. Proceedings of the National Academy of Sciences of the United States of America. 1969;64:304–310. doi: 10.1073/pnas.64.1.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Frasca JM, Parks VR. A Routine Technique for Double-Staining Ultrathin Sections Using Uranyl and Lead Salts. J Cell Biol. 1965;25:157–161. doi: 10.1083/jcb.25.1.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhao FQ, Craig R. Capturing time-resolved changes in molecular structure by negative staining. Journal of structural biology. 2003;141:43–52. doi: 10.1016/s1047-8477(02)00546-4. [DOI] [PubMed] [Google Scholar]
- 76.Quintarelli G, Zito R, Cifonelli JA. On phosphotungstic acid staining. I. J Histochem Cytochem. 1971;19:641–647. doi: 10.1177/19.11.641. [DOI] [PubMed] [Google Scholar]
- 77.Seidel D, Agostini B, Muller P. Structure of an Abnormal Plasma Lipoprotein (Lp-X) Characterizing Obstructive-Jaundice. Biochimica Et Biophysica Acta. 1972;260:146. doi: 10.1016/0005-2760(72)90082-3. [DOI] [PubMed] [Google Scholar]
- 78.Knight DP. Negative staining of rat tail tendon collagen fibrils with uranyl formate. Tissue Cell. 1975;7:651–654. doi: 10.1016/0040-8166(75)90033-6. [DOI] [PubMed] [Google Scholar]
- 79.Dash S, Kamruddin M, Bera S, Ajikumar PK, Tyagi AK, Narasimhan SV, Raj B. Temperature programmed decomposition of uranyl nitrate hexahydrate. Journal of Nuclear Materials. 1999;264:271–282. [Google Scholar]
- 80.Harris JR, Gebauer W, Guderian FU, Markl J. Keyhole limpet hemocyanin (KLH), I: Reassociation from Immucothel followed by separation of KLH1 and KLH2. Micron. 1997;28:31–41. doi: 10.1016/s0968-4328(97)00012-7. [DOI] [PubMed] [Google Scholar]
- 81.Harris JR, Gebauer W, Sohngen SM, Nermut MV, Markl J. Keyhole limpet hemocyanin (KLH), II: Characteristic reassociation properties of purified KLH1 and KLH2. Micron. 1997;28:43–56. doi: 10.1016/s0968-4328(97)00011-5. [DOI] [PubMed] [Google Scholar]
- 82.HMA . Principles and Techniques of Electron Microscopy: Biological Applications. Cambridge University Press; Cambridge: 2000. [Google Scholar]
- 83.Jiang ZG, Simon MN, Wall JS, McKnight CJ. Structural analysis of reconstituted lipoproteins containing the N-terminal domain of apolipoprotein B. Biophys J. 2007;92:4097–4108. doi: 10.1529/biophysj.106.101105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Barter PJ, Brewer HB, Jr, Chapman MJ, Hennekens CH, Rader DJ, Tall AR. Cholesteryl ester transfer protein: a novel target for raising HDL and inhibiting atherosclerosis. Arterioscler Thromb Vasc Biol. 2003;23:160–167. doi: 10.1161/01.atv.0000054658.91146.64. [DOI] [PubMed] [Google Scholar]
- 85.Camejo G, Waich S, Quintero G, Berrizbeitia ML, Lalaguna F. The affinity of low density lipoproteins for an arterial macromolecular complex. A study in ischemic heart disease and controls. Atherosclerosis. 1976;24:341–354. doi: 10.1016/0021-9150(76)90126-x. [DOI] [PubMed] [Google Scholar]
- 86.Gordon T, Castelli WP, Hjortland MC, Kannel WB, Dawber TR. High density lipoprotein as a protective factor against coronary heart disease. The Framingham Study. The American journal of medicine. 1977;62:707–714. doi: 10.1016/0002-9343(77)90874-9. [DOI] [PubMed] [Google Scholar]
- 87.Hayek T, Azrolan N, Verdery RB, Walsh A, Chajek-Shaul T, Agellon LB, Tall AR, Breslow JL. Hypertriglyceridemia and cholesteryl ester transfer protein interact to dramatically alter high density lipoprotein levels, particle sizes, and metabolism. Studies in transgenic mice. The Journal of clinical investigation. 1993;92:1143–1152. doi: 10.1172/JCI116683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Brown ML, Inazu A, Hesler CB, Agellon LB, Mann C, Whitlock ME, Marcel YL, Milne RW, Koizumi J, Mabuchi H, et al. Molecular basis of lipid transfer protein deficiency in a family with increased high-density lipoproteins. Nature. 1989;342:448–451. doi: 10.1038/342448a0. [DOI] [PubMed] [Google Scholar]
- 89.Inazu A, Brown ML, Hesler CB, Agellon LB, Koizumi J, Takata K, Maruhama Y, Mabuchi H, Tall AR. Increased high-density lipoprotein levels caused by a common cholesteryl-ester transfer protein gene mutation. The New England journal of medicine. 1990;323:1234–1238. doi: 10.1056/NEJM199011013231803. [DOI] [PubMed] [Google Scholar]
- 90.Niesor EJ. Different effects of compounds decreasing cholesteryl ester transfer protein activity on lipoprotein metabolism. Current opinion in lipidology. 2011;22:288–295. doi: 10.1097/MOL.0b013e3283475e00. [DOI] [PubMed] [Google Scholar]
- 91.Miyares MA. Anacetrapib and dalcetrapib: two novel cholesteryl ester transfer protein inhibitors. The Annals of pharmacotherapy. 2011;45:84–94. doi: 10.1345/aph.1P446. [DOI] [PubMed] [Google Scholar]
- 92.Kappelle PJ, van Tol A, Wolffenbuttel BH, Dullaart RP. Cholesteryl ester transfer protein inhibition in cardiovascular risk management: ongoing trials will end the confusion. Cardiovascular therapeutics. 2011;29:e89–99. doi: 10.1111/j.1755-5922.2010.00201.x. [DOI] [PubMed] [Google Scholar]
- 93.Zhang L, Kaspar A, Woodnutt G, Ren G. Monitoring the Structural Changes of Conjugated Antibodies by High-Resolution Electron Microscopy and Individual-Particle Electron Tomography. Biophysical Journal. 2010;98:440a–441a. [Google Scholar]
- 94.Zhang L, Ren G. Determining the Dynamic Protein Structure by Individual-Particle Electron Tomography: An Individual Antibody Structure at a Nanometer Resolution. Biophysical Journal. 2010;98:441a. [Google Scholar]
- 95.Ren G, Zhang L. Asymmetric Small Protein Structure Determination by Individual Particle Electron Tomography. Biophysical Journal. 2012;102:394a. [Google Scholar]
- 96.Zhang L, Ren G. Structural Determination of Heterogeneous Protein by Individual-Particle Electron Tomography - Combination of Electron Tomography and Local Refinement Reconstruction Method for High-Resolution Structural Determination of Each Individual Protein Particle. Biophysical Journal. 2010;98:441a. [Google Scholar]