Abstract
Protein disorder remains an intrinsically fuzzy concept. Its role in protein function is difficult to conceptualize and its experimental study is challenging. Although a wide variety of roles for protein disorder have been proposed, establishing that disorder is functionally important, particularly in vivo, is not a trivial task. Several molecular chaperones have now been identified as conditionally disordered proteins; fully folded and chaperone-inactive under non-stress conditions, they adopt a partially disordered conformation upon exposure to distinct stress-conditions. This disorder appears to be vital for their ability to bind multiple aggregation-sensitive client proteins and to protect cells against the stressors. The study of these conditionally disordered chaperones should prove useful in understanding the functional role for protein disorder in molecular recognition.
Protein disorder: A fuzzy concept
Our view of proteins has been strongly shaped by the many beautiful structures that have been solved by X-ray crystallography. Looking at these structures, it is easy to forget that proteins often have very dynamic properties and regions that show considerable flexibility. Indeed, only 25% of crystal structures represent >95% of the complete molecule; all others have missing electron density for more than 5% of their sequence, usually because these regions take on multiple conformations [1]. Moreover, the crystallographic portion of the protein database is almost certainly biased toward those proteins that fold readily into a single or, at most, a few distinct conformations and are thus crystallizable. However, proteins possess a wide range of stabilities and degrees of (dis)order [2]. Globally ‘intrinsically disordered’ proteins lie at one extreme part of a continuous spectrum of structural states, which ranges from very flexible to static, and can involve either part of a protein or the complete polypeptide chain (Figure 1). Thus it is difficult to summarize the degree of protein flexibility with a single term, and not surprisingly, a number of descriptors have been proposed including intrinsically disordered, natively unfolded, or partially folded [3]. For the purpose of this review, we will define intrinsically disordered proteins as those that show substantial disordered regions at least in vitro. Furthermore, we would like to introduce the term conditionally disordered (see Glossary) to describe those proteins that are intrinsically disordered under some circumstances and gain order under others, such as in the presence of their physiological binding partners. These proteins make up a large percentage and perhaps even the majority of intrinsically disordered proteins [4].
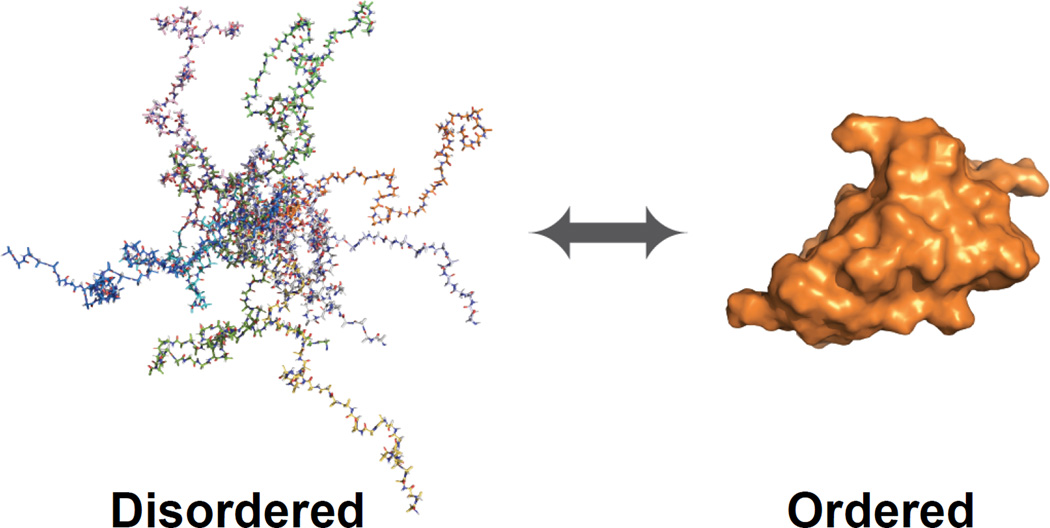
Figure 1.
Proteins have wide ranges of flexibility that span a continuous spectrum including those that are globally intrinsically disordered (shown as an ensemble of structures generated by the program flexible-meccano) [77] to proteins that show well-folded, stable structures (shown in orange). Many proteins are conditionally disordered and can transition between order and disorder depending on the presence or absence of binding partners or other conditions (shown by the double headed arrow). Proteins can also show portions that are well folded and other portions that are disordered. This figure was generated by Loic Salmon.
Intrinsic disorder appears to be very common within proteins. Between 30 and 50% of eukaryotic proteins are estimated to contain regions of >30 amino acids that do not adopt a defined secondary structure in vitro, and many proteins have been predicted to be completely unstructured [5–7]. Computational methods to predict disorder are increasingly becoming more sophisticated [8–10]. However, verifying the (un)folding status of proteins, particularly within the context of the cell, remains challenging. It is currently unclear how many of the proteins that have been experimentally shown to be partially or fully unfolded in vitro are indeed unstructured in the cell. Molecular crowding and the presence of appropriate binding partners are thought to shift the equilibrium of many disordered proteins towards their folded state, suggesting that the percentage of globally intrinsically disordered proteins estimated from in vitro and bioinformatics-based approaches might actually be significantly lower in the cell [11]. This conclusion is supported by the observation that although intrinsically disordered proteins are inherently sensitive to proteolytic degradation in vitro, they do not generally exhibit reduced half-lives in vivo, possibly because they are involved in stabilizing interactions in the cell that might decrease the extent of their disorder [12, 13].
Thus, although tremendous progress has been made in computationally distinguishing intrinsically disordered regions (IDRs) from ordered regions [8–10] careful in vitro and in vivo studies are still needed on an individual protein basis to verify computational predictions of the order/disorder status of proteins. Various approaches can provide experimental information on protein disorder [14], with nuclear magnetic resonance (NMR) being arguably unrivaled in its ability to provide detailed residue-by-residue information on the extent of disorder through chemical shift, residual dipolar coupling and paramagnetic resonance enhancement measurements [15, 16]. Significant progress has also been made in developing in-cell NMR spectroscopy techniques, culminating recently in the first de novo protein structure determinations within living Escherichia coli cells [17]. These and other in-cell techniques, such as SUPREX (stability of unpurified proteins from rates of H/D exchange) [18] will very likely be instrumental in establishing the true in vivo extent of disorder within proteins.
Is protein disorder the default?
In viewing well-ordered crystal structures of proteins, it is easy to forget that disorder is the mathematical default for peptide sequences. Lau and Dill calculated that a maximum of only about 1 in 1010 random sequences are expected to fold into a defined structure [19]. The observation that the majority of proteins show at least some regions of ordered structure indicates that order is strongly selected for in evolution. This finding has led to the ‘form dictates function’ axiom, which has guided biological research for many decades. Based on the same rationale, however, regions of proteins that are not functionally or structurally important can be expected to rapidly devolve into disorder. Most mutations are destabilizing, therefore one possibility is that protein disorder is simply a negative consequence of the random mutations that proteins are subjected to during their evolution [20]. Thus, even the frequent occurrence of disordered regions within proteins does not necessarily establish their relevance for the function of proteins. However, it has long been recognized that disorder might provide functional advantages by allowing the atomic fluctuations that can enhance binding plasticity, enzymatic catalysis, and even allosteric coupling [4, 21, 22]. As such, disorder might play a role in molecular recognition and cellular signaling [21–23]. Moreover, disorder also decreases stability and increases conformational entropy and flexibility [15]. Thus, protein disorder might play a helpful role in in vivo regulation, for instance, by facilitating proteolysis. Finally, recent molecular dynamics and simulation studies on aggregation-prone plant hydrophobins also suggested that the conformational entropy conferred by disordered regions decreases the propensity of proteins to self-aggregate [24]. It is hypothesized that intrinsically disordered regions can prevent unwanted aggregation processes within the crowded environment of the cell while fostering controlled fibril formation in distinct environments, such as at air-water interfaces [24]. These studies and many other examples discussed in recent reviews [2, 25] argue that disorder is not simply the result of the strong destabilizing tendency of mutations. However, perhaps because of the difficulties in studying disorder, direct demonstrations of functional roles for disorder in proteins are less numerous than inferred roles for disorder.
Conditionally disordered proteins
In our view, the concept of conditional disorder provides an important way to reconcile the increasing evidence for the various roles that disorder plays in biological systems with the difficulty in understanding how a completely disordered protein can carry out a specific biological function. Conditionally disordered proteins can exist in a least two states, one that shows a high degree of flexibility and a second state where the protein is more ordered (Figure 2). The ability to study proteins in both states provides a valuable opportunity to establish cause and effect relationships between disorder and function. Many disordered proteins refold once they bind their partners, such as DNA, proteins, or membranes [4, 26], which is likely guided by thermodynamic principles that dictate that binding will stabilize both interaction partners in direct proportion to the strength of the binding interaction [27]. In addition, order-to-disorder-to-order transitions can occur as part of the catalytic cycle of enzymes [28–31]. Proteins that display a single interface that engages multiple binding partners provide particularly attractive example of the concept of conditional disorder. Binding surfaces that are disordered prior to binding seem much better poised to fold into multiple distinct conformations upon binding to different partners than binding surfaces that are well-organized [4]. Showing that a disordered protein acquires multiple distinct conformations upon binding to different partners would thus provide convincing evidence that disorder (or at least the ability to exist in an ensemble of conformations) is functionally important. Two somewhat related and potentially synergistic models have been proposed to describe how a partially unfolded surface regains structure upon binding to different partners (Figure 2). The ‘conformational selection hypothesis’ proposes that by binding, different partners are able to stabilize different members of the conformational ensemble [4, 22, 32]. Alternatively, the ‘folding upon binding’ model proposes that proteins may fold into different conformations upon binding different partners.
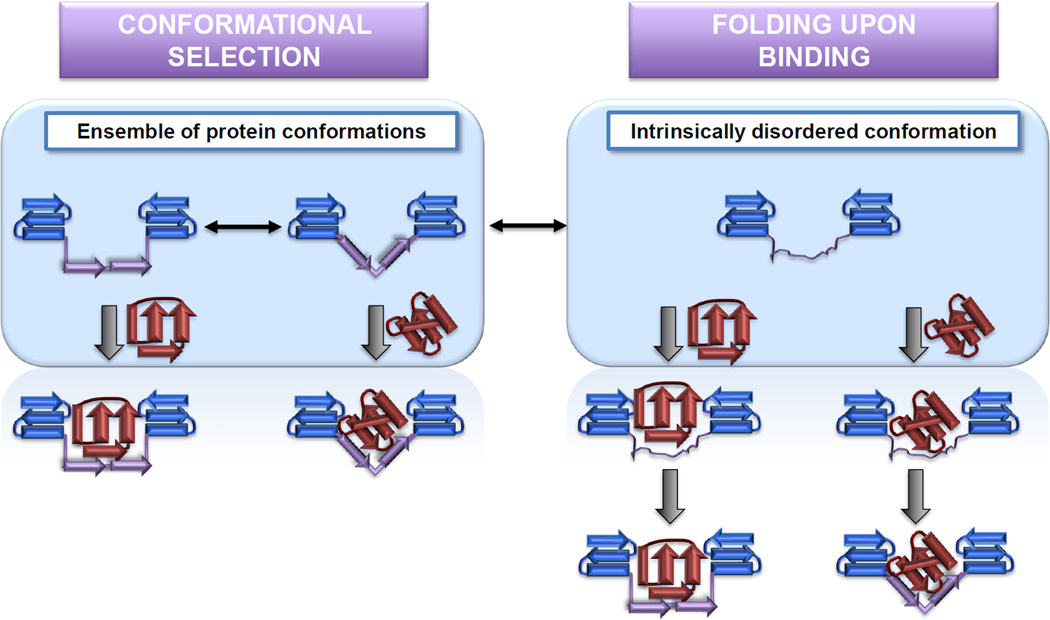
Figure 2.
Molecular recognition mechanisms in conditionally disordered proteins. Two models, ‘conformational selection’ and ‘folding upon binding’, are currently being discussed to explain the observation that intrinsically disordered proteins regain structure in the presence of their binding partners. The conformational selection theory proposes that a small proportion of the intrinsically disordered protein population (shown in blue and purple) is in an appropriate configuration to interact with specific binding partners (shown in red). This interaction then shifts the equilibrium towards the binding-competent conformation. The folding upon binding model proposes that the intrinsically disordered region first binds to the partner and then subsequently refolds. As indicated by the central arrow, components of both mechanisms might contribute to the coupled folding/binding events of conditionally disordered proteins [25].
Various short peptides derived from the disordered C terminus of p53 take on distinct conformations, in a chameleon-like fashion, when bound to various clients [33]. This result is consistent with a functional role for the disorder in the unliganded C terminus of p53, which is known to interact with over 40 different binding partners [33]. However, at this point it is unclear whether the C-terminal region of p53 will adopt the same set of distinct conformations in the context of the full-length protein. Moreover, true chameleon-like behavior for intrinsically disordered proteins still appears to be rare [26]. That only a few examples have been discovered so far might be due to the multiple challenges involved: (1) the identification of structurally different binding partners for the same intrinsically disordered protein, and (2) the solution of the structures of these complexes.
Molecular chaperones: prototypes of proteins with multiple binding partners
Proteins that engage in multiple mutually exclusive transient interactions have on average a higher degree of disorder than non-hub proteins [34, 35]. This also applies to molecular chaperones, which act to bind to a large variety of different protein folding intermediates to prevent their non-specific protein aggregation and facilitate protein folding both in vitro and in vivo. Their predicted degree of disorder ranges from 24–100% [36, 37]. Folding chaperones (i.e., foldases), such as Hsp70 (E. coli DnaK), Hsp60 (E. coli GroEL), and Hsp90 (E. coli HtpG), undergo large conformational rearrangements that modulate client-protein interactions, which are driven by ATP-binding and -hydrolysis as well as co-chaperones [38–40]. Some of these conformational changes have been associated with intrinsically disordered regions. In Hsp70, for instance, the linker between the nucleotide binding domain and the client-binding domain is highly flexible, allowing extensive inter-domain conformational changes [38, 41, 42]. The C terminus of Hsp70 is also disordered, but its exact role in chaperone activity remains controversial. Smock et al. presented evidence that the C terminus of Hsp70 encodes a weak auxiliary peptide binding motif that is important for chaperone activity in vitro [43]. However, Aponte et al. performed an in vivo selection for enhanced chaperone activity and found that truncation of 35 residues from the disordered C terminus of Hsp70 actually increased in vivo and in vitro chaperone activity [44]. One explanation for these apparently conflicting results is that chaperones bind their own disordered regions and thus might treat themselves as substrates [43]. Disordered regions in chaperones like Hsp70 would thus be acting as covalently linked inhibitors. The GroEL (Hsp60) 14-mer contains C termini that are disordered in the ATP-free apo-GroEL but are more ordered in the ATP-bound form [40]. The functional consequences of this apparent disorder-to-order transition remain to be elucidated.Hsp90 harbors several unstructured regions that appear to play important roles in its chaperone activity by providing interdomain flexibility and by conferring solubility to Hsp90-client complexes [45, 46]. Moreover, several of the phosphorylation sites on Hsp90 are located in regions that are disordered and potentially involved in order-to-disorder transitions during its ATP binding and hydrolysis cycle [47].
It is evident from these studies that intrinsic disorder contributes to the chaperone function of ATP-dependent chaperones by promoting the dynamic conformational rearrangements necessary for client protein maturation. It remains to be determined, however, whether the disordered regions in these ATP-dependent chaperones are also directly involved in chaperone-client interactions. In summary, despite extensive research spanning almost 25 years, a clear role for the disordered regions in ATP-dependent chaperones has been difficult to establish. This is likely due to their often very complex quaternary structure (many function as very high molecular weight oligomers) and because interactions between chaperones and their partially-folded client proteins are technically challenging to resolve. Only recently, however, a group of small, ATP-independent, highly promiscuous chaperones have been identified, whose stress-specific activation converts fully folded proteins into intrinsically disordered chaperones. Their study might now open the door to elucidate the role of intrinsic disorder in protein function.
ATP-independent chaperones: activation by stress-induced unfolding
Chaperones that need to work independently of ATP, either because they work in ATP-free cellular compartments (e.g., periplasm) or they function specifically under ATP-depleted stress conditions (e.g., oxidative stress) [48], require alternative means to control substrate binding and release. Recent work has revealed several ATP-independent chaperones that use large-scale order-to-disorder transitions to trigger activation and client binding and employ disorder-to-order transitions to control client release. These chaperones make particularly attractive models for studying the relationships between disorder, client specificity, and protein activity.
Extreme examples of chaperones that undergo activation by protein unfolding are the acid-activated chaperone HdeA [49] and the redox-regulated chaperone Hsp33 [50]. The stress conditions that activate these chaperones (low pH and severe oxidative stress, respectively) cause widespread protein unfolding and lead to the inactivation of numerous proteins. HdeA and Hsp33 are also unfolded by these stress conditions, yet in a clever twist, their unfolding triggers the activation of their chaperone function. This mode of posttranslational regulation is orders of magnitude faster than transcription or translation, which are reduced under both low pH [51] and oxidative stress conditions [52]. The ability to utilize stress-induced order-to-disorder transitions as a mode of activation makes these chaperones thus uniquely suited to serve as a first line of defense against stress conditions that lead to widespread protein unfolding.
HdeA: an acid-activated conditionally disordered chaperone
When E. coli is ingested by humans, the pH of its periplasm rapidly equilibrates with the pH of the stomach (~pH 2). This would cause the irreversible acid denaturation and inactivation of most proteins were it not for the action of the periplasmic chaperone HdeA [53]. At pH 7, HdeA is an abundant, well-folded dimer with no discernable chaperone function. Upon shifting to pH 2, however, HdeA very rapidly (0.125 seconds) partially unfolds and monomerizes, and within less than 2 seconds it becomes active as a chaperone [49, 54]. The partially disordered nature of HdeA at low pH is thought to be vital for its ability to flexibly interact with a wide variety of substrates, which protects proteins against irreversible damage and thereby shields E. coli from the harmful effects of stomach acid.
HdeA binds tightly to its client proteins during the entire time E. coli is exposed to stomach acid because of a disordered hydrophobic surface that becomes exposed upon activation. Using Förster resonance energy transfer (FRET) to monitor HdeA-protein complexes, it was shown that acid-unfolded HdeA flexibly adopts different conformations when bound to different substrate proteins [49]. The importance of conformational flexibility in HdeA-client protein interactions suggests that HdeA could indeed have chameleon-like binding properties. Upon transition from the low pH encountered in the stomach to the neutral pH in the small intestine, HdeA very slowly releases its client proteins in a folding-competent form [54]. The slow release appears to keep the concentration of aggregation-sensitive folding intermediates low, allowing its client proteins to passively refold and disfavoring aggregation (Figure 3). HdeA is a 10 kDa protein and hence well-suited to a variety of spectroscopic techniques, providing a unique opportunity to monitor conformational changes in both HdeA and its clients in response to simple pH shifts; these studies should provide crucial clues about the role of order-to-disorder transitions in both client binding and chaperone action.
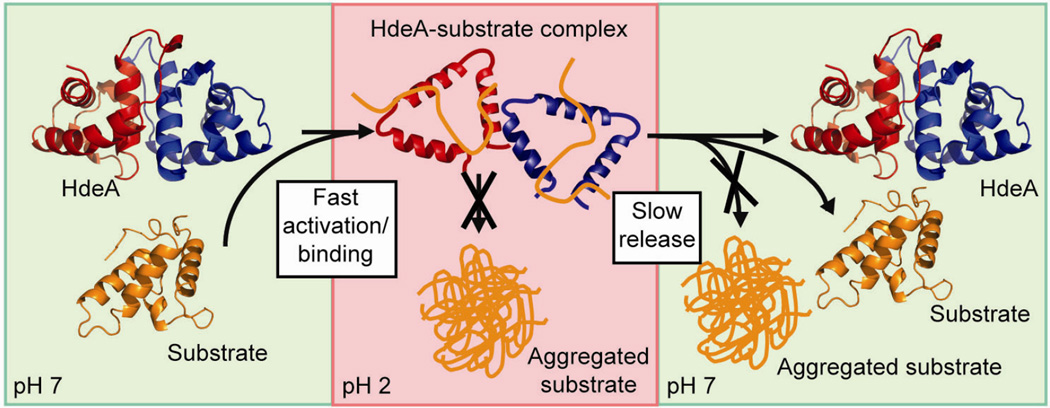
Figure 3.
Mechanism of action of the flexible protein HdeA. At neutral pH (pH 7, left panel), HdeA exists as an inactive dimer. When exposed to acid (pH 2, center panel), HdeA, like many other proteins, unfolds and becomes more flexible. Although this unfolding process inactivates most other proteins, it triggers the activation of HdeA. The flexibility of HdeA allows it to mold itself to fit other proteins that have become aggregation-prone by acid-induced unfolding, thereby preventing their aggregation. HdeA slowly releases proteins after shifting to neutral pH (pH 7, right panel), keeping the concentration of aggregation-sensitive folding intermediates low.
Hsp33: an oxidative stress-activated intrinsically disordered chaperone
Like HdeA, the redox-regulated chaperone Hsp33 is compactly folded under non-stress conditions. In contrast to HdeA, however, inactive Hsp33 is a monomeric two-domain protein. Moreover, it contains four absolutely conserved cysteines in the far C terminus that are engaged in the tetrahedral, high-affinity binding of a single zinc ion (Figure 4). Zinc binding stabilizes both the far C terminus of Hsp33 and an adjacent meta-stable linker region, which folds on top of the N-terminal domain [55]. Exposure to oxidizing conditions that cause protein unfolding (i.e., bleach stress, oxidative heat stress) leads to the rapid formation of two intramolecular disulfide bonds, concomitant with zinc release; destabilization of the zinc binding domain; unfolding of the adjacent linker region; and dimerization [48, 55]. Unfolding of the linker region appears to be crucial for activation, as a single mutation that exclusively alters the folding status of the linker is sufficient to constitutively activate the chaperone function of Hsp33 [56]. Once activated, Hsp33 protects hundreds of cytosolic client proteins against stress-induced protein aggregation and shields bacteria, including E. coli and Vibrio cholerae, against the antimicrobial oxidant bleach [48].
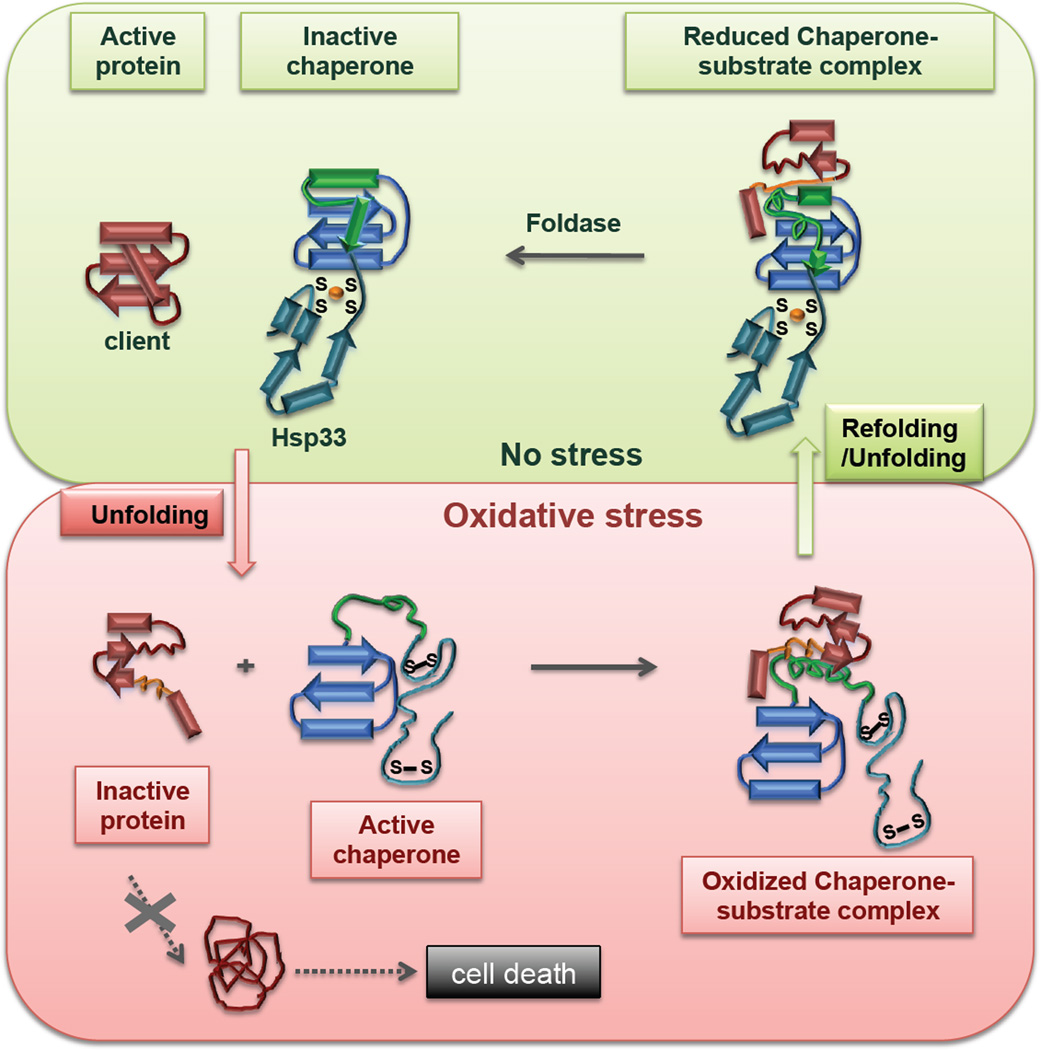
Figure 4.
Mechanism of Hsp33, a redox-regulated chaperone. Under non-stress conditions, Hsp33 is a chaperone-inactive, zinc-binding monomer (upper left). Upon exposure to oxidative stress conditions that cause protein unfolding (e.g., bleach stress, oxidative heat stress), Hsp33 undergoes massive conformational rearrangements, triggered by the formation of two intramolecular disulfide bonds and zinc release (orange ball). This leads to unfolding of the C-terminal redox switch domain (cyan and green), and the activation of Hsp33 as chaperone (dimerization of Hsp33 has been omitted for simplicity). The linker region (green) of Hsp33 appears to directly interact with secondary structure elements of newly unfolded client proteins (red), adopting a more stabilized conformation in this process (lower right). Return to reducing, non-stress conditions causes refolding of the Hsp33 zinc-binding domain (cyan), which appears to trigger conformational changes in the client proteins (orange), consistent with increased client unfolding (upper right). Client proteins are then released to cellular foldases (e.g., the DnaK-system), which are known to bind unstructured folding intermediates, and support the refolding of client proteins to their native state.
Recent hydrogen/deuterium exchange experiments, combined with peptic digests and mass spectrometric analysis, suggested that Hsp33 uses its intrinsically disordered linker region to interact with protein folding intermediates that contain substantial amounts of secondary structure [57]. Stabilization of the intrinsically disordered region of Hsp33 upon binding to secondary structure elements in client proteins appears to contribute to its high affinity for structured folding intermediates and provides insight into how chaperones might select their client proteins. It is currently unclear how Hsp33 distinguishes between native proteins and partially-unfolded proteins, both of which obviously contain secondary structure motifs. Recognition of hydrophobic motifs is likely to be involved. Whether this additional binding occurs at sites distinct from the intrinsically disordered regions or via small ordered segments (i.e., recognition modules) interspersed in the natively disordered C-terminal redox switch domain (analogous to the natively disordered ubiquitin ligase San1 [58]) remains to be tested.
Structural analysis of the client proteins in complex with Hsp33 during preparation for substrate release (i.e., return to reducing non-stress conditions) revealed extensive conformational changes in both the chaperone and client protein, consistent with the partial refolding of the zinc binding domain in Hsp33 and further destabilization of the client proteins [57]. These results are reminiscent of the ‘entropy transfer model’ [36], which proposes that the entropic costs of refolding the intrinsically disordered regions in chaperones upon client binding might be used to unfold client proteins. Yet in the case of Hsp33, unfolding of the client proteins appears to be triggered primarily by the refolding of Hsp33 upon return to non-stress conditions. Destabilization of the client proteins might either be due to an active ratchet-like unfolding process, which would imply very stable interactions between Hsp33 and parts of the client proteins, or more likely, by a passive capturing mechanism, in which the more stabilized linker region captures a more unfolded state of the client protein. Unfolding of the client proteins likely decreases the affinity of Hsp33 for them, hence facilitating their release and preparing them for transfer to ATP-dependent chaperone foldases such as the Hsp70 (DnaK) systems (Figure 4), which are known to preferentially bind to extended polypeptide chains [57, 59]. Thus although the role of disorder in the mechanistic cycle of ATP-dependent chaperones is still unclear, disorder clearly plays an important role in the function of the ATP-independent chaperones. Other examples include the chaperone activity of small heat shock proteins as well as the chaperone-like activity of several globally disordered proteins as outlined in the next section.
The role of intrinsic disorder in small heat shock proteins
Yeast Hsp26 is another example of an ATP-independent chaperone that utilizes specific stress conditions to trigger the conformational changes needed to activate its chaperone function. Hsp26, a member of the small heat shock protein (sHsp)/alpha-crystallin family, is chaperone-inactive at 25°C but readily interacts with client proteins upon incubation at heat shock temperatures [60]. Biophysical studies revealed that Hsp26 uses the folding status of a unique thermo-sensing middle domain, whose heat shock-induced order-to-disorder transition appears to confer conformational changes in Hsp26 that are necessary for its improved chaperone function [61]. Yet, unlike HdeA and Hsp33, this region does not appear to be directly involved in client binding. Instead, photo-and chemical crosslinking studies using different members of the sHsp family indicate that the intrinsically disordered N terminus of small heat shock proteins interacts with client proteins, allowing these chaperones to present a diverse assembly of hydrophobic residues for their promiscuous interaction with different client proteins [62]. These N-terminal regions are, however, not the exclusive interaction sites of sHsps, as other regions on sHsps/alpha-crystallins have also been implicated in client binding [63]. The role of disorder in the sHSPs has been reviewed very recently [64].
Globally intrinsically disordered chaperones and molecular shields
Three other proteins with anti-aggregation activity (casein [65], late embryogenesis abundant (LEA) dehydration proteins [66], and α-synuclein [67]) appear to be globally disordered in vitro. Both casein and LEA proteins have been proposed to function as small ‘cageless’ chaperones, which act via transient hydrophobic interactions to shield aggregation-prone surfaces and increase refolding rates [68]. The LEA proteins are small highly hydrophilic proteins that appear to use physical interference to reduce intermolecular cohesion rates. They protect against dehydration-mediated protein inactivation and, potentially via the same mechanism, against temperature-mediated protein inactivation and aggregation in vitro [66]. Intriguingly, although disordered under standard buffer conditions, LEA proteins have a strong tendency to adopt a structured, α-helical configuration when dehydrated [69, 70]; that is, under one of the conditions in which they are typically active as chaperones. Some LEA proteins have chaperone activity approaching that of the well-established chaperone Hsp90, whereas others have activities comparable to that of BSA, a protein which, though often used as chaperone-inactive control in chaperone assays, has been reported to have measurable chaperone activity when used at very high concentrations [71]. α- and β-casein, long considered prototypes of natively unfolded proteins, inhibit the aggregation of a variety of different proteins in vitro [72]. It is postulated that caseins form soluble, high molecular weight micellar complexes with client proteins, which helps them prevent client protein precipitation. Caseins, however, cannot be considered folding chaperones as they actively inhibit lysozyme refolding and are unable to prevent heat-induced activity loss of catalase or alcohol dehydrogenase [73]. A third globally intrinsically disordered protein, α-synuclein [74] inhibits heat-induced aggregation of several model substrates in vitro, albeit with efficiencies that are about 160- to 350-fold lower than that of the well-established small heat shock protein Hsp27 [75].
Although the stress-mediated induction of LEA proteins points to an in vivo role in protein folding, it is unclear at this point if casein or α-synuclein play any role as chaperones in vivo. We speculate that the relative inefficiency of α-synuclein, casein, and the LEA proteins as molecular chaperones might reflect a common mode of action, inhibiting aggregation by physically surrounding folding intermediates with a molecular shield that prevents them from interacting with other aggregation-sensitive entities. Chaperones like GroEL appear to function in the same basic way, surrounding folding intermediates with a relatively passive cage that inhibits aggregation [76]. However, GroEL has also evolved highly regulated cycles of client binding and release that dramatically increase its efficiency as a chaperone. LEA proteins and casein might compensate for their relative inefficiency by being present in high concentrations.
Concluding remarks
Establishing specific roles for disorder in protein function is experimentally very challenging. However, demonstrating that the disordered regions of a protein can assume multiple distinct conformations upon binding to different partner proteins would provide convincing evidence of an important functional role for disorder. The recent discovery of conditionally disordered molecular chaperones provide us now with excellent test cases, as these folding helper proteins can be studied in both their ordered and disordered states and must be able to interact with multiple partners in order to function. Results obtained from the chaperone studies described here have shown that conditional disorder is important for client recognition. Accumulating further evidence that disordered regions within these chaperones are both directly and indirectly involved in substrate binding will provide an important route to establishing a functional role for protein disorder (Box 1).
BOX 1: Outstanding Questions.
When is disorder selected for by function and when is it the inevitable result of random destabilizing mutations?
What is the extent of protein disorder within the cell?
Are all intrinsically disordered proteins actually conditionally disordered?
Does chaperone function generally depend upon conditional disorder?
Acknowledgements
We are indebted to Drs. Dana Reichmann, Linda Foit and Loic Salmon for helping us preparing some of the figures of this manuscript, and for many useful comments on the manuscript. This work was supported by a National Institutes of Health RO1 GM065318 award (to U.J.). J.C.A.B. is a Howard Hughes Medical Institute Investigator.
Glossary
- BSA
bovine serum albumin, a commonly used negative control protein in chaperone assays due to its very weak chaperone activity.
- Chaperone
a protein that assists the non-covalent folding of proteins and the assembly or disassembly of other macromolecular structures. Its most common characteristic is its ability to dramatically reduce non-specific protein aggregation, a side reaction of protein folding and unfolding processes.
- Conditionally disordered proteins
proteins that can exist in a least two states, one that shows a high degree of flexibility and a second state where the protein is more ordered.
- Conformational selection
molecular recognition mechanism based on the assumption that a proportion of the population of a protein is in an appropriate configuration to interact with its binding partner.
- DnaK (Hsp70)
the eubacterial heat shock protein chaperone homologous to eukaryotic Hsp70, which uses ATP binding and hydrolysis to support the folding of newly synthesized proteins.
- Folding upon binding
molecular recognition mechanism that assumes an initial interaction between a flexible protein and its binding partner, followed by a folding reaction that increases the strength of the interaction.
- FRET
Förster resonance energy transfer, a commonly used method to measure molecular distances within and between proteins.
- GroEL(Hsp60)
a bacterial heat shock protein chaperone homologous to eukaryotic Hsp60, one of the best-characterized ATP-dependent protein folding machines.
- HdeA
a small bacterial acid-activated molecular chaperone, which protects proteins against low pH-induced protein aggregation and bacteria against acid-stress.
- Hsp26
a member of the small heat shock protein (sHsp) chaperone family, which functions as ATP-independent chaperone holdase.
- Hsp33
a ~33 kDa redox regulated heat shock protein chaperones, which is rapidly activated by oxidants such as hypochlorous acid (i.e., bleach) and protects proteins against oxidative unfolding.
- Hsp90
a family of eukaryotic heat shock protein chaperones that have an average molecular weight of ~90 kDa. This class of chaperones interacts with a large number of partially folded proteins, including kinases, steroid receptors and p53.
- HtpG
the bacterial heat shock protein chaperone homologous to eukaryotic Hsp90.
- IDR
intrinsically disordered region.
- Intrinsically disordered proteins
proteins that lack a stable structure when studied as an isolated polypeptide chain under reasonably physiological conditions in vitro.
- LEA
late embryogenesis abundant dehydration proteins in plants and animals, which protect other proteins against desiccation or osmotic stresses.
- p53
a tumor suppressor protein that is involved in cell cycle regulation.
- SH3 domain
Src homology domain 3, a small ~60 amino acid domain to which proline-rich sequences bind.
- sHsp
a family of small heat shock protein chaperones with molecular weights ranging from 12 to 26 kDa.
- SUPREX
stability of unpurified proteins from rates of H/D exchange; an H/D exchange method typically combined with mass spectrometric analysis, which is used to determine relative thermodynamic stabilities of proteins or protein-ligand complexes.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Le Gall T, et al. Intrinsic disorder in the Protein Data Bank. Journal of biomolecular structure & dynamics. 2007;24:325–342. doi: 10.1080/07391102.2007.10507123. [DOI] [PubMed] [Google Scholar]
- 2.Uversky VN. Intrinsically disordered proteins from A to Z. Int J Biochem Cell Biol. 2011;43:1090–1103. doi: 10.1016/j.biocel.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 3.Cortese MS, et al. Intrinsic disorder in scaffold proteins: getting more from less. Progress in biophysics and molecular biology. 2008;98:85–106. doi: 10.1016/j.pbiomolbio.2008.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tompa P, Fuxreiter M. Fuzzy complexes: polymorphism and structural disorder in protein-protein interactions. Trends Biochem Sci. 2008;33:2–8. doi: 10.1016/j.tibs.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 5.Tompa P, et al. Prevalent structural disorder in E. coli and S. cerevisiae proteomes. J Proteome Res. 2006;5:1996–2000. doi: 10.1021/pr0600881. [DOI] [PubMed] [Google Scholar]
- 6.Dunker AK, et al. Intrinsic protein disorder in complete genomes. Genome informatics. Workshop on Genome Informatics. 2000;11:161–171. [PubMed] [Google Scholar]
- 7.Ward JJ, et al. Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. Journal of molecular biology. 2004;337:635–645. doi: 10.1016/j.jmb.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 8.Deng X, et al. A comprehensive overview of computational protein disorder prediction methods. Mol Biosyst. 2012;8:114–121. doi: 10.1039/c1mb05207a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dosztanyi Z, et al. Bioinformatical approaches to characterize intrinsically disordered/unstructured proteins. Brief Bioinform. 2010;11:225–243. doi: 10.1093/bib/bbp061. [DOI] [PubMed] [Google Scholar]
- 10.Ghalwash MF, et al. Uncertainty analysis in protein disorder prediction. Mol Biosyst. 2012;8:381–391. doi: 10.1039/c1mb05373f. [DOI] [PubMed] [Google Scholar]
- 11.Szasz CS, et al. Protein disorder prevails under crowded conditions. Biochemistry. 2011;50:5834–5844. doi: 10.1021/bi200365j. [DOI] [PubMed] [Google Scholar]
- 12.Yen HC, et al. Global protein stability profiling in mammalian cells. Science. 2008;322:918–923. doi: 10.1126/science.1160489. [DOI] [PubMed] [Google Scholar]
- 13.Suskiewicz MJ, et al. Context-dependent resistance to proteolysis of intrinsically disordered proteins. Protein Sci. 2011;20:1285–1297. doi: 10.1002/pro.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Receveur-Brechot V, et al. Assessing protein disorder and induced folding. Proteins. 2006;62:24–45. doi: 10.1002/prot.20750. [DOI] [PubMed] [Google Scholar]
- 15.Brzovic PS, et al. The acidic transcription activator Gcn4 binds the mediator subunit Gal11/Med15 using a simple protein interface forming a fuzzy complex. Mol Cell. 2011;44:942–953. doi: 10.1016/j.molcel.2011.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Salmon L, et al. Measurement and analysis of NMR residual dipolar couplings for the study of intrinsically disordered proteins. Methods in molecular biology. 2012;895:115–125. doi: 10.1007/978-1-61779-927-3_9. [DOI] [PubMed] [Google Scholar]
- 17.Sakakibara D, et al. Protein structure determination in living cells by in-cell NMR spectroscopy. Nature. 2009;458:102–105. doi: 10.1038/nature07814. [DOI] [PubMed] [Google Scholar]
- 18.Ghaemmaghami S, Oas TG. Quantitative protein stability measurement in vivo. Nature structural biology. 2001;8:879–882. doi: 10.1038/nsb1001-879. [DOI] [PubMed] [Google Scholar]
- 19.Lau KF, Dill KA. Theory for protein mutability and biogenesis. Proc Natl Acad Sci U S A. 1990;87:638–642. doi: 10.1073/pnas.87.2.638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.DePristo MA, et al. Missense meanderings in sequence space: a biophysical view of protein evolution. Nat Rev Genet. 2005;6:678–687. doi: 10.1038/nrg1672. [DOI] [PubMed] [Google Scholar]
- 21.Hilser VJ, Thompson EB. Intrinsic disorder as a mechanism to optimize allosteric coupling in proteins. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:8311–8315. doi: 10.1073/pnas.0700329104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen J. Towards the physical basis of how intrinsic disorder mediates protein function. Archives of biochemistry and biophysics. 2012;524:123–131. doi: 10.1016/j.abb.2012.04.024. [DOI] [PubMed] [Google Scholar]
- 23.Sanchez IE, et al. Point mutations in protein globular domains: contributions from function, stability and misfolding. Journal of Molecular Biology. 2006;363:422–432. doi: 10.1016/j.jmb.2006.08.020. [DOI] [PubMed] [Google Scholar]
- 24.De Simone A, et al. Intrinsic disorder modulates protein self-assembly and aggregation. Proc Natl Acad Sci U S A. 2012;109:6951–6956. doi: 10.1073/pnas.1118048109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dyson HJ. Expanding the proteome: disordered and alternatively folded proteins. Quarterly reviews of biophysics. 2011;44:467–518. doi: 10.1017/S0033583511000060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hsu WL, et al. Intrinsic protein disorder and protein-protein interactions. Pac Symp Biocomput. 2012:116–127. [PubMed] [Google Scholar]
- 27.Shriver JW, Edmondson SP. Ligand-binding interactions and stability. Methods Mol Biol. 2009;490:135–164. doi: 10.1007/978-1-59745-367-7_6. [DOI] [PubMed] [Google Scholar]
- 28.Bahar I, et al. Intrinsic dynamics of enzymes in the unbound state and relation to allosteric regulation. Curr Opin Struct Biol. 2007;17:633–640. doi: 10.1016/j.sbi.2007.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bemporad F, et al. Biological function in a non-native partially folded state of a protein. EMBO J. 2008;27:1525–1535. doi: 10.1038/emboj.2008.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Olsson U, Wolf-Watz M. Overlap between folding and functional energy landscapes for adenylate kinase conformational change. Nature Communications. 2010;1:111. doi: 10.1038/ncomms1106. [DOI] [PubMed] [Google Scholar]
- 31.Tsou CL. Active site flexibility in enzyme catalysis. Annals of the New York Academy of Sciences. 1998;864:1–8. doi: 10.1111/j.1749-6632.1998.tb10282.x. [DOI] [PubMed] [Google Scholar]
- 32.Boehr DD, Wright PE. Biochemistry. How do proteins interact? Science. 2008;320:1429–1430. doi: 10.1126/science.1158818. [DOI] [PubMed] [Google Scholar]
- 33.Oldfield CJ, et al. Flexible nets: disorder and induced fit in the associations of p53 and 14-3-3 with their partners. BMC Genomics. 2008;9(Suppl 1):S1. doi: 10.1186/1471-2164-9-S1-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Singh GP, et al. Role of intrinsic disorder in transient interactions of hub proteins. Proteins. 2007;66:761–765. doi: 10.1002/prot.21281. [DOI] [PubMed] [Google Scholar]
- 35.Higurashi M, et al. Identification of transient hub proteins and the possible structural basis for their multiple interactions. Protein Sci. 2008;17:72–78. doi: 10.1110/ps.073196308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tompa P, Csermely P. The role of structural disorder in the function of RNA and protein chaperones. FASEB J. 2004;18:1169–1175. doi: 10.1096/fj.04-1584rev. [DOI] [PubMed] [Google Scholar]
- 37.Tompa P, Kovacs D. Intrinsically disordered chaperones in plants and animals. Biochem Cell Biol. 2010;88:167–174. doi: 10.1139/o09-163. [DOI] [PubMed] [Google Scholar]
- 38.Zhuravleva A, Gierasch LM. Allosteric signal transmission in the nucleotide-binding domain of 70-kDa heat shock protein (Hsp70) molecular chaperones. Proc Natl Acad Sci U S A. 2011;108:6987–6992. doi: 10.1073/pnas.1014448108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Genest O, et al. Heat shock protein 90 from Escherichia coli collaborates with the DnaK chaperone system in client protein remodeling. Proc Natl Acad Sci U S A. 2011;108:8206–8211. doi: 10.1073/pnas.1104703108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Clare DK, et al. ATP-triggered conformational changes delineate substrate-binding and -folding mechanics of the GroEL chaperonin. Cell. 2012;149:113–123. doi: 10.1016/j.cell.2012.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bertelsen EB, et al. Solution conformation of wild-type E. coli Hsp70 (DnaK) chaperone complexed with ADP and substrate. Proc Natl Acad Sci U S A. 2009;106:8471–8476. doi: 10.1073/pnas.0903503106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zuiderweg ER, et al. Allostery in the Hsp70 Chaperone Proteins. Top Curr Chem. 2012 doi: 10.1007/128_2012_323. epubl. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smock RG, et al. Conserved, disordered C terminus of DnaK enhances cellular survival upon stress and DnaK in vitro chaperone activity. J Biol Chem. 2011;286:31821–31829. doi: 10.1074/jbc.M111.265835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aponte RA, et al. Directed evolution of the DnaK chaperone: mutations in the lid domain result in enhanced chaperone activity. J Mol Biol. 2010;399:154–167. doi: 10.1016/j.jmb.2010.03.060. [DOI] [PubMed] [Google Scholar]
- 45.Bron P, et al. Apo-Hsp90 coexists in two open conformational states in solution. Biol Cell. 2008;100:413–425. doi: 10.1042/BC20070149. [DOI] [PubMed] [Google Scholar]
- 46.Wayne N, Bolon DN. Charge-rich regions modulate the anti-aggregation activity of Hsp90. J Mol Biol. 401:931–939. doi: 10.1016/j.jmb.2010.06.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Soroka J, et al. Conformational switching of the molecular chaperone Hsp90 via regulated phosphorylation. Mol Cell. 2012;45:517–528. doi: 10.1016/j.molcel.2011.12.031. [DOI] [PubMed] [Google Scholar]
- 48.Winter J, et al. Bleach activates a redox-regulated chaperone by oxidative protein unfolding. Cell. 2008;135:691–701. doi: 10.1016/j.cell.2008.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tapley TL, et al. Structural plasticity of an acid-activated chaperone allows promiscuous substrate binding. Proc Natl Acad Sci U S A. 2009;106:5557–5562. doi: 10.1073/pnas.0811811106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jakob U, et al. Chaperone activity with a redox switch. Cell. 1999;96:341–352. doi: 10.1016/s0092-8674(00)80547-4. [DOI] [PubMed] [Google Scholar]
- 51.Arnold CN, et al. Global analysis of Escherichia coli gene expression during the acetate-induced acid tolerance response. J Bacteriol. 2001;183:2178–2186. doi: 10.1128/JB.183.7.2178-2186.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shenton D, Grant CM. Protein S-thiolation targets glycolysis and protein synthesis in response to oxidative stress in the yeast Saccharomyces cerevisiae. Biochem J. 2003;374:513–519. doi: 10.1042/BJ20030414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hong W, et al. Chaperone-dependent mechanisms for acid resistance in enteric bacteria. Trends in Microbiology. 2012;20:328–335. doi: 10.1016/j.tim.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 54.Tapley TL, et al. Protein refolding by pH-triggered chaperone binding and release. Proc Natl Acad Sci U S A. 2010;107:1071–1076. doi: 10.1073/pnas.0911610107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ilbert M, et al. The redox-switch domain of Hsp33 functions as dual stress sensor. Nat Struct Mol Biol. 2007;14:556–563. doi: 10.1038/nsmb1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cremers CM, et al. Unfolding of metastable linker region is at the core of Hsp33 activation as a redox-regulated chaperone. J Biol Chem. 2010;285:11243–11251. doi: 10.1074/jbc.M109.084350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Reichmann D, et al. Order out of disorder: working cycle of an intrinsically unfolded chaperone. Cell. 2012;148:947–957. doi: 10.1016/j.cell.2012.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rosenbaum JC, et al. Disorder targets misorder in nuclear quality control degradation: a disordered ubiquitin ligase directly recognizes its misfolded substrates. Mol Cell. 2011;41:93–106. doi: 10.1016/j.molcel.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hoffmann JH, et al. Identification of a redox-regulated chaperone network. EMBO J. 2004;23:160–168. doi: 10.1038/sj.emboj.7600016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Franzmann TM, et al. The activation mechanism of Hsp26 does not require dissociation of the oligomer. Journal of Molecular Biology. 2005;350:1083–1093. doi: 10.1016/j.jmb.2005.05.034. [DOI] [PubMed] [Google Scholar]
- 61.Franzmann TM, et al. Activation of the chaperone Hsp26 is controlled by the rearrangement of its thermosensor domain. Mol Cell. 2008;29:207–216. doi: 10.1016/j.molcel.2007.11.025. [DOI] [PubMed] [Google Scholar]
- 62.Jaya N, et al. Substrate binding site flexibility of the small heat shock protein molecular chaperones. Proc Natl Acad Sci U S A. 2009;106:15604–15609. doi: 10.1073/pnas.0902177106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Basha E, et al. Small heat shock proteins and alpha-crystallins: dynamic proteins with flexible functions. Trends in Biochemical Sciences. 2012;37:106–117. doi: 10.1016/j.tibs.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sudnitsyna MV, et al. The role of intrinsically disordered regions in the structure and functioning of small heat shock proteins. Current protein & peptide science. 2012;13:76–85. doi: 10.2174/138920312799277875. [DOI] [PubMed] [Google Scholar]
- 65.Bhattacharyya J, Das KP. Molecular chaperone-like properties of an unfolded protein, alpha(s)-casein. J Biol Chem. 1999;274:15505–15509. doi: 10.1074/jbc.274.22.15505. [DOI] [PubMed] [Google Scholar]
- 66.Kovacs D, et al. Chaperone activity of ERD10 and ERD14, two disordered stress-related plant proteins. Plant physiology. 2008;147:381–390. doi: 10.1104/pp.108.118208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rekas A, et al. The chaperone activity of alpha-synuclein: Utilizing deletion mutants to map its interaction with target proteins. Proteins. 2012;80:1316–1325. doi: 10.1002/prot.24028. [DOI] [PubMed] [Google Scholar]
- 68.Chakrabortee S, et al. Intrinsically disordered proteins as molecular shields. Mol Biosyst. 2012;8:210–219. doi: 10.1039/c1mb05263b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Haaning S, et al. An unusual intrinsically disordered protein from the model legume Lotus japonicus stabilizes proteins in vitro. The Journal of Biological Chemistry. 2008;283:31142–31152. doi: 10.1074/jbc.M805024200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hundertmark M, et al. Influence of drying on the secondary structure of intrinsically disordered and globular proteins. Biochemical and biophysical research communications. 2012;417:122–128. doi: 10.1016/j.bbrc.2011.11.067. [DOI] [PubMed] [Google Scholar]
- 71.Finn TE, et al. Serum albumin prevents protein aggregation and amyloid formation and retains chaperone-like activity in the presence of physiological ligands. The Journal of biological chemistry. 2012;287:21530–21540. doi: 10.1074/jbc.M112.372961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yong YH, Foegeding EA. Caseins: utilizing molecular chaperone properties to control protein aggregation in foods. Journal of Agricultural and Food Chemistry. 2010;58:685–693. doi: 10.1021/jf903072g. [DOI] [PubMed] [Google Scholar]
- 73.Treweek TM, et al. The chaperone action of bovine milk alphaS1- and alphaS2-caseins and their associated form alphaS-casein. Archives of biochemistry and biophysics. 2011;510:42–52. doi: 10.1016/j.abb.2011.03.012. [DOI] [PubMed] [Google Scholar]
- 74.Fauvet B, et al. Characterization of semisynthetic and naturally Nalpha-acetylated alpha-synuclein in vitro and in intact cells: implications for aggregation and cellular properties of alpha-synuclein. The Journal of biological chemistry. 2012 doi: 10.1074/jbc.M112.383711. epubl. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Souza JM, et al. Chaperone-like activity of synucleins. FEBS letters. 2000;474:116–119. doi: 10.1016/s0014-5793(00)01563-5. [DOI] [PubMed] [Google Scholar]
- 76.Horwich AL, et al. The GroEL/GroES cis cavity as a passive anti-aggregation device. FEBS letters. 2009;583:2654–2662. doi: 10.1016/j.febslet.2009.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ozenne V, et al. Flexible-meccano: a tool for the generation of explicit ensemble descriptions of intrinsically disordered proteins and their associated experimental observables. Bioinformatics. 2012;28:1463–1470. doi: 10.1093/bioinformatics/bts172. [DOI] [PubMed] [Google Scholar]