Summary
Fur (ferric uptake regulator) is the master regulator of iron homeostasis in many bacteria, but how it responds specifically to Fe(II) in vivo is not clear. Biochemical analyses of Bacillus subtilis Fur (BsFur) reveal that in addition to Fe(II), both Zn(II) and Mn(II) allosterically activate BsFur-DNA binding. Dimeric BsFur co-purifies with site 1 structural Zn(II) (Fur2Zn2) and can bind four additional Zn(II) or Mn(II) ions per dimer. Metal ion binding at previously described site 3 occurs with highest affinity, but the Fur2Zn2:Me2 form has only a modest increase in DNA binding affinity (~7-fold). Metallation of site 2 (Fur2Zn2:Me4) leads to a ~150-fold further enhancement in DNA binding affinity. Fe(II) binding studies indicate that BsFur buffers the intracellular Fe(II) concentration at ~1 μM. Both Mn(II) and Zn(II) are normally buffered at levels insufficient for metallation of BsFur site 2, thereby accounting for the lack of crosstalk observed in vivo. However, in a perR mutant, where the BsFur concentration is elevated, BsFur may now use Mn(II) as a co-repressor and inappropriately repress iron uptake. Since PerR repression of fur is enhanced by Mn(II), and antagonized by Fe(II), PerR may co-regulate Fe(II) homeostasis by modulating BsFur levels in response to the Mn(II)/Fe(II) ratio.
Keywords: Fur, metalloregulation, Fe(II) homeostasis
Introduction
Iron is an essential nutrient for nearly all living organisms. However, in aerobic environments, the predominant ferric form is extremely insoluble and thus largely unavailable to most cells. Cells have evolved a variety of iron uptake systems with high affinity and specificity (Andrews et al., 2003). However, the intracellular concentration of iron must be tightly controlled due to the ability of ferrous ion to participate in the Fenton reaction and generate highly toxic hydroxyl radicals (Imlay, 2008).
The master regulator of iron homeostasis in many bacteria, Fur (ferric uptake regulator), was first identified in Escherichia coli as a result of a selection with high Mn(II) three decades ago (Hantke, 1981; Lee and Helmann, 2007). It was proposed that Fur is the primary target of Mn(II) toxicity and Fur can bind Mn(II) as a co-repressor to repress iron uptake regardless of the intracellular Fe(II) level in E. coli. Although it was shown in early studies that Fur responds to Fe(II) to repress gene expression (Bagg and Neilands, 1987), the molecular details of this key sensing step have remained obscure. Bacillus subtilis contains a Fur ortholog (BsFur) that maintains iron homeostasis through the transcriptional repression of iron uptake systems under iron sufficient conditions. BsFur selectively responds to Fe(II) but not Mn(II) in vivo (Baichoo et al., 2002; Gaballa et al., 2008). As a result, selection for Mn(II) resistance does not result in the recovery of fur mutations (unpublished observations).
The ability of metalloregulatory proteins to respond specifically to their cognate metal ions can be governed by chemical or biological factors. Chemically, different metal ions interact with protein binding sites with widely varying affinities that reflect the nature and geometry of the available ligands. Biologically, metal-responsiveness may be governed at the protein level due to ability of some metals, but not others, to trigger the relevant allosteric changes in protein structure. Responsiveness may also be determined at the cellular level by the limited intracellular availability of metal ions. Collectively, these factors (which can be summarized as affinity, allostery, and access) are responsible for metal specificity (Ma et al., 2009; Waldron and Robinson, 2009).
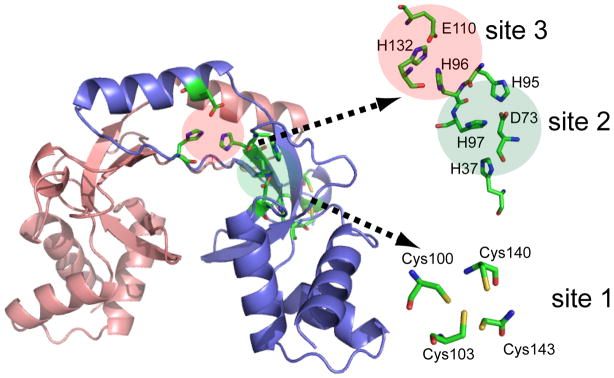
Despite numerous structural studies, the location and affinity of the metal binding site(s) for Fur proteins have been controversial. Crystal structures of Fur and its homologs have revealed up to three distinct metal binding sites (Fig. 1): i) site 1, a Cys4-Zn(II) structural site that is located close to the carboxyl-terminus and consists of two Cys-X-X-Cys motifs; ii) site 2, at the junction of the DNA binding domain and the dimerization domain, and iii) site 3, adjacent to site 2 and also close to the dimer interface (Pohl et al., 2003; Lucarelli et al., 2007; Dian et al., 2011; Shin et al., 2011). While the role of site 1 as a structural site has been widely appreciated (Lee and Helmann, 2006a), the involvement of site 2 and 3 in metal binding and activation of DNA binding is unresolved. It was initially proposed that site 3 was the primary Fe-sensing site, based on structural studies of Pseudomonas aeruginosa Fur (which lacks site 1) (Pohl et al., 2003), but site 2 is the sensing site for the homologous proteins BsZur and BsPerR (Lee and Helmann, 2006b; Ma et al., 2011a). E. coli Fur has been reported to bind Fe(II) with a dissociation constant of 1.2 μM as monitored by intrinsic protein fluorescence, but neither the stoichiometry nor the site(s) of binding were determined (Mills and Marletta, 2005).
Figure 1.
Homology model of BsFur using ScZur structure as template (pdb: 3mwm; sequence identity: 33.3%) (Shin et al., 2011). Three metal binding sites are highlighted with conserved putative metal binding ligands shown in stick configuration. Putative metal binding ligands that are not conserved between ScZur and BsFur are not shown.
Metal sensing sites have been defined both structurally and biochemically for some Fur homologs. B. subtilis encodes PerR and Zur, involved in peroxide and Zn(II) sensing, respectively (Bsat et al., 1998; Gaballa and Helmann, 1998). For both proteins, site 2 is the metal binding site, and no metal binding was detected at site 3 (Ma et al., 2011b; Ma et al., 2011a). In contrast, structural studies of other Fur family proteins have revealed metal occupancy at both sites 2 and 3 (Pohl et al., 2003; Lucarelli et al., 2007; Dian et al., 2011; Shin et al., 2011). However, high and non-physiologically relevant concentrations of metal ions are present during crystallization and the functional relevance of these metal binding sites has not been tested. Recent results with Helicobacter pylori Fur and Streptomyces coelicolor Zur indicate that both site 2 and 3 bind metal ions, with site 2 playing a primary role in metal sensing (Dian et al., 2011; Shin et al., 2011). In these cases, however, neither the total metal binding stoichiometry in solution nor the connection between metal occupancy and activation of DNA binding was determined.
Here, we demonstrate that BsFur, unlike the other two Fur paralogs in B. subtilis, binds metal ions with physiologically relevant affinities at both sites 2 and 3. Further, the full activation of DNA binding corresponds to metal occupancy at both sites, with site 2 likely playing the primary role in metal sensing. The metal selectivity of gene regulation mediated by BsFur is primarily governed by fine-tuning of metal ion binding affinities so that non-cognate metal ions, which may be potent agonists, are normally maintained at levels below those that interfere with regulation by the cognate metal. The regulation of Fur protein level by PerR appears to play a critical role in this process.
Results and Discussion
Metal-dependent DNA-binding by BsFur in vitro
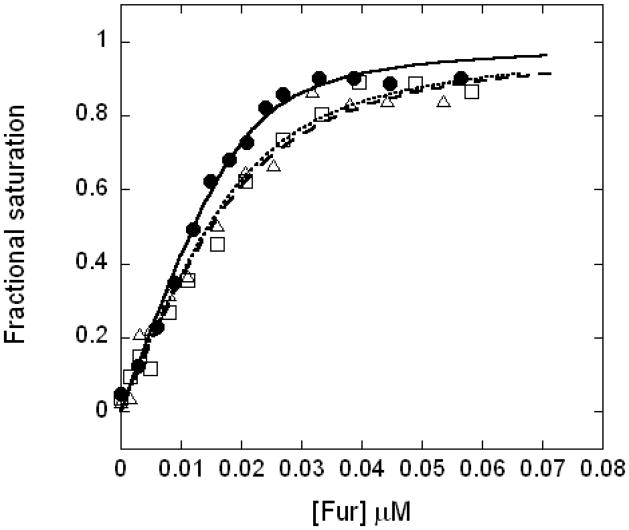
Fe(II), Mn(II), and Zn(II) all activate BsFur-DNA interaction to the same degree as monitored by a fluorescence anisotropy (FA)-based DNA-binding assay (1 mM sodium dithionite was presented to maintain the Fe(II) form in the experiments with iron; see Experimental Procedures). A DNA probe containing the previously characterized Fur box operator site from the feuABC-ybbA operon was used (Baichoo et al., 2002). This operator conforms to the 7-1-7 consensus sequence (with one mismatch) but lacks the extended sequence of the 19 bp Fur box which binds two dimers to two overlapping operators, as previously described (Baichoo and Helmann, 2002). Data fitting revealed DNA binding affinities (Ka) of 9.7 × 108 M−1 with Fe(II), 4.0 × 108 M−1 with Mn(II), and 4.4 × 108 M−1 with Zn(II) (Fig. 2). We conclude that both Mn(II) and Zn(II) are effective at triggering the allosteric changes needed for DNA-binding and are therefore reasonable surrogates for Fe(II) for analyses of metal binding properties. Since Fe(II) is a redox active metal ion, and requires strict anaerobic conditions for in vitro experiments, nearly all reported biochemical studies of Fur proteins have used Mn(II) as the activating cofactor.
Figure 2.
Normalized binding curves of BsFur binding to DNA in the presence of Mn(II) (open square, dashed curve), Zn(II) (open triangle, dotted curve), or Fe(II) (filled circle, solid curve) are shown. Curves represent the best fit to the data.
The fact that DNA binding by BsFur can be activated by either Mn(II) or Zn(II) raises the question of how BsFur achieves an Fe(II)-specific response in vivo. Coordination number and geometry are known to play important roles in dictating the metal specificity of some metalloregulatory proteins (Ma et al., 2009). Mn(II) can often adopt the same coordination geometry as Fe(II) and is therefore commonly used as a surrogate. In contrast, Zn(II) sites are often tetrahedral, although five- and six-coordinated Zn(II) has been observed in crystal structures of Fur proteins (Dian et al., 2011; Pohl et al., 2003). We hypothesized that since coordination chemistry fails to distinguish these metal ions for activation of BsFur in vitro, binding affinity and access to metals within the cellular milieu are likely to be the major determinants of metal specificity in vivo. We have therefore measured the stoichiometry and affinity of the binding interactions between BsFur and metal ions.
Zn(II) binding studies indicate that a BsFur dimer binds a total of six metal ions
Previous structural studies have revealed that Fur family proteins often (but not always) contain a structural Zn(II) site (site 1), and as many as two additional metal ion binding sites: sites 2 and 3 (Fig. 1) (Pohl et al., 2003; Dian et al., 2011; Shin et al., 2011; Lucarelli et al., 2007). Although this suggests a possible stoichiometry of as many as three metals per monomer, this stoichiometry has never been confirmed via direct metal titration in solution. Indeed, BsPerR and BsZur each contain a structural (site 1) Zn(II) and can only bind one additional metal ion at site 2, resulting in a total stoichiometry of two metals per monomer (Ma et al., 2011b; Ma et al., 2011a). Since the metal binding ligands of sites 2 and 3 are also largely conserved in BsFur (Fig. 1), we first sought to address the metal binding stoichiometry and affinity of BsFur.
After purification in the presence of 1 mM EDTA, BsFur contained ~0.9 to 1.0 Zn(II) per monomer. This corresponds to the non-exchangeable structural Zn(II) (site 1), consistent with previous studies indicating that metal at this site cannot be easily removed by chelators such as EDTA and is only removed upon protein unfolding (Ma et al., 2011a; Lee and Helmann, 2006a; Outten et al., 1999). The dimeric protein as purified is designated Fur2Zn2. Correspondingly, Fur2Zn2:Men, [where Me can be Zn(II), Mn(II) or Fe(II)] is used to describe the products of subsequent metal binding events.
We used competition binding experiments with Magfura-2 (Mf2) to monitor the binding of Zn(II) and Mn(II) to BsFur. The UV absorption of Mf2 at both 325 nm and 366 nm changes upon formation of 1:1 complex with Zn(II) or Mn(II) with affinities under our buffer conditions (0.1 mM TCEP, 20 mM Tris, pH 8.0, 0.1 M NaCl for Mn(II) binding or 0.4 M NaCl for Zn(II) binding) of 5.0 × 106 M−1 and 1.9 × 106 M−1, respectively (Fig. S1). Note that these values differ from the previously reported affinities of 5.0 × 107 M−1 for Zn(II) and 1.0 × 106 M−1 for Mn(II), presumably due to different solution conditions (Golynskiy et al., 2006; Timothy J.B, 1993; VanZile et al., 2000).
To monitor Zn(II) binding, we titrated Zn(II) into a mixture of 3.2 μM BsFur dimer and 10.1 μM Mf2. The Mf2 absorbance did not change significantly until about 7 μM Zn(II) was added as indicated by the plateau at the first part of the binding curve (Fig. 3A). This indicates that BsFur can bind 2.0 mol Zn(II) per dimer with much higher affinity than Mf2 (KZn1, KZn2 ≥109 M−1) to generate Fur2Zn2:Zn2. The significant curvature in the second part of the binding curve suggested competition for Zn(II) binding between Fur2Zn2:Zn2 and Mf2. Analysis of these curves revealed that the competition was due to two independent Zn(II) binding events to Fur2Zn2:Zn2, both with 1.0 Zn(II) per dimer stoichiometry. These latter binding events had strong negative cooperativity with affinities of KZn3 = 2.0 × 108 M−1 and KZn4 = 6.9 × 105 M−1, respectively (Table 1). Thus, under these conditions we can sequentially form the Fur2Zn2:Zn2, Fur2Zn2:Zn3, and Fur2Zn2:Zn4 forms of the Fur protein.
Figure 3.
Zn(II) binding to BsFur in competition with Mf2 or Quin-2. (A) Zn(II) titrated into 10.1 μM Mf2 and 3.2 μM BsFur dimer. Changes in absorbance at 325 (circles) and 366 nm (squares) are plotted against Zn(II) concentrations. (B) Zn(II) titrated into 8.0 μM Quin-2 and 2.4 μM BsFur dimer. Changes in absorbance at 261 nm are plotted against Zn(II) concentrations. All curves represent the best fit with parameters compiled in Table 1.
Table 1.
| Fur | Site 3 (Zn) | Site 2 (Zn) | Site 3 (Mn) | Site 2 (Mn) | Site 2 (Fe) | ||
|---|---|---|---|---|---|---|---|
| KZn1 (×1010 M−1) | KZn2 (×1010 M−1) | KZn3 (×106 M−1) | KZn4 (×106 M−1) | KMn1 (×104 M−1) | KMn2 (×104 M−1) | KFed (×104 M−1) | |
| WT | 710 ± 250 | 5.4 ± 1.0 | 200 ± 40 | 0.69 ± 0.08 | 620 ± 40 | 4.2 ± 0.4 | 120 ± 10 |
| H37A (site 2) | 510 ± 210 | 34 ± 7 | 3.5 ± 0.2 | 3.5 ± 0.2 | 230 ± 20 | 0.13 ± 0.04 | 0.6 |
| H95A (site 2) | 1300 ± 900 | 2.3± 0.8 | 190 ± 30 | 3.3 ± 0.2 | 250 ± 30 | n.d. c | n.d. |
| H97A (site 2) | 300 ± 40 | 11 ± 1 | 0.67± 0.08 | 0.1 | 110 ± 10 | n.d. | n.d. |
| H96A (site 3) | 51 ± 4 | 6.4± 0.8 | 15000 ± 3000 | 2.6 ± 0.2 | 31 ± 2 | n.d. | n.d. |
| H132A(site3) | 4.4± 0.7 | 0.023 ± 0.004 | 31 ± 2 | 1.8 ± 0.1 | 22 ± 2 | 5.7 ± 0.2 | 99 ± 20 |
All Zn(II) binding affinities were determined using competition assays with both Mf2 and Quin2. Representative result from multiple experiments (N≥3) is shown. Errors generated by Dynafit are shown to indicate the goodness of fit.
KMn1 was determined by Mf2 competition, while KMn2 was determined by FA-based DNA binding activation assay.
n.d.: not determined
KFe is determined by FA-based assay in 20 mM MOPS, pH 7.5, 0.4 M NaCl, 0.1 mM NAC.
The binding affinities of the first two binding events (KZn1 and KZn2) were further determined using a higher affinity competitor, Quin-2 (KZn = 2.7 × 1011 M−1) (Jefferson et al., 1990) to reveal binding constants of 7.1 × 1012 M−1 and 5.4 × 1010 M−1 for KZn1 and KZn2, respectively (Fig. 3B, Table 1). The binding affinities of the latter two events (KZn3 and KZn4) are below the lower detection limit of this assay and are therefore not observed. Since purified BsFur dimer contained two structural Zn(II) (Fur2Zn2), these data indicate that BsFur can bind a total of 6.0 Zn(II) per dimer (3.0 per monomer) to form Fur2Zn2:Zn4, a stoichiometry consistent with most previous crystal structures of Fur family members. These data support a model in which each BsFur protomer binds Zn(II) at both sites 2 and 3 with negative cooperativity (homotropic negative cooperativity), resulting in four distinct binding affinities as observed (Table 1). Such homotropic negative cooperativity was also noted for Zn(II) binding at site 2 of BsZur dimer (Ma et al., 2011a).
When Mn(II) was titrated into a mixture of BsFur and Mf2, a simpler binding curve resulted. These data were best fit to a competition model with only two identical Mn(II) binding events per dimer with an affinity (KMn1) of 7.3 × 106 M−1 (Fig. 4, Table 1). The homotropic negative cooperativity noted for Zn(II) binding was not observed in the case of Mn(II), for reasons not fully understood. This may be due to a decreased magnitude of cooperativity, which cannot be confidently resolved by the assay, or a total loss of cooperativity. Importantly, the apparent binding stoichiometry of Mn(II) in this assay was distinct from that for Zn(II). We reasoned that since the affinities of the last two Zn(II) binding events to BsFur dimer (KZn3 and KZn4) were relatively low, the affinities of Mn(II) for these same sites were likely to be even lower, in accord with the Irving-William series. In this case, these binding affinities would be below the lower detection limit (~105 M−1) of this competition assay. The presence of such low affinity Mn(II) binding sites was subsequently confirmed by an FA-based assay (see below).
Figure 4.
Mn(II) binding to WT (filled circles, solid curve) or H132A BsFur (open circles, dashed curve) in competition with Mf2. Mn(II) was titrated into 6.8 μM Mf2 mixed with either 5.0 μM H132A or 3.2 μM WT BsFur dimer. Changes in absorbance at 325 (circles) and 366 nm (squares) are plotted. Curves represent the best fit with parameters compiled in Table 1.
Both site 2 and 3 are involved in metal binding
Based on the reported structures of Fur family proteins and our observed metal binding stoichiometry, we hypothesized that BsFur binds metal ions at both site 2 and 3. Metal bound at site 3 was shown to be less easily exchanged in the case of P. aeruginosa Fur (Pohl et al., 2003), consistent with a generally higher affinity for site 3 than site 2 (Fig. 1). In the case of Zn(II), it binds to the equivalent sites in each protomer with negative cooperativity, resulting in four distinct binding affinities (KZn1 to KZn4) (Table 1). BsFur mutants with amino acid substitutions of putative metal binding residues at site 2 (H37A, H95A and H97A) and site 3 (H96A and H132A) (Fig. 1) were purified and characterized to test this hypothesis.
Since we proposed that the Mn(II) binding we observed in the Mf2 competition assay was at the high affinity site, we expected that site 3 but not site 2 mutations would affect this binding. Indeed, the affinity of Mn(II) binding to BsFur was significantly decreased in both site 3 mutant proteins, H96A and H132A. The binding affinities (KMn1) were 3.1 × 105 M−1 and 2.2 × 105 M−1, or ~20 and ~30-fold decreased comparing to WT BsFur, respectively (Fig. 4, Table 1). In contrast, all three site 2 mutants only moderately affected the Mn(II) binding affinity, with the largest decrease of ~6-fold observed with H97A (Table 1). These minor changes are expected since site 2 and 3 are physically close and contain metal binding ligands from a shared loop region (Fig. 1).
We next determined the Zn(II) binding affinities of these mutants using a combination of Quin2 and Mf2 as competitors. As expected, the high affinity binding events (KZn1 and KZn2) of both site 3 mutants were significantly weakened, while the low affinity ones (KZn3 and KZn4) were not. In contrast, only the low affinity Zn(II) binding of some site 2 mutants were substantially weakened (Table 1). This further supports our model that purified BsFur (Fur2Zn2) can bind four additional Zn(II) with distinct affinities in a stepwise manner. These binding studies imply the presence of multiple species of Fur dimer that can be indicated as Fur2Zn2:Men, where n=0 to 4 and represents different metal binding status at sites 2 and 3. Site 3 binds metal ions with generally higher affinity than site 2, and is therefore occupied first.
Further examination of the Zn(II) binding data in Table 1 reveals several other interesting features. Firstly, the Zn(II) binding of the H95A protein was not affected, although H95 was predicted to be a ligand in site 2 (Fig. 1). Therefore, it is possible that H95 is not an important metal binding ligand, at least for Zn(II). Alternatively, a reorganization or recruitment of additional ligands to the coordination sphere may occur to accommodate Zn(II) which thereby results in unchanged affinity. Secondly, some mutations appeared to affect the Zn(II) binding to the two protomers unequally and reduce the magnitude of homotropic negative cooperativity. For instance, site 2 mutation H37A more significantly affected the first binding event at site 2 in a dimer (KZn3) than the second (KZn4), resulting in a loss of negative cooperativity of Zn(II) binding (KZn3≈KZn4). Similar results characterize the site 3 mutant H96A. In contrast, the H97A or H132A mutations appear to perturb the metal binding energy in each protomer equally, resulting in no change in the magnitude of homotropic negative cooperativity (Table 1). Although the detailed mechanism of negative cooperativity is not clear, it is possibly caused by an energetic penalty of the second binding event in the dimer once the equivalent site in the other protomer is occupied. Loss of metal binding ligand H37 or H96 affects the inter-protomer communication, suggesting that they may play roles in this process. Lastly, substitution of a site 3 residue H96 with alanine not only decreased Zn(II) binding at site 3, but also unexpectedly increased the affinity at site 2, at least for KZn3. One possible explanation is that H96 is in between two metal binding ligands (H95 and H97) of site 2 (Fig. 1). Some restraints imposed on this loop region from coordinating metal ion at site 3 are absent in H96A, which could in turn favor metal binding at site 2.
High affinity BsFur-DNA interaction requires metal binding at both site 2 and 3
Next, we sought to determine which of the metal binding events resolved above, involving sites 2 and 3, are required for activation of BsFur-DNA binding. We therefore monitored the DNA binding affinity of WT, site 2, and site 3 mutant proteins using FA under conditions that allow the formation of distinct metallated complexes.
The effect of saturating metal ions on BsFur-DNA binding was first addressed. With 1 mM Mn(II) in solution, BsFur binds the feuA operator DNA with an affinity (KMn4) of 4.0 × 108 M−1, or ~1,000-fold higher than in the absence of Mn(II) (Kapo) (Figs. 2, 5A, and Table 2). Interestingly, all BsFur mutant proteins characterized here can bind DNA with a high affinity similar to WT in the presence of saturating Mn(II) concentrations to form Fur2Zn2:Mn4 (Table 2). This suggests that none of the mutations cause any gross defects in allosteric regulation, although higher metal ion concentrations (10 mM Mn(II) was used in the case of H37A and H97A) may be required. Saturating Zn(II) in the solution led to similar results for WT and some BsFur mutants, but the high concentrations of Zn(II) required to saturate the H37A and H97A proteins caused quenching of the fluorescent probe (data not shown), and therefore the DNA-binding properties of these mutant proteins with Zn(II) were not determined.
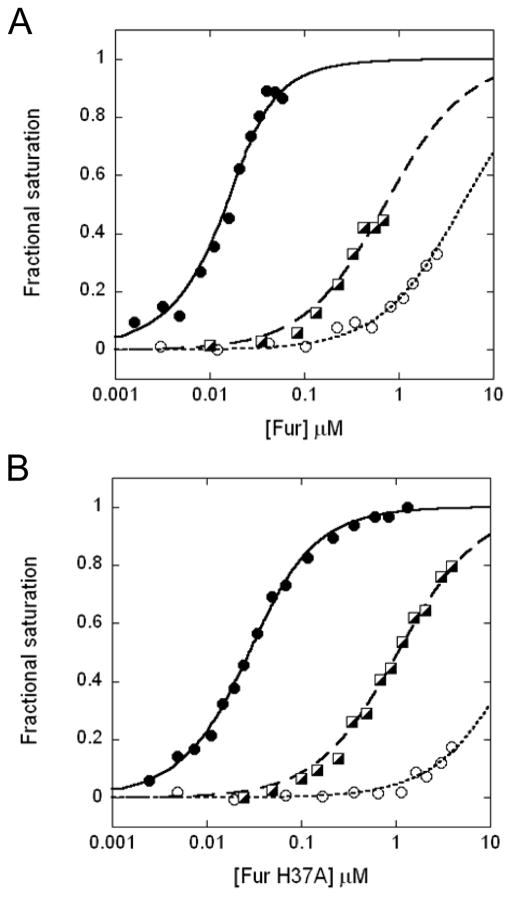
Figure 5.
DNA binding of (A) WT and (B) H37A BsFur. Normalized DNA binding curves of BsFur in presence of saturating Mn(II) conditions (filled circles, solid curve) or 1 mM EDTA (open circles, dotted curve) are shown. The DNA binding of the Fur2Zn2:Zn2 form of both WT and H37A BsFur are also shown (square, dashed curve). Curves represent the best fit with parameters compiled in Table 2.
Table 2.
DNA binding affinities of BsFur WT and mutants.a
| Fur | KMn4 (Fur2Zn2:Mn4) (×106 M−1) | KZn2 (Fur2Zn2:Zn2) (×106 M−1) | Kapo (Fur2Zn2) (×106 M−1) |
|---|---|---|---|
| WT | 400 ±50 | 2.8 ±0.2 | 0.42 ±0.02 |
| H37A (site 2) | 110 ±10 | 1.9 ±0.1 | 0.095 ±0.007 |
| H95A (site 2) | 650 ±40 | n.d. b | 0.67 ±0.02 |
| H97A (site 2) | 210 ±20 | 0.98 ±0.06 | 0.19 ±0.01 |
| H96A (site 3) | 870 ±80 | n.d. | 0.42 ±0.02 |
| H132A(site3) | 160 ±20 | n.d. | 1.3 ±0.1 |
Representative result from multiple FA experiments (N≥3) is shown. Errors generated by Dynafit are shown to indicate the goodness of fit.
n.d.: not determined
We next determined the DNA binding activity of the partially metallated state (Fur2Zn2:Me2), where only site 3 but not site 2 was occupied. Since the affinity of Zn(II) binding was best characterized, a Fur2Zn2:Zn2 form was used. We also included 10 μM NTA (KZn = 8.7 × 108 M−1 at pH 8.0; Martell and Smith, 1979–1989) in the solution to assure that site 2 would not be affected by any trace Zn(II) contamination. Under these conditions, the DNA binding of WT BsFur was only partially activated with KZn2 = 2.8 × 106 M−1, or ~7-fold higher affinity than that of the Fur2Zn2 form (Kapo) (Fig. 5A, Table 2). This suggests that metal binding at site 3 does have a measurable effect on DNA binding. It can also be inferred that metal binding at site 2 further increases the DNA binding affinity by ~150-fold.
The metal binding at site 2 was significantly perturbed in two site 2 mutants, H37A and H97A (Table 1), which further facilitates the specific metal loading at only site 3 in these mutants. Therefore, the DNA-binding affinities of the Fur2Zn2:Zn2 form, with Zn(II) bound at site 3 but not site 2, of both the H37A and H97A proteins were measured. They both bound DNA with intermediate affinity, consistent with the observations with WT BsFur (Fig. 5B, Table 2). We therefore conclude that metal binding at both site 2 and 3 are important for DNA binding, with site 2 playing the predominant regulatory role, consistent with its physical location at the inter-domain hinge region (Fig. 1).
Primary sensing role of site 2 and implications for metal specific responsiveness
In the competition assay with Mf2, we noted different stoichiometries of metal binding with Zn(II) and Mn(II). We hypothesized that the second Mn(II) binding event was not observed due to the low affinity. With the FA-based DNA binding assay, we were able to examine this low affinity Mn(II) binding.
We titrated Mn(II) into a solution containing 10 nM labeled DNA and 50 nM BsFur dimer, where no BsFur:DNA complex should form in the absence of any activating metal ions (Fig. 5B). Under these conditions, the overall equilibrium constant reflects BsFur binding to Mn(II) and DNA to form a ternary complex. Since the affinity of Mn(II)-bound BsFur-DNA interaction (KMn4) is known (Table 2), the affinity of Mn(II)-BsFur interaction can then be resolved. In this assay, we also Chelex-treated both BsFur and the buffer to avoid contamination of trace metal ions and especially the potent agonist Zn(II) (see Experimental Procedures). Due to the high affinity of the Zn(II)-BsFur interaction (Table 1), trace Zn(II) present in the solution may partially activate DNA binding and interfere with the assay. Indeed, Zn(II) contamination likely explains our previous report that Fur binds DNA in the absence of added Fe(II) (Bsat and Helmann, 1999). After Chelex-treatment, no change in FA was observed upon mixing 50 nM BsFur dimer with DNA. Therefore, the subsequent change in FA is fully dependent on Mn(II).
When Mn(II) was titrated into the solution of BsFur and DNA, we noticed a gradual increase of FA, indicating activation of DNA binding by Mn(II). More than 5 μM Mn(II) was needed to fully activate the DNA binding (Fig. 6A). Further data fitting using a model including both Mn(II) binding and DNA binding (with a fixed affinity of 4.0 × 108 M−1, Table 2) revealed a Mn(II) binding affinity (KMn2) of 4.2 (±0.4) × 104 M−1, clearly different from the one (KMn1 = 6.2 × 106 M−1) determined by Mf2 competition (Table 1). We conclude that this affinity corresponds to the low affinity Mn(II) binding to BsFur site 2. It was not observed in Mf2 competition since it is lower than the detection limit for this assay, as initially hypothesized. More importantly, these data suggest that a high concentration of Mn(II) is required to activate BsFur-DNA interaction under these conditions.
Figure 6.
Activation of BsFur-DNA interaction by Mn(II) and Fe(II). (A) Mn(II) and (B) Fe(II) concentration dependence of DNA binding of WT (circles, solid curve), H132A (squares, dashed curve) and H37A (triangles, dotted curve) are shown. Inset: the full Mn(II)-dependent DNA binding curve of H37A. All curves represent the best fit with parameters compiled in Table 1.
This FA-based assay also provides a way to monitor Fe(II) binding and determine the Fe(II) binding affinity of BsFur. For these experiments, chelex-treated MOPS buffer (20 mM, pH 7.5) containing 0.4 M NaCl and 0.1 mM N-acetyl-cysteine (NAC), which has a lower reduction potential than dithiothreitol (Noszal et al., 2000), is used to minimize Fe(II) oxidation. NAC does not appear to compete with BsFur in Fe(II) binding, because increasing NAC concentration to 1 mM resulted in similar binding isotherms. The DNA binding affinity of BsFur in the presence of saturating metal ion is similar to the previous results (data not shown). However, despite extensive treatment, when 50 nM Fur dimer is mixed with DNA, a slight increase in anisotropy was observed, suggesting minor metal contamination in the buffer. We further decreased the protein concentration to 25 nM dimer to minimize the increase of FA and assure the Fe(II)-dependent activation of DNA binding. Titration of Fe(II) into a solution containing DNA and BsFur resulted in activation of DNA binding. Subsequent data analysis revealed BsFur binds Fe(II) with an affinity of 1.2 × 106 M−1 (Table 1, Fig. 6B). This affinity gives the first estimation of the bioavailable (buffered) Fe(II) concentration of ~1 μM in B. subtilis.
To further verify that metal binding at site 2 is responsible for the observed activation of DNA binding, we performed the same assay with two representative mutant proteins, H37A (site 2) and H132A (site 3) (Fig. 1). As expected, both the Fe(II) and Mn(II) concentration dependence of DNA binding of H132A were similar to that of WT BsFur, with binding affinities determined to be similar to WT (Fig. 6, Table 1). However, much higher Fe(II) and Mn(II) concentrations were needed to activate DNA binding by the site 2 mutant protein H37A (Fig. 6, Table 1).
These experiments reveal that metal binding at site 2 activates DNA binding and this is therefore assigned as the metal-sensing event. The metal binding affinity of site 2 thus determines the concentration of metal ions required to activate BsFur-DNA binding. These results predict that site 2 mutant proteins should be defective in sensing Fe(II) in vivo. Indeed, previous studies have shown that a Fur H97A mutant protein is defective in the Fe(II)-mediated repression of siderophore biosynthesis despite accumulating to levels comparable to or greater than wild-type Fur in the cell (Bsat and Helmann, 1999). Zn(II) binds at site 2 with affinities in the sub- to low-micromolar range (Table 1), while the intracellular bioavailable Zn(II) concentration, as controlled at least in part by BsZur, is in the pM-fM range (Ma et al., 2011a). This is similar to reported values in E. coli (Outten and O’Halloran, 2001) and S. coelicolor (Shin et al., 2011). Therefore, Zn(II) levels are normally insufficient to activate BsFur-DNA binding in vivo.
The intracellular level of free Mn(II) is likely determined by MntR, the Mn(II)-sensing repressor of high affinity Mn(II) uptake systems (Que and Helmann, 2000). Two different affinities, obtained with different methods and conditions, have been previously reported: Kd = 1 and 10 μM corresponding to the two Mn(II) binding to MntR monomer (site A and C) at pH 8.0 as measured by ITC (Kliegman et al., 2006), and Kd = 92 μM at pH 7.2 determined by competition with ANS (Golynskiy et al., 2006). The difference in pH could be one reason for the significant difference in these observed affinities. To gain insight into the likely levels of free Mn(II) in the cell we measured the Mn(II) affinity of MntR using a competition assay with Mf2 at pH 8.0. Our results indicate that the MntR monomer binds two Mn(II) with an identical Kd of 6.3 μM (Fig. S2). This result is consistent with the previous ITC study, although no difference in metal binding to site A and C was discerned in our analysis. This measured affinity is slightly higher than the Mn(II) binding affinity of BsFur at site 2 (4.2 × 104 M−1, or Kd = 24 μM), which suggests the intracellular level of free Mn(II) is normally buffered at levels just below those needed to activate BsFur-DNA binding. Taken together, these results can explain why BsFur selectively responds to Fe(II) in vivo.
BsFur can respond to Mn(II) when Fur protein level is elevated in perR mutant
Although the above results are consistent with the observation that BsFur normally responds only to Fe(II) in vivo, the affinity of MntR for Mn(II) is only marginally lower than that required to activate Fur-DNA binding. This suggests that Mn(II)-dependent activation of Fur might occur under conditions where the normal levels of the binding partners are perturbed, such as a sudden influx of Mn(II) or an increase in BsFur protein levels.
The effect of increasing Mn(II) has previously been investigated with an mntR mutant, where the Mn(II) level is increased due to the constitutive expression of Mn(II) uptake systems (Guedon et al., 2003). As confirmed here, adding Mn(II) to metal limiting minimal medium (MLMM) does cause repression of a Fur-regulated dhbA promoter-lacZ fusion. However, this effect seems to be indirect because the observed repression is still sensitive to treatment with dipyridyl (DP), an Fe(II) specific chelator known to prevent repression by Fe(II)-bound, but not Mn(II)-bound, Fur (Fig. 7) (Guedon et al., 2003). We therefore suggest that in this case an increase in the intracellular Mn(II) levels has resulted in an increase in bioavailable Fe(II) (Guedon et al., 2003).
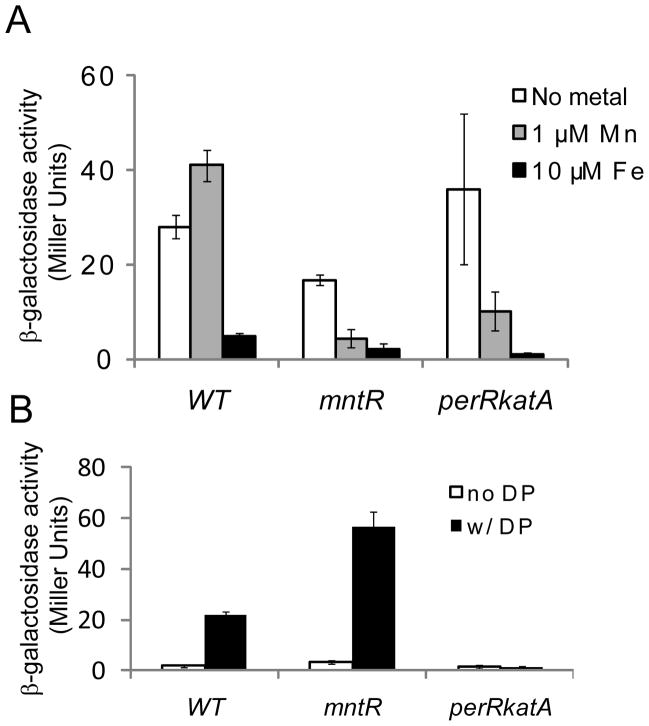
Figure 7.
Repression of Pdhb-lacZ fusion in different strains. (A) Cells grown in MLMM with the indicated metal ions added. (B) Cells grown in LB medium with or without dipyridyl (DP).
Expression of Fur is regulated by PerR in B. subtilis, and the Fur protein level is elevated by ~2.2 fold in perR mutant (Faulkner et al., 2012). We have previously shown that under these conditions Fur appears to be constitutively active as a repressor thereby leading to iron starvation. Therefore, we sought to test whether the increase of Fur protein may now allow Fur to respond to Mn(II) directly. Since a perR null mutant grows slowly and accumulates suppressor mutants at a high frequency, we used perRkatA strain instead to assess the metal dependency of Fur repression (Faulkner et al., 2012). By eliminating the abundant heme-requiring enzyme KatA, the poor growth of the perR mutant is largely alleviated in this strain (Faulkner et al., 2012). When grown in MLMM supplemented with Mn(II) or Fe(II), the expression of the dhbA reporter gene was only repressed by Fe(II) in WT cells, suggesting an Fe(II) specific response as expected (Fig. 7A). Interestingly, gene expression was also repressed by Mn(II) in perRkatA cells (Fig. 7A). Moreover, when wild-type cells are grown in a relatively metal-rich medium (LB), dhbA is normally repressed and this repression is relieved by DP treatment consistent with the use of Fe(II) as corepressor. A similar result was observed with an mntR mutant strain (Fig. 7B), as previously reported for studies in minimal medium (Guedon et al., 2003). In contrast, in the perRkatA cells repression of the dhbA reporter was insensitive to DP treatment (Fig. 7B). These results suggest that, in perRkatA cells, Fur does indeed respond directly to Mn(II).
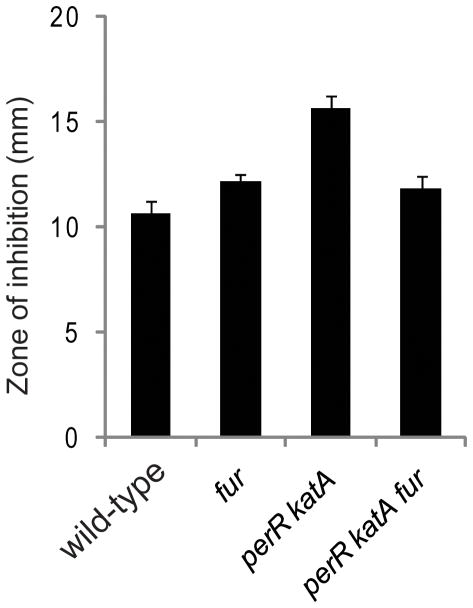
Together, these findings indicate that regulation of Fur protein levels by PerR (Fuangthong et al., 2002) avoids crosstalk between Mn(II) and Fur. Such crosstalk can be harmful to the cell, as evidenced by the fact that Fur is a target of Mn(II) toxicity in E. coli (Hantke, 1981). Interestingly, BsFur is not a target of Mn(II) toxicity in WT cells because fur mutant cells show the same level of sensitivity to Mn(II) as WT in a zone of inhibition assay (Fig. 8). However, the presence of Fur in perRkatA cells increased the Mn(II) sensitivity comparing to perRkatAfur cells (Fig. 8). These results suggest Fur is a target of Mn(II) toxicity when it is derepressed, likely due to the ability of Mn(II)-Fur to repress gene expression under these conditions.
Figure 8.
Fur is a target of Mn(II) toxicity in perRkatA cells as tested by zone of inhibition assay (the diameter of the inhibition zone is shown).
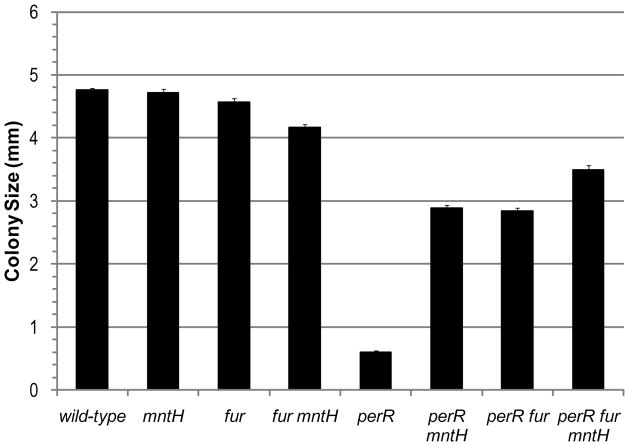
We recently reported that severe iron starvation, due to the constitutive repression of iron uptake genes, causes the slow growth phenotype of the perR mutant. This phenotype is largely alleviated in perRfur double mutant (Faulkner et al., 2012). Based on the results above, we further hypothesized that Mn(II) might be acting as an agonist of Fur thereby leading to the inappropriate repression of iron uptake genes. Consistent with this, an mntHperR double mutant grew much faster than a perR mutant as judged by the colony size after 24 hrs of growth (Fig. 9). Mutation of mntH leads to a significant (~50%) decrease in total Mn(II) levels as measured by ICP-MS (data not shown), which may be sufficient to prevent the activation of Fur. However, the mntHperRfur triple mutant grows somewhat better than the mntHperR double mutant (Fig. 9). It is possible that the mntH mutation does not lower Mn(II) sufficiently to completely eliminate Fur activity. Alternatively, reduced Mn(II) levels may alleviate the consequence of iron starvation, perhaps by avoiding mis-loading Mn(II) into Fe(II) requiring proteins.
Figure 9.
Effect of mntH and fur mutations on the growth of perR mutant cells as measured by the colony size (diameter) after 24 hrs of growth.
Involvement of PerR in metal ion homeostasis
PerR is a metal-dependent regulator that can normally utilize either Fe(II) or Mn(II) as co-repressor. However, only the Fe(II)-bound PerR can mediate metal catalyzed oxidation in response to peroxide (Lee and Helmann, 2006b). Moreover, while Mn(II)-bound PerR appears to efficiently repress all of the PerR regulated genes, the Fe(II)-bound form does not (Fuangthong et al., 2002). In fact, Fur and ZosA (a P-type ATPase previously proposed to be involved in Zn(II) uptake; Gaballa and Helmann, 2002) can only be repressed by Mn(II)-bound PerR (Fuangthong et al., 2002). This regulation pattern of Fur, along with the data presented above, suggest that PerR may unexpectedly be involved in regulating iron homeostasis in response to the intracellular Mn(II) level. Under normal conditions where Mn(II) is sufficient, Fur is repressed by PerR, which prevents the crosstalk between Mn(II) and Fur as shown above. During Mn(II) deficiency, Fur is then de-repressed but, since Mn(II) levels are low, this is unlikely to interfere with Fur-mediated regulation. This response could serve normally to reduce Fe levels by increasing sensitivity to Fe(II) and thereby coordinate Fe(II) and Mn(II) homeostasis. For example, elevated levels of Fur protein could decrease the set-point for intracellular bioavailable Fe(II) concentration. We therefore speculate that PerR regulation of Fur may help to maintain an optimal Mn(II)/Fe(II) ratio: under Mn(II) deficient conditions, elevated Fur protein level can lower the set-point for Fe(II); When Mn(II) is sufficient, the set-point for Fe(II) is higher due to the repression of Fur by PerR. Notably, such regulation of iron homeostasis by Mn(II) levels has previously been shown in Bradyrhizobium japonicum, where the stability of the regulatory protein Irr is affected by both Mn(II) and heme (Puri et al., 2010). As a result, Mn(II) deficiency leads to iron deficiency in that organism, a trend similar to what we hypothesize in B. subtilis. This model is currently being investigated in our laboratory. This type of regulatory circuit may be important to allow the correct metallation of metalloproteins under various conditions.
Experimental Procedures
Protein overexpression purification
pET-17b plasmids containing the coding sequence of BsFur between the NdeI and KpnI restriction sites were used for protein overexpression. BsFur mutants were constructed using site-directed mutagenesis. WT and mutant BsFur proteins were overexpressed and purified using previously described protocols with some modifications (Bsat and Helmann, 1999; Ma et al., 2011a). Briefly, the expression plasmids were transformed into E. coli BL21 pLysS cells. A single colony was grown overnight in 50 mL LB supplemented with 100 μg/mL ampicillin and 35 μg/mL chloramphenicol at 37 °C with rigorous shaking (200 rpm). A 10 mL overnight culture was diluted into 1 L fresh LB containing the same antibiotics and grown to mid-log phase. A final concentration of 1 mM IPTG was added for induction, cells were incubated for 2 hours, and harvested by centrifugation. Cell pellets were resuspended in buffer A (20 mM Tris, pH 8.0, 0.1 M NaCl, 2 mM EDTA, and 2 mM DTT) and lysed by sonication. BsFur proteins remained in the supernatant and were subjected to a three-step chromatography purification procedure using a heparin column, a Superdex 200 column, and a Mono-Q anion exchange column as described (Bsat and Helmann, 1999; Ma et al., 2011a). The resultant BsFur proteins were more than 90% pure judging by coomassie blue stained SDS-PAGE gel. Purified BsFur proteins were dialyzed against buffer B (20 mM Tris, pH 8.0, 0.1 M NaCl, 1 mM TCEP) and stored at −80 °C. Protein concentrations were determined using a calculated extinction coefficient at 280 nm (ε = 11,460 M−1cm−1) and the Zn(II) contents were determined by PAR assay (Ma et al., 2011a). All purified WT and mutant BsFur proteins contained ~0.9 to 1.0 Zn(II) per monomer, suggesting that the structural Zn(II) was retained during purification, as expected. No Fe(II) was detected by a ferrozine assay (Fish, 1988). All BsFur proteins appeared to be dimers by Superdex G75 chromatography (~24 mL column volume) when 200 μL of 2 μM monomer were loaded (data not shown). Under these same conditions, dimerization defects were visualized in some BsZur mutants (Ma et al., 2011a).
MntR was overexpressed and purified using the same protocol as BsFur except that no reductant was used in the purification. Purified MntR was finally dialyzed against Buffer C (20 mM Tris, pH 8.0, 0.4 M NaCl). MntR concentration was determined using a calculated extinction coefficient at 280 nm (ε = 18,916 M−1cm−1).
Metal binding experiments
All Zn(II) and Mn(II) binding experiments of BsFur were carried out in buffer B containing only 0.1 mM TCEP, which did not interfere with the Magfura-2 (Mf2) or Quin2 probes used here. The salt concentration was increased to 0.4 M NaCl in all the Zn(II) binding experiments to avoid protein precipitation. Under these buffer conditions, Mf2 binds Mn(II) with an affinity of 1.9 × 106 M−1 (pH 8.0, 0.1M NaCl) (Fig. S1A), close to the reported value (KMn = 1.0 × 106 M−1) (Golynskiy et al., 2006). However, Mf2 binds Zn(II) with an affinity of 5.0 × 106 M−1 (pH 8.0, 0.4M NaCl), much lower than the reported value (KZn = 5.0 × 107 M−1, pH 7.0, 0.1 M NaCl) (Fig. S1B) (Timothy J.B, 1993; VanZile et al., 2000). These values were used in subsequent data analysis. The reported value of the binding affinity of Zn(II) to Quin2 was (KZn = 2.7 × 1011 M−1) was used (Jefferson et al., 1990). For Mn(II) binding to MntR, experiments were carried out in Buffer C and the Mn(II)-Mf2 binding affinity was determined to be 3.0 × 105 M−1 (Fig. S2).
A typical metal binding experiment was carried out by titrating Zn(II) or Mn(II) into a mixture of BsFur protein and probe, either Mf2 or Quin2. UV-vis spectra were recorded after 10 min, a time sufficient to reach equilibrium as determined in initial experiments comparing the spectra as a function of time. The absorbance (325 and 366 nm for Mf2; 261 nm for Quin2) was plotted against metal ion concentration for data fitting by Dynafit (Kuzmic, 1996). In the competition experiment with Mf2, both changes in 325 and 366 nm absorbance were simultaneously fitted. A competition model involving four independent Zn(II) binding (KZn1 to KZn4) to BsFur dimer was used. For Mn(II) binding, a model with only one Mn(II) binding to one BsFur monomer (KMn1) was used (see text). Multiple experiments varying the concentration of BsFur or the probe were performed and representative data are reported here. The errors of the fitting determined by Dynafit are typically less then 20% (Tables 1 and 2). Results from replicate determinations of metal ion affinity using the FA and competition assays were similar (<30% variation) for Mn(II) and Fe(II). Larger variations in individual binding constants were observed for Zn(II), depending largely on the computed value of the cooperativity factor (determined by the shape of the binding curve), but the overall affinities (i.e. KZn1KZn2 for site 3 or KZn3KZn4 for site 2) were consistent between replicas. The data of Mn(II)-MntR binding were fit using a model involving two identical Mn(II) binding affinities for each MntR monomer (sites A and C).
Fluorescence anisotropy (FA)
All FA-based experiments were performed in buffer B containing 0.4 M NaCl. A 6-FAM (6-carboxyfluorescein)-labeled DNA (5′-6-FAM-CCAATTGATAATAGTTATCAATTGA-3′ and its complement) derived from the feuA promoter region including the fur-box (in bold). 1 mM EDTA, 1 mM Mn(II) or 10 μM Zn(II) was added to determine the binding affinity BsFur to DNA in the absence or presence of activating metal ions. For some BsFur mutants with defective metal binding capabilities, up to 10 mM Mn(II) was used to ensure saturation of all metal binding sites in solution. For FA experiments with Fe(II), 100 μM ferrous ammonium sulfate (Fe(NH4)2(SO4)2) was added from a freshly prepared stock solution in 0.1% HCl (no significant change of pH observed). 1 mM sodium dithionite was included in the solution to prevent Fe(II) oxidation. Fluorescence anisotropy (FA; λex = 495 nm; λem = 520 nm) was measured after each addition of BsFur into the DNA and 2 min equilibration time. An average of at least 5 continuous FA reading was reported for each data point and plotted against BsFur concentration. A 1:1 non-dissociable dimer to DNA binding model was used for data fitting by Dynafit (Kuzmic, 1996). This model was justified by the fact that a stoichiometry of ~1.1 BsFur dimer per DNA was observed when the same FA assay was carried out with 1 μM DNA, a concentration where stoichiometric binding was expected. These results indicate that the protein used was at least 90% active. A similar stoichiometry was determined for BsFur mutants, suggesting the purified proteins had no significant differences in the percentage of active protein (data not shown). In cases where saturation was not achieved within the protein concentration range examined, the total change of anisotropy was fixed in the fitting to the same value of WT BsFur under saturating metal ion conditions. For Mn(II) and Fe(II) binding assays monitored by FA, both protein and buffer were further chelex-treated (Chelex-100, BioRad) overnight at 4 °C to remove trace metal contamination. The structural Zn(II) was not affected by such treatment as determined by ICP-MS (data not shown). The Chelex-treated protein bound DNA with similar affinity as untreated BsFur in the presence of saturating metal ions (data not shown). For Fe(II) binding experiments, buffer (20 mM MOPS, pH 7.5, 0.4 M NaCl) containing 0.1 mM N-acetyl-cysteine was used to minimize Fe(II) oxidation.
β-galactosidase assay
All B. subtilis strains used in this study are listed in Table S1. Heat induction was used to obtain SPβ lysates from HB606 for transduction. For experiments in metal limiting minimal medium (MLMM) (Ma et al., 2011b), cells were grown in MLMM with supplement of 1 μM Mn(II), Fe(II) and Zn(II) (in the form of ultrapure MnCl2, FeCl2 and ZnSO4) overnight. FeCl2 solution was prepared with 0.1% HCl but its oxidation to Fe(III) was expected once mixed with MLMM. Cells were harvested and washed with MLMM, and then inoculated (1:25 dilution) into MLMM with no metal, Mn(II) or Fe(II) added. Mid-log phase cells were collected for β-galactosidase assay. For experiments with LB medium, overnight cultures were inoculated (1:100 dilution) into fresh LB medium with or without addition of 100 μM dipyridyl and grown to mid-log phase. β-galactosidase assays were carried out as reported (Miller, 1972; Ma et al., 2011a).
Zone of inhibition assay
HB14123 was constructed by transforming chromosomal DNA from HB2501 into HB14109 and select for kanamycin resistance (Table S1). Zone of inhibition assays were carried out as reported (Que and Helmann, 2000) with the modification that 100 μL cells at OD600~0.4 were mixed with 4 mL soft agar. The paper disks contained 5 μL of 2.0 M MnCl2 were used in this assay.
Colony size measurement
HB14106 and HB14127 were constructed by chromosomal DNA transformation. Colony size measurement was carried out on LB-agar plates exactly as reported (Faulkner et al., 2012). Twenty-five colonies were measured for each strain to determine its average colony size.
Supplementary Material
Acknowledgments
We thank Mr. Jihong Song for assistance with purification of a BsFur mutant. This work was supported by National Institute of Health grant GM059323 to J.D.H.
References
- Andrews SC, Robinson AK, Rodriguez-Quinones F. Bacterial iron homeostasis. FEMS microbiol Rev. 2003;27:215–237. doi: 10.1016/S0168-6445(03)00055-X. [DOI] [PubMed] [Google Scholar]
- Bagg A, Neilands JB. Ferric uptake regulation protein acts as a repressor, employing iron(II) as a cofactor to bind the operator of an iron transport operon in Escherichia coli. Biochemistry. 1987;26:5471–5477. doi: 10.1021/bi00391a039. [DOI] [PubMed] [Google Scholar]
- Baichoo N, Helmann JD. Recognition of DNA by Fur: a Reinterpretation of the Fur Box Consensus Sequence. J Bacteriol. 2002;184:5826–5832. doi: 10.1128/JB.184.21.5826-5832.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baichoo N, Wang T, Ye R, Helmann JD. Global analysis of the Bacillus subtilis Fur regulon and the iron starvation stimulon. Mol Microbiol. 2002;45:1613–1629. doi: 10.1046/j.1365-2958.2002.03113.x. [DOI] [PubMed] [Google Scholar]
- Bsat N, Helmann JD. Interaction of Bacillus subtilis Fur (Ferric Uptake Repressor) with the dhb Operator In Vitro and In Vivo. J Bacteriol. 1999;181:4299–4307. doi: 10.1128/jb.181.14.4299-4307.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bsat N, Herbig A, Casillas-Martinez L, Setlow P, Helmann JD. Bacillus subtilis contains multiple Fur homologues: identification of the iron uptake (Fur) and peroxide regulon (PerR) repressors. Mol Microbiol. 1998;29:189–198. doi: 10.1046/j.1365-2958.1998.00921.x. [DOI] [PubMed] [Google Scholar]
- Dian C, Vitale S, Leonard GA, Bahlawane C, Fauquant C, Leduc D, Muller C, de Reuse H, Michaud-Soret I, Terradot L. The structure of the Helicobacter pylori ferric uptake regulator Fur reveals three functional metal binding sites. Mol Microbiol. 2011;79:1260–1275. doi: 10.1111/j.1365-2958.2010.07517.x. [DOI] [PubMed] [Google Scholar]
- Faulkner MJ, Ma Z, Fuangthong M, Helmann JD. Derepression of the Bacillus subtilis PerR peroxide stress response leads to iron deficiency. J Bacteriol. 2012;194:1226–1235. doi: 10.1128/JB.06566-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fish WW. Rapid colorimetric micromethod for the quantitation of complexed iron in biological samples. Methods Enzymol. 1988;158:357–364. doi: 10.1016/0076-6879(88)58067-9. [DOI] [PubMed] [Google Scholar]
- Fuangthong M, Herbig AF, Bsat N, Helmann JD. Regulation of the Bacillus subtilis fur and perR Genes by PerR: Not All Members of the PerR Regulon Are Peroxide Inducible. J Bacteriol. 2002;184:3276–3286. doi: 10.1128/JB.184.12.3276-3286.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaballa A, Antelmann H, Aguilar C, Khakh SK, Song KB, Smaldone GT, Helmann JD. The Bacillus subtilis iron-sparing response is mediated by a Fur-regulated small RNA and three small, basic proteins. Proc Natl Acad Sci USA. 2008;105:11927–11932. doi: 10.1073/pnas.0711752105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaballa A, Helmann JD. Identification of a Zinc-Specific Metalloregulatory Protein, Zur, Controlling Zinc Transport Operons in Bacillus subtilis. J Bacteriol. 1998;180:5815–5821. doi: 10.1128/jb.180.22.5815-5821.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaballa A, Helmann JD. A peroxide-induced zinc uptake system plays an important role in protection against oxidative stress in Bacillus subtilis. Mol Microbiol. 2002;45:997–1005. doi: 10.1046/j.1365-2958.2002.03068.x. [DOI] [PubMed] [Google Scholar]
- Golynskiy MV, Gunderson WA, Hendrich MP, Cohen SM. Metal binding studies and EPR spectroscopy of the manganese transport regulator MntR. Biochemistry. 2006;45:15359–15372. doi: 10.1021/bi0607406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guedon E, Moore CM, Que Q, Wang T, Ye RW, Helmann JD. The global transcriptional response of Bacillus subtilis to manganese involves the MntR, Fur, TnrA and σB regulons. Mol Microbiol. 2003;49:1477–1491. doi: 10.1046/j.1365-2958.2003.03648.x. [DOI] [PubMed] [Google Scholar]
- Hantke K. Regulation of ferric iron transport in Escherichia coli K12: Isolation of a constitutive mutant. MGG. 1981;182:288–292. doi: 10.1007/BF00269672. [DOI] [PubMed] [Google Scholar]
- Imlay JA. Cellular defenses against superoxide and hydrogen peroxide. Annu Rev Biochem. 2008;77:755–776. doi: 10.1146/annurev.biochem.77.061606.161055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson JR, Hunt JB, Ginsburg A. Characterization of indo-1 and quin-2 as spectroscopic probes for Zn2+-protein interactions. Anal Biochem. 1990;187:328–336. doi: 10.1016/0003-2697(90)90465-l. [DOI] [PubMed] [Google Scholar]
- Kliegman JI, Griner SL, Helmann JD, Brennan RG, Glasfeld A. Structural basis for the metal-selective activation of the manganese transport regulator of Bacillus subtilis. Biochemistry. 2006;45:3493–3505. doi: 10.1021/bi0524215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmic P. Program DYNAFIT for the analysis of enzyme kinetic data: application to HIV proteinase. Anal Biochem. 1996;237:260–273. doi: 10.1006/abio.1996.0238. [DOI] [PubMed] [Google Scholar]
- Lee JW, Helmann JD. Biochemical characterization of the structural Zn2+ site in the Bacillus subtilis peroxide sensor PerR. J Biol Chem. 2006a;281:23567–23578. doi: 10.1074/jbc.M603968200. [DOI] [PubMed] [Google Scholar]
- Lee JW, Helmann JD. The PerR transcription factor senses H2O2 by metal-catalysed histidine oxidation. Nature. 2006b;440:363–367. doi: 10.1038/nature04537. [DOI] [PubMed] [Google Scholar]
- Lee JW, Helmann JD. Functional specialization within the Fur family of metalloregulators. Biometals. 2007;20:485–499. doi: 10.1007/s10534-006-9070-7. [DOI] [PubMed] [Google Scholar]
- Lucarelli D, Russo S, Garman E, Milano A, Meyer-Klaucke W, Pohl E. Crystal structure and function of the zinc uptake regulator FurB from Mycobacterium tuberculosis. J Biol Chem. 2007;282:9914–9922. doi: 10.1074/jbc.M609974200. [DOI] [PubMed] [Google Scholar]
- Ma Z, Gabriel SE, Helmann JD. Sequential binding and sensing of Zn(II) by Bacillus subtilis Zur. Nucleic Acids Res. 2011a;39:9130–9138. doi: 10.1093/nar/gkr625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z, Jacobsen FE, Giedroc DP. Coordination Chemistry of Bacterial Metal Transport and Sensing. Chem Rev. 2009;109:4644–4681. doi: 10.1021/cr900077w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z, Lee JW, Helmann JD. Identification of altered function alleles that affect Bacillus subtilis PerR metal ion selectivity. Nucleic Acids Res. 2011b;39:5036–5044. doi: 10.1093/nar/gkr095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martell AE, Smith RM. Critical Stability Constants. Plenum Press; New York: 1979–1989. [Google Scholar]
- Miller JH. Expermiments in molecular genetics. Cold Spring Harbor Laboratory; Cold Spring Harbor, N.Y: 1972. [Google Scholar]
- Mills SA, Marletta MA. Metal binding characteristics and role of iron oxidation in the ferric uptake regulator from Escherichia coli. Biochemistry. 2005;44:13553–13559. doi: 10.1021/bi0507579. [DOI] [PubMed] [Google Scholar]
- Noszal B, Visky D, Kraszni M. Population, Acid-Base, and Redox Properties of N-Acetylcysteine Conformers. J Med Chem. 2000;43:2176–2182. doi: 10.1021/jm9909600. [DOI] [PubMed] [Google Scholar]
- Outten CE, O’Halloran TV. Femtomolar sensitivity of metalloregulatory proteins controlling zinc homeostasis. Science. 2001;292:2488–2492. doi: 10.1126/science.1060331. [DOI] [PubMed] [Google Scholar]
- Outten CE, Olson KE, Cao H, O’Halloran TV. The Ferric Uptake Regulation (Fur) Repressor Is a Zinc Metalloprotein. Biochemistry. 1999;38:6559–6569. doi: 10.1021/bi982788s. [DOI] [PubMed] [Google Scholar]
- Pohl E, Haller JC, Mijovilovich A, Meyer-Klaucke W, Garman E, Vasil ML. Architecture of a protein central to iron homeostasis: crystal structure and spectroscopic analysis of the ferric uptake regulator. Mol Microbiol. 2003;47:903–915. doi: 10.1046/j.1365-2958.2003.03337.x. [DOI] [PubMed] [Google Scholar]
- Puri S, Hohle TH, O’Brian MR. Control of bacterial iron homeostasis by manganese. Proc Natl Acad Sci USA. 2010;107:10691–10695. doi: 10.1073/pnas.1002342107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Que Q, Helmann JD. Manganese homeostasis in Bacillus subtilis is regulated by MntR, a bifunctional regulator related to the diphtheria toxin repressor family of proteins. Mol Microbiol. 2000;35:1454–1468. doi: 10.1046/j.1365-2958.2000.01811.x. [DOI] [PubMed] [Google Scholar]
- Shin JH, Jung HJ, An YJ, Cho YB, Cha SS, Roe JH. Graded expression of zinc-responsive genes through two regulatory zinc-binding sites in Zur. Proc Natl Acad Sci USA. 2011;108:5045–5050. doi: 10.1073/pnas.1017744108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timothy JBS. Measurement of free Zn2+ ion concentration with the fluorescent probe mag-fura-2 (furaptra) J Biochem Biophys Methods. 1993;27:25–37. doi: 10.1016/0165-022x(93)90065-v. [DOI] [PubMed] [Google Scholar]
- VanZile ML, Cosper NJ, Scott RA, Giedroc DP. The zinc metalloregulatory protein Synechococcus PCC7942 SmtB binds a single zinc ion per monomer with high affinity in a tetrahedral coordination geometry. Biochemistry. 2000;39:11818–11829. doi: 10.1021/bi001140o. [DOI] [PubMed] [Google Scholar]
- Waldron KJ, Robinson NJ. How do bacterial cells ensure that metalloproteins get the correct metal? Nat Rev Microbiol. 2009;7:25–35. doi: 10.1038/nrmicro2057. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.