Highlights
► Epigenetic control is involved in stress signaling and stress responses. ► Stress can modify epigenetic regulation at many different levels. ► Epigenetic and genetic components of stress responses are connected. ► Epigenetic diversity might be an important factor in stress adaptation and evolution.
Abstract
Stressful conditions for plants can originate from numerous physical, chemical and biological factors, and plants have developed a plethora of survival strategies including developmental and morphological adaptations, specific signaling and defense pathways as well as innate and acquired immunity. While it has become clear in recent years that many stress responses involve epigenetic components, we are far from understanding the mechanisms and molecular interactions. Extending our knowledge is fundamental, not least for plant breeding and conservation biology. This review will highlight recent insights into epigenetic stress responses at the level of signaling, chromatin modification, and potentially heritable consequences.
Current Opinion in Plant Biology 2012, 15:568–573
This review comes from a themed issue on Cell signaling and gene regulation
Edited by Jerzy Paszkowski and Robert A. Martienssen
For a complete overview see the Issue and the Editorial
Available online 7th September 2012
1369-5266/$ – see front matter, © 2012 Elsevier Ltd. All rights reserved.
Introduction
Plants in the field are permanently exposed to stress. Limitations originate from many different factors: too high or too low temperatures/water supply/light intensity, non-optimal mineral composition or soil contamination, mechanic inhibition, pathogen infestation, lack of symbiotic partners, interactions with other plants, parasites or herbivores. These conditions can activate defense responses by which plants can minimize detrimental consequences of stressful conditions for their survival, growth, or propagation. These mechanisms allow plants to occupy even extreme habitats, despite their sedentariness. Under field conditions, different stress types usually occur concomitantly, like heat and drought, and the molecular responses can be difficult to separate. Response to one stress type can be also antagonistic to another, as adaptation to nutrient-poor substrates might reduce stress by competition. Therefore, mechanistic studies of stress effects are mostly performed under controlled laboratory conditions, applying one stress type and distinguishing biotic from abiotic stress. While this resulted in information about signaling cascades, transcription factors, and defense compounds, interference of stress with epigenetic factors became evident only recently. Stress effects on the epigenetic level are expected to allow more permanent changes of gene expression and potentially long-term adaptation that could have evolutionary impact, as chromatin modifications can be mitotically or meiotically heritable. This review will focus on the connection of stress with epigenetic regulation, separated in three levels.
Level 1: epigenetic components of stress signaling
Investment into stress defense, alongside constitutive morphological and metabolic survival equipment and seasonal adaptations, expends plants’ general resources and therefore should be restricted to the actual occurrence of stressful situations. Plants use a range of different sensing and signaling mechanisms to induce dynamic stress responses only when challenged. Signaling includes mainly the hormones salicylic acid (SA), jasmonic acid (JA) and ethylene upon biotic stress, and abscisic acid (ABA) in case of abiotic stress (reviewed in [1]). Nevertheless, there is growing evidence that noncoding and siRNAs and the proteins generating or binding them are involved in stress-signaling and can subsequently induce transcriptional or posttranscriptional gene silencing (TGS or PTGS, respectively). These principles are described in the review by Wierzbicki (this issue).
There are many examples of differential siRNA, miRNA or ncRNA expression upon stress [2]. The recent identification of a mutated NRPD2 gene responsible for constitutive overexpression of a SA-inducible gene provided a mechanistic link between stress signaling and elements of the RNA directed DNA methylation (RdDM) pathway, excluding only PolIV [3•]. Some RdDM mutants had a compromised immune response to pathogenic fungi correlated with a lack of gene induction by JA. In contrast, resistance to a bacterial pathogen was increased, corresponding with elevated levels of salicylic acid-related defense genes and enriched activating chromatin marks H3K4me3 and H3K9ac at the promoter of the SA-inducible gene PR-1. This argues for the overlap between targets of RdDM and SA-signaling and a role of RdDM to relay stress signals to the nucleus.
Stress can certainly induce the production of siRNAs, either via antisense transcription of protein-coding or non-protein-coding sequences, or from inverted repeats. An early example of naturally occurring antisense was the discovery that induction of the Arabidopsis SRO5 expression under salt stress creates a transcript partially complementary to that of the constitutively expressed P5CDH, resulting in dsRNA as a substrate for Dicer-like proteins and siRNAs that are only present and effective under stress [4]. Among 76 long non-protein-coding RNAs (npcRNA) in the Arabidopsis genome, 26 had altered expression levels upon low phosphate, salt or drought stress [5]. Some of them gave rise to 24 bp siRNAs, and npc536 conferred improved root growth under salt stress. By now, many pairs of potential antisense transcripts are identified [6], and natural cis-antisense siRNAs (nat-siRNAs) are defined as a separate class of the small RNA family. Many of them are found exclusively or enriched upon specific stress conditions [7]. One example role of siRNAs in extreme stress tolerance is a dehydration- and ABA-inducible retroelement-derived siRNA that regulates an adaptive response in the resurrection plant Craterostigma plantagineum [8].
Applying tailor-made transcripts of inverted repeats (IRs) is a routine technique to interfere with transcription of target genes with homology to the resulting siRNAs, but similar siRNA can also originate from transcripts of endogenous IRs. Two Arabidopsis repeats, IR71 and IR2039 [9••], produce siRNA of different size classes, of which some can silence a GFP-reporter in trans. Endogenous IRs are highly variable in the genome of different ecotypes of Arabidopsis, indicating fast evolution and rapid adaptive changes [9••]. However, the actual response of IR-derived siRNA to environmental factors and contribution to stress-adaptation is yet to be demonstrated.
Responses of transposable elements (TE) to stress are the topic of reviews by Lisch (this issue) and Bucher et al. (this issue), but TE activation raises the interesting potential of TE-derived small RNAs that target stress-related protein-coding genes [10] and thereby represent an indirect stress signaling pathway.
Level 2: stress etching on chromatin
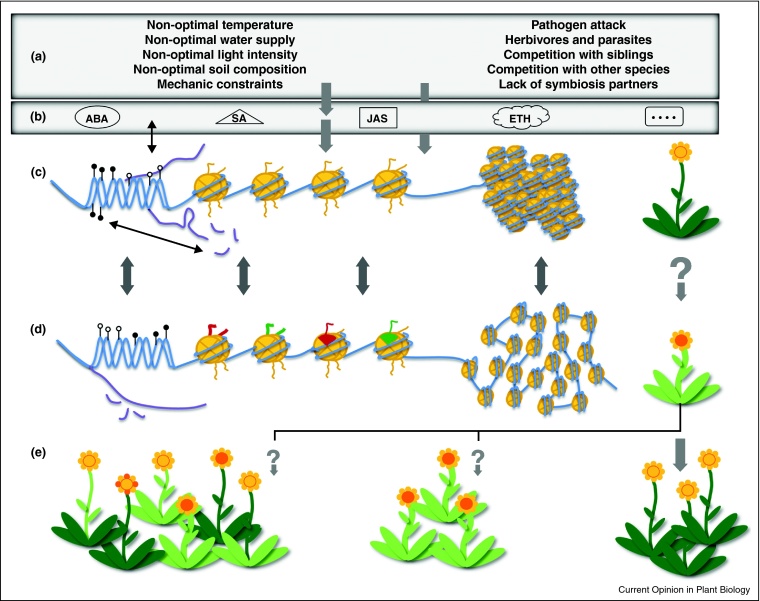
Stress signaling leads to stress-adapted gene expression and also affects chromatin structure at responsive genes, directly or indirectly [11–14]. The changes can affect DNA methylation, histone tail modifications, exchange of histone variants, or nucleosome occupancy and larger chromatin configuration, as documented in the following examples (Figure 1).
Figure 1.
Abiotic and biotic stress conditions (a) can change gene expression with and without involving plant stress hormones (b). Transcription changes, or stress factors directly, can affect chromatin via DNA methylation, histone tail modifications, histone variant replacements, or nucleosome loss and chromatin de-condensation (c, d). These changes are largely reversible but can modify metabolic or morphologic plant features under stress conditions. Usually, the new phenotypes are not transmitted to progeny. However, chromatin-associated changes have the potential to be heritable and might result in uniform maintenance of new features or new combination and epigenetic diversity (e).
DNA methylation is an important defense strategy against infections by DNA-viruses. Double-stranded viral transcripts can induce methylation at homologous sequences, which led to the discovery of the RdDM phenomenon [15]. Arabidopsis RdDM mutants are hypersensitive to gemini viruses infection [16]. Some siRNAs associated with virus infection can be complementary to regions of the host genome, especially to regulatory sequences containing retro-elements evolved from ancient integration events, and can exert stable gene silencing on such ‘off-targets’. The same principle could apply to other endogenous siRNAs that match with promoter regions and could affect stress-related genes [17]. For example, repeat elements in the promoter of a sodium transporter gene are methylation targets and determine salt stress tolerance [18]. A plausible correlation is also the DNA methylation of two genes controlling stomata development, their transcriptional repression, and a reduced number of stomata in Arabidopsis plants grown under low humidity [19••]. Methylation changes that appear non-targeted or not yet associated with stress gene targets are induced by salt stress in Arabidopsis [20], or in roots of rice plants, to different degree depending on the salt sensitivity of the cultivars [21]. These could be secondary effects due to stress-induced activity changes of epigenetic regulators. Multiple methylation changes were also observed associated with herbivore and pathogen attack in dandelions [22], and between mangroves or rubber trees growing in different habitats [23,24]. Higher resistance to pathogens of Arabidopsis mutants with defects in DNA methyltransferases inspired a genome-wide analysis after infection with a bacterial pathogen, revealing methylation changes in all sequence contexts and at multiple loci, including defense-related genes, transposons and repeats [25•].
There are several reports of stress-induced histone tail modifications and altered stress resistance in mutants lacking histone-modifying enzymes. Installation of activating marks can result in increased transcription levels of stress genes but may also simply poise target genes for faster or stronger response upon a more serious attack later [26,27]. ABA and salt change H3 phosphorylation and H4 acetylation in cell cultures of tobacco and Arabidopsis [28]. Activation of a gene for an immune receptor protein (R) gene in Arabidopsis depends on a histone methyltransferase installing H3K36me3 [29•]. A prominent target is also the PR-1 (Pathogen-Related 1) gene involved in initiating defense or stress adaptation where H3K4me2, H3K4me3 and H3ac levels increase upon SA treatment or pathogens [30,31]. PR1 expression is negatively regulated by two WRKY transcription factors, which both interact with histone deacetylase 19 (HDA19) [32]. Deacetylation reduces active marks, and the induction of HDA19 upon bacterial infection could be a chromatin-based way to weaken the plant defense system, among many other strategies [13]. Another histone deacetylase, HDA6, is required for freezing tolerance in Arabidopsis, although the link to histone acetylation at a specific target gene is missing [33,34]. A histone ubiquitin ligase determines the resistance to necrotrophic fungi [35], and the growing number of identified modification types makes it likely that more of them will also be connected with stress regulation.
The histone methylation at H3K27 under control of the Polycomb/Trithorax complexes and its response to environmental signals are described in detail elsewhere [36]. MSI1, a member of the PRC2 as well as the CAF1 complex, contributes to drought stress resistance by controlling the expression of ABA-responsive genes and proline synthesis [37]. Lower H3K27me3 levels at abscisic acid-responsive genes upon downregulation of the cell cycle regulator RETINOBLASTOMA-RELATED (RBR), and induction of several stress-related genes before deregulation of the cell cycle machinery [38,39] indicate a tight and complex connection between stress factors that modify chromatin features and developmental regulation. Three other histone variants are so far also connected with stress responses. Histone variant H2A.Z associated with the 5′end of many genes is relevant for repression of pathogen response, as mutants lacking subunits of the SWR1 complex that installs H2A.Z show increased resistance to bacterial infections [40]. A clear connection of this variant also with abiotic stress is evident from its eviction by heat exposure [41••]. Lack of ARP6, one SWR1 subunit, mimics the transcriptional changes even at ambient temperatures. Further, a plant-specific linker histone H1 variant is expressed under drought stress in Arabidopsis, and its downregulation in tomato revealed its importance in water stress response [42]. DNA damaging stress is followed by incorporation of H2A.X, which becomes phosphorylated and attracts DNA repair complexes and transcriptional activators [43]. However, many more chromatin components might be involved in enabling DNA repair and recombination.
Lastly, the association of nucleosomes with DNA can be modified in response to stress. Indirect evidence came from analyzing mutants with defects in putative or proven chromatin remodeling factors in response to stress (e.g. [44]). A protein phosphatase 2C and ABA co-receptor, HYPERSENSITIVE TO ABA1 (HAB1) can interact with a subunit of the ATP-dependent chromatin remodeling complex SWI3B, and swi3b mutants show reduced ABA responses [45]. The deregulation of epigenetically controlled genes and repetitive sequences upon heat stress [46–48] is associated with transient loss of DNA-bound nucleosomes at transcribed and non-transcribed genomic regions, and with substantial heterochromatin de-condensation [47]. A role for nucleosome occupancy is further suggested by delayed re-silencing of heat stress-activated repeats in mutants with impaired CHROMATIN ASSEMBLY FACTOR 1 (CAF-1) subunits [47]. Nucleosome eviction, together with histone tail modifications were also observed at target genes of stress-induced transcription factors in the green alga Chlamydomonas [49•].
Level 3: potential for lasting adaptation
Extreme stress can result in a complete growth arrest, but, after limited damage, plants can recover and resume growth and development. This is in part exerted by activation of replication- and cell cycle checkpoints that prohibit amplification and transmission of damaged DNA [50]. It is tempting to propose the existence of a similar epigenetic checkpoint control that senses detrimental alterations in the epigenome and halts growth until these alterations are repaired. However, plants tolerate loss of DNA damage checkpoint components much better than animals [50]. Similarly, epigenetic mutations with lethal consequences in animals are often less severe in plants, which could indicate a less stringent ‘quality’ control of epigenetic integrity. Many plants proliferate partially or even exclusively by vegetative propagation, circumventing epigenetic reprogramming during gamete formation and sexual reproduction as observed in mammals. However, if such ‘resetting’ occurs in plants, it seems to be at least not as extensive [51], and mitotic and meiotic transmission of stress-induced epigenetic disruptions is therefore conceivable. Indeed, there is growing interest in ‘memory’ effects in stressed plants. These could even be beneficial if they make exposed plants and/or their progeny more resistant upon recapitulated challenges. The concept of priming plants with a certain stress, resulting in a faster and more pronounced response later, is exemplified by Systemic Acquired Resistance (SAR), in which soluble and/or volatile signaling molecules can spread within an individual or be transmitted between plants. Several stress types were reported to induce a priming effect that can be assayed also in subsequent generations [52–57]. In some cases, the lack of epigenetic components, mainly in the RdDM pathway, was shown to reduce or eliminate the transmission to the progeny [52,54] or mimic the primed state [53]. However, the effects did not last more than one or two generations, and concomitant chromatin changes were connected only in one case with the primed state of stress-specific genes [53]. Seemingly transgenerational responses can also originate from parental stress affecting embryo development, seed germination or early growth of the progeny, independent or dependent from chromatin-regulated components.
Although a role of chromatin in priming is widely assumed [58–60], proof of strong causal epigenetic changes affecting traits with adaptive values, without concomitant genetic changes or long-living signaling components transmitted through the cytoplasm of gametes, would be desirable [51,61]. This is not meant to discourage further studies: on the contrary, the wealth of interesting phenomena that could indicate transgenerational epigenetic inheritance [62,63], together with the growing toolbox for thorough genome-wide genetic and epigenetic analysis, make it a very interesting field of research. Analysis of DNA methylation in individuals, between generations or populations from different habitats indicates a range of epigenetic diversity that might by far exceed genetic diversity, as described in the contribution of Becker and Weigel (this issue). But it is also clear that many epigenetic changes are triggered by genetic mutations, especially transposon movements, in the vicinity. So far, these genetically triggered epigenetic changes are those with the most relevant and drastic consequences, as exemplified by the development of genetic incompatibility [64•]. On the other hand, many transposons are under epigenetic control, as reviewed by Bucher et al. (this issue), which makes the question of a direct or indirect epigenetic stress memory an interesting, but difficult-to-investigate chicken-and-egg problem.
Conclusions
Stress and epigenetic regulation meet at many different levels from which only a few aspects are documented so far. Current information about the connection resembles a few fragments of a jigsaw puzzle for which neither the number of parts nor the dimensions of the picture are known. It is very likely that epigenetic variation contributes to the adaptation potential of plants and, like genetic diversity, is under selection by environmental conditions. Whether epigenetic responses to stress can serve as adaptive traits remains a matter of debate. Surprisingly few primary publications have addressed the issue, while many reviews discuss this possibility. Adding another one will not shift this imbalance but we hope to stimulate more work in the field.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
Acknowledgements
We thank Thomas Friese for editing the manuscript. The space limitation required omitting references to many interesting publications, for which we apologize. Our research is supported by the Austrian Academy of Sciences and a grant from the Austrian Science Fund (FWF I489) to O.M.S.
References
- 1.Robert-Seilaniantz A., Grant M., Jones J.D. Hormone crosstalk in plant disease and defense: more than just jasmonate-salicylate antagonism. Annu Rev Phytopathol. 2011;49:317–343. doi: 10.1146/annurev-phyto-073009-114447. [DOI] [PubMed] [Google Scholar]
- 2.Khraiwesh B., Zhu J.-K., Zhu J. Role of miRNAs and siRNAs in biotic and abiotic stress responses of plants. Biochim Biophys Acta. 2012;1819:137–148. doi: 10.1016/j.bbagrm.2011.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3•.Lopez A., Ramirez V., Garcia-Andrade J., Flors V., Vera P. The RNA silencing enzyme RNA polymerase v is required for plant immunity. PLoS Genetics. 2011;7:e1002434. doi: 10.1371/journal.pgen.1002434. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study links stress signaling to the RdDM pathway, by demonstrating that a subunit of PolV and several RdDM pathway genes determine plant immune responses. RdDM mutants, with the exception of those lacking Pol IV, showed also an enhanced salicylic acid-mediated defense.
- 4.Borsani O., Zhu J.H., Verslues P.E., Sunkar R., Zhu J.K. Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell. 2005;123:1279–1291. doi: 10.1016/j.cell.2005.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ben Amor B., Wirth S., Merchan F., Laporte P., d’Aubenton-Carafa Y., Hirsch J., Maizel A., Mallory A., Lucas A., Deragon J.M. Novel long non-protein coding RNAs involved in Arabidopsis differentiation and stress responses. Genome Res. 2009;19:57–69. doi: 10.1101/gr.080275.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen D., Yuan C., Zhang J., Zhang Z., Bai L., Meng Y., Chen L.-L., Chen M. PlantNATsDB: a comprehensive database of plant natural antisense transcripts. Nucleic Acids Res. 2012;40:D1187–D1193. doi: 10.1093/nar/gkr823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang X., Xia J., Lii Y., Barrera-Figueroa B., Zhou X., Gao S., Lu L., Niu D., Liang W., Chen Z. Genome-wide analysis of plant nat-siRNAs reveals insights into their distribution, biogenesis and function. Genome Biol. 2012;13:R20. doi: 10.1186/gb-2012-13-3-r20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hilbricht T., Varotto S., Sgaramella V., Bartels D., Salamini F., Furini A. Retrotransposons and siRNA have a role in the evolution of desiccation tolerance leading to resurrection of the plant Craterostigma plantagineum. New Phytol. 2008;179:877–887. doi: 10.1111/j.1469-8137.2008.02480.x. [DOI] [PubMed] [Google Scholar]
- 9••.Dunoyer P., Brosnan C.A., Schott G., Wang Y., Jay F., Alioua A., Himber C., Voinnet O. An endogenous, systemic RNAi pathway in plants. EMBO. 2010;29:1699–1712. doi: 10.1038/emboj.2010.65. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]; This study demonstrates very elegantly that siRNAs originating from two endogenous inverted repeats can move throughout the plant and silence target loci in trans via RdDM. Presence of the inverted repeats, their stress response, and processing of transcripts might be under selection and highly variable between ecotypes.
- 10.McCue A.D., Nuthikattu S., Reeder S.H., Slotkin R.K. Gene expression and stress response mediated by the epigenetic regulation of a transposable element small RNA. PLoS Genetics. 2012;8:e1002474. doi: 10.1371/journal.pgen.1002474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berr A., Menard R., Heitz T., Shen W.H. Chromatin modification and remodelling: a regulatory landscape for the control of Arabidopsis defence responses upon pathogen attack. Cell Microbiol. 2012;14:829–839. doi: 10.1111/j.1462-5822.2012.01785.x. [DOI] [PubMed] [Google Scholar]
- 12.Grativol C., Hemerly A.S., Ferreira P.C.G. Genetic and epigenetic regulation of stress responses in natural plant populations. Biochim Biophys Acta. 2012;1819:502–512. doi: 10.1016/j.bbagrm.2011.08.010. [DOI] [PubMed] [Google Scholar]
- 13.Ma K.-W., Flores C., Ma W. Chromatin configuration as a battlefield in plant-bacteria interactions. Plant Physiol. 2011;157:535–543. doi: 10.1104/pp.111.182295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mirouze M., Paszkowski J. Epigenetic contribution to stress adaptation in plants. Curr Opin Plant Biol. 2011;14:267–274. doi: 10.1016/j.pbi.2011.03.004. [DOI] [PubMed] [Google Scholar]
- 15.Wassenegger M., Heimes S., Riedel L., Sanger H.L. RNA-directed de novo methylation of genomic sequences in plants. Cell. 1994;76:567–576. doi: 10.1016/0092-8674(94)90119-8. [DOI] [PubMed] [Google Scholar]
- 16.Raja P., Sanville B.C., Buchmann R.C., Bisaro D.M. Viral genome methylation as an epigenetic defense against geminiviruses. J Virol. 2008;82:8997–9007. doi: 10.1128/JVI.00719-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baev V., Naydenov M., Apostolova E., Ivanova D., Doncheva S., Minkov I., Yahubyan G. Identification of RNA-dependent DNA-methylation regulated promoters in Arabidopsis. Plant Physiol Biochem. 2010;48:393–400. doi: 10.1016/j.plaphy.2010.03.013. [DOI] [PubMed] [Google Scholar]
- 18.Baek D., Jiang J., Chung J.-S., Wang B., Chen J., Xin Z., Shi H. Regulated AtHKT1 gene expression by a distal enhancer element and DNA methylation in the promoter plays an important role in salt tolerance. Plant Cell Physiol. 2011;52:149–161. doi: 10.1093/pcp/pcq182. [DOI] [PubMed] [Google Scholar]
- 19••.Tricker P., Gibbings J., Rodríguez López C., Hadley P., Wilkinson M. Low relative humidity triggers RNA-directed de novo DNA methylation and suppression of genes controlling stomatal development. J Exp Bot. 2012;63:3799–3813. doi: 10.1093/jxb/ers076. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study links stress-induced epigenetic modifications to a developmental program. Low humidity correlates with hypermethylation at genes of two transcription factors necessary for stomata differentiation, resulting in fewer stomata. Neither stress-dependent methylation nor reduced stomata density was observed in DNA methylation pathway mutants.
- 20.Bilichak A., Ilnystkyy Y., Hollunder J., Kovalchuk I. The progeny of Arabidopsis thaliana plants exposed to salt exhibit changes in DNA methylation, histone modifications and gene expression. PLoS One. 2012;7:e30515. doi: 10.1371/journal.pone.0030515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang W., Zhao X., Pan Y., Zhu L., Fu B., Li Z. DNA methylation changes detected by methylation-sensitive amplified polymorphism in two contrasting rice genotypes under salt stress. J Genet Genomics. 2011;38:419–424. doi: 10.1016/j.jgg.2011.07.006. [DOI] [PubMed] [Google Scholar]
- 22.Verhoeven K.J.F., Jansen J.J., van Dijk P.J., Biere A. Stress-induced DNA methylation changes and their heritability in asexual dandelions. New Phytol. 2010;185:1108–1118. doi: 10.1111/j.1469-8137.2009.03121.x. [DOI] [PubMed] [Google Scholar]
- 23.Lira-Medeiros C.F., Parisod C., Fernandes R.A., Mata C.S., Cardoso M.A., Ferreira P.C. Epigenetic variation in mangrove plants occurring in contrasting natural environment. PLoS One. 2010;5:e10326. doi: 10.1371/journal.pone.0010326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Uthup T.K., Ravindran M., Bini K., Thakurdas S. Divergent DNA methylation patterns associated with abiotic stress in Hevea brasiliensis. Mol Plant. 2011;4:996–1013. doi: 10.1093/mp/ssr039. [DOI] [PubMed] [Google Scholar]
- 25•.Dowen R.H., Pelizzola M., Schmitz R.J., Lister R., Dowen J.M., Nery J.R., Dixon J.E., Ecker J.R. Widespread dynamic DNA methylation in response to biotic stress. Proc Natl Acad Sci USA. 2012;109:E2183–E2191. doi: 10.1073/pnas.1209329109. [DOI] [PMC free article] [PubMed] [Google Scholar]; The first genome-wide DNA methylation analysis in infected plants revealed changes at cytosines of all sequence contexts, partially connected with expression changes of stress related genes.
- 26.Berr A., McCallum E.J., Alioua A., Heintz D., Heitz T., Shen W.H. Arabidopsis histone methyltransferase SET DOMAIN GROUP8 mediates induction of the jasmonate/ethylene pathway genes in plant defense response to necrotrophic fungi. Plant Physiol. 2010;154:1403–1414. doi: 10.1104/pp.110.161497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jaskiewicz M., Conrath U., Peterhaensel C. Chromatin modification acts as a memory for systemic acquired resistance in the plant stress response. EMBO Rep. 2011;12:50–55. doi: 10.1038/embor.2010.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sokol A., Kwiatkowska A., Jerzmanowski A., Prymakowska-Bosak M. Up-regulation of stress-inducible genes in tobacco and Arabidopsis cells in response to abiotic stresses and ABA treatment correlates with dynamic changes in histone H3 and H4 modifications. Planta. 2007;227:245–254. doi: 10.1007/s00425-007-0612-1. [DOI] [PubMed] [Google Scholar]
- 29•.Palma K., Thorgrimsen S., Malinovsky F.G., Fiil B.K., Nielsen H.B., Brodersen P., Hofius D., Petersen M., Mundy J. Autoimmunity in Arabidopsis acd11 is mediated by epigenetic regulation of an immune receptor. PLoS Pathog. 2010;6:e1001137. doi: 10.1371/journal.ppat.1001137. [DOI] [PMC free article] [PubMed] [Google Scholar]; A forward genetic screen for mutants suppressing the autoimmunity phenotype of acd11 mutants identified a histone H3K36me3 methyltransferase (SDG8) and an immune receptor. The expression of the immune receptor depends on SDG8 activity, hence connecting histone modification with autoimmunity and plant resistance to biotic stress.
- 30.de-la-Peña C., Rangel-Cano A., Alvarez-Venegas R. Regulation of disease-responsive genes mediated by epigenetic factors: interaction of Arabidopsis-Pseudomonas. Mol Plant Pathol. 2012;13:388–398. doi: 10.1111/j.1364-3703.2011.00757.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mosher R.A., Durrant W.E., Wang D., Song J., Dong X. A comprehensive structure–function analysis of Arabidopsis SNI1 defines essential regions and transcriptional repressor activity. Plant Cell. 2006;18:1750–1765. doi: 10.1105/tpc.105.039677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim K.C., Lai Z., Fan B., Chen Z. Arabidopsis WRKY38 and WRKY62 transcription factors interact with histone deacetylase 19 in basal defense. Plant Cell. 2008;20:2357–2371. doi: 10.1105/tpc.107.055566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim J., To T.K., Seki M. An epigenetic integrator: New insights into genome regulation, environmental stress responses and developmental controls by histone deacetylase 6. Plant Cell Physiol. 2012;53:794–800. doi: 10.1093/pcp/pcs004. [DOI] [PubMed] [Google Scholar]
- 34.To T.K., Nakaminami K., Kim J.M., Morosawa T., Ishida J., Tanaka M., Yokoyama S., Shinozaki K., Seki M. Arabidopsis HDA6 is required for freezing tolerance. Biochem Biophys Res Commun. 2011;406:414–419. doi: 10.1016/j.bbrc.2011.02.058. [DOI] [PubMed] [Google Scholar]
- 35.Dhawan R., Luo H., Foerster A.M., AbuQamar S., Du H.-N., Briggs S.D., Mittelsten Scheid O., Mengiste T. Histone monoubiquitination1 interacts with a subunit of the mediator complex and regulates defense against necrotrophic fungal pathogens in Arabidopsis. Plant Cell. 2009;21:1000–1019. doi: 10.1105/tpc.108.062364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim D.H., Sung S. Environmentally coordinated epigenetic silencing of FLC by protein and long noncoding RNA components. Curr Opin Plant Biol. 2012;15:51–56. doi: 10.1016/j.pbi.2011.10.004. [DOI] [PubMed] [Google Scholar]
- 37.Alexandre C., Moller-Steinbach Y., Schonrock N., Gruissem W., Hennig L. Arabidopsis MSI1 is required for negative regulation of the response to drought stress. Mol Plant. 2009;2:675–687. doi: 10.1093/mp/ssp012. [DOI] [PubMed] [Google Scholar]
- 38.Borghi L., Gutzat R., Futterer J., Laizet Y., Hennig L., Gruissem W. Arabidopsis retinoblastoma-related is required for stem cell maintenance, cell differentiation, and lateral organ production. Plant Cell. 2010;22:1792–1811. doi: 10.1105/tpc.110.074591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gutzat R., Borghi L., Futterer J., Bischof S., Laizet Y., Hennig L., Feil R., Lunn J., Gruissem W. Retinoblastoma-related protein controls the transition to autotrophic plant development. Development. 2011;138:2977–2986. doi: 10.1242/dev.060830. [DOI] [PubMed] [Google Scholar]
- 40.March-Diaz R., Garcia-Dominguez M., Lozano-Juste J., Leon J., Florencio F.J., Reyes J.C. Histone H2A.Z. and homologues of components of the SWR1 complex are required to control immunity in Arabidopsis. Plant J. 2008;53:475–487. doi: 10.1111/j.1365-313X.2007.03361.x. [DOI] [PubMed] [Google Scholar]
- 41••.Kumar S.V., Wigge P.A. H2A.Z-containing nucleosomes mediate the thermosensory response in Arabidopsis. Cell. 2010;140:136–147. doi: 10.1016/j.cell.2009.11.006. [DOI] [PubMed] [Google Scholar]; A screen for mutants with an aberrant thermosensory response identified a subunit of the SWR1 chromatin remodeling complex that is necessary for deposition of the histone variant H2A.Z. The study demonstrates that eviction of H2A.Z. is crucial for the thermosensory response and could provide a direct mechanism for chromatin-based temperature sensing.
- 42.Scippa G.S., Di Michele M., Onelli E., Patrignani G., Chiatante D., Bray E.A. The histone-like protein H1-S and the response of tomato leaves to water deficit. J Exp Bot. 2004;55:99–109. doi: 10.1093/jxb/erh022. [DOI] [PubMed] [Google Scholar]
- 43.Lang J., Smetana O., Sanchez-Calderon L., Lincker F., Genestier J., Schmit A.-C., Houlné G., Chabouté M.-E. Plant γH2AX foci are required for proper DNA DSB repair responses and colocalize with E2F factors. New Phytol. 2012;194:353–363. doi: 10.1111/j.1469-8137.2012.04062.x. [DOI] [PubMed] [Google Scholar]
- 44.Walley J.W., Rowe H.C., Xiao Y., Chehab E.W., Kliebenstein D.J., Wagner D., Dehesh K. The chromatin remodeler splayed regulates specific stress signaling pathways. PLoS Pathog. 2008;4:e1000237. doi: 10.1371/journal.ppat.1000237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saez A., Rodrigues A., Santiago J., Rubio S., Rodriguez P.L. HAB1–SWI3B interaction reveals a link between abscisic acid signaling and putative SWI/SNF chromatin-remodeling complexes in Arabidopsis. Plant Cell. 2008;20:2972–2988. doi: 10.1105/tpc.107.056705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lang-Mladek C., Popova O., Kiok K., Berlinger M., Rakic B., Aufsatz W., Jonak C., Hauser M.T., Luschnig C. Transgenerational inheritance and resetting of stress-induced loss of epigenetic gene silencing in Arabidopsis. Mol Plant. 2010;3:594–602. doi: 10.1093/mp/ssq014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pecinka A., Dinh H.Q., Baubec T., Rosa M., Lettner N., Mittelsten Scheid O. Epigenetic regulation of repetitive elements is attenuated by prolonged heat stress in Arabidopsis. Plant Cell. 2010;22:3118–3129. doi: 10.1105/tpc.110.078493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tittel-Elmer M., Bucher E., Broger L., Mathieu O., Paszkowski J., Vaillant I. Stress-induced activation of heterochromatic transcription. PLoS Genet. 2010;6:e1001175. doi: 10.1371/journal.pgen.1001175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49•.Strenkert D., Schmollinger S., Sommer F., Schulz-Raffelt M., Schroda M. Transcription factor-dependent chromatin remodeling at heat shock and copper-responsive promoters in Chlamydomonas reinhardtii. Plant Cell. 2011;23:2285–2301. doi: 10.1105/tpc.111.085266. [DOI] [PMC free article] [PubMed] [Google Scholar]; This is a comprehensive study of stress-induced chromatin dynamics at two stress-responsive transcription factors in a model organism suitable for large-scale biochemical analysis.
- 50.Cools T., De Veylder L. DNA stress checkpoint control and plant development. Curr Opin Plant Biol. 2009;12:23–28. doi: 10.1016/j.pbi.2008.09.012. [DOI] [PubMed] [Google Scholar]
- 51.Paszkowski J., Grossniklaus U. Selected aspects of transgenerational epigenetic inheritance and resetting in plants. Curr Opin Plant Biol. 2011;14:195–203. doi: 10.1016/j.pbi.2011.01.002. [DOI] [PubMed] [Google Scholar]
- 52.Boyko A., Blevins T., Yao Y., Golubov A., Bilichak A., Ilnytskyy Y., Hollander J., Meins F., Jr., Kovalchuk I. Transgenerational adaptation of Arabidopsis to stress requires DNA methylation and the function of Dicer-like proteins. PLoS One. 2010;5:e9514. doi: 10.1371/journal.pone.0009514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Luna E., Bruce T.J., Roberts M.R., Flors V., Ton J. Next-generation systemic acquired resistance. Plant Physiol. 2012;158:844–853. doi: 10.1104/pp.111.187468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rasmann S., De Vos M., Casteel C.L., Tian D., Halitschke R., Sun J.Y., Agrawal A.A., Felton G.W., Jander G. Herbivory in the previous generation primes plants for enhanced insect resistance. Plant Physiol. 2012;158:854–863. doi: 10.1104/pp.111.187831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Slaughter A., Daniel X., Flors V., Luna E., Hohn B., Mauch-Mani B. Descendants of primed Arabidopsis plants exhibit resistance to biotic stress. Plant Physiol. 2012;158:835–843. doi: 10.1104/pp.111.191593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Verhoeven K.J., van Gurp T.P. Transgenerational effects of stress exposure on offspring phenotypes in apomictic dandelion. PLoS One. 2012;7:e38605. doi: 10.1371/journal.pone.0038605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Whittle C.A., Otto S.P., Johnston M.O., Krochko J.E. Adaptive epigenetic memory of ancestral temperature regime in Arabidopsis thaliana. Botany-Botanique. 2009;87:650–657. [Google Scholar]
- 58.Conrath U. Molecular aspects of defence priming. Trends Plant Sci. 2011;16:524–531. doi: 10.1016/j.tplants.2011.06.004. [DOI] [PubMed] [Google Scholar]
- 59.van den Burg H.A., Takken F.L.W. Does chromatin remodeling mark systemic acquired resistance? Trends Plant Sci. 2009;14:286–294. doi: 10.1016/j.tplants.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 60.Birney E. Chromatin and heritability: how epigenetic studies can complement genetic approaches. Trends Genet. 2011;27:172–176. doi: 10.1016/j.tig.2011.02.005. [DOI] [PubMed] [Google Scholar]
- 61.Pecinka A., Mittelsten Scheid O. Stress-induced chromatin changes: a critical view on their heritability. Plant Cell Physiol. 2012;53:801–808. doi: 10.1093/pcp/pcs044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Herman J., Se S. Adaptive transgenerational plasticity in plants: case studies, mechanisms, and implications for natural populations. Front Plant Sci. 2011;2 doi: 10.3389/fpls.2011.00102. Article 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jablonka E., Raz G. Transgenerational epigenetic inheritance: prevalence, mechanisms, and implications for the study of heredity and evolution. Q Rev Biol. 2009;84:131–176. doi: 10.1086/598822. [DOI] [PubMed] [Google Scholar]
- 64•.Durand S., Bouche N., Perez Strand E., Loudet O., Camilleri C. Rapid establishment of genetic incompatibility through natural epigenetic variation. Curr Biol: CB. 2012;22:326–331. doi: 10.1016/j.cub.2011.12.054. [DOI] [PubMed] [Google Scholar]; The authors present an example of a natural occurring epiallele that is responsible for genetic incompatibility between different Arabidopsis ecotypes. This epiallel is stable over several generations and can trigger RdDM in trans.