Abstract
Sulfur is both crucial to life and a potential threat to health. While colonic sulfur metabolism mediated by eukaryotic cells is relatively well studied, much less is known about sulfur metabolism within gastrointestinal microbes. Sulfated compounds in the colon are either of inorganic (e.g., sulfates, sulfites) or organic (e.g., dietary amino acids and host mucins) origin. The most extensively studied of the microbes involved in colonic sulfur metabolism are the sulfate-reducing bacteria (SRB), which are common colonic inhabitants. Many other microbial pathways are likely to shape colonic sulfur metabolism as well as the composition and availability of sulfated compounds, and these interactions need to be examined in more detail. Hydrogen sulfide is the sulfur derivative that has attracted the most attention in the context of colonic health, and the extent to which it is detrimental or beneficial remains in debate. Several lines of evidence point to SRB or exogenous hydrogen sulfide as potential players in the etiology of intestinal disorders, inflammatory bowel diseases (IBDs) and colorectal cancer in particular. Generation of hydrogen sulfide via pathways other than dissimilatory sulfate reduction may be as, or more, important than those involving the SRB. We suggest here that a novel axis of research is to assess the effects of hydrogen sulfide in shaping colonic microbiome structure. Clearly, in-depth characterization of the microbial pathways involved in colonic sulfur metabolism is necessary for a better understanding of its contribution to colonic disorders and development of therapeutic strategies.
Keywords: sulfur, hydrogen sulfide, colonic microbiota, sulfate-reducing bacteria, inflammatory bowel disease, colorectal cancer, irritable bowel syndrome
Introduction
The crucial role of the microbiome in digestive processes and intestinal health is now fully integrated in gastroenterology research (Gordon et al., 2007). The immense number of microbes resident in the small intestine and colon form complex communities and metabolic interactions, which impact the host beneficially or detrimentally. Accordingly, the intestinal microbiota plays a central role in digestive biochemistry and element cycling. Whereas abundant data are available regarding microbial carbon metabolism (carbohydrate or fiber degradation), less is known about microbial metabolism of other elements in the human intestine.
Historically, the sulfur cycle was one of the first to be well documented in environmental microbiology (Kertesz, 2000; Canfield and Farquhar, 2012). Chemolithoautotrophic and metabolic conversion of sulfur compounds were first described in the late 19th century by Beijerinck (Kelly et al., 1997) and are of central interest in the context of environmental pollution (Kellogg et al., 1972). Many sulfur compounds are also recognized for their toxicity and impact on human health (Ware et al., 1981). While there is no evidence for a microbial sulfur cycle per se in digestive systems, direct and indirect evidence points to a greater diversity and influence of microbial sulfur metabolism in the human intestine than previously recognized.
The sulfur present in the human body is provided exclusively by diet and either converted to sulfated compounds, assimilated by host cells or excreted. Sulfur is generally acquired in the human diet through protein (Stipanuk, 2004). It is noteworthy that one essential and another conditionally essential amino acid (methionine and cysteine, respectively) are sulfated, and thus, sulfur acquisition is crucial to humans. Cysteine is considered a conditionally essential amino acid as it can be synthesized from methionine via transsulfuration (Dominy and Stipanuk, 2004). As reviewed elsewhere, in addition to dietary inputs, host sulfur amino acids are actively recycled through a wide array of metabolic pathways (Stipanuk, 1986, 2004; Stipanuk and Dominy, 2006). Similar to the host, gut microbes require sulfur inputs and, because of their active metabolism and tremendous number, are likely to play a major role in the metabolism of luminal sulfur. Consequently, gut microbes are key players in determining the balance of beneficial to detrimental effects of sulfur-containing compounds (Table 1). This review summarizes microbial pathways influencing sulfur metabolism and their recognized and potential contributions to colonic health and disease.
Table 1.
Microbial taxa involved in pathways of colonic sulfur metabolism*.
| Sulfur source | Sulfur-containing substrate | Microbial genus | References |
|---|---|---|---|
| Inorganic | Sulfate (SO42−) | Desulfovibrio spp. (Desulfomonas spp) | Gibson et al., 1988; Fite et al., 2004; Marchesi et al., 2009; Nava et al., 2012 |
| Desulfobacter spp. | Gibson et al., 1988; Nava et al., 2012 | ||
| Desulfobulbus spp. | Gibson et al., 1988; Nava et al., 2012 | ||
| Desulfotomaculum spp. | Gibson et al., 1988; Nava et al., 2012 | ||
| Sulfite (SO32−) | Bilophila wadsworthia | Baron et al., 1989 | |
| Campylobacter jejuni | Kelly and Myers, 2005 | ||
| Organic | Cysteine | Escherichia coli | Metaxas and Delwiche, 1955; Shatalin et al., 2011 |
| Staphylococcus aureus | Shatalin et al., 2011 | ||
| Salmonella thyphimurium | Kredich et al., 1972 | ||
| Mycobacterium tuberculosis | Wheeler et al., 2005 | ||
| Helicobacter pylori | Kim et al., 2006 | ||
| Leishmania major (protozoa) | Nowicki et al., 2010 | ||
| Prevotella intermedia | Igarashi et al., 2009 | ||
| Fusobacterium nucleatum | Yoshida et al., 2010 | ||
| Streptococcus anginosus | Yoshida et al., 2011 | ||
| Clostridium spp. | Genomic cysteine desulfhydrase | ||
| Enterobacter spp. | Genomic cysteine desulfhydrase | ||
| Klebsiella spp. | Genomic cysteine desulfhydrase | ||
| Desulfovibrio spp. | Genomic cysteine desulfhydrase | ||
| Sulfomucin | Prevotella strain RS2 | Roberton et al., 2000 | |
| Bacteroides fragilis | Roberton et al., 2000 | ||
| Helicobacter pylori | Slomiany et al., 1992 | ||
| Akkermansia spp. | Genomic glycosulfatase | ||
| Taurine | Bilophila wadsworthia | Laue et al., 1997 | |
| Sulfated bile acids (rat model) | Clostridium perfringens | Huijghebaert and Eyssen, 1982; Huijghebaert et al., 1982; Robben et al., 1986 | |
| Estrogen-3-sulfates and phenylsulfates | Peptococcus niger | Vaneldere et al., 1991 |
Assimilatory sulfate reduction, widespread and present in virtually all microbes, has not been included.
Intestinal sources of sulfur
Food sources of inorganic sulfate include commercial breads, dried fruits, vegetables, nuts, fermented beverages, and brassica vegetables (Florin et al., 1991). Diets supplemented with inorganic sulfate stimulate hydrogen sulfide (H2S) production within the colon (Christl et al., 1992; Lewis and Cochrane, 2007). Furthermore, in vitro incubation studies using human feces indicate that organic sulfur-containing compounds including cysteine, taurocholic acid, and mucin provide a more efficient source for sulfide production than inorganic sulfate (Florin, 1991; Levine et al., 1998), with meat being a particularly important source (Magee et al., 2000). In particular red meat, eggs, and milk contain elevated concentrations of cysteine. Concentrations of both free cysteine and methionine in colon are relatively low (Ahlman et al., 1993), indicating efficient metabolism by the microbiota and host epithelial cells. An additional source of colonic sulfur includes the sulfomucins. Mucins, consisting of a peptide backbone, are largely responsible for the viscous properties of the colonic mucus layer and can be broadly classified as sialomucins or sulfomucins based on the presence of terminal sialic acid or sulfate groups, respectively, on the oligosaccharide chain (Verdugo, 1990; Jass and Roberton, 1994; Croix et al., 2011).
Many other trace sulfated compounds are provided by dietary elements. In particular, fermented foodstuffs contain a wide array of volatile sulfur compounds (Landaud et al., 2008). Microbial and host cell metabolism also produce a large variety of simple to complex sulfated compounds available for further microbial utilization or degradation.
Microbial pathways involved in colonic sulfur metabolism
Assimilatory sulfate reduction
Among microbes, assimilatory sulfate reduction is the most widespread and essential biochemical process linked to sulfur and provides a mechanism by which microorganisms can reduce sulfate to elemental sulfur in order to satisfy physiological requirements (Peck, 1961). This process is mediated by phosphoadenosine-5′-phosphosulfate (PAPS) reductases (Figure 1), which are widely distributed among microbes and other living organisms except animals (for example 2924 hits for annotated nucleotide sequences and 10,402 for protein sequences were found in the NCBI database). Assimilatory sulfate reduction can also be performed by other enzymatic pathways (Vaneldere et al., 1991; Seitz and Leadbetter, 1995) and likely represents a crucial step in sulfur cycling as it allows for significant disposal of sulfate and conversion to necessary organic sulfated compounds.
Figure 1.
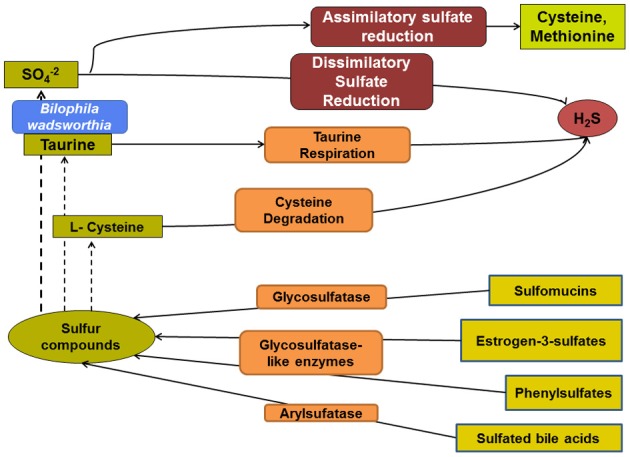
Overview of the microbial pathways involved in colonic sulfur metabolism. The sulfate remaining from assimilatory sulfate reduction is available for sulfate-reducing bacteria and, thus, H2S production. Taurine and cysteine are additional potentially important substrates for microbial production of H2S. Various microbial metabolic pathways influence the composition and relative abundance of organic sulfur compounds.
Dissimilatory sulfate reduction
While the process of assimilatory sulfate reduction is widespread among microbes, only restricted microbial groups are capable of dissimilatory sulfate reduction (Figure 2). The sulfate-reducing bacteria (SRB) are notable as the only microbes in intestinal ecosystems that rely on inorganic sulfate for conservation of energy. This sulfate respiration pathway has been conserved in five bacterial and two archaeal phyla (Wagner et al., 1998). Most of the SRB belong to the Deltaproteobacteria with more than 25 genera, followed by the Gram-positive SRB within the Clostridia (Desulfotomaculum, Desulfosporosinus, and Desulfosporomusa genera). The SRB use sulfate as a terminal electron acceptor for respiration, with the concomitant production of H2S (Peck, 1961). In the colon, sulfate reduction is generally associated with dihydrogen oxidation (Carbonero et al., 2012), but the electrons may also be provided from the oxidation of organic compounds, such as lactate (Flint et al., 2009). SRB are ubiquitously present in the human intestinal mucosa (Nava et al., 2012) and have been enumerated by cultivation-dependent methods from human stool in numbers ranging from 103 to 1011/g (Leclerc et al., 1980; Gibson et al., 1988). In a culture-based study by Gibson et al., the principal SRB were lactate- and hydrogen-utilizing Desulfovibrio spp. (64–81%), acetate-utilizing Desulfobacter spp. (9–16%), propionate- and hydrogen-utilizing Desulfobulbus spp. (5–8%), lactate-utilizing Desulfomonas spp (reclassified within the genus Desulfovibrio) (3–10%), and acetate- and butyrate-utilizing Desulfotomaculum spp. (2%). However, these observations are based on cultivation methods, which underestimate true bacterial diversity. More recently molecular-based techniques have been applied successfully to describing SRB diversity in various environments. SRB have thus far been detected and quantified from stool and colonic mucosa samples (Zinkevich and Beech, 2000; Fite et al., 2004; Nava et al., 2012). The genus Desulfovibrio has generally been found in higher numbers (Fite et al., 2004; Marchesi et al., 2009), with lower abundances of Desulfobacter, Desulfobulbus, and Desulfotomaculum (Nava et al., 2012). The biochemistry of dissimilatory sulfate reduction has been investigated most extensively within species of Desulfovibrio. In the environment, SRB are able to utilize a wide range of substrates as electron donors and acceptors and can even adopt non-sulfidogenic lifestyles (Plugge et al., 2011). For example Desulfobulbus spp. can ferment organic matter under certain conditions (Kuever et al., 2005). It is not known if colonic SRB possess the ability to switch from sulfidogenic to non-sulfidogenic lifestyles in the colon, but an ability to adapt to varying sulfate levels would confer a competitive advantage.
Figure 2.
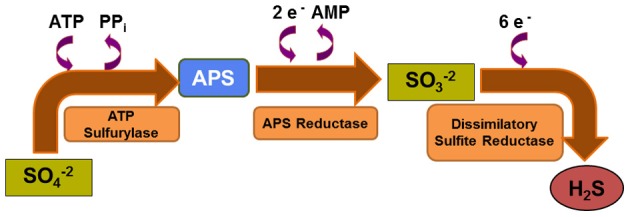
Dissimilatory sulfate reduction pathway. Microbial sulfate reduction relies on sequential catalytic reactions in which reduction of sulfate is coupled with oxidation of H2 or simple organic molecules. This anaerobic respiration pathway is less favorable thermodynamically than aerobic respiration.
Cysteine degradation
Cysteine degradation to H2S mediated by L-cysteine desulfhydrase (Figure 3) was described in the 1950s in the Escherichia coli model, a well-known inhabitant of the human gut (Metaxas and Delwiche, 1955). Since that time, L- or D-cysteine desulfhydrase have been described in diverse taxa (Kumagai et al., 1975) including a few intestinal pathogens such as Salmonella thyphimurium (Kredich et al., 1972), Mycobacterium tuberculosis (Wheeler et al., 2005), H. pylori (Kim et al., 2006), and the protozoan Leishmania major (Nowicki et al., 2010). L- or D-cysteine desulfhydrase have also been studied in various oral microorganisms as a source of malodor and abscess formation (Igarashi et al., 2009; Yoshida et al., 2010, 2011). Those genera found in the oral cavity, namely Streptococcus, Prevotella and Fusobacterium, are also common inhabitants of the human colon. A search in the GenBank and EBI databases revealed that cysteine desulfhydrase-encoding genes have been annotated in several common colonic genera such as Clostridium, Enterobacter, Klebsiella, Streptococcus, and, surprisingly, the SRB genus Desulfovibrio. Although desulfhydrase activity has not been quantified in intestinal ecosystems and the prevalence of the gene among intestinal taxa remains undefined, it appears likely that desulfhydrase-harboring microbes are important producers of H2S in the human colon.
Figure 3.
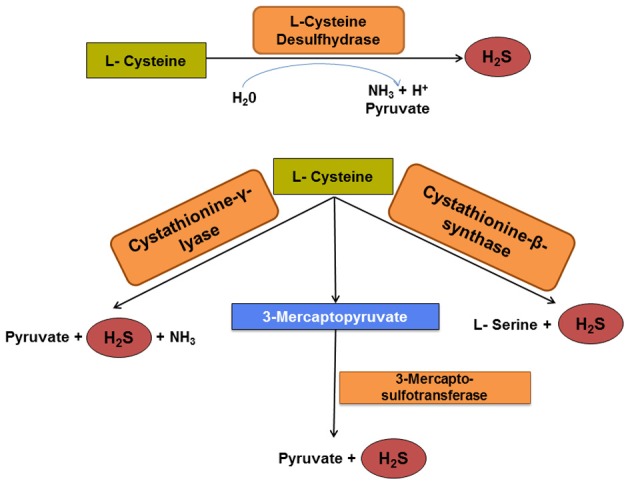
Pathways of cysteine degradation to H2S. Cysteine desulfhydrase is a key enzyme for initial microbial cysteine fermentation and pyruvate production. However, recent evidence indicates that three other enzymes orthologous to eukaryotic enzymes may catalyze similar reactions. The generation of H2S via these four enzymatic pathways may surpass that by dissimilatory sulfate reduction, especially in hosts consuming a protein-rich diet.
Other microbial enzymes possess cysteine desulfhydrase activity, and, more generally, the ability to convert cysteine or cysteine analogs is widespread. Cysteine desulfidases, that carry out reactions similar to other desulfhydrases, have been described in environmental archaea (Tchong et al., 2005). Beta C-S lyases or cystathionases mediates the cleavage of cystathionine to homocysteine in E. coli (Zdych et al., 1995) and Lactobacillus spp. (De Angelis et al., 2002; Irmler et al., 2008). Another example is the SRB Desulfovibrio desulfuricans, which is able to obtain sulfate from cysteine degradation (Forsberg, 1980). Conversely, the prevalent O-acetyl-L-serine sulfhydrylase (OASS) and homocysteine synthase mediate the microbial synthesis of cysteine from O-acetyl-L-serine and sulfide in bacteria and archaea (Borup and Ferry, 2000; Tai and Cook, 2001).
A recent paper provided evidence that a high proportion of bacterial genomes harbor orthologs of mammalian cystathionine β-synthase (CBS), cystathionine γ-lyase (CSE), and 3-mercaptopyruvate sulfurtransferase (3MST), three enzymes involved in H2S production (Shatalin et al., 2011). The authors confirmed that pathogenic strains, including E. coli and Staphylococcus aureus, converted cysteine to H2S through activities of one of those enzymes. Thus, it may be hypothesized that many gut microbes are capable of cysteine conversion to H2S. Further, the authors demonstrated that sulfide production reduces oxidative stress, thereby increasing antibiotic resistance potential (Shatalin et al., 2011). Indeed, those microbial metabolic pathways are very poorly documented in gut commensals, and characterization of these enzymes and associated genes will be crucial to understanding if microbial-generated sulfide plays a role in shaping the colonic ecosystem.
Taurine and sulfite respiration
Only a few genera are able to gain energy through sulfite respiration rather than direct sulfate respiration (Pukall et al., 1999; Soulimane et al., 2011). Among those, the only intestinal resident characterized to date is Bilophila wadsworthia, a close relative of Desulfovibrio spp. (Baron et al., 1989). This unique bacterial species has attracted much attention because of its links with various disease symptoms (Finegold et al., 1992; Arzese et al., 1997; Aucher et al., 1998; Nakayama et al., 2000; Devkota et al., 2012). It reduces sulfite after an initial degradation of taurine and harbors a different dissimilatory sulfite reductase than other SRB (Laue et al., 2001) (Figure 4). A sulfite oxidation metabolic pathway has also been demonstrated in the food-borne human pathogen Campylobacter jejuni (Kelly and Myers, 2005); however, the existence of similar pathways in intestinal commensal microbes has not been described.
Figure 4.
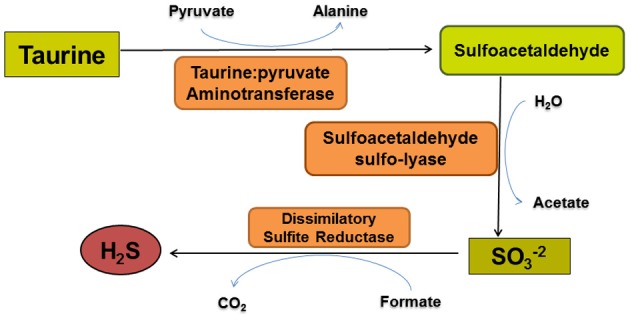
The taurine degradation pathway of Bilophila wadsworthia. Bilophila wadsworthia is the only known intestinal microbe that uses taurine as an electron acceptor for anaerobic respiration. The first two enzymatic reactions result in sulfite production. Sulfite is subsequently converted to H2S by a dissimilatory sulfite reductase that differs structurally from those used by sulfate-reducing bacteria.
Taurine, a sulfated compound secreted by the eukaryotic host that is crucial for chelating bile acids in addition to contributing to osmoregulation, cardiac function, and retinal development is degraded by B. wadsworthia using a taurine:pyruvate aminotransferase to obtain sulfite (Laue et al., 1997; Laue and Cook, 2000) (Figure 4). An intriguing link between dietary fat and the utilization of taurochloric acid as a source of taurine for B. wadsworthia is suggested by two recent studies (Swann et al., 2011; Devkota et al., 2012). Compared to mice fed a low fat or high polyunsaturated fat diet, mice fed a high milk fat diet demonstrated an overrepresentation of B. wadsworthia, which was determined to be mediated by taurine-conjugated bile acids (Devkota et al., 2012). Furthermore, milk fat was essential for colonization by B. wadsworthia in the germ-free mouse colon. Swann et al. (2011) demonstrated that the lack of an established microbiota results in a dominance of taurine-conjugated bile acids over unconjugated or glycine-conjugated bile acids, suggesting that bile acids, whose composition is modulated by the gut microbiota, may serve as signaling molecules in liver and other tissues (Swann et al., 2011). Although taurine has also been demonstrated to be a substrate for SRB in marine microbial mats (Visscher et al., 1999), it is not known if colonic SRB are capable of taurine utilization.
Arylsulfatases
Bile acids can be sulfated by the action of host sulfotransferases, which allow their subsequent detoxification and disposal (Alnouti, 2009). It was demonstrated in a rat model that the opposite conversion, bile acid desulfation, can be mediated by microbial enzymes with arylsulfatase activity (Robben et al., 1988). Arylsulfatases were purified from Clostridium perfringens (Huijghebaert and Eyssen, 1982; Huijghebaert et al., 1982; Robben et al., 1986) inhabiting the rat intestine and are also widespread in several abundant colon genera. Little arylsulfatase activity was reported in human stool (McBain and Macfarlane, 1998), which does not preclude a more extensive activity of microbial arylsulfatase in the colonic ecosystem where significant concentrations of sulfated bile acids are found (Palmer, 1967; Hofmann et al., 2008).
Glycosulfatases
Some colonic microbes have evolved to degrade colonic mucins (Roberton and Stanley, 1982; Derrien et al., 2004, 2008). In particular, microbial glycosulfatases catalyze the release of sulfate from sulfomucins (Corfield et al., 1992; Roberton and Wright, 1997). Glycosulfatase enzymes have been described primarily in the intestinal resident Prevotella strain RS2 and Bacteroides fragilis (Roberton et al., 2000), but also in the stomach resident Helicobacter pylori (Slomiany et al., 1992). A suite of glycosulfatase-like enzymes were also described in the intestinal Peptococcus niger, using less abundant substrates such as estrogen-3-sulfates and phenylsulfates (Vaneldere et al., 1991). Relative abundance and activity of glycosulfatase-harboring bacteria may greatly influence colonic sulfur metabolism; however, little is known about prevalence, abundance, and activity of bacterial glycosulfatases. A search in the GenBank and EBI databases confirmed that glycosulfatases encoding genes have been annotated in important colonic genera such as Prevotella, Bacteroides (Arumugam et al., 2011), and Akkermansia (Belzer and De Vos, 2012).
Links with colonic health and disease
Hydrogen sulfide
Toxicity
Hydrogen sulfide is the most suspect sulfated compound in the etiology of colonic disorders and inflammation (Nakamura et al., 2010; Danese and Fava, 2011; Medani et al., 2011). In addition, it is among the most hazardous gases in industrial applications, exhibiting toxicity to different organs at low concentrations, with increasing concentrations being fatal (Guidotti, 1996). There is ample evidence that H2S at physiological concentrations is genotoxic and induces DNA damage as well as inflammatory responses (Attene-Ramos et al., 2006, 2007, 2010). Exogenous hydrogen sulfide is highly toxic to colonocytes and impairs their metabolic function, especially butyrate oxidation (Roediger et al., 1993; Babidge et al., 1998). In the human colon, sulfide exists largely in the volatile, highly toxic undissociated form (H2S), which is quickly absorbed by the mucosa or passes as flatus (Suarez et al., 1997). Over 90% of sulfate disappears during passage through the colon of individuals lacking a sulfate-reducing microbiota, indicating that additional colonic processes compete for sulfate (Strocchi et al., 1993).
Detoxification
Hydrogen sulfide is oxidized in many tissues including colonic mucosa (Bartholomew et al., 1980; Furne et al., 2001), which presumably is exposed frequently to bacterial-derived sulfide, given the persistent colonization of the human colonic mucosa by SRB (Nava et al., 2012). The first step in the sulfide oxidation pathway is catalyzed by sulfide quinone reductase (SQR), a mitochondrial flavoprotein that oxidizes H2S to protein-bound persulfide. Subsequently, a sulfur dioxygenase and a sulfur transferase are thought to sequentially convert SQR-bound persulfide into sulfite and thiosulfate, respectively (Wilson et al., 2008; Kabil and Banerjee, 2010). Given that most of the colonic disorders for which a potential pathogenic role for sulfide exists are effectively gene-environment disorders, it is straight-forward to envision how common polymorphisms in host genes encoding components of the sulfide oxidation pathway might underlie ineffective epithelial sulfide detoxification and thereby predispose certain individuals to chronic sulfide-generated inflammation or genotoxicity.
H2S as an endogenous signaling molecule
Despite its potential inflammatory, toxic and genotoxic properties, there is also evidence that human cells may be able to use H2S as a inorganic substrate for mitochondrial energization (Bouillaud et al., 2007). It is postulated that H2S may be an important endogenous signaling molecule (Abe and Kimura, 1996; Distrutti et al., 2006). In addition, H2S has been suggested to be an effective drug against various GI diseases in a rodent model (Wallace et al., 2007, 2009; Wallace, 2010). However, the potential therapeutic properties of exogenous H2S still remain controversial (Bouillaud and Blachier, 2011; Olson, 2011). Intriguingly, a recent report demonstrated that inhibition of H2S synthesis in healthy rats resulted in mucosal injury and inflammation in the small intestine and colon, while intracolonic administration of H2S donors significantly reduced the severity of trinitrobenzene sulfonic acid-induced colitis (Wallace et al., 2009). These data indicate that the outright assumption that colonic H2S is only deleterious may be challenged and justify additional study of both bacterial and endogenous sources of H2S in the human colon.
H2S as an architect of microbiome structure
In the context of culture-based microbiology, sulfide in different forms has been used as a strong reducing agent, allowing maintenance of anoxia in media. However, sulfide concentrations must be kept low to avoid toxicity in microbial cells. For example, in microcosms simulating anaerobic digesters, lactose fermentation and methanogenesis were inhibited by sulfide in a dose-dependent manner. Interestingly, it was also observed that SRB activities were inhibited by increased sulfide concentrations (Hilton and Oleszkiewicz, 1988). In surface sediments of sandy intertidal flats, high sulfide concentrations may reduce functional diversity (Freitag et al., 2003). Finally, sulfide production was shown to provide cytoprotection from oxidative stress (Shatalin et al., 2011). As a start, in vitro studies with representative colonic strains are needed to better understand the potential for sulfide as well as sulfated compounds to influence the structure of colonic ecosystem.
Inflammatory bowel disease
The two major forms of inflammatory bowel disease (IBD), Crohn's disease (CD), and ulcerative colitis (UC), afflict 0.1–0.5% of individuals in Western countries (Podolsky, 2002; Hanauer, 2006). It is commonly accepted that IBD results from multifactorial interactions among genetic and environmental factors that lead to a dysregulation of the innate immune response to the intestinal microbiota in genetically predisposed individuals (Podolsky, 2002).
Substantial evidence exists for a potential pathogenic role of H2S in IBD, particularly in UC (Pitcher and Cummings, 1996). Healthy colonic epithelial cells depend on the availability of short-chain fatty acids such as butyrate for nutrition. Butyrate is produced during colonic fermentation and oxidized by colonocytes via the enzyme acyl-CoA dehydrogenase. Because this enzyme is inhibited by H2S, oxidation of butyrate is impaired by H2S (Babidge et al., 1998). Patients with UC were shown to have reduced breath excretion of CO2 after administration of butyrate intrarectally, reflecting reduced colonic butyrate oxidation (Den Hond et al., 1998). UC patients with reduced colonic butyrate oxidation also exhibit increased intestinal permeability (Den Hond et al., 1998). Indeed, as ion absorption, mucin synthesis, membrane lipid synthesis, and detoxification processes in colonocytes depend on the oxidation of butyrate (Roediger et al., 1997), decreases of butyrate oxidation would be expected to compromise epithelial barrier function (Ramakrishna et al., 1991).
Patients with UC have been shown to consume more protein and, thereby, more sulfur amino acids than control subjects (Tragnone et al., 1995). Removing foods such as milk, cheese, and eggs improved symptoms in UC patients (Truelove, 1961). Further, the numbers of SRB and rate of sulfidogenesis were reported greater in UC patients than control cases (Gibson et al., 1991; Pitcher et al., 2000). In a study comparing patients with IBD to healthy individuals or patients with other gastrointestinal symptoms, the prevalence of Desulfovibrio piger detected via PCR was significantly higher in the IBD patients (Loubinoux et al., 2002). Levine et al. (1998) found that production of H2S from feces of patients with UC was 3–4 times greater than from feces of healthy controls. However, this difference in H2S production may not have been due to colonization by a greater number of SRB, as qPCR did not show that patients with active UC harbored more SRB in stool or rectal mucosa than healthy controls (Fite et al., 2004). A common treatment regimen for patients with UC may contribute to conflicting results regarding the density of SRB populations in UC patients. Specifically, 5-aminosalicylic acid (5-ASA), an anti-inflammatory medication commonly prescribed for UC, also inhibits SRB growth and production of H2S (Pitcher et al., 2000; Edmond et al., 2003). For example, stool sulfide concentrations between patients with UC and non-colitic controls did not differ when the use of salicylates in colitic patients was not controlled (Moore et al., 1998); in those patients with UC who were not administered 5-ASA, fecal sulfide concentrations were significantly greater (Pitcher et al., 2000).
Additional evidence for the role of H2S in UC is the observation that SRB were found in surgically constructed ileo-anal pouches of UC patients but not in pouches of patients with familial adenomatous polyposis (FAP). Furthermore, H2S production in UC pouches was 10 times greater than that in FAP pouches (Duffy et al., 2002). Finally, in a later study, the severity of pouchitis was positively correlated with fecal concentrations of H2S (Ohge et al., 2005), possibly reflecting a pathogenic role for this gas. Coffey et al. (2009) proposed that the surgical creation of ileo-anal pouches in UC patients might lead to colonic metaplasia, which, in turn, could result in increased production of sulfomucin and, thus, higher colonization by SRB. The adverse consequences of such colonization would be greater exposure to potentially proinflammatory concentrations of H2S (Coffey et al., 2009).
Increased activity of mucin sulfatase, an enzyme belonging to the glycosulfatase group, was observed in patients with active UC but not CD (Tsai et al., 1995). In most patients, fluctuations in fecal sulfatase activity corresponded with severity of clinical disease, suggesting that the increased fecal sulfatase activity contributed to perpetuation of the disease. In this scenario, individuals genetically predisposed to a high SRB carriage rate who then experience increased sulfatase activity might be at increased risk of UC due to the increased availability of endogenous sulfate for SRB sulfide production. Similarly, diets high in exogenous sources of sulfate would represent the greatest risk for those genetically predisposed to a higher abundance of SRB.
Recently, a culture-dependent study demonstrated that mucosal biopsies from IBD patients were more often colonized by Fusobacterium spp. than those from matched healthy controls (Strauss et al., 2011). It was further demonstrated that Fusobacterium isolates from IBD patients trigger inflammatory pathways in colonic cells (Dharmani et al., 2011). Fusobacterium spp. produce H2S through cysteine desulfhydrase activity. It also appears possible that direct consumption of cysteine can reduce in some instances sulfate availability for SRB, thereby possibly explaining the conflicting reports of SRB association with IBD. The recent demonstration of selection for B. wadsworthia in the gut microbiota of mice fed a high-saturated fat diet that was associated with higher occurrence and severity of colitis compared to those fed high-unsaturated fat (Devkota et al., 2012) provides a potential mechanistic model for the development of IBD in susceptible individuals. Notably, this effect was mediated by an increase of specific bile acids, particularly taurocholic acid, a potential source of taurine for B. wadsworthia.
Colorectal cancer
Colorectal cancer is the third most frequent cancer worldwide and the fourth most common cause of cancer death, with responsibility for 4,92,000 deaths annually (Weitz et al., 2005; Ferlay et al., 2010). Genetic and environmental factors play a significant role in the development of colorectal cancer (Kinzler and Vogelstein, 1996; Rhodes and Campbell, 2002; de la Chapelle, 2004). Although etiologically divided into sporadic (90% of the cases), hereditary (5–10%), and IBD-associated (2%), all colorectal cancers show multistep development with several mutations (Kinzler and Vogelstein, 1996; Rhodes and Campbell, 2002; de la Chapelle, 2004). Doll and Peto (1981) estimated that over 90% of gastrointestinal cancers are determined by environmental factors such as diet. It has been suggested that environmental cancer risk is determined by the interaction between diet and colonic microbial metabolism (O'Keefe et al., 2007). Particularly, there is strong epidemiologic and experimental evidence that diets with high animal fat and protein (meat) are associated with increased risk of colorectal cancer (Willett et al., 1990; Sandhu et al., 2001; Norat et al., 2002). As discussed earlier, meat contains relatively high dietary sulfur, which can promote microbial sulfate reduction in the colon.
Kanazawa and colleagues (1996) demonstrated that H2S concentrations were significantly greater in patients who had previously undergone surgery for sigmoid colon cancer and who later developed new epithelial neoplasia of the colon, compared to individuals of similar age with a healthy colon. The ability of the colon to detoxify H2S is also reduced in patients with colon cancer (Ramasamy et al., 2006). The association of H2S with colon cancer is further supported by the finding that H2S induces colonic mucosal hyperproliferation, with this effect reversed by butyrate (Christl et al., 1996). The mucosal hyperproliferation effect of H2S may be mediated by mitogen-activated protein kinase (MAPK)-mediated proliferation (Deplancke and Gaskins, 2003). Hydrogen sulfide is also a potent genotoxin that induces direct radical-associated DNA damage (Attene-Ramos et al., 2006, 2007). Colon cancer in UC and, perhaps, sporadic colon cancer in general may reflect genomic instability resulting from exposure to H2S (Attene-Ramos et al., 2007). As the number of SRB was reported to be not significantly different (Balamurugan et al., 2008) or reduced in colorectal cancer patients when compared to healthy controls (Scanlan et al., 2009), impaired detoxification of H2S may be critical to the role of this compound in colon cancer.
Increased detection of Fusobacterium spp. in CRC tumors has been reported in two independent studies (Kostic et al., 2011; Castellarin et al., 2012). As described previously, Fusobacterium spp. produce H2S through cysteine desulfhydrase activity. As H2S has been demonstrated to be genotoxic, it can be hypothesized that it may be one causative metabolite in the etiology of CRC. It also appears possible that direct bacterial consumption of cysteine would limit sulfate availability for SRB, thereby possibly explaining the conflicting reports of SRB association with CRC.
Irritable bowel syndrome
Irritable bowel syndrome (IBS) is a functional bowel disorder characterized by chronic abdominal pain, bloating, and abnormal bowel habits (Mayer, 2008). Diarrhea or constipation may predominate or these symptoms may alternate. Microbial dysbioses have been described, and as microbial gases excreted in breath are higher in IBS patients, abnormal microbial pathways are a possible cause (King et al., 1998).
Consistent with this possibility, it was demonstrated recently that exogenous H2S (NaHS) inhibits in vitro motor patterns in the human, rat and mouse colon and jejunum mainly through an action on multiple potassium channels (Gallego et al., 2008). Exogenous H2S (NaHS) inhibited nociception induced by colorectal distension in both healthy and post-colitic rats (Distrutti et al., 2006) but, conversely, triggered visceral nociceptive behavior when administered intracolonically in the mouse (Matsunami et al., 2009). Recently, a functional dysbiosis was described in constipation-predominant IBS patients with, in particular, a significant increase of SRB and fecal sulfide, indicating a potential role for sulfide in the development of IBS symptoms (Chassard et al., 2012).
Conclusions
Microbial sulfur metabolism in the human colon is likely more extensive than has been previously recognized. For example, H2S, the sulfated compound with the highest potential influence on digestive health, can be generated from cysteine degradation as well as via dissimilatory sulfate reduction. The extent to which organic sulfur metabolism is operative in the human colon, how diet influences relative activities of the enzymatic pathways involved, and which organisms carry out these processes are poorly understood. In addition, the importance of the microbiota and the metabolism of sulfated bile acids are now established, and further work is needed to understand how dietary fat intake influences these pathways. With clear evidence that the human colonic mucosa is persistently colonized by SRB, the beneficial versus toxic effects of H2S need to be delineated. One possibility is that the mucosal microbiome is shaped in part through the differential susceptibility of mutualistic microbes to sulfide toxicity. Clearly, extensive work is needed to understand the primary pathways for the production of H2S and how their activities may influence both host and microbial components of the colonic ecosystem. This new body of knowledge may identify relatively innocuous approaches for the prevention or treatment of chronic colonic disorders.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Abe K., Kimura H. (1996). The possible role of hydrogen sulfide as an endogenous neuromodulator. J. Neurosci. 16, 1066–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahlman B., Leijonmarck C. E., Lind C., Vinnars E., Wernerman J. (1993). Free amino acids in biopsy specimens from the human colonic mucosa. J. Surg. Res. 55, 647–653 10.1006/jsre.1993.1198 [DOI] [PubMed] [Google Scholar]
- Alnouti Y. (2009). Bile acid sulfation: a pathway of bile acid elimination and detoxification. Toxicol. Sci. 108, 225–246 10.1093/toxsci/kfn268 [DOI] [PubMed] [Google Scholar]
- Arumugam M., Raes J., Pelletier E., Le Paslier D., Yamada T., Mende D. R., et al. (2011). Enterotypes of the human gut microbiome. Nature 473, 174–180 10.1038/nature09944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arzese A., Mercuri F., Trevisan R., Menozzi M. G., Botta G. A. (1997). Recovery of Bilophila wadsworthia from clinical specimens in Italy. Anaerobe 3, 219–224 10.1006/anae.1997.0076 [DOI] [PubMed] [Google Scholar]
- Attene-Ramos M. S., Nava G. M., Muellner M. G., Wagner E. D., Plewa M. J., Gaskins H. R. (2010). DNA damage and toxicogenomic analyses of hydrogen sulfide in human intestinal epithelial FHs 74 Int cells. Environ. Mol. Mutagen. 51, 304–314 10.1002/em.20546 [DOI] [PubMed] [Google Scholar]
- Attene-Ramos M. S., Wagner E. D., Gaskins H. R., Plewa M. J. (2007). Hydrogen sulfide induces direct radical-associated DNA damage. Mol. Cancer Res. 5, 455–459 10.1158/1541-7786.MCR-06-0439 [DOI] [PubMed] [Google Scholar]
- Attene-Ramos M. S., Wagner E. D., Plewa M. J., Gaskins H. R. (2006). Evidence that hydrogen sulfide is a genotoxic agent. Mol. Cancer Res. 4, 9–14 10.1158/1541-7786.MCR-05-0126 [DOI] [PubMed] [Google Scholar]
- Aucher P., Richard L., Grollier G., Moinard N., Mioche L., Babin P., et al. (1998). Bilophila wadsworthia isolated from acute pancreatitis. Medecine Et Maladies Infectieuses 28, 260–263 [Google Scholar]
- Babidge W., Millard S., Roediger W. (1998). Sulfides impair short chain fatty acid beta-oxidation at acyl-CoA dehydrogenase level in colonocytes: implications for ulcerative colitis. Mol. Cell. Biochem. 181, 117–124 [DOI] [PubMed] [Google Scholar]
- Balamurugan R., Rajendiran E., George S., Samuel G. V., Ramakrishna B. S. (2008). Real-time polymerase chain reaction quantification of specific butyrate-producing bacteria, Desulfovibrio and Enterococcus faecalis in the feces of patients with colorectal cancer. J. Gastroenterol. Hepatol. 23, 1298–1303 10.1111/j.1440-1746.2008.05490.x [DOI] [PubMed] [Google Scholar]
- Baron E. J., Summanen P., Downes J., Roberts M. C., Wexler H., Finegold S. M. (1989). Bilophila wadsworthia, gen-nov and sp-nov, a unique Gram-negative anaerobic rod recovered from appendicitis specimens and human feces. J. Gen. Microbiol. 135, 3405–3411 [DOI] [PubMed] [Google Scholar]
- Bartholomew T. C., Powell G. M., Dodgson K. S., Curtis C. G. (1980). Oxidation of sodium sulphide by rat liver, lungs and kidney. Biochem. pharmacol. 29, 2431–2437 10.1016/0006-2952(80)90346-9 [DOI] [PubMed] [Google Scholar]
- Belzer C., De Vos W. M. (2012). Microbes inside - from diversity to function: the case of Akkermansia. ISME J. 6, 1449–1458 10.1038/ismej.2012.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borup B., Ferry J. G. (2000). O-acetylserine sulfhydrylase from Methanosarcina thermophila. J. Bacteriol. 182, 45–50 10.1128/JB.182.1.45-50.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouillaud F., Blachier F. (2011). Mitochondria and sulfide: a very old story of poisoning, feeding, and signaling? Antioxid. Redox Signal. 15, 379–391 10.1089/ars.2010.3678 [DOI] [PubMed] [Google Scholar]
- Bouillaud F., Goubern M., Andriamihaja M., Nubel T., Blachier F. (2007). Sulfide, the first inorganic substrate for human cells. FASEB J. 21, 1699–1706 10.1096/fj.06-7407com [DOI] [PubMed] [Google Scholar]
- Canfield D. E., Farquhar M. (2012). The global sulfur cycle, in Fundamentals of Geobiology, eds Knoll A. H., Canfield D. E., Konhauser K. O. (West Sussex: Wiley-Blackwell; ). [Google Scholar]
- Carbonero F., Benefiel A. C., Gaskins H. R. (2012). Contributions of the microbial hydrogen economy to colonic homeostasis. Nat. Rev. Gastroenterol. Hepatol. 9, 504–518 10.1038/nrgastro.2012.85 [DOI] [PubMed] [Google Scholar]
- Castellarin M., Warren R. L., Freeman J. D., Dreolini L., Krzywinski M., Strauss J., et al. (2012). Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 22, 299–306 10.1101/gr.126516.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chassard C., Dapoigny M., Scott K. P., Crouzet L., Del'Homme C., Marquet P., et al. (2012). Functional dysbiosis within the gut microbiota of patients with constipated-irritable bowel syndrome. Aliment. Pharmacol. Ther. 35, 828–838 10.1111/j.1365-2036.2012.05007.x [DOI] [PubMed] [Google Scholar]
- Christl S. U., Eisner H. D., Dusel G., Kasper H., Scheppach W. (1996). Antagonistic effects of sulfide and butyrate on proliferation of colonic mucosa - a potential role for these agents in the pathogenesis of ulcerative colitis. Dig. Dis. Sci. 41, 2477–2481 [DOI] [PubMed] [Google Scholar]
- Christl S. U., Gibson G. R., Cummings J. H. (1992). Role of dietary sulphate in the regulation of methanogenesis in the human large intestine. Gut 33, 1234–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffey J. C., Rowan F., Burke J., Dochery N., Kirwan W. O., O'Connell P. R. (2009). Pathogenesis of and unifying hypothesis for idiopathic pouchitis. Am. J. Gastroenterol. 104, 1013–1023 10.1038/ajg.2008.127 [DOI] [PubMed] [Google Scholar]
- Corfield A. P., Wagner S. A., Clamp J. R., Kriaris M. S., Hoskins L. C. (1992). Mucin degradation in the human colon - production of sialidase, sialate O-acetylesterase, N-acetylneuraminate lyase, arylesterase, and glycosulfatase activities by strains of fecal bacteria. Infect. Immun. 60, 3971–3978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croix J. A., Carbonero F., Nava G. M., Russell M., Greenberg E., Gaskins H. R. (2011). On the relationship between sialomucin and sulfomucin expression and hydrogenotrophic microbes in the human colonic mucosa. PLoS ONE 6:e24447 10.1371/journal.pone.0024447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danese S., Fava F. (2011). Intestinal microbiota in inflammatory bowel disease: friend or foe? World J. Gastroenterol. 17, 557–566 10.3748/wjg.v17.i5.557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Angelis M., Curtin A. C., McSweeney P. L. H., Faccia M., Gobbetti M. (2002). Lactobacillus reuteri DSM 20016: purification and characterization of a cystathionine gamma-lyase and use as adjunct starter in cheesemaking. J. Dairy Res. 69, 255–267 [DOI] [PubMed] [Google Scholar]
- de la Chapelle A. (2004). Genetic predisposition to colorectal cancer. Nat. Rev. Cancer 4, 769–780 10.1038/nrc1453 [DOI] [PubMed] [Google Scholar]
- Den Hond E., Hiele M., Evenepoel P., Peeters M., Ghoos Y., Rutgeerts P. (1998). In vivo butyrate metabolism and colonic permeability in extensive ulcerative colitis. Gastroenterology 115, 584–590 [DOI] [PubMed] [Google Scholar]
- Deplancke B., Gaskins H. R. (2003). Hydrogen sulfide induces serum-independent cell cycle entry in nontransformed rat intestinal epithelial cells. FASEB J. 17, 1310–1312 10.1096/fj.02-0883fje [DOI] [PubMed] [Google Scholar]
- Derrien M., Collado M. C., Ben-Amor K., Salminen S., De Vos W. M. (2008). The mucin degrader Akkermansia muciniphila is an abundant resident of the human intestinal tract. Appl. Environ. Microbiol. 74, 1646–1648 10.1128/AEM.01226-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien M., Vaughan E. E., Plugge C. M., De Vos W. M. (2004). Akkermansia muciniphila gen. nov., sp nov., a human intestinal mucin-degrading bacterium. Int. J. Syst. Evol. Microbiol. 54, 1469–1476 10.1099/ijs.0.02873-0 [DOI] [PubMed] [Google Scholar]
- Devkota S., Wang Y., Musch M. W., Leone V., Fehlner-Peach H., Nadimpalli A., et al. (2012). Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10-/- mice. Nature 487, 104–108 10.1038/nature11225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmani P., Strauss J., Ambrose C., Allen-Vercoe E., Chadee K. (2011). Fusobacterium nucleatum infection of colonic cells stimulates MUC2 mucin and tumor necrosis factor-alpha. Infect. Immun. 79, 2597–2607 10.1128/IAI.05118-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Distrutti E., Sediari L., Mencarelli A., Renga B., Orlandi S., Antonelli E., et al. (2006). Evidence that hydrogen sulfide exerts antinociceptive effects in the gastrointestinal tract by activating K-ATP channels. J. Pharmacol. Exp. Ther. 316, 325–335 10.1124/jpet.105.091595 [DOI] [PubMed] [Google Scholar]
- Doll R., Peto R. (1981). The causes of cancer: quantitative estimates of avoidable risks of cancer in the United States today. J. Natl. Cancer Inst. 66, 1191–1308 [PubMed] [Google Scholar]
- Dominy J. E., Stipanuk M. H. (2004). New roles for cysteine and transsulfuration enzymes: production of H2S, a neuromodulator and smooth muscle relaxant. Nutr. Rev. 62, 348–353 10.1111/j.1753-4887.2004.tb00060.x [DOI] [PubMed] [Google Scholar]
- Duffy M., O'Mahony L., Coffey J. C., Collins J. K., Shanahan F., Redmond H. P., et al. (2002). Sulfate-reducing bacteria colonize pouches formed for ulcerative colitis but not for familial adenomatous polyposis. Dis. Colon Rectum 45, 384–388 [DOI] [PubMed] [Google Scholar]
- Edmond L. M., Hopkins M. J., Magee E. A., Cummings J. H. (2003). The effect of 5-aminosalicylic acid-containing drugs on sulfide production by sulfate-reducing and amino acid-fermenting bacteria. Inflamm. Bowel Dis. 9, 10–17 [DOI] [PubMed] [Google Scholar]
- Ferlay J., Shin H. R., Bray F., Forman D., Mathers C., Parkin D. M. (2010). GLOBOCAN (2008). v1.2, Cancer Incidence and Mortality Worldwide: IARC CancerBase No. 10 [Online], (Lyon, France: World Health Organization, International Agency for Research on Cancer; ). Available online at: http://globocan.iarc.fr [Accessed July 31 2012]. [Google Scholar]
- Finegold S., Summanen P., Gerardo S. H., Baron E. (1992). Clinical importance of Bilophila wadsworthia. Eur. J. Clin. Microbiol. Infect. Dis. 11, 1058–1063 [DOI] [PubMed] [Google Scholar]
- Fite A., Macfarlane G. T., Cummings J. H., Hopkins M. J., Kong S. C., Furrie E., et al. (2004). Identification and quantitation of mucosal and faecal desulfovibrios using real time polymerase chain reaction. Gut 53, 523–529 10.1136/gut.2003.031245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint H. J., Marquet P., Duncan S. H., Chassard C., Bernalier-Donadille A. (2009). Lactate has the potential to promote hydrogen sulphide formation in the human colon. FEMS Microbiol. Lett. 299, 128–134 10.1111/j.1574-6968.2009.01750.x [DOI] [PubMed] [Google Scholar]
- Florin T., Neale G., Gibson G. R., Christl S. U., Cummings J. H. (1991). Metabolism of dietary sulfate - absorption and excretion in humans. Gut 32, 766–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florin T. H. J. (1991). Hydrogen sulfide and total acid-volatile sulfide in feces, determined with a direct spectrophotometric method. Clin. Chim. Acta 196, 127–134 [DOI] [PubMed] [Google Scholar]
- Forsberg C. W. (1980). Sulfide production from cysteine by Desulfovibrio desulfuricans. Appl. Environ. Microbiol. 39, 453–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitag T. E., Klenke T., Krumbein W. E., Gerdes G., Prosser J. I. (2003). Effect of anoxia and high sulphide concentrations on heterotrophic microbial communities in reduced surface sediments (Black Spots) in sandy intertidal flats of the German Wadden Sea. FEMS Microbiol. Ecol. 44, 291–301 10.1016/S0168-6496(03)00076-X [DOI] [PubMed] [Google Scholar]
- Furne J., Springfield J., Koenig T., Demaster E., Levitt M. D. (2001). Oxidation of hydrogen sulfide and methanethiol to thiosulfate by rat tissues: a specialized function of the colonic mucosa. Biochem. Pharmacol. 62, 255–259 10.1016/S0006-2952(01)00657-8 [DOI] [PubMed] [Google Scholar]
- Gallego D., Clave P., Donovan J., Rahmati R., Grundy D., Jimenez M., et al. (2008). The gaseous mediator, hydrogen sulphide, inhibits in vitro motor patterns in the human, rat and mouse colon and jejunum. Neurogastroenterol. Motil. 20, 1306–1316 10.1111/j.1365-2982.2008.01201.x [DOI] [PubMed] [Google Scholar]
- Gibson G. R., Cummings J. H., Macfarlane G. T. (1991). Growth and activities of sulphate-reducing bacteria in gut contents of healthy subjects and patients with ulcerative colitis. FEMS Microbiol. Lett. 86, 103–111 [Google Scholar]
- Gibson G. R., Macfarlane G. T., Cummings J. H. (1988). Occurrence of sulphate-reducing bacteria in human faeces and the relationship of dissimilatory sulphate reduction to methanogenesis in the large gut. J. Appl. Bacteriol. 65, 103–111 [DOI] [PubMed] [Google Scholar]
- Gordon J. I., Turnbaugh P. J., Ley R. E., Hamady M., Fraser-Liggett C. M., Knight R. (2007). The Human microbiome project. Nature 449, 804–810 10.1038/nature06244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guidotti T. L. (1996). Hydrogen sulphide. Occup. Med. (Lond.) 46, 367–371 [DOI] [PubMed] [Google Scholar]
- Hanauer S. B. (2006). Inflammatory bowel disease: epidemiology, pathogenesis, and therapeutic opportunities. Inflamm. Bowel Dis. 12Suppl. 1, S3–S9 [DOI] [PubMed] [Google Scholar]
- Hilton B. L., Oleszkiewicz J. A. (1988). Sulfide-induced inhibition of anaerobic digestion. J. Environ. Eng. (New York) 114, 1377–1391 [Google Scholar]
- Hofmann A. F., Loening-Baucke V., Lavine J. E., Hagey L. R., Steinbach J. H., Packard C. A., et al. (2008). Altered bile acid metabolism in childhood functional constipation: inactivation of secretory bile acids by sulfation in a subset of patients. J. Pediatr. Gastroenterol. Nutr. 47, 598–606 10.1097/MPG.0b013e31816920a6 [DOI] [PubMed] [Google Scholar]
- Huijghebaert S. M., Eyssen H. J. (1982). Specificity of bile salt sulfatase activity from Clostridium sp. strain-S1. Appl. Environ. Microbiol. 44, 1030–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huijghebaert S. M., Mertens J. A., Eyssen H. J. (1982). Isolation of a bile-salt sulfatase-producing Clostridium strain from rat intestinal microflora. Appl. Environ. Microbiol. 43, 185–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igarashi T., Yano T., Fukamachi H., Yamamoto M. (2009). Characterization of L-cysteine desulfhydrase from Prevotella intermedia. Oral Microbiol. Immunol. 24, 485–492 10.1111/j.1399-302X.2009.00546.x [DOI] [PubMed] [Google Scholar]
- Irmler S., Raboud S., Beisert B., Rauhut D., Berthoud H. (2008). Cloning and characterization of two Lactobacillus casei genes encoding a cystathionine lyase. Appl. Environ. Microbiol. 74, 99–106 10.1128/AEM.00745-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jass J. R., Roberton A. M. (1994). Colorectal mucin histochemistry in health and disease - a critical review. Pathol. Int. 44, 487–504 [DOI] [PubMed] [Google Scholar]
- Kabil O., Banerjee R. (2010). Redox biochemistry of hydrogen sulfide. J. Biol. Chem. 285, 21903–21907 10.1074/jbc.R110.128363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanazawa K., Konishi F., Mitsuoka T., Terada A., Itoh K., Narushima S., et al. (1996). Factors influencing the development of sigmoid colon cancer: bacteriologic and biochemical studies. Cancer 77, 1701–1706 [DOI] [PubMed] [Google Scholar]
- Kellogg W. W., Allen E. R., Cadle R. D., Lazrus A. L., Martell E. A. (1972). Sulfur cycle. Science 175, 587 10.1126/science.175.4022.587 [DOI] [PubMed] [Google Scholar]
- Kelly D. J., Myers J. D. (2005). A sulphite respiration system in the chemoheterotrophic human pathogen Campylobacter jejuni. Microbiology 151, 233–242 10.1099/mic.0.27573-0 [DOI] [PubMed] [Google Scholar]
- Kelly D. P., Shergill J. K., Lu W. P., Wood A. P. (1997). Oxidative metabolism of inorganic sulfur compounds by bacteria. Antonie Van Leeuwenhoek 71, 95–107 [DOI] [PubMed] [Google Scholar]
- Kertesz M. A. (2000). Riding the sulfur cycle–metabolism of sulfonates and sulfate esters in gram-negative bacteria. FEMS Microbiol. Rev. 24, 135–175 [DOI] [PubMed] [Google Scholar]
- Kim Y. K., Lee H., Kho H. S., Chung J. W., Chung S. C. (2006). Volatile sulfur compounds produced by Helicobacter pylori. J. Clin. Gastroenterol. 40, 421–426 [DOI] [PubMed] [Google Scholar]
- King T. S., Elia M., Hunter J. O. (1998). Abnormal colonic fermentation in irritable bowel syndrome. Lancet 352, 1187–1189 10.1016/S0140-6736(98)02146-1 [DOI] [PubMed] [Google Scholar]
- Kinzler K. W., Vogelstein B. (1996). Lessons from hereditary colorectal cancer. Cell 87, 159–170 10.1016/S0092-8674(00)81333-1 [DOI] [PubMed] [Google Scholar]
- Kostic A. D., Gevers D., Pedamallu C. S., Michaud M., Duke F., Earl A. M., et al. (2011). Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 22, 292–298 10.1101/gr.126573.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kredich N. M., Keenan B. S., Foote L. J. (1972). Purification and subunit structure of cysteine desulfhydrase from Salmonella typhimurium. J. Biol. Chem. 247, 7157 [PubMed] [Google Scholar]
- Kuever J., Rainey F. A., Widdel F. (2005). Family II. Desulfobublaceae fam. nov., in Bergey's Manual of Systematic Bacteriology: VolumeTwo: The Proteobacteria, eds Staley J. T., Brenner D. J., Krieg I. R. (East Lansing, MI: Bergey's Manual Trust; ), 992–998 [Google Scholar]
- Kumagai H., Choi Y. J., Sejima S., Yamada H. (1975). Formation of cysteine desulfhydrase by bacteria. Agric. Biol. Chem. 39, 387–392 [Google Scholar]
- Landaud S., Helinck S., Bonnarme P. (2008). Formation of volatile sulfur compounds and metabolism of methionine and other sulfur compounds in fermented food. Appl. Microbiol. Biotechnol. 77, 1191–1205 10.1007/s00253-007-1288-y [DOI] [PubMed] [Google Scholar]
- Laue H., Cook A. M. (2000). Biochemical and molecular characterization of taurine: pyruvate aminotransferase from the anaerobe. Eur. J. Biochem. 267, 6841–6848 10.1046/j.1432-1033.2000.01782.x [DOI] [PubMed] [Google Scholar]
- Laue H., Denger K., Cook A. M. (1997). Taurine reduction in anaerobic respiration of Bilophila wadsworthia RZATAU. Appl. Environ. Microbiol. 63, 2016–2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laue H., Friedrich M., Ruff J., Cook A. M. (2001). Dissimilatory sulfite reductase (desulfoviridin) of the taurine-degrading, non-sulfate-reducing bacterium Bilophila wadsworthia RZATAU contains a fused DsrB-DsrD subunit. J. Bacteriol. 183, 1727–1733 10.1128/JB.183.5.1727-1733.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leclerc H., Oger C., Beerens H., Mossel D. A. A. (1980). Occurrence of sulfate reducing bacteria in the human intestinal flora and in the aquatic environment. Water Res. 14, 253–256 [Google Scholar]
- Levine J., Ellis C. J., Furne J. K., Springfield J., Levitt M. D. (1998). Fecal hydrogen sulfide production in ulcerative colitis. Am. J. Gastroenterol. 93, 83–87 10.1111/j.1572-0241.1998.083_c.x [DOI] [PubMed] [Google Scholar]
- Lewis S., Cochrane S. (2007). Alteration of sulfate and hydrogen metabolism in the human colon by changing intestinal transit rate. Am. J. Gastroenterol. 102, 624–633 10.1111/j.1572-0241.2006.01020.x [DOI] [PubMed] [Google Scholar]
- Loubinoux J., Bronowicki J. P., Pereira I. A., Mougenel J. L., Faou A. E. (2002). Sulfate-reducing bacteria in human feces and their association with inflammatory bowel diseases. FEMS Microbiol. Ecol. 40, 107–112 10.1111/j.1574-6941.2002.tb00942.x [DOI] [PubMed] [Google Scholar]
- Magee E. A., Richardson C. J., Hughes R., Cummings J. H. (2000). Contribution of dietary protein to sulfide production in the large intestine: an in vitro and a controlled feeding study in humans. Am. J. Clin. Nutr. 72, 1488–1494 [DOI] [PubMed] [Google Scholar]
- Marchesi J. R., Scanlan P. D., Shanahan F. (2009). Culture-independent analysis of desulfovibrios in the human distal colon of healthy, colorectal cancer and polypectomized individuals. FEMS Microbiol. Ecol. 69, 213–221 10.1111/j.1574-6941.2009.00709.x [DOI] [PubMed] [Google Scholar]
- Matsunami M., Tarui T., Mitani K., Nagasawa K., Fukushima O., Okubo K., et al. (2009). Luminal hydrogen sulfide plays a pronociceptive role in mouse colon. Gut 58, 751–761 10.1136/gut.2007.144543 [DOI] [PubMed] [Google Scholar]
- Mayer E. A. (2008). Clinical practice. Irritable bowel syndrome. N. Engl. J. Med. 358, 1692–1699 10.1056/NEJMcp0801447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBain A. J., Macfarlane G. T. (1998). Ecological and physiological studies on large intestinal bacteria in relation to production of hydrolytic and reductive enzymes involved in formation of genotoxic metabolites. J. Med. Microbiol. 47, 407–416 [DOI] [PubMed] [Google Scholar]
- Medani M., Collins D., Docherty N. G., Baird A. W., O'Connell P. R., Winter D. C. (2011). Emerging role of hydrogen sulfide in colonic physiology and pathophysiology. Inflamm. Bowel Dis. 17, 1620–1625 10.1002/ibd.21528 [DOI] [PubMed] [Google Scholar]
- Metaxas M. A., Delwiche E. A. (1955). The L-cysteine desulfhydrase of Escherichia coli. J. Bacteriol. 70, 735–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore J., Babidge W., Millard S., Roediger W. (1998). Colonic luminal hydrogen sulfide is not elevated in ulcerative colitis. Dig. Dis. Sci. 43, 162–165 [DOI] [PubMed] [Google Scholar]
- Nakamura N., Lin H. C., McSweeney C. S., Mackie R. I., Gaskins H. R. (2010). Mechanisms of microbial hydrogen disposal in the human colon and implications for health and disease. Ann. Rev. Food Sci. Tech. 1, 363–395 10.1146/annurev.food.102308.124101 [DOI] [PubMed] [Google Scholar]
- Nakayama I., Akieda Y., Yamaji E., Suzuki T., Anraku N., Watanabe T., et al. (2000). Is Bilophila wadsworthia the cause of acute gangrenous appendicitis? Clin. Infect. Dis. 31, 239–239 [Google Scholar]
- Nava G. M., Carbonero F., Croix J. A., Greenberg E., Gaskins H. R. (2012). Abundance and diversity of mucosa-associated hydrogenotrophic microbes in the healthy human colon. ISME J. 6, 57–70 10.1038/ismej.2011.90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norat T., Lukanova A., Ferrari P., Riboli E. (2002). Meat consumption and colorectal cancer risk: dose-response meta-analysis of epidemiological studies. Int. J. Cancer 98, 241–256 10.1002/ijc.10126 [DOI] [PubMed] [Google Scholar]
- Nowicki C., Marciano D., Santana M., Mantilla B. S., Silber A. M., Marino-Buslje C. (2010). Biochemical characterization of serine acetyltransferase and cysteine desulfhydrase from Leishmania major. Mol. Biochem. Parasitol. 173, 170–174 10.1016/j.molbiopara.2010.06.004 [DOI] [PubMed] [Google Scholar]
- Ohge H., Furne J. K., Springfield J., Rothenberger D. A., Madoff R. D., Levitt M. D. (2005). Association between fecal hydrogen sulfide production and pouchitis. Dis. Colon Rectum 48, 469–475 10.1007/s10350-004-0820-8 [DOI] [PubMed] [Google Scholar]
- O'Keefe S. J., Chung D., Mahmoud N., Sepulveda A. R., Manafe M., Arch J., et al. (2007). Why do African Americans get more colon cancer than Native Africans? J. Nutr. 137, 175S–182S [DOI] [PubMed] [Google Scholar]
- Olson K. R. (2011). The therapeutic potential of hydrogen sulfide: separating hype from hope. Am. J. Physiol. Regul. Integr. Comp. Physiol. 301, R297–R312 10.1152/ajpregu.00045.2011 [DOI] [PubMed] [Google Scholar]
- Palmer R. H. (1967). The formation of bile acid sulfates: a new pathway of bile acid metabolism in humans. Proc. Natl. Acad. Sci. U.S.A. 58, 1047–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peck H. D. (1961). Enzymatic basis for assimilatory and dissimilatory sulfate reduction. J. Bacteriol. 82, 933–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitcher M. C. L., Beatty E. R., Cummings J. H. (2000). The contribution of sulphate reducing bacteria and 5-aminosalicylic acid to faecal sulphide in patients with ulcerative colitis. Gut 46, 64–72 10.1136/gut.46.1.64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitcher M. C. L., Cummings J. H. (1996). Hydrogen sulphide: a bacterial toxin in ulcerative colitis? Gut 39, 1–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plugge C. M., Zhang W., Scholten J. C., Stams A. J. (2011). Metabolic flexibility of sulfate-reducing bacteria. Front. Microbio. 2:81 10.3389/fmicb.2011.00081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podolsky D. K. (2002). Inflammatory bowel disease. N. Engl. J. Med. 347, 417–429 10.1056/NEJMra020831 [DOI] [PubMed] [Google Scholar]
- Pukall R., Stackebrandt E., Buntefuss D., Fruhling A., Rohde M., Kroppenstedt R. M., et al. (1999). Sulfitobacter mediterraneus sp. nov., a new sulfite-oxidizing member of the alpha-Proteobacteria. Int. J. Syst. Bacteriol. 49, 513–519 10.1099/00207713-49-2-513 [DOI] [PubMed] [Google Scholar]
- Ramakrishna B. S., Robertsthomson I. C., Pannall P. R., Roediger W. E. W. (1991). Impaired sulfation of phenol by the colonic mucosa in quiescent and active ulcerative colitis. Gut 32, 46–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramasamy S., Singh S., Taniere P., Langman M. J., Eggo M. C. (2006). Sulfide-detoxifying enzymes in the human colon are decreased in cancer and upregulated in differentiation. Am. J. Physiol. Gastrointest. Liver Physiol. 291, G288–G296 10.1152/ajpgi.00324.2005 [DOI] [PubMed] [Google Scholar]
- Rhodes J. M., Campbell B. J. (2002). Inflammation and colorectal cancer: IBD-associated and sporadic cancer compared. Trends Mol. Med. 8, 10–16 10.1016/S1471-4914(01)02194-3 [DOI] [PubMed] [Google Scholar]
- Robben J., Caenepeel P., Vaneldere J., Eyssen H. (1988). Effects of intestinal microbial bile-salt sulfatase activity on bile-salt kinetics in gnotobiotic rats. Gastroenterology 94, 494–502 [DOI] [PubMed] [Google Scholar]
- Robben J., Parmentier G., Eyssen H. (1986). Isolation of a rat intestinal Clostridium strain producing 5-alpha and 5-beta bile salt 3-alpha-sulfatase activity. Appl. Environ. Microbiol. 51, 32–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberton A. M., Stanley R. A. (1982). In vitro utilization of mucin by Bacteroides fragilis. Appl. Environ. Microbiol. 43, 325–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberton A. M., Wright D. P. (1997). Bacterial glycosulphatases and sulphomucin degradation. Can. J. Gastroenterol. 11, 361–366 [DOI] [PubMed] [Google Scholar]
- Roberton A. M., Wright D. P., Rosendale D. I. (2000). Prevotella enzymes involved in mucin oligosaccharide degradation and evidence for a small operon of genes expressed during growth on mucin. FEMS Microbiol. Lett. 190, 73–79 [DOI] [PubMed] [Google Scholar]
- Roediger W. E., Moore J., Babidge W. (1997). Colonic sulfide in pathogenesis and treatment of ulcerative colitis. Dig. Dis. Sci. 42, 1571–1579 [DOI] [PubMed] [Google Scholar]
- Roediger W. E. W., Duncan A., Kapaniris O., Millard S. (1993). Reducing sulfur-compounds of the colon impair colonocyte nutrition - implications for ulcerative-colitis. Gastroenterology 104, 802–809 [DOI] [PubMed] [Google Scholar]
- Sandhu M. S., White I. R., McPherson K. (2001). Systematic review of the prospective cohort studies on meat consumption and colorectal cancer risk: a meta-analytical approach. Cancer Epidemiol. Biomarkers Prev. 10, 439–446 [PubMed] [Google Scholar]
- Scanlan P. D., Shanahan F., Marchesi J. R. (2009). Culture-independent analysis of desulfovibrios in the human distal colon of healthy, colorectal cancer and polypectomized individuals. FEMS Microbiol. Ecol. 69, 213–221 10.1111/j.1574-6941.2009.00709.x [DOI] [PubMed] [Google Scholar]
- Seitz A. P., Leadbetter E. R. (1995). Microbial assimilation and dissimilation of sulfonate sulfur, in Geochemical Transformations of Sedimentary Sulfur. American Chemical Society Symposium Series, no. 612, eds Vairavamurthy M. A., Schoonen A. A. (Washington, DC: American Chemical Society; ), 365–376 [Google Scholar]
- Shatalin K., Shatalina E., Mironov A., Nudler E. (2011). H2S: a universal defense against antibiotics in bacteria. Science 334, 986–990 10.1126/science.1209855 [DOI] [PubMed] [Google Scholar]
- Slomiany B. L., Murty V. L. N., Piotrowski J., Grabska M., Slomiany A. (1992). Glycosulfatase activity of Helicobacter pylori toward human gastric mucin - effect of sucralfate. Am. J. Gastroenterol. 87, 1132–1137 [PubMed] [Google Scholar]
- Soulimane T., Robin S., Arese M., Forte E., Sarti P., Giuffre A. (2011). A sulfite respiration pathway from Thermus thermophilus and the key role of newly identified cytochrome c(550). J. Bacteriol. 193, 3988–3997 10.1128/JB.05186-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stipanuk M. H. (1986). Metabolism of sulfur-containing amino-acids. Annu. Rev. Nutr. 6, 179–209 10.1146/annurev.nu.06.070186.001143 [DOI] [PubMed] [Google Scholar]
- Stipanuk M. H. (2004). Sulfur amino acid metabolism: pathways for production and removal of homocysteine and cysteine. Annu. Rev. Nutr. 24, 539–577 10.1146/annurev.nutr.24.012003.132418 [DOI] [PubMed] [Google Scholar]
- Stipanuk M. H., Dominy J. E. (2006). Surprising insights that aren't so surprising in the modeling of sulfur amino acid metabolism. Amino Acids 30, 251–256 10.1007/s00726-005-0288-4 [DOI] [PubMed] [Google Scholar]
- Strauss J., Kaplan G. G., Beck P. L., Rioux K., Panaccione R., Devinney R., et al. (2011). Invasive potential of gut mucosa-derived Fusobacterium nucleatum positively correlates with IBD status of the host. Inflamm. Bowel Dis. 17, 1971–1978 10.1002/ibd.21606 [DOI] [PubMed] [Google Scholar]
- Strocchi A., Ellis C. J., Levitt M. D. (1993). Use of metabolic-inhibitors to study H2 consumption by human feces - evidence for a pathway other than methanogenesis and sulfate reduction. J. Lab. Clin. Med. 121, 320–327 [PubMed] [Google Scholar]
- Suarez F., Furne J., Springfield J., Levitt M. (1997). Insights into human colonic physiology obtained from the study of flatus composition. Am. J. Physiol. Gastro. Liver Physiol. 272, G1028–G1033 [DOI] [PubMed] [Google Scholar]
- Swann J. R., Want E. J., Geier F. M., Spagou K., Wilson I. D., Sidaway J. E., et al. (2011). Systemic gut microbial modulation of bile acid metabolism in host tissue compartments. Proc. Natl. Acad. Sci. U.S.A. 108Suppl. 1, 4523–4530 10.1073/pnas.1006734107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai C. H., Cook P. F. (2001). Pyridoxal 5′-phosphate dependent alpha, beta-elimination reactions: mechanism of O-acetylserine sulfhydrylase. Acc. Chem. Res. 34, 49–59 10.1021/ar990169l [DOI] [PubMed] [Google Scholar]
- Tchong S. I., Xu H. M., White R. H. (2005). L-cysteine desulfidase: an [4Fe-4S] enzyme isolated from Methanocaldococcus jannaschii that catalyzes the breakdown of L-cysteine into pyruvate, ammonia, and sulfide. Biochemistry 44, 1659–1670 10.1021/bi0484769 [DOI] [PubMed] [Google Scholar]
- Tragnone A., Valpiani D., Miglio F., Elmi G., Bazzocchi G., Pipitone E., et al. (1995). Dietary habits as risk factors for inflammatory bowel disease. Eur. J. Gastroenterol. Hepatol. 7, 47–51 [PubMed] [Google Scholar]
- Truelove S. C. (1961). Ulcerative colitis provoked by milk. Br. Med. J. 1, 154–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai H. H., Dwarakanath A. D., Hart C. A., Milton J. D., Rhodes J. M. (1995). Increased faecal mucin sulphatase activity in ulcerative colitis: a potential target for treatment. Gut 36, 570–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaneldere J., Parmentier G., Asselberghs S., Eyssen H. (1991). Partial characterization of the steroidsulfatases in Peptococcus niger H4. Appl. Environ. Microbiol. 57, 69–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdugo P. (1990). Goblet cells secretion and mucogenesis. Annu. Rev. Physiol. 52, 157–176 10.1146/annurev.ph.52.030190.001105 [DOI] [PubMed] [Google Scholar]
- Visscher P. T., Gritzer R. F., Leadbetter E. R. (1999). Low-molecular-weight sulfonates, a major substrate for sulfate reducers in marine microbial mats. Appl. Environ. Microbiol. 65, 3272–3278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner M., Roger A. J., Flax J. L., Brusseau G. A., Stahl D. A. (1998). Phylogeny of dissimilatory sulfite reductases supports an early origin of sulfate respiration. J. Bacteriol. 180, 2975–2982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace J. L. (2010). Physiological and pathophysiological roles of hydrogen sulfide in the gastrointestinal tract. Antioxid. Redox Signal. 12, 1125–1133 10.1089/ars.2009.2900 [DOI] [PubMed] [Google Scholar]
- Wallace J. L., Dicay M., McKnight W., Martin G. R. (2007). Hydrogen sulfide enhances ulcer healing in rats. FASEB J. 21, 4070–4076 10.1096/fj.07-8669com [DOI] [PubMed] [Google Scholar]
- Wallace J. L., Vong L., McKnight W., Dicay M., Martin G. R. (2009). Endogenous and exogenous hydrogen sulfide promotes resolution of colitis in rats. Gastroenterology 137, 569–578 10.1053/j.gastro.2009.04.012 [DOI] [PubMed] [Google Scholar]
- Ware J. H., Thibodeau L. A., Speizer F. E., Colome S., Ferris B. G. (1981). Assessment of the health effects of atmospheric sulfur-oxides and particulate matter - evidence from observational studies. Environ. Health Perspect. 41, 255–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weitz J., Koch M., Debus J., Hohler T., Galle P. R., Buchler M. W. (2005). Colorectal cancer. Lancet 365, 153–165 10.1016/S0140-6736(05)17706-X [DOI] [PubMed] [Google Scholar]
- Wheeler P. R., Coldham N. G., Keating L., Gordon S. V., Wooff E. E., Parish T., et al. (2005). Functional demonstration of reverse transsulfuration in the Mycobacterium tuberculosis complex reveals that methionine is the preferred sulfur source for pathogenic mycobacteria. J. Biol. Chem. 280, 8069–8078 10.1074/jbc.M412540200 [DOI] [PubMed] [Google Scholar]
- Willett W. C., Stampfer M. J., Colditz G. A., Rosner B. A., Speizer F. E. (1990). Relation of meat, fat, and fiber intake to the risk of colon cancer in a prospective study among women. N. Engl. J. Med. 323, 1664–1672 10.1056/NEJM199012133232404 [DOI] [PubMed] [Google Scholar]
- Wilson K., Mudra M., Furne J., Levitt M. (2008). Differentiation of the roles of sulfide oxidase and rhodanese in the detoxification of sulfide by the colonic mucosa. Dig. Dis. Sci. 53, 277–283 10.1007/s10620-007-9854-9 [DOI] [PubMed] [Google Scholar]
- Yoshida A., Takahashi Y., Nagata E., Hoshino T., Oho T., Awano S., et al. (2011). Streptococcus anginosusl-cysteine desulfhydrase gene expression is associated with abscess formation in BALB/c mice. Mol. Oral Microbiol. 26, 221–227 10.1111/j.2041-1014.2010.00599.x [DOI] [PubMed] [Google Scholar]
- Yoshida Y., Ito S., Kamo M., Kezuka Y., Tamura H., Kunimatsu K., et al. (2010). Production of hydrogen sulfide by two enzymes associated with biosynthesis of homocysteine and lanthionine in Fusobacterium nucleatum subsp nucleatum ATCC 25586. Microbiology 156, 2260–2269 10.1099/mic.0.039180-0 [DOI] [PubMed] [Google Scholar]
- Zdych E., Peist R., Reidl J., Boos W. (1995). MalY of Escherichia coli is an enzyme with the activity of a beta-C-S lyase (cystathionase). J. Bacteriol. 177, 5035–5039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinkevich V., Beech I. B. (2000). Screening of sulfate-reducing bacteria in colonoscopy samples from healthy and colitic human gut mucosa. FEMS Microbiol. Ecol. 34, 147–155 10.1111/j.1574-6941.2000.tb00764.x [DOI] [PubMed] [Google Scholar]


