SUMMARY
The lack of small-molecule inhibitors for anion-selective transporters and channels has impeded our understanding of the complex mechanisms that underlie ion passage. The ubiquitous CLC “Chloride Channel” family represents a unique target for biophysical and biochemical studies because its distinctive protein fold supports both passive chloride channels and secondary-active chloride-proton transporters. Here, we describe the synthesis and characterization of the first specific small-molecule inhibitor directed against a CLC antiporter (ClC-ec1). This compound, 4,4′-octanamidostilbene-2,2′-disulfonate (OADS), inhibits ClC-ec1 with low micromolar affinity and has no specific effect on a CLC channel (ClC-1). Inhibition of ClC-ec1 occurs by binding to two distinct intracellular sites. The location of these sites and the lipid-dependence of inhibition suggest potential mechanisms of action. The discovery of this compound will empower research to elucidate differences between antiporter and channel mechanisms and to develop treatments for CLC-mediated disorders.
INTRODUCTION
Small-molecule modulators of ion channels and transporters have a rich history as tools for uncovering mechanisms of ion permeation and as therapeutic agents in treatment of disease (Gupta, 2011; Yan, 2010). However, the availability of small-molecule modulators and the resulting scientific advances have overwhelmingly favored the broad family of cation-selective channels and transporters. The study of their anion-selective counterparts—and the treatment of diseases related to anion transport (Planells-Cases and Jentsch, 2009; Verkman and Galietta, 2009)—have been hindered by a general lack of anion-transport modulators. Here we describe a new small-molecule inhibitor for one family of anion-transport proteins, the CLC “Chloride Channel” family. The availability of such compounds will enable further studies to understand one of the most intriguing aspects of ion transport in the CLC family – the ability of the same general structure to support either passive Cl− transport or active Cl−/H+ antiport. Our findings provide a foundation for the design of new pharmacological agents for studying CLC-mediated physiological processes, including epithelial ion transport, neuronal and skeletal muscle excitability, hippocampal neuroprotection, cardiac pacemaker activity, endocytosis, and lysosomal acidification (Graves, et al., 2008; Huang, et al., 2009; Jentsch, 2008; Jeworutzki, et al., 2012; Lossin and George, 2008; Nighot, et al., 2009; Ratte and Prescott, 2011) and for treating CLC-related disorders including osteoporosis, neurodegeneration, and cardiovascular disease (Duan, 2011; Jentsch, 2008; Liantonio, et al., 2012; Zhao, et al., 2009).
In expanding the list of CLC modulators, we focused on finding inhibitors of the CLC antiporter from E. coli, ClC-ec1 (Dutzler, 2006; Maduke, et al., 1999). We chose this particular protein because it is an excellent structural model for other CLC members (Engh and Maduke, 2005; Estevez, et al., 2003; Lin and Chen, 2003) and it can be produced and purified in large quantities for analysis by multiple biophysical and biochemical techniques. To date, the only known inhibitor of a CLC antiporter was 4,4′-diisothiocyano-2,2′-stilbenedisulfonic acid (DIDS) (Matulef and Maduke, 2005), which inhibits many different transporters and channels. DIDS is of limited practical utility due to its low-affinity, promiscuous binding (Cabantchik and Greger, 1992), and instability in aqueous solution (Jakobsen and Horobin, 1989). This instability leads to the formation of polythiourea compounds, which are more stable than DIDS and are more potent inhibitors of ClC-ec1 (Matulef, et al., 2008). These compounds, however, possess two major disadvantages that limit their usefulness. First, they are large, conjugated molecules that are impractical to synthesize and modify for structure-activity investigations. Second, the polythiourea inhibitors cause aggregation of detergent-solubilized protein, thus precluding crystallographic or spectroscopic characterization of the binding site. Here, we introduce a new ClC-ec1 inhibitor design, 4,4′-octanamidostilbene-2,2′-disulfonate or “OADS”, and provide a comprehensive series of experiments to elucidate the details of its molecular interactions with the protein.
In the context of the CLC family, OADS offers a unique opportunity to study differences and similarities between two major classes of membrane proteins. Although CLCs originally were thought to be solely voltage-gated Cl− channels, many are Cl−/H+ antiporters (Accardi and Miller, 2004; Picollo and Pusch, 2005; Scheel, et al., 2005). Selective CLC inhibitors thus offer the potential to become tools to probe how these two ostensibly different ion transport processes can be supported by closely related protein structures (as deduced by sequence similarity together with compelling structure-function analyses (Engh and Maduke, 2005; Estevez, et al., 2003; Lin and Chen, 2003; Lisal and Maduke, 2009; Miller, 2006)). The classic “chloride-channel inhibitors” have broad specificity, inhibiting many types of Cl− channels and transporters, and even cation channels (Alper, 2009; Chen, et al., 2007; Evanko, et al., 2004; Kasai, et al., 1999; Li, et al., 2008). OADS is highly effective against the Cl−/H+ antiporter ClC-ec1, but has little activity against a CLC from the channel branch of the family—suggesting that the differences between permeation and active transport may be teased apart using molecular probes. OADS also is the first inhibitor of the CLC antiporters that is reversible, has low molecular weight (<1000 Da), and is amenable to structural modification. We capitalize on these properties to characterize inhibitor binding, thus providing a basis for developing future generations of CLC-specific inhibitors that eventually may have therapeutic application in treating osteoporosis, and neurodegenerative and cardiovascular diseases (Duan, 2011; Liantonio, et al., 2012; Zhao, et al., 2009).
RESULTS
Chemical combination of generic Cl−-channel inhibitors yields a potent CLC antiporter inhibitor
In our search for modulators of CLC function, we tested known CLC channel inhibitors as lead compounds. This panel included flufenamic acid, the clofibric acid derivative 2-(p-chlorophenoxy)-3-(4-chlorophenyl)propionic acid, 9-anthracene carboxylate (9AC), 5-nitro-2-(3-phenylpropylamino)benzoic acid (NPPB), niflumic acid, octanoic acid, and 4,4′-diaminostilbene-2,2′-disulfonate (DADS) (Estevez, et al., 2003; Gradogna and Pusch, 2010; Venglarik, et al., 1994; Zhang and Chen, 2009). None of these compounds inhibited ClC-ec1; however, a modified compound (OADS) combining DADS with two aliphatic side chains mimicking the hydrophobic element of octanoic acid (Figure 1A) inhibited ClC-ec1 at low-micromolar concentrations.
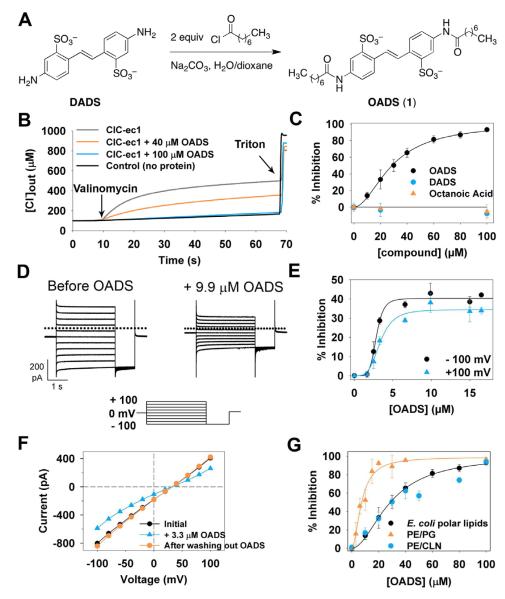
Figure 1. OADS is a CLC antiporter inhibitor.
A) Synthesis of OADS from the generic chloride channel inhibitors octanoic acid and DIDS.
B) Raw data from Cl− efflux assays. ClC-ec1 is reconstituted into liposomes, and efflux is initiated by the addition of valinomycin; Triton X-100 is added at the end of each experiment to release all of the chloride from the liposomes (see Experimental Procedures for more details).
| equation |
D) Currents carried by ClC-ec1 in planar lipid bilayers in response to the voltage protocol shown below before (Left) and after (Right) addition of 9.9 μM OADS to one side (the cis chamber) of the bilayer.
E) Concentration dependence of the inhibition by OADS in bilayers. Each data point represents the mean inhibition ± SEM from 3-7 bilayers with OADS added to the cis chamber. At +100 mV, Imax = 34%, Ki = 3.4 μM, and n = 3.9. At −100 mV, Imax = 40%, Ki = 2.9 μM, and n = 6.1.
F) Reversibility of OADS inhibition in bilayers. Current-voltage curves before (black); and after adding 3.3 μM OADS to the trans chamber (blue squares), and after perfusion with OADS-free solution (red triangles). Similar results were obtained with two other bilayers, and in all cases reversal was complete within 4-10 min (with 4 min being the minimum amount of time required for full perfusion).
G) Effect of liposome composition on ClC-ec1 sensitivity to OADS in Cl− efflux assays. Data for E. coli polar lipids are identical to those in panel (C). For PE/PG liposomes, Ki = 7.3 μM and n =1.9; for PE/CLN , Ki = 32 μM and n = 1.5.
We first assessed OADS’ inhibition of ClC-ec1 using a Cl− efflux assay (Walden, et al., 2007), where ClC-ec1 is reconstituted into liposomes containing a high internal chloride concentration relative to the extraliposomal buffer, and activity is measured as the rate at which the protein passes Cl− along this steep concentration gradient (Figure 1B). OADS inhibited ClC-ec1 with a concentration for half-maximal inhibition (Ki) of 29 μM (Figure 1C, circles), whereas neither parent compound detectably inhibited ClC-ec1 (Figure 1C, squares and triangles) at concentrations up to 1 mM (not shown). Thus, the chemical combination of two conventional Cl− channel inhibitors (octanoic acid and DADS) results in an increase in potency compared to either parent compound by well over 100-fold.
We also examined ClC-ec1 inhibition by OADS using bilayer recordings, which allow for precise control of transmembrane voltage and solutions on both sides of the membrane (Miller, 1983). In these experiments, ClC-ec1-reconstituted liposomes were fused to planar lipid bilayers, and the ionic currents carried by ClC-ec1 were measured under voltage clamp. OADS was then perfused onto one side of the bilayer, and the degree of inhibition was recorded (Figure 1D). We observe maximal inhibition of ~40% when OADS is added to the cis chamber (Figures 1E). Because CLC-ec1 inserts into bilayers with random orientation (Matulef and Maduke, 2005) and OADS is a side-specific inhibitor (as discussed below and shown in Figures 5 and S4), the maximal inhibition possible in these experiments is ~50%. In contrast, in the Cl− efflux assays OADS is allowed access to both sides of the membrane through freeze fracture of liposomes during their preparation, thereby allowing nearly complete inhibition (Figure 1C). In bilayers, the modest increase in potency from Ki ~3.6 μM at +100 mV to ~2.7 μM at −100 mV (Figure 1E) is consistent with the negatively charged inhibitor sensing part of the transmembrane electric field on its way to the inhibitory binding site (Woodhull, 1973). Inhibition by OADS is completely reversible within 4-10 minutes (with 4 minutes being the minimum amount of time required for full solution exchange of the bilayer chamber) (Figure 1F).
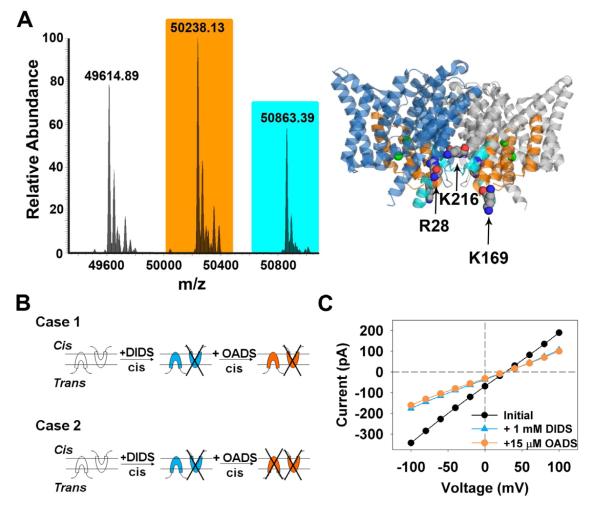
Figure 5. Binding and inhibition by OADS occurs at two distinct intracellular sites.
A) “Top-down” mass spectrometry was used to identify the OADS-binding regions in ClC-ec1. The ClC-ec1 structure illustrates the identified region of labeling of the singly-modified protein in orange (residues 109-185) and an additional site identified in the doubly modified protein in cyan (residues 15-40). The singly modified protein was modified only at the 109-185 site and not at the 15-40 site; the doubly modified protein was modified at both locations. The two subunits are depicted in blue and grey, and Cl− ions are space-filled in green.
B) Schematic cartoons depicting side-specific inhibition of ClC-ec1 by OADS. ClC-ec1 protein is randomly oriented in bilayers. Case I shows that if OADS, like DIDS, acts from the intracellular side, adding OADS to the cis chamber after addition of DIDS will have no effect. Case 2 shows that if OADS acts from the extracellular side, adding OADS to the cis chamber after addition of DIDS should cause further inhibition.
C) Current-voltage curves obtained in bilayer recording experiments before (black circles) and after adding 1 mM DIDS to the cis chamber (blue triangles), and after subsequent addition of 15 μM OADS to the cis chamber (orange squares). That no additional inhibition was observed upon addition of OADS is consistent with Case 1, with OADS inhibition occurring at the intracellular side of ClC-ec1.
The Ki values measured in bilayers (~3 μM, Figure 1E) are an order of magnitude lower than the values determined using the Cl− efflux assay (~30 μM, Figure 1C, circles). Given the sensitivity of ClC-ec1 activity to membrane composition (Robertson, et al., 2010) (see also Figure S1), we hypothesized that the different Ki values could be due to the presence of an additional negatively charged lipid (cardiolipin) in the E. coli lipid extract used for the efflux assays, which was not present in the bilayer experiments. To test whether the presence of cardiolipin (CLN) affects OADS potency, we did Cl− efflux assays on ClC-ec1 reconstituted into liposomes composed of a defined lipid mixture identical to that used in the bilayer experiments (with phosphatidylethanolamine (PE) and phosphatidylglycerol (PG) head groups and lacking CLN) (PE/PG liposomes). Consistent with our hypothesis, the change in lipid composition decreased Ki from 29 to 7.3 μM, (Figure 1G, triangles), close to that observed in the planar bilayer experiments. Substituting CLN for PG restores the Ki to that observed in the original E.coli polar lipid liposomes (Figure 1G, blue and black circles). These data indicate that inhibition of ClC-ec1 by OADS is sensitive to the lipid bilayer composition.
OADS effect on protein activity is not solely mediated by bilayer modification
With a Ki as low as 3 μM (in PE/PG membranes, Figure 1E), OADS is among the most potent CLC inhibitors, as well as the only well-behaved small-molecule inhibitor of any CLC antiporter. But the dependence of ClC-ec1 activity and OADS potency on bilayer composition (Figures S1, 1G) raises the question whether the inhibition results from a specific interaction with the protein, from a more general modification of the lipid bilayer, or from some combination of these mechanisms (Figure 2A).
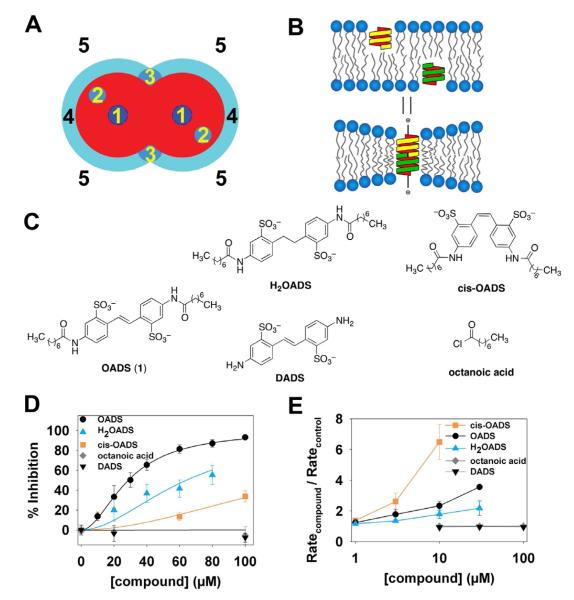
Figure 2. Inhibition of ClC-ec1 by OADS results predominantly from drug-protein interactions.
A) Mechanisms by which drugs can inhibit transporter function: 1) Occluding the transport path to block ion movement; 2) binding to sites formed by the protein, to alter function by altering the free energy difference between different conformations; 3) binding to selective sites at the protein/bilayer interface, which may also alter the bilayer deformation energy contribution to the free energy difference between different conformations; 4) non-specific accumulation at the protein/bilayer interface to alter the local lipid packing and 5) partitioning at the bilayer/solution interface to alter lipid bilayer material properties, which both will alter the bilayer contribution to the membrane protein’s conformational equilibrium (Modified after (Andersen, 2008).
B) Changes in bilayer properties can be determined by measuring changes in the gramicidin (gA) monomer↔dimer equilibrium, e.g., (Lundbaek, et al., 2010) because the hydrophobic length of the bilayer-spanning, ion conducting, dimeric gA channel is less than the hydrophobic length of the host bilayer. gA channel formation therefore leads to a local membrane thinning, with an associated energetic cost that varies with changes in the bilayer properties, thus making gA channels suitable probes for changes in bilayer properties.
C) Panel of OADS derivatives tested for ClC-ec1 potency and bilayer modification: OADS, cis-OADS, and H2OADS, DADS, and octanoic acid.
D) Dose-response curves for inhibition of ClC-ec1 activity by OADS derivatives, as determined by Cl−-efflux assays on liposomes formed from E. coli polar lipids (as in Figure 1B,C.) Each data point represents the mean inhibition ± SEM for 3-5 experiments at each concentration, with the exception of H2OADS points in which represent the average of 2 experiments ± range.
E) Dose-response curves for the OADS derivatives’ effects on bilayer properties, as reported by increases in gA activity. gA activity is determined from the time course of Tl+-induced quenching of a fluorophore encapsulated in gA-doped large unilamellar liposomes (see Experimental Procedures for more details). The quench rate in the presence of a compound normalized to the rate in the absence of added compound reflects the degree of bilayer modification (Ingolfsson and Andersen, 2010)
To address these questions, we employed an assay that exploits gramicidin channels to distinguish between inhibition that occurs solely through modification of bilayer properties and inhibition that involves a direct effect on the protein (Figure 2B). Briefly, the sensitivity of gramicidin channel activity to lipid bilayer properties (Lundbaek, et al., 2010) is used to report on bilayer modification by an exogenously added compound (Ingolfsson and Andersen, 2010). On its own, a positive result in the gramicidin assay may not be remarkable because many drug-like compounds are amphiphiles. Such compounds will therefore also have bilayer modifying activity (at some concentration) in addition to their target-specific activity. The power of the assay lies in its application to a panel of (related) compounds. If a compound inhibits activity of a membrane protein target solely through a bilayer-modification mechanism, the target-specific and the bilayer effects should occur at similar concentrations; moreover, the bilayer-modifying potency of a panel of related compounds will be positively correlated with their relative inhibitory potency. Conversely, if the compound has specific interactions with the target, the relative inhibitory potency for the set of compounds will deviate from such a correlation (Ingolfsson and Andersen, 2010). To implement this analysis, we synthesized and tested a panel of OADS derivatives (Figure 2C-E). There is an unmistakable break in the correlation between the two assays: the cis isomer of OADS is several-fold less potent against ClC-ec1 than its trans counterpart (Figure 2D), yet it is the more potent bilayer-modifying compound (Figure 2E). Thus, inhibition is not due solely to a modification of bilayer properties (mechanisms 4 and 5 in Figure 2A), but rather the predominant mechanism of OADS inhibition of ClC-ec1 occurs through specific interactions with the protein (mechanisms 1, 2, or 3 as in Figure 2A).
OADS selectively inhibits the CLC antiporter ClC-ec1 over the CLC channel ClC-1
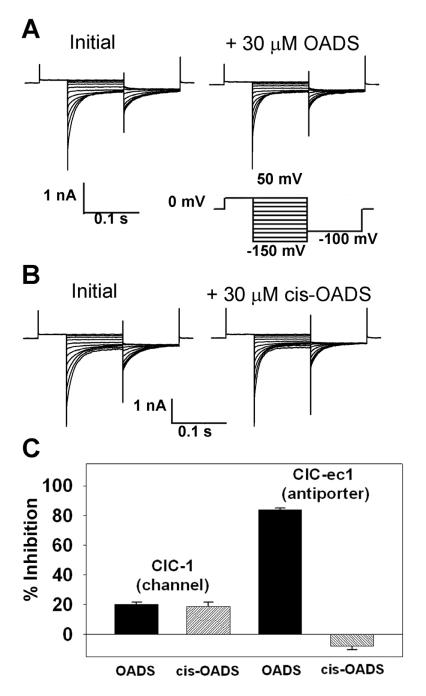
Having established that OADS inhibits ClC-ec1 through specific interactions with the protein, we next sought to address whether OADS (like other stilbene derivatives) also acts as a broad-spectrum inhibitor of chloride channels or whether it displays selectivity towards the chloride-proton antiporter ClC-ec1. OADS was tested for activity against the CLC chloride channel ClC-1 using inside-out patches excised from Xenopus oocytes expressing human ClC-1. As shown in Figures 3A and C, 30 μM OADS (which inhibits ClC-ec1 by 84% in PE/PG liposomes) inhibits ClC-1 by only 20%. This low potency of OADS towards ClC-1 could be due solely to the lipid composition of the oocyte membrane, given the previously noted influence of lipids on inhibition by OADS (Figure 1). Accordingly, it is informative to compare the cis and trans isomers of OADS within each environment. In contrast to inhibition of ClC-ec1, similar degrees of ClC-1 inhibition are observed with both cis and trans OADS isomers (Figures 3B, 3C). This result suggests that the cis isomer of OADS acts through a more generic mechanism (such as bilayer modification, as discussed above), whereas the trans isomer (“OADS”, Figure 2B) acts through more specific molecular contact with ClC-ec1.
Figure 3. OADS is a selective CLC inhibitor.
A) Representative current families from inside-out patches excised from Xenopus oocytes expressing human ClC-1. Currents before (left), and after (right) steady-state inhibition in 30 μM OADS were elicited by the voltage protocol shown below the traces.
B) ClC-1 currents as in (A) but with 30 μM cis-OADS.
C) Summary data comparing inhibition of ClC-1 by OADS and cis-OADS (left) to inhibition of ClC-ec1 by OADS and cis-OADS (right). ClC-ec1 was reconstituted in PE/PG liposomes and measured in Cl− efflux assays. For each experiment, data represent the mean ± SEM for 3 experiments.
OADS binds to and inhibits ClC-ec1 with a 2:1 stoichiometry
In the chloride efflux assays, the inhibition curves are best fit with a Hill coefficient of 1.9, which suggests that ClC-ec1 inhibition requires at least 2 OADS to bind per protein subunit (Figure 1C). (ClC-ec1 is a homodimer, with each monomer acting as an independent transporting unit (Robertson, et al., 2010)). In bilayer recordings, the Hill coefficient for OADS inhibition ranges from 3.9 at +100 mV to 6.1 at −100 mV (Figure 1E). Given that OADS has some bilayer-modifying activity (Figure 2D), the larger Hill coefficient in the latter experiments could be due to OADS interacting with the bilayer, and not directly with the protein. To focus on the protein-specific aspect of OADS binding to and inhibition of ClC-ec1, we investigated the covalent binding of a photo-reactive OADS derivative with detergent-solubilized protein. Covalent modification of ClC-ec1 by this reagent allows for use of mass spectrometry to identify the number of OADS molecules bound per protein subunit. By quantifying the Cl− flux activity of the modified protein, we then could determine how many bound OADS molecules are necessary and sufficient for inhibition.
To synthesize an OADS analog suitable for photoaffinity labeling, we chose a diazirine photophore, as this functional group only slightly perturbs the OADS structure, its photoactivation generates an electrophilic carbene that can react with any nearby C– H, N–H, or O–H bond, and it is activated at 365 nm, a wavelength that will not induce photolytic protein damage (Dorman and Prestwich, 2000). The synthesis of a diazirine-OADS photoaffinity-labeling reagent (“PAL-OADS”) is shown in Figure S2A. The Ki of PAL-OADS for ClC-ec1 is 80 μM as determined by the Cl− efflux assay in E coli polar lipids (Figure S2B), a 2-3-fold increase compared to OADS (Figure 1C).
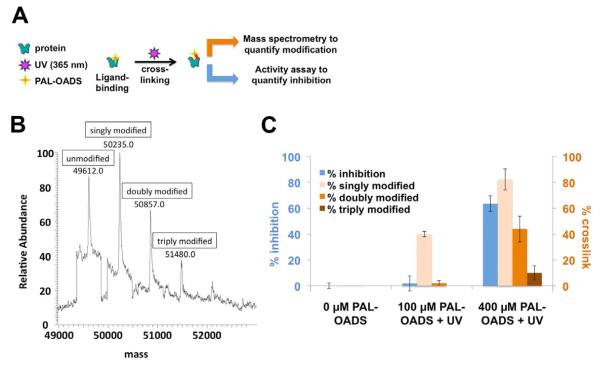
In liposomes, irreversible inhibition by PAL-OADS was observed only when the sample was irradiated with 365-nm light, thus confirming that PAL-OADS acts as a light-dependent covalent inhibitor (Figure S2C). To quantify the relationship between photo-incorporation and inhibition, solubilized ClC-ec1 was incubated with PAL-OADS and irradiated with 365 nm light. These samples were then split into aliquots for either analysis by mass spectrometry or reconstitution into liposomes for activity analysis (Figure 4A-C). The sample labeled with the lower concentration of PAL-OADS showed no inhibition relative to unmodified ClC-ec1, though 40% of the protein was modified by a single OADS molecule (Figure 4C, middle); this result indicates that binding of one equivalent of OADS per ClC-ec1 is not sufficient to inhibit function. The sample labeled with the higher concentration of PAL-OADS (400 μM) was inhibited 62%, which best correlates to the amount of protein that is at least doubly labeled (44%), but not to the amount that is triply labeled (10%). These results are consistent with the Hill coefficient of ~2 determined in the Cl− efflux assays (Figure 1B) and demonstrate that the binding of two OADS molecules per subunit is necessary and sufficient for inhibition.
Figure 4. Binding two molecules of OADS per ClC-ec1 protein subunit is necessary and sufficient to inhibit ClC-ec1 function.
A) Experimental schematic: cross-linked protein was subjected to mass spectrometry analysis (as in panel B) and functional studies (reconstitution and Cl−-efflux assays, as in Figure 1 B, C).
B) Covalent modification of ClC-ec1 with 400 μM PAL-OADS detected by mass spectrometry.
C) Comparison of % inhibition of ClC-ec1 activity measured using Cl− efflux (blue bars) to % ClC-ec1 that is singly, doubly and triply modified (light orange, medium orange, and brown bars, respectively). ClC-ec1 in the presence of 0, 100 or 400 μM PAL-OADS was irradiated with 365-nm light UV and then split into two samples as illustrated in panel A. “Singly modified” corresponds to percent ClC-ec1 covalently linked to at least one equivalent of PAL-OADS (sum of singly, doubly, and triply modified protein), detected by mass spectrometry as in panel B. “Doubly modified” = % ClC-ec1 covalently linked to at least two equivalents of PAL-OADS (sum of doubly and triply modified protein). “Triply modified” = % ClC-ec1 covalently linked to three equivalents of OADS. The combined results demonstrate that (1) binding of one OADS molecule to ClC-ec1 does not inhibit ClC-ec1 (middle, results with 100 uM PAL-OADS), (2) binding of two OADS molecules correlates well with inhibition (right, results with 400 μM PAL-OADS), and (3) binding of three OADS molecules is not required for inhibition (right, results with 400 μM OADS). Results represent the mean of three experiments (± SEM).
Inhibition by OADS is mediated by binding to 2 distinct intracellular sites
Having established that direct binding of OADS to ClC-ec1 is responsible for inhibition, we sought to obtain structural details of the inhibitor-binding sites. ClC-ec1 crystals grown in the presence of (or soaked with) OADS failed to reveal density corresponding to the small molecule. An alternative strategy using PAL-OADS in combination with ‘top-down’ mass spectrometry (MS) was, therefore, explored (Robinette, et al., 2006). Top-down MS uses high-resolution Fourier-transform mass spectrometry to measure the intact mass of the protein in combination with fragmentation either by collisionally activated or electron-capture dissociation (CAD/ECD), producing product ions for tandem mass spectrometry whose masses are sequence and modification specific (Kelleher, 2004). Analysis of the single-labeled protein revealed a binding site localized within residues 109 and 185 of ClC-ec1; analysis of the double-labeled protein revealed a second locale within the first 40 residues of the N-terminus (Figures 5A, S3, Tables S1, S2).
The MS data show that both OADS-binding sites are localized to the intracellular side of the membrane. This result cannot be compared directly with our functional assays because reconstituted ClC-ec1 adopts mixed orientations in liposomes and bilayers. We can achieve “functional orientation”, however, by taking advantage of the fact that DIDS inhibits ClC-ec1 specifically from the intracellular side, as demonstrated in bilayer recordings (Matulef and Maduke, 2005). Figure 5B shows a schematic representation of transporters oriented within a bilayer in opposite directions relative to the two bilayer chambers (called cis and trans). If OADS inhibits exclusively from the intracellular side, addition of OADS to the cis chamber after addition of DIDS to the same chamber should have no further effect (Figure 5B, case 1). In contrast, if OADS inhibits from the extracellular side, addition of OADS to the cis chamber after addition of DIDS to the same chamber should cause further inhibition (Figure 5B, case 2). We observe that application of OADS following addition of DIDS to the same chamber has no effect, consistent with Case 1 (Figure 5C). Likewise, application of DIDS following addition of OADS to the same chamber has no effect (Figure S4A). In further support of this model, application of OADS to the opposite chamber from DIDS results in significant additional inhibition (Figure S6B, D), as does application of DIDS to the opposite chamber from OADS (Figure S6C, D). Hence, OADS inhibits ClC-ec1 from the intracellular side, in agreement with the mass spectrometry results.
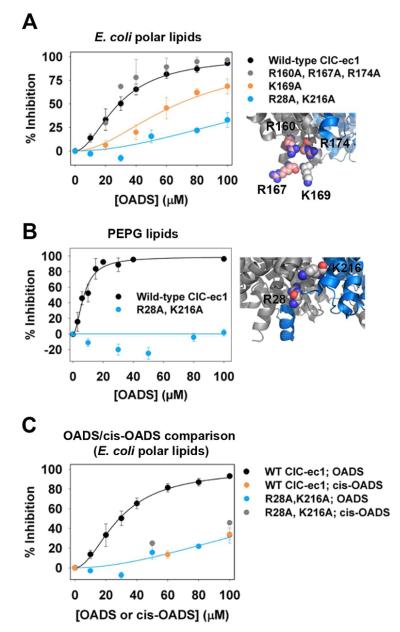
To confirm and further refine the location of the OADS-binding sites, we tested the functional effect of mutations in each of the regions identified through mass spectrometry. We targeted cationic amino acid residues, reasoning that neutralization of such residues near the binding pockets would decrease affinity of the di-anionic OADS molecule. Figure S5 shows a summary of the mutations that had no (or very little) effect on Ki for OADS. Of the residues tested, two sites were found in which neutralization disrupted OADS inhibition. Within the first region identified by MS (residues 109-185, Figure 5A), the K169A mutant exhibited a two-fold decrease in sensitivity to OADS (Ki = 68 μM), whereas mutating the three nearby arginine residues (R160, R167 and R174, located within 5 Å of K169) had no influence (Figures 6A, S5A). This result indicates that a mutation at K169 changes OADS affinity through disruption of a specific interaction rather than through a more general electrostatic effect. Within the second region identified by MS (residues 15-40, Figure 5A), we found that R28A in combination with K216A (which is within 9 Å in 3-dimensional space) eliminated all detectable inhibition in PE/PG lipids (Figure 6B) and reduced the inhibition in E. coli lipids several fold (Figure 6A). The residual inhibition in E. coli lipids is similar to the inhibition observed with cis-OADS (Figure 6C). Ki values could not be determined for the R28A/K216A mutant due to the solubility limit of OADS (100 μM). This decrease in OADS affinity for mutant ClC-ec1 is specific to R28/K216, as mutation of R23 and K30 or R17, R18, and R19 had no effect on OADS inhibition (Figure S5).
Figure 6. K169 and R28/K216 constitute functionally relevant OADS binding sites.
A) Cl− efflux assays reveal decreased sensitivity of K169 and R28/K216 mutants to OADS. Simultaneous mutation of three other cationic residues R164, R160, and R172, all within 5 Å of K169 (inset), has no effect on inhibition. Assays were performed on liposomes formed from E. coli polar lipids. Error bars indicate SEM for 3-7 experiments at each concentration.
B) Inhibition of R28A/K216A by OADS is not detectable in Cl− efflux assays with PE/PG liposomes. Data for WT are identical to that shown in Figure 1G. For R28A/K216A, error bars indicate SEM for 5 experiments at each concentration.
C) In contrast to WT ClC-ec1, cis-OADS inhibits R28A/K216A just as potently as trans-OADS (“OADS”). Data are from Cl− efflux assays on liposomes formed from E. coli polar lipids.
Simulations of OADS:ClC-ec1 binding in a lipid bilayer
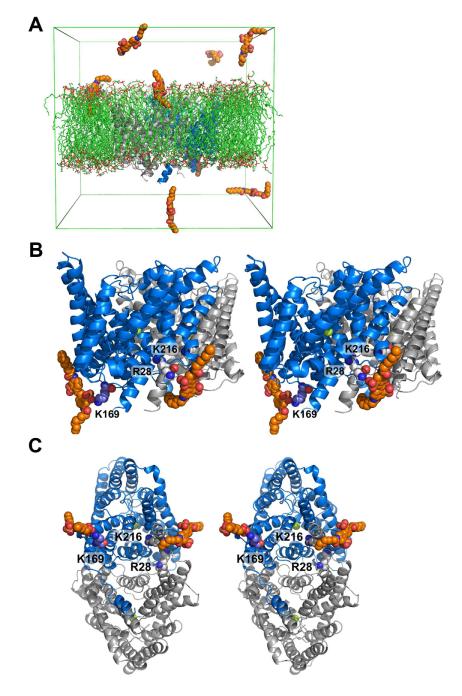
We also studied the binding of OADS to ClC-ec1 by atomistic molecular dynamics simulations in POPE/POPG (3:1) membranes, which were done without prior knowledge of any experimental data concerning the OADS binding sites. Figure 7A shows the simulation system. In five simulations (each 30-85 ns, with a total simulation time of 265 ns), six to eight OADS molecules were inserted in the aqueous phase near the membrane. Most OADS molecules rapidly moved to the membrane/solution interface (not all molecules had entered the interfacial region at the time the snapshot in Figure 7a was taken), with the OADS sulfate groups interacting with the phospholipid head groups. Contacts between OADS and K169 were observed in two simulations; contacts between OADS and the region near R28 and K216 were observed in three simulations. Both interactions were observed in one simulation, where two OADS molecules associated with the intracellular side of ClC-ec1: one of them formed a tight ionic interaction between K169 and the anionic sulfate on OADS; the second interacted with a larger region of the adjoining ClC-ec1 subunit near K216 and R28, involving a contact to the backbone of R28 (Figure 7B, C). The limited time-scale of the simulations does not allow conclusions about the thermodynamic stability of these binding modes, but the results lend further support to our experimental data implicating K169 and R28/K216 as specific, functionally relevant sites of OADS binding to ClC-ec1.
Figure 7. OADS binding to ClC-ec1 at K169 and R28/K216 as observed by atomistic molecular dynamics simulations.
A) Simulation system showing ClC-ec1 (blue/grey) in a lipid bilayer (green) with OADS (orange). Lower compartment corresponds to the intracellular side of the protein (see Experimental Procedures for more details of simulation system).
B) Stereo view of ClC-ec1 showing that OADS associates with K169 through an ionic interaction and with R28/K216 through direct contact with the backbone of R28. For reference, chloride ions bound to the chloride permeation pathway of ClC-ec1 are shown space-filled in green
C) Rotated view, showing perspective from the cytoplasm.
DISCUSSION
Despite recent progress in the generation of CLC-specific inhibitors of certain homologs, (including a scorpion venom toxin specific for ClC-2 (Thompson, et al., 2009) and small-molecule inhibitors of the ClC-K channels (Gradogna and Pusch, 2010; Liantonio, et al., 2008)), the CLCs lack the extensive array of small-molecule modulators available for other membrane proteins. We have developed and characterized a novel CLC inhibitor, “OADS” which is the first small-molecule inhibitor of CLC antiporters that is of sufficient affinity and stability to be used in both functional and biochemical studies. OADS was created through the combination of two generic chloride-channel inhibitors, each of which shows no efficacy against ClC-ec1. The maximal observed affinity of OADS for ClC-ec1 (Ki ~ 3 μM) is well within the range of typical lead compounds, and we anticipate that further iterative modification (performed in tandem with functional studies), will result in improved next-generation inhibitors that will facilitate atomistic studies of these important membrane proteins.
The information we have gathered on OADS binding and inhibition of ClC-ec1 provide insight into possible mechanisms of inhibition. Functional and mass spectrometric analysis of the singly-modified protein (Figure 4C, D) shows that binding at a single site between residues 109 and 185 occurs first (since no modification at the second site is observed in the singly-modified protein.) This site involves K169, as evident from our mutational (Figure 6A, C, D) and modeling (Figure 7B, C) studies. Binding to this site alone is not sufficient to inhibit function, however; inhibition is observed only when OADS is bound to two discrete locations (Figure 4D). Because binding to the second (R28/K216) site is not observed in the absence of binding to the K169 site (Figure 5A), we conclude that binding of OADS at the K169 site is required for binding to the second site.
Mutating R28 and K216 to alanine eliminates all detectable inhibition of ClC-ec1 reconstituted in PE/PG and decreases inhibition in E. coli lipids several fold (Figure 6A, B). The residual inhibition in E. coli lipids may be due to nonspecific bilayer effects, rather than specific OADS-protein interactions. The magnitude of such an effect is likely to depend on the bilayer composition, which could account for the differences between the inhibition observed in PE/PG versus E. coli lipids. Non-specific binding, however, cannot be the primary mechanism of action of OADS on the wild-type protein (as discussed above, Figure 2), but may account for the residual inhibition observed in the R28A/K216A mutant. In support of this hypothesis, cis-OADS (which is more bilayer-modifying but a less effective inhibitor of wild-type ClC-ec1 Figure 2C, D, 3D) inhibits the R28A/K216A mutant with similar potency to the trans-OADS isomer (Figure 6C). Likewise, ClC-1 (which lacks positively charged residues at the corresponding positions) is inhibited only 20% by OADS, and is equally inhibited by both the cis and trans-OADS isomers (Figure 3). Together, these data further support our model for specific OADS-R28/K216 interactions in the inhibition of ClC-ec1 and illustrate the importance of testing for bilayer effects.
Does OADS inhibit eukaryotic CLC antiporters? Addressing this question is challenging given the low currents exhibited by eukaryotic CLCs in inside-out patches and the fact that eukaryotic CLCs have not been functionally reconstituted for use in either bilayer recordings or efflux assays. Sequence alignments around the regions that contribute to OADS potency in ClC-ec1 are too wobbly to reliably predict which homologs contain positively charged residues in positions equivalent to R28, K169, and K216 (and thus to predict which homologs are sensitive to OADS). Nevertheless, it is intriguing that in some alignments position 169 (at the site of initial binding) contains a positively charged residue in most CLC antiporters and a negatively charged residue in all of the known channels (Figure S5C). This observation suggests that OADS might be generally specific for CLC antiporters over channels.
The effect of cardiolipin (CLN) on both OADS potency (Figure 1G) and ClC-ec1 activity (Figure S1) suggests a functionally important CLN-protein interaction in ClC-ec1, perhaps similar to those observed in other proton-pumping proteins. In the cytochrome bc1 complex, for example, CLN is thought to serve as a local buffer capable of directly feeding protons into the ion permeation path (Haines and Dencher, 2002; Lange, et al., 2001). Consistent with CLN playing such a role in H+ transfer by ClC-ec1, Cl− efflux at pH 4.5 (slightly above the pH 4.0 used in the assays reported here) is robust in PE/CLN liposomes but severely reduced in PE/PG (in the absence of CLN) (Figure S1). OADS (Figure 1A) shares some basic structural features with CLN (including the presence of two central negative charges flanked by hydrocarbon chains). Thus, owing to the higher potency of OADS the absence of CLN (Figure 1E, G), it is tempting to speculate that OADS and CLN compete for a binding site on ClC-ec1 (consistent with mechanism 3 in Figure 2A). A direct competition is unlikely, however, given that the R28A/K216A mutant has similar activity to wild-type ClC-ec1.
How does OADS binding impact ClC-ec1 function? The location of the OADS binding sites – away from the Cl−-permeation pathway (Figures 5A, 7B) – shows that OADS is not a classic “pore blocker”. This conclusion is supported by mutagenesis of the intracellular vestibule leading to the Cl−-permeation pathway, as simultaneous neutralization of three charged residues in this region has no effect on inhibition by OADS (Figure S5A). We can further conclude that OADS does not indirectly cause occlusion of the chloride permeation pathway because ITC experiments reveal no significant effect on Cl− binding to ClC-ec1 (Figure S6), Together, these results strongly suggest that OADS leaves the Cl−-permeation pathway intact and acts either by disrupting the H+-permeation pathway or by hindering the transition between protein conformations involved in the Cl−/H+ transport cycle (hence slowing transport). The CLN influence on OADS affinity could thus arise from several possible mechanisms including a specific interaction of CLN with the H+-permeation pathway, a general membrane effect (Lundbaek, et al., 2010), and/or a propensity for ClC-ec1 to adopt a conformation more favorable to OADS binding without CLN present. If OADS does indeed act by “trapping” ClC-ec1 in a particular conformation, then OADS will be a powerful tool for investigating CLC conformational states that have been undetected by crystallography but glimpsed by NMR (Elvington, et al., 2009). In any case, our findings set the stage for inhibitor-driven investigations to elucidate function in this widespread and distinctive family of channels and transporters.
SIGNIFICANCE
CLC “chloride channel” proteins are essential for a wide variety of physiological processes and are distinctive in that only half of the CLC family members are true chloride channels, whereas the other half are chloride-proton antiporters. Investigations of this unique family of proteins thus hold promise for generating insight into fundamental mechanistic distinctions between channels and transporters. The study of CLCs has been hindered, however, by the lack of an established pharmacology. Here, we begin to fill this void by identifying and characterizing a novel compound that inhibits a CLC antiporter (ClC-ec1), and has little effect on a CLC channel (ClC-1). This newly developed molecule, OADS, was generated through combination of two generic anion-channel inhibitors that alone show no efficacy against ClC-ec1. OADS is the first small-molecule inhibitor of the CLC antiporters that is both stable in solution and amenable to structural modification. These traits have allowed us to go beyond pure functional analysis to characterize details of inhibitor-protein binding. We first used a new gramicidin-based assay to demonstrate that inhibition is due to protein binding rather than bilayer modification. This novel technology has broad applicability and vast potential for streamlining drug discovery efforts. We next used top-down mass spectrometry analysis and a photoaffinity-labeled OADS derivative to identify inhibitor binding sites, thus establishing the feasibility of these methods with large (50-kD) integral membrane proteins. Finally, we use molecular dynamics simulations, mutagenesis, and activity assays were used to substantiate the relevance and specificity of the identified OADS-binding sites. The identification of these sites will aid in further development and refinement of next-generation CLC inhibitors.
EXPERIMENTAL PROCEDURES
Chemical Syntheses
OADS and its analogs were synthesized as described in the Supplemental Experimental Procedures.
Expression, purification, and reconstitution of ClC-ec1
ClC-ec1 containing a C-terminal polyhistidine tag was overexpressed in E. coli, purified, and reconstituted as described (Accardi, et al., 2004; Walden, et al., 2007). Reconstituted liposomes were formed from either E. coli polar lipid extract or synthetic lipids (65% POPE/ 35% POPG or 65% POPE and 35% bovine cardiolipin), where POPE is 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine and POPG is 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1′-rac-glycerol). All lipids were purchased from Avanti Polar Lipids.
Chloride efflux assays
Cl− efflux assays were performed as previously described (Matulef, et al., 2008). The selected inhibitor was diluted from a 2–10 mM stock solution in H2O to ClC-ec1-containing liposomes (in 300 mM KCl, 25 mM Na+-citrate, pH 4.0). An equal volume of H2O was added to liposomes containing no inhibitor. OADS solubility is limited in salt solution (unlike in pure H2O), and thus a final OADS concentration of no more than 100 μM was used in these experiments. (In contrast, PAL-OADS remains soluble up to mM concentrations in salt solution, and thus higher concentrations could be used in the mass spectrometry experiments described below.) Intra- and extravesicular solutions were equalized by freezing and thawing samples four times prior to extrusion through a 400 nm filter. Extravesicular solution was exchanged for low-chloride flux buffer (150 mM K2SO4, 50 mM KCl, 25 mM Na+-citrate, pH 4.0) immediately prior to performing the experiment. Cl− efflux was initiated by the addition of the potassium-selective ionophore valinomycin, which allows bulk electroneutral efflux of KCl, and monitored using an extravesicular AgCl electrode. The turnover rate is defined as the initial rate of Cl− efflux minus the initial efflux through control liposomes containing no ClC-ec1, divided by the number of ClC-ec1 molecules in the assay (Cl− s−1 ClC-ec1−1) (quantified as described by Walden et al, 2007, using an extinction coefficient of 0.85 cm2/mg) . Triton X-100 (0.1%) was added at the end of each assay as a control to reveal the total chloride.
Electrophysiological Recordings
Bilayer and patch-clamp recording were performed as described previously (Lisal and Maduke, 2009; Matulef and Maduke, 2005). See Supplemental Experimental Procedures for details.
Measurement of bilayer modification by OADS and its derivatives
Large unilamellar vesicles (LUVs) were made of 1,2-dierucoyl-sn-glycero-3-phosphocholine (DC22:1 PC), loaded with the water-soluble fluorophore 8-aminonaphthalene-1,3,6-trisulfonic acid, disodium salt (ANTS) using a four-part process of sonication, freeze-thaw cycles, extrusion, and elution. The LUVs were doped with gramicidin (gA) from B brevis and incubated for 20-24 hours at 12.5°C. Alternatively, gA containing vesicles were prepared by adding gA directly to the lipid. Both methods of vesicle preparation were subjected to the same four-step process and produced consistent control rates. The fluorescence produced by ANTS is quenched by thallium (Tl+). Tl+ and ANTS cross the bilayer slowly, whereas the conducting gA channels are very Tl+ permeable, such that the Tl+ influx rate (the rate of fluorescence quenching) becomes a measure of the number of conducting gA channels in the LUV membranes. To measure the fluorescence quench rate, the ANTS-loaded LUVs were mixed with the Tl+ quencher using a stopped-flow spectrofluorometer (Applied Photophysics, SX.20) and the fluorescence quench rate was determined from a stretched-exponential fit to the fluorescence time course, as described previously (Ingolfsson and Andersen, 2011; Ingolfsson and Andersen, 2010). The stilbene derivatives are fluorescent, and background fluorescence was corrected by measuring the fluorescence time-course in the absence and presence of quencher, thus enabling correction for the background fluorescence and photobleaching of the OADS derivatives.
Binding stoichiometry determination by mass spectrum analysis
Samples were analyzed by LC-MS on an Agilent 1100 binary pump HPLC and Thermo Fisher LTQ XL ion trap mass spectrometer. A 75 × 2.1 mm Zorbax 300SB-C3 Poroshell column was employed with initial conditions of 98% A (0.1% formic acid in water)/2% B (0.1% formic acid in acetonitrile) ramped to 95% B in 10 min at a flow of 250 μL/min. Ionization was in positive ESI with mass range 400-2000 m/z. The injection volume was 10 μL of a 100× diluted in H2O sample solution. Promass deconvolution software was used to plot the data as mass vs. intensity.
Top-down mass-spectrometry of covalently modified ClC-ec1
ClC-ec1 covalently modified with 500 μM PAL-OADS were precipitated with chloroform/methanol and dissolved in 100 μL of 90% formic acid (ACS grade, Fisher). The sample was immediately injected into a size-exclusion chromatography system previously equilibrated with CHCl3/MeOH/1% aqueous formic acid (4:4:1, v/v), as previously described for lactose permease (a protein that weighs close to 50 kDa and has twelve transmembrane helices) (Whitelegge, et al., 1999). Column eluent was directed to a standard electrospray source (IonMax, Thermo Scientific) attached to a 7 Tesla hybrid linear ion trap Fourier-transform ion cyclotron resonance mass spectrometer (LTQ-FT Ultra, Thermo Scientific). The column (SW2000 XL, Tosoh Biosciences) was eluted with a flow rate of 250 μL/min, which was reduced to 5 μL/min immediately prior to elution of the protein peak 7 minutes into the run. This ‘peak parking’ allows extended data collection on OADS/ClC-ec1 (10 – 90 minutes). The FT-MS was operated at 400,000 resolution (at 400 m/z) in order to achieve unit resolution of 13C isotopomers. The mass spectrum was then transformed to the zero charge state using Xtract software (Xcaliber, Thermo Scientific). Collisionally activated dissociation (CAD) of the ion at m/z 1571 gave a complex mass spectrum that was analyzed using ProSightPC (Version 1.0; Thermo Scientific) to match CAD product ions to the primary structure of ClC-ec1 with 1 or 2 OADS molecules placed at different positions.
Molecular dynamics simulations
The ClC-ec1 dimer (PDB entry 1OTS)(Dutzler, et al., 2003) was inserted in a hydrated and equilibrated POPE/POPG(3:1) bilayer, set up with CHARMM-GUI (Jo, et al., 2008), using g_membed (Wolf, et al., 2010). The system contained 240 POPE lipids and 39 POPG lipids, 29509 TIP3P water molecules, and KCl salt at 150 mM concentration. The CHARMM36 force field was used together with the TIP3P water model (Klauda, et al., 2010). After energy minimization and equilibration for ~40 ns, eight molecules of OADS were inserted into the aqueous phase near the membrane. OADS was assumed to be in a dianionic state and the parameters for OADS were derived using CgenFF (Vanommeslaeghe, et al., 2010). See Supplemental Experimental Procedures for further details.
Supplementary Material
HIGHLIGHTS.
 OADS is the first known small-molecule inhibitor of a CLC antiporter
OADS is the first known small-molecule inhibitor of a CLC antiporter OADS specifically inhibits the ClC-ec1 antiporter but not the ClC-1 channel
OADS specifically inhibits the ClC-ec1 antiporter but not the ClC-1 channel Photoaffinity labeling and mass spectrometry have localized OADS binding to two discrete sites
Photoaffinity labeling and mass spectrometry have localized OADS binding to two discrete sites The unique binding mode and lipid-dependence of OADS suggest potential mechanisms of action
The unique binding mode and lipid-dependence of OADS suggest potential mechanisms of action
ACKNOWLEDGEMENTS
We thank Martin Prieto for comments on the manuscript, David Kopfer for help with the parameterization of OADS, Hyun-Ho Lim and Chris Miller for help with crystallization attempts, Ricky Cheng for remedial Pymol and assistance with figures, and Thomas Jentsch for providing the hClC-1 DNA. This work was supported by NIH grants GM070773 (M.M.) and GM021342 (O.S.A.), ARRA Supplements GM070773-05S1 (M.M.) and GM021342-35S1 (O.S.A), AHA grant 1137885-100-UADJU (M.M.), the Mathers Foundation (M.M.), a Stanford University SPARK award (M.M. and J.D.B.) Stanford University (J.D.B.), a Research Corporation for Science Advancement Cottrell College Science Award (K.M.), start-up funds from the University of San Diego (K.M.), Scottish Universities Physics Alliance (U.Z.), and the National Physical Laboratory (U.Z.). Top-down mass spectrometry was performed on a 7T LTQ-FT Ultra purchased with National Institutes of Health support (S10 RR023045). A.H. was supported by an Amgen Graduate Fellowship and a Roche Graduate Fellowship. S.P. was supported in part by a Summer Undergraduate Research Experience grant from the University of San Diego. K.T. was supported by a Stanford BioX Fellowship. T.C. was supported by the Stanford Institutes of Medicine Summer Research Program.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Accardi A, Kolmakova-Partensky L, Williams C, Miller C. Ionic currents mediated by a prokaryotic homologue of CLC Cl− channels. J. Gen. Physiol. 2004;123:109–119. doi: 10.1085/jgp.200308935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Accardi A, Miller C. Secondary active transport mediated by a prokaryotic homologue of ClC Cl− channels. Nature. 2004;427:803–807. doi: 10.1038/nature02314. [DOI] [PubMed] [Google Scholar]
- Alper SL. Molecular physiology and genetics of Na+-independent SLC4 anion exchangers. J. Exp. Biol. 2009;212:1672–1683. doi: 10.1242/jeb.029454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen OS. Perspectives on how to drug an ion channel. J. Gen. Physiol. 2008;131:395–397. doi: 10.1085/jgp.200810012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabantchik ZI, Greger R. Chemical probes for anion transporters of mammalian cell membranes. Am. J. Physiol. 1992;262:C803–827. doi: 10.1152/ajpcell.1992.262.4.C803. [DOI] [PubMed] [Google Scholar]
- Chen B, Nicol G, Cho WK. Role of calcium in volume-activated chloride currents in a mouse cholangiocyte cell line. J. Membr. Biol. 2007;215:1–13. doi: 10.1007/s00232-007-9000-9. [DOI] [PubMed] [Google Scholar]
- Dorman G, Prestwich GD. Using photolabile ligands in drug discovery and development. Trends Biotechnol. 2000;18:64–77. doi: 10.1016/s0167-7799(99)01402-x. [DOI] [PubMed] [Google Scholar]
- Duan DD. The ClC-3 chloride channels in cardiovascular disease. Acta Pharmacol. Sin. 2011;32:675–684. doi: 10.1038/aps.2011.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutzler R. The ClC family of chloride channels and transporters. Curr. Opin. Struct. Biol. 2006;16:439–446. doi: 10.1016/j.sbi.2006.06.002. [DOI] [PubMed] [Google Scholar]
- Dutzler R, Campbell EB, MacKinnon R. Gating the selectivity filter in ClC chloride channels. Science. 2003;300:108–112. doi: 10.1126/science.1082708. [DOI] [PubMed] [Google Scholar]
- Elvington SM, Liu CW, Maduke MC. Substrate-driven conformational changes in ClC-ec1 observed by fluorine NMR. EMBO J. 2009;28:3090–3102. doi: 10.1038/emboj.2009.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engh AM, Maduke M. Cysteine accessibility in ClC-0 supports conservation of the ClC intracellular vestibule. J. Gen. Physiol. 2005;125:601–617. doi: 10.1085/jgp.200509258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estevez R, Schroeder BC, Accardi A, Jentsch TJ, Pusch M. Conservation of Chloride Channel Structure Revealed by an Inhibitor Binding Site in ClC-1. Neuron. 2003;38:47–59. doi: 10.1016/s0896-6273(03)00168-5. [DOI] [PubMed] [Google Scholar]
- Evanko DS, Zhang Q, Zorec R, Haydon PG. Defining pathways of loss and secretion of chemical messengers from astrocytes. Glia. 2004;47:233–240. doi: 10.1002/glia.20050. [DOI] [PubMed] [Google Scholar]
- Gradogna A, Pusch M. Molecular Pharmacology of Kidney and Inner Ear CLC-K Chloride Channels. Front Pharmacol. 2010;1:130. doi: 10.3389/fphar.2010.00130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves AR, Curran PK, Smith CL, Mindell JA. The Cl-/H+ antiporter ClC-7 is the primary chloride permeation pathway in lysosomes. Nature. 2008;453:788–792. doi: 10.1038/nature06907. [DOI] [PubMed] [Google Scholar]
- Gupta SP, editor. Ion channels and their inhibitors. 1Edition Springerlink; Berlin: 2011. [Google Scholar]
- Haines TH, Dencher NA. Cardiolipin: a proton trap for oxidative phosphorylation. FEBS Lett. 2002;528:35–39. doi: 10.1016/s0014-5793(02)03292-1. [DOI] [PubMed] [Google Scholar]
- Huang ZM, Prasad C, Britton FC, Ye LL, Hatton WJ, Duan D. Functional role of CLC-2 chloride inward rectifier channels in cardiac sinoatrial nodal pacemaker cells. J. Mol. Cell. Cardiol. 2009;47:121–132. doi: 10.1016/j.yjmcc.2009.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolfsson HI, Andersen OS. Alcohol’s effects on lipid bilayer properties. Biophys. J. 2011;101:847–855. doi: 10.1016/j.bpj.2011.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolfsson HI, Andersen OS. Screening for small molecules’ bilayer-modifying potential using a gramicidin-based fluorescence assay. Assay Drug Dev. Technol. 2010;8:427–436. doi: 10.1089/adt.2009.0250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsen P, Horobin RW. Preparation and characterization of 4-acetamido-4′-isothiocyanostilbene-2,2′-disulfonic acid (SITS) and related stilbene disulfonates. Stain Technol. 1989;64:301–313. doi: 10.3109/10520298909107022. [DOI] [PubMed] [Google Scholar]
- Jentsch TJ. CLC chloride channels and transporters: from genes to protein structure, pathology and physiology. Crit. Rev. Biochem. Mol. Biol. 2008;43:3–36. doi: 10.1080/10409230701829110. [DOI] [PubMed] [Google Scholar]
- Jeworutzki E, Lopez-Hernandez T, Capdevila-Nortes X, Sirisi S, Bengtsson L, Montolio M, Zifarelli G, Arnedo T, Muller CS, Schulte U, et al. GlialCAM, a Protein Defective in a Leukodystrophy, Serves as a ClC-2 Cl(-) Channel Auxiliary Subunit. Neuron. 2012;73:951–961. doi: 10.1016/j.neuron.2011.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo S, Kim T, Iyer VG, Im W. CHARMM-GUI: a web-based graphical user interface for CHARMM. J Comput Chem. 2008;29:1859–1865. doi: 10.1002/jcc.20945. [DOI] [PubMed] [Google Scholar]
- Kasai M, Kawasaki T, Yamaguchi N. Regulation of the ryanodine receptor calcium release channel: a molecular complex system. Biophys. Chem. 1999;82:173–181. doi: 10.1016/s0301-4622(99)00117-9. [DOI] [PubMed] [Google Scholar]
- Kelleher NL. Top-down proteomics. Anal. Chem. 2004;76:197A–203A. [PubMed] [Google Scholar]
- Klauda JB, Venable RM, Freites JA, O’Connor JW, Tobias DJ, Mondragon-Ramirez C, Vorobyov I, MacKerell AD, Jr., Pastor RW. Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. J. Phys. Chem. B. 2010;114:7830–7843. doi: 10.1021/jp101759q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange C, Nett JH, Trumpower BL, Hunte C. Specific roles of protein-phospholipid interactions in the yeast cytochrome bc1 complex structure. EMBO J. 2001;20:6591–6600. doi: 10.1093/emboj/20.23.6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Ma KT, Zhao L, Si JQ, Zhang ZS, Zhu H, Li J. Niflumic acid hyperpolarizes smooth muscle cells via calcium-activated potassium channel in spiral modiolar artery of guinea pigs. Acta Pharmacol. Sin. 2008;29:789–799. doi: 10.1111/j.1745-7254.2008.00803.x. [DOI] [PubMed] [Google Scholar]
- Liantonio A, Gramegna G, Camerino GM, Dinardo MM, Scaramuzzi A, Potenza MA, Montagnani M, Procino G, Lasorsa DR, Mastrofrancesco L, et al. In-vivo administration of CLC-K kidney chloride channels inhibitors increases water diuresis in rats: a new drug target for hypertension? J. Hypertens. 2012;30:153–167. doi: 10.1097/HJH.0b013e32834d9eb9. [DOI] [PubMed] [Google Scholar]
- Liantonio A, Picollo A, Carbonara G, Fracchiolla G, Tortorella P, Loiodice F, Laghezza A, Babini E, Zifarelli G, Pusch M, et al. Molecular switch for CLC-K Cl− channel block/activation: optimal pharmacophoric requirements towards high-affinity ligands. Proc. Natl. Acad. Sci. U. S. A. 2008;105:1369–1373. doi: 10.1073/pnas.0708977105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CW, Chen TY. Probing the pore of ClC-0 by substituted cysteine accessibility method using methane thiosulfonate reagents. J. Gen. Physiol. 2003;122:147–159. doi: 10.1085/jgp.200308845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisal J, Maduke M. Review. Proton-coupled gating in chloride channels. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2009;364:181–187. doi: 10.1098/rstb.2008.0123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lossin C, George AL., Jr. Myotonia congenita. Adv. Genet. 2008;63:25–55. doi: 10.1016/S0065-2660(08)01002-X. [DOI] [PubMed] [Google Scholar]
- Lundbaek JA, Collingwood SA, Ingolfsson HI, Kapoor R, Andersen OS. Lipid bilayer regulation of membrane protein function: gramicidin channels as molecular force probes. J. R. Soc. Interface. 2010;7:373–395. doi: 10.1098/rsif.2009.0443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maduke M, Pheasant DJ, Miller C. High-level expression, functional reconstitution, and quaternary structure of a prokaryotic ClC-type chloride channel. J. Gen. Physiol. 1999;114:713–722. doi: 10.1085/jgp.114.5.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matulef K, Howery AE, Tan L, Kobertz WR, Du Bois J, Maduke M. Discovery of potent CLC chloride channel inhibitors. ACS Chem. Biol. 2008;3:419–428. doi: 10.1021/cb800083a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matulef K, Maduke M. Side-dependent inhibition of a prokaryotic ClC by DIDS. Biophys. J. 2005;89:1721–1730. doi: 10.1529/biophysj.105.066522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller C. ClC chloride channels viewed through a transporter lens. Nature. 2006;440:484–489. doi: 10.1038/nature04713. [DOI] [PubMed] [Google Scholar]
- Miller C. Integral membrane channels: studies in model membranes. Physiol. Rev. 1983;63:1209–1242. doi: 10.1152/physrev.1983.63.4.1209. [DOI] [PubMed] [Google Scholar]
- Nighot PK, Moeser AJ, Ryan KA, Ghashghaei T, Blikslager AT. ClC-2 is required for rapid restoration of epithelial tight junctions in ischemic-injured murine jejunum. Exp. Cell Res. 2009;315:110–118. doi: 10.1016/j.yexcr.2008.10.001. [DOI] [PubMed] [Google Scholar]
- Picollo A, Pusch M. Chloride/proton antiporter activity of mammalian CLC proteins ClC-4 and ClC-5. Nature. 2005;436:420–423. doi: 10.1038/nature03720. [DOI] [PubMed] [Google Scholar]
- Planells-Cases R, Jentsch TJ. Chloride channelopathies. Biochim. Biophys. Acta. 2009;1792:173–189. doi: 10.1016/j.bbadis.2009.02.002. [DOI] [PubMed] [Google Scholar]
- Ratte S, Prescott SA. ClC-2 channels regulate neuronal excitability, not intracellular chloride levels. J. Neurosci. 2011;31:15838–15843. doi: 10.1523/JNEUROSCI.2748-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson JL, Kolmakova-Partensky L, Miller C. Design, function and structure of a monomeric ClC transporter. Nature. 2010;468:844–847. doi: 10.1038/nature09556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinette D, Neamati N, Tomer KB, Borchers CH. Photoaffinity labeling combined with mass spectrometric approaches as a tool for structural proteomics. Expert Rev. Proteomics. 2006;3:399–408. doi: 10.1586/14789450.3.4.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheel O, Zdebik AA, Lourdel S, Jentsch TJ. Voltage-dependent electrogenic chloride/proton exchange by endosomal CLC proteins. Nature. 2005;436:424–427. doi: 10.1038/nature03860. [DOI] [PubMed] [Google Scholar]
- Thompson CH, Olivetti PR, Fuller MD, Freeman CS, McMaster D, French RJ, Pohl J, Kubanek J, McCarty NA. Isolation and characterization of a high affinity peptide inhibitor of ClC-2 chloride channels. J. Biol. Chem. 2009 doi: 10.1074/jbc.M109.031724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanommeslaeghe K, Hatcher E, Acharya C, Kundu S, Zhong S, Shim J, Darian E, Guvench O, Lopes P, Vorobyov I, et al. CHARMM general force field: A force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J Comput Chem. 2010;31:671–690. doi: 10.1002/jcc.21367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venglarik CJ, Singh AK, Bridges RJ. Comparison of -nitro versus - amino 4,4′-substituents of disulfonic stilbenes as chloride channel blockers. Mol. Cell. Biochem. 1994;140:137–146. doi: 10.1007/BF00926752. [DOI] [PubMed] [Google Scholar]
- Verkman AS, Galietta LJ. Chloride channels as drug targets. Nat. Rev. Drug Discov. 2009;8:153–171. doi: 10.1038/nrd2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walden M, Accardi A, Wu F, Xu C, Williams C, Miller C. Uncoupling and turnover in a Cl-/H+ exchange transporter. J. Gen. Physiol. 2007;129:317–329. doi: 10.1085/jgp.200709756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitelegge JP, le CJ, Lee JC, Engel CK, Prive GG, Faull KF, Kaback HR. Toward the bilayer proteome, electrospray ionization-mass spectrometry of large, intact transmembrane proteins. Proc. Natl. Acad. Sci. U.S.A. 1999;96:10695–10698. doi: 10.1073/pnas.96.19.10695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf MG, Hoefling M, Aponte-Santamaria C, Grubmuller H, Groenhof G. g_membed: Efficient insertion of a membrane protein into an equilibrated lipid bilayer with minimal perturbation. J Comput Chem. 2010;31:2169–2174. doi: 10.1002/jcc.21507. [DOI] [PubMed] [Google Scholar]
- Woodhull AM. Ionic Blockage of Sodium in Nerve. The Journal of General Physiology. 1973;61:687–708. doi: 10.1085/jgp.61.6.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Q, editor. Membrane transporters in drug discovery and development. Humana; New York: 2010. [Google Scholar]
- Zhang XD, Chen TY. Amphiphilic blockers punch through a mutant CLC-0 pore. J. Gen. Physiol. 2009;133:59–68. doi: 10.1085/jgp.200810005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q, Wei Q, He A, Jia R, Xiao Y. CLC-7: a potential therapeutic target for the treatment of osteoporosis and neurodegeneration. Biochem. Biophys. Res. Commun. 2009;384:277–279. doi: 10.1016/j.bbrc.2009.04.088. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.