Summary
The origins and developmental mechanisms of coronary arteries are incompletely understood. We showed here by fate mapping, clonal analysis and immunohistochemistry that endocardial cells generate the endothelium of coronary arteries. Dye tracking, live imaging, and tissue transplantation also revealed that ventricular endocardial cells are not terminally differentiated; instead, they are angiogenic and form coronary endothelial networks. Myocardial Vegf-a or endocardial Vegfr-2 deletion inhibited coronary angiogenesis and arterial formation by ventricular endocardial cells. In contrast, lineage and knockout studies showed that endocardial cells make a small contribution to the coronary veins, the formation of which is independent of myocardial-to-endocardial Vegf signaling. Thus, contrary to the current view of a common source for the coronary vessels, our findings indicate that the coronary arteries and veins have distinct origins and are formed by different mechanisms. This information may help develop better cell therapies for coronary artery disease.
Introduction
Despite the medical importance of coronary arteries, their embryonic origins and developmental mechanisms remain unclear. These arteries are the loci for coronary artery disease, the most widespread disease in western societies. Elucidating mechanisms of coronary artery formation may help recapitulate this developmental process for coronary artery regeneration.
Coronary arteries have 3 tissue layers: the inner layer of endothelium, the middle layer of smooth muscle cells, and the outer layer of fibroblasts. The endothelium is the first layer formed during coronary artery formation. Primitive coronary vessels (or coronary plexuses) consist of one endothelial cell layer. The plexuses then recruit smooth muscle cells and fibroblasts to assemble mature arteries. Endothelium is also the first site where coronary artery disease occurs in adults. Thus identifying the cellular origins of coronary endothelium is essential to elucidate mechanisms of coronary artery development or regeneration.
The heart is made of three major tissue layers: the endocardium, myocardium, and epicardium. The myocardium is the central layer, and the coronary vasculature forms within this layer during development. The epicardium is the outermost epithelial layer of the heart; it is derived from the proepicardium outside the heart (Komiyama et al., 1987; Viragh and Challice, 1981). Studies have shown that epicardial cells generate coronary vascular smooth muscle cells (Cai et al., 2008; Dettman et al., 1998; Mikawa and Fischman, 1992; Mikawa and Gourdie, 1996; Vrancken Peeters et al., 1999; Zhou et al., 2008). It is less clear whether proepicardial/epicardial cells make any significant contribution to coronary endothelial cells, although some coronary endothelial cells in avian species are derived from proepicardial cells (Mikawa et al., 1992; Perez-Pomares et al., 2002). Fate-mapping studies in mice have suggested the sinus venosus as a common origin of the endothelium of coronary arteries and veins (Red-Horse et al., 2010) while a subset of proepicardial cells also contribute to some coronary endothelial cells (Katz et al., 2012).
The endocardium is the internal epithelial layer of the heart. Endocardial cells are one of the earliest endothelial populations acquired in development, differentiating from multi-potent progenitors in the cardiac field (Misfeldt et al., 2009; Sugi and Markwald, 1996; Yamashita et al., 2000; Yang et al., 2008). They form an endocardial tube by vasculogenesis and later become the endocardium of the heart (Drake and Fleming, 2000). Endothelial cells of coronary vessels arise later in development and form coronary vessels in the myocardium (Lavine and Ornitz, 2009; Luttun and Carmeliet, 2003; Majesky, 2004; Olivey and Svensson, 2010; Wada et al., 2003). Ventricular endocardial cells have been thought to be terminally differentiated without a significant role in coronary vessel formation.
Here we showed that ventricular endocardial cells are a major origin of coronary artery endothelium. Myocardial Vegf-a to endocardial Vegfr-2 signaling is required for these cells to differentiate into coronary endothelium. The information may have implications for engineering better vessels for coronary artery regeneration.
Results
Characterization of Nfatc1 expression during coronary vessel development
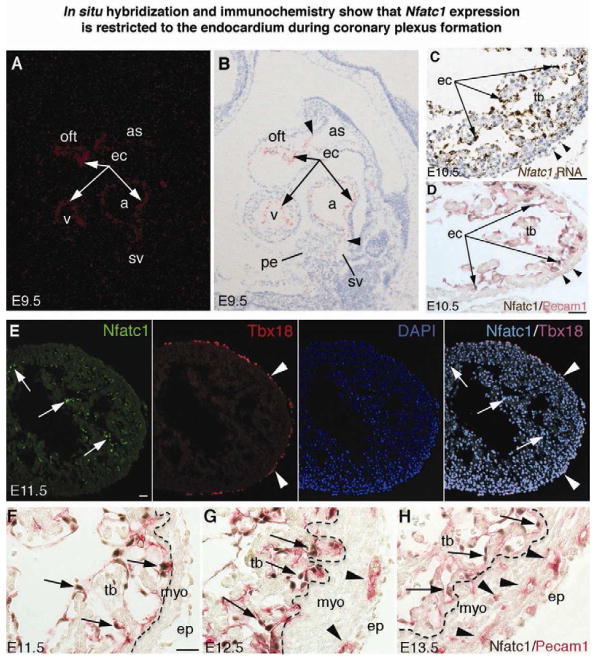
Cardiac endocardial cells comprise a unique endothelial cell population that expresses Nfatc1 during development, while vascular endothelial cells do not express Nfatc1 (Chang et al., 2004; de la Pompa et al., 1998; Ranger et al., 1998; Zhou et al., 2005). In this study, we further characterized Nfatc1 expression in embryonic tissues relative to coronary development. We confirmed by in situ hybridization that Nfatc1 transcripts demarcated endocardium at embryonic day (E) 9.5, since the endothelium of aortic sac, sinus venosus, and the rest of the peripheral vasculature was negative for Nfatc1 transcripts (Figure 1A, 1B). Nfatc1 transcripts were not found in the proepicardium either. At E10.5 Nfatc1 transcripts were similarly restricted to the endocardium (Figure 1C). Likewise, double immunostaining of Nfatc1 and Pecam1 (pan-endothelial marker) revealed that Nfatc1 proteins were confined to the endocardium (Figure 1D). Neither Nfatc1 transcripts nor proteins were detected in the forming epicardium. Furthermore, co-immunostaining of Nfatc1 and Tbx18 (epicardial marker) (Kraus et al., 2001) confirmed that epicardial cells did not express Nfatc1 at E11.5 (Figure 1E).
Figure 1. In situ hybridization and immunochemistry show that Nfatc1 expression is restricted to the endocardium during coronary plexus formation.
(A,B) E9.5 heart sections show Nfatc1 transcripts in the endocardium (ec, arrows). Nfatc1 transcript signals separate the positive endocardium from the negative endothelium of aortic sac (as) and sinus venosus (sv) (arrowheads). Background signals are seen in the proepicardium (pe). a, atrium; oft, outflow tract; v, ventricle
(C) E10.5 ventricular sections show Nfatc1 transcripts in the endocardium (arrows) but not epicardium (arrowheads). tb, trabeculae
(D) E10.5 heart sections co-immunostained with antibodies against Nfatc1 (brown nuclear staining) and Pecam1 (red membrane staining) show Nfatc1 proteins in the endocardium (arrows) but not epicardium (arrowheads).
(E) E11.5 heart sections show Nfatc1 proteins in the endocardium (arrows) but not in the Tbx18-positive epicardium (arrowheads).
(F–H) E11.5–E13.5 heart sections stained with antibodies against Nfatc1 and Pecam1 show that Nfatc1 proteins are restricted to the endocardium (arrows). They are not present in the endothelium of coronary plexuses or Pecam1-positive cells in the myocardium (myo, arrowheads) or the mature coronary vessels (see also Figure S1). Dashed line separates the trabeculae from the compact wall. ep, epicardium. All bars = 25 μm.
When coronary plexuses developed from E11.5 to E13.5, Nfatc1 transcripts were downregulated in the ventricular endocardium (data not shown) while Nfatc1 proteins remained in a subset of endocardial cells (Figure 1F–1H). Neither Nfatc1 transcripts (data not shown) nor Nfatc1 proteins were found in the endothelium of coronary plexuses (Figure 1G, 1H). Likewise, the endothelium of developed coronary vessels from E14.5 to E16.5 did not have detectable Nfatc1 transcripts or Nfatc1 proteins, which were found only in the valve endocardium (Figure S1, A–F; Supplemental Information available online). These findings showed that Nfatc1 expression is restricted to the endocardium during coronary development. Nfatc1 is not expressed in the proepicardium/epicardium, sinus venosus, or developing coronary vessels.
Nfatc1+ endocardial precursors generate coronary plexuses
To study the developmental fate and function of endocardial cells, we generated a Cre knock-in mouse strain, Nfatc1Cre, in which Cre cDNA with an internal ribosomal entry site was inserted downstream of the stop codon of the mouse Nfatc1 (Zhou et al., 2002) (Figure S2, A–C). Nfatc1Cre mice developed normally and bred to the RCEfsEGFP (Miyoshi et al., 2010; Sousa et al., 2009) or R26fslz mice (Soriano, 1999). Cre expression was restricted to the endocardium of Nfatc1Cre embryos; no expression was seen in the sinus venosus, liver, pharyngeal arch, proepicardium/epicardium, myocardium at E9.5–E10.5 (Figure S2, D–F), or developing coronary vessels (data not shown).
The fate of endocardial cells was then tracked by the inherited expression of enhanced green fluorescent protein (EGFP) or beta-galactosidase (β-gal hereafter) (Figure S3, A). Nfatc1Cre-mediated EGFP expression began at E9.0 in the endocardium of Nfatc1Cre;RCEfsEGFP embryos (Figure S3, B). At E10.5, EGFP expression was limited to the heart proper of Nfatc1Cre;RCEfsEGFP embryos (Figure S3, C). Sectional examination of E9.5–E10.5 Nfatc1Cre;RCEfslz embryos confirmed restricted β-gal expression in the endocardium (Figure S3, D,E) and in the cushion mesenchyme derived from endocardial cells (Figure S3, F,G). Neither Cre reporter gene was expressed in the proepicardium/epicardium, myocardium, pharyngeal arch, and liver bud (Figure S3, B–G). Although Nfatc1 expression was not found in the sinus venous endothelium (Figure S3, H), Nfatc1Cre-mediated β-gal or EGFP expression was observed in some sinus venous endothelial cells at E10.5–E11.5 (Figure S3, I–J), suggesting that Nfatc1+ endocardial cells contribute to a subset of sinus venous endothelial population.
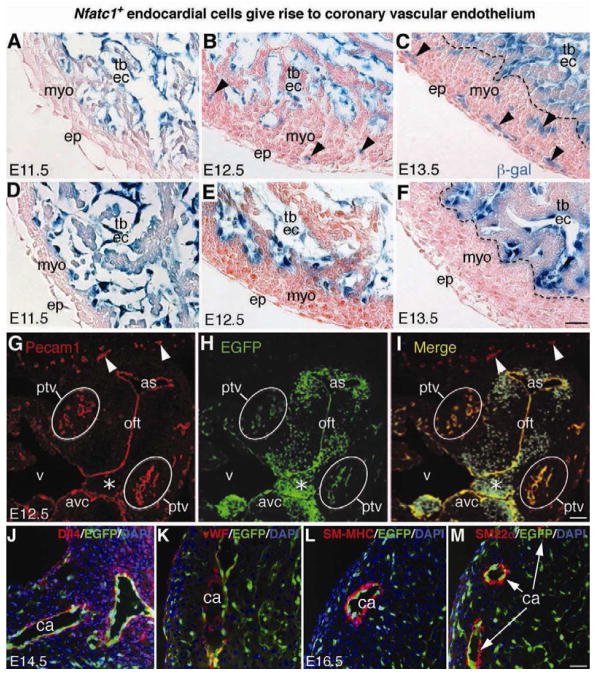
We then examined the lineage contribution of endocardial cells to coronary vessels in Nfatc1Cre;R26fslz embryos and found that cells of the emerging coronary plexuses at E11.5–E13.5 uniformly expressed β-gal (Figure 2). The β-gal+ cells, derived from the Nfatc1+ precursors at E11.5 (Figure 2A), invaded the myocardium at E12.5 (Figure 2B) and formed coronary plexuses throughout the myocardium at E13.5 (Figure 2C). Conversely, in control Nfatc1lacZ-BAC embryos that expressed β-gal driven by the Nfatc1 promoter/enhancer (Misfeldt et al., 2009), β-gal expression (an indicator of the Nfatc1Cre activities) was restricted to the endocardium and absent in coronary plexuses (Figure 2D–2F). This observation is consistent with the finding that coronary plexuses do not express Nfatc1 at these stages, thus further eliminating the possibility of ectopic Cre expression in coronary plexuses.
Figure 2. Fate-mapping analysis reveals that Nfatc1+ endocardial cells generate coronary vascular endothelium.
(A–C) X-gal stained E11.5–E13.5 heart sections of Nfatc1Cre;R26fslz embryos show that the β-gal tagged Nfatc1+ endocardial cells reside in the endocardium of the myocardial wall and trabeculae at E11.5 (A), they begin to invade the myocardium at E12.5 (B, arrowheads) and generate networks of coronary plexuses at E13.5 (C, arrowheads). The descendants of Nfatc1+ cells are not present in the epicardium and myocardium (see also Figures S2 and S3).
(D–F) X-gal stained E11.5–E13.5 heart sections of control Nfatc1lacZ-BAC embryos show that the β-gal activities directed by the Nfatc1 promoter/enhancer are restricted to the endocardium and not present in the myocardium and epicardium. Unlike Nfatc1Cre, Nfatc1lacZ does not label the coronary plexuses.
(G–I) Dual fluorescent sections through ventricular (v) outflow tract (oft) of E12.5 Nfatc1Cre;RCEfsEGFP embryos show the EGFP+ endothelial descendants of Nfatc1+ endocardial cells in the Pecam1+ coronary plexuses in the peritruncal region (ptv). The peripheral vessels (arrowheads) expressing Pecam1 but not EGFP are not derived from the Nfatc1+ endocardial cells (see also Figure S4). Mesenchyme of atrioventricular canal (avc, asterisk), derived from the cushion endocardial cells, is EGFP positive but Pecam1 negative.
(J–K) Dual fluorescent sections of E14.5 Nfatc1Cre;RCEfsEGFP heart show EGFP+ endothelial descendants of endocardial cells in the Dll4+ or vWF+ endothelium (red) of the main coronary arteries (ca). The trabecular endocardium is negative for vWF.
(L–M) Dual fluorescent sections of E16.5 Nfatc1Cre;RCEfsEGFP heart show EGFP+ endothelial descendants of endocardial cells in the inner layer of the coronary arteries (ca) surrounded by smooth muscle cells positive for SM-MHC or SM22a (see also Figure S5 and S6). All Bars = 25 μm.
To verify the cell identity of the endocardial-derived coronary cells, we co-labeled the EGFP+ descendants of Nfatc1+ endocardial cells with Pecam1 antibodies in Nfatc1Cre;RCEfsEGFP embryos. We found that most Pecam1-positive cells in the peritruncal coronary vessels expressed EGFP at E12.5, whereas the vessels outside the heart were labeled only by Pecam1 antibodies (Figure 2G–2I). Also, all coronary plexuses arising at E11.5–E13.5 in the ventricular wall co-expressed Pecam1 and EGFP (Figure S4). We then stained E14.5 or E16.5 Nfatc1Cre;RCEfsEGFP heart sections with multiple cardiovascular markers and found that the EGFP+ descendants of Nfatc1+ endocardial cells were present in the endothelium of coronary arteries expressing arterial endothelial markers Dll4 (Figure 2J and Figure S5, A–C), Ephb2 and Jagged1 (data not shown). We also noted that while endocardial cells were negative for vWF, their descendants in coronary arteries acquired vWF expression (Figure 2K). Thus, the acquisition of vWF and loss of Nfatc1 expression in coronary endothelial cells may serve as makers for endocardial to coronary endothelial differentiation. Further staining of smooth muscle myosin heavy chain (SM-MHC) and SM22α showed that the EGFP+ endocardial descendants contributed to the endothelium of developed coronary arteries, but they did not become vascular smooth muscle cells of the cognate vessels (Figure 2L, 2M). In contrast to their prominent arterial presence, few EGFP+ descendants of endocardial cells were found in coronary veins (Figure S5, D–F). EGFP+ descendants were also absent in lymphatic vessels (Figure S5, G–I) and did not become cardiomyocytes (Figure S5, J–L).
We further compared the fate mapping of Nfatc1Cre-marked endocardial cells in coronary vessels to that of Tie1Cre-labeled pan-endothelial (arterial and venous) cells (Gustafsson et al., 2001). Unlike Tie1+ endothelial descendants which formed vascular networks on the surface of E14.5 hearts, Nfatc1+ endocardial descendants did not form the networks but contributed to the intramyocardial vessels (Figure S6, A,B). Pecam1 staining of E16.5 Nfatc1Cre;RCEfsEGFP hearts confirmed that Nfatc1+ endocardial descendants contributed to most intramyocardial vessels (including the main arteries), with much less presence in the subepicardial vessels (Figure S6, C,D). Coronary arteriograms of E16.5 Nfatc1Cre;RCEfsEGFP hearts validated that EGFP+ descendants of Nfatc1+ endocardial cells contributed to the entire coronary artery network, including the main coronary arteries and their branches, arterioles, and capillaries (Figure S6, E–J).
Quantitative analysis of Nfatc1+ endocardial descendants in the coronary vessels of E16.5 hearts showed that they contributed to 72%, 81%, or 37% endothelial population of major coronary arteries, intramyocardial, or subepicardial vessels, respectively (Figure S6, K). These findings establish that endocardial cells are a major source of endothelial cells of the intramyocardial vessels including major coronary arteries and suggest that the majority of subepicardial vascular endothelium arises from a different origin.
Clonal analysis establishes spatiotemporal differentiation of endocardial cells into coronary endothelial cells
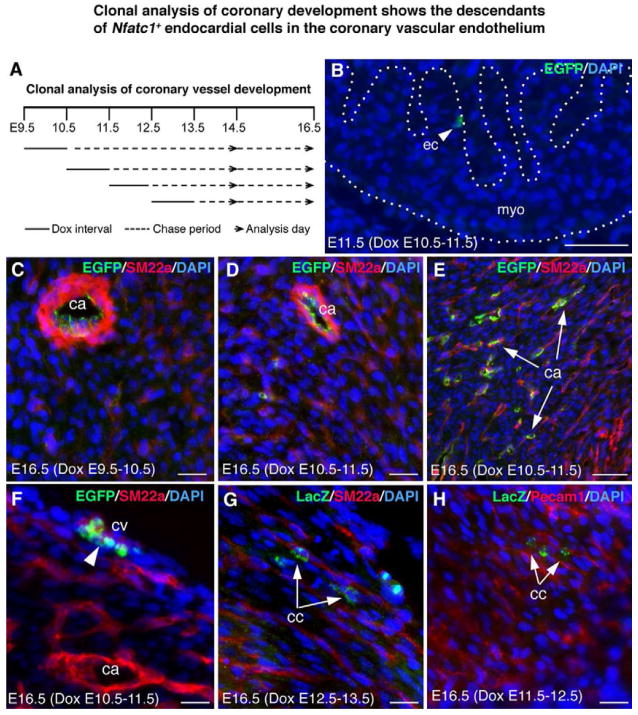
To confirm that the endocardium is a major source of coronary arterial endothelial cells and determine when endocardial cells are committed to a coronary endothelial fate, we generated Nfatc1nrtTA-BAC mice using an Nfatc1-BAC DNA (Figure S7, A,B), and bred them to tetO-Cre;RCEfsEGFP or tetO-Cre;R26fslz mice. When induced with doxycycline (Dox) at E7.5–E11.5, the tetO-Cre-mediated EGFP expression was found to mark endocardial cells and their mesenchymal descendants (Figure S7, C–F). Limited Dox dose was then applied for 24 hours, at E9.5, E10.5, E11.5, or E12.5 (Figure 3A), to induce rare recombination events in individual cells in the endocardium (Figure 3B). The expansion of EGFP+ or β-gal+ single cells were chased for 1–7 days until E14.5 or E16.5, and the clonality and coronary location of cell clusters were determined by expression of the reporters, vascular markers, and histology. We analysed 39 whole hearts by evaluating serial sections (Table S2, Figure 3C–3H). Single or separated EGFP+ or β-gal + cell clusters were found in the endothelium of major or small coronary arteries (Figure 3C–3E), veins (Figure 3F), or capillaries (Figure 3G, 3H). Some cells near developing arteries exhibited filopodia (Figure S8, A), suggesting that they were migrating cells. Cell clusters were also found in heart valves (Figure S8, B–D). However, they were not detected in the sinus venosus.
Figure 3. Clonal analysis of coronary development shows the descendants of Nfatc1+ endocardial cells in the coronary vascular endothelium.
(A) A schematic diagram showing clonal analysis of coronary development using the Nfatc1nrtTA;tetO-Cre system induced by doxycycline (Dox).
(B) Heart section of Nfatc1nrtTA;tetO-Cre;RCEfsEGFP E11.5 embryo induced at E10.5 with a limited Dox dose shows single EGFP-labeled cells in the endocardium (arrowhead) (see also Figure S7).
(C–E) Sections of E16.5 Nfatc1nrtTA;tetO-Cre;RCEfsEGFP heart induced at E9.5 (C) or E10.5 (D,E) show the EGFP+ cell clusters in the large (C, D) or small coronary arteries (arrows) expressing SM22a.
(F) Sections of E16.5 Nfatc1nrtTA;tetO-Cre;RCEfsEGFP heart induced at E10.5 shows the EGFP+ cell clusters (arrowhead) in the subepicardial vessels alongside the SM22a+ cells.
(G) Sections of E16.5 Nfatc1nrtTA;tetO-Cre;R26fslz heart induced at E12.5 shows the LacZ-labeled capillary cells (cc) cluster (green; arrows) alongside the SM22a-expressing small arteries (red).
(H) Sections of E16.5 Nfatc1nrtTA;tetO-Cre;R26fslz heart induced at E11.5 shows the LacZ+ capillary cells cluster (arrows) alongside the Pecam1+ small vessels (red). See also Figures S8, S9, and Table S2. All bars = 20 μm.
Quantitative analysis of clones showed that over 86% of the Nfatc1+ endothelial descendants that derived at E9.5–E13.5 were in the intramyocardial vessels at E14.5 or E16.5; the remainders were in the subepicardial vessels (Figure S9, A,B). The endocardial precursors labeled around E11.5 generated most coronary endothelial cells; their ability to differentiate into endothelial cells was greatly reduced after E12.5. Consistent with their predominant presence in the endothelium of intramyocardial vessels, the number of Nfatc1+ endothelial descendants found in the main coronary arteries at E14.5 or E16.5 was 13 or 5 times greater than that in the main veins (Figure S9, C). Furthermore, the numbers of labeled endocardial cells across the 39 analyzed hearts correlated significantly with the numbers of their sister endothelial cells in both intramyocardial and subepicardial vessels. However, the correlation between the endocardial and intramyocardial numbers (Pearson’s correlation coefficient r = 0.86, p = 4.0 × 10−12) was higher than that between the endocardial and subepicardial numbers (r = 0.59, p = 8.2 × 10−5) (Figure S9, D).
Thus, clonal analysis suggests that endocardial cells commit to a coronary endothelial fate right before coronary plexus formation and supports the conclusion from the Nfatc1Cre fate mapping that the endocardium is a major source of the endothelium of intramyocardial vessels and major coronary arteries.
Ventricular endocardial cells form coronary plexuses by angiogenesis
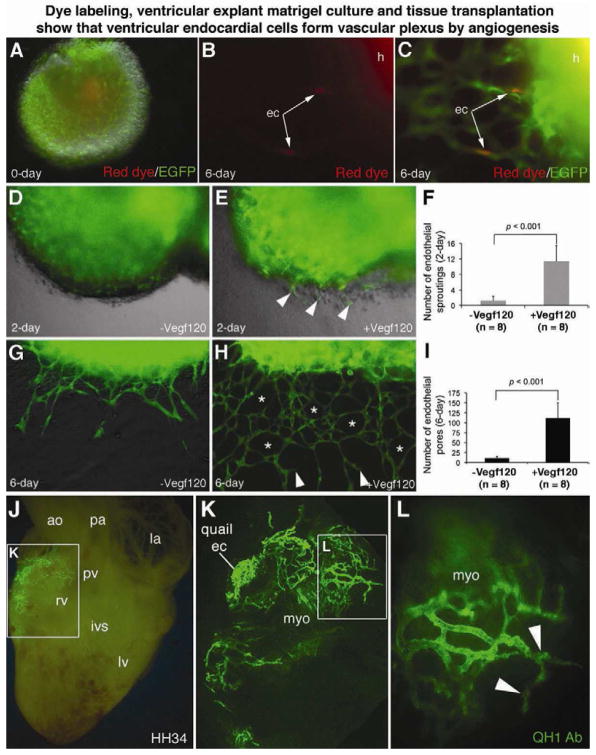
To visualize how ventricular endocardial cells generated coronary plexuses, we isolated hearts from E10.5 or E11.5 embryos and labeled endocardial cells with Red Fluorescent CMTPX. The ventricles were used in a Matrigel endothelial tube assay. We found single labeled endocardial cells that had migrated through the myocardial wall into Matrigel in the presence of Vegf120 (Figure S10, A–C). When using the red dye to label Nfatc1Cre;RCEfsEGFP embryonic ventricles, we observed dye-labeled individual cells integrated into an endothelial network generated by EGFP+ descendants of Nfatc1+ ventricular endocardial cells (Figure 4, A–C).
Figure 4. Dye labeling, ventricular explant culture and tissue transplantation show that ventricular endocardial cells form the vascular plexus by angiogenesis.
(A–C) Dye labeling and explant matrigel cultures (see also Figure S10, A to C) using ventricles from E11.5 Nfatc1Cre;RCEfsEGFP hearts (h) show red fluorescent CMTPX-labeled individual endocardial cells (arrows) that had migrated through the ventricular wall and integrated into an EGFP+ endothelial network upon Vegf120 treatment.
(D–I) Matrigel angiogenesis assays show that Vegf120 significantly promotes transmural migration, sprouting (arrowheads), and endothelial networking (asterisks) by Nfatc1+ descendants. The error bars represent SD. See also Movies S1 and S2.
(J–L) Images of QH1 antibody stained HH34 quail-chick chimerical heart sliced through the right ventricle at the implanted site show that the QH1+ descendants of engrafted HH15 quail endocardial cells invade chick myocardium and generate intramyocardial vessels. Quail endocardial cells, implanted at the surface of the atrioventricular junction of the chick heart (K), generate intensive myocardial vascular networks by angiogenic sprouting and branching (L, arrowheads). See also Figure S10, D–F, and Table S3.
The angiogenic function of endocardial cells was further characterized by time-lapse fluorescence live imaging and quantified. Without Vegf120, Nfatc1+ precursors invaded the ventricular wall and sprouted to form endothelial tubes, which then fused into distinct vessels in the myocardium (Movie S1). With Vegf120, the Nfatc1+ precursors readily migrated through the ventricular wall (Movie S2) and underwent angiogenic sprouting (Figure 4D, 4E). Within 6 days of Vegf120 exposure, the descendants of Nfatc1+ precursors generated sophisticated endothelial networks (Figure 4G, 4H). In contrast, angiogenic sprouting and networking were significantly limited without Vegf120 (Figure 4F, 4I). Besides Vegf120, other Vegf-a isoforms such as Vegf164 and Vegf188 were capable of promoting coronary angiogenesis, whereas Vegf-c, Vegf-d, Fgf2, and Fgf9 had partial or no effects (data not shown). These data showed that endocardial cells can develop into coronary endothelial networks and suggested that Vegf-a is a major factor involved in coronary plexus formation by ventricular endocardial cells.
To further test coronary angiogenesis by ventricular endocardial cells, we conducted quail-to-chick transplantation experiments in which we isolated ventricular endocardium or proepicardium from quail embryonic hearts at the Hamburger and Hamilton (HH) stage 15 (prior to coronary plexus formation) and implanted them at the inner curvature of the atrioventricular junction of HH15 chick embryonic hearts (Figure S10, D). Using the QH1 antibody that labels quail, but not host chick, endothelial cells, we detected the contribution of implanted quail cells to coronary arteries of chick hearts at HH34 (Figure S10, E,F). Quail endocardial cells invaded chick ventricular wall and developed into an extensive endothelial network in the myocardium (Figure 4J–4L), while quail proepicardium largely generated subepicardial vessels (Table S3 and data not shown).
Together, the results from four distinct experimental approaches (Nfatc1Cre fate-mapping, clonal analysis, dye labeling, and cross-species transplantation) all demonstrate that ventricular endocardial cells generate endothelial cells of coronary arteries. The endocardial descendants comprise the majority of endothelial cells in intramyocardial vessels and coronary arteries. They further show that endocardial cells are not terminally differentiated; they are angiogenic and are activated by Vegf-a to generate coronary plexuses.
Myocardial Vegf-a is required for coronary angiogenesis and artery formation
Vegf-a is known to be produced by the developing myocardium (Giordano et al., 2001; Miquerol et al., 2000; Tomanek et al., 2006) and required for vasculogenesis and angiogenesis in development (Carmeliet et al., 1996; Ferrara et al., 1996). Therefore, we asked if myocardial Vegf-a was necessary in vivo for endocardial cells to differentiate into coronary endothelial cells and form coronary arteries. We used the Tnnt2 promoter-Cre (Tnnt2Cre)(Chen et al., 2006; Jiao et al., 2003) and Vegf-af/f mice (Gerber et al., 1999) to disrupt Vegf-a in the myocardium (data not shown). Wholemount or sectional Pecam1 staining of E12.5 Tnnt2Cre;Vegf-a+/+ (Control) and Tnnt2Cre;Vegf-af/f (Vegf-a null) hearts showed that angiogenic sprouts or coronary plexuses were present in the peritruncal area or ventricular septum of control (Figure 5, A–D) but not Vegf-a null hearts (Figure 5, E–H). Thus myocardial Vegf-a is necessary for coronary plexus formation.
Figure 5. Disruption of Vegf-a in the myocardium reveals that myocardial Vegf-a is required for coronary angiogenesis and artery formation.
(A–D) Pictures of E12.5 Tnnt2Cre;Vegf-a+/+ (Control) heart stained with Pecam1 antibodies exhibit coronary plexuses (arrows) in the peritruncal region and interventricular septum (ivs). Arrowheads indicate the branching endothelial tubes. Dashed lines separate the Pecam1+ vessels in the septum from the Pecam1+ trabeculae.
(E–H) Pictures of E12.5 Tnnt2Cre;Vegf-af/f (Vegf-a null) heart show no plexus formation in the peritruncal and septal myocardium.
(I–K) Pictures of E14.5 control heart show the Pecam1+ coronary arteries (ca, arrows) and subepicardial vessels (arrowheads).
(L–N) Pictures of E14.5 Vegf-a null heart show less and immaturely formed coronary arteries (arrows) but numerous and dilated subepicardial vessels (arrowheads). The null heart also has necrotic peritruncal and septal myocardium (L, arrows) (see also Figures S11, S12, and Table S4). Bar in I,L = 100 μm; rest = 20 μm.
From E12.5 to E13.5 when coronary plexuses had developed into a well-organized intramyocardial endothelial network in control hearts (Figure S11, A,C), the angiogenic defect persisted in all Vegf-a null hearts leading to myocardial necrosis (Figure S11, B,D). By E14.5 when control hearts developed intramyocardial coronary arteries and subepicardial veins with distinct patterns (Figure 5, I–K), Vegf-a null hearts had only few immature myocardial coronary arteries and developed dilated subepicardial veins (Figure 5, L–N). Quantitative analysis confirmed an 88% reduction in the number of Pecam1+ intramyocardial endothelial cells in E14.5 Vegf-a null hearts, but only a 37% decrease in the subepicardial endothelial cells (Figure S11, E). These findings indicated that myocardial Vegf-a triggers coronary angiogenesis and is required for arterial formation. The fact that coronary veins formed indicated that they have an embryonic origin independent of myocardial Vegf-a, although the vein defect might be a consequence of myocardial Vegf-a deficiency and/or they might be secondary to the arterial defect.
Vegf-a null hearts also exhibited cardiac phenotypes, including thin ventricular walls, necrotic septa, cardiac hemorrhages (Figure S12, B,D), and ruptured septa at E15.5 (Figure S12, F). All Vegf-a null embryos were runted and died after E15.5 (Table S4). To rule out early myocardial defects that might affect coronary angiogenesis, we examined myocardial function and structure but found no myocardial apoptosis, abnormal cardiac gene expression, and alterations in the ultrastructure of sarcomeres, mitochondria, or Golgi apparatus in E11.5–E12.5 Vegf-a null hearts (data not shown). These results suggest that the cardiac phenotypes are a result of the vascular defects.
Endocardial Vegfr-2 is required for coronary angiogenesis and artery formation
Vegfr-2 is a major Vegf receptor required for vessel formation during development (Shalaby et al., 1995). We thus investigated if Vegfr-2 could transduce Vegf-a signals to trigger coronary angiogenesis by Nfatc1+ endocardial cells. We used Nfatc1Cre and Vegfr-2f/f (Haigh et al., 2003) to remove Vegfr-2 in the endocardium. Vegfr-2 antibody staining of E10.5 Vegfr-2+/+;Nfatc1Cre (Control) and Vegfr-2f/f;Nfatc1Cre embryos (Vegfr-2 null) showed that the deletion was restricted to the endocardium (Figure S13, A vs. A′), whereas Vegfr-2 expression in the vasculature outside the heart was not affected (Figure S13, B–E vs. B′-E′). Similar to E12.5 Vegf-a null embryos, E12.5 Vegfr-2 null embryos did not develop coronary plexuses in the peritruncal/coronary sulcus area or ventricular septum (Figure 6, A,B vs. C,D). The early angiogenic defect resulted in severely diminished or no coronary arteries in all E14.5 Vegfr-2 null embryos (Figure 6, E vs. H). In contrast, coronary veins developed in these embryos (Figure 6, F vs. I). The arterial specific defect was consistent with Vegfr-2 deletion in the Nfatc1+ endocardial precursors, as Vegfr-2 expression in the endothelium of coronary veins was not affected (Figure 6, G vs. J).
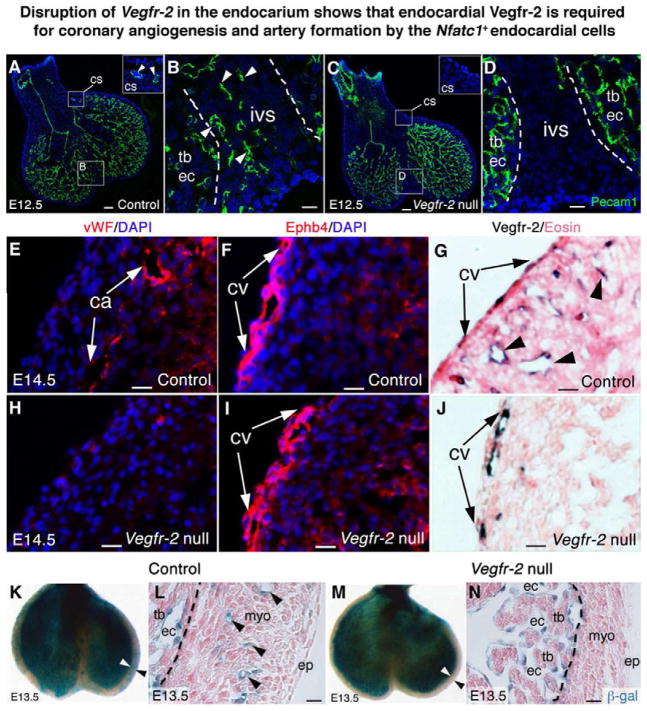
Fig. 6. Disruption of Vegfr-2 in the endocardium shows that endocardial Vegfr-2 is required for coronary angiogenesis and artery formation by endocardial cells.
(A,B) E12.5 Nfatc1Cre;Vegfr-2+/+ (Control) heart sections stained with Pecam1 antibodies show coronary endothelial tubes in the myocardium of the coronary sulcus (cs) (A, inset) and interventricular septum (B, arrowheads).
(C,D) E12.5 Nfatc1Cre;Vegfr-2f/f (Vegfr-2 null) heart sections stained with Pecam1 antibodies show no coronary endothelial tubes in the coronary sulcus (C, inset) and interventricular septum (D). Dashed lines separate the septum from the Pecam1+ endocardium of trabeculae.
(E–G) E14.5 control heart sections show that mature coronary arteries are positive for vWF staining (E, arrows) while subepicardial coronary veins (cv) are positive for Ephb4 staining (F, arrows). Vegfr-2 proteins are present in arteries (G, arrowheads) and veins (G, arrows).
(H–J) E14.5 Vegfr-2 null heart sections show no vWF+ coronary arteries in the myocardium (H), but presence of Ephb4+ subepicardial coronary veins (I, arrows). Vegfr-2 proteins are only present in the coronary veins (J, arrows).
(K,L) X-gal stained E13.5 Nfatc1Cre;Vegfr-2+/+;R26fslz (Control) heart show that the β-gal+ descendants of Nfatc1+ endocardial cells generate the coronary vessels in the ventricular wall (arrowheads). Dashed lines distinguish the β-gal+ coronary plexuses in the compact wall from the β-gal+ endocardium of trabeculae.
(M,N) X-gal stained E13.5 Nfatc1Cre;Vegfr-2f/f;R26fslz (Vegfr-2 null) heart show no β-gal+ coronary plexuses derived from endocardial cells in the ventricular wall. See also Figures S14, S15, and Table S5. Bar in A,C = 100 μm; rest = 20 μm.
Like Vegf-a null embryos, all Vegfr-2 null embryos developed cardiac hemorrhages by E14.5 (Figure S14, A–D), they were runted thereafter, and died in utero (Table S5). Quantitative analysis of immunostaining confirmed a 74% reduction in the number of Pecam1+ intramyocardial endothelial cells in E14.5 Vegfr-2 null hearts and only a 24% decrease in subepicardial endothelial cells (Figure S14, E–I). These results demonstrate that Vegfr-2 function is necessary for differentiation of endocardial cells into endothelial cells to form coronary arteries.
To track the fate of Vegfr-2 null endocardial cells during coronary angiogenesis, we generated Nfatc1Cre;Vegfr-2f/f;R26fslz and Nfatc1Cre;Vegfr-2f/f;RCEfsEGFP mice to simultaneously delete Vegfr-2 and activate reporter gene expression in endocardial cells. We found that the β-gal+ descendants of Vegfr-2 null endocardial cells failed to generate intramyocardial coronary arteries in Nfatc1Cre;Vegfr-2f/f;R26fslz embryos (Figure 6K,L vs. 6M,N). Consistently, coronary angiogenesis assays with ventricles of E11.5 Nfatc1Cre;Vegfr-2f/f;RCEfsEGFP embryos showed that EGFP+ Vegfr-2 null endocardial cells could not respond to Vegf120, they failed to migrate, sprout and form endothelial networks (Figure S15, B,E vs. C,F), which was confirmed by quantitative analysis (Figure S15, G,H). Thus these results again indicated that the initial coronary arteries arise from the Nfatc1+ precursors in the endocardium and that Vegf signaling is important for this process.
Taken together, the data from the studies of Vegf-a and Vegfr-2 null hearts demonstrate that myocardially produced Vegf-a signals through endocardial Vegfr-2 to stimulate ventricular endocardial cells to undergo angiogenesis that generates coronary arteries. They provide a fifth, independently derived line of functional evidence that endocardial cells are the progenitors of coronary arteries, whereas the endothelium of coronary veins has a different origin.
Discussion
These studies demonstrated that the endocardium is a major source of endothelial cells of the coronary vasculature. The endocardium generates the precursor cells that form coronary plexuses and develop into endothelial cells of coronary arteries, arterioles and capillaries. Such angiogenic functions of endocardial cells appear evolutionarily conserved, as revealed in transplantation studies in which avian endocardial cells are capable of generating coronary arterial network.
Our results show that ventricular endocardial cells are not terminally differentiated but instead are angiogenic for coronary arteries. The angiogenic sprouting and migration of endocardial cells into the myocardium is induced by myocardial Vegf-a to endocardial Vegfr-2 signaling. This is consistent with previous morphological analysis suggesting that the earliest vessels develop when encasing endocardial cells penetrate the myocardium and proliferate into an interconnected coronary network (Viragh and Challice, 1981). In contrast to the requirement for myocardial Vegf-a to endocardial Vegfr2 signaling, neither endocardial deletion of Vegf-a nor proepicardial/epicardial deletion of Vegfr-2 blocked intramyocardial coronary angiogenesis (Zhang et al. unpublished data).
A hypoxia-dependent Vegf-a concentration gradient exists in the myocardium of the developing heart (Wikenheiser et al., 2006), and our results indicate that this gradient provides a cue through Vegfr-2 in the endocardium to trigger migration of angiogenic precursor cells from the endocardium into the myocardium (Figure 7A). Once in the myocardium, these cells predominantly differentiate into arterial endothelial cells and populate intramyocardial arteries, arterioles, and capillaries.
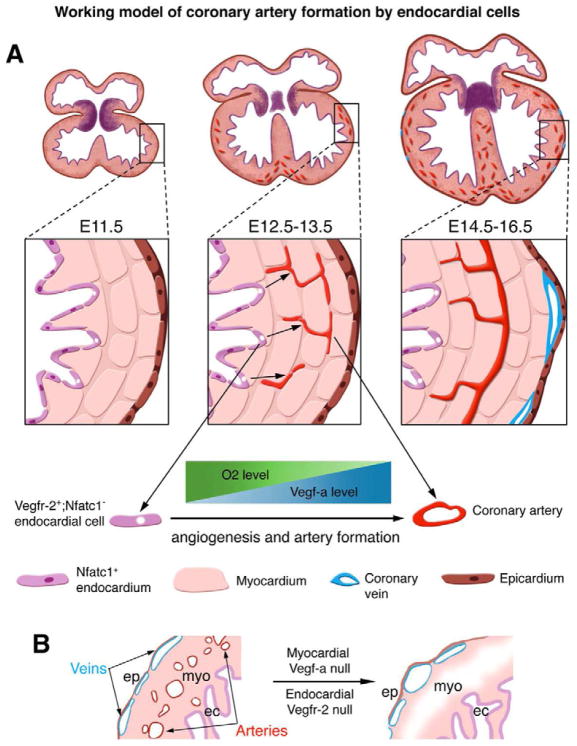
Figure 7. Working model of coronary artery formation by endocardial cells.
(A) Diagram shows an ontogenic role for endocardial cells in generation of the coronary arteries. Between E11.5 and E13.5 of mouse embryogenesis, myocardial proliferation generates a Vegf-a gradient across the thickening ventricular wall, possibly regulated by a reverse gradient of myocardial O2 content. At the same time, some ventricular endocardial cells turn off nuclear Nfatc1 expression (purple nuclei). The Vegf-a gradient induces these Nfatc1− endocardial cells (white nuclei) to invade the myocardium and proliferate into coronary plexuses by angiogenesis via their expression of Vegfr-2. The plexuses then develop into coronary arteries.
(B) Knockout of Vegf-a in the myocardium or Vegfr-2 in the endocardium prevents coronary angiogenesis and artery formation, but does not block coronary vein formation, suggesting that coronary veins arise from non-endocardial origins, independent of myocardial Vegf-a to endocardial Vegfr-2 signaling.
Our analysis showed that Nfatc1 transcription occurs in the endocardium, but not in the descendant endothelial cells of coronary vessels. Thus, while the Nfatc1 promoter is first active in the endocardial cells, it is then shut off as those cells develop into endothelial cells of the coronary plexuses. Previous studies have found that spatiotemporally regulated Nfatc1 expression maintains progenitor cell status and that its down-regulation promotes the differentiation of progenitor cells in the developing heart valves (Wu et al., 2011) or hair follicles (Horsley et al., 2008). Whether Nfatc1 expression in ventricular endocardial cells maintains their progenitor status and/or primes them for later angiogenesis, and whether scheduled Nfatc1 shut down has a role in coronary angiogenesis are some of important questions currently under investigation.
While the endocardium is the source of most arterial endothelial cells, the endocardium provides a lesser contribution to coronary veins. Therefore, the coronary veins have their major origin outside the endocardium and arise by a different mechanism than the arterial cells. Previous fate mapping using an inducible Cre (VE-cadherin-CreERT2) (Monvoisin et al., 2006) showed that the vast majority of the endothelial cells in the coronary veins arise during development by E9.5 from the sinus venosus (Red-Horse et al., 2010). A small number of coronary venous endothelial descendants became coronary artery endothelial cells. While this led to the conclusion that venous endothelial cells are reprogrammed into arterial endothelial cells, the fraction of coronary arterial endothelial cells arising from the sinus venosus was far less than the fraction of coronary venous endothelial cells. Additionally, a recent study using Scx and Sema3D Cre lines suggested that a subset of proepicardial/epicardial cells expressing these genes at E9.5 is able to differentiate into multiple cardiovascular cells, mostly epicardial cells and/or fibroblasts as well as a small fractions of coronary endothelial and smooth muscle cells (Katz et al., 2012). Our fate-mapping experiments also showed that a fraction of the sinus venous endothelial cells are derived from the endocardium, suggesting that endocardial cells may contribute to a minor fraction of the coronary venous endothelium via the sinus venosus route.
Collectively, these results suggest a mechanism for coronary vessel development in which the arterial and venous portions mainly arise from largely distinct embryonic endothelial cell populations at different anatomical sites and during distinct developmental windows. Our results showed that around E11.5, endocardial precursor cells, through Vegfr-2, respond to Vegf-a signaling from the myocardium and migrate into the myocardium to form coronary plexuses where they mature into arteries, arterioles, and capillaries. In contrast, the coronary venous endothelial cells arise mostly from sinus venous endothelial cells before E9.5 (Red-Horse et al., 2010). We propose that these processes act in parallel to form the arterial and venous endothelial populations of the complete coronary vasculature.
Given the importance of coronary artery disease, generation of coronary arteries by tissue engineering is obviously a highly desirable goal. Regenerative approaches that recapitulate normal development may represent an especially attractive path for development of such therapies. Our results imply that programmed transient Nfatc1 expression couple with sustained Vegfr-2-dependent Vegf-a signaling in the progenitor cells of coronary arterial endothelium might be useful strategy for regenerating coronary arteries.
Experimental Procedures
Animals
To generate the Nfatc1Cre mice, an IRES-Cre/PGK-hygromycin cassette was inserted at the 3′ untranslational region of Nfatc1 (Figure S2). To disrupt Vegf-a in the myocardium, Tnnt2Cre mice (Chen et al., 2006; Jiao et al., 2003) were mated to Vegf-af/f mice (Gerber et al., 1999). Inactivation of Vegfr-2 expression in the endocardium was achieved by crossing Nfatc1Cre with Vegfr-2f/f mice. The maintenance of mice and mouse experiments were performed according to protocols approved by the Institutional Animal Care and Use Committee of Albert Einstein College of Medicine and Vanderbilt University. Noontime on the day of detecting vaginal plugs was designated as E0.5.
Immunostainings and RNA in situ hybridization
Primary and secondary antibodies used in this study are listed in Table S1. For immunostainings, mouse embryos from E9.5 to E12.5 or hearts from E13.5 to E16.5 were fixed in 4% PFA. A 15%–30% sucrose gradient was applied to those for frozen sections. Immunohistochemistry was carried out using the ABC-HRP or ABC-AK method (Vector Laboratories). Fluorescent avidin kit (Vector Laboratories) was used for immunofluorescence visualization. Whole mount or section RNA in situ hybridization was performed to detect Nfatc1 or Cre transcripts in E9.5 to E15.5 embryos or hearts using probes for Nfatc1 or Cre mRNA.
Whole mount X-gal Staining
Embryos from E9.5 to E12.5 or hearts isolated at E13.5 to E14.5 were fixed in 4% PFA on ice for 30 minutes to 2 hours depending on the developmental stage, washed in PBS, and then stained with freshly prepared X-gal solution for 2 hours at 37°C or overnight at room temperature. The stained samples were washed by PBS, sectioned, and photographed.
Mouse ventricular explant culture and coronary angiogenesis assay
Ventricles without the atria and sinus venosus were isolated from E10.5 or E11.5 embryos, rinsed with PBS to remove circulating cells and placed in the Matrigel (growth factor reduced, BD Biosciences) in Nunc 4-well plates. The Matrigel was prepared by addition of an equal volume of M199 medium plus 10% FBS with or without a testing growth factor including Vegf120, 164, 189, Vegf-c, Vegf-d, Fgf2 or Fgf9 (R&D Systems). The final concentration of each growth factor was 10 ng/ml. Explants were cultured for 6 days and angiogenesis by the ventricular endocardial cells was photographed.
Dye tracing of ventricular endocardial cells in ventricular explant cultures
E11.5 hearts were microinjected with the Red Fluorescent CMTPX (CellTracker™, Invitrogen) through their ventricles. After a 2-minute incubation on ice, atria and sinuses were removed. Ventricles were then rinsed with PBS and placed in the Matrigel with Vegf120. The transmural migration of the labeled individual endocardial cells and their integration into an endothelial network was photographed.
Quail-to-chick transplantation assays
Quail ventricular endocardium or ventricular apex without the epicardium were isolated from the Hamburger and Hamilton stages (HH) 15/16 quail embryos, labeled with carbon particles for visualization, and implanted at the inner curvature of the atrioventricular junction of HH15/16 chick embryonic hearts. The implanted chick embryos were incubated at 37°C until HH34 or HH42. The chimerical hearts were isolated and fixed with 3.7% formaldehyde, cut into anterior and posterior halves through the right ventricle to expose intramyocardial vasculature, followed by incubation with a mouse monoclonal anti-QH1 antibody to visualize the contribution of quail endocardial cells to the developing chick coronary vasculature.
Supplementary Material
Highlights.
Ventricular endocardial cells are a major origin of the coronary arteries
Ventricular endocardial cells generate coronary arteries by angiogenesis
Myocardial Vegf-a to endocardial Vegfr-2 signaling regulates coronary angiogenesis
The coronary arteries and veins arise largely from different origins and mechanisms
Acknowledgments
We are grateful to Napoleone Ferrara, Janet Rossant and Kyunghee Choi, and Gordon Fishell for Vegf-af/f, Vegfr-2f/f, or RCEfsEGFP mice. We also thank Drs. Bernice Morrow and Richard Kitsis for critical reading of the manuscript. B.Z. was supported by NIH (HL078881), AHA (0435128N) and Tuner-Hazinski Research Award. H.S.B. and C.P.C. were supported by NIH (HL100398 and HL85345, respectively).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Cai CL, Martin JC, Sun Y, Cui L, Wang L, Ouyang K, Yang L, Bu L, Liang X, Zhang X, et al. A myocardial lineage derives from Tbx18 epicardial cells. Nature. 2008;454:104–108. doi: 10.1038/nature06969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmeliet P, Ferreira V, Breier G, Pollefeyt S, Kieckens L, Gertsenstein M, Fahrig M, Vandenhoeck A, Harpal K, Eberhardt C, et al. Abnormal blood vessel development and lethality in embryos lacking a single VEGF allele. Nature. 1996;380:435–439. doi: 10.1038/380435a0. [DOI] [PubMed] [Google Scholar]
- Chang CP, Neilson JR, Bayle JH, Gestwicki JE, Kuo A, Stankunas K, Graef IA, Crabtree GR. A field of myocardial-endocardial NFAT signaling underlies heart valve morphogenesis.[see comment] Cell. 2004;118:649–663. doi: 10.1016/j.cell.2004.08.010. [DOI] [PubMed] [Google Scholar]
- Chen JW, Zhou B, Yu QC, Shin SJ, Jiao K, Schneider MD, Baldwin HS, Bergelson JM. Cardiomyocyte-specific deletion of the coxsackievirus and adenovirus receptor results in hyperplasia of the embryonic left ventricle and abnormalities of sinuatrial valves. Circ Res. 2006;98:923–930. doi: 10.1161/01.RES.0000218041.41932.e3. [DOI] [PubMed] [Google Scholar]
- de la Pompa JL, Timmerman LA, Takimoto H, Yoshida H, Elia AJ, Samper E, Potter J, Wakeham A, Marengere L, Langille BL, et al. Role of the NF-ATc transcription factor in morphogenesis of cardiac valves and septum.[see comment] Nature. 1998;392:182–186. doi: 10.1038/32419. [DOI] [PubMed] [Google Scholar]
- Dettman RW, Denetclaw W, Jr, Ordahl CP, Bristow J. Common epicardial origin of coronary vascular smooth muscle, perivascular fibroblasts, and intermyocardial fibroblasts in the avian heart. Dev Biol. 1998;193:169–181. doi: 10.1006/dbio.1997.8801. [DOI] [PubMed] [Google Scholar]
- Drake CJ, Fleming PA. Vasculogenesis in the day 6.5 to 9.5 mouse embryo. Blood. 2000;95:1671–1679. [PubMed] [Google Scholar]
- Ferrara N, Carver-Moore K, Chen H, Dowd M, Lu L, O’Shea KS, Powell-Braxton L, Hillan KJ, Moore MW. Heterozygous embryonic lethality induced by targeted inactivation of the VEGF gene. Nature. 1996;380:439–442. doi: 10.1038/380439a0. [DOI] [PubMed] [Google Scholar]
- Gerber HP, Hillan KJ, Ryan AM, Kowalski J, Keller GA, Rangell L, Wright BD, Radtke F, Aguet M, Ferrara N. VEGF is required for growth and survival in neonatal mice. Development. 1999;126:1149–1159. doi: 10.1242/dev.126.6.1149. [DOI] [PubMed] [Google Scholar]
- Giordano FJ, Gerber HP, Williams SP, VanBruggen N, Bunting S, Ruiz-Lozano P, Gu Y, Nath AK, Huang Y, Hickey R, et al. A cardiac myocyte vascular endothelial growth factor paracrine pathway is required to maintain cardiac function. Proc Natl Acad Sci U S A. 2001;98:5780–5785. doi: 10.1073/pnas.091415198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafsson E, Brakebusch C, Hietanen K, Fassler R. Tie-1-directed expression of Cre recombinase in endothelial cells of embryoid bodies and transgenic mice. J Cell Sci. 2001;114:671–676. doi: 10.1242/jcs.114.4.671. [DOI] [PubMed] [Google Scholar]
- Haigh JJ, Morelli PI, Gerhardt H, Haigh K, Tsien J, Damert A, Miquerol L, Muhlner U, Klein R, Ferrara N, et al. Cortical and retinal defects caused by dosage-dependent reductions in VEGF-A paracrine signaling. Dev Biol. 2003;262:225–241. doi: 10.1016/s0012-1606(03)00356-7. [DOI] [PubMed] [Google Scholar]
- Horsley V, Aliprantis AO, Polak L, Glimcher LH, Fuchs E. NFATc1 balances quiescence and proliferation of skin stem cells. Cell. 2008;132:299–310. doi: 10.1016/j.cell.2007.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao K, Kulessa H, Tompkins K, Zhou Y, Batts L, Baldwin HS, Hogan BL. An essential role of Bmp4 in the atrioventricular septation of the mouse heart. Genes Dev. 2003;17:2362–2367. doi: 10.1101/gad.1124803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz TC, Singh MK, Degenhardt K, Rivera-Feliciano J, Johnson RL, Epstein JA, Tabin CJ. Distinct compartments of the proepicardial organ give rise to coronary vascular endothelial cells. Developmental cell. 2012;22:639–650. doi: 10.1016/j.devcel.2012.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komiyama M, Ito K, Shimada Y. Origin and development of the epicardium in the mouse embryo. Anat Embryol (Berl) 1987;176:183–189. doi: 10.1007/BF00310051. [DOI] [PubMed] [Google Scholar]
- Kraus F, Haenig B, Kispert A. Cloning and expression analysis of the mouse T-box gene Tbx18. Mech Dev. 2001;100:83–86. doi: 10.1016/s0925-4773(00)00494-9. [DOI] [PubMed] [Google Scholar]
- Lavine KJ, Ornitz DM. Shared circuitry: developmental signaling cascades regulate both embryonic and adult coronary vasculature. Circ Res. 2009;104:159–169. doi: 10.1161/CIRCRESAHA.108.191239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luttun A, Carmeliet P. De novo vasculogenesis in the heart. Cardiovascular Research. 2003;58:378–389. doi: 10.1016/s0008-6363(03)00258-x. [DOI] [PubMed] [Google Scholar]
- Majesky MW. Development of coronary vessels. Curr Top Dev Biol. 2004;62:225–259. doi: 10.1016/S0070-2153(04)62008-4. [DOI] [PubMed] [Google Scholar]
- Mikawa T, Borisov A, Brown AM, Fischman DA. Clonal analysis of cardiac morphogenesis in the chicken embryo using a replication-defective retrovirus: I. Formation of the ventricular myocardium. Dev Dyn. 1992;193:11–23. doi: 10.1002/aja.1001930104. [DOI] [PubMed] [Google Scholar]
- Mikawa T, Fischman DA. Retroviral analysis of cardiac morphogenesis: discontinuous formation of coronary vessels. Proc Natl Acad Sci U S A. 1992;89:9504–9508. doi: 10.1073/pnas.89.20.9504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikawa T, Gourdie RG. Pericardial mesoderm generates a population of coronary smooth muscle cells migrating into the heart along with ingrowth of the epicardial organ. Dev Biol. 1996;174:221–232. doi: 10.1006/dbio.1996.0068. [DOI] [PubMed] [Google Scholar]
- Miquerol L, Langille BL, Nagy A. Embryonic development is disrupted by modest increases in vascular endothelial growth factor gene expression. Development. 2000;127:3941–3946. doi: 10.1242/dev.127.18.3941. [DOI] [PubMed] [Google Scholar]
- Misfeldt AM, Boyle SC, Tompkins KL, Bautch VL, Labosky PA, Baldwin HS. Endocardial cells are a distinct endothelial lineage derived from Flk1+ multipotent cardiovascular progenitors. Dev Biol. 2009;333:78–89. doi: 10.1016/j.ydbio.2009.06.033. [DOI] [PubMed] [Google Scholar]
- Miyoshi G, Hjerling-Leffler J, Karayannis T, Sousa VH, Butt SJ, Battiste J, Johnson JE, Machold RP, Fishell G. Genetic fate mapping reveals that the caudal ganglionic eminence produces a large and diverse population of superficial cortical interneurons. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2010;30:1582–1594. doi: 10.1523/JNEUROSCI.4515-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monvoisin A, Alva JA, Hofmann JJ, Zovein AC, Lane TF, Iruela-Arispe ML. VE-cadherin-CreERT2 transgenic mouse: a model for inducible recombination in the endothelium. Dev Dyn. 2006;235:3413–3422. doi: 10.1002/dvdy.20982. [DOI] [PubMed] [Google Scholar]
- Olivey HE, Svensson EC. Epicardial-myocardial signaling directing coronary vasculogenesis. Circ Res. 106:818–832. doi: 10.1161/CIRCRESAHA.109.209197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivey HE, Svensson EC. Epicardial-myocardial signaling directing coronary vasculogenesis. Circulation Research. 2010;106:818–832. doi: 10.1161/CIRCRESAHA.109.209197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Pomares JM, Carmona R, Gonzalez-Iriarte M, Atencia G, Wessels A, Munoz-Chapuli R. Origin of coronary endothelial cells from epicardial mesothelium in avian embryos. Int J Dev Biol. 2002;46:1005–1013. [PubMed] [Google Scholar]
- Ranger AM, Grusby MJ, Hodge MR, Gravallese EM, de la Brousse FC, Hoey T, Mickanin C, Baldwin HS, Glimcher LH. The transcription factor NF-ATc is essential for cardiac valve formation.[see comment] Nature. 1998;392:186–190. doi: 10.1038/32426. [DOI] [PubMed] [Google Scholar]
- Red-Horse K, Ueno H, Weissman IL, Krasnow MA. Coronary arteries form by developmental reprogramming of venous cells. Nature. 2010;464:549–553. doi: 10.1038/nature08873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalaby F, Rossant J, Yamaguchi TP, Gertsenstein M, Wu XF, Breitman ML, Schuh AC. Failure of blood-island formation and vasculogenesis in Flk-1-deficient mice. Nature. 1995;376:62–66. doi: 10.1038/376062a0. [DOI] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nature Genetics. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Sousa VH, Miyoshi G, Hjerling-Leffler J, Karayannis T, Fishell G. Characterization of Nkx6-2-derived neocortical interneuron lineages. Cerebral cortex. 2009;19(Suppl 1):i1–10. doi: 10.1093/cercor/bhp038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugi Y, Markwald RR. Formation and early morphogenesis of endocardial endothelial precursor cells and the role of endoderm. Developmental Biology. 1996;175:66–83. doi: 10.1006/dbio.1996.0096. [DOI] [PubMed] [Google Scholar]
- Tomanek RJ, Ishii Y, Holifield JS, Sjogren CL, Hansen HK, Mikawa T. VEGF family members regulate myocardial tubulogenesis and coronary artery formation in the embryo. Circ Res. 2006;98:947–953. doi: 10.1161/01.RES.0000216974.75994.da. [DOI] [PubMed] [Google Scholar]
- Viragh S, Challice CE. The origin of the epicardium and the embryonic myocardial circulation in the mouse. Anat Rec. 1981;201:157–168. doi: 10.1002/ar.1092010117. [DOI] [PubMed] [Google Scholar]
- Vrancken Peeters MP, Gittenberger-de Groot AC, Mentink MM, Poelmann RE. Smooth muscle cells and fibroblasts of the coronary arteries derive from epithelial-mesenchymal transformation of the epicardium. Anat Embryol (Berl) 1999;199:367–378. doi: 10.1007/s004290050235. [DOI] [PubMed] [Google Scholar]
- Wada AM, Willet SG, Bader D. Coronary vessel development: a unique form of vasculogenesis. Arterioscler Thromb Vasc Biol. 2003;23:2138–2145. doi: 10.1161/01.ATV.0000098645.38676.CC. [DOI] [PubMed] [Google Scholar]
- Wikenheiser J, Doughman YQ, Fisher SA, Watanabe M. Differential levels of tissue hypoxia in the developing chicken heart. Dev Dyn. 2006;235:115–123. doi: 10.1002/dvdy.20499. [DOI] [PubMed] [Google Scholar]
- Wu B, Wang Y, Lui W, Langworthy M, Tompkins KL, Hatzopoulos AK, Baldwin HS, Zhou B. Nfatc1 coordinates valve endocardial cell lineage development required for heart valve formation. Circulation Research. 2011;109:183–192. doi: 10.1161/CIRCRESAHA.111.245035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita J, Itoh H, Hirashima M, Ogawa M, Nishikawa S, Yurugi T, Naito M, Nakao K. Flk1-positive cells derived from embryonic stem cells serve as vascular progenitors.[see comment] Nature. 2000;408:92–96. doi: 10.1038/35040568. [DOI] [PubMed] [Google Scholar]
- Yang L, Soonpaa MH, Adler ED, Roepke TK, Kattman SJ, Kennedy M, Henckaerts E, Bonham K, Abbott GW, Linden RM, et al. Human cardiovascular progenitor cells develop from a KDR+ embryonic-stem-cell-derived population. Nature. 2008;453:524–528. doi: 10.1038/nature06894. [DOI] [PubMed] [Google Scholar]
- Zhou B, Cron RQ, Wu B, Genin A, Wang Z, Liu S, Robson P, Baldwin HS. Regulation of the murine Nfatc1 gene by NFATc2. Journal of Biological Chemistry. 2002;277:10704–10711. doi: 10.1074/jbc.M107068200. [DOI] [PubMed] [Google Scholar]
- Zhou B, Ma Q, Rajagopal S, Wu SM, Domian I, Rivera-Feliciano J, Jiang D, von Gise A, Ikeda S, Chien KR, et al. Epicardial progenitors contribute to the cardiomyocyte lineage in the developing heart. Nature. 2008;454:109–113. doi: 10.1038/nature07060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B, Wu B, Tompkins KL, Boyer KL, Grindley JC, Baldwin HS. Characterization of Nfatc1 regulation identifies an enhancer required for gene expression that is specific to pro-valve endocardial cells in the developing heart. Development. 2005;132:1137–1146. doi: 10.1242/dev.01640. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.