Abstract
Plants synthesize several classes of small (15- to 30-kD monomer) heat-shock proteins (sHSPs) in response to heat stress, including a nuclear-encoded, chloroplast-localized sHSP (HSP21). Cytosolic sHSPs exist as large oligomers (approximately 200–800 kD) composed solely or primarily of sHSPs. Phosphorylation of mammalian sHSPs causes oligomer dissociation, which appears to be important for regulation of sHSP function. We examined the native structure and phosphorylation of chloroplast HSP21 to understand this protein's basic properties and to compare it with cytosolic sHSPs. The apparent size of native HSP21 complexes was > 200 kD and they did not dissociate during heat stress. We found no evidence that HSP21 or the plant cytosolic sHSPs are phosphorylated in vivo. A partial HSP21 complex purified from heat-stressed pea (Pisum sativum L.) leaves contained no proteins other than HSP21. Mature recombinant pea and Arabidopsis thaliana HSP21 were expressed in Escherichia coli, and purified recombinant Arabidopsis HSP21 assembled into homo-oligomeric complexes with the same apparent molecular mass as HSP21 complexes observed in heat-stressed leaf tissue. We propose that the native, functional form of chloroplast HSP21 is a large, oligomeric complex containing nine or more HSP21 subunits, and that plant sHSPs are not regulated by phosphorylation-induced dissociation.
sHSPs with a monomeric molecular mass of 15 to 30 kD are among the most abundant proteins synthesized by plants in response to heat stress (Vierling, 1991). sHSPs are members of a large family of proteins that includes the vertebrate eye lens α-crystallins (Ingolia and Craig, 1982; Arrigo and Landry, 1994). Members of this family share a conserved carboxy-terminal sequence and are found in high-molecular-mass complexes in vivo (approximately 200–800 kD). The plant sHSPs can be divided into several classes based on DNA-sequence analysis, immunological cross-reactivity, and intracellular localization. There are two classes of cytosolic sHSP (classes I and II), as well as distinct types of sHSP localized to the ER, the mitochondrion, and the chloroplast (Waters et al., 1996). Compared with other plant sHSPs, the chloroplast-localized sHSP is most closely related to the cytosolic class I sHSPs, but it also has unique features, including a Met-rich domain and an amino-terminal transit peptide (Chen and Vierling, 1991). The diversity of sHSPs is unique to plants; other eukaryotes produce fewer sHSPs and do not have organelle-localized sHSPs.
In plants and other organisms, sHSPs are believed to protect cells from heat-induced damage (Nagao et al., 1986; Vierling, 1991). A number of studies have correlated sHSP expression in plants with thermotolerance. Induction of HSPs in Arabidopsis thaliana via a constitutively active heat-shock transcription factor resulted in constitutive expression of at least one cytosolic sHSP and a substantial increase in thermotolerance (Lee et al., 1995b). Overexpression of the Drosophila or mammalian sHSP confers thermotolerance in cultured rodent cells (Landry et al., 1989; Rollet et al., 1992; Lavoie et al., 1993; Lee et al., 1995b). However, little is known about the mechanisms by which the sHSPs may confer thermotolerance. Recent evidence indicates that cytosolic sHSPs from plants, as well as the α-crystal-lins and the mammalian sHSPs, function as ATP-independent chaperones in vitro (Horwitz, 1992; Jakob et al., 1993; Lee et al., 1995a, 1997). The in vitro studies suggest that sHSPs prevent the aggregation of substrate proteins caused by heat or chemical denaturation and facilitate the reactivation of denatured substrate proteins.
Studies of mammalian cells indicate that phosphorylation of sHSPs may be an important regulator of sHSP function. The mammalian sHSPs are phosphorylated via a mitogen-activated protein kinase-activated protein kinase pathway at three conserved R-X-X-S sites (Gaestel et al., 1991). Phosphorylation of mammalian sHSPs results in a dissociation of the sHSP oligomer from approximately 400 kD to less than 70 kD (Kato et al., 1994) or from 200 and 250 kD to 150 and 125 kD (Lavoie et al., 1995), respectively, depending on the method of size determination used. sHSP phosphorylation is proposed to affect interaction of sHSPs with actin and, in some cases, also thermotolerance (Benndorf et al., 1994; Knauf et al., 1994; Lavoie et al., 1995).
Although members of the sHSP family are present in high-molecular-mass complexes in vivo, the structure and composition of these complexes are not yet fully defined. Recombinant forms of the mammalian sHSPs have been estimated as oligomers of 16 (Ehrnsperger et al., 1997) or 32 subunits (Behlke et al., 1996). The α-crystallins have been isolated in complexes ranging in size from 300 to 800 kD (Stevens and Augusteyn, 1993), corresponding to approximately 15 to 40 subunits, respectively. A 9-subunit sHSP oligomer has been isolated from Mycobacterium tuberculosis (Chang et al., 1996). Several different plant sHSPs have also been studied. A partially purified soybean (Glycine max L.) cytosolic class I sHSP complex is reported to contain at least 15 polypeptides ranging from 15 to 18 kD, all of which react with antibodies raised against a soybean sHSP, and may also contain higher-molecular-mass HSPs as minor components (Jinn et al., 1995). The same study reports that the class I complexes in rice, mung bean, and pea (Pisum sativum L.) contain at least 11, 15, and 8 sHSP polypeptides, respectively. Overexpression of a recombinant class I or class II pea cytosolic sHSP in Escherichia coli leads to the formation of complexes that have been isolated and shown to be homo-oligomeric dodecamers (Lee et al., 1995a). sHSP complexes have not been purified to homogeneity directly from plants. However, in total, the data suggest that the large sHSP-containing complexes seen in vivo represent oligomers of the sHSPs.
To gain information that will further our understanding of the diverse plant sHSPs, we have investigated the structure and phosphorylation state of HSP21, the chloroplast sHSP. HSP21 is a nuclear-encoded protein targeted to the chloroplast by an amino-terminal transit peptide that is removed after import to yield the mature 21-kD subunit. HSP21 is not constitutively expressed but becomes detectable in both leaves and roots after heat stress. In heat-stressed leaves, HSP21 has been estimated to represent approximately 0.05% of soluble chloroplast protein (Chen et al., 1994), making HSP21 much less abundant than the cytosolic sHSPs, which represent 1% of total plant protein after heat stress (DeRocher et al., 1991).
Like the mammalian and plant cytosolic sHSPs, mature HSP21 is detected in a soluble, high-molecular-mass complex. The chloroplast-localized sHSP (HSP21) was found in complexes of approximately 230 and 300 kD in pea and Arabidopsis, respectively, when examined in plant extracts by N-PAGE (Chen et al., 1994; Osteryoung and Vierling, 1994). Additionally, in pea a 42-kD form of HSP21 was detected when in vitro-translation products were imported into isolated chloroplasts (Chen et al., 1994). Previous work has not ruled out the possibility that proteins other than HSP21 are in the HSP21-containing complexes.
The work reported here examines the dynamics, phosphorylation, and composition of chloroplast HSP21 in pea and Arabidopsis. We provide evidence from in vivo studies indicating that HSP21 exists primarily in large complexes that do not dissociate during heat stress and recovery. In addition, unlike the mammalian sHSPs, HSP21 and other plant sHSPs do not appear to be phosphorylated in response to heat stress. Expression of mature pea and Arabidopsis HSP21 in E. coli yields homo-oligomers with the same apparent molecular masses as the HSP21 complexes seen in heat-stressed leaves. These results suggest that the chloroplast HSP21-containing complex is an oligomer of HSP21 subunits in vivo.
MATERIALS AND METHODS
Plant Growth and Heat-Shock Treatment
Pea (Pisum sativum L. cv Little Marvel) seeds were germinated and grown in vermiculite under a 16-h photoperiod for 9 d as described previously (Chen et al., 1994). For heat-stress treatment intact plants were taken from 22 to 38°C at a rate of 4°C h−1, this maximum temperature was maintained for 4 h, and then the temperature was decreased at 4°C h−1 back to 22°C (Chen et al., 1990).
Arabidopsis thaliana plants, which constitutively overexpress Arabidopsis HSP21 (Osteryoung et al., 1993), were grown in soil for 4 weeks under a 16-h photoperiod. Heat treatment was as described for pea, except that the temperature was increased or decreased at a rate of 6°C h−1 and the maximum temperature reached was 42°C.
Electrophoresis and Immunoblotting
Samples for N-PAGE were prepared by homogenizing leaf tissue directly in nondenaturing sample buffer (60 mm Tris-HCl, pH 8.0, 5 mm ε-amino-n-caproic acid, 1 mm benzamidine, and 15% Suc) using a ground-glass homogenizer. The concentration of tissue in buffer was 100 mg mL−1. Samples were microcentrifuged at 16,250g for 10 min and applied directly to 4 to 22% gradient N-PAGE. Electrophoresis was carried out as previously described (Chen et al., 1994), except that gels were run at 90 V for 48 h in 50 mm Tris-Gly buffer. Native molecular mass standards were: thyroglobulin, 669 kD; ferritin, 440 kD; catalase, 232 kD; lactate dehydrogenase, 147 kD; and BSA, 67 kD (Pharmacia).
Samples for SDS-PAGE were prepared as described above, except that SDS to 2% and DTT at 60 mm were added to the sample buffer and samples were heated for 2 min at 100°C after homogenization. SDS-PAGE was carried out on 10 to 16% or 12.5% gels using the method of Laemmli (1970). After electrophoresis, gels were either stained with Coomassie brilliant blue R or transferred to nitrocellulose for western-blot analysis. Rabbit antiserum against a pea HSP21 fusion protein was used at a dilution of 1:3000 as described previously (Vierling et al., 1989). Anti-pea HSP18.1 (DeRocher et al., 1991), HSP17.7 (Helm et al., 1997), and HSP70 (DeRocher and Vierling, 1995) antisera were used as described previously. Bound antibodies were detected using goat anti-rabbit horseradish peroxidase and an enhanced chemiluminescent system (Amersham). Molecular mass standards were: mysosin H-chain, 200 kD; phosphorylase B, 97 kD; BSA, 67 kD; ovalbumin, 43 kD; carbonic anhydrase, 29 kD; β-lactoglobin, 18 kD; and lysozyme, 14 kD (Life Technologies).
Size-Exclusion Chromatography
Pea extracts were prepared by homogenizing 200 mg of leaf tissue in 1.5 mL of column buffer (10 mm Tris-HCl, pH 8.0, 150 mm NaCl) with the addition of ε-amino-n-caproic acid to 10 mm and benzamidine to 1 mm. The insoluble material was removed by microcentrifugation at 16,250g for 15 min at 4°C. The resulting supernatant was combined with 200 μL of 1 mg mL−1 protein standards: thyroglobulin, 669 kD (Pharmacia); catalase, 232 kD (Pharmacia); β-amylase 200 kD (Sigma); BSA, 67 kD (Sigma); chymo-trypsinogen, 25 kD (Pharmacia); and Dextran Blue 2000 (Pharmacia). The sample was applied to a Sephadex G-200 column (1.5 cm i.d. × 100 cm) equilibrated with column buffer and 0.01% sodium azide. Elution was carried out at 0.4 mL min−1 and 2.25-mL fractions were collected. SDS-PAGE and immunoblotting were used to identify fractions containing HSP21. The relative amount of HSP21 in each fraction was determined by scanning the x-ray film using a laser densitometer and ImageQuant 3.1 software (Molecular Dynamics, Sunnyvale, CA). Purified pea HSP21 was analyzed on size-exclusion chromatography using the same methods with the following changes: samples were dialyzed into the appropriate column buffer, and additional buffers were used to test the effect of various salts on the elution of HSP21 (150 and 300 mm KCl, and 500 mm [NH4]2SO4).
The variability in the apparent molecular mass of proteins determined by size-exclusion chromatography with Sephadex G-200 was estimated as follows: Four of the five standard proteins were used to generate a linear equation relating the log of the molecular mass to the elution volume. This line was then used to calculate the apparent molecular mass of the fifth standard. The apparent molecular masses for three replications varied by approximately 12.5%.
Heat-stressed pea leaf tissue samples were also analyzed on an HPLC size-exclusion column (model G3000, TosoHaas, Montgomeryville, PA). The relative amounts of HSP21 in the column fractions, as examined by SDS-PAGE, were analyzed using a Macintosh computer and the public-domain NIH Image program (developed at the National Institutes of Health and available on the Internet at http://rsb.info.nih.gov/nih-image/). The size standards used for the HPLC size-exclusion column were: thyroglobulin, 669 kD (Pharmacia); ferritin, 440 kD (Pharmacia); aldolase, 158 kD (Pharmacia); ovalbumin, 42 kD (Pharmacia); and Cyt c, 12.3 kD (Sigma). A plot of the log of the molecular mass against the elution time for thyroglobulin, Rubisco, and ferritin produced a linear relationship (r2 = 0.996), which was used to calculate the apparent molecular mass of the HSP21 complex. The smaller molecular mass standards were used to ensure that the column was run for sufficient time to elute species the size of monomers or dimers of HSP21.
In Vivo Labeling
Nine-day-old pea seedlings were labeled with 0.1 to 0.8 mCi of H332PO4 (carrier free, 5 mCi mL−1, DuPont-NEN) in 2 mm Tris-HCl, pH 8.0, for 1 or 2 h. Specific samples, as indicated, were also incubated with 5 μm okadaic acid or 50 μm cantharidin (Sigma). Seedlings were heat treated as described above, and labeled at the time points indicated in the figure legend. At the start of the heat stress, seedlings in damp vermiculite were placed in the growth chamber. To initiate labeling, seedlings were removed from the vermiculite and the roots were excised under water approximately 1.5 cm below the expanded leaves. The cut ends were placed in 200 μL of labeling solution, and seedlings were then returned to the growth chamber for the described treatments.
At the end of the labeling period seedlings were processed by one of the following three methods: (a) Leaf tissue (200 mg) was homogenized in a glass homogenizer in SDS sample buffer or in SDS sample buffer containing 10 mm NaF. Samples were heated for 2 min at 100°C, microcentrifuged, and analyzed by gradient SDS-PAGE and immunoblotting as described above. (b) Labeled seedlings were crushed in liquid nitrogen using a mortar and pestle and approximately 0.2 g was transferred to a ground-glass homogenizer. Samples were then treated as described above, except the sample buffer also contained 50 mm NaF, 100 mm EDTA, and 100 mm EGTA. (c) Seedlings were homogenized in 2 mL of 20% TCA and 80% acetone using a glass homogenizer immersed in a bath of dry ice and acetone. Samples were transferred to a microcentrifuge tube and the precipitated protein was pelleted at 16,250g at 4°C for 30 min. These samples were then resuspended in SDS sample buffer, pH 10.0, and boiled for 2 min. Gels containing radiolabeled proteins were dried and exposed to x-ray film for 4 to 20 h. 32P-labeled samples that had been transferred to nitrocellulose were also exposed to film.
Purification of Pea HSP21
HSP21 accumulation was induced in 9-d-old peas using the gradual 38°C heat-stress regimen described above. Chloroplasts were isolated from approximately 1 kg of heat-stressed leaf tissue using methods described previously (Vierling et al., 1986), except that the final Percoll gradient step was omitted. Washed chloroplasts were diluted to 5 mg mL−1 chlorophyll and lysed in 10 mm Hepes, pH 8.0, containing 10 mm ε-amino-n-caproic acid and 1 mm benzamidine. The membranes were removed by centrifugation for 45 min at 12,000g in a fixed-angle rotor. A 40% (NH4)2SO4 fractionation was carried out by adding 0.1 m citrate buffer, pH 5.0 (equal to one-half that of the volume of the supernatant), and a saturated solution of (NH4)2SO4. After incubation at 4°C for 1.5 h, precipitated protein was removed by centrifugation at 4°C for 45 min at 12,000g. The soluble fraction containing HSP21 was filtered through a 0.2-μm filter and applied to an HPLC hydrophobic-interaction column (21.4 × 100 mm, 300-Å pore size; model 83-B23-E, Dynamax Hydropore, Rainin, Woburn, MA). HSP21 was eluted from the column with a gradient from 2 to 0 m (NH4)2SO4 in 100 mm phosphate buffer, pH 7.0. The flow rate was 5 mL min−1 and 5-mL fractions were collected. Samples were dialyzed against 10 mm Tris, pH 8.0, and fractions containing HSP21 were identified by SDS-PAGE and immunoblotting. Amino-terminal sequencing of purified pea HSP21 was carried out at the Arizona Research Laboratory Biotech Facility (Tucson).
Construction of Escherichia coli Expression Vectors
The mature amino terminus of HSP21 resulting from the removal of the transit peptide was determined for Arabidopsis and pea by amino-terminal sequencing of processed, mature, radiolabeled HSP21. The methods used for production of chloroplast protein precursors and their import into isolated chloroplasts were basically the same as those described previously (Vierling et al., 1988). The Arabidopsis HSP21 cDNA (Osteryoung et al., 1993) was transcribed in vitro to produce RNA coding for the HSP21 precursor protein. The RNA was then translated in vitro in the presence of [3H]Ile (DuPont-NEN, 89.6 Ci/mmol) and [3H]Gln (DuPont-NEN, 43.98 Ci/mmol), and the labeled HSP21 precursor protein was imported into isolated chloroplasts. The chloroplasts were lysed, membranes were removed by microcentrifugation, and the supernatant containing the processed, labeled HSP21 was amino-terminally sequenced (Arizona Research Laboratories). Radioactivity was monitored in 21 cycles in a scintillation counter (model LS6000 IC, Beckman), and the pattern of labeled residues was aligned with Ile and Gln residues in the deduced amino acid sequence of the Arabidopsis HSP21 precursor. The amino-terminal residue of mature Arabidopsis HSP21 was determined to be Gln-45. The amino terminus of pea HSP21 was also determined by the same method, but using [3H]Asp (DuPont-NEN, 10 Ci/mmol) for precursor protein labeling, and monitoring radioactivity for 15 cycles. The amino terminus of mature pea HSP21 was found to be Gln-50, consistent with Edman degradation of purified mature pea HSP21 (see Results). This result is further evidence of the amino terminus determined for mature Arabidopsis HSP21. In addition, Arabidopsis HSP21 residue Gln-45 and pea HSP21 residue Gln-50 both occur immediately after the amino acids ArgAla in the precursor sequence.
To replace the amino-terminal transit peptide with a start Met in both Arabidopsis and pea HSP21, PCR mutagenesis was used. Plasmid AZ311, the Arabidopsis HSP21 cDNA cloned into Bluescript (Osteryoung et al., 1994), was used as the PCR template, and the universal T3 primer and the custom 5′ primer CATATGCAAGACCAGAGAGAAAAC were used to generate the PCR product. This DNA was ligated into the pCR II vector using the Original TA Cloning kit (Invitrogen, San Diego, CA) and amplified. A NdeI-XhoI partial digest was used to obtain a segment of DNA that included the entire HSP21 cDNA. The segment was cloned into the expression vector pJC20 (Clos and Brandau, 1994) to produce pAZ376.
The mature pea HSP21 construct was synthesized as follows: The appropriate PCR fragment was generated from the plasmid AZ043 using the 5′ primer 3004 (GGCCGGATCCCATATGCAGGCTGGTGGTGATGG) and the 3′ primer 3005 (CGGAATTCCCTATCACTGAATTTGAAC). The PCR fragment was cloned into the pCR II vector as described above. A NdeI/EcoRI fragment corresponding to mature pea HSP21 and a start Met was then subcloned into pJC20 to make plasmid AZ315. After transformation into E. coli BL21(DE3) cells, induction of pea or Arabidopsis HSP21 was directed by T7 polymerase, which was induced by 1 mm isopropyl-β-d-thiogalactopyranoside for 6 h.
Purification of Recombinant Arabidopsis HSP21 from E. coli
Cells were harvested by centrifugation and washed with 50 mm Tris-HCl and 1 mm EDTA, pH 7.5 (buffer T50E1). Cells were sonicated in the presence of 1 mm benzamidine, 10 mm ε-amino-n-caproic acid and centrifuged at 17,500g for 30 min. The supernatant containing HSP21 and a wash of the pellet fraction were pooled. (NH4)2SO4 precipitation was carried out and the 40 to 70% pellet was found to be enriched for HSP21. This pellet was resolubilized in 25 mm Tris-HCl, 1 mm EDTA, pH 7.5 (T25E1), dialyzed against T25E1 at 4°C, and separated onto 0.2 to 0.8 m Suc gradients for 3 h at 45,000 rpm in a rotor (model VTi50, Beckman). Fractions containing HSP21 were pooled and applied to a DEAE-Sepharose column equilibrated with T25E1. HSP21 was eluted with a 0 to 0.15 m NaCl gradient. Fractions containing HSP21 were pooled, dialyzed, brought to 2 m (NH4)2SO4, and passed through a 0.2-μm syringe filter (Schleicher & Schuell). Samples were then separated on a hydrophobic-interaction column (Rainin) as described above for isolation of pea HSP21.
RESULTS
HSP21 Complexes Do Not Dissociate during Heat Stress
In leaf extracts analyzed by N-PAGE after heat stress, pea HSP21 is detected in complexes with apparent molecular masses of 230 and 200 kD (Chen et al., 1994). We were interested in determining if HSP21-containing complexes changed in size or relative abundance during heat shock or recovery in vivo, as seen for mammalian cytosolic sHSPs. Samples were taken at hourly intervals during a gradual heat stress and recovery. When analyzed by N-PAGE and western blotting, immunoreactive species were detected at 230, 200, and 42 kD at all time points after HSP21 was induced (data not shown). The 42-kD immunoreactive species was not seen in plant extracts in previous studies (Chen and Vierling, 1991), although a 42-kD form of HSP21 has been observed upon import of radiolabeled HSP21 into isolated chloroplasts (Chen et al., 1994). The relative abundance of the large forms and the 42-kD form could not be compared because their relative reactivity with the HSP21 antibody is not known.
To obtain an estimate of the size of the HSP21-containing complexes by an independent method, and to determine the relative abundance of the 42-kD form of HSP21, samples taken during a gradual heat stress were analyzed by size-exclusion chromatography using an HPLC size-exclusion column followed by western blotting. Native extracts of pea leaves were prepared during a gradual heat stress in which the temperature was increased from 22 to 38°C, maintained at 38°C for 4 h, and then gradually returned to 22°C. Samples were analyzed from the following temperature points: (a) at 34°C during HSP induction, (b) as the temperature reached 38°C, (c) after 4 h at 38°C, (d) as the temperature decreased to 27°C, (e) after 1 h of recovery at 22°C, and (f) after 2 h of recovery at 22°C followed by 30 min of abrupt stress at 38°C. Samples were also taken from plants that were allowed to gradually return to 22°C and were then abruptly restressed at 38°C. As shown in Figure 1, the highest concentration of HSP21 occurred in a fraction corresponding to a molecular mass of 550 ± 50 kD. The appearance of HSP21-containing complexes occurred in parallel to accumulation of the HSP21 polypeptide, and the protein was not consistently detected in any fraction corresponding to 42-kD or other low-molecular-mass species.
Figure 1.
Comparison of pea HSP21 complex size during heat stress and recovery. Leaf samples were taken at the temperature points indicated as discussed in the text and fractionated using an HPLC size-exclusion column, and the fractions were analyzed by immunoblotting with pea HSP21 antibodies. Lane numbers correspond to time of elution in minutes. The elution times of thyroglobulin (669 kD), aldolase (158 kD), and Cyt c (12 kD) are indicated. R indicates the elution time of endogenous pea Rubisco.
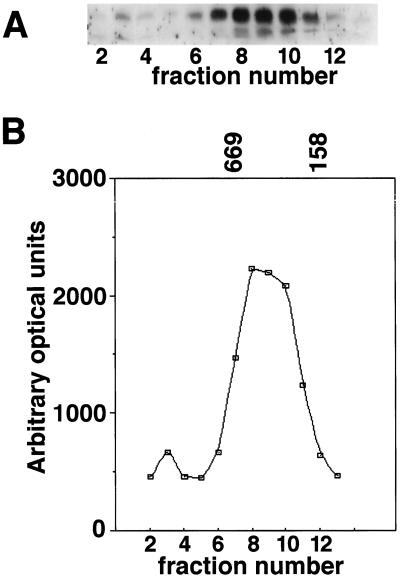
The size of the HSP21-containing complex in extracts of pea leaves exposed to a 12-h gradual heat stress was also analyzed using a gravity-loaded size-exclusion column with a different matrix (Sephadex G-200) followed by SDS-PAGE, western blotting, and laser densitometry. HSP21 eluted in a single peak corresponding to 420 ± 50 kD, as shown in Figure 2. The resolution of size-exclusion chromatography is less than that of N-PAGE, so we cannot rule out the possibility that the single large HSP21 complex detected with size-exclusion chromatography is actually composed of two species of very similar size, as was seen by N-PAGE. A small amount of the HSP21 signal was detected in a fraction corresponding to an apparent molecular mass > 700 kD. The nature of this minor form is unclear. No HSP21 was detected by western-blot analysis in the 42-kD range, so these fractions were not analyzed by laser densitometry.
Figure 2.
Estimation of pea HSP21 complex size in heat-stressed leaf tissue by size-exclusion chromatography. A, Total leaf extracts were analyzed on a Sephadex G-200 size-exclusion column and pea HSP21 was detected in column fractions by immunoblotting. B, The western-blot signal was quantified and plotted to identify the elution point of the majority of HSP21 relative to standard proteins. The elution times of thyroglobulin (669 kD) and aldolase (158 kD) are indicated.
The absence of the 42-kD form of HSP21 in heat-stressed leaf tissue analyzed using the two size-exclusion chromatography columns suggests that the 42-kD form of HSP21 is substantially less abundant than the larger forms in vivo. Alternatively, the 42-kD band appearing with N-PAGE of tissue extracts may be an artifact produced during electrophoresis. The fact that HSP21 is detected in a large complex throughout a gradual heat stress and recovery suggests that, unlike the sHSPs in mammalian systems, chloroplast sHSP function during heat stress may not be regulated by dissociation.
Phosphorylation of HSP21 Is Not Detected in Vivo
Phosphorylation has a significant influence on the structure and activity of many proteins, including the mammalian sHSPs, in which phosphorylation correlates with dissociation of the sHSP complex, as well as a decrease in thermoresistance in some cell lines (Landry et al., 1991; Kato et al., 1994). Although changes in the native size of HSP21 were not observed during heat shock or recovery, it is still possible that phosphorylation might modulate HSP21 function. To determine if HSP21 is phosphorylated in vivo, pea seedlings were labeled with 32Pi as described in Methods. Samples were taken at several time points during heat stress and recovery to detect transient protein phosphorylation. Seedlings were labeled in several separate experiments and were then processed in the presence of phosphatase inhibitors using three alternative techniques designed to minimize phosphatase activity (see Methods).
Seedlings were labeled for 1 or 2 h under the following conditions: (a) during induction of HSP21 expression, starting when leaf tissue reached 30°C, (b) at 22°C in the absence of heat stress, (c) during the final 2 h at the maximum temperature of 38°C, (d) during recovery from heat stress as the temperature returned to 22°C, and (e) after samples had returned to 22°C following the gradual heat stress. In addition, plants that had been heat stressed and allowed to recover at 22°C were then incubated with label and returned abruptly to 38°C (data not shown).
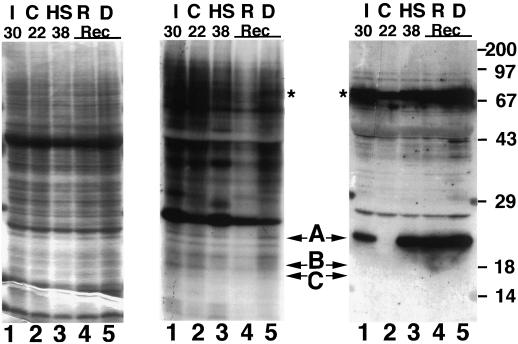
In the representative experiment documented in Figure 3, radiolabeled samples extracted from equal weights of tissue were analyzed. No incorporation of 32P was detected in proteins corresponding to HSP21 or to any of the other plant sHSPs (Fig. 3). Strong incorporation of 32P by other proteins indicates that abundant label was present and that protein phosphorylation occurred. Samples transferred to nitrocellulose blots were reacted with antibodies to pea HSP21 and HSP70 (Fig. 3), or with pea cytosolic class I HSP18.1 or class II HSP17.7 (data not shown) to confirm that sHSPs accumulated during the treatment. Comparison of the western blots and the autoradiograms indicates that neither HSP21 nor the cytosolic sHSPs were phosphorylated in vivo under any of the conditions tested. Addition of the protein phosphatase inhibitors cantharidin or okadaic acid during in vivo labeling did not alter the labeling pattern (data not shown). Analysis of samples containing equal counts per minute (rather than from equal weight of tissue) was also performed, and did not change the data interpretation. These data indicate that neither the chloroplast nor the cytosolic plant sHSPs are phosphorylated in vivo.
Figure 3.
Phosphorylation of pea HSP21 is not detectable in vivo. Leaf samples prepared after in vivo labeling with 32Pi, as described in the text, were separated by SDS-PAGE and gels were either stained (left panel), autoradiographed (center panel), or subjected to western blotting using anti-pea HSP21 or HSP70 antibodies (right panel). Lanes I, Leaf sample taken at 30°C during gradual temperature increase; lanes C, nonstressed control leaf tissue; lanes HS, sample taken after 2 h at 38°C; lanes R, sample taken at 22°C after gradual heat stress; and lanes D, sample taken at 38°C at the end of the heat stress, as temperature began to decline. Arrows at A, B, and C indicate the positions of HSP21, HSP18.1, and HSP17.7, respectively. The positions of HSP18.1 and HSP17.7 were determined on separate blots. The positions of heat-shock cognate 70 and HSP70, which are not separable in this analysis, are indicated by asterisks. Molecular mass standards in kilodaltons are indicated at the right of the figure. Rec, Recovery.
Purification of an HSP21 Complex from Heat-Stressed Pea Leaves
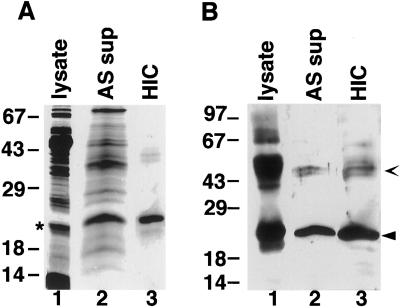
To examine the composition of the HSP21-containing complexes, steps were taken toward purification of HSP21 from pea leaves. As described in “Materials and Methods,” chloroplasts were lysed and soluble proteins processed through (NH4)2SO4 precipitation and hydrophobic-interaction chromatography to yield a preparation of nearly homogenous HSP21 polypeptide (Fig. 4A). As shown in Figure 4B, western blotting confirmed that the 21-kD polypeptide corresponded to HSP21, and an immunoreactive 42-kD species is likely to correspond to a dimer of HSP21. Edman degradation of the purified 21-kD polypeptide yielded the following amino acids: Q-A-G/D-G-D-G/N-D/K-N-K-D, with amino acids separated by a slash indicating sequencing cycles in which two amino acids were detected. Amino acids in boldface are the amino acids predicted from the HSP21 cDNA previously isolated from pea (Vierling et al., 1988). These data further confirm the identity of the purified polypeptide and locate the mature amino terminus at Gln-50. The presence of amino acids not predicted by the HSP21 cDNA may represent a sequence derived from an HSP21 isomer. Two distinct forms of HSP21 that differ slightly in their pI values have been observed, and Southern-blot analysis indicates that there is probably more than one HSP21 gene in pea (Vierling et al., 1988). Approximately 1 mg of HSP21 was recovered per kilogram of heat-stressed pea leaf tissue. The low abundance of HSP21 is consistent with previous estimates (Chen et al., 1994). The level of HSP21 in pea is also comparable to the amount of HSP21 found in heat-stressed Arabidopsis leaf tissue (approximately 0.05% of soluble leaf protein), determined using a rabbit anti-Arabidopsis HSP21 antibody (Vierling et al., 1989) and purified, recombinant Arabidopsis HSP21 as the standard (data not shown).
Figure 4.
Purification of pea HSP21 from heat-stressed leaf tissue. A, Coomassie blue-stained gel. B, Western blot. Lanes 1, Total soluble chloroplast lysate proteins (lysate); lanes 2, 40% (NH4)2SO4 supernatant (AS sup); and lanes 3, purified HSP21 after hydrophobic-interaction chromatography (HIC). Closed arrowhead indicates HSP21, and open arrowhead indicates dimer of HSP21 that does not dissociate on SDS-PAGE (Chen et al., 1994). The asterisk next to lane 1 in A represents a chloroplast protein that comigrates with HSP21 in the chloroplast lysate, but is separated from HSP21 during the (NH4)2SO4 precipitation and does not react with anti-HSP21 antibodies. Numbers indicate molecular mass standards in kilodaltons.
The size of purified pea HSP21 was compared with the size of the HSP21-containing complex observed in leaf extracts. The purified HSP21 complexes could not be resolved into discrete bands by N-PAGE for reasons that remain unclear. However, the HSP21 complexes had an apparent molecular mass of 300 kD when analyzed by size- exclusion chromatography on Sepharose G-200, significantly smaller than the apparent molecular mass of 420 ± 50 kD observed for HSP21 in homogenates of whole-leaf tissue analyzed using the same size-exclusion column. As shown in Table I, the apparent molecular mass of the purified complex was not altered in the presence of 500 mm NaCl, indicating that the decrease in apparent size was not caused by an interaction between the HSP21 complexes and the column matrix. The fact that no proteins other than HSP21 were identified in the complex is consistent with HSP21 forming a homo-oligomer. The difference in apparent molecular mass between purified HSP21 and HSP21 in leaf extracts may be attributable to the loss of subunits during the low-pH (NH4)2SO4 precipitation (data not shown). Although variations in the purification were tried, we were unsuccessful in isolating a larger stable complex containing pea HSP21.
Table I.
Analysis of the pea HSP21 complex by size-exclusion chromatography
| Sample | Salt Concentrationa | Apparent Molecular Massb |
|---|---|---|
| kD | ||
| Chloroplast lysate | 150 mm NaCl | 420 |
| 40% (NH4)2SO4 supernatant | 150 mm KCl | 300 |
| Purified complex | 150 mm NaCl or 150 mm KCl | 310 |
| Purified complex | 500 mm (NH4)2SO4 | 340 |
Concentration and salt used during size-exclusion chromatography with the Sephadex G-200 matrix.
Estimated molecular masses are ± 12.5%, as described in Methods.
Expression of Pea and Arabidopsis HSP21 in E. coli
The low abundance of HSP21 and the difficulties in purification of the native-sized complex prompted us to undertake expression of HSP21 in E. coli as an avenue for further study of this protein. As noted above, cytosolic sHSPs from plants have been successfully overexpressed in E. coli and found to assemble into oligomeric complexes similar to those observed in vivo (Lee et al., 1995a; Helm et al., 1997). We were interested in determining if HSP21 would exhibit similar properties. Because HSP21 is synthesized as a precursor, to express HSP21 in E. coli, plasmids were constructed in which the transit peptide was replaced by a single Met residue (see Methods).
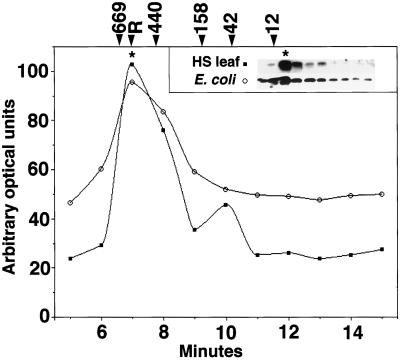
To determine if recombinant pea HSP21 assembled into complexes comparable to those observed in heat-stressed leaf tissue, recombinant pea HSP21 was compared with HSP21 in heat-stressed pea leaves by N-PAGE. In both samples, HSP21 was detected in complexes with apparent molecular masses of approximately 230 and 200 kD (data not shown). When analyzed using an HPLC size-exclusion column, recombinant pea HSP21 and HSP21 from heat-stressed leaves had the same apparent molecular mass (Fig. 5). Rubisco (molecular mass, 550 kD) eluted at 7 min on this column, and HSP21 in samples that did not contain Rubisco (E. coli lysates and purified HSP21) also consistently eluted at 7 min. This information, combined with the elution times of thyroglobulin (660 kD) and ferritin (440 kD), was used to calculate an apparent molecular mass of 550 ± 50 kD for the recombinant pea HSP21 complex.
Figure 5.
HSP21 complexes from heat-stressed pea tissue and E. coli extracts are the same size. Shown are the results of size analysis of HSP21 from heat-stressed pea tissue (HS leaf) and E. coli extracts size-exclusion chromatography using an HPLC size-exclusion column. HSP21 was detected by immunoblotting (inset), and the resulting signal was quantified and plotted to identify the fraction containing the most HSP21 (indicated by asterisks). ○, Signal from recombinant HSP21 expressed in E. coli; ▪, signal from HSP21 in heat-stressed leaf tissue. R indicates the elution point of Rubisco in leaf homogenates, and numbers indicate the elution point of molecular mass standards (in kilodaltons) listed in Methods.
Similar results were obtained with Arabidopsis HSP21. Recombinant Arabidopsis HSP21 migrated as three species with very similar molecular masses, the largest of which had an apparent molecular mass of 300 kD when analyzed by N-PAGE (Fig. 6A). This 300-kD complex comigrated with the single 300-kD complex detected in heat-stressed leaf samples, as shown in Figure 6B. The largest 300-kD complex also comigrated with the HSP21 complex formed in transgenic Arabidopsis (Osteryoung et al., 1993), which constitutively overexpressed HSP21 in the absence of heat stress (Fig. 6B). Recombinant Arabidopsis HSP21 and HSP21 from the transgenic Arabidopsis also coeluted with an apparent molecular mass of 550 kD when analyzed using the HPLC column (data not shown). In total, these results suggest that either HSP21-containing complexes are homo-oligomeric, or that HSP21-containing complexes include other factors common to both E. coli and the chloroplast.
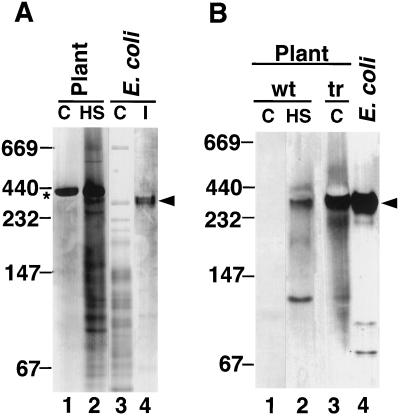
Figure 6.
Arabidopsis HSP21 expressed in E. coli comigrates with HSP21 from heat-stressed wild-type and transgenic Arabidopsis plants. A, Coomassie blue-stained N-PAGE. Lane 1, Nonstressed Arabidopsis leaf tissue; lane 2, heat-stressed leaf tissue; lane 3, lysate from control E. coli; and lane 4, E. coli lysate containing HSP21. B, Western blot of N-PAGE. Lane 1, Nonstressed Arabidopsis leaf tissue; lane 2, heat-stressed leaf tissue; lane 3, transgenic Arabidopsis (tr) constitutively expressing HSP21; and lane 4, lysate from E. coli containing HSP21. The asterisk indicates the position of Rubisco, and the arrowheads indicate the position of the HSP21 oligomer. C, Control; HS, heat stressed; I, induced; tr, transgenic; and wt, wild type.
Purification of Recombinant Arabidopsis HSP21 Complexes
To confirm that the recombinant HSP21 complexes were composed solely of HSP21 subunits, we undertook further purification of the recombinant complexes. Although highly expressed, the pea complexes formed in E. coli could not be isolated as stable oligomers. The E. coli-expressed Arabidopsis HSP21 complexes proved to be more stable and were purified as detailed in Methods. Analysis of the purified HSP21 complex by SDS-PAGE revealed that it was composed solely of the HSP21 polypeptide (Fig. 7A). When analyzed by N-PAGE, the purified recombinant HSP21 was observed in three similarly sized, higher-molecular-mass forms that are also present in the initial E. coli lysate (Fig. 7B). The largest purified recombinant Arabidopsis HSP21 complex co-migrates with the complex identified in heat-stressed leaf tissue. The fact that no proteins other than HSP21 were identified in the E. coli-expressed HSP21 complexes indicates that HSP21 is a homo-oligomer in vivo that does not contain other protein components.
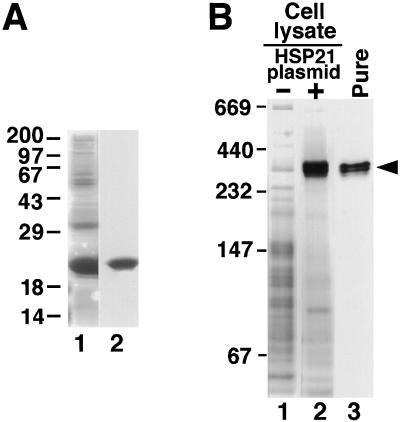
Figure 7.
Purified recombinant Arabidopsis HSP21 is a large homo-oligomer. A, Coomassie blue-stained SDS-PAGE of recombinant HSP21 before and after purification. Lane 1, Lysate of E. coli cells expressing HSP21; lane 2, purified HSP21. B, Recombinant Arabidopsis HSP21 forms a 300-kD complex that comigrates with HSP21 from an E. coli lysate. Lane 1, Lysate of E. coli cells that do not express HSP21 (−); lane 2, lysate of E. coli cells expressing HSP21 (+); and lane 3, assembled HSP21 after purification (Pure) (indicated by arrowhead). Samples were separated by N-PAGE and the gel was stained with Coomassie blue. Numbers at left indicate molecular mass standards (in kilodaltons).
To determine the possible origin of the recombinant Arabidopsis complexes that migrated slightly faster than the major 300-kD complex, each of the high-molecular-mass bands separated by N-PAGE was excised and subjected to SDS-PAGE. Western blotting indicated that the two faster-migrating forms of the HSP21 complex contain HSP21 that had been proteolytically cleaved to remove 1 to 2 kD (data not shown). The extent of proteolysis was not reduced by the inclusion of protease inhibitors and did not increase during the course of purification, suggesting that proteolysis may be occurring in E. coli before lysis.
DISCUSSION
We have investigated the native structure of the chloroplast-localized sHSP, HSP21. Our data indicate that the protein is a large oligomer composed of HSP21 subunits, which appear not to be phosphorylated and do not dissociate during heat stress and recovery. These findings contrast with what has been observed for mammalian cytosolic sHSPs, which also exist as large oligomers, but are known to undergo regulated phosphorylation and dissociation during heat stress (Lavoie et al., 1995). The dissociation of the mammalian sHSPs is believed to alter their interaction with other proteins in vivo (Lavoie et al., 1995) and in vitro (Benndorf et al., 1994).
Although both size-exclusion chromatography and N-PAGE indicate that HSP21 is in a high-molecular-mass complex during stress, the two methods give different absolute values for the apparent molecular mass of the HSP21 complexes. In lysates of heat-stressed pea leaves, HSP21 is 420 ± 50 kD when measured by Sephadex G-200 size-exclusion chromatography, and 550 ± 50 kD when analyzed on an HPLC size-exclusion column. When the same samples are analyzed by N-PAGE, pea HSP21 is detected in species with apparent molecular masses of 230, 200, and 42 kD. Arabidopsis HSP21 also migrates as a 550-kD complex on the HPLC size-exclusion column, but has an apparent molecular mass of 300 kD on N-PAGE. Similar discrepancies have been observed when the mammalian sHSP, HSP27, is analyzed by size-exclusion chromatography (apparent molecular mass, 400 kD) or N-PAGE (apparent molecular mass, 230 kD) (Lavoie et al., 1995). Both methods of analysis are secondary methods, and differences in the shape of the HSP21 complexes relative to the standard proteins may give anomalous estimates of molecular mass. However, by either method, HSP21 appears to form a stable complex of at least 200 kD in vivo.
The absence of the 42-kD form of HSP21 in heat-stressed pea leaf tissue analyzed by size-exclusion chromatography suggests that this form of HSP21 is substantially less abundant than the larger forms or that it is an artifact of the N-PAGE technique. In addition, a 42-kD form of HSP21 is not observed in Arabidopsis samples analyzed by N-PAGE and western blotting. In mammalian cell lines the 70- to 150-kD HSP oligomers that result from the dissociation of the large oligomers are the major form of sHSP after heat stress (Kato et al., 1994; Lavoie et al., 1995). In contrast, in those experiments in which the 42-kD form of HSP21 has been observed, it is a minor component. It has been suggested that the 42-kD form may represent an intermediate assembly that subsequently assembles into the large oligomers (Chen et al., 1994).
When purified from heat-stressed pea leaves, HSP21 was found in a complex of 300 kD, as measured by size-exclusion chromatography using a Sephadex G-200 matrix. The purified protein did not yield discrete complexes on N-PAGE. HSP21 complexes may have been less disrupted by size-exclusion chromatography than by N-PAGE. The 300-kD HSP21 complex was an oligomer of HSP21 subunits and may represent a stable core of the larger (> 400 kD) HSP21 complex seen in whole-leaf homogenates. Similar suggestions have been made for the α-crystallins, which have been purified as complexes with molecular masses ranging from 300 to 800 kD (Walsh et al., 1991). The absence of measurable decreases in the size of the HSP21 complex during heat stress is significant because the dissociation of the mammalian sHSPs is believed to be important for their function in vivo.
The dissociation of the mammalian sHSP complexes is regulated by phosphorylation of S at conserved R-X-X-S sites (Gaestel et al., 1991; Kato et al., 1994). In some mammalian cell lines, phosphorylatable sites are required for the increased thermotolerance that accompanies the overexpression of the mammalian sHSPs, but these sites are not required in other cell lines (E. Hickey, personal communication; Knauf et al., 1994; Lavoie et al., 1995). Plants are known to have a mitogen-activated protein kinase-activated protein-like kinase cascade (Jonak et al., 1994; Pöpping et al., 1996). However, neither HSP21 nor the cytosolic sHSPs contain this conserved R-X-X-S motif, and our results indicate that plant sHSPs are not phosphorylated in vivo and do not undergo the dissociation that accompanies phosphorylation of the sHSPs in mammals. These results indicate that the plant sHSP oligomeric structure is unlikely to be regulated by phosphorylation, as seen for mammalian sHSPs. It has been suggested that the large sHSP oligomers are the active species in vivo (Leroux et al., 1997). If so, then the plant sHSPs may be regulated by other mechanisms, such as control of the rate of sHSP degradation.
Lack of sHSP phosphorylation in plants has been suggested by previous studies. Phosphorylation of sHSPs was not detected in cultured tomato cells (Nover et al., 1989), and work by Clarke and Critchley (1992) indicated that a 260-kD, nuclear-encoded, chloroplast-localized, sHSP-containing complex in barley (Hordeum vulgare L.) was also not phosphorylated in vivo. The latter authors suggest that the barley complex represents an octamer of a 32-kD sHSP, although they noted that the presence of additional nuclear-encoded proteins could not be ruled out. The sequence of this 32-kD protein has not been determined, so it is unknown whether this protein is a member of the sHSP family. In sorghum and millet, small heat-induced, chloroplast-localized proteins have been found in complexes of approximately 380 kD. These chloroplast proteins also do not appear to be phosphorylated in vivo (Clarke and Critchley, 1994). Our experiments using antibodies specific to defined chloroplast and cytosolic sHSPs confirm and extend these earlier observations regarding sHSP phosphorylation.
In vitro-translated HSP21 precursor, when imported into chloroplasts isolated from heat-stressed leaves, is incorporated into the same HSP21 complexes observed in heat- stressed leaves alone (Chen et al., 1994). However, the same precursor does not form high-molecular-mass complexes when imported into chloroplasts isolated from leaves that have not been heat stressed. It was suggested that the failure of imported HSP21 to assemble in the latter case might be the result of the low concentration of in vitro- translation product or a requirement for heat-induced factors (Chen et al., 1990). However, HSP21 overexpressed in transgenic Arabidopsis in the absence of heat stress assembles into a complex with the same apparent molecular mass as the complexes seen in heat-stressed tissue, suggesting that heat stress is not required for assembly (Osteryoung et al., 1994). The fact that a substantial portion of Arabidopsis HSP21 expressed in E. coli assembles into large oligomers with the same apparent molecular mass as HSP21 complexes in heat-stressed leaves suggests that HSP21 complexes in vivo do not contain chloroplast proteins other than HSP21. In addition, recombinant HSP21, which co-migrates with HSP21 complexes from heat-stressed leaves, has been purified and shown to be homo-oligomeric. These data also indicate that no chloroplast factors are strictly required for oligomerization. A small proportion of HSP21 was found in complexes slightly smaller than those observed in heat-stressed leaf tissue, and it is possible that, although not strictly required, plant factors may increase the yield of correctly assembled HSP21.
If chloroplast HSP21 acts as a chaperone, as is proposed for the cytosolic sHSPs, it is perhaps surprising that we saw no evidence of an increase in HSP21 oligomer size during heat stress in the plant. However, we cannot rule out the possibility that HSP21 oligomers associate with substrates or other chaperones, because such interactions may be too labile to be observed in tissue homogenates using these techniques. Alternatively, these interactions may be too dynamic to be identified by the nonequilibrium methods used. In addition, small changes in apparent molecular mass (≤ 30 kD) are not easily observed using size-exclusion chromatography and cannot be ruled out.
Plant sHSPs are present in the cytoplasm, chloroplast, mitochondrion, and ER, whereas mammalian and yeast cells express only cytosolic sHSPs. The work presented here suggests that the size, stability, and regulation of the plant sHSPs may differ from what is observed in other organisms. We conclude that native HSP21 is a nonphosphorylated oligomer of nine or more HSP21 subunits in vivo that do not dissociate during heat stress. Additional work will be required to identify the substrates of the sHSPs and to determine how they function in vivo.
ACKNOWLEDGMENT
We thank Dr. Ricardo Azpiroz for stimulating scientific discussion and critical reading of the manuscript.
Abbreviations:
- HSP
heat-shock protein
- N-PAGE
nondenaturing, pore-exclusion PAGE
- sHSP
small HSP
Footnotes
This work was supported by National Institutes of Health grant no. RO1-GM42762, by University of Arizona Experiment Station funds, and by American Cancer Society Faculty Research Award no. FRA-420 to E.V.
LITERATURE CITED
- Arrigo A-P, Landry J (1994) Expression and function of the low-molecular-weight heat shock proteins. In R Morimoto, A Tissieres, C Georgopoulus, eds, Cold Spring Harbor Monograph Series: The Biology of Heat Shock Proteins and Molecular Chaperones, Ed 26. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp 335–373
- Behlke J, Lutsch G, Gaestel M, Bielka J. Supramolecular structure of the recombinant murine small heat shock protein HSP25. FEBS Lett. 1996;288:119–122. doi: 10.1016/0014-5793(91)81016-2. [DOI] [PubMed] [Google Scholar]
- Benndorf R, Hayess K, Ryazantsev S, Wieske M, Behlke J. Phosphorylation and supramolecular organization of murine small heat shock protein HSP25 abolish its actin polymerization-inhibiting activity. J Biol Chem. 1994;269:20780–20784. [PubMed] [Google Scholar]
- Chang Z, Primm TP, Jakana J, Lee IH, Serysheva I. Mycobacterium tuberculosis 16-kDa antigen (Hsp16.3) functions as an oligomeric structure in vitro to suppress thermal aggregation. J Biol Chem. 1996;271:7218–7223. [PubMed] [Google Scholar]
- Chen Q, Lauzon LM, DeRocher AE, Vierling E. Accumulation, stability, and localization of a major chloroplast heat-shock protein. J Cell Biol. 1990;110:1873–1883. doi: 10.1083/jcb.110.6.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Osteryoung K, Vierling E. J Biol Chem. 1994;269:13216–13223. [PubMed] [Google Scholar]
- Chen Q, Vierling E. Analysis of conserved domains identifies a unique structural feature of a chloroplast heat shock protein. Mol Gen Genet. 1991;226:425–431. doi: 10.1007/BF00260655. [DOI] [PubMed] [Google Scholar]
- Clark AK, Critchley C. The identification of a heat shock protein complex in chloroplasts of barley leaves. Plant Physiol. 1992;100:2081–2089. doi: 10.1104/pp.100.4.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke AK, Critchley C. Characterisation of chloroplast heat shock proteins in young leaves of C4 monocots. Physiol Plant. 1994;92:118–130. [Google Scholar]
- Clos J, Brandau S. Protein Exp Purif. 1994;5:133–137. doi: 10.1006/prep.1994.1020. [DOI] [PubMed] [Google Scholar]
- DeRocher A, Vierling E. Cytoplasmic HSP70 homologues of pea: differential expression in vegetative and embryonic organs. Plant Mol Biol. 1995;27:441–456. doi: 10.1007/BF00019312. [DOI] [PubMed] [Google Scholar]
- DeRocher AE, Helm KW, Lauzon LM, Vierling E. Expression of a conserved family of cytoplasmic low molecular weight heat shock proteins during heat stress and recovery. Plant Physiol. 1991;96:1038–1047. doi: 10.1104/pp.96.4.1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrnsperger M, Gräber S, Gaestel M, Buchner J. Binding of nonnative protein to HSP25 during heat shock creates a reservoir of folding intermediates for reactivation. EMBO J. 1997;16:221–229. doi: 10.1093/emboj/16.2.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaestel M, Schroder W, Benndorf R, Lippman C, Buchner J. Identification of the phosphorylation sites of the murine small heat shock protein HSP25. J Biol Chem. 1991;266:14721–14724. [PubMed] [Google Scholar]
- Helm K, Lee G, Vierling E. Expression and structure of cytosolic class II heat-shock proteins. Plant Physiol. 1997;114:1477–1485. doi: 10.1104/pp.114.4.1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horwitz J. Alpha-crystallin can function as a molecular chaperone. Proc Natl Acad Sci USA. 1992;89:10449–10453. doi: 10.1073/pnas.89.21.10449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia TD, Craig EA. Four small Drosophila heat shock proteins are related to each other and to mammalian α-crystallin. Proc Natl Acad Sci USA. 1982;79:2360–2364. doi: 10.1073/pnas.79.7.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakob U, Gaestel M, Engel K, Buchner J. Small heat shock proteins are molecular chaperones. J Biol Chem. 1993;268:1517–1520. [PubMed] [Google Scholar]
- Jinn T-L, Chen Y-M, Lin C-Y. Characterization and physiological function of class I low-molecular-mass, heat-shock protein complex in soybean. Plant Physiol. 1995;108:693–701. doi: 10.1104/pp.108.2.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonak C, Heberle-Bors E, Hirt H. MAP kinases: universal multi-purpose signaling tools. Plant Mol Biol. 1994;24:407–416. doi: 10.1007/BF00024109. [DOI] [PubMed] [Google Scholar]
- Kato K, Hasegawa K, Goto S, Inaguma Y. Dissociation as a result of phosphorylation of an aggregated form of the small stress protein, hsp27. J Biol Chem. 1994;269:11274–11278. [PubMed] [Google Scholar]
- Knauf U, Jakob U, Engel K, Buchner J, Gaestel M. Stress- and mitogen-induced phosphorylation of the small heat shock protein HSP25 by MAPKAP kinase 2 is not essential for chaperone properties and cellular thermoresistance. EMBO J. 1994;13:54–60. doi: 10.1002/j.1460-2075.1994.tb06234.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Landry J, Chrétien P, Lambert H, Hickey E, Weber LA. Heat shock resistance conferred by expression of the human HSP27 gene in rodent cells. J Cell Biol. 1989;109:7–15. doi: 10.1083/jcb.109.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry J, Chrétien P, Laszlo A, Lambert H. Phosphorylation of HSP27 during development and decay of thermotolerance in Chinese hamster cells. J Cell Physiol. 1991;147:93–101. doi: 10.1002/jcp.1041470113. [DOI] [PubMed] [Google Scholar]
- Lavoie J, Lambert H, Hickey E, Weber L, Landry J. Modulation of cellular thermoresistance and actin filament stability accompanies phosphorylation-induced changes in the oligomeric structure of heat shock protein 27. Mol Cell Biol. 1995;15:505–516. doi: 10.1128/mcb.15.1.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavoie JN, Gingras-Breton G, Tanguay R, Landry J. Induction of Chinese hamster HSP27 gene expression in mouse cells confers resistance to heat shock. J Biol Chem. 1993;268:3420–3429. [PubMed] [Google Scholar]
- Lee GJ, Pokala N, Vierling E. Structure and in vitro molecular chaperone activity of cytosolic small heat shock proteins from pea. J Biol Chem. 1995a;270:10432–10438. doi: 10.1074/jbc.270.18.10432. [DOI] [PubMed] [Google Scholar]
- Lee GJ, Roseman AM, Sabil HR, Vierling E. A small heat shock protein stably binds heat-denatured model substrates and can maintain a substrate in a folding-competent state. EMBO J. 1997;16:659–671. doi: 10.1093/emboj/16.3.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Hubel A, Schöffl F. Derepression of the activity of genetically engineered heat shock factor causes constitutive synthesis of heat shock proteins and increased thermotolerance in transgenic Arabidopsis. Plant J. 1995b;8:603–612. doi: 10.1046/j.1365-313x.1995.8040603.x. [DOI] [PubMed] [Google Scholar]
- Leroux MR, Melki R, Gordon B, Batelier G, Candido EPM. Structure-function studies on small heat shock protein oligomeric assembly and interaction with unfolded peptides. J Biol Chem. 1997;272:24646–24656. doi: 10.1074/jbc.272.39.24646. [DOI] [PubMed] [Google Scholar]
- Nagao R, Kimpel J, Vierling E, Key J. The heat shock response: a comparative analysis. Oxf Surv Plant Mol Cell Biol. 1986;3:384–438. [Google Scholar]
- Nover L, Scharf K-D, Neumann D. Cytoplasmic heat shock granules are formed from precursor particles and are associated with a specific set of mRNAs. Mol Cell Biol. 1989;9:1298–1308. doi: 10.1128/mcb.9.3.1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osteryoung K, Pipes B, Wehmeyer N, Vierling E (1994) Studies of a chloroplast-localized small heat shock protein in Arabidopsis. In J Cherry, ed, Biochemical and Cellular Mechanisms of Stress Tolerance in Plants, Vol 86. Springer-Verlag, Berlin, pp 97–113
- Osteryoung K, Sundberg H, Vierling E. Poly(A) tail length of a heat shock protein RNA is increased by severe heat stress, but intron splicing is unaffected. Mol Gen Genet. 1993;239:323–333. doi: 10.1007/BF00276930. [DOI] [PubMed] [Google Scholar]
- Osteryoung K, Vierling E. Dynamics of small heat shock protein distribution within the chloroplasts of higher plants. J Biol Chem. 1994;269:28676–28682. [PubMed] [Google Scholar]
- Pöpping B, Gibbons T, Watson MD. The Pisum sativum MAP kinase homologue (PsMAP) rescues the Saccharomyces cerevisiae hog1 deletion mutant under conditions of high osmotic stress. Plant Mol Biol. 1996;31:355–363. doi: 10.1007/BF00021795. [DOI] [PubMed] [Google Scholar]
- Rollet E, Lavioe J, Landry J, Tanguay R. Expression of Drosophila's 27 kDa heat shock protein into rodent cells confers thermal resistance. Biochem Biophys Res Commun. 1992;185:116–120. doi: 10.1016/s0006-291x(05)80963-5. [DOI] [PubMed] [Google Scholar]
- Stevens A, Augusteyn RC. Acid-induced dissociation of αA- and αB-crystallin homopolymers. Biophys J. 1993;65:1648–1655. doi: 10.1016/S0006-3495(93)81219-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierling E. The heat shock response in plants. Annu Rev Plant Physiol Plant Mol Biol. 1991;42:579–620. [Google Scholar]
- Vierling E, Harris L, Chen Q. The major low-molecular weight heat shock protein in chloroplasts shows antigenic conservation among diverse higher plant species. Mol Cell Biol. 1989;9:461–468. doi: 10.1128/mcb.9.2.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierling E, Mishkind ML, Schmidt GW, Key JL. Specific heat shock proteins are transported into chloroplasts. Proc Natl Acad Sci USA. 1986;83:361–365. doi: 10.1073/pnas.83.2.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierling E, Nagao RT, DeRocher AE, Harris LM. A heat shock protein localized to chloroplasts is a member of a eukaryotic superfamily of heat shock proteins. EMBO J. 1988;7:575–581. doi: 10.1002/j.1460-2075.1988.tb02849.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh M, Sen A, Chakrabarti B. Micellar subunit assembly in a three-layer model of oligomeric alpha crystallin. J Biol Chem. 1991;266:20079–20084. [PubMed] [Google Scholar]
- Waters ER, Lee GJ, Vierling E. Evolution, structure and function of the small heat shock proteins in plants. J Exp Bot. 1996;47:325–338. [Google Scholar]