Abstract
Virus-based vectors are widely used in hematopoietic stem cell (HSC) gene therapy, and have the ability to integrate permanently into genomic DNA, thus driving long-term expression of corrective genes in all hematopoietic lineages. To date, HSC gene therapy has been successfully employed in the clinic for improving clinical outcomes in small numbers of patients with X-linked severe combined immunodeficiency (SCID-X1), adenosine deaminase deficiency (ADA-SCID), adrenoleukodystrophy (ALD), thalassemia, chronic granulomatous disease (CGD), and Wiskott-Aldrich syndrome (WAS). However, adverse events were observed during some of these HSC gene therapy clinical trials, linked to insertional activation of proto-oncogenes by integrated proviral vectors leading to clonal expansion and eventual development of leukemia. Numerous studies have been performed to understand the molecular basis of vector-mediated genotoxicity, with the aim of developing safer vectors and lower-risk gene therapy protocols. This review will summarize current information on the mechanisms of insertional mutagenesis in hematopoietic stem and progenitor cells due to integrating gene transfer vectors, discuss the available assays for predicting genotoxicity and mapping vector integration sites, and introduce newly-developed approaches for minimizing genotoxicity as a way to further move HSC gene therapy forward into broader clinical application.
Keywords: gene therapy, hematopoietic stem cells, insertional mutagenesis, genotoxicity, induced pluripotent stem cell
Introduction
Over the past two decades, hematopoietic stem cells (HSC) based gene therapy has been used in clinical trials for severe inherited diseases such as X-linked severe combined immunodeficiency (SCID-X1) [1,2], adenosine deaminase deficiency (ADA-SCID) [3,4], X-linked chronic granulomatous disease (X-CGD) [5,6], X-linked adrenoleukodystrophy (X-ALD) [7] and Wiskott-Aldrich syndrome (WAS) [8]. Initial disappointment with results in the 1990s, due to low HSC gene transfer efficiency, was replaced by optimism beginning around 2000, as improved transduction conditions and vectors began to result in evidence for reversal of clinical immunodeficiency syndromes. However, serious safety concerns were raised just a few years later, when in 2003 clonal vector-associated leukemias were reported in several patients enrolled in the pioneering SCID-X1 trial [9,10]. The development of methods for evaluating viral vector genotoxicity and design of lower risk integrating vectors is critical for further use of HSC genetic modification to treat inherited and acquired diseases. In this review, we will summarize what has been learned regarding vector-related genotoxicity from human clinical trials (Table 1), in vitro studies, and animal models, and suggest ways to reduce this risk, in order to move HSC gene therapy forward safely.
Table 1.
Reported genotoxic events in HSC gene therapy clinical trials
| Diseases | Patients | Vector backbone |
Transgene | Follow-up | Severe adverse effects |
Insertion sites (clonal dominance) |
Clinical outcome |
References | |
|---|---|---|---|---|---|---|---|---|---|
| SCID-X1 | 9 | MFG | γC | 9 years | T-ALL | No.1 | LMO2 | Died despite chemotherapy | [1,2,10,15] |
| γ-retroviral | (range, 8 to 11) | in four patients | No.2 | LMO2 | Remission with chemotherapy | ||||
| vector | No.3 | CCND2 | Remission with chemotherapy | ||||||
| No.4 | LMO2, BMI1 | Remission with chemotherapy | |||||||
| Other 5 | No clonal dominance | Alive, improved immune function | |||||||
| SCID-X1 | 10 | MFG | γC | 5 years | T-ALL | No.1 | LMO2 | Remission with chemotherapy | [14,16] |
| γ-retroviral | in one patient | Other 9 | No clonal dominance | Alive with improved immune | |||||
| vector | function | ||||||||
| ADA-SCID | 10 | GIADAl | ADA | 4.0 years | None | Integration hotspots near | All alive with improved | [3,4] | |
| MLV-based | (range, | DYRK1A,BLM,LMO2, | immune and metabolic | ||||||
| retroviral vector | 1.8 to 8.0) | CCND2 and BCL2 no clonal | parameters | ||||||
| selection ) | |||||||||
| X-CGD | 2 | pSF7, SFFV- | gp91phox | 4 years | MDS | No.1 | MDS1/Evi1, PRDM16, and | Died from sepsis | [5,9] |
| based retrovirus | SETBP1 | 27 months | |||||||
| vector | MDS | No.2 | MDS1/Evi1, PRDM16, and | Underwent an allogeneic | |||||
| SETBP1 | HSC transplantation at month 45 | ||||||||
| X-CGD | 3 | MFGS | gp91phox | 3 years | None | No clonal dominance | Clinical improvement | [6] | |
| retroviral vector | |||||||||
| X-ALD | 2 | HIV-1-derived | ABCD1 | 24 to 30 months | None | No clonal dominance | Alive with decreased progression | [7] | |
| lentiviral vector | of neurologic phenotype | ||||||||
| β-thalassemia | 1 | HIV-1-derived | β-globin | 33 months | None | Dominant HMGA2 expression | Alive and transfusion- | [26] | |
| lentiviral vector | clone | independent | |||||||
| WAS | 2 | CMMP | WASP | 3 years | T-ALL in one | LMO2, CCND2, and BMI1 | T-ALL patient with ongoing | [8] | |
| retroviral vector | patient(LMO2) | in T-cells, and MDS1 | chemotherapy, the other alive | ||||||
| /EVI1, PRDM16, and | with improved immune | ||||||||
| SETBP1 in granulocytes | function and decreased cytopenias |
Results of pivotal HSC gene therapy human clinical trials
HSC gene therapy trials for SCID-X1
SCID-X1 is an X-linked inherited disorder caused by inactivating mutations of the γC cytokine receptor common subunit gene, located on the X-chromosome. Patients with SCID-X1 lack mature T and NK cells, and die in early childhood due to severe infections resulting from profound immunodeficiency [11,12]. In 2000, the first report of successful gene therapy for SCID-X1 provided a tremendous boost for the field [1]. In this trial, autologous bone marrow CD34+ cells were collected and transduced with a replication-defective γC Moloney retrovirus containing the corrective gene, and reinfused into the patients without any myeloablation. By 10 months post-infusion, the T and NK compartments had been filled by γC transgene-expressing cells. T, B, and NK cell counts and function, including antigen-specific responses, were comparable to those of healthy children of the same age [1], and at a median of 9 years follow-up, a recent paper documented complete correction of the immunodeficiency associated with SCID-X1 in these patients [2].
However, almost 3 years after gene therapy, uncontrolled exponential clonal proliferation of vector-containing T cells was observed in the two youngest patients. Remarkably, the leukemic cells in both patients were shown to have proviral insertions activating aberrant LMO2 gene expression [10]. From 1999 to 2009, a total of 20 patients with SCID-X1 underwent HSC gene therapy with corrective γ-retroviral vectors in trials in France and England [13,14]. To date, five of 20 have developed T cell leukemias between 23 and 68 months after receiving transduced CD34+ cells, with one death, and successful treatment of the other four with chemotherapy and/or allogeneic transplantation [15,16]. Activation of the proto-oncogene LMO2 via the proviral enhancer was documented in all four cases and the CCND2 proto-oncogene in the fifth (Table 1).
These findings raised concerns about the safety of gene therapy and promoted in-depth analysis of the genotoxicity of viral vectors. Many theories were initially put forward suggesting that the risk of leukemia was unique to the particulars of the SCID-X1 trial in France, and would not extend to other diseases being treated with HSC gene therapy, or even to other SCID-X1 trials using slightly different vectors, transduction conditions or patient populations. Factors proposed to contribute to the high apparent risk included the underlying severity of the immunodeficiency in X-SCID, constitutive activation of the signaling molecule transgene, the young age of the patients, very high doses of CD34+ cells, the transduction conditions or particular vector backbone utilized in France but not other SCID-X1 trials, expanded target lymphoid progenitors, rapid T cell expansion in vivo following correction, or some unique synergy between the γC transgene and LMO2 activation [17,18]. However, the eventual occurrence of similar leukemias in a separate British trial, utilizing a slightly different vector and transduction conditions, along with occurrence of leukemia linked to vector insertions in other clinical situations and in animal models as described below, silenced most of this speculation, and suggested that there were significant inherent risks resulting from vector integration into the genome of HSCs.
HSC gene therapy trials for ADA-SCID patients
After several studies completed in the 1990s did not provide evidence for significant immunologic improvement or clinical benefit, likely due to insufficient HSC transduction efficiency and/or poor transgene expression [19,20], a more successful gene therapy trial for ADA-SCID was performed in Milan, utilizing γ-retroviral Moloney murine leukemia virus (MLV) vectors to deliver a normal human ADA gene [3]. Patients received nonmyeloablative conditioning with busulfan before infusion [3], since it appears that in ADA deficiency, corrected T cell progenitors do not robustly outcompete uncorrected cells without some advantage supplied by at least partial myeloablation. Immune reconstitution and normal T cell function were observed in nine of ten patients [4]. As opposed to the SCID-X1 trial, none of the 19 patients with ADA deficiency (median follow-up period, 3 years; range, 0.5–9 years) showed any adverse effects, with no progression to clonal hematopoietic or any evidence for hematopoietic malignancies [13,21].
Insertion site analysis in genetically corrected CD34+ cells and their multilineage progeny before and up to 47 months after transplantation into 5 patients with ADA-SCID revealed that retroviral insertions sites (IS) were in gene-dense regions, promoters, and transcriptionally active genes, both in preinfusion transduced CD34+ cells and in vivo cells after gene therapy [22]. Insertion sites were identified close to or within proto-oncogenes or genes controlling cell growth and self-renewal including LMO2. But in this group of patients, the T cells carrying LMO2 IS did not clonally expand and did not progress to leukemia [22]. These observations indicate that the properties of the transduced progenitor, its in vivo proliferative history, the underlying disease and transgene, or specific vector and transduction characteristics could impact on the likelihood that potentially “dangerous clones” progress to clonal dominance and leukemia. Insertions in potentially dangerous genomic sites are not sufficient per se to induce a proliferative advantage in T cells in vivo, and oncogenic transformation likely requires multiple cooperating events beyond proviral-linked insertional mutagenesis.
HSC gene therapy trials for X-CGD patients
X-linked chronic granulomatous disease (CGD) is caused by mutations of the CYBB gene, encoding gp91phox required by neutrophils to produce microbicidal oxidants. Patients develop recurrent life threatening bacterial and fungal infections [23]. Several trials were performed in the 1990s without the use of conditioning prior to infusion of transduced cells, and failed to show long-term engraftment of corrected neutrophils or any clinical benefit. In a more recent trial, two patients received corrected CD34+ cells after busulfan conditioning [5]. A monocistronic gamma retroviral vector pSF71 was used to express gp91phox. Significant levels of corrected neutrophils and monocytes and notable clinical improvements with clearance of serious chronic infections were observed in both patients. However, marked expansion of vector-containing myeloid cells occurred in both patients beginning several months following infusion, and was linked to expansion of multiple clones with proviral activation of insertional activation of MDS1/EVI1, SETBP1 or PRDM16 genes, and silencing of the gp91phox transgene. Both patients eventually developed monosomy 7 in an MDS1/EVI1 dominant clone and progressed to myelodysplasia/acute myeloid leukemia at 15 or 28 months post-infusion. One patient died 27 months after gene therapy of sepsis [9]. In another gene therapy trial for X-CGD conducted at the NIH [6], three patients received CD34+ cells transduced with an MFGS retroviral vector encoding gp91phox also following busulfan conditioning. Sustained long-term correction of neutrophils with clinical improvement occurred in two patients, with no evidence for clonal dominance or progression to myelodysplastic syndrome (MDS) during 3 years follow-up [6]. The different degree of insertional genotoxicity observed in these two otherwise very similar trials may be related to the more potent proviral enhancer contained in the SF71 vector backbone compared to the MFGS backbone, more likely to activate nearby proto-oncogenes.
HSC gene therapy trials for WAS patients
A German group recently published results on two patients entered into a gene therapy trial for Wiscott-Aldrich Syndrome (WAS), an X-linked recessive primary immunodeficiency-thrombocytopenia disorder [8]. The CMMP retroviral vector backbone expressed the WAS protein, and was pseudotyped with the gibbon ape leukemia virus (GALV) envelope protein. Moderate dose busulfan was given before reinfusion of CD34+ cells. The clinical condition of both patients markedly improved, and functional improvement of T cells, B cells, NK cells, and monocytes was documented, along with an increase in the platelet count. During the initial follow-up period of 2 and a half years reported in the paper, the clonal distribution and fate of gene-corrected cells in vivo were monitored by large-scale analyses of retroviral vector insertion sites and a highly polyclonal reconstitution pattern was found. However, both patients had clones with IS in the LMO2, CCND2, and BMI1 genes detected in T cells, all genes shown to trigger malignant transformation of CD3+ T cells when activated by vector insertions in patients with SCIDX1. Clones with insertions in MDS1/EVI1, PRDM16, and SETBP1 were detected in granulocytes, genes associated with myeloid clonal expansion in patients with CGD [8]. However, in unpublished data reported at the 2010 American Society of Hematology Meeting, Dr. Christoph Klein, the lead trial investigator, reported that one patient had developed T-ALL linked to a clone with vector-activated LMO2, similar to the T-ALLs observed in the SCID-X1 trial. This occurrence clearly indicates that T-ALLs, even those linked to LMO2 activation, are not confined to SCID-X1 trials and do not require participation of the γC receptor transgene or rapid expansion of T cells into an empty T cell compartment, since none of these conditions existed in the WAS trial.
HSC gene therapy trial for X-ALD patients
X-linked adrenoleukodystrophy (X-ALD) is a fatal neurodegenerative disease caused by ABCD1 gene mutations, resulting in a shortage (deficiency) of adrenoleukodystrophy protein (ALDP) and an inability to process very long chain fatty acids [24]. Replacement of brain HSC-derived microglial cells via HSC gene therapy was hypothesized to be a potential therapeutic approach, and a pilot study using a replication-defective lentiviral vector expressing the ABCD1 gene to transducer autologous CD34+ cells enrolled two patients and was reported in 2009 [7]. Reconstitution with 9% to 14% of granulocytes, monocytes, and Tand B lymphocytes expressing the corrective protein was observed for the 24 to 30 months of follow-up. The progressive cerebral demyelination characteristic of ALD appeared to slow in the two patients [7]. Large-scale analysis of lentivirus IS revealed a high number of distinct ISs, indicating a consistently polyclonal distribution of lentivirally-corrected hematopoietic cells over time. As expected for lentiviruses, insertions were distributed mainly in gene coding regions without a particular preference for transcriptional start sites. There was no evidence for clonal expansion over time in either patient.
HSC gene therapy trial for β-thalassemia patients
The β-thalassemias are inherited disorders caused by mutations in the β-globin gene or its promoter/enhancer elements, characterized by absent or severely reduced β-globin protein production, and complete transfusion dependence for survival in β-thalassemia major. Gene therapy for hemoglobin disorders has been pursued for decades in the laboratory, because it conceptually holds great promise for these disorders, via permanent production of functional red blood cells from genetically-modified HSCs [25]. A clinical trial using an HIV-based lentiviral vector to express the corrective β-globin was initiated in 2007 [26]. An 18-year-old male patient with β(E)/β(o)-thalassemia received autologous transduced CD34+ cells following busulfan conditioning. One year after the treatment, the patient became transfusion-independent with increased levels of β-globin. This improvement was stable for at least 33 months of follow-up. Approximately 11% of circulating nucleated cells contained the vector; however unlike the polyclonal pattern seen in ALD following HIV-based gene transfer, in this patient clonal dominance of myeloid/erythroid cells containing an insertion in the HMGA2 developed. Most of the therapeutic benefit resulted from erythropoiesis originating from this dominant clone. Although abnormal HMGA2 expression has been implicated as a potential oncogenic stimulus, the authors note that increased levels of the protein were present in only 5% of all circulating hematopoietic cells, and that there was no evidence of further expansion or a malignant or pre-malignant state. A splicing event between the vector globin transcript and the HMGA2 locus resulted in increased expression of a truncated HMGA2 transcript insensitive to normal let-7 microRNA-mediated degradation. In a recent murine model, the truncated Hmga2 transcript has been shown to confer a clonal growth advantage [27]. It remains unclear whether this was an extraordinarily rare outcome of a single integration and splicing event in a single myeloid progenitor that will be unlikely to progress to leukemia, and unlikely to occur in additional patients, or whether this is more concerning event that indicates these safety-modified lentiviruses will also result in an unacceptable rate of genotoxicity.
Methodology for tracking virus insertion sites
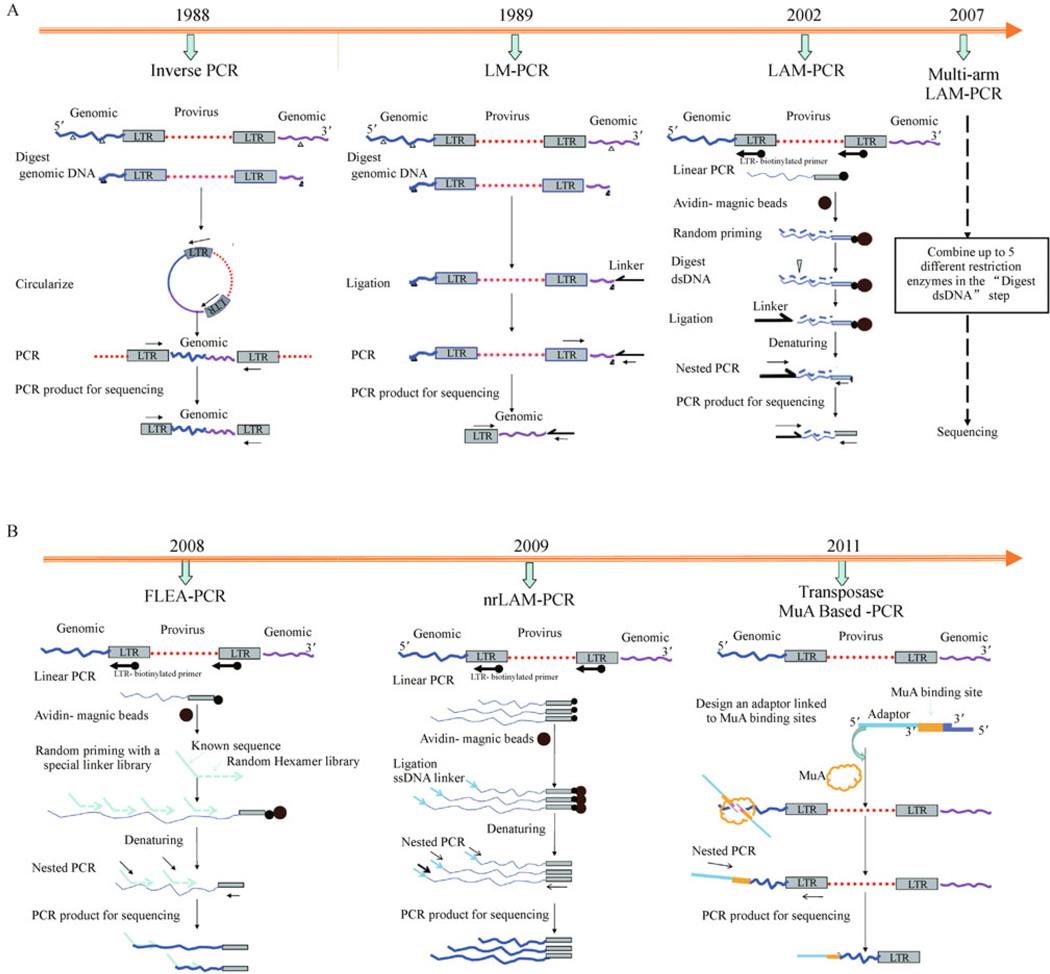
Several different classes of integrating viral vectors have been used in experimental and clinical gene therapy studies to achieve stable expression of corrective genes in HSCs and their progeny. However, an integrated vector provirus can influence the expression of adjacent genes, up to 50–100 kb away, and confer an altered phenotype on the gene-modified target cell. As summarized above, clinical gene therapy protocols utilizing integrating vectors have already been shown to result in both clinical improvement, but also in insertional activation of proto-oncogenes and resulting malignant or premalignant uncontrolled clonal expansions in several patients with SCID-X1, WAS, and X-CGD [5,10,16]. It is thus very important to be able to identify and track proviral integration sites in individual transduced clones after transduction and following engraftment with the transduced HSCs. It is even more important to identify IS in the setting of clonal dominance or transformation to vector-associated malignancies, in order to gain deeper insights into vector-host cell interactions and to understand and potentially avoid genotoxicity [22]. Prior to the sequencing of the entire human and murine genomes, identification of integration sites had been difficult and not particularly fruitful, since integrating retroviruses do not appear to target a particular sequence motif or a small set of genes. However, the availability of the draft complete human genome sequence in 2001 opened the door to detailed IS analysis. Several methods for identifying and tracking vector IS have been developed over the past two decades. Here we summarize the different methods (as shown in Fig. 1) and discuss their efficiency, sensitivity and biases. All rely on working out from known sequences in the proviral integrated genome into the unknown adjacent eukaryotic genomic DNA, isolating the junction fragment, amplifying it and sequencing it.
Fig. 1.
Schematic diagrams for summarizing the different methods available to identify proviral insertion sites. (A)Methods requiring restriction enzyme digestion adjacent to insertion sites, including inverse PCR, LM-PCR, LAM-PCR and multi-arm optimized LAMPCR. (B) Methods without restriction enzyme digestion, including FLEA-PCR, nrLAM-PCR and transposase MuA based-PCR. (Timeline shows the year when each method was first described.)
Inverse polymerase chain reaction (IPCR)
In 1988, Howard et al. published a method for in vitro amplification of unknown DNA sequences that flank a region of known sequence [28]. A frequent cutting restriction enzyme is used to cleave genomic DNA into small fragments, which are then ligated into circles. In contrast to conventional PCR used to amplify known DNA fragments, in inverse PCR primers annealing to known sequences in the LTR are oriented in the reverse direction, away from each other, and used to amplify the circularized DNA, including the LTR-genomic DNA junction. Amplified DNA can then be sequenced. This was the first reported approach for isolating genomic integration sites; however, it has a number of drawbacks. It is inefficient and insensitive, since the vast majority of fragmented DNA does not contain any LTR sequences and is not amplified, but acts as a sink for ligase and other reaction components. Some fragments may be too small or too large to be efficiently ligated into circles, and many circles may be too large to allow amplification. Differences in the size of the circles, and thus the size of the amplified fragment, greatly impact on the likelihood of detecting any specific insertion site. The dependence on restriction enzyme digestion also imparts bias, via all the issues listed above in terms of highly variable fragment lengths. However, the technique is quite simple, and has been successfully utilized for sophisticated IS retrieval following HSC gene transfer in non-human primates and murine tumors [29,30].
Ligation-mediated PCR (LM-PCR)
In 1989, ligation-mediated PCR (LM-PCR) was first described [31]. Genomic DNA is digested with frequent-cutting restriction enzymes, similar to the first step in inverse PCR. However, instead of circularizing the DNA, a linker is ligated to the genomic end of the cleaved DNA, and PCR amplification using one primer annealing to the linker and another to the vector LTR results in amplification of the vector-genome junction, followed by sequencing. This method has been adopted and modified by Kustikova et al. in 2008 for analyzing retroviral IS in HSCs [32].This method has restriction enzyme bias, and the linker ligation step is very inefficient, since the vast majority of DNA “ends” are not on fragments containing vector. This methodology has worked well to identify IS in clonal or oligoclonal samples, with vector copy numbers of 0.10–0.20 or higher [33], but most clinical samples have much lower copy numbers and it is not sufficiently sensitive or efficient to retrieve IS effectively in this setting.
Linear amplification-mediated polymerase chain reaction (LAM-PCR)
A major advance in IS retrieval occurred one decade ago, when LM-PCR was modified by von Kalle and coworkers to greatly increase sensitivity and efficiency in a method termed “linear amplification mediated PCR” or LAM-PCR [34–36]. Efficiency and sensitivity was greatly increased by adding in a linear amplification step, using a biotinylated primer annealing to the LTR in the orientation out toward the junction with genomic DNA, and then purifying these amplified fragments away from non-vector-containing DNA via strepavidin-coated magnetic beads. These purified fragments are converted to dsDNA by random hexamer priming, digested with a restriction enzyme(s), typically frequent-cutting enzymes with a 4 bp recognition site, and then a double-stranded linker is ligated to the ends. Exponential nested PCR using primers annealing to the linker and to the LTR (identical to LM-PCR) is followed by direct sequencing, shotgun cloning into bacterial plasmids and sequencing or pyrosequencing. The technique is technically challenging and labor intensive.
LAM-PCR allows isolation and identification of large numbers of IS in highly polyclonal samples, as demonstrated in numerous publications reporting IS data from clinical trials and preclinical primate studies [5,9,15,16,37]. It is very sensitive, down to close to a single cell level. It is much more efficient, in terms of “signal to noise (PCR and cloning artifacts) when the overall level of vector copy number is at least 0.01, in our experience. Relatively small amounts of DNA are sufficient for most analyses (100–500 ng), allowing analysis of sorted cell populations for tracking of clones in specific lineages over time. We would stress that simply running out LAM-PCR products on a thin gel and seeing multiple bands is not sufficient to document polyclonality or come to other conclusions regarding IS, but that shotgun sequencing or pyrosequencing and actual identification of IS is required for rigorous analysis.
But LAM-PCR also has limitations. It is impossible to detect the entire constellation of vector integration sites using any method that relies on restriction enzyme cutting, since some IS occur either too close or too far from any specific restriction enzyme site, resulting in fragments that are too small to resolve, or alternatively, too long to be amplified, thus limiting the analysis to a subset of clones in a mix [38,39]. For instance, frequent-cutting enzymes recognizing AATT motifs can only access to 54.5% of all possible integrations in the human genome. A combination of the 5 most potent four cutter restriction enzymes gives access to 88.7% of the analyzable genome; however, performing LAM-PCR with 5 different enzymes is even more labor intensive, expensive, and impractical in terms of the amounts of DNA required [39]. Like inverse PCR, there is little evidence that LAM-PCR is sufficiently quantitative to draw conclusions regarding the relative frequencies of individual clonal contributions to a population of cells, short of very marked skewing, usually confirmed by Southern blot or allele-specific PCR. This is due to differences in efficiency of ligation and amplification depending on fragment length and potentially chromatin characteristics. In other words, if an individual IS clone represents 20% of contigs identified by shotgun cloning following LAM-PCR, this does not mean that the clone is truly present in 20% of the starting cell population.
The power of any IS retrieval methodology has been greatly increased by replacing shotgun cloning and then sequencing of inverse PCR or LAM-PCR products with high throughput direct sequencing methodologies such as 454 pyrosequencing [40], Instead of at most hundreds of IS being able to be isolated and identified by an army of laboratory members over a time period of months to years, these new sequencing approaches can generate thousands of IS in a day or two [41].
FLEA-PCR
Another nonrestrictive method was reported by Pule et al. [42], termed flanking-sequence exponential anchored-polymerase chain reaction (FLEA-PCR). In contrast to standard LAM-PCR, following the linear PCR step, primers containing a known sequence and then a random sequence library are used for priming the single-stranded DNA, followed by exponential nested PCR and sequencing. This method should be able to decrease bias and allow more consistent and possibly quantitative detection of vector integration sites. But the efficiency and sensitivity of these methods need to be tested, in combination with high-throughput 454 pyrosequencing.
Nonrestrictive linear-amplification-mediated PCR (nrLAM-PCR)
To avoid the biases related to restriction enzymes cutting in the methods described above, a nonrestrictive linear-amplification-mediated PCR (nrLAM-PCR) was recently developed by von Kalle’s group [34,36,39]. This method starts identically to LAM-PCR, using a biotinylated LTR-specific primer for the linear amplification step. But instead of converting ssDNA to dsDNA after linear PCR and bead enrichment, and then cutting with a restriction enzyme, the single-stranded DNA fragments containing the provirus-genome junctions are ligated to a linker oligonucleotide without any restriction enzyme cutting, and exponential nested PCR is then followed by high-throughput pyrosequencing. Because the single-stranded ligation step in nrLAM-PCR is less efficient than ligation to dsDNA in standard LAM-PCR, the sensitivity of nrLAM-PCR is lower, and larger DNA samples are required.
Transposase MuA based method
Recently, Brady et al. reported a new method based on phage Mu transposition for tracking virus integration sites [43]. This method uses the bacterial transposase MuA to introduce adaptors into genomic DNA to allow PCR amplification. There are no restriction enzyme or ligation steps and it is quick and simple. It appears to recover integration sites in a near random fashion, and provides at least a rough measure of clonal abundance.
Distribution of integration sites for different classes of vectors
Human gene therapy clinical studies provide important information about gene therapy efficiency and genotoxicity of viral vectors. However, information is limited due to the small number and size of the clinical studies, and potential impact of the underlying disease on the clonal reconstitution pattern. Therefore, integration analyses of viral vectors in cell lines and in animal models have been very helpful to assess vector genotoxicity preclinically, and to investigate IS distribution of different vectors. Before the sequence of the human genome became available a decade ago, it was impossible to study IS distribution. Soon after the genome was published, the first large scale analysis of HIV lentivirus IS in a human T cell line was published by using LM-PCR [44]. There was an unexpectedly strong bias for HIV IS within genes, particularly transcribed genes that were activated in cells after infection by HIV-1. Comparative studies of IS patterns for other viral vectors such as MLV, avian sarcoma leukosis virus (ASLV) and foamy viruses were then performed in various cell lines [45–48]. In a landmark study from Wu et al., 903 MLV integrations and 379 HIV-1 integrations were mapped after transduction and short-term culture of a human cell line, demonstrating that MLV retroviruses integrated near the start of transcriptional units whereas HIV-1 was again shown to preferentially integrated anywhere within the transcriptional unit, but not upstream of the transcriptional start site [47]. Avian sarcoma leukosis virus and foamy viruses were shown to have the weakest preferences for transcriptional units [45,46].
Both vector and target cell characteristics can impact on integration patterns. Retroviral vector (RV) long-terminal repeat (RV-LTR) strong promoter/enhancer elements can change the expression levels of nearby host genes, The mechanisms of the RV-LTR in oncogenesis were reviewed recently by Dr. Trobridge [49]. For MLVand HIV vectors, the pattern of gene activation influences the loci most likely to be preferred integration sites, therefore the activation status of the cell, the target cell type (HSC versus lymphocyte for instance), and cell culture conditions during transduction, would be expected to influence integration profiles via epigenetic and transcriptional pathways [45,50–52]. Viral factors including the gag component of the integrase and interaction of the provirus or viral proteins with target cell proteins such as lens epithelium-derived growth factor (LEDGF) also play crucial roles in controlling integration profiles [53–57]. The interactions of the host cell genome and integrating viruses still require further investigation to fully characterize the factors influencing viral vector integration patterns.
One common approach for evaluating genotoxicity in vitro has been developed based on high level transduction of primary murine bone marrow cells, expansion, and then limiting dilution plating to detect immortalized clones [58]. This technique was developed based on the earlier observation by Du et al. that immortalized myeloid cells arose following transduction with MLV vectors containing only a marker gene, and that these clones contained vector insertions activating expression of Evi1 or related proto-oncogenes [59]. Using this approach, the authors showed that self-inactivating (SIN) MLV vectors using a strong internal enhancer/promoter may also transform cells by insertional mutagenesis, but that the transforming capacity was significantly reduced compared with standard LTR containing vectors [58]. This method was also used to compare genotoxicity of other classes of various viral vectors [60,61].
Insights from HSC gene transfer animal models
As early as 1992, there was evidence that integration of proviruses into the genome of hematopoietic stem and progenitor cells could result in tumors. Rhesus macaques transplanted with CD34+ cells transduced with MLV vectors developed T cell leukemia/lymphoma, but these animals were found to be viremic with replication-competent recombinant retroviruses that arose in the producer cell line, and the tumors were attributed to scores of integration events arising in this unusual situation [62]. Regulatory agencies and investigators focused on ensuring that producer cell lines were not contaminated with replication-competent recombinant viruses, and the assumption was that the risk of insertional mutagenesis was extremely low. In 2002, the first report was published regarding a murine leukemia arising in an animal receiving HSCs transduced with a replication-defective retroviral vector. The leukemic insertion site was near the Evi1 gene [33,63]. However, the short lifespan of mice and relatively low mutagenesis risk detected using clinical vectors hampered studies of genotoxicity in normal mice. Tumorprone Cdkn2a−/− mice, which are particularly susceptible to cancer-triggering genetic lesions due to the presence of predisposing genetic lesions in all somatic cells, proved very useful to study genotoxicity of retroviral and lentiviral vectors [64,65]. The results from this mouse model suggested that retroviral vectors triggered leukemia/lymphomas contingent on LTR enhancer activity in a dose dependent manner; in contrast, lentiviral vectors seem relatively safe even with a higher integration load. IS enrichment in oncogenes and cell cycle related genes was found in retroviral vectors but not in lentiviral vector insertion patterns. This mouse model was also used to evaluate genotoxicity of vectors with removal of enhancer elements (SIN γ-retroviral vectors), and greatly reduced genotoxicity of these modified vectors was confirmed [66].
The downside to all murine models is the fact that mice are short-lived animals, and transplantation of cells transduced with most vectors containing therapeutic genes into normal mice has generally not resulted in tumors within their life span. One approach to accelerate genotoxicity in the mouse has been to perform serial transplants, with progression to clonal hematopoiesis and leukemia in secondary and tertiary transplant recipients, presumably due to much more intense selective pressure for activated proto-oncogenes in a setting requiring rapid HSC expansion [33]. However, there are numerous differences between murine and human hematopoiesis, and the serial transplant murine models still require almost a year of follow-up. Even though there are a lot of disadvantages of using mouse models for preclinical gene therapy development, they can help to assess HSC gene therapy efficacy in disease models, and give relevant insights into safety. Many reports have used a humanized mouse model to engraft transduced human long-term repopulating cells as another relevant approach to optimize gene therapy technology or test gene therapy efficiency [67–70].
However, we believe that large animal models will also be desirable to fully evaluate genotoxicity in preclinical gene therapy studies. Dogs and non-human primates have been investigated and appear to be predictive models [71]. Dogs are relatively easy to handle, and inbreeding has produced a number of models for human inherited genetic diseases, including α-l-iduronidase deficiency, SCID-X1, canine leukocyte adhesion deficiency (CLAD) and pyruvate kinase deficiency [72–75]. HSC gene therapy has been tested in dog models showing phenotypic correction of canine SCID-X1 and CLAD, which provided important information for clinical studies [76,77]. In a dog model comparing IS of long-term repopulating cells transduced with γ-retroviral vectors, lentiviral vectors or foamy viral vectors [78]. γ-retroviral vectors showed a high frequency of IS close to transcription start sites. Also, γ-retroviral proviruses were found more frequently within and close to proto-oncogene transcription sites than lentiviral or foamy vectors. These data confirm that this retroviral system may be the most prone to risky gene activation.
Compared with dog models, nonhuman primates have a closer genetic relationship to humans, and results from these models better predict outcome in human gene therapy trials [71]. Genotoxicity related study results from nonhuman primates are the most relevant data available to help assess the risk of insertional mutagenesis associated with viral vector gene transfer in humans. We followed 42 rhesus macaques, 23 baboons, and 17 dogs with significant levels of gene transfer for a median of 3.5 years with marker or drug-resistance genes containing retroviral vectors transduced CD34+ cells. In this study, no evidence of progression toward oligoclonal or monoclonal hematopoiesis was observed [79]. However, 5 years after transplantation, one rhesus macaque developed a fatal myeloid sarcoma, a type of acute myeloid leukemia. Analysis of the tumor showed two clonal vector insertions, and one was in the anti-apoptotic gene BCL2-A1 [80].
Our laboratory also compared genomic integration sites of the widely used γ-retroviral vector MLV and a lentiviral vector simian immunodeficiency virus (SIV) vector in nonhuman primates. MLV or SIV transduced CD34+ cells were transplanted and recipients followed for 6 months to 6 years. MLV integrants were located predominantly around transcription start sites while SIV integrants strongly favored transcription units and gene-dense regions of the genome [37]. Insertions in the MDS1/EVI1 region were detected at a very high frequency with MLV but not SIV following HSC transduction in primates, although thus far we have not observed progression to abnormal hematopoiesis or leukemia resulting from in vivo clonal expansion of the MDS1/EVI1 populations [81,82]. In vitro expansion of transduced cells prior to transplantation resulted in more marked MDS1/EVI1 clonal dominance [83].
Our group also investigated the use of ASLV vectors in rhesus long-term repopulating cells. Compared with MLV and SIV vectors, ASLV vector integration was non-clustered, did not favor gene-rich regions or transcription start sites, despite a weak preference for gene-coding regions [46]. No insertions close to or within the MDS1/EVI1 locus were found in vivo utilizing ASLV vectors. Moreover, ASLV LTRs do not have detectable promoter and enhancer activity [84,85] in mammalian cells. These data suggests that optimized vectors based on ASLV could be useful and safe for gene therapy applications.
Impact of target cell characteristics
Compared with HSC targets, gene transfer into mature T cells appears less risky at least in regards to genotoxicity. In human clinical trials utilizing MLV vectors to transducer mature T cells, malignant transformation has not been observed even with 10 years follow-up [86–88]. These observations suggest target cell characteristics may influence the genotoxicity of integrating viral vectors. To clarify this issue, Newrzela et al. directly compared susceptibility of mature murine T cells and HSCs to transformation after retroviral gene transfer of potent T cell oncogenes [89]. Mice receiving transduced HSCs all developed Tcell lymphoma/leukemia; in contrast, none of the mice that received T cell transplants transduced with the same vectors developed leukemia/lymphoma, despite persistence of gene modified cells. The difference might be explained by the dependence of pre-malignant mature T cell clones on major histocompatibility complex (MHC) self-peptide interactions for survival, which would be restricted by the size of the corresponding MHC/self-peptide niche. Proto-oncogenes responsible for self-renewal in HSCs may be accessible to vector insertion in HSCs, in contrast to T cells.
Further directions for minimizing the risks of genotoxicity
Vector design and modification
In most human gene therapy trials, γ-retroviral vectors were used to transfer therapeutic genes to autologous hematopoietic cells. However, serious adverse events, especially 5 cases of leukemia in the SCID-X1 trial and 2 cases of MDS in the X-CGD trial, have raised strong reservations regarding the further use of these vectors, due to their apparent high genotoxic risk [13]. Powerful enhancer elements within the γ-retroviral long-terminal repeats (LTRs) of these vectors can activate the transcription of nearby proto-oncogenes. There is hope that lentiviral vectors may be less genotoxic [64], based on their integration patterns: lentiviral vectors integrate randomly in entire active transcribed genes while γ-retroviral vectors prefer promoter and enhancer regions and many lentiviruses therefore may be more likely to inactivate than activate genes [37,47]. All lentiviral vectors under clinical development have their LTR enhancers deleted, in order to decrease the risk of recombination with wild-type HIV in a patient, but the added benefit of this design may be much lower risk for activation of adjacent genes. However, as detailed above, despite these safety features and the encouraging highly polyclonal integration pattern of lentiviral vectors in the X-ALD trial, the single patient in the β-thalassemia trial developed marked clonal dominance of cells with lentiviral vector-induced aberrant splicing and activation of the HMGA2 gene [26]. No integrating vector can be considered completely safe regarding insertional activation of proto-oncogenes or inactivation of tumor suppressor genes or mirRNAs. All vectors need to be assessed for their relative risk, and as many safety modifications as possible incorporated based on results in preclinical testing.
Ongoing trials for SCID-X1 utilize SIN γ-retroviral vectors with removal of LTR promoter/enhancers, and instead use of an internal elongation factor (EF)-1α cellular promoter to direct transgene expression. A second trial has been proposed utilizing a SIN lentiviral vector that incorporates an insulator element to decrease activation of adjacent genes, and also exploits the EF-1α promoter to drive gene expression. The comparison of clinical outcomes and insertion-site profile in these two trials will provide very instructive safety information on these two safety-modified vectors [90].
There are also several alternative virus vectors under development. The ASLV vector is replication-incompetent in mammalian cells, with a promoter and enhancer in the LTR selected for optimal expression in avian cells. Genome-wide analyses of ASLV integration sites were done both in cell lines and in vivo in our rhesus macaques HSC transplantation model. ASLV integration sites are distributed broadly in the human genome [48]. Despite a weak preference toward gene-coding regions [48,84], ASLV integration is non-clustered, and does not favor gene-rich regions, transcription start sites nor CpG islands. There was no propensity for ASLV insertions within or near proto-oncogenes, and most importantly, no insertions close to or within the MDS1/EVI1 locus, which is in contrast to the significant over-representation of this insertion site for MLV vectors in the same transplantation model [48]. The combination of these features is unique for ASLVand suggests that optimized vectors based on this virus could be useful and safe for gene transfer to HSCs and progenitor cells.
Another alternative vector is based on the Foamy virus (FV). Analysis of FV vector integration sites in vitro and in hematopoietic repopulating cells of dogs demonstrated a unique integration profile and lack of preferential integration within genes, despite a modest preference for integration near transcription start sites and a significant preference for CpG islands. The genome wide distribution of FV vector proviruses was nonrandom, with both clusters and gaps. Transcriptional profiling showed that gene expression had little influence on integration site selection [45,91], suggesting that FV vector may be safer alternatives to γ-retroviruses or lentiviral vectors. Several recent reviews summarize these and other approaches to reducing genotoxicity via redesign of current vectors or development of completely novel vectors [49,92].
Optimization of ex vivo cell culture conditions and cytokine combinations
A very important step in HSC gene therapy is the required ex vivo cell culture of target cells during transduction. The ex vivo cell culture conditions and cytokine combinations are critical to maintain hematopoietic stem/progenitor cell activity and can affect critical behaviors including homing, engraftment and risk of genotoxicity. It has been reported that murine and human cells cultured for prolonged period ex vivo in the presence of hematopoietic cytokines which are necessary to induce cell proliferation have decreased stem cell activity in vivo [93,94]. A previous in vitro study with rhesus CD34+ cells indicated that after 4 days of stimulatory culture in stem cell factor (SCF), megakaryocyte growth and development factor (MDGF), and flt3 ligand (FLT), transfer of the cells to SCF alone on retronectin (FN) support resulted in decreased active cycling and a halt to proliferation, without a loss of viability or induction of apoptosis, and improved engraftment [95]. Our group’s recent studies showed that prolonged ex vivo expansion of retrovirally transduced primate CD34+ cells resulted in overrepresentation of clones with MDS1/EVI1 insertion sites in the myeloid lineage after transplantation which indicated that prolonged ex vivo expansion of transduced cells may increase the risk of genotoxicity [83]. In vitro and in vivo studies for optimizing ex vivo cell culture conditions and cytokine combinations are important for modified HSC gene therapy clinical protocol to reduce genotoxicity.
Co-expression of suicide gene in therapeutic vector transduced HSC
Including an inducible suicide gene in a therapeutic vector has been proposed as a strategy for reducing risk by providing a means to eliminate vector-containing cells that are producing toxicities, including neoplastic transformation [96]. A number of suicide gene systems have been investigated, all relying on specific killing of vector-containing cells following administration of an activator drug, which is not toxic in the absence of the transgene, and the best developed system utilized the herpes thymidine kinase (TK) gene, which converts ganciclovir to a DNA replication-terminator, and preferentially kills cells expressing the suicide gene. This suicide gene has been used successfully to allow deletion of graft-versus-host disease producing vector-containing donor T cells in the allogeneic transplantation setting [97–99]. However, drug administration may select for cells harboring a suicide gene that has undergone an inactivating mutation, resulting in ganciclovir-resistant cells. Tumor cells could escape suicide gene toxicity by genetic and epigenetic instability, i.e. gene silencing or loss of the suicide gene resulting from recombination, chromosomal deletion, or chromosomal loss [100]. An in vivo tumor cell model showed that a double suicide gene strategy, with a vector containing both thymidine kinase and cytosine deaminase genes, were required for reliable elimination of tumor cells; neither single suicide gene system was effective alone [101]. A very potent new suicide gene is the inducible Caspase 9 system, which can be activated by a small molecule specific chemical inducer of dimerization to result in apoptosis [102–104]. Recently, a study has shown that an inducible suicide gene system including both Caspase 9 and yeast cytosine deaminase (YCD) can efficiently and specifically induce apoptosis of transduced induced pluripotent stem cells (iPSC) [105]. After optimizing, a suicide gene strategy may provide an added measure of safety for HSC-directed gene therapy utilizing integrating vectors, since no integrating system can be guaranteed safe regarding insertional mutagenesis.
Gene and cell therapy using safe harbor iPSC clone
Induced pluripotent stem cell (iPSC) technology has the potential to revolutionize regenerative medicine, disease modeling and drug discovery. Yamanaka and coworkers first succeeded in generating iPSC by reprogramming fibroblasts with four transcription factors conferring pluripotency and an embryonic phenotype, Oct4, Sox2, Klf4, and c-Myc [106,107]. Instead of gene therapy targeting HSC, it is possible that iPSC could be an alternative target for correction and production of therapeutic hematopoietic progeny cells. Recent studies have explored the potential of iPSC generation combined with gene and cell therapy for disease treatment in mice and humans [108,109]. In a humanized sickle cell anemia mouse model, hematopoietic progenitors obtained in vitro from autologous iPSC and containing a normal human sickle hemoglobin allele could correct the disease phenotype after transplantation [108]. Another in vitro study showed that corrected Fanconi-anemia-specific iPSC can give rise to disease-free hematopoietic progenitors of the myeloid and erythroid lineages [109]. There is concern about genotoxicity related to using integrating retroviruses carrying proto-oncogenes for reprogramming, due to insertional mutagenesis of retro- or lenti-viral reprogramming methods. A recent report showed that iPSC-like cells could be generated simply by very high level transduction of target cells, even without utilization of transcription factors [110], although IS profiles of iPSCs have not shown concerning common integration sites [111]. Even transient expression of the transcription factor transgenes may predispose to genetic or epigenetic changes predisposing to malignancy, like c-Myc, one of the factors used for reprogramming is a proto-oncogene which is a tumor inducer and also epigenetic hotspots exist in iPSCs [110,112,113]. Non-integrating or excisable reprogramming methods have been developed to avoid some of these problems [111, 114–119]. Recently, Papapetrou et al. describe a strategy to genetically modify human iPSC at “safe harbor” sites in the genome. They used an excisable single polycistronic vector co-expressing Oct4, Klf4, c-Myc and Sox2 to generate β-thalassemia-patient iPSCs, and then transduced these cells with a corrective lentiviral β-globin vector. Five “safe harbor” criteria based on integration position relative to contiguous coding genes, microRNAs and ultra conserved regions were defined. They found about 10% of integrations of a lentivirally encoded β-globin transgene in β-thalassemia-patient iPSC clones met the “safe harbor” criteria and permitted high-level β-globin expression upon erythroid differentiation, with little risk of insertional mutagenesis [119]. This study defined a feasible strategy for the potential safe use of patient-specific iPSC combined with corrective gene therapy or introduction of a suicide gene.
Another gene therapy study using X-CGD patient specific iPSC cell with gp91phox deficiency was reported by Zou et al. [120]. Instead of using viruses to correct the X-CGD mutation, zinc finger nuclease-mediated gene targeting was used. A single-copy gp91phox therapeutic minigene was mediated into one allele of the “safe harbor” AAVS1 locus in X-CGD iPSCs, and resulted in sustained expression of gp91phox and substantially restored neutrophil ROS production with functional correction of neutrophils differentiated from the iPSCs. This study showed non-viral high-efficiency gene transfer/targeting represents alternative and promising approach for iPSC-based gene therapy.
Conclusions
Great progress has been made in the development of HSC mediated gene therapy over the past two decades. Understanding the factors that influence vector-mediated genotoxicity will allow for the future development of safer vectors and protocols, extending applications to diseases that are not immediately fatal. Appropriate models including in vitro cell lines, mouse models, and large animal models continue to be developed and refined in their predictive capabilities. Numerous methods for insertion site retrieval have been developed, and now permit large-scale rapid assessment of insertion site patterns. All these efforts should benefit patients who can best be cured with HSC gene therapy. The possibilities for utilizing iPSCs to generate corrected hematopoietic cells are an exciting recent development that could circumvent problems associated with HSC targets; however, there are numerous hurdles to overcome, most importantly inefficient production of hematopoietic cells from iPSCs, and ongoing safety concerns.
Footnotes
Competing interests: The authors have no conflict of interest to declare.
References
- 1.Cavazzana-Calvo M, Hacein-Bey S, de Saint Basile G, Gross F, Yvon E, Nusbaum P, Selz F, Hue C, Certain S, Casanova JL, Bousso P, Deist FL, Fischer A. Gene therapy of human severe combined immunodeficiency (SCID)-X1 disease. Science. 2000;288(5466):669–672. doi: 10.1126/science.288.5466.669. [DOI] [PubMed] [Google Scholar]
- 2.Hacein-Bey-Abina S, Hauer J, Lim A, Picard C, Wang GP, Berry CC, Martinache C, Rieux-Laucat F, Latour S, Belohradsky BH, Leiva L, Sorensen R, Debré M, Casanova JL, Blanche S, Durandy A, Bushman FD, Fischer A, Cavazzana-Calvo M. Efficacy of gene therapy for X-linked severe combined immunodeficiency. N Engl J Med. 2010;363(4):355–364. doi: 10.1056/NEJMoa1000164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aiuti A, Slavin S, Aker M, Ficara F, Deola S, Mortellaro A, Morecki S, Andolfi G, Tabucchi A, Carlucci F, Marinello E, Cattaneo F, Vai S, Servida P, Miniero R, Roncarolo MG, Bordignon C. Correction of ADA-SCID by stem cell gene therapy combined with nonmyeloablative conditioning. Science. 2002;296(5577):2410–2413. doi: 10.1126/science.1070104. [DOI] [PubMed] [Google Scholar]
- 4.Aiuti A, Cattaneo F, Galimberti S, Benninghoff U, Cassani B, Callegaro L, Scaramuzza S, Andolfi G, Mirolo M, Brigida I, Tabucchi A, Carlucci F, Eibl M, Aker M, Slavin S, Al-Mousa H, Al Ghonaium A, Ferster A, Duppenthaler A, Notarangelo L, Wintergerst U, Buckley RH, Bregni M, Marktel S, Valsecchi MG, Rossi P, Ciceri F, Miniero R, Bordignon C, Roncarolo MG. Gene therapy for immunodeficiency due to adenosine deaminase deficiency. N Engl J Med. 2009;360(5):447–458. doi: 10.1056/NEJMoa0805817. [DOI] [PubMed] [Google Scholar]
- 5.Ott MG, Schmidt M, Schwarzwaelder K, Stein S, Siler U, Koehl U, Glimm H, Kühlcke K, Schilz A, Kunkel H, Naundorf S, Brinkmann A, Deichmann A, Fischer M, Ball C, Pilz I, Dunbar C, Du Y, Jenkins NA, Copeland NG, Lüthi U, Hassan M, Thrasher AJ, Hoelzer D, von Kalle C, Seger R, Grez M. Correction of Xlinked chronic granulomatous disease by gene therapy, augmented by insertional activation of MDS1-EVI1, PRDM16 or SETBP1. Nat Med. 2006;12(4):401–409. doi: 10.1038/nm1393. [DOI] [PubMed] [Google Scholar]
- 6.Kang EM, Choi U, Theobald N, Linton G, Long Priel DA, Kuhns D, Malech HL. Retrovirus gene therapy for X-linked chronic granulomatous disease can achieve stable long-term correction of oxidase activity in peripheral blood neutrophils. Blood. 2010;115(4):783–791. doi: 10.1182/blood-2009-05-222760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cartier N, Hacein-Bey-Abina S, Bartholomae CC, Veres G, Schmidt M, Kutschera I, Vidaud M, Abel U, Dal-Cortivo L, Caccavelli L, Mahlaoui N, Kiermer V, Mittelstaedt D, Bellesme C, Lahlou N, Lefrère F, Blanche S, Audit M, Payen E, Leboulch P, l’Homme B, Bougnères P, Von Kalle C, Fischer A, Cavazzana-Calvo M, Aubourg P. Hematopoietic stem cell gene therapy with a lentiviral vector in X-linked adrenoleukodystrophy. Science. 2009;326(5954):818–823. doi: 10.1126/science.1171242. [DOI] [PubMed] [Google Scholar]
- 8.Boztug K, Schmidt M, Schwarzer A, Banerjee PP, Díez IA, Dewey RA, Böhm M, Nowrouzi A, Ball CR, Glimm H, Naundorf S, Kühlcke K, Blasczyk R, Kondratenko I, Maródi L, Orange JS, von Kalle C, Klein C. Stem-cell gene therapy for the Wiskott-Aldrich syndrome. N Engl J Med. 2010;363(20):1918–1927. doi: 10.1056/NEJMoa1003548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stein S, Ott MG, Schultze-Strasser S, Jauch A, Burwinkel B, Kinner A, Schmidt M, Krämer A, Schwäble J, Glimm H, Koehl U, Preiss C, Ball C, Martin H, Göhring G, Schwarzwaelder K, Hofmann WK, Karakaya K, Tchatchou S, Yang R, Reinecke P, Kühlcke K, Schlegelberger B, Thrasher AJ, Hoelzer D, Seger R, von Kalle C, Grez M. Genomic instability and myelodysplasia with monosomy 7 consequent to EVI1 activation after gene therapy for chronic granulomatous disease. Nat Med. 2010;16(2):198–204. doi: 10.1038/nm.2088. [DOI] [PubMed] [Google Scholar]
- 10.Hacein-Bey-Abina S, Von Kalle C, Schmidt M, McCormack MP, Wulffraat N, Leboulch P, Lim A, Osborne CS, Pawliuk R, Morillon E, Sorensen R, Forster A, Fraser P, Cohen JI, de Saint Basile G, Alexander I, Wintergerst U, Frebourg T, Aurias A, Stoppa-Lyonnet D, Romana S, Radford-Weiss I, Gross F, Valensi F, Delabesse E, Macintyre E, Sigaux F, Soulier J, Leiva LE, Wissler M, Prinz C, Rabbitts TH, Le Deist F, Fischer A, Cavazzana-Calvo M. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302(5644):415–419. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- 11.Leonard WJ. The molecular basis of X-linked severe combined immunodeficiency: defective cytokine receptor signaling. Annu Rev Med. 1996;47:229–239. doi: 10.1146/annurev.med.47.1.229. [DOI] [PubMed] [Google Scholar]
- 12.Sugamura K, Asao H, Kondo M, Tanaka N, Ishii N, Ohbo K, Nakamura M, Takeshita T. The interleukin-2 receptor gamma chain: its role in the multiple cytokine receptor complexes and T cell development in XSCID. Annu Rev Immunol. 1996;14(1):179–205. doi: 10.1146/annurev.immunol.14.1.179. [DOI] [PubMed] [Google Scholar]
- 13.Fischer A, Hacein-Bey-Abina S, Cavazzana-Calvo M. 20 years of gene therapy for SCID. Nat Immunol. 2010;11(6):457–460. doi: 10.1038/ni0610-457. [DOI] [PubMed] [Google Scholar]
- 14.Gaspar HB, Parsley KL, Howe S, King D, Gilmour KC, Sinclair J, Brouns G, Schmidt M, Von Kalle C, Barington T, Jakobsen MA, Christensen HO, Al Ghonaium A, White HN, Smith JL, Levinsky RJ, Ali RR, Kinnon C, Thrasher AJ. Gene therapy of X-linked severe combined immunodeficiency by use of a pseudotyped gammaretroviral vector. Lancet. 2004;364(9452):2181–2187. doi: 10.1016/S0140-6736(04)17590-9. [DOI] [PubMed] [Google Scholar]
- 15.Hacein-Bey-Abina S, Garrigue A, Wang GP, Soulier J, Lim A, Morillon E, Clappier E, Caccavelli L, Delabesse E, Beldjord K, Asnafi V, MacIntyre E, Dal Cortivo L, Radford I, Brousse N, Sigaux F, Moshous D, Hauer J, Borkhardt A, Belohradsky BH, Wintergerst U, Velez MC, Leiva L, Sorensen R, Wulffraat N, Blanche S, Bushman FD, Fischer A, Cavazzana-Calvo M. Insertional oncogenesis in 4 patients after retrovirus-mediated gene therapy of SCID-X1. J Clin Invest. 2008;118(9):3132–3142. doi: 10.1172/JCI35700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Howe SJ, Mansour MR, Schwarzwaelder K, Bartholomae C, Hubank M, Kempski H, Brugman MH, Pike-Overzet K, Chatters SJ, de Ridder D, Gilmour KC, Adams S, Thornhill SI, Parsley KL, Staal FJ, Gale RE, Linch DC, Bayford J, Brown L, Quaye M, Kinnon C, Ancliff P, Webb DK, Schmidt M, von Kalle C, Gaspar HB, Thrasher AJ. Insertional mutagenesis combined with acquired somatic mutations causes leukemogenesis following gene therapy of SCID-X1 patients. J Clin Invest. 2008;118(9):3143–3150. doi: 10.1172/JCI35798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davé UP, Jenkins NA, Copeland NG. Gene therapy insertional mutagenesis insights. Science. 2004;303(5656):333. doi: 10.1126/science.1091667. [DOI] [PubMed] [Google Scholar]
- 18.Woods NB, Bottero V, Schmidt M, von Kalle C, Verma IM. Gene therapy: therapeutic gene causing lymphoma. Nature. 2006;440(7088):1123. doi: 10.1038/4401123a. [DOI] [PubMed] [Google Scholar]
- 19.Blaese RM, Culver KW, Miller AD, Carter CS, Fleisher T, Clerici M, Shearer G, Chang L, Chiang Y, Tolstoshev P, Greenblatt JJ, Rosenberg SA, Klein H, Berger M, Mullen CA, Ramsey WJ, Muul L, Morgan RA, Anderson WF. T lymphocyte-directed gene therapy for ADA- SCID: initial trial results after 4 years. Science. 1995;270(5235):475–480. doi: 10.1126/science.270.5235.475. [DOI] [PubMed] [Google Scholar]
- 20.Kohn DB, Hershfield MS, Carbonaro D, Shigeoka A, Brooks J, Smogorzewska EM, Barsky LW, Chan R, Burotto F, Annett G, Nolta JA, Crooks G, Kapoor N, Elder M, Wara D, Bowen T, Madsen E, Snyder FF, Bastian J, Muul L, Blaese RM,Weinberg K, Parkman R. T lymphocytes with a normal ADA gene accumulate after transplantation of transduced autologous umbilical cord blood CD34+ cells in ADA-deficient SCID neonates. Nat Med. 1998;4(7):775–780. doi: 10.1038/nm0798-775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cassani B, Montini E, Maruggi G, Ambrosi A, Mirolo M, Selleri S, Biral E, Frugnoli I, Hernandez-Trujillo V, Di Serio C, Roncarolo MG, Naldini L, Mavilio F, Aiuti A. Integration of retroviral vectors induces minor changes in the transcriptional activity of T cells from ADA-SCID patients treated with gene therapy. Blood. 2009;114(17):3546–3556. doi: 10.1182/blood-2009-02-202085. [DOI] [PubMed] [Google Scholar]
- 22.Aiuti A, Cassani B, Andolfi G, Mirolo M, Biasco L, Recchia A, Urbinati F, Valacca C, Scaramuzza S, Aker M, Slavin S, Cazzola M, Sartori D, Ambrosi A, Di Serio C, Roncarolo MG, Mavilio F, Bordignon C. Multilineage hematopoietic reconstitution without clonal selection in ADA-SCID patients treated with stem cell gene therapy. J Clin Invest. 2007;117(8):2233–2240. doi: 10.1172/JCI31666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heyworth PG, Cross AR, Curnutte JT. Chronic granulomatous disease. Curr Opin Immunol. 2003;15(5):578–584. doi: 10.1016/s0952-7915(03)00109-2. [DOI] [PubMed] [Google Scholar]
- 24.Moser HW, Mahmood A, Raymond GV. X-linked adrenoleukodystrophy. Nat Clin Pract Neurol. 2007;3(3):140–151. doi: 10.1038/ncpneuro0421. [DOI] [PubMed] [Google Scholar]
- 25.Yannaki E, Emery DW, Stamatoyannopoulos G. Gene therapy for β-thalassaemia: the continuing challenge. Expert Rev Mol Med. 2010;12:e31. doi: 10.1017/S1462399410001626. [DOI] [PubMed] [Google Scholar]
- 26.Cavazzana-Calvo M, Payen E, Negre O, Wang G, Hehir K, Fusil F, Down J, Denaro M, Brady T, Westerman K, Cavallesco R, Gillet-Legrand B, Caccavelli L, Sgarra R, Maouche-Chrétien L, Bernaudin F, Girot R, Dorazio R, Mulder GJ, Polack A, Bank A, Soulier J, Larghero J, Kabbara N, Dalle B, Gourmel B, Socie G, Chrétien S, Cartier N, Aubourg P, Fischer A, Cornetta K, Galacteros F, Beuzard Y, Gluckman E, Bushman F, Hacein-Bey-Abina S, Leboulch P. Transfusion independence and HMGA2 activation after gene therapy of human β-thalassaemia. Nature. 2010;467(7313):318–322. doi: 10.1038/nature09328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ikeda K, Mason PJ, Bessler M. 3′UTR-truncated Hmga2 cDNA causes MPN-like hematopoiesis by conferring a clonal growth advantage at the level of HSC in mice. Blood. 2011;117(22):5860–5869. doi: 10.1182/blood-2011-02-334425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ochman H, Gerber AS, Hartl DL. Genetic applications of an inverse polymerase chain reaction. Genetics. 1988;120(3):621–623. doi: 10.1093/genetics/120.3.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim HJ, Tisdale JF, Wu T, Takatoku M, Sellers SE, Zickler P, Metzger ME, Agricola BA, Malley JD, Kato I, Donahue RE, Brown KE, Dunbar CE. Many multipotential gene-marked progenitor or stem cell clones contribute to hematopoiesis in nonhuman primates. Blood. 2000;96(1):1–8. [PubMed] [Google Scholar]
- 30.Suzuki T, Minehata K, Akagi K, Jenkins NA, Copeland NG. Tumor suppressor gene identification using retroviral insertional mutagenesis in Blm-deficient mice. EMBO J. 2006;25(14):3422–3431. doi: 10.1038/sj.emboj.7601215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mueller PR, Wold B. In vivo footprinting of a muscle specific enhancer by ligation mediated PCR. Science. 1989;246(4931):780–786. doi: 10.1126/science.2814500. [DOI] [PubMed] [Google Scholar]
- 32.Kustikova OS, Baum C, Fehse B. Retroviral integration site analysis in hematopoietic stem cells. Methods Mol Biol. 2008;430:255–267. doi: 10.1007/978-1-59745-182-6_18. [DOI] [PubMed] [Google Scholar]
- 33.Kustikova O, Fehse B, Modlich U, Yang M, Düllmann J, Kamino K, von Neuhoff N, Schlegelberger B, Li Z, Baum C. Clonal dominance of hematopoietic stem cells triggered by retroviral gene marking. Science. 2005;308(5725):1171–1174. doi: 10.1126/science.1105063. [DOI] [PubMed] [Google Scholar]
- 34.Schmidt M, Schwarzwaelder K, Bartholomae CC, Glimm H, von Kalle C. Detection of retroviral integration sites by linear amplification-mediated PCR and tracking of individual integration clones in different samples. Methods Mol Biol. 2009;506:363–372. doi: 10.1007/978-1-59745-409-4_24. [DOI] [PubMed] [Google Scholar]
- 35.Schmidt M, Zickler P, Hoffmann G, Haas S, Wissler M, Muessig A, Tisdale JF, Kuramoto K, Andrews RG, Wu T, Kiem HP, Dunbar CE, von Kalle C. Polyclonal long-term repopulating stem cell clones in a primate model. Blood. 2002;100(8):2737–2743. doi: 10.1182/blood-2002-02-0407. [DOI] [PubMed] [Google Scholar]
- 36.Schmidt M, Schwarzwaelder K, Bartholomae C, Zaoui K, Ball C, Pilz I, Braun S, Glimm H, von Kalle C. High-resolution insertionsite analysis by linear amplification-mediated PCR (LAM-PCR) Nat Methods. 2007;4(12):1051–1057. doi: 10.1038/nmeth1103. [DOI] [PubMed] [Google Scholar]
- 37.Hematti P, Hong BK, Ferguson C, Adler R, Hanawa H, Sellers S, Holt IE, Eckfeldt CE, Sharma Y, Schmidt M, von Kalle C, Persons DA, Billings EM, Verfaillie CM, Nienhuis AW, Wolfsberg TG, Dunbar CE, Calmels B. Distinct genomic integration of MLV and SIV vectors in primate hematopoietic stem and progenitor cells. PLoS Biol. 2004;2(12):e423. doi: 10.1371/journal.pbio.0020423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harkey MA, Kaul R, Jacobs MA, Kurre P, Bovee D, Levy R, Blau CA. Multiarm high-throughput integration site detection: limitations of LAM-PCR technology and optimization for clonal analysis. Stem Cells Dev. 2007;16(3):381–392. doi: 10.1089/scd.2007.0015. [DOI] [PubMed] [Google Scholar]
- 39.Gabriel R, Eckenberg R, Paruzynski A, Bartholomae CC, Nowrouzi A, Arens A, Howe SJ, Recchia A, Cattoglio C, Wang W, Faber K, Schwarzwaelder K, Kirsten R, Deichmann A, Ball CR, Balaggan KS, Yáñez-Muñoz RJ, Ali RR, Gaspar HB, Biasco L, Aiuti A, Cesana D, Montini E, Naldini L, Cohen-Haguenauer O, Mavilio F, Thrasher AJ, Glimm H, von Kalle C, Saurin W, Schmidt M. Comprehensive genomic access to vector integration in clinical gene therapy. Nat Med. 2009;15(12):1431–1436. doi: 10.1038/nm.2057. [DOI] [PubMed] [Google Scholar]
- 40.Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z, Dewell SB, Du L, Fierro JM, Gomes XV, Godwin BC, He W, Helgesen S, Ho CH, Irzyk GP, Jando SC, Alenquer ML, Jarvie TP, Jirage KB, Kim JB, Knight JR, Lanza JR, Leamon JH, Lefkowitz SM, Lei M, Li J, Lohman KL, Lu H, Makhijani VB, McDade KE, McKenna MP, Myers EW, Nickerson E, Nobile JR, Plant R, Puc BP, Ronan MT, Roth GT, Sarkis GJ, Simons JF, Simpson JW, Srinivasan M, Tartaro KR, Tomasz A, Vogt KA, Volkmer GA, Wang SH, Wang Y, Weiner MP, Yu P, Begley RF, Rothberg JM. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437(7057):376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang GP, Garrigue A, Ciuffi A, Ronen K, Leipzig J, Berry C, Lagresle-Peyrou C, Benjelloun F, Hacein-Bey-Abina S, Fischer A, Cavazzana-Calvo M, Bushman FD. DNA bar coding and pyrosequencing to analyze adverse events in therapeutic gene transfer. Nucleic Acids Res. 2008;36(9):e49. doi: 10.1093/nar/gkn125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pule MA, Rousseau A, Vera J, Heslop HE, Brenner MK, Vanin EF. Flanking-sequence exponential anchored-polymerase chain reaction amplification: a sensitive and highly specific method for detecting retroviral integrant-host-junction sequences. Cytotherapy. 2008;10(5):526–539. doi: 10.1080/14653240802192636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brady T, Roth SL, Malani N, Wang GP, Berry CC, Leboulch P, Hacein-Bey-Abina S, Cavazzana-Calvo M, Papapetrou EP, Sadelain M, Savilahti H, Bushman FD. A method to sequence and quantify DNA integration for monitoring outcome in gene therapy. Nucleic Acids Res. 2011;39(11):e72. doi: 10.1093/nar/gkr140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schröder AR, Shinn P, Chen H, Berry C, Ecker JR, Bushman F. HIV-1 integration in the human genome favors active genes and local hotspots. Cell. 2002;110(4):521–529. doi: 10.1016/s0092-8674(02)00864-4. [DOI] [PubMed] [Google Scholar]
- 45.Trobridge GD, Miller DG, Jacobs MA, Allen JM, Kiem HP, Kaul R, Russell DW. Foamy virus vector integration sites in normal human cells. Proc Natl Acad Sci USA. 2006;103(5):1498–1503. doi: 10.1073/pnas.0510046103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mitchell RS, Beitzel BF, Schroder AR, Shinn P, Chen H, Berry CC, Ecker JR, Bushman FD. Retroviral DNA integration: ASLV, HIV, MLV show distinct target site preferences. PLoS Biol. 2004;2(8):e234. doi: 10.1371/journal.pbio.0020234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu X, Li Y, Crise B, Burgess SM. Transcription start regions in the human genome are favored targets for MLV integration. Science. 2003;300(5626):1749–1751. doi: 10.1126/science.1083413. [DOI] [PubMed] [Google Scholar]
- 48.Narezkina A, Taganov KD, Litwin S, Stoyanova R, Hayashi J, Seeger C, Skalka AM, Katz RA. Genome-wide analyses of avian sarcoma virus integration sites. J Virol. 2004;78(21):11656–11663. doi: 10.1128/JVI.78.21.11656-11663.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Trobridge GD. Genotoxicity of retroviral hematopoietic stem cell gene therapy. Expert Opin Biol Ther. 2011;11(5):581–593. doi: 10.1517/14712598.2011.562496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Biasco L, Ambrosi A, Pellin D, Bartholomae C, Brigida I, Roncarolo MG, Di Serio C, von Kalle C, Schmidt M, Aiuti A. Integration profile of retroviral vector in gene therapy treated patients is cell-specific according to gene expression and chromatin conformation of target cell. EMBO Mol Med. 2011;3(2):89–101. doi: 10.1002/emmm.201000108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang GP, Ciuffi A, Leipzig J, Berry CC, Bushman FD. HIV integration site selection: analysis by massively parallel pyrosequencing reveals association with epigenetic modifications. Genome Res. 2007;17(8):1186–1194. doi: 10.1101/gr.6286907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brady T, Agosto LM, Malani N, Berry CC, O’Doherty U, Bushman F. HIV integration site distributions in resting and activated CD4+ T cells infected in culture. AIDS. 2009;23(12):1461–1471. doi: 10.1097/QAD.0b013e32832caf28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lewinski MK, Yamashita M, Emerman M, Ciuffi A, Marshall H, Crawford G, Collins F, Shinn P, Leipzig J, Hannenhalli S, Berry CC, Ecker JR, Bushman FD. Retroviral DNA integration: viral and cellular determinants of target-site selection. PLoS Pathog. 2006;2(6):e60. doi: 10.1371/journal.ppat.0020060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cherepanov P, Maertens G, Proost P, Devreese B, Van Beeumen J, Engelborghs Y, De Clercq E, Debyser Z. HIV-1 integrase forms stable tetramers and associates with LEDGF/p75 protein in human cells. J Biol Chem. 2003;278(1):372–381. doi: 10.1074/jbc.M209278200. [DOI] [PubMed] [Google Scholar]
- 55.Llano M, Delgado S, Vanegas M, Poeschla EM. Lens epitheliumderived growth factor/p75 prevents proteasomal degradation of HIV-1 integrase. J Biol Chem. 2004;279(53):55570–55577. doi: 10.1074/jbc.M408508200. [DOI] [PubMed] [Google Scholar]
- 56.Emiliani S, Mousnier A, Busschots K, Maroun M, Van Maele B, Tempé D, Vandekerckhove L, Moisant F, Ben-Slama L, Witvrouw M, Christ F, Rain JC, Dargemont C, Debyser Z, Benarous R. Integrase mutants defective for interaction with LEDGF/p75 are impaired in chromosome tethering and HIV-1 replication. J Biol Chem. 2005;280(27):25517–25523. doi: 10.1074/jbc.M501378200. [DOI] [PubMed] [Google Scholar]
- 57.Ciuffi A, Llano M, Poeschla E, Hoffmann C, Leipzig J, Shinn P, Ecker JR, Bushman F. A role for LEDGF/p75 in targeting HIV DNA integration. Nat Med. 2005;11(12):1287–1289. doi: 10.1038/nm1329. [DOI] [PubMed] [Google Scholar]
- 58.Modlich U, Bohne J, Schmidt M, von Kalle C, Knöss S, Schambach A, Baum C. Cell-culture assays reveal the importance of retroviral vector design for insertional genotoxicity. Blood. 2006;108(8):2545–2553. doi: 10.1182/blood-2005-08-024976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Du Y, Jenkins NA, Copeland NG. Insertional mutagenesis identifies genes that promote the immortalization of primary bone marrow progenitor cells. Blood. 2005;106(12):3932–3939. doi: 10.1182/blood-2005-03-1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zychlinski D, Schambach A, Modlich U, Maetzig T, Meyer J, Grassman E, Mishra A, Baum C. Physiological promoters reduce the genotoxic risk of integrating gene vectors. Mol Ther. 2008;16(4):718–725. doi: 10.1038/mt.2008.5. [DOI] [PubMed] [Google Scholar]
- 61.Modlich U, Navarro S, Zychlinski D, Maetzig T, Knoess S, Brugman MH, Schambach A, Charrier S, Galy A, Thrasher AJ, Bueren J, Baum C. Insertional transformation of hematopoietic cells by self-inactivating lentiviral and gammaretroviral vectors. Mol Ther. 2009;17(11):1919–1928. doi: 10.1038/mt.2009.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Donahue RE, Kessler SW, Bodine D, McDonagh K, Dunbar C, Goodman S, Agricola B, Byrne E, Raffeld M, Moen R. Helper virus induced T cell lymphoma in nonhuman primates after retroviral mediated gene transfer. J Exp Med. 1992;176(4):1125–1135. doi: 10.1084/jem.176.4.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li Z, Düllmann J, Schiedlmeier B, Schmidt M, von Kalle C, Meyer J, Forster M, Stocking C, Wahlers A, Frank O, Ostertag W, Kühlcke K, Eckert HG, Fehse B, Baum C. Murine leukemia induced by retroviral gene marking. Science. 2002;296(5567):497. doi: 10.1126/science.1068893. [DOI] [PubMed] [Google Scholar]
- 64.Montini E, Cesana D, Schmidt M, Sanvito F, Ponzoni M, Bartholomae C, Sergi Sergi L, Benedicenti F, Ambrosi A, Di Serio C, Doglioni C, von Kalle C, Naldini L. Hematopoietic stem cell gene transfer in a tumor-prone mouse model uncovers low genotoxicity of lentiviral vector integration. Nat Biotechnol. 2006;24(6):687–696. doi: 10.1038/nbt1216. [DOI] [PubMed] [Google Scholar]
- 65.Lund AH, Turner G, Trubetskoy A, Verhoeven E, Wientjens E, Hulsman D, Russell R, DePinho RA, Lenz J, van Lohuizen M. Genome-wide retroviral insertional tagging of genes involved in cancer in Cdkn2a-deficient mice. Nat Genet. 2002;32(1):160–165. doi: 10.1038/ng956. [DOI] [PubMed] [Google Scholar]
- 66.Montini E, Cesana D, Schmidt M, Sanvito F, Bartholomae CC, Ranzani M, Benedicenti F, Sergi LS, Ambrosi A, Ponzoni M, Doglioni C, Di Serio C, von Kalle C, Naldini L. The genotoxic potential of retroviral vectors is strongly modulated by vector design and integration site selection in a mouse model of HSC gene therapy. J Clin Invest. 2009;119(4):964–975. doi: 10.1172/JCI37630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Drake AC, Khoury M, Leskov I, Iliopoulou BP, Fragoso M, Lodish H, Chen J. Human CD34+ CD133+ hematopoietic stem cells cultured with growth factors including Angptl5 efficiently engraft adult NOD-SCID Il2rγ−/− (NSG) mice. PLoS ONE. 2011;6(4):e18382. doi: 10.1371/journal.pone.0018382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Frecha C, Fusil F, Cosset FL, Verhoeyen E. In vivo gene delivery into hCD34+ cells in a humanized mouse model. Methods Mol Biol. 2011;737:367–390. doi: 10.1007/978-1-61779-095-9_15. [DOI] [PubMed] [Google Scholar]
- 69.Joseph A, Zheng JH, Chen K, Dutta M, Chen C, Stiegler G, Kunert R, Follenzi A, Goldstein H. Inhibition of in vivo HIV infection in humanized mice by gene therapy of human hematopoietic stem cells with a lentiviral vector encoding a broadly neutralizing anti- HIV antibody. J Virol. 2010;84(13):6645–6653. doi: 10.1128/JVI.02339-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Spraul CW, Roth HJ, Gäckle H, Lang GE, Lang GK. Influence of special-effect contact lenses (Crazy Lenses) on visual function. CLAO J. 1998;24(1):29–32. [PubMed] [Google Scholar]
- 71.Trobridge GD, Kiem HP. Large animal models of hematopoietic stem cell gene therapy. Gene Ther. 2010;17(8):939–948. doi: 10.1038/gt.2010.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Felsburg PJ, Somberg RL, Perryman LE. Domestic animal models of severe combined immunodeficiency: canine X-linked severe combined immunodeficiency and severe combined immunodeficiency in horses. Immunodefic Rev. 1992;3(4):277–303. [PubMed] [Google Scholar]
- 73.Spellacy E, Shull RM, Constantopoulos G, Neufeld EF. A canine model of human alpha-L-iduronidase deficiency. Proc Natl Acad Sci USA. 1983;80(19):6091–6095. doi: 10.1073/pnas.80.19.6091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kijas JM, Bauer TR, Jr, Gäfvert S, Marklund S, Trowald-Wigh G, Johannisson A, Hedhammar A, Binns M, Juneja RK, Hickstein DD, Andersson L. A missense mutation in the beta-2 integrin gene (ITGB2) causes canine leukocyte adhesion deficiency. Genomics. 1999;61(1):101–107. doi: 10.1006/geno.1999.5948. [DOI] [PubMed] [Google Scholar]
- 75.Beard BC, Kiem HP. Canine models of gene-modified hematopoiesis. Methods Mol Biol. 2009;506:341–361. doi: 10.1007/978-1-59745-409-4_23. [DOI] [PubMed] [Google Scholar]
- 76.Ting-De Ravin SS, Kennedy DR, Naumann N, Kennedy JS, Choi U, Hartnett BJ, Linton GF, Whiting-Theobald NL, Moore PF, Vernau W, Malech HL, Felsburg PJ. Correction of canine X-linked severe combined immunodeficiency by in vivo retroviral gene therapy. Blood. 2006;107(8):3091–3097. doi: 10.1182/blood-2005-10-4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bauer TR, Jr, Hai M, Tuschong LM, Burkholder TH, Gu YC, Sokolic RA, Ferguson C, Dunbar CE, Hickstein DD. Correction of the disease phenotype in canine leukocyte adhesion deficiency using ex vivo hematopoietic stem cell gene therapy. Blood. 2006;108(10):3313–3320. doi: 10.1182/blood-2006-03-006908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Beard BC, Keyser KA, Trobridge GD, Peterson LJ, Miller DG, Jacobs M, Kaul R, Kiem HP. Unique integration profiles in a canine model of long-term repopulating cells transduced with gammaretrovirus, lentivirus, or foamy virus. Hum Gene Ther. 2007;18(5):423–434. doi: 10.1089/hum.2007.011. [DOI] [PubMed] [Google Scholar]
- 79.Kiem HP, Sellers S, Thomasson B, Morris JC, Tisdale JF, Horn PA, Hematti P, Adler R, Kuramoto K, Calmels B, Bonifacino A, Hu J, von Kalle C, Schmidt M, Sorrentino B, Nienhuis A, Blau CA, Andrews RG, Donahue RE, Dunbar CE. Long-term clinical and molecular follow-up of large animals receiving retrovirally transduced stem and progenitor cells: no progression to clonal hematopoiesis or leukemia. Mol Ther. 2004;9(3):389–395. doi: 10.1016/j.ymthe.2003.12.006. [DOI] [PubMed] [Google Scholar]
- 80.Seggewiss R, Pittaluga S, Adler RL, Guenaga FJ, Ferguson C, Pilz IH, Ryu B, Sorrentino BP, Young WS, 3rd, Donahue RE, von Kalle C, Nienhuis AW, Dunbar CE. Acute myeloid leukemia is associated with retroviral gene transfer to hematopoietic progenitor cells in a rhesus macaque. Blood. 2006;107(10):3865–3867. doi: 10.1182/blood-2005-10-4108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Calmels B, Ferguson C, Laukkanen MO, Adler R, Faulhaber M, Kim HJ, Sellers S, Hematti P, Schmidt M, von Kalle C, Akagi K, Donahue RE, Dunbar CE. Recurrent retroviral vector integration at the Mds1/Evi1 locus in nonhuman primate hematopoietic cells. Blood. 2005;106(7):2530–2533. doi: 10.1182/blood-2005-03-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kim YJ, Kim YS, Larochelle A, Renaud G, Wolfsberg TG, Adler R, Donahue RE, Hematti P, Hong BK, Roayaei J, Akagi K, Riberdy JM, Nienhuis AW, Dunbar CE, Persons DA. Sustained high-level polyclonal hematopoietic marking and transgene expression 4 years after autologous transplantation of rhesus macaques with SIV lentiviral vector-transduced CD34+ cells. Blood. 2009;113(22):5434–5443. doi: 10.1182/blood-2008-10-185199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sellers S, Gomes TJ, Larochelle A, Lopez R, Adler R, Krouse A, Donahue RE, Childs RW, Dunbar CE. Ex vivo expansion of retrovirally transduced primate CD34+ cells results in overrepresentation of clones with MDS1/EVI1 insertion sites in the myeloid lineage after transplantation. Mol Ther. 2010;18(9):1633–1639. doi: 10.1038/mt.2010.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hu J, Renaud G, Gomes TJ, Ferris A, Hendrie PC, Donahue RE, Hughes SH, Wolfsberg TG, Russell DW, Dunbar CE. Reduced genotoxicity of avian sarcoma leukosis virus vectors in rhesus long-term repopulating cells compared to standard murine retrovirus vectors. Mol Ther. 2008;16(9):1617–1623. doi: 10.1038/mt.2008.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hu J, Ferris A, Larochelle A, Krouse AE, Metzger ME, Donahue RE, Hughes SH, Dunbar CE. Transduction of rhesus macaque hematopoietic stem and progenitor cells with avian sarcoma and leukosis virus vectors. Hum Gene Ther. 2007;18(8):691–700. doi: 10.1089/hum.2006.175. [DOI] [PubMed] [Google Scholar]
- 86.Deeks SG, Wagner B, Anton PA, Mitsuyasu RT, Scadden DT, Huang C, Macken C, Richman DD, Christopherson C, June CH, Lazar R, Broad DF, Jalali S, Hege KM. A phase II randomized study of HIV-specific T-cell gene therapy in subjects with undetectable plasma viremia on combination antiretroviral therapy. Mol Ther. 2002;5(6):788–797. doi: 10.1006/mthe.2002.0611. [DOI] [PubMed] [Google Scholar]
- 87.Mitsuyasu RT, Anton PA, Deeks SG, Scadden DT, Connick E, Downs MT, Bakker A, Roberts MR, June CH, Jalali S, Lin AA, Pennathur-Das R, Hege KM. Prolonged survival and tissue trafficking following adoptive transfer of CD4zeta gene-modified autologous CD4(+) and CD8(+) T cells in human immunodeficiency virus-infected subjects. Blood. 2000;96(3):785–793. [PubMed] [Google Scholar]
- 88.Walker RE, Bechtel CM, Natarajan V, Baseler M, Hege KM, Metcalf JA, Stevens R, Hazen A, Blaese RM, Chen CC, Leitman SF, Palensky J, Wittes J, Davey RT, Jr, Falloon J, Polis MA, Kovacs JA, Broad DF, Levine BL, Roberts MR, Masur H, Lane HC. Long-term in vivo survival of receptor-modified syngeneic T cells in patients with human immunodeficiency virus infection. Blood. 2000;96(2):467–474. [PubMed] [Google Scholar]
- 89.Newrzela S, Cornils K, Li Z, Baum C, Brugman MH, Hartmann M, Meyer J, Hartmann S, Hansmann ML, Fehse B, von Laer D. Resistance of mature T cells to oncogene transformation. Blood. 2008;112(6):2278–2286. doi: 10.1182/blood-2007-12-128751. [DOI] [PubMed] [Google Scholar]
- 90.Persons DA. Lentiviral vector gene therapy: effective and safe? Mol Ther. 2010;18(5):861–862. doi: 10.1038/mt.2010.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Nowrouzi A, Dittrich M, Klanke C, Heinkelein M, Rammling M, Dandekar T, von Kalle C, Rethwilm A. Genome-wide mapping of foamy virus vector integrations into a human cell line. J Gen Virol. 2006;87(Pt 5):1339–1347. doi: 10.1099/vir.0.81554-0. [DOI] [PubMed] [Google Scholar]
- 92.Nienhuis AW, Dunbar CE, Sorrentino BP. Genotoxicity of retroviral integration in hematopoietic cells. Mol Ther. 2006;13:1031–1049. doi: 10.1016/j.ymthe.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 93.Dorrell C, Gan OI, Pereira DS, Hawley RG, Dick JE. Expansion of human cord blood CD34(+)CD38( – ) cells in ex vivo culture during retroviral transduction without a corresponding increase in SCID repopulating cell (SRC) frequency: dissociation of SRC phenotype and function. Blood. 2000;95(1):102–110. [PubMed] [Google Scholar]
- 94.Williams DA. Ex vivo expansion of hematopoietic stem and progenitor cells—robbing Peter to pay Paul? Blood. 1993;81(12):3169–3172. [PubMed] [Google Scholar]
- 95.Dunbar CE, Takatoku M, Donahue RE. The impact of ex vivo cytokine stimulation on engraftment of primitive hematopoietic cells in a non-human primate model. Ann N Y Acad Sci. 2001;938:236–244. doi: 10.1111/j.1749-6632.2001.tb03594.x. discussion 244–245. [DOI] [PubMed] [Google Scholar]
- 96.Spencer DM. Developments in suicide genes for preclinical and clinical applications. Curr Opin Mol Ther. 2000;2(4):433–440. [PubMed] [Google Scholar]
- 97.Lupo-Stanghellini MT, Provasi E, Bondanza A, Ciceri F, Bordignon C, Bonini C. Clinical impact of suicide gene therapy in allogeneic hematopoietic stem cell transplantation. Hum Gene Ther. 2010;21(3):241–250. doi: 10.1089/hum.2010.014. [DOI] [PubMed] [Google Scholar]
- 98.Ciceri F, Bonini C, Gallo-Stampino C, Bordignon C. Modulation of GvHD by suicide-gene transduced donor T lymphocytes: clinical applications in mismatched transplantation. Cytotherapy. 2005;7(2):144–149. doi: 10.1080/14653240510018136. [DOI] [PubMed] [Google Scholar]
- 99.Bonini C, Bondanza A, Perna SK, Kaneko S, Traversari C, Ciceri F, Bordignon C. The suicide gene therapy challenge: how to improve a successful gene therapy approach. Mol Ther. 2007;15(7):1248–1252. doi: 10.1038/sj.mt.6300190. [DOI] [PubMed] [Google Scholar]
- 100.Frank O, Rudolph C, Heberlein C, von Neuhoff N, Schröck E, Schambach A, Schlegelberger B, Fehse B, Ostertag W, Stocking C, Baum C. Tumor cells escape suicide gene therapy by genetic and epigenetic instability. Blood. 2004;104(12):3543–3549. doi: 10.1182/blood-2004-03-0852. [DOI] [PubMed] [Google Scholar]
- 101.Uckert W, Kammertöns T, Haack K, Qin Z, Gebert J, Schendel DJ, Blankenstein T. Double suicide gene (cytosine deaminase and herpes simplex virus thymidine kinase) but not single gene transfer allows reliable elimination of tumor cells in vivo. Hum Gene Ther. 1998;9(6):855–865. doi: 10.1089/hum.1998.9.6-855. [DOI] [PubMed] [Google Scholar]
- 102.Straathof KC, Pulè MA, Yotnda P, Dotti G, Vanin EF, Brenner MK, Heslop HE, Spencer DM, Rooney CM. An inducible caspase 9 safety switch for T-cell therapy. Blood. 2005;105(11):4247–4254. doi: 10.1182/blood-2004-11-4564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Tey SK, Dotti G, Rooney CM, Heslop HE, Brenner MK. Inducible caspase 9 suicide gene to improve the safety of allodepleted T cells after haploidentical stem cell transplantation. Biol Blood Marrow Transplant. 2007;13(8):913–924. doi: 10.1016/j.bbmt.2007.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Quintarelli C, Vera JF, Savoldo B, Giordano Attianese GM, Pule M, Foster AE, Heslop HE, Rooney CM, Brenner MK, Dotti G. Co-expression of cytokine and suicide genes to enhance the activity and safety of tumor-specific cytotoxic T lymphocytes. Blood. 2007;110(8):2793–2802. doi: 10.1182/blood-2007-02-072843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zhong B, Watts KL, Gori JL, Wohlfahrt ME, Enssle J, Adair JE, Kiem HP. Safeguarding nonhuman primate iPS cells with suicide genes. Mol Ther. 2011;19:1667–1675. doi: 10.1038/mt.2011.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126(4):663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 107.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131(5):861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 108.Hanna J, Wernig M, Markoulaki S, Sun CW, Meissner A, Cassady JP, Beard C, Brambrink T, Wu LC, Townes TM, Jaenisch R. Treatment of sickle cell anemia mouse model with iPS cells generated from autologous skin. Science. 2007;318(5858):1920–1923. doi: 10.1126/science.1152092. [DOI] [PubMed] [Google Scholar]
- 109.Raya A, Rodríguez-Pizà I, Guenechea G, Vassena R, Navarro S, Barrero MJ, Consiglio A, Castellà M, Río P, Sleep E, González F, Tiscornia G, Garreta E, Aasen T, Veiga A, Verma IM, Surrallés J, Bueren J, Izpisúa Belmonte JC. Disease-corrected haematopoietic progenitors from Fanconi anaemia induced pluripotent stem cells. Nature. 2009;460(7251):53–59. doi: 10.1038/nature08129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kane NM, Nowrouzi A, Mukherjee S, Blundell MP, Greig JA, Lee WK, Houslay MD, Milligan G, Mountford JC, von Kalle C, Schmidt M, Thrasher AJ, Baker AH. Lentivirus-mediated reprogramming of somatic cells in the absence of transgenic transcription factors. Mol Ther. 2010;18(12):2139–2145. doi: 10.1038/mt.2010.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Winkler T, Cantilena A, Métais JY, Xu X, Nguyen AD, Borate B, Antosiewicz-Bourget JE, Wolfsberg TG, Thomson JA, Dunbar CE. No evidence for clonal selection due to lentiviral integration sites in human induced pluripotent stem cells. Stem Cells. 2010;28(4):687–694. doi: 10.1002/stem.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lister R, Pelizzola M, Kida YS, Hawkins RD, Nery JR, Hon G, Antosiewicz-Bourget J, O’Malley R, Castanon R, Klugman S, Downes M, Yu R, Stewart R, Ren B, Thomson JA, Evans RM, Ecker JR. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature. 2011;471(7336):68–73. doi: 10.1038/nature09798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hussein SM, Nagy K, Nagy A. Human induced pluripotent stem cells: the past, present, and future. Clin Pharmacol Ther. 2011;89(5):741–745. doi: 10.1038/clpt.2011.37. [DOI] [PubMed] [Google Scholar]
- 114.Okita K, Nakagawa M, Hyenjong H, Ichisaka T, Yamanaka S. Generation of mouse induced pluripotent stem cells without viral vectors. Science. 2008;322(5903):949–953. doi: 10.1126/science.1164270. [DOI] [PubMed] [Google Scholar]
- 115.Stadtfeld M, Nagaya M, Utikal J, Weir G, Hochedlinger K. Induced pluripotent stem cells generated without viral integration. Science. 2008;322(5903):945–949. doi: 10.1126/science.1162494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Sommer CA, Stadtfeld M, Murphy GJ, Hochedlinger K, Kotton DN, Mostoslavsky G. Induced pluripotent stem cell generation using a single lentiviral stem cell cassette. Stem Cells. 2009;27(3):543–549. doi: 10.1634/stemcells.2008-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kaji K, Norrby K, Paca A, Mileikovsky M, Mohseni P, Woltjen K. Virus-free induction of pluripotency and subsequent excision of reprogramming factors. Nature. 2009;458(7239):771–775. doi: 10.1038/nature07864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Warren L, Manos PD, Ahfeldt T, Loh YH, Li H, Lau F, Ebina W, Mandal PK, Smith ZD, Meissner A, Daley GQ, Brack AS, Collins JJ, Cowan C, Schlaeger TM, Rossi DJ. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell. 2010;7(5):618–630. doi: 10.1016/j.stem.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Papapetrou EP, Lee G, Malani N, Setty M, Riviere I, Tirunagari LM, Kadota K, Roth SL, Giardina P, Viale A, Leslie C, Bushman FD, Studer L, Sadelain M. Genomic safe harbors permit high β-globin transgene expression in thalassemia induced pluripotent stem cells. Nat Biotechnol. 2011;29(1):73–78. doi: 10.1038/nbt.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zou J, Sweeney CL, Chou BK, Choi U, Pan J, Wang H, Dowey SN, Cheng L, Malech HL. Oxidase-deficient neutrophils from Xlinked chronic granulomatous disease iPS cells: functional correction by zinc finger nuclease-mediated safe harbor targeting. Blood. 2011;117(21):5561–5572. doi: 10.1182/blood-2010-12-328161. [DOI] [PMC free article] [PubMed] [Google Scholar]