Abstract
The characterization of post-transcriptional gene regulation by small regulatory (20–30 nt) RNAs, particularly miRNAs and piRNAs, has become a major focus of research in recent years. A prerequisite for characterizing small RNAs is their identification and quantification across different developmental stages, and in normal and disease tissues, as well as model cell lines. Here we present a step-by-step protocol for generating barcoded small RNA cDNA libraries compatible with Illumina HiSeq sequencing, thereby facilitating miRNA and other small RNA profiling of large sample collections.
Introduction
MicroRNAs (miRNAs) are short (20–23 nucleotide (nt)) RNAs that guide sequence-specific posttranscriptional gene regulation and are encoded in the genomes of animals, plants, and some viruses [1–6]. These genetic regulators are expressed in tissue-, cell-type- and developmental-stage-specific patterns and dysregulation or mutation of miRNA genes causes or contributes to several human diseases [5, 7–15]. To assess miRNA expression in multiple biological or clinical samples, it is essential to develop reliable, accurate, and efficinet methods for measuring miRNA abundance.
Standard methods for miRNA profiling comprise microarray analysis [16–20] or quantitative RT-PCR (qRT-PCR) [21]. While cost-effective and allowing for relatively high sample-throughput, these approaches are limited to the study of previously identified miRNAs deposited at miRbase (www.mirbase.org) [22, 23]. Microarray assays can be hampered by cross-hybridization preventing the identification of individual members of miRNAs sequence families or mutant variants of miRNAs, whereas qRT-PCR methods are limited to a pre-selected subset of miRNAs. Here we provide a step-by-step protocol to generate miRNA expression profiles from deep sequencing of small RNA cDNA libraries. In addition to generating information on miRNA abundance, sequencing-based methods allow for the discovery of new or mutated miRNAs as well as novel families of small RNAs [24–34]. Deep sequencing of a limited set of tissues is sufficient to detect virtually every annotated miRNA species, albeit most of them in low frequency [31, 35]. Typically, a small subset of miRNAs (30 to 50 in mammalian tissues) represents the majority (>90%) of all sequence reads annotated as miRNAs [36–39]. Given that each miRNA potentially regulates hundreds of transcripts [38, 40], it is those abundantly expressed miRNAs that have been shown to control gene expression in a quantifiable manner [41–43].
The sequencing depth (currently more than 150 million sequence reads) obtained in a standard Illumina HiSeq sequencing is more than enough to record the relative abundance of miRNAs engaged in gene regulation. At the same time, prototypic small RNA cDNA library preparation and sequencing is time-consuming and more expensive on a per sample basis than microarray or PCR-based assays. These disadvantages are alleviated by the introduction of a barcoding approach to allow multiplexing of several samples; the introduction of a barcode at the first step of small RNA cDNA library preparation followed by pooling of multiple samples reduces processing time and sequencing costs [37, 44–49].
Overview of the method
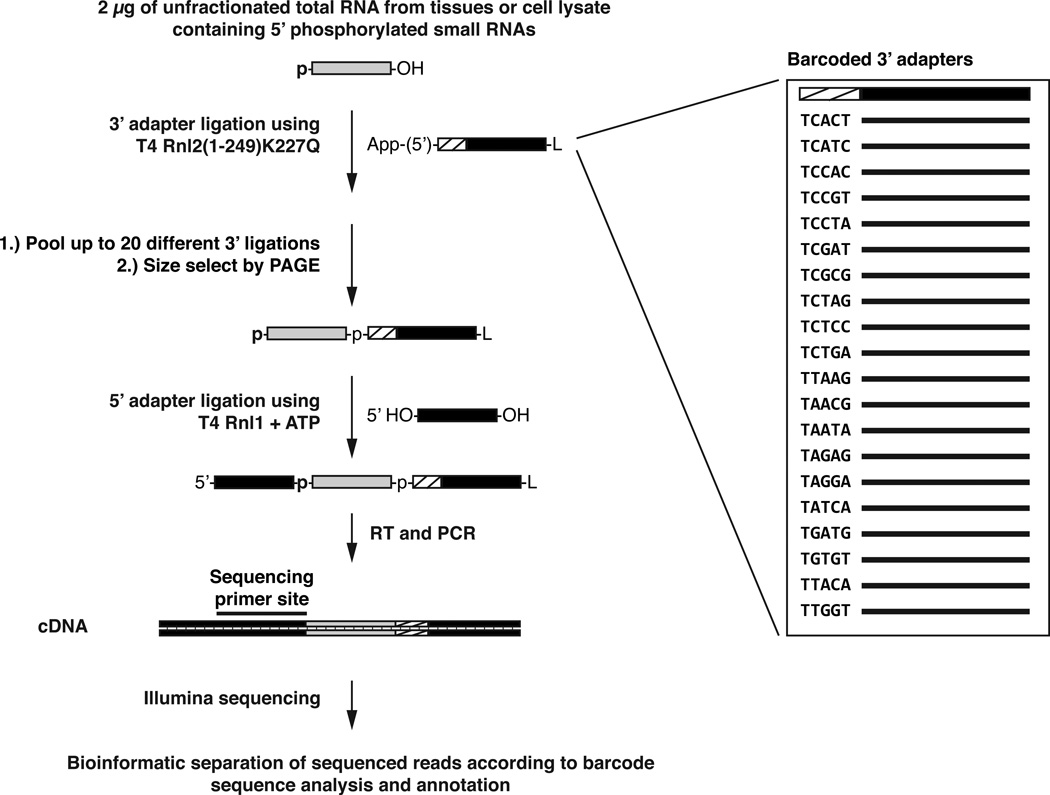
The experimental process (Fig. 1) of small RNA cDNA library generation consists of small RNA isolation, ligation of barcoded 3’ adapters to up to 20 individual samples, pooling of samples, ligation of a 5’ adapter, reverse transcription and PCR to generate the cDNA library, and sequencing. In contrast to a number of RNA cDNA library generation protocols for RNAseq applications, ligation of 3’ and 5’ adapters of different sequence to the small RNA preserves the orientation of the RNA insert and allows for the determination of its origin from the sense or antisense strand of the genome after cDNA sequencing. Downstream bioinformatic analyses start with the separation of sequence reads according to the barcode sequence, followed by mapping and annotation of the extracted sequence reads as described in an accompanying protocol (accompanying Methods manuscript by Farazi et al.) [50].
Fig. 1.
Schematic representation of barcoded small RNA cDNA library preparation. The barcode represents a 5-nt unique sequence at the 5’ end of the 3’ adapter oligodeoxynucleotides. Illumina HiSeq sequencing at its smallest scale yields more than 150 million sequence reads per sequencing lane. Most (>70%) sequence reads contain recognizable barcode sequences [37, 44], resulting in more than 3 million sequence reads per sample when using the full set of 20 barcoded adapters. L, 3’ aminohexyl blocking group that prevents adapter circularization.
Small regulatory RNAs bound by Argonaute and PIWI proteins are characterized by the presence of a 5’ monophosphate and a 3’ hydroxyl group. In contrast, most RNA turnover and hydrolysis products generally carry a 5’ hydroxyl group and a 2’,3’ cyclic phosphate or 2’ or 3’ monophosphate. Our protocol is designed to enrich for RNAs with 5’ monophosphate and 3’ hydroxyl termini. Due to the use of RNA ligases for joining adapters, circularization of small 5’ phosphorylated RNAs as well as adapters carrying 5’ phosphate and 3’ hydroxyl termini during those steps has to be prevented [44, 51, 52]. (1) Our 3’ adapter oligodeoxynucleotides are blocked at their 3’ hydroxyl terminus and the 5’ adapter lacks a 5’ monophosphate preventing their circularization. (2) To minimize adenylation and subsequent circularization of the input RNA during 3’ adapter ligation, we use pre-adenylated 3’ adapters together with a truncated and mutated form of T4 RNA ligase 2, Rnl2(1–249)K227Q, in the absence of ATP.
Some classes of small RNAs, such as piRNAs or plant miRNAs may carry 3’ terminal 2’-O-methyl modifications. This modification does not prevent but may reduce the efficiency of the 3’ adapter ligation step [53, 54]. 2’-O-methylated RNA containing samples may be pretreated with periodate under basic conditions to oxidize and eliminate unmodified 2’,3’ hydroxyl 3’ termini, yielding 3’ monophosphate ends, which cannot be joined to the 3’ adapter [55, 56]. These libraries are then specifically depleted for miRNAs.
Some reports indicate that the overall abundance of miRNAs might be altered under certain conditions [57–59]. To capture global variations in miRNA content, we add a cocktail of oligoribonucleotides of distinct sequence and known concentration as internal standards [37]. Based on the the ratio of calibrator to total miRNA sequence reads the absolute amount of miRNAs in the total RNA input can be calculated.
Materials
All reagents need to be RNase-free. RNA samples should be stored at −20°C or below and kept on ice while the reactions are set up to minimize hydrolysis. Importantly, use siliconized tubes for all manipulations of small RNAs after the recovery of the 3’ adapter ligation products, because oligonucleotides in the nanomolar concentration range readily adsorb to surfaces of non-siliconized tubes and pipette tips. For the same reason, homogenize reaction mixtures by gentle vortexing rather than pipetting up and down.
Oligonucleotides
Size Marker Oligoribonucleotides
We add trace amounts of 32P-end-labeled 19 and 24 nt size markers (or 35 nt when sequencing includes piRNAs) to the samples of total RNA to monitor the yield of adapter ligation and to guide the recovery of the expected length fraction of RNA ligation products.
| Size | Sequence |
|---|---|
| 19 nt | 5’-CGUACGCGGGUUUAAACGA |
| 24 nt | 5’-CGUACGCGGAAUAGUUUAAACUGU |
| 35 nt | 5’- CUCAUCUUGGUCGUACGCGGAAUAGUUUAAACUGU |
The RNA size markers contain a PmeI restriction endonuclease recognition site (underlined). After PCR-amplification the cDNA libraries are digested with PmeI to avoid sequencing of the size markers.
Calibrator Oligoribonucleotide Sequences
The calibrator oligoribonucleotides have no match to the human or mouse genome [16]. We recommend the addition of 0.5 fmol each of the ten following calibrator oligoribonucleotides to 2 µg of total RNA. The preparation of a calibrator cocktail requires the use of carrier oligonucleotide to prevent surface adsorption in the nanomolar concentration range (we use an 11-nt oligodeoxynucleotide, 5’-TCGAAGTATTC).
| Name | Sequence |
|---|---|
| Cal01 | pGUCCCACUCCGUAGAUCUGUUC |
| Cal02 | pGAUGUAACGAGUUGGAAUGCAA |
| Cal03 | pUAGCAUAUCGAGCCUGAGAACA |
| Cal04 | pCAUCGGUCGAACUUAUGUGAAA |
| Cal05 | pGAAGCACAUUCGCACAUCAUAU |
| Cal06 | pUCUUAACCCGGACCAGAAACUA |
| Cal07 | pAGGUUCCGGAUAAGUAAGAGCC |
| Cal08 | pUAACUCCUUAAGCGAAUCUCGC |
| Cal09 | pAAAGUAGCAUCCGAAAUACGGA |
| Cal10 | pUGAUACGGAUGUUAUACGCAGC |
p, 5’ monophosphate, sequences are RNA.
Pre-adenylated 3’ Adapters for Illumina HiSeq sequencing
We use the following set of 20 pre-adenylated 3’ adapter oligodeoxynucleotides, each containing a unique pentamer barcode sequence at the 5’ end (bold and underlined).
| 3’ Adapter | Sequence |
|---|---|
| Ad01 | rAppTCACTTCGTATGCCGTCTTCTGCTTG-L |
| Ad02 | rAppTCATCTCGTATGCCGTCTTCTGCTTG-L |
| Ad03 | rAppTCCACTCGTATGCCGTCTTCTGCTTG-L |
| Ad04 | rAppTCCGTTCGTATGCCGTCTTCTGCTTG-L |
| Ad05 | rAppTCCTATCGTATGCCGTCTTCTGCTTG-L |
| Ad06 | rAppTCGATTCGTATGCCGTCTTCTGCTTG-L |
| Ad07 | rAppTCGCGTCGTATGCCGTCTTCTGCTTG-L |
| Ad08 | rAppTCTAGTCGTATGCCGTCTTCTGCTTG-L |
| Ad09 | rAppTCTCCTCGTATGCCGTCTTCTGCTTG-L |
| Ad10 | rAppTCTGATCGTATGCCGTCTTCTGCTTG-L |
| Ad11 | rAppTTAAGTCGTATGCCGTCTTCTGCTTG-L |
| Ad12 | rAppTAACGTCGTATGCCGTCTTCTGCTTG-L |
| Ad13 | rAppTAATATCGTATGCCGTCTTCTGCTTG-L |
| Ad14 | rAppTAGAGTCGTATGCCGTCTTCTGCTTG-L |
| Ad15 | rAppTAGGATCGTATGCCGTCTTCTGCTTG-L |
| Ad16 | rAppTATCATCGTATGCCGTCTTCTGCTTG-L |
| Ad17 | rAppTGATGTCGTATGCCGTCTTCTGCTTG-L |
| Ad18 | rAppTGTGTTCGTATGCCGTCTTCTGCTTG-L |
| Ad19 | rAppTTACATCGTATGCCGTCTTCTGCTTG-L |
| Ad20 | rAppTTGGTTCGTATGCCGTCTTCTGCTTG-L |
L, 3’ aminohexyl blocking group; rApp, 5’ terminal adenosine residue connected via a 5’,5’-diphosphate bridge to the 5’OH of the 5’ nucleotide, which activates the adapter for ligation. The synthesis and purification of the pre-adenylated 3’ adapters has been described previously [32].
Oligoribonucleotide 5’ Adapter Compatible with Illumina Sequencing
5’ adapter: 5’-GUUCAGAGUUCUACAGUCCGACGAUC
Primers for Amplification of the Barcoded cDNA Library
The primers are compatible with the Illumina small RNA sequencing workflow and allow for the clonal amplification of the cDNA on HiSeq flowcells in preparation for sequencing.
Forward primer: 5’-AATGATACGGCGACCACCGACAGGTTCAGAGTTCTACAGTCCGA Underlined sequence corresponds to the Illumina sequencing primer.
Reverse primer: 5’-CAAGCAGAAGACGGCATACGA
Enzymes
-
-
T4 polynucleotide kinase (PNK; NEB)
-
-
Truncated and mutated T4 RNA ligase 2; Rnl2(1–249)K227Q (NEB). The plasmid for expression of recombinant, His-tagged T4 Rnl2(1–249)K227Q can also be obtained from www.addgene.org.
-
-
RNA ligase, T4 Rnl1 (Fermentas or NEB).
-
-
SuperScript III reverse transcriptase (Invitrogen)
-
-
Taq DNA polymerase (any supplier)
-
-
PmeI (NEB).
Buffers and Solutions
| Buffer | Composition |
|---|---|
| 10x Ligation buffer without ATP | 0.5 M Tris-HCl, pH 7.6, 0.1 M MgCl2, 0.1 M 2-mercaptoethanol, 1 mg/ml acetylated BSA (Sigma-Aldrich, B-8894) |
| Denaturing PAA gel loading solution | 98.8% Formamide, 1% (v/v) 0.5 M Na2H2EDTA, pH 8.0, 0.2% Bromophenol blue |
| 5x TBE buffer | 0.45 M Tris base, 40 mM Boric acid, 10 mM Na2H2EDTA |
| 10x Ligation buffer with ATP | 0.5 M Tris-HCl, pH 7.6, 0.1 M MgCl2, 0.1 M 2-mercaptoethanol 1 mg/ml acetylated BSA (Sigma-Aldrich, B-8894), 2 mM ATP |
| 5x First strand buffer (supplied with SuperScript III reverse transcriptase) | 250 mM Tris-HCl, pH 8.3, 375 mM KCl, 15 mM MgCl2, 100 mM DTT |
| 10x dNTP mix | 2 mM dATP, 2 mM dCTP, 2 mM dGTP, 2 mM dTTP |
| 10x PCR buffer | 100 mM Tris-HCl, pH 8.0, 500 mM KCl, 1% Triton-X100, 20 mM MgCl2, 10 mM 2-mercapthoethanol |
| 5x Agarose gel loading solution | 0.2% (w/v) Bromophenol blue, 0.2% (w/v) Xylene cyanol FF, 50 mM Na2H2EDTA, pH 8.0, 20% Ficoll type 400 |
Other Materials
-
-
Siliconized 1.5 ml reaction tubes (BioPlas)
-
-
QiaQuick gel extraction kit (Qiagen)
-
-
TRIzol® reagent (Invitrogen)
-
-
25 bp DNA Ladder (Invitrogen)
Procedure
The following step-by-step procedure describes the generation of a small RNA cDNA library for Illumina sequencing. We use 2 µg of total RNA as input per biological or clinical sample and pool up to 20 samples directly after the 3’ adapter ligation step that introduced the pentameric barcode sequence. We have also generated miRNA profiles from smaller quantities of total RNA (minimum 10 ng) from body fluids, exosome preparations, or serum, adjusting calibrator oligoribonucleotide concentrations as required.
Isolation of total RNA
Total RNA is isolated from either freshly collected cultured cells or tissues, flash-frozen samples stored below −70°C, or formalin-fixed paraffin embedded tissues [60]. Typically, 1 mg of tissue or 105 cells yield about 1 µg of total RNA. We recommend isolation of total RNA from tissue or cultured cells either using the guanidinium isothiocyanate (GITC)/phenol method [61] or the commercial TRIzol reagent (Invitrogen). We avoid aqueous LiCl precipitation, as small RNAs, including tRNAs, will not precipitate. Protocols and kits designed to enrich for small RNAs, such as the Qiagen miRNeasy kit, can be used for small RNA isolation, but may complicate the determination of the ratio of miRNA to total RNA by addition of calibrator oligoribonucleotides.
Preparation of Calibrator Oligoribonucleotide Cocktail
-
1
Prepare 7 ml of carrier solution containing 500 nM 11-nt carrier oligodeoxyribonucleotide in water. The carrier is necessary to prevent surface adsorption during dilution and storage of low concentrations of calibrator oligoribonucleotides.
-
2
Prepare 50 µl of a calibrator cocktail containing 1 µM of each calibrator oligoribonucleotide.
-
3
Dilute the calibrator cocktail 1:10 in carrier solution resulting in a calibrator concentration of 0.1 µM each.
-
4
Further dilute the calibrator cocktail dilution 1:100 in carrier solution resulting in a final oligoribonucleotide calibrator concentration of 1 nM each.
Preparation of Radiolabeled Size Markers
-
5
Radiolabel the size markers individually in a 10 µl reaction containing 1 µM RNA, 10 U T4 polynucleotide kinase and 50 µCi [-32P]ATP (6,000 Ci/mmol) at 37°C for 15 min.
-
6
Quench the reactions by adding 10 µl of denaturing PAA gel loading solution.
-
7
Incubate the sample at 90 C for 1 min.
-
8
Load samples on a 15% denaturing polyacrylamide (PAA) gel (15 cm wide, 17 cm long, 0.5 mm thick; 30 ml gel volume). Run the gel approx. 50 min at 30 W using 0.5×TBE buffer until the bromophenol blue dye is close to the bottom of the gel.
-
9
Dismantle the gel, leaving it mounted on one glass plate. Wrap the gel in plastic film (e.g. Saran wrap), place it in a cassette and align against one corner, align an X-ray film against the same corner and expose for 1 min; develop the film.
-
10
Align the gel on glass plate to the X-ray film. Cut out the radioactive bands corresponding to the length markers and transfer the gel slices into 1.5 ml siliconized tubes (one for each marker). Also collect and store some moderately radioactive gel pieces from the gel running front. These pieces will be needed later for placement in gels to facilitate alignment with phosphorimager printouts in order to excise 3’ and 5’ adapter ligation products.
-
11
Add 300 µl of 0.3 M NaCl and elute the RNA from the gel by incubating the tube overnight at 4°C under constant agitation.
-
12
Collect the supernatant and add 900 µl (3 volumes) of absolute ethanol. Keep the sample on ice for 1 h or overnight at −20°C.
-
13
Centrifuge in a tabletop centrifuge at 4°C and at maximum speed (approx. 14,000 g) for 15 min.
-
14
Discard the supernatant. Collect residual ethanol by centrifugation at 14,000 g for 1 min and discard residual liquid. Air-dry the RNA pellets for 10 min.
-
15
Dissolve each pellet in 10 µl of H2O.
-
16
Combine length marker solutions to obtain the size marker cocktail.
-
17
Dilute the combined length markers 1:50 in carrier solution prepared in step 1.
3’ Adapter Ligation
-
18
Provide 2 µg of total RNA in 8 µl of H2O in a siliconized reaction tube.
-
19
Prepare a Mastermix for ligating the 3’ adapter, multiplying the volume for one reaction (9 µl) by the number of samples being processed (up to 20 samples plus one control for the combined length markers); increase the volume of the Mastermix by another 10% to compensate for dispensing errors. Each 3’ adapter ligation reaction requires 2 µl of 10x RNA ligase buffer without ATP, 6 µl 50% aqueous DMSO, 0.5 µl of a 10 nM calibrator cocktail solution, and 0.5 µl of 5’-32P-labeled length marker oligoribonucleotide mix.
-
20
Add 9 µl of the Mastermix to each RNA sample prepared in step 18 (for the length marker control reaction add Mastermix to 8 µl H2O and 2 µl of radioactive size marker solution)
-
21
Add 2 µl of 50 µM adenylated 3’ adapter with a unique barcode to each sample.
-
22
Incubate the reaction mixture at 90°C for 1 min to denature the RNA and immediately place on ice for 2 min.
-
23
Add 1 µl T4 Rnl2(1–249)K227Q (1 µg/µl), mix gently, and incubate overnight on ice.
-
24
Place 2 µl of 3 M NaCl multiplied by the number of samples into a new 2-ml reaction tube (i.e. 40 µl for 20 samples). Add 3 times the total volume of the combined 3’ adapter ligation reactions of absolute ethanol (i.e. 1.3 ml ethanol for 20 samples). Transfer the individual adapter ligation reactions into the ethanol-containing tube, which stops the ligation reaction and at the same time combines the barcoded libraries.
-
25
Precipitate the ligation products by incubation on ice for 1 h and collect the pellet by centrifugation in a tabletop centrifuge at 4°C and maximum speed (approx. 14,000 g) for 15 min.
-
26
Discard the supernatant. Collect residual ethanol by centrifugation at 14,000 g for 1 min. Air-dry the RNA pellet for 10 min.
-
27
Dissolve the RNA pellet in 20 µl H2O.
-
28
Add 20 µl of denaturing PAA gel loading solution and load the samples in two adjacent wells in the center of a 20-well 15% denaturing PAA gel (15 cm wide, 17 cm long, 0.5 mm thick; 30 ml gel solution). When processing more than one barcoded library (i.e. more than 20 individual small RNA samples at once), space samples by a two-well distance to avoid cross contamination. Load the length marker ligation reaction separated by one blank lane into the lanes flanking the leftmost and the rightmost of the samples loaded in the center of the gel.
-
29
Run the gel at 30 W for approximately 45 min in 0.5×TBE buffer until the bromophenol blue dye reaches the lower third of the gel. Do not run the gel much further in order to contain the ligation products within a gel area as small as possible for efficient elution.
-
30
Dismantle the gel, leaving it mounted on one glass plate.
-
31
To facilitate the alignment of the gel to the phosphorimager paper printout, excise small triangles (approx. 5 mm size) at three of the four corners of the gel and implant into each a tiny radioactive gel pieces collected in step 10.
-
32
Cover the gel in plastic film (e.g. Saran wrap) to avoid contamination and expose to a phosphorimaging screen for 45 min.
-
33
Print out a 100%-scaled image of the phosphorimaged gel, align the gel on top of the printout according to the positions of the three radioactive gel pieces. For gel excision in sample lanes, use the positions of the 19-nt/3’ adapter and 24-nt/3’ adapter (35-nt/3’adapter, when sequencing piRNAs) ligation products as margins. Transfer the gel piece(s) into a siliconized 1.5 ml tube. Also excise the ligation products for the length marker control and place into a separate tube.
-
34
Add 350 µl of 0.3 M NaCl to each tube and elute the ligated RNAs from the gel slices by incubating overnight at 4°C under constant agitation.
-
35
Collect the supernatant and add 3 volumes absolute ethanol to precipitate the RNA as described in steps 26 and 27.
5’ Adapter Ligation
-
36
Dissolve the RNA pellets in 9 µl H2O. This also pertains to the control length marker ligation product.
-
37
Prepare a Mastermix as outlined in step 19, containing the following components per reaction: 1 µl of 100 µM 5’ adapter, 2 µl of 10x RNA ligase buffer with ATP, and 6 µl 50% aqueous DMSO.
-
38
Add 9 µl of the Mastermix to each RNA sample from step 37. Incubate the tube at 90°C for 1 min to denature the RNA and immediately place on ice for 2 min.
-
39
Add 2 µl of Rnl1 (1 mg/ml), mix gently, and incubate at 37°C for 1 h.
-
40
Add 20 µl of denaturing PAA gel loading solution and load the samples in two adjacent wells of a 20-well 12% denaturing PAA gel (15 cm wide, 17 cm long, 0.5 mm thick; 30 ml gel solution). Load samples as in step 28. Run the gel at 30 W using 0.5×TBE buffer until the bromophenol blue dye is close to the bottom of the gel. Image the gel as described in steps 30–33 and excise the new ligation products.
-
41
Add 350 µl of 0.3 M NaCl and 1 µl of 100 µM reverse PCR primer as carrier and elute the ligated RNAs from the gel by incubating the tube overnight at 4°C under constant agitation. The carrier facilitates the recovery of the ligation product.
-
42
Add 3 volumes absolute ethanol to precipitate and collect the RNA as described in steps 26 and 27.
Reverse Transcription (RT)
-
43
Dissolve pellets in 5.6 µl H2O. This also pertains to the control length marker ligation product.
-
44
Prepare an RT Mastermix (as outlined in step 19) containing the following components per reaction: 1.5 µl 0.1 M DTT, 3 µl 5x first-strand buffer, and 4.2 µl 10x dNTPs.
-
45
Denature the RNA from step 44 by incubating the tube at 90°C for 1 min, followed by transfer of the tube to a 50°C incubator.
-
46
Add 8.7 µl of the RT Mastermix to each sample and incubate for 3 min at 50°C. Add 0.75 µl of Superscript III reverse transcriptase and incubate at 42°C for 30 min.
-
47
To hydrolyze the RNA template, add 40 µl of 150 mM KOH/20 mM Tris base and incubate at 90°C for 10 min.
-
48
Neutralize the solution by addition of 40 µl 150 mM HCl and adjust the pH to a value between 7.5 and 9.0. Monitor the pH change by spotting 1 µl of cDNA solution on a pH paper. The pH of the solution should be slightly alkaline to not inhibit the subsequent PCR.
PCR amplification
-
49
Prepare a PCR mix containing the following components: 10 µl of the cDNA solution, 0.5 µl 100 µM forward primer, 0.5 µl 100 µM reverse primer, 10 µl 10×dNTP mix, 10 µl 10×PCR buffer, 68 µl H2O, and 1 µl Taq DNA polymerase (5 U/µl).
Also prepare a no-template control PCR reaction (H2O instead of cDNA) to check for DNA contamination.
-
50
Program the following cycle conditions: 45 s at 94°C, 85 s at 50°C, 60 s at 72°C. Remove 12 µl aliquots every other cycle following cycle number 8 by temporarily putting the PCR cycler on hold at the end of the 72°C step. This is done to determine the necessary number of cycles for amplifying the cDNA library. In our experience, with a pool of 20 sample of 2 µg input RNA, it is usually not necessary to amplify for more than 15 cycles.
-
51
Analyze the PCR products on a 2.5 % agarose gel. These products might appear as a double band with a higher band running at the expected length of 90–95 bp (90–106 bp when generating piRNA libraries) and a 70 bp band corresponding to 3’-adapter-to-5’-adapter ligation side products without insert. Carryover from unligated 3’ adapter into the 5’ adapter ligation reaction is responsible for this byproduct.
-
52
Define the optimal cycle number for cDNA amplification (in our experience not more than 15 cycles), which has to be within the exponential amplification phase of the PCR, i.e. approx. 5 cycles away from reaching the saturation level of PCR amplification. It is important to limit the PCR to the exponential phase, otherwise sequence-specific distortions will be introduced into the small RNA profiles, a process commonly referred to as clonal amplification. The excess of primers over cDNA in the exponential phase also prevents reannealing of products with different inserts, which leads to incomplete removal of length markers in the PmeI digestion step (steps 62–64).
-
53
Perform a 300 µl PCR according to steps 49 and 50 with the previously determined optimal cycle number by distributing the volume over 3 PCR tubes. After the reaction, remove 5 µl and combine with 5 µl agarose loading dye solution to verify product formation on a 2.5% agarose gel. If the PCR product is visible, proceed to the next step, otherwise add 3 cycles of PCR or repeat the entire amplification experiment, including pilot (steps 50/51) and large-scale PCR (this step).
-
54
Combine the aliquoted PCR products in a 1.5 ml tube, add 30 µl of 3 M NaCl and extract with 330 µl of phenol/chloroform and vortex for 20 s.
-
55
Separate phases by centrifugation at 14,000 g in a tabletop centrifuge for 2 min.
-
56
Take off the upper, aqueous phase and transfer to a new tube. Make sure not to take up the interphase where insoluble material accumulates.
-
57
Re-extract the aqueous phase with 330 µl of chloroform to remove residual phenol and vortex for 20 s, then separate phases by centrifugation at 14,000 g in a tabletop centrifuge for 2 min.
-
58
Take off the upper, aqueous phase and transfer to a new tube.
-
59
Add 1 ml of absolute ethanol and incubate on ice for 1 h or overnight at −20°C.
-
60
Collect the pellet by centrifugation in a tabletop centrifuge at 4°C at maximum speed (14,000 g) for 15 min.
-
61
Discard the supernatant. Collect residual liquid by centrifugation at 14,000 g for 1 min. Remove all of the supernatant but do not dry the pellet as this will cause the DNA to denature and thereby inhibit the following restriction enzyme digestion. Immediately proceed to the next step.
PmeI digestion
This step cleaves PCR products originating from the radiolabeled length markers used during small RNA cDNA library preparation. Be careful not to denature the double-stranded PCR product before or during the PmeI digestion. Denaturation and subsequent re-annealing of a complex sequence pool will result in imperfect rehybridization and formation of DNA duplexes with internal bulges that might compromise PmeI digestion. As control, the PCR product obtained from the ligation of adapters to the marker oligonucleotides alone (marker control sample) must be digested completely.
-
62
Prepare a PmeI digestion cocktail containing the following components per reaction: 2 µl 10x PmeI buffer (NEB), 0.2 µl of 100x BSA (10 mg/ml, NEB), 17.3 µl of H2O, and 0.5 µl (5 U) of PmeI (NEB).
-
63
Dissolve the DNA pellet from step 62 in the 20 µl PmeI digestion mixture and incubate at 37°C for at least 2 h. Do not vortex vigorously to avoid denaturing the restriction enzyme.
-
64
Next, to separate small RNA insert-containing adapter ligation products from shorter 5’-adapter-to-3’-adapter ligation side products, prepare a 2.5% agarose gel containing 0.4 µg/ml of ethidium bromide. Add 20 µl of agarose gel loading solution and load all of the restriction digest into two wells of the agarose gel, as well as the 25-bp DNA ladder in a separate well. Run the gel in 0.5×TBE buffer for approximately 1.5 h at 180 V until the 25-bp ladder is sufficiently resolved.
-
65
Visualize the DNA in the gel using a 360 nm UV transilluminator and excise the band of approx. 90–95 bp (90–106 bp for piRNA libraries) in size. Avoid excising below 90 bp to make sure the 5’-to-3’-adapter ligation product is not recovered.
-
66
Transfer the gel slice to a pre-weighed 1.5 ml reaction tube and weigh it again. Elute the DNA from the gel using the QiaQuick gel extraction kit or a comparable kit according to the manufacturer’s instructions. Recover the DNA in 30 µl of 1×TE buffer.
-
67
The DNA is then ready for Illumina sequencing.
Troubleshooting
-
-
No PCR product: A) Adapter ligation failed. Take care to cool the reaction mixture after heat-shock and before addition of ligase. Rnl2(1–249)K227Q in particular is inactive at temperatures above 37°C and may be irreversibly inactivated at higher temperatures. B) After precipitation take care that residual ethanol is evaporated before addition of reaction mix, as residual ethanol will inhibit ligases and reverse transcriptases. C) Revisit reverse transcription.
-
-
No full-length, but only 5’-to-3’-adapter ligation products of 70 bp are visible after PCR: A) Make sure adapter concentrations in the ligation reactions are correct. B) Make sure to only excise ligation products between 19 and 24 nt (or 35 nt) length. Never cut below the 19 nt marker.
Acknowledgments
M.H. is supported by a fellowship of the Charles Revson, Jr. Foundation. N.R. is supported through a K08 award (NS072235) from the National Institute of Neurological Disorders and Stroke. T.A.F. is supported by the RUCCTS Grant #UL1RR024143. T.T. is an HHMI investigator, and work in his laboratory was supported by NIH grants MH08442 and the Starr Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bhattacharyya SN, Filipowicz W. Argonautes and company: sailing against the wind. Cell. 2007;128:1027–1028. doi: 10.1016/j.cell.2007.02.033. [DOI] [PubMed] [Google Scholar]
- 2.Bartel DP. MicroRNAs: Target Recognition and Regulatory Functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stefani G, Slack FJ. Small non-coding RNAs in animal development. Nat. Rev. Mol. Cell. Biol. 2008;9:219–230. doi: 10.1038/nrm2347. [DOI] [PubMed] [Google Scholar]
- 4.Voinnet O. Origin, Biogenesis, and Activity of Plant MicroRNAs. Cell. 2009;136:669–687. doi: 10.1016/j.cell.2009.01.046. [DOI] [PubMed] [Google Scholar]
- 5.Skalsky RL, Cullen BR. Viruses, microRNAs, and Host Interactions. Annual Review of Microbiology. 2010;64:123–141. doi: 10.1146/annurev.micro.112408.134243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cullen BR. Viruses and microRNAs: RISCy interactions with serious consequences. Genes Dev. 2011;25:1881–1894. doi: 10.1101/gad.17352611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hebert SS, de Strooper B. Molecular biology. miRNAs in neurodegeneration. Science. 2007;317:1179–1180. doi: 10.1126/science.1148530. [DOI] [PubMed] [Google Scholar]
- 8.Ryan BM, Robles AI, Harris CC. Genetic variation in microRNA networks: the implications for cancer research. Nat. Rev. Cancer. 2010;10:389–402. doi: 10.1038/nrc2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nature Reviews Genetics. 2009;10:704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat. Rev. Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 11.Latronico MV, Condorelli G. MicroRNAs and cardiac pathology. Nature Reviews Cardiology. 2009;6:419–429. doi: 10.1038/nrcardio.2009.56. [DOI] [PubMed] [Google Scholar]
- 12.Inui M, Martello G, Piccolo S. MicroRNA control of signal transduction. Nat. Rev. Mol. Cell. Biol. 2010;11:252–263. doi: 10.1038/nrm2868. [DOI] [PubMed] [Google Scholar]
- 13.Zhang C. Novel functions for small RNA molecules. Current Opinion in Molecular Therapeutics. 2009;11:641–651. [PMC free article] [PubMed] [Google Scholar]
- 14.Poy MN, Spranger M, Stoffel M. microRNAs and the regulation of glucose and lipid metabolism. Diabetes, Obesity and Metabolism. 2007;9:67–73. doi: 10.1111/j.1463-1326.2007.00775.x. [DOI] [PubMed] [Google Scholar]
- 15.Rottiers V, Naar AM. MicroRNAs in metabolism and metabolic disorders. Nat. Rev. Mol. Cell. Biol. 2012;13:239–250. doi: 10.1038/nrm3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bissels U, Wild S, Tomiuk S, Holste A, Hafner M, Tuschl T, et al. Absolute quantification of microRNAs by using a universal reference. Rna. 2009;15:2375–2384. doi: 10.1261/rna.1754109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bissels U, Wild S, Tomiuk S, Hafner M, Scheel H, Mihailovic A, et al. Combined Characterization of microRNA and mRNA Profiles Delineates Early Differentiation Pathways of CD133(+) and CD34(+) Hematopoietic Stem and Progenitor Cells. Stem Cells. 2011;29:847–857. doi: 10.1002/stem.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barad O, Meiri E, Avniel A, Aharonov R, Barzilai A, Bentwich I, et al. MicroRNA expression detected by oligonucleotide microarrays: system establishment and expression profiling in human tissues. Genome Res. 2004;14:2486–2494. doi: 10.1101/gr.2845604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nelson PT, Baldwin DA, Scearce LM, Oberholtzer JC, Tobias JW, Mourelatos Z. Microarray-based, high-throughput gene expression profiling of microRNAs. Nat. Meth. 2004;1:155–161. doi: 10.1038/nmeth717. [DOI] [PubMed] [Google Scholar]
- 20.Baskerville S, Bartel DP. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. Rna. 2005;11:241–247. doi: 10.1261/rna.7240905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fiedler SD, Carletti MZ, Christenson LK. Quantitative RT-PCR methods for mature microRNA expression analysis. Methods Mol. Biol. 2010;630:49–64. doi: 10.1007/978-1-60761-629-0_4. [DOI] [PubMed] [Google Scholar]
- 22.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucl. Acids Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deepsequencing data. Nucl. Acids Res. 2011;39:D152–D157. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aravin AA, Lagos-Quintana M, Yalcin A, Zavolan M, Marks D, Snyder B, et al. The small RNA profile during Drosophila melanogaster development. Dev. Cell. 2003;5:337–350. doi: 10.1016/s1534-5807(03)00228-4. [DOI] [PubMed] [Google Scholar]
- 25.Chen PY, Manninga H, Slanchev K, Chien M, Russo JJ, Ju J, et al. The developmental miRNA profiles of zebrafish as determined by small RNA cloning. Genes Dev. 2005;19:1288–1293. doi: 10.1101/gad.1310605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aravin AA, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442:203–207. doi: 10.1038/nature04916. [DOI] [PubMed] [Google Scholar]
- 27.Lau NC, Seto AG, Kim J, Kuramochi-Miyagawa S, Nakano T, Bartel DP, et al. Characterization of the piRNA Complex from Rat Testes. Science. 2006;313:363–367. doi: 10.1126/science.1130164. [DOI] [PubMed] [Google Scholar]
- 28.Saito K, Sakaguchi Y, Suzuki T, Suzuki T, Siomi H, Siomi MC. Pimet, the Drosophila homolog of HEN1, mediates 2“-O-methylation of Piwi- interacting RNAs at their 3” ends. Genes Dev. 2007;21:1603–1608. doi: 10.1101/gad.1563607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carmell MA, Girard A, van de Kant HJG, Bourc'his D, Bestor TH, de Rooij DG, et al. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev. Cell. 2007;12:503–514. doi: 10.1016/j.devcel.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 30.Ruby JG, Jan CH, Player C, Axtell MJ, Lee W, Nusbaum C, et al. Large-Scale Sequencing Reveals 21U-RNAs and Additional MicroRNAs and Endogenous siRNAs in C. elegans. Cell. 2006;127:1193–1207. doi: 10.1016/j.cell.2006.10.040. [DOI] [PubMed] [Google Scholar]
- 31.Chiang HR, Schoenfeld LW, Ruby JG, Auyeung VC, Spies N, Baek D, et al. Mammalian microRNAs: experimental evaluation of novel and previously annotated genes. Genes Dev. 2010;24:992–1009. doi: 10.1101/gad.1884710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hafner M, Landgraf P, Ludwig J, Rice A, Ojo T, Lin C, et al. Identification of microRNAs and other small regulatory RNAs using cDNA library sequencing. Methods. 2008;44:3–12. doi: 10.1016/j.ymeth.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bar M, Wyman SK, Fritz BR, Qi J, Garg KS, Parkin RK, et al. MicroRNA Discovery and Profiling in Human Embryonic Stem Cells by Deep Sequencing of Small RNA Libraries. Stem Cells. 2008;26:2496–2505. doi: 10.1634/stemcells.2008-0356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Friedländer MR, Adamidi C, Han T, Lebedeva S, Isenbarger TA, Hirst M, et al. High-resolution profiling and discovery of planarian small RNAs. PNAS. 2009;106:11546–11551. doi: 10.1073/pnas.0905222106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Friedländer MR, Chen W, Adamidi C, Maaskola J, Einspanier R, Knespel S, et al. Discovering microRNAs from deep sequencing data using miRDeep. Nat. Biotech. 2008;26:407–415. doi: 10.1038/nbt1394. [DOI] [PubMed] [Google Scholar]
- 36.Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Farazi TA, Horlings HM, Ten Hoeve JJ, Mihailovic A, Halfwerk H, Morozov P, et al. MicroRNA Sequence and Expression Analysis in Breast Tumors by Deep Sequencing. Cancer Research. 2011;71:4443–4453. doi: 10.1158/0008-5472.CAN-11-0608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, et al. Transcriptome-wide Identification of RNA-Binding Protein and MicroRNA Target Sites by PAR-CLIP. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ugras S, Brill E, Jacobsen A, Hafner M, Socci ND, DeCarolis PL, et al. Small RNA Sequencing and Functional Characterization Reveals MicroRNA-143 Tumor Suppressor Activity in Liposarcoma. Cancer Research. 2011;71:5659–5669. doi: 10.1158/0008-5472.CAN-11-0890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Friedman RC, Farh KK-H, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 42.Baek D, Villén J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Krutzfeldt J, Kuwajima S, Braich R, Rajeev KG, Pena J, Tuschl T, et al. Specificity, duplex degradation and subcellular localization of antagomirs. Nucl. Acids Res. 2007;35:2885–2892. doi: 10.1093/nar/gkm024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hafner M, Renwick N, Brown M, Mihailovic A, Holoch D, Lin C, et al. RNAligase- dependent biases in miRNA representation in deep-sequenced small RNA cDNA libraries. Rna. 2011;17:1697–1712. doi: 10.1261/rna.2799511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu Q, Schlabach MR, Hannon GJ, Elledge SJ. Design of 240,000 orthogonal 25mer DNA barcode probes. PNAS. 2009;106:2289–2294. doi: 10.1073/pnas.0812506106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Parameswaran P, Jalili R, Tao L, Shokralla S, Gharizadeh B, Ronaghi M, et al. A pyrosequencing-tailored nucleotide barcode design unveils opportunities for large-scale sample multiplexing. Nucl. Acids Res. 2007;35:e130. doi: 10.1093/nar/gkm760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schulte JH, Marschall T, Martin M, Rosenstiel P, Mestdagh P, Schlierf S, et al. Deep sequencing reveals differential expression of microRNAs in favorable versus unfavorable neuroblastoma. Nucl. Acids Res. 2010;38:5919–5928. doi: 10.1093/nar/gkq342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hamady M, Walker JJ, Harris JK, Gold NJ, Knight R. Error-correcting barcoded primers for pyrosequencing hundreds of samples in multiplex. Nat. Meth. 2008;5:235–237. doi: 10.1038/nmeth.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vigneault F, Sismour AM, Church GM. Efficient microRNA capture and bar-coding via enzymatic oligonucleotide adenylation. Nat. Meth. 2008;5:777–779. doi: 10.1038/nmeth.1244. [DOI] [PubMed] [Google Scholar]
- 50.Berninger P, Gaidatzis D, van Nimwegen E, Zavolan M. Computational analysis of small RNA cloning data. Methods. 2008;44:13–21. doi: 10.1016/j.ymeth.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 51.Pfeffer S, Lagos-Quintana M, Tuschl T. Cloning of small RNA molecules. Curr. Prot. Mol. Biol. 2003:26.4.1–26.4.18. doi: 10.1002/0471142727.mb2604s72. [DOI] [PubMed] [Google Scholar]
- 52.Lau NC, Lim LP, Weinstein EG, Bartel DP. An Abundant Class of Tiny RNAs with Probable Regulatory Roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 53.Ebhardt HA, Thi EP, Wang M-B, Unrau PJ. Extensive 3' modification of plant small RNAs is modulated by helper component-proteinase expression. PNAS. 2005;102:13398–13403. doi: 10.1073/pnas.0506597102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Munafo DB, Robb GB. Optimization of enzymatic reaction conditions for generating representative pools of cDNA from small RNA. Rna. 2010;16:2537–2552. doi: 10.1261/rna.2242610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vagin VV, Sigova A, Li C, Seitz H, Gvozdev V, Zamore PD. A distinct small RNA pathway silences selfish genetic elements in the germline. Science. 2006;313:320–324. doi: 10.1126/science.1129333. [DOI] [PubMed] [Google Scholar]
- 56.Alefelder S, Patel BK, Eckstein F. Incorporation of terminal phosphorothioates into oligonucleotides. Nucl. Acids Res. 1998;26:4983–4988. doi: 10.1093/nar/26.21.4983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Krol J, Busskamp V, Markiewicz I, Stadler MB, Ribi S, Richter J, et al. Characterizing Light-Regulated Retinal MicroRNAs Reveals Rapid Turnover as † a Common Property of Neuronal MicroRNAs. Cell. 2010;141:618–631. doi: 10.1016/j.cell.2010.03.039. [DOI] [PubMed] [Google Scholar]
- 58.Kumar MS, Lu J, Mercer KL, Golub TR, Jacks T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat. Genet. 2007;39:673–677. doi: 10.1038/ng2003. [DOI] [PubMed] [Google Scholar]
- 59.Hwang H-W, Wentzel EA, Mendell JT. Cell-cell contact globally activates microRNA biogenesis. PNAS. 2009;106:7016–7021. doi: 10.1073/pnas.0811523106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Masuda N, Ohnishi T, Kawamoto S, Monden M, Okubo K. Analysis of chemical modification of RNA from formalin-fixed samples and optimization of molecular biology applications for such samples. Nucl. Acids Res. 1999;27:4436–4443. doi: 10.1093/nar/27.22.4436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]