Abstract
With the increased focus on healthy eating and consuming raw vegetables, this study assessed the extent of contamination of fresh vegetables by Pseudomonas aeruginosa in Jamaica and examined the antibiotic susceptibility profiles and the presence of various virulence associated determinants of P. aeruginosa. Analyses indicated that vegetables from retail markets and supermarkets were widely contaminated by P. aeruginosa; produce from markets were more frequently contaminated, but the difference was not significant. Lettuce and carrots were the most frequently contaminated vegetables, while tomatoes were the least. Pigment production (Pyoverdine, pyocyanin, pyomelanin and pyorubin), fluorescein and alginate were common in these isolates. Imipenem, gentamicin and ciprofloxacin were the most inhibitory antimicrobial agents. However, isolates were resistant or showed reduced susceptibility to ampicillin, chloramphenicol, sulphamethoxazole/trimethoprim and aztreonam, and up to 35% of the isolates were resistant to four antimicrobial agents. As many as 30% of the isolates were positive for the fpv1 gene, and 13% had multiple genes. Sixty-four percent of the isolates harboured an exoenzyme gene (exoS, exoT, exoU or exoY), and multiple exo genes were common. We conclude that P. aeruginosa is a major contaminant of fresh vegetables, which might be a source of infection for susceptible persons within the community.
1. Introduction
Pseudomonas aeruginosa is an oxidase-positive, nonfermentative, motile, and gram-negative bacterium that is ubiquitous and very versatile. While P. aeruginosa is considered an opportunistic pathogen, several reports indicate that the organism can also cause infections in healthy hosts [1–4]. Further, evidence has suggested that there are no major differences in virulence between clinical and environmental isolates, for example, clone and pilin-type distributions [5], pilin genes [6], flagellin genes [7], genes for multidrug efflux and type III secretion system, the porin gene oprD [8], haemolytic and proteolytic activities, and invasion of epithelial cells [9]. Consequently, consuming raw vegetables that have been contaminated by this organism could have serious implication on human health.
Although many of the organisms associated with food plants are nonpathogenic, many, including vegetables, may be contaminated through insufficiently-treated water and fertilizers or may be compromised by the use of biocides during cultivation. Contamination can occur in the field, during harvest, processing, distribution, and even at use [10, 11].
The intrinsic and acquired resistance of P. aeruginosa to many structurally-unrelated antibiotics is due to several adaptations, including active efflux systems, reduced cell wall permeability, plasmid acquisition, expression of various enzymes, or by biofilm formation [12, 13]. Pathogenesis involves production of both extracellular and cell-associated virulence factors [14]. Many virulence factors are expressed through a cell density-dependent mechanism known as quorum sensing [14]. These additional virulence factors include elastase, lipase, protease, and several cytotoxins, encoded by exo genes. Elastase and alkaline protease are known to degrade a large variety of tissue components such as proteinaceous elements of connective tissue and cleave the cell surface receptors on neutrophils [15].
In the present study, we examined the prevalence and antibiotic-resistance profiles of the organism and the presence of various virulence factors in P. aeruginosa isolated from vegetables obtained from various markets and supermarkets across Jamaica.
2. Materials and Methods
2.1. Microbial Isolation and Identification
Seventeen individual retail market and supermarket outlets, including two canteens serving lunches, located in several parishes in Jamaica were selected for the study. A total of 95 vegetable samples including lettuce (n = 15), white cabbage (n = 15), red cabbage (n = 3), carrots (n = 15), sweet pepper (n = 15), cucumber (n = 15), tomatoes (n = 15), and mixed vegetable salad (n = 2) were collected in plastic sample bags and stored 4°C for a maximum of 48 hr. Approximately seven samples were collected from each outlet. Prior to testing, sample bags were wiped with 70% alcohol, and a sterile knife was used to cut samples in sterile trays. Samples were trimmed of any spoiled parts, and their outer leaves removed. Samples were cut into pieces, and 25 g portions placed into sterile 225 mL Tryptic Soy Broth (TSB). Further decimal dilutions were made using physiological saline, and aliquots were plated onto Cetrimide base agar. Plates were incubated at 42°C for 48 hr, and organisms were purified on blood agar and MacConkey agar plates. Further, gram stain, motility, oxidase, and catalase tests were carried out. The criteria for identifying an isolate as Pseudomonas aeruginosa were oxidase positivity, catalase positivity, growth at 42°C, and pigment production [16]. P. aeruginosa PAO1 and PAOR1 (gifts from B. Iglewski) and ATCC 27853 (gift from R. Khan) were used as the reference strains in all tests completed. P. aeruginosa isolates were grown on blood agar and MacConkey agar plates to assess purity and on Mueller-Hinton agar plates to assess pigment production. Pyoverdine and pyocyanin production was assessed on Pseudomonas B and Pseudomonas A media, respectively. Escherichia coli isolates were purified on MacConkey agar and then inoculated on Triple iron agar, Urea agar, Simmons citrate agar, and Motility Indole-lysine media. Isolates that showed similar biochemical reactions to the E. coli standard strain ATCC25922 were selected for further study. Unless otherwise noted, bacteria were grown in Luria-Bertani medium (Oxoid, Basingstoke, UK, USA) or in Mueller-Hinton broth or agar (Becton Dickinson, Cockeysville, MD) at 37°C for 18 h.
2.2. Antimicrobial Susceptibility Testing
Susceptibility testing was performed by the standard CLSI (formerly known as NCCLS) disk diffusion method [17] using common antipseudomonad antibiotics: ampicillin (10 μg), aztreonam (30 μg), ceftazidime (30 μg), chloramphenicol (30 μg), ciprofloxacin (5 μg), gentamicin (10 μg), imipenem (10 μg), sulfamethoxazole-trimethoprim (1.25/23.75 μg), and tetracycline (30 μg) (BD Biosciences, MD, USA). Inocula were prepared by suspending growth from LB broth to a starting concentration of 5 × 105 cfu/mL. Mueller-Hinton plates were incubated at 35°C for 16 to 18 hours after inoculation with organisms and placement of the disks, and zones of inhibition were measured.
2.3. Multiplex PCR for Detection of Virulence Genes
Eight primers were used to assess the distribution of the Type III secretion (TTS) genes, exoS, exoT, exoU, and exoY [18]. Bacteria were grown overnight at 37°C in lauryl tryptose broth, and DNA extracted using the Wizard genomic DNA purification kit (Promega Corp.) according to the manufacturer's instructions. The PCR was set up as follows: 1 μL of DNA template (100 to 200 ng), 1 μL of each PCR primer (IDT, USA), (a final 200 mM concentration of each primer), 10 μL of GoTaq MasterMix (Promega), and 1 μL of sterile water. The negative control contained 1 μL sterile water instead of DNA. The PCR was carried out as follows: initial denaturation at 94°C for 2 min; 36 cycles of 94°C for 30 s, 58°C for 30 s, and 68°C for 1 min; a final extension step at 68°C for 7 min. Reaction products were separated in a 2% agarose gel and stained with 0.5 mg/mL of ethidium bromide.
2.4. Multiplex PCR for the Identification of fpvAI, fpvAII, and fpvAIII
Six primers were used for the simultaneous amplification of the different fpvA genes, which code for the three types of pyoverdine receptors produced by P. aeruginosa [19]. The PCR was set up as follows: 1 μL of DNA template (100 to 200 ng), 1 μL of each PCR primer (IDT), (a final 200 mM concentration of each primer), 10 μL of GoTaq MasterMix (Promega), and 3 μL of sterile water. The negative control contained 1 μL of sterile water instead of DNA. The following conditions were used: initial denaturation at 94°C for 3 min, followed by 30 cycles with denaturation at 94°C for 30 s, annealing at 55°C for 30 s and elongation at 72°C for 30 s, and terminating with a last cycle at 72°C for 10 min.
2.5. Statistical Analysis
Statistical analysis was performed using the χ 2 test. Differences were considered significant at P < 0.05.
3. Results
Samples of vegetables from markets (72.3%; range 50–100%) were more frequently contaminated than those from supermarkets (55.6%; range 14–86%). This difference was, however, not significant. Lettuce (89% in supermarkets; 100% in markets; P > 0.05) and carrots (67% in supermarkets; 100% in markets; P < 0.05) were the most frequently contaminated vegetable (Table 1), while tomatoes had the lowest level of contamination (22% in supermarkets; 50% in markets; P < 0.05) with counts of nil to 1.5 × 103 cfu/g. One supermarket in the eastern region had lettuce as the only sample being contaminated by P. aeruginosa with a count of >3.0 × 102 cfu/g. With the exception of tomatoes and lettuce, all other samples were contaminated with high levels of P. aeruginosa. Both samples of mixed vegetable salad were contaminated, with counts of 2.6 × 102 cfu/g and >6.0 × 103 cfu/g.
Table 1.
Level of contamination by Pseudomonas aeruginosa of vegetable samples from nine supermarkets and six markets investigated in this study.
| Vegetable sample | Supermarket (%) | Markets (%) |
|---|---|---|
| Cabbage, white | 6/9 (67%) | 3/6 (50%) |
| Cabbage, red | 2/3 (67%) | 0 |
| Carrots | 6/9 (67%) | 6/6 (100%) |
| Cucumbers | 5/9 (56%) | 4/6 (67%) |
| Lettuce | 8/9 (89%) | 6/6 (100%) |
| Sweet potatoes | 5/9 (56%) | 3/6 (50%) |
| Tomatoes | 2/9 (22%) | 3/6 (50%) |
|
| ||
| Total | 34/57 (60%) | 26/36 (72%) |
A total of 88 isolates of P. aeruginosa and three isolates of E. coli were recovered from 95 vegetable samples, and two vegetable salads included in this study (Table 2). Isolates were recovered from all vegetable samples, and several samples showed high viable count of bacteria (2.6 × 102 up to >1.2 × 106 cfu/g).
Table 2.
Frequency of contamination by Pseudomonas aeruginosa of vegetable samples from supermarkets, markets, and canteens in the three regions investigated in this study.
| Region | Supermarkets | Markets | Canteens |
|---|---|---|---|
| Eastern | 11 | 21 | 2 |
| Central | 14 | 12 | 0 |
| Western | 16 | 12 | 0 |
|
| |||
| Total | 41 | 45 | 2 |
Imipenem (100%), gentamicin (97%), ciprofloxacin (93%), and ceftazidime (79%) were the most inhibitory antibiotics found in this study based on susceptibility results (Figure 1). However, all isolates were resistant to ampicillin, and 84% and 83% were resistant to chloramphenicol and sulfamethoxazole/trimethoprim, respectively. However, it was of concern that reduced susceptibilities were observed to aztreonam (41%) and tetracycline (29%), given that these are environmental isolates. In the case of these latter antimicrobial agents, these values were in addition to resistance frequencies of 55% observed. Multidrug resistance (defined as resistance to three or more antimicrobial agents) was observed in isolates from all regions: 23%, 35%, and 20% of isolates were resistant to three, four, and five antimicrobial agents, respectively. Two isolates were resistant to six antimicrobials. Both of these isolates were from lettuce samples in the eastern region. Carrots, cabbage, cucumbers, and lettuce were the vegetables frequently associated with multidrug resistant isolates. Resistance to chloramphenicol, ampicillin, and trimethoprim/sulfamethoxazole was most frequently observed.
Figure 1.
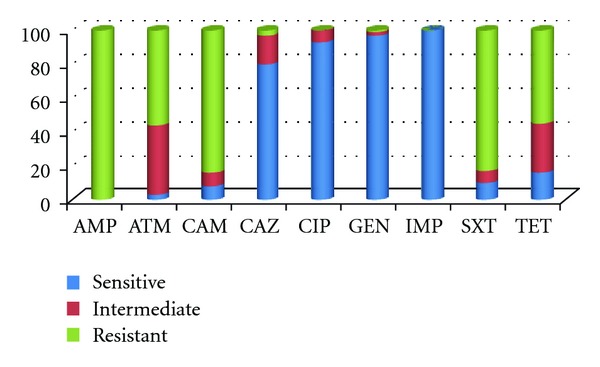
Frequency of resistance among P. aeruginosa isolates to common several antimicrobial agents. AMP: ampicillin; ATM: aztreonam; CAM: chloramphenicol; CAZ: ceftazidime; CIP: ciprofloxacin; GEN: gentamicin; IMP: imipenem; SXT: trimethoprim/sulfamethoxazole; TET: tetracycline.
Isolates produced pyoverdine, pyorubin, pyocyanin, pyomelanin, and fluorescein pigments. However, it was apparent that more pigmented isolates were obtained from sources in western Jamaica. While some pigments occurred in isolates from all regions (pyoverdine, pyomelanin, and fluorescein), others only occurred in isolates from the central and western (pyorubin) or western and eastern (pyocyanin) regions. In all, 42% isolates produced fluorescein while 68% isolates produced pyoverdine. Several isolates (n = 24) were nonpigmented and mucoid due to the overproduction of alginate, and were observed in the all regions, and associated with all vegetable samples. One of the isolates from the vegetable salad was non-pigmented, while the other produced pyoverdine and fluorescein.
The distributions of the pyoverdine receptor genes are illustrated in Figure 2. While a large proportion of isolates had the fpvI gene only (22%), a significant proportion (13%) had multiple genes. The total frequencies of isolates harbouring fpv1, fpv2, and fpv3 genes were 30%, 20%, and 23%, respectively. This correlated well with the observation that 68% of isolates produced pyoverdine pigment. Pyoverdine receptor genes were identified in all regions, and multiple genes were associated with cabbage and carrot samples. The two isolates that had all three FPV receptors were from cabbage samples: one was from a white cabbage sample from the central region and the other was a red cabbage from the eastern region.
Figure 2.
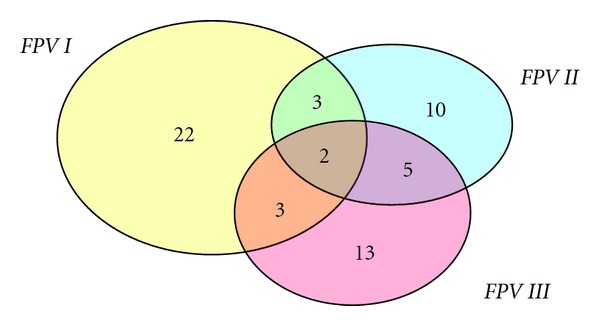
Distribution of pyoverdine receptor genes detected by PCR in P. aeruginosa isolates in this study.
Seventy-three (64%) isolates were PCR-positive for exoenzyme genes: 6% was positive for exoS (Figure 3); exoT, exoU, and exoY were identified in 40%, 33%, and 30% of isolates, respectively. While no isolate expressed exoS in conjunction with any other exoenzyme gene, 14% of isolates were positive for exoT, exoU, and exoY. These exo genes occurred in isolates from all regions, and were more frequently associated with carrots, lettuce, and cabbage samples.
Figure 3.
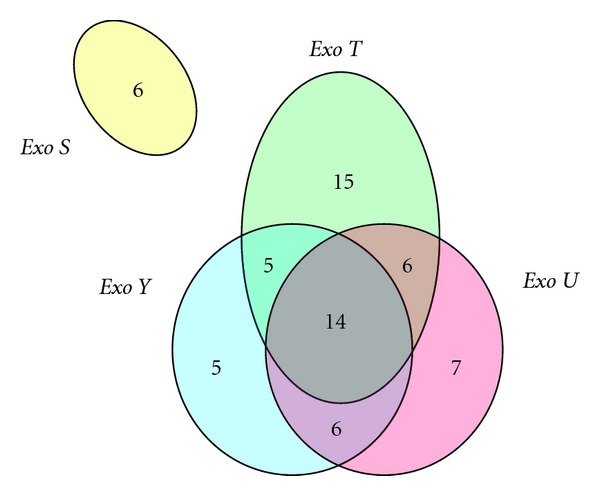
Distribution of exoenzyme genes detected by PCR in P. aeruginosa isolates in this study.
We identified 19 isolates with multidrug resistance, multiple exoenzyme genes, and at least one FPV receptor gene. There was no apparent association noted between pigment production and multiple drug resistance. However, non-pigment producers (alginate producers) were less likely to have genes for exoenzyme or FPV receptor genes, with the exception of two isolates. One was from a red cabbage in the eastern region, which was resistant to five antimicrobial agents, and had exoT and U and fpv1, fpv2, and fpv3. The other was from sweet pepper in the western region and was resistant to four antimicrobials, had exoU, exoY, fpv2, and fpv3. One isolate from a vegetable salad sample was resistant to four antimicrobials, was pigmented and had exoT, exoU, and exoY in addition to the fpv3 gene.
4. Discussion
Observations showed considerable bacteriological load carried by vegetable samples with total viable counts ranging from 102 to more than 106 cfu/g. The presence of high numbers of viable bacteria, an indicator of the expected shelf life of the vegetable, increases the likelihood of spoilage, as well as the possibility of produce-associated outbreaks [20]. In recent decades, public health promotion of healthier lifestyles has led to increased demand for fresh produce in many countries, including developing countries like Jamaica. Due to the fact that these products are often consumed raw or with very little cooking, consumers face an increased risk of infection from contaminating microorganisms. Despite the risk management strategies instituted in many countries, the number of reported illnesses linked to contaminated produce have increased in the USA [21] and in Europe [22]. While most reports have identified Salmonella and E. coli as the main produce-associated pathogens, we have to remain vigilant about the potential risks from others such as Pseudomonas aeruginosa.
In this study, we sought to identify the prevalence of P. aeruginosa contaminating fresh vegetables in markets and supermarkets in Jamaica. We noted that vegetables from both markets and supermarkets were contaminated, with lettuce and carrots being the most frequently contaminated. Numerous studies have investigated the potential sources of contamination in the supply chain at the pre- and postharvest stages. For example, during the preharvest phase, pathogen populations can establish themselves on growing crops, and the risk can be amplified after harvest either by further direct contamination or by proliferation of existing pathogen populations during processing and postharvest handling procedures. Water is likely to be an important source of contamination in the field, including runoff from nearby animal pastures and irrigation from contaminated sources [23, 24]. Further, use of water in postharvest processing has also played a role, for example, in trying to prevent importation of fruit flies [25]. It is well established that pathogens may be disseminated in the environment via the use of inadequately composted or raw animal manures or sewage [26, 27], via the faeces of wild animals [28], or via flies [29, 30]. Post-harvest processes, ranging from storage and rinsing to cutting, are also possible sources of contamination [31], and the use of inadequately decontaminated water in hydrocoolers, which are used to store and process large quantities of fresh produce, can lead to contamination of an entire lot [32]. Given that there were few isolates of E. coli relative to P. aeruginosa, we concluded that contamination was more likely from nonmanure sources, particularly soil, flies, cockroaches, or rodents. In other words, the organism's presence in the environment and increased temperature and humidity, such as those normally experienced in tropical countries, may be predisposing factors for growth or colonization of vegetables. In fact, frank unsanitary conditions were not observed in the markets included in the study, so it is likely that the growth medium used is selected for P. aeruginosa rather than E. coli. Further, Shigeharu and coworkers [33] noted that contamination was not markedly decreased after disinfection with sodium hypochlorite, a common agent used to provide protection against pathogenic organisms. Historically, Correa et al. [34] reported that 19% of the fresh vegetable samples fed to oncology patients were found to be contaminated by P. aeruginosa even though 1% hypochlorite was used as a disinfectant. Similar to this study, those authors reported that lettuce was among the vegetables that yielded the highest frequency of isolation. Further, the authors reported that some of the pyocin typing and serotyping of vegetable isolates were found to be identical to those recovered from clinical sources.
Lettuce and carrots were the two vegetables that were mainly associated with P. aeruginosa contamination in all regions analyzed in this study. An increased growth of this organism on lettuce may be as a result of its wide surface area. A combination of factors may have contributed to the high contamination yield obtained from all supermarkets and markets across the island as suggested previously. There were no major differences with isolation of P. aeruginosa from the various regions assessed as all seem to have been equally implicated by this organism. There was no major difference in counts between vegetables obtained.
The inner tissues of vegetables are supposed to be free of microorganisms [35]. For this study, even though the outer leaves were removed from cabbage and lettuce, the recovery of P. aeruginosa remained considerably high. The market samples as expected were comparatively more contaminated by P. aeruginosa than those of the supermarkets. Considering that poorer persons buy produce in the markets, it is likely that these are at increased risk with the often additional burden of no or inadequate health coverage. In addition to supermarkets and markets, it was considered that consumers may conduct additional washing before use. Through washing, more nutrients become available, and potential pathogenic organisms can spread from contaminated parts to uncontaminated areas [36]. Further, ready-to-eat mixed vegetable salads are frequently contaminated and may be directly related to the quality of the inputs from market or supermarket sources.
Although healthy individuals require >105 cfu/mL P. aeruginosa, the ingestion of <103 cfu/g may colonize the intestine of susceptible individuals and may lead to a gastrointestinal infection, bacteraemia, and haematogenous spread [37]. Pseudomonas septicaemia in infants manifested as necrotizing bowel lesions with a history of diarrhoea is also possible [38]. While bacteria in the gastrointestinal tract have been regarded as important in immunocompromised patients, such as those undergoing anticancer chemotherapy, it is of direct concern that the pathogen can also infect persons from the community and might cause fatal bacteraemia [2] and community-acquired pneumonia [1]. Other reports showed incidences of community-acquired Pseudomonas infection of the gastrointestinal tract causing diarrhoea in infants [39].
Further, we noted that several of these isolates had multiple pyoverdine receptors genes. Pyoverdine acts as a siderophore and enables P. aeruginosa to acquire more iron from the host cells to carry out its metabolic functions and would therefore increase the virulence of this organism. Pyoverdine also has cytotoxic effect because of its ability to stimulate the production of reactive oxygen and has been determined to be an essential component in biofilm production [14]. Pyocyanin also contributes to the virulence of this organism. Like pyoverdine, it is toxic to both bacterial and eukaryotic cells, this is due to the reactive oxygen that intermediates the organism generates [40]. In a previous study [41], we noted that isolates that produced either pyoverdine or pyocyanin were more likely to produce additional virulence factors. It was therefore not surprising to observe the extent of multiple exo genes and/or FPV receptor genes in these organisms. Expression of these genes is governed by quorum sensing [42] and is known to be involved in the pathogenesis of P. aeruginosa [43].
Alginate production by P. aeruginosa is an important virulence factor as its expression allows the bacteria to resist phagocytosis. This expression contributes to the development and persistence of the bacteria in patients with chronic pulmonary infections, including cystic fibrosis patients [44–46]. Some of these alginate producing strains had multiple exo and fpv genes.
Many of the isolates were found to have several genes that are expressed and secreted by type 3 secretion systems (TTSS). In this study, we noted that the exoS gene did not occur with any other exoenzyme gene and suggest that exoS might be mutually exclusive to other exo genes. Exoenzyme S inhibits RHO GTPase family signalling, paralyzing macrophages, and inhibiting phagocytosis [47, 48]. ExoS and ExoT are highly related and have dual functions. ExoT contributes to actin cytoskeleton disruption and inhibition of internalization of the bacteria [49, 50]. ExoU has the ability to lyse host cell membrane and has cytotoxic effects [51], while ExoY acts as adenylate cyclase that increases the levels of intracellular cAMP [52].
P. aeruginosa is one of the leading causes of hospital- and community-acquired infections due to its ability to cause a variety of diseases and its high-level resistance to several antibiotics. It was originally believed that this ability was due to the low outer membrane (OM) permeability. However, it is now known to be as a result of the combined action of multidrug resistance pumps in association with OM permeability [53]. However, crop irrigation and application of pesticides with contaminated water can be a primary source of resistant bacteria in a field, and antimicrobial resistance could also originate from bacterial adaptation to heavy metals and plant metabolites, resulting in the formation of multidrug efflux systems [54]. It is also likely that these adaptations could result in increased expression of existing efflux systems. From the isolates studied, many showed multiple resistance to the antimicrobials tested. While most of the isolates were susceptible to ceftazidime, ciprofloxacin, gentamicin, and imipenem, it is of concern that these environmental isolates exhibited significant frequencies of resistance or reduced susceptibilities to so many antimicrobial agents including ampicillin, chloramphenicol, sulfamethoxazole/trimethoprim, tetracycline, and aztreonam. Of note was that many of these isolates expressed pyoverdine pigment and fluorescein and were also alginate-producing strains. While antimicrobial resistance does not appear to be associated with the production of pigment [41], more work might be warranted in environmental isolates, particularly those associated with produce. As has been expressed elsewhere, contaminated vegetables may be an important reservoir or source of community- and hospital-acquired strains of P. aeruginosa.
5. Conclusions
P. aeruginosa contamination of raw vegetables was found to be prevalent in Jamaica as contamination was observed in samples collected from markets and supermarkets in all regions (eastern, western, and central locations). Many of these isolates were resistant or had reduced susceptibilities to multiple antimicrobial agents, produced pigments or alginate, and had multiple exoenzyme or pyoverdine receptor genes. Because of the increased focus on healthy eating and consumption of vegetables, this is a major health concern as the possibility of innocuous spread of pathogenic organisms can be facilitated among unsuspecting and vulnerable individuals. Consequently, the application of proper hygiene practices along the food production/supply chain is essential.
Conflict of Interests
The authors declare that they have no conflict of interests.
Acknowledgments
This work was supported by a grant from the Bureau of Standards Jamaica (to K. Allydice-Francis). The authors thank the Northern Caribbean University and the University of the West Indies at Mona, Jamaica for facilitating this research and a number of other persons for their advice.
References
- 1.Hatchette TF, Gupta R, Marrie TJ. Pseudomonas aeruginosa community-acquired pneumonia in previously healthy adults: case report and review of the literature. Clinical Infectious Diseases. 2000;31(6):1349–1356. doi: 10.1086/317486. [DOI] [PubMed] [Google Scholar]
- 2.Kang CI, Kim SH, Park WB, et al. Clinical features and outcome of patients with community-acquired Pseudomonas aeruginosa bacteraemia. Clinical Microbiology and Infection. 2005;11(5):415–418. doi: 10.1111/j.1469-0691.2005.01102.x. [DOI] [PubMed] [Google Scholar]
- 3.Lo WT, Wang CC, Hsu ML, Chu ML. Pyogenic liver abscess caused by Pseudomonas aeruginosa in a previously healthy child: report of one case. Acta Paediatrica Taiwanica. 2000;41(2):98–100. [PubMed] [Google Scholar]
- 4.McCallum SJ, Gallagher MJ, Corkill JE, Hart CA, Ledson MJ, Walshaw MJ. Spread of an epidemic Pseudomonas aeruginosa strain from a patient with cystic fibrosis (CF) to non-CF relatives. Thorax. 2002;57(6):559–560. doi: 10.1136/thorax.57.6.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Römling U, Wingender J, Muller H, Tümmler B. A major Pseudomonas aeruginosa clone common to patients and aquatic habitats. Applied and Environmental Microbiology. 1994;60(6):1734–1738. doi: 10.1128/aem.60.6.1734-1738.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Spangenberg C, Fislage R, Sierralta W, Tümmler B, Römling U. Comparison of type IV-pilin genes of Pseudomonas aeruginosa of various habitats has uncovered a novel unusual sequence. FEMS Microbiology Letters. 1995;127(1-2):p. 158. doi: 10.1111/j.1574-6968.1995.tb07466.x. [DOI] [PubMed] [Google Scholar]
- 7.Morgan JAW, Bellingham NF, Winstanley C, Ousley MA, Hart CA, Saunders JR. Comparison of flagellin genes from clinical and environmental Pseudomonas aeruginosa isolates. Applied and Environmental Microbiology. 1999;65(3):1175–1179. doi: 10.1128/aem.65.3.1175-1179.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pirnay JP, De Vos D, Mossialos D, Vanderkelen A, Cornelis P, Zizi M. Analysis of the Pseudomonas aeruginosa oprD gene from clinical and environmental isolates. Environmental Microbiology. 2002;4(12):872–882. doi: 10.1046/j.1462-2920.2002.00281.x. [DOI] [PubMed] [Google Scholar]
- 9.Alonso A, Rojo F, Martínez JL. Environmental and clinical isolates of Pseudomonas aeruginosa show pathogenic and biodegradative properties irrespective of their origin. Environmental Microbiology. 1999;1(5):421–430. doi: 10.1046/j.1462-2920.1999.00052.x. [DOI] [PubMed] [Google Scholar]
- 10.Beuchat LR. Pathogenic microorganisms associated with fresh produce. Journal of Food Protection. 1996;59(2):204–216. doi: 10.4315/0362-028X-59.2.204. [DOI] [PubMed] [Google Scholar]
- 11.Garg N, Churey JJ, Splittstoesser DF. Effect of processing conditions on the microflora of fresh-cut vegetables. Journal of Food Protection. 1990;53:701–773. doi: 10.4315/0362-028X-53.8.701. [DOI] [PubMed] [Google Scholar]
- 12.Deplano A, Denis O, Poirel L, et al. Molecular characterization of an epidemic clone of panantibiotic-resistant Pseudomonas aeruginosa . Journal of Clinical Microbiology. 2005;43(3):1198–1204. doi: 10.1128/JCM.43.3.1198-1204.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Köhler T, Van Delden C, Curty LK, Hamzehpour MM, Pechere JC. Overexpression of the MexEF-OprN multidrug efflux system affects cell-to-cell signaling in Pseudomonas aeruginosa . Journal of Bacteriology. 2001;183(18):5213–5222. doi: 10.1128/JB.183.18.5213-5222.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wagner VE, Filiatrault MJ, Picardo KF, Iglewski BH. Pseudomonas: Genomics and Molecular Biology. Norfolk, UK: Caister Academic Press; 2008. Pseudomonas aeruginosa: virulence and pathogenesis issues; pp. 129–158. [Google Scholar]
- 15.Lomholt JA, Poulsen K, Kilian M. Epidemic population structure of Pseudomonas aeruginosa: evidence for a clone that is pathogenic to the eye and that has a distinct combination of virulence factors. Infection and Immunity. 2001;69(10):6284–6295. doi: 10.1128/IAI.69.10.6284-6295.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kiska DL, Gilligan PH. Pseudomonas. In: Murray PR, editor. Manual of Clinical Microbiology. 8th edition. Vol. 47. Washington, DC, USA: ASM Press; 2003. pp. 719–728. [Google Scholar]
- 17.National Committee for Clinical Laboratory Standards. Twelfth Informational Supplement. M100-S12. Wayne, Pa, USA: NCCLS; 2002. Performance standards for antimicrobial susceptibility testing. [Google Scholar]
- 18.Ajayi T, Allmond LR, Sawa T, Wiener-Kronish JP. Single-nucleotide-polymorphism mapping of the Pseudomonas aeruginosa type III secretion toxins for development of a diagnostic multiplex PCR system. Journal of Clinical Microbiology. 2003;41(8):3526–3531. doi: 10.1128/JCM.41.8.3526-3531.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Chial M, Ghysels B, Beatson SA, et al. Identification of type II and type III pyoverdine receptors from Pseudomonas aeruginosa . Microbiology. 2003;149(4):821–831. doi: 10.1099/mic.0.26136-0. [DOI] [PubMed] [Google Scholar]
- 20.Berger CN, Sodha SV, Shaw RK, et al. Fresh fruit and vegetables as vehicles for the transmission of human pathogens. Environmental Microbiology. 2010;12(9):2385–2397. doi: 10.1111/j.1462-2920.2010.02297.x. [DOI] [PubMed] [Google Scholar]
- 21.Sivapalasingam S, Friedman CR, Cohen L, Tauxe RV. Fresh produce: a growing cause of outbreaks of foodborne illness in the United States, 1973 through 1997. Journal of Food Protection. 2004;67(10):2342–2353. doi: 10.4315/0362-028x-67.10.2342. [DOI] [PubMed] [Google Scholar]
- 22.Westrell T, Ciampa N, Boelaert F, et al. Zoonotic infections in Europe in 2007: a summary of the EFSA-ECDC annual report. Eurosurveillance. 2009;14:p. 19100. [PubMed] [Google Scholar]
- 23.Hamilton AJ, Stagnitti F, Premier R, Boland AM, Hale G. Quantitative microbial risk assessment models for consumption of raw vegetables irrigated with reclaimed water. Applied and Environmental Microbiology. 2006;72(5):3284–3290. doi: 10.1128/AEM.72.5.3284-3290.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tyrrel SF, Knox JW, Weatherhead EK. Microbiological water quality requirements for salad irrigation in the United Kingdom. Journal of Food Protection. 2006;69(8):2029–2035. doi: 10.4315/0362-028x-69.8.2029. [DOI] [PubMed] [Google Scholar]
- 25.Sivapalasingam S, Barrett E, Kimura A, et al. A multistate outbreak of Salmonella enterica serotype newport infection linked to mango consumption: impact of water-dip disinfestation technology. Clinical Infectious Diseases. 2003;37(12):1585–1590. doi: 10.1086/379710. [DOI] [PubMed] [Google Scholar]
- 26.Natvig EE, Ingham SC, Ingham BH, Cooperband LR, Roper TR. Salmonella enterica serovar typhimurium and Escherichia coli contamination of root and leaf vegetables grown in soils with incorporated bovine manure. Applied and Environmental Microbiology. 2002;68(6):2737–2744. doi: 10.1128/AEM.68.6.2737-2744.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Santamaría J, Toranzos GA. Enteric pathogens and soil: a short review. International Microbiology. 2003;6(1):5–9. doi: 10.1007/s10123-003-0096-1. [DOI] [PubMed] [Google Scholar]
- 28.Ackers ML, Mahon BE, Leahy E, et al. An outbreak of Escherichia coli O157:H7 infections associated with leaf lettuce consumption. Journal of Infectious Diseases. 1998;177(6):1588–1593. doi: 10.1086/515323. [DOI] [PubMed] [Google Scholar]
- 29.Sela S, Nestel D, Pinto R, Nemny-Lavy E, Bar-Joseph M. Mediterranean fruit fly as a potential vector of bacterial pathogens. Applied and Environmental Microbiology. 2005;71(7):4052–4056. doi: 10.1128/AEM.71.7.4052-4056.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Talley JL, Wayadande AC, Wasala LP, et al. Association of Escherichia coli O157:H7 with filth flies (Muscidae and Calliphoridae) captured in leafy greens fields and experimental transmission of E. coli O157:H7 to spinach leaves by house flies (diptera: Muscidae) Journal of Food Protection. 2009;72(7):1547–1552. doi: 10.4315/0362-028x-72.7.1547. [DOI] [PubMed] [Google Scholar]
- 31.Wachtel MR, Charkowski AO. Cross-contamination of Lettuce with Escherichia coli O157:H7. Journal of Food Protection. 2002;65(3):465–470. doi: 10.4315/0362-028x-65.3.465. [DOI] [PubMed] [Google Scholar]
- 32.Gagliardi JV, Millner PD, Lester G, Ingram D. On-farm and postharvest processing sources of bacterial contamination to melon rinds. Journal of Food Protection. 2003;66(1):82–87. doi: 10.4315/0362-028x-66.1.82. [DOI] [PubMed] [Google Scholar]
- 33.Shigeharu O, Kiyonaga H, Matsuzaka Y, et al. Microbial contamination of fruit and vegetables and their disinfection. Biological and Pharmaceutical Bulletin. 2008;31(10):1902–1905. doi: 10.1248/bpb.31.1902. [DOI] [PubMed] [Google Scholar]
- 34.Correa CMC, Tibana A, Gontijo Filho PP. Vegetables as a source of infection with Pseudomonas aeruginosa in a university and oncology hospital of Rio de Janeiro. Journal of Hospital Infection. 1991;18(4):301–306. doi: 10.1016/0195-6701(91)90187-d. [DOI] [PubMed] [Google Scholar]
- 35.Lund BM. Ecosystems in vegetable foods. Journal of Applied Bacteriology Symposium Supplement. 1992;(21) doi: 10.1111/j.1365-2672.1992.tb03631.x. [DOI] [PubMed] [Google Scholar]
- 36.European Union Scientific Committee on Food. Risk Profile on the Microbiological Contamination of Fruits and Vegetables Eaten Raw, 2002, http://europa.eu.int/comm/food/fs/sc/scf/index_en.html.
- 37.Kominos SD, Copeland CE, Grosiak B. Mode of transmission of Pseudomonas aeruginosa in a burn unit and an intensive care unit in a general hospital. Applied microbiology. 1972;23(2):309–312. doi: 10.1128/am.23.2.309-312.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tsai MJ, Teng CJ, Teng RJ, Lee PI, Chang MH. Necrotizing bowel lesions complicated by Pseudomonas septicaemia in previously healthy infants. European Journal of Pediatrics. 1996;155(3):216–218. doi: 10.1007/BF01953941. [DOI] [PubMed] [Google Scholar]
- 39.Yeung CK, Lee KH. Community acquired fulminant Pseudomonas infection of the gastrointestinal tract in previously healthy infants. Journal of Paediatrics and Child Health. 1998;34(6):584–587. doi: 10.1046/j.1440-1754.1998.00290.x. [DOI] [PubMed] [Google Scholar]
- 40.Wilson R, Dowling RB. Pseudomonas aeruginosa and other related species. Thorax. 1998;53(3):213–219. doi: 10.1136/thx.53.3.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Finlayson EA, Brown PD. Comparison of antibiotic resistance and virulence factors in pigmented and non-pigmented Pseudomonas aeruginosa . West Indian Medical Journal. 2011;60:24–32. [PubMed] [Google Scholar]
- 42.Mavrodi DV, Bonsall RF, Delaney SM, Soule MJ, Phillips G, Thomashow LS. Functional analysis of genes for biosynthesis of pyocyanin and phenazine-1-carboxamide from Pseudomonas aeruginosa PAO1. Journal of Bacteriology. 2001;183(21):6454–6465. doi: 10.1128/JB.183.21.6454-6465.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reszka KJ, O’Malley Y, McCormick ML, Denning GM, Britigan BE. Oxidation of pyocyanin, a cytotoxic product from Pseudomonas aeruginosa, by microperoxidase 11 and hydrogen peroxide. Free Radical Biology and Medicine. 2004;36(11):1448–1459. doi: 10.1016/j.freeradbiomed.2004.03.011. [DOI] [PubMed] [Google Scholar]
- 44.Gacesa P. Bacterial alginate biosynthesis-recent progress and future prospects. Microbiology. 1998;144(5):1133–1143. doi: 10.1099/00221287-144-5-1133. [DOI] [PubMed] [Google Scholar]
- 45.Pier GB, Coleman F, Grout M, Franklin M, Ohman DE. Role of alginate O acetylation in resistance of mucoid Pseudomonas aeruginosa to opsonic phagocytosis. Infection and Immunity. 2001;69(3):1895–1901. doi: 10.1128/IAI.69.3.1895-1901.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stapper AP, Narasimhan G, Ohman DE, et al. Alginate production affects Pseudomonas aeruginosa biofilm development and architecture, but is not essential for biofilm formation. Journal of Medical Microbiology. 2004;53(7):679–690. doi: 10.1099/jmm.0.45539-0. [DOI] [PubMed] [Google Scholar]
- 47.Krueger KM, Barbieri JT. The family of bacterial ADP-riboscylating exotoxins. Clinical Microbiology Reviews. 1995;8(1):34–47. doi: 10.1128/cmr.8.1.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ganesan AK, Vincent TS, Olson JC, Barbieri JT. Pseudomonas aeruginosa exoenzyme S disrupts Ras-mediated signal transduction by inhibiting guanine nucleotide exchange factor-catalyzed nucleotide exchange. Journal of Biological Chemistry. 1999;274(31):21823–21829. doi: 10.1074/jbc.274.31.21823. [DOI] [PubMed] [Google Scholar]
- 49.Krall R, Schmidt G, Aktories K, Barbieri JT. Pseudomonas aeruginosa ExoT is a Rho GTPase-activating protein. Infection and Immunity. 2000;68(10):6066–6068. doi: 10.1128/iai.68.10.6066-6068.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Garrity-Ryan L, Kazmierczak B, Kowal R, Comolli J, Hauser A, Engel JN. The arginine finger domain of ExoT contributes to actin cytoskeleton disruption and inhibition of internalization of Pseudomonas aeruginosa by epithelial cells and macrophages. Infection and Immunity. 2000;68(12):7100–7113. doi: 10.1128/iai.68.12.7100-7113.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Finck-Barbançon V, Frank DW. Multiple domains are required for the toxic activity of Pseudomonas aeruginosa ExoU. Journal of Bacteriology. 2001;183(14):4330–4344. doi: 10.1128/JB.183.14.4330-4344.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yahr TL, Vallis AJ, Hancock MK, Barbieri JT, Frank DW. ExoY, an adenylate cyclase secreted by the Pseudomonas aeruginosa type III system. Proceedings of the National Academy of Sciences of the United States of America. 1998;95(23):13899–13904. doi: 10.1073/pnas.95.23.13899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ma D, Cook DN, Hearst JE, Nikaido H. Efflux pumps and drug resistance in gram-negative bacteria. Trends in Microbiology. 1994;2(12):489–493. doi: 10.1016/0966-842x(94)90654-8. [DOI] [PubMed] [Google Scholar]
- 54.Schwaiger K, Hölzel CH, Bauer J. Detection of the macrolide-efflux protein A gene mef(A) in Enterococcus faecalis . Microbial Drug Resistance. 2011;17(3):429–432. doi: 10.1089/mdr.2010.0192. [DOI] [PubMed] [Google Scholar]


