Abstract
Recent studies support the notion that the pre-translocation (PRE) ribosomal complex functions, at least in part, as a Brownian machine, stochastically fluctuating among multiple conformations and transfer RNA (tRNA) binding configurations. Apart from the relatively more energetically stable conformational states of the PRE complex, termed macrostate I (MS I) and macrostate II (MS II), several additional intermediate states have been recently discovered. Structural and kinetic analyses of these states, made possible by cryogenic electron microscopy (cryo-EM), X-ray crystallography, and single-molecule fluorescence resonance energy transfer (smFRET), have provided important insights into the translocation process, which is now understood to proceed, at least in the first step of the process, as a Brownian machine that is transiently stabilized in the “productive” MS II conformation by the binding of the translocase elongation factor G (EF-G).
INTRODUCTION
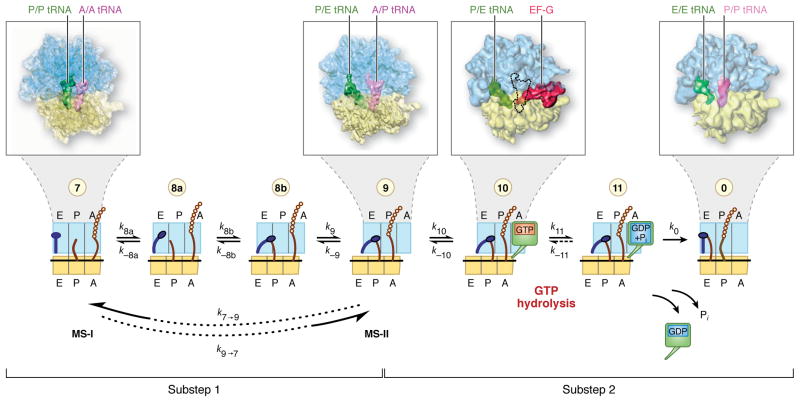
Translocation is the step during the elongation cycle, immediately following peptidyl transfer, in which mRNA and the tRNAs are advanced by the span of one codon, preparing the ribosome for the selection and incorporation of the next aminoacyl-tRNA (Fig. 1). Conceptually, this step is divided into two substeps: in the first substep (Fig. 1, states denoted as 7–9), the small subunit rotates relative to the large subunit and the tRNAs advance from the A (aminoacyl) site to the P (peptidyl) site, and from the P to the E (exit) site, respectively, relative to the large subunit to form hybrid (A/P, P/E) configurations [1]. In the second substep (Fig. 1, states denoted as 9-0), following the binding of EF-G and GTP hydrolysis, the small subunit rotates back, and mRNA and tRNAs bound to it advance relative to the small subunit by the span of one codon.
Fig. 1.
The mRNA-tRNA translocation process, and definition of translocation substeps 1 and 2. This diagram presents a sequence of distinct steps during the process of mRNA-tRNA translocation, as characterized by biochemical and structural studies. The blue feature on the left symbolizes the L1 stalk in its “open” and “closed” conformations. The green box on the right in states 10 and 11 stands for EF-G in GTP and GDP forms. Substep 1 requires the rotation of the small subunit body (symbolized by a shift of the yellow box with respect to the blue box) and effects the translocation of the tRNA on the large subunit, through the formation of hybrid A/P and P/E configurations. Substep 2, reached upon binding of EF-G followed by GTP hydrolysis, effects the translocation of mRNA and the tRNAs bound to it with respect to the small subunit. This review focuses on substep 1, which involves several structural intermediates. (Note: In state 10, it has not been possible to trap a complex containing P/E-tRNA, A/P-tRNA and EF-G all in one complex for cryo-EM. For illustration of the general binding position of EF-G on the ribosome, the cryo-EM map depicted here shows a complex that lacks the A-site tRNA and thus allows EF-G to bind in the GTP form. The predicted position of A/P-tRNA in a hypothetical authentic EF-G-bound pretranslocational complex is indicated by an outline. In such a complex, the steric conflict with domain IV of EF-G is presumably resolved by the positional flexibility of this domain known for the free form of EF-G [13]) (Adapted from [2]).
A growing body of studies focusing on the first substep strongly suggest that the ribosome is a Brownian machine that performs a major part of its work by harnessing thermal energy from its surroundings [2•,3,4,5,6•], though a detailed discussion of ribosome dynamics must include a consideration of the energy set free by peptide bond formation [7]. As such, the mechanism of ribosome-catalyzed protein synthesis in the cell can be analyzed and described within the framework of statistical mechanics [8,9]. Necessary ingredients that enable a Brownian ratchet machine to undergo a processive motion are the “pawl” and the expenditure of chemical energy, which are responsible for directionally biasing what are otherwise random, thermally driven conformational fluctuations [10]. What distinguishes the ribosome from other processive, thermally powered biomolecular machines are the large length scales of its motions – relative to the molecule’s size – and the extensive number of non-covalent interactions that must be remodeled during these motions, both of which are difficult to conceive of without postulating the existence of structural intermediates, or conformational states with moderate stability, along the reaction pathway. While it is likely that the Brownian machine principle is at work at every stage of translation, an idea that has spurred molecular dynamics (MD) simulation studies of ribosomal components [11,12], individual factors [13], and the ribosome in its entirety [14,15•], this article focuses on the first substep defined above.
Intersubunit (“ratchet-like”) rotation of the pre-translocation (PRE) ribosomal complex (i.e., after peptidyl transfer but before translocation) was first recognized by cryogenic electron microscopy (cryo-EM) [16], and is a requirement for translocation, as was established by an experiment in which the two ribosomal subunits were cross-linked across the intersubunit interface, preventing the rotation [17]. In fact, the idea that the two ribosomal subunits are likely to move relative to each other during translation was spelled out early on independently by Bretscher (1968) [18]and Spirin (1968)[19]. Bulk [20] as well as single-molecule fluorescence resonance energy transfer (FRET) studies [21,22] indicated that at physiological (~3.5 mM) concentrations of Mg++, in the absence of EF-G, the PRE ribosome undergoes random intersubunit rotations, as it oscillates between the two macrostates (MS I and II, see ref [23], also known as “global states” *22+). Indeed, structural analysis of such a sample with cryo-EM revealed that the PRE complex exists in MS I and MS II in a mixture of roughly 30%:70%, respectively [24], though this ratio has been recently revised based on a more detailed analysis of the same dataset [25••], as will be seen below. Julian et al. (2008) [26] came to similar conclusions. The fact that the ribosome binding configuration of the tRNAs depends strongly on the concentration of Mg++, with [Mg++] > 5 mM favoring the classical tRNA configuration [27], explains why earlier cryo-EM studies using high Mg++ concentrations failed to find structural evidence of spontaneous interconversion between different states of the PRE complex.
The absence of EF-G in the bulk and single-molecule FRET (smFRET) experiments, and thus the absence of the required unidirectional rectification of the Brownian motion, permits the PRE complex to idle back and forth unproductively between MS I and MS II but prevents the full translocation reaction from being efficiently carried out. The frequency with which the PRE complex oscillates between MS I and MS II in the absence of EF-G exhibits the temperature-dependence that would be expected for a thermally driven Brownian process, as has been demonstrated by recent smFRET experiments [6•].
EXPERIMENTAL EVIDENCE SUPPORTING THE EXISTENCE OF INTERMEDIATE STATES BEYOND MS I and MS II
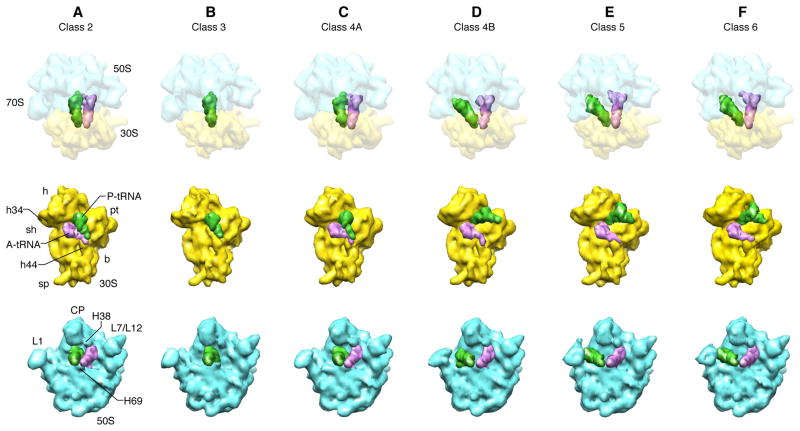
The first indications suggesting the existence of structural intermediates beyond MS I and MS II sampled by the bacterial PRE complex came from smFRET studies of a PRE complex harboring a single-point mutation within the so-called P-loop of the 23S ribosomal RNA (rRNA) component of the large ribosomal subunit, which is responsible for directly base pairing to the aminoacyl acceptor stem of the P/P configured deacylated tRNA within MS I or the A/P configured peptidyl-tRNA within MS II [28]. The assumption underlying this experiment is that an intermediate state that is sampled along the translocation pathway of a wild-type PRE complex, but that is too short-lived to be observed given the typical time resolution of smFRET experiments (~40 ms), is sufficiently stabilized in the mutant PRE complex such that it becomes long-lived enough to be detectable by smFRET. In fact, a hidden Markov modeling in the analysis of an analogous wild-type experiment detected evidence of the mutation-induced intermediates [28], though these results could not be independently confirmed by another lab [29]. A subsequent cryo-EM study of this mutant PRE complex [30•] showed that the ribosome within the mutant PRE complex indeed assumes at least two intermediate angles of intersubunit rotation along with two intermediate configurations of the P-site tRNA that had not been previously observed. The discovery of these two mutation-stabilized intermediates prompted a re-examination of the wild-type dataset that was previously classified into MS I and II [24] with a more powerful method of classification, a maximum likelihood method developed by the Carazo group [31]. This re-examination resulted in the discovery of at least two additional intermediate states of the wild-type PRE complex (Fig. 2) [25••] which have similarity to, but are not identical with, the two states discovered in the mutant PRE complex [30•]. Not surprisingly, given the degree of similarity in ribosome core structure and functional mechanism, analogous states with intermediate rotations and tRNA positions have also been found in a cryo-EM study of the eukaryotic PRE complex (Fig. 3) [32••], although a detailed comparison with bacterial intermediates is not yet available. At any rate, these authors do note significant differences in the classical A/A, P/P configurations when comparing eukaryotic with bacterial ribosome complexes.
Fig. 2.
Inventory of states in a pre-translocational sample of wild-type E. coli ribosomes.
First row: reconstructions of the 70S ribosome from six classes found by classification, shown in top view to aid in the comparison with Fig. 4. Second and third row: interface of small and large subunit, after computational removal of the opposite subunit, respectively. Class 3 depicts a population of ribosomes in which no A-site tRNA is bound. Class 2 represents the unrotated (MS I) state, with tRNAs in the classical A/A, P/P configuration. Classes 5 and 6 are very similar, representing the rotated MS II state, with tRNAs in the fully hybrid A/P, P/E configuration. Classes 4A and 4B are two distinct novel intermediates. (Landmarks on small subunit: h, head; h34, helix 34; sh, shoulder; h44, helix 44; sp, spur; b, body; pt, platform. Landmarks on large subunit: L1, L1 stalk; H69, helix 69; L7/L12, L7/L12 stalk; H38, helix 38; CP, central protuberance.
(Middle and bottom rows reproduced from [25]; with permission from Proceedings of the National Academy of Sciences).
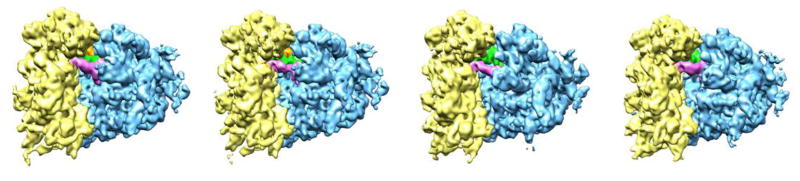
Fig. 3.
Inventory of states in a pre-translocational sample of rabbit liver ribosomes. From left to right: states are termed classical-1, classical-2, rotated-1 and rotated-2. A-, P- and E-site tRNA are painted pink, green, and orange, respectively. Yellow = 40S subunit, blue = 60S subunit. The PRE-translocation complex bearing deacylated tRNAPhe in the P site and N-acetyl-Lys-tRNA Lys3 in the A site was prepared by stepwise addition of purified tRNAs to reassociated 80S ribosomes (0.8 μM) programmed with MFK-mRNA. (Reproduced from [32]; with permission by Cell Press).
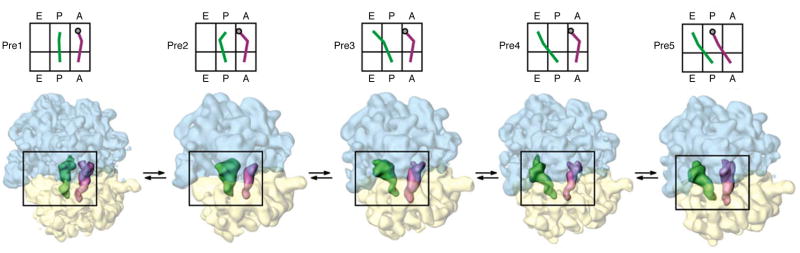
Another process, reverse translocation, was studied by an exhaustive analysis of a large cryo-EM dataset (>2 million particles) from Escherichia coli, yielding a number of intermediate states with distinct ribosome conformations and tRNA configurations [33••]. At the end of the slow process, however, the pool of ribosomes is left in the pre-translocational mixture of fast-equilibrating states (Fig. 4), which should be directly comparable to the pools analyzed by Agirrezabala et al. (2012) [25••] and Budkevich et al. (2011) [32••]. Thus even though the sample preparation, and the path that brought the ribosome population to the pre-translocational state is quite different, the results (in terms of the structures of intermediate states defined by ensemble averages of subpopulations) must be very similar. This expectation is borne out by a comparison between the relevant class reconstructions of Agirrezabala et al. (2012) [25••] and Fischer et al. (2010) [33••] (Figs. 2 and 4): the states denoted as pre1, pre2, pre3, pre4, and pre5 of Fischer et al. (Fig. 4) are comparable, respectively, to states captured by Agirrezabala et al. in classes 2, 4A, 4B, 5 and 6 (Fig. 2), though this has yet to be analyzed in detail. The biggest step, in terms of reconfiguration of tRNA positions, goes from pre2 to pre3 (or 4A to 4B) where the P/P-tRNA swings to the P/E position. Other more subtle changes between successive states are characterized as reorientations of the tRNAs, around an axis linking anticodon with CCA, that result in different positions of the elbow (e.g., the P/E-tRNA in pre4 versus pre5).
Fig. 4.
Relevant states (Pre1 through Pre5) corresponding to the pool of pre-translocational ribosomes generated by a back-translocation. Cartoons on top depict the tRNA positions, with Pre1 and Pre2 being two variants of the classical configuration, and Pre3–Pre5 being three variants of the hybrid configuration. As in the other experiments depicted in Figs. 2 and 3, intersubunit rotation proceeds from the classical, nonrotated state MS I to the rotated state MS II (on each panel from left to right).
(Reproduced from [33]; with permission from Nature Publishing Company)
X-ray crystallography has provided atomic-resolution structures of ribosome conformations that are captured by happenstance due to crystal packing, allowing an exhaustive analysis of the remodeling of intersubunit interactions and, in some cases, ribosome-tRNA interactions that are probably related to the remodeling that has been observed at lower resolution in the functional PRE complex intermediates that have been identified by cryo-EM [34,35••,36,37]. For a detailed description of these structures, the reader is referred to the companion article by Jamie Cate in this issue. Quantification of the intersubunit rotation angle, the translational movement of various intrasubunit structural domains, and the tRNA configurations, using a method developed by Klaus Schulten’s group in another context *38•], has enabled a quantitative comparison of all of the structural intermediates that have been studied thus far for which atomic coordinates are available, either from X-ray crystallography or by fitting atomic-resolution X-ray crystallographic structures to cryo-EM-derived electron density [25••]. Notably, the agreement between cryo-EM and X-ray values is not always close. For instance, while cryo-EM values for the angle that is formed between the “platform” domain and the “body” domain of the small ribosomal subunit change via a smooth trajectory as a function of intersubunit rotation, the corresponding measurements in the X-ray structures are widely scattered.
ENSEMBLE METHODS IN CRYO-EM
The discovery of multiple structural intermediate states within the same PRE complex sample, for both forward- and reverse translocation processes, has put a spotlight on novel methods recently developed in cryo-EM. It has been the initial premise of single-particle reconstruction [39] that a large ensemble of molecules exhibits the same structure, making it possible that it can be characterized by a single average. Hence, in the usual practice of visualizing a molecular machine, one would make an attempt to stabilize a single conformational state, for example by the use of an antibiotic [40]. More recently, through the use of sophisticated classification methods, it has now become possible to deal with a mixture of states, and extract individual three-dimensional structures for each of these states [41,42•,43,44,45,46,31,47,48,49]. Although the relationship between these states needs to be established by different experimental techniques, particularly smFRET, the ability to recover multiple structures of a molecule in thermal equilibrium is nevertheless a significant step in the continuing development of the cryo-EM methodology, and carries great potential for future studies of dynamic processes. This approach has been successfully employed in the cases of ribosome biogenesis [50], reverse translocation [33••], and translocation [25••,32••,30•,51••]. However, as the development of classification methods is still in flux, careful validation is required to ascertain the exact number of classes in a pool of molecules, and the structures they represent.
THE STRUCTURAL BASIS FOR INTERMEDIATE STATES
In attempting to understand the structural basis for intermediate states, we must look at the way the small and large subunits interact with each other via intersubunit bridges [52,53]. The spatial distribution and composition of the bridges (relatively tight RNA-RNA associations in the center, relatively loose noncovalent RNA-protein or protein-protein interactions at the periphery) lend themselves to rotational mobility, with the rotation center being situated at the tip of helix 27 of the 16S rRNA component of the small ribosomal subunit [54]. Peripheral bridges, which absorb up to 20Å relative motions of the subunits, may play the role of control elements that ensure ordered structural transitions. Two peripheral bridges, both involving small-subunit ribosomal protein S13, have been singled out for detailed study: bridge B1a, where 23S rRNA helix 38 (the so-called “A-site finger”) is the large-subunit component of the bridge, and bridge B1b, where large-subunit ribosomal protein L5 plays that role. A control function of both bridges is indicated by the fact that ΔS12/ΔS13 mutants exhibit an increased rate of EF-G-independent translocation, albeit with increased error rate [55], reinforcing Woese’s view *56+ that the ribosome was initially an RNA-based machine that acquired proteins at a late stage for improved accuracy and processivity.
An analysis of the long, curved helix 38 by MD simulations showed that this element has anisotropic elastic properties, lending itself to the steering and stabilization of the path of intersubunit rotation [11]. Bridge B1b, on the other hand, is formed by interaction between L5 and S13. Protein S13 glides along L5 as the small subunit rotates with respect to the large subunit, forming salt bridges at certain intervals [35••,37] that are likely to be instrumental in defining the intermediates that have been observed by cryo-EM. Computation of the electrostatic surface charges along S13 and L5 suggests that the gliding contacts that comprise bridge B1b are stabilized or modulated by juxtaposition of opposite or equal polarities [23,57].
THE FREE-ENERGY LANDSCAPE OF INTERSUBUNIT ROTATION AND THE COUPLING OF tRNA MOTION TO RIBOSOME DYNAMICS
It is remarkable that cryo-EM, by furnishing population counts for the occupancy of co-existing states, is able to give us information about the topography of the free-energy landscape that governs dynamic processes such as intersubunit rotation [25••,33••]. Predictably, differences in the free energies of the various conformational states which have been observed by cryo-EM, corresponding to differences in the minima of the wells representing the conformational states in the free-energy landscape, are very small, on the order of 1 kcal/mol or less. On the other hand, the size of the energetic barriers, or hurdles, separating these minima cannot be gauged by cryo-EM, since the appropriate conformational transition states are not sufficiently populated to allow computational separation by classification and separate structural reconstruction. This information is potentially available from smFRET experiments; however, for various technical reasons, smFRET has failed thus far to find intermediate states in wild-type PRE ribosomes beyond the well-characterized states MS I and MS II. The only hints come from the results of hidden-Markov modeling [28] and from signals derived from L1-tRNA distance fluctuations [58].
Studies employing smFRET have looked at the question of how movements of the tRNAs between their classical and hybrid configurations might be coupled to intersubunit rotation -- kinetically, or thermodynamically. Cornish et al. (2008) [21] observed that the rate of intersubunit motion was 10-fold slower than the rate of tRNA fluctuations between classical and hybrid configurations. A three-wavelength study has concluded, similarly, that closing and opening of the L1 stalk of the large ribosomal subunit is only loosely coupled to fluctuations of the P-site tRNA between its classical and hybrid configurations [58]. It is plausible to assume [25••] that the binding configurations of the tRNAs on the ribosome are influenced by the more slowly changing molecular environment presented by the ribosome; in other words, that tRNA movement is thermodynamically, not kinetically coupled with intersubunit motion. This argument, incidentally, underlies the presentation of conformational changes of intrasubunit structural domains of the ribosome and the binding configurations of the tRNAs as a function of the intersubunit rotation angle as an effective “independent variable” in refs. *25••,33••].
CONCLUSIONS
The study of ribosome structure and dynamics, made possible by the exhaustive analysis of cryo-EM data collected on samples of functionally active ribosomal complexes, by a close examination of X-ray crystallographic structures of ribosomes in a variety of conformational states, as well as several bulk FRET and smFRET studies, has highlighted several fascinating aspects of ribosome dynamics which were not initially appreciated when the first X-ray structures of the ribosome appeared. First, it was recognized that intersubunit (“ratchet-like”) rotation occurs, second, that this rotation is a functional requirement for translocation, and third, that the gross ribosome architecture lends itself to this mode of motion when thermal energy is supplied from the surroundings. Forth, it was established that intersubunit rotation occurs even in the absence of factors. Thus, the ribosome exhibits all expected attributes of a Brownian machine. The point of the most recent studies reviewed here is that, moreover, there is an intricate fine structure in the subunit interface that produces coarse granularity in the rotation, leading to the formation of intermediate states, beyond MS I and MS II, in which the ribosome and the ribosome-bound tRNAs adopt conformations that are intermediate between those observed in MS I and MS II. These additional intermediate states must be seen as “safe” conduits leading processively from MS I to MS II -- the state that must apparently be reached for binding of EF-G and productive completion of the translocation process. By necessity, the free-energy wells corresponding to these newly identified intermediate states must be shallow enough to permit rapid interconversions between them at physiological temperatures, and this is indeed the conclusion from cryo-EM experiments conducted on samples engaged in forward and reverse translocation.
A recent article by Peter Moore (2012) [7] suggested that ribosome research, thanks to the seminal contributions of X-ray crystallography and cryo-EM combined, has now reached a “poststructural” era in which the dynamic aspect will take center stage. In some sense this is true, particularly as far as the contributions by X-ray crystallography are concerned, but, on the other hand, the specter of having to track multiple conformational transitions in a dynamic model of translation, as this literature review has tabulated, would seem to suggest that a lot of structural work still needs to be done.
Highlights.
During the elongation cycle, mRNA-tRNA translocation entails large-scale relative movements of the ribosomal subunits, mRNA, and two tRNAs.
Increasing evidence from cryo-electron microscopy (cryo-EM), X-ray crystallography, and, to some extent, single-molecule FRET (smFRET) point to the existence of intermediate states characterized by intermediate intersubunit rotations and tRNA (hybrid) positions.
Identification and visualization of such intermediates equilibrating in a factor-free pre-translocational sample has now become possible by cryo-EM with the aid of classification, as demonstrated by recent studies in several labs.
Analysis of atomic structures shows that one of the peripheral intersubunit bridges, B1b, may be responsible for the observed “granularity” of the intersubunit motion, and may have an important control function in translocation.
The model of the ribosome as a Brownian machine is supported by evidence of temperature-dependent interconversion between states reported by smFRET, and the coexistence of ribosome subpopulations with distinct structures related by intersubunit rotation, as visualized by cryo-EM.
Acknowledgments
I’m indebted to Ruben Gonzalez for an extensive discussion of the manuscript of this review and for many helpful suggestions. I thank Melissa Thomas, Wen Li, and Xabier Agirrezabala for assistance with the preparation of the illustrations, and Wen Li for a discussion of the mechanism of EF-G entry into the ribosome. This work was supported by HHMI and by NIH grants R01 GM29169 and GM55440.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Moazed D, Noller HF. Intermediate states in the movement of transfer RNA in the ribosome. Nature. 1989;342:142–148. doi: 10.1038/342142a0. [DOI] [PubMed] [Google Scholar]
- 2•.Frank J, Gonzalez RL., Jr Structure and dynamics of a processive Brownian motor: the translating ribosome. Annu Rev Biochem. 2010;79:381–412. doi: 10.1146/annurev-biochem-060408-173330. This review, as the earlier one by Munro et al. (2009), makes the point that the ribosome should be viewed as a Brownian machine. It sets out to integrate two aspects of ribosomal dynamics as reflected in the experiment: cryo-EM depicting three-dimensional ensemble averages of molecules in different states, and smFRET depicting the transitions between these states and dwell times in these states. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Munro JB, Sanbonmatsu KY, Spahn CMT, Blanchard SC. Navigating the ribosome’s metastable energy landscape. Trends Biochem Sci. 2009;34:390–400. doi: 10.1016/j.tibs.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Spirin AS. The ribosome as a conveying thermal ratchet machine. J Biol Chem. 2009;284:21103–21119. doi: 10.1074/jbc.X109.001552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Spirin AS, Finkelstein AV. The ribosome as a Brownian ratchet machine. In: Frank J, editor. Molecular Machines in Biology–Workshop of the Cell. Cambridge University Press; New York: 2011. [Google Scholar]
- 6•.Wang B, Ho J, Fei J, Gonzalez RL, Jr, Lin Q. A microfluidic approach for investigating the temperature dependence of biomolecular activity with single-molecule resolution. Lab Chip. 2011;11:274–281. doi: 10.1039/c0lc00157k. A device is described enabling the measurement of smFRET signals as a function of temperature. Temperature dependence of the MS I → II (or Global states I → II) transition was found in agreement with theoretical predictions, corroborating the view that the ribosome is a machine driven by Brownian motion. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moore PB. How should we think about the ribosome? Ann Rev Biophys. 2012;41:1–19. doi: 10.1146/annurev-biophys-050511-102314. [DOI] [PubMed] [Google Scholar]
- 8.Tinoco I, Wen JD. Simulation and analysis of single-ribosome translation. Phys Biol. 2009;6:025006. doi: 10.1088/1478-3975/6/2/025006. [DOI] [PubMed] [Google Scholar]
- 9.Chowdhury D. Statistical mechanical treatment of molecular machines. In: Frank J, editor. Molecular Machines in Biology–Workshop of the Cell. Cambridge University Press; New York: 2011. [Google Scholar]
- 10.Astumian RD. Thermodynamics and kinetics of a Brownian motor. Science. 1997;276:917–922. doi: 10.1126/science.276.5314.917. [DOI] [PubMed] [Google Scholar]
- 11.Réblová K, Rázga F, Li W, Gao H, Frank J, Sponer J. Dynamics of the base of ribosomal A-site finger revealed by molecular dynamics simulations and cryo-EM. Nucl Acids Res. 2010;38:1325–1340. doi: 10.1093/nar/gkp1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Réblová K, Sponer J, Lankas F. Structure and mechanical properties of the ribosomal L1 stalk three-way junction. Nucl Acid Res. 2012 doi: 10.1093/nar/gks258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li W, Trabuco LG, Schulten K, Frank J. Molecular dynamics of EF-G during translocation. Proteins. 2011;79:1478–1486. doi: 10.1002/prot.22976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sanbonmatsu KY, Joseph S, Tung CS. Simulating movement of tRNA into the ribosome during decoding. Proc Natl Acad Sci USA. 2005;102:15854–15859. doi: 10.1073/pnas.0503456102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15•.Whitford PC, Ahmed A, Yu Y, Hennelly SP, Tama F, Spahn CMT, Onuchic JN, Sanbonmatsu KY. Excited states of ribosome translocation revealed through integrative molecular modeling. Proc Natl Acad Sci USA. 2011;108:18943–18948. doi: 10.1073/pnas.1108363108. The authors present a methodology (MDfit) that utilizes molecular dynamics simulations to generate configurations of excited, poorly occupied states that are consistent with available biophysical and biochemical measurements, including structural information obtained from cryo-EM and X-ray crystallography. This study can make predictions that may be verified by future 3D visualization experiments. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frank J, Agrawal RK. A ratchet-like intersubunit reorganization of the ribosome during translocation. Nature. 2000;406:318–322. doi: 10.1038/35018597. [DOI] [PubMed] [Google Scholar]
- 17.Horan LH, Noller HF. Intersubunit movement is required for ribosomal translocation. Proc Natl Acad Sci USA. 2007;104:4881–4885. doi: 10.1073/pnas.0700762104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bretscher MS. Translocation in protein synthesis: a hybrid structure model. Nature. 1968;218:675–677. doi: 10.1038/218675a0. [DOI] [PubMed] [Google Scholar]
- 19.Spirin AS. How does the ribosome work? A hypothesis based on the two subunit construction of the ribosome. Cur Modern Biol. 1968;2:115–127. doi: 10.1016/0303-2647(68)90017-8. [DOI] [PubMed] [Google Scholar]
- 20.Ermolenko DN, Majumdar ZK, Hickerson RP, Spiegel PC, Clegg RM, Noller HF. Observation of intersubunit movement of the ribosome in solution using FRET. J Mol Biol. 2007;370:530–540. doi: 10.1016/j.jmb.2007.04.042. [DOI] [PubMed] [Google Scholar]
- 21.Cornish PV, Ermolenko DN, Noller HF, Ha T. Spontaneous intersubunit rotation in single ribosomes. Mol Cell. 2008;30:578–588. doi: 10.1016/j.molcel.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fei J, Kosuri P, Macdougall DD, Gonzalez RL. Coupling of ribosomal L1 stalk and tRNA dynamics during translation elongation. Mol Cell. 2008;30:348–359. doi: 10.1016/j.molcel.2008.03.012. [DOI] [PubMed] [Google Scholar]
- 23.Frank J, Gao H, Sengupta J, Gao N, Taylor DJ. The process of mRNA-tRNA translocation. Proc Natl Acad Sci USA. 2007;104:19671–19678. doi: 10.1073/pnas.0708517104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Agirrezabala X, Lei J, Brunelle JL, Ortiz-Meoz RF, Green R, Frank J. Visualization of the hybrid state of tRNA binding promoted by spontaneous ratcheting of the ribosome. Mol Cell. 2008;32:190–197. doi: 10.1016/j.molcel.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25••.Agirrezabala X, Liao HY, Schreiner E, Fu J, Ortiz-Meoz RF, Schulten K, Green R, Frank J. Structural characterization of mRNA-tRNA translocation intermediates. Proc Natl Acad Sci USA. 2012;109:6094–6099. doi: 10.1073/pnas.1201288109. In this study, more than 200,000 cryo-EM images of wild-type Escherichia coli ribosomes in the PRE state were analyzed by classification. Five distinct subpopulations containing A- and P-site tRNA were found. The corresponding reconstructions show the ribosome in different incrementally varying conformations, ranging from macrostate I to macrostate II, distinguished by intersubunit rotation and tRNA positions. This article also contains a quantitative analysis of the conformations of intermediate states of the PRE ribosome reported in the literature thus far. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Julián P, Konevega AL, Scheres SH, Lázaro M, Gil D, Wintermeyer W, Rodnina MV, Valle M. Structure of ratcheted ribosomes with tRNAs in hybrid states. Proc Natl Acad Sci USA. 2008;105:16924–16927. doi: 10.1073/pnas.0809587105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim HD, Puglisi JD, Chu S. Fluctuations of transfer RNAs between classical and hybrid states. Biophys J. 2007;93:3575–3582. doi: 10.1529/biophysj.107.109884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Munro JB, Altman RB, O’Connor N, Blanchard SC. Identification of two distinct hybrid state intermediates on the ribosome. Mol Cell. 2007;25:505–517. doi: 10.1016/j.molcel.2007.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen C, Stevens B, Kaur J, Cabral D, Liu H, Wang Y, Zhang H, Rosenblum G, Smilansky Z, Goldman YE, Cooperman BS. Single-molecule fluorescence measurements of ribosomal translocation dynamics. Mol Cell. 2011;42:367–77. doi: 10.1016/j.molcel.2011.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30•.Fu J, Munro JB, Blanchard SC, Frank J. Cryoelectron microscopy structures of the ribosome complex in intermediate states during tRNA translocation. Proc Natl Acad Sci USA. 2011;108:4817–4821. doi: 10.1073/pnas.1101503108. In this study, a PRE sample of a P-loop mutant of the E. coli ribosome, previously characterized by smFRET, was imaged by cryo-EM and the resulting images analyzed by classification and 3D reconstructions of classes obtained. Two intermediate states were found, one of which is characterized by an unusual position of the E-site tRNA in contact with the L1 stalk. The findings were interpreted in terms of the L1 stalk’s role in translocation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scheres SHW, Gao H, Valle M, Herman GT, Eggermont PPB, Frank J, Carazo JM. Disentangling conformational states of macromolecules in 3D-EM through likelihood optimization. Nat Methods. 2007;4:27–29. doi: 10.1038/nmeth992. [DOI] [PubMed] [Google Scholar]
- 32••.Budkevich T, Giesebrecht J, Altman RB, Munro JB, Mielke T, Nierhaus KH, Blanchard SC, Spahn CMT. Structure and dynamics of the mammalian ribosomal pretranslocation complex. Mol Cell. 2011;44:214–224. doi: 10.1016/j.molcel.2011.07.040. A pre-translocational sample of 80S ribosomes from rabbit liver was imaged by cryo-EM. 300,000 particles were separated into four groups by classification. Although the ribosome in the PRE complex was from rabbit liver, the tRNAs are of bovine origin (for the cryo-EM study) and of E. coli origin for the accompanying smFRET experiments. Reconstructions from these groups show the 80S ribosome in four distinct conformations and tRNA binding configurations, denoted by “classical 1, classical 2, hybrid 1, and hybrid 2”. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33••.Fischer N, Konevega AL, Wintermeyer W, Rodnina MV, Stark H. Ribosome dynamics and tRNA movement by time-resolved electron cryomicroscopy. Nature. 2010;466:329–333. doi: 10.1038/nature09206. This study is based on an exhaustive analysis of a large dataset collected from a sample of a back-translocating ribosome at different temperatures. Because of the length of the process (~20 mins), time resolution is achieved by collecting samples in few-minute intervals. Many clusters are found depicting the ribosome in multiple states during retro-translocation. As the pool of post-translocational ribosomes decreases, a pool of pre-translocational ribosomes forms, which are in rapid equilibrium. As later found in authentic forward translocation in bacteria (see Agirrezabala et al., 2012) and eukaryotes (see Budkevich et al., 2011), the pre-translocational data are clustered, revealing intermediate ribosome conformations and associated tRNA constellations. Significantly, the number and definition of observed states changes strongly with temperature. [DOI] [PubMed] [Google Scholar]
- 34.Berk V, Zhang W, Pai RD, Cate JH. Structural basis for mRNA and tRNA positioning on the ribosome. Proc Natl Acad Sci USA. 2006;103:15830–15834. doi: 10.1073/pnas.0607541103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35••.Dunkle JA, Wang L, Feldman MB, Pulk A, Chen VB, Kapral GJ, Noeske J, Richardson JS, Blanchard SC, Cate JH. Structures of the bacterial ribosome in classical and hybrid states of tRNA binding. Science. 2011;332:981–984. doi: 10.1126/science.1202692. The authors use the ribosome recyling factor to capture the E. coli ribosome in the 30S body-rotated (‘fully rotated’) conformation. This 3.2-Å crystallographic structure shows a tRNA in a ‘highly bent’ P/E hybrid position. The conformation resembles a mirror image of the A/T conformation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjurjo A, Holton JM, Cate JH. Structures of the bacterial ribosome at 3.5Å resolution. Science. 2005;310:827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- 37.Zhang W, Dunkle JA, Cate JHD. Structures of the ribosome in intermediate states of ratcheting. Science. 2009;325:1014–1017. doi: 10.1126/science.1175275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38•.Agirrezabala X, Schreiner E, Trabuco LG, Lei J, Ortiz-Meoz RF, Schulten K, Green R, Frank J. Structural insights into cognate versus near-cognate discrimination during decoding. EMBO J. 2011;30:1497–1507. doi: 10.1038/emboj.2011.58. A novel quantitative characterization of tRNA and ribosome domain positions and movements is for the first time employed in this study, using a tensor analysis. This analysis was subsequently employed in the characterization of intermediate states in Agirrezabala et al. (2012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Frank J. Single-particle reconstruction of biological macromolecules in electron microscopy—30 years. Quart Rev Biophys. 2009;42:139–158. doi: 10.1017/S0033583509990059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Valle M, Zavialov A, Sengupta J, Rawat U, Ehrenberg M, Frank J. Locking and unlocking of ribosomal motions. Cell. 2003;114:123–134. doi: 10.1016/s0092-8674(03)00476-8. [DOI] [PubMed] [Google Scholar]
- 41.Elmlund D, Elmlund H. Ab initio structure determination from electron microscopic images of single molecules coexisting in different functional states. Structure. 2010;14:777–786. doi: 10.1016/j.str.2010.06.001. [DOI] [PubMed] [Google Scholar]
- 42•.Frank J. The ribosome comes alive. Isr J Chem. 2010;50:95–98. doi: 10.1002/ijch.201000010. This article espouses the idea of “Story in a Sample” as a new paradigm of the single-particle cryo-EM approach to the study of molecular machines. In case where several states of a molecular machine are in free equilibrium, as in the pre-translocational ribosome, cryo-EM aided by powerful classification techniques can identify the corresponding subpopulations and reconstruct the molecule in these states. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Frank J. Visualization of molecular machines by cryo-electron microscopy. In: Frank J, editor. Molecular Machines in Biology – Workshop of the Cell. Cambridge University Press; New York: 2011. [Google Scholar]
- 44.Liao HY, Frank J. Classification by bootstrapping in single particle methods. Proc IEEE Int Symp Biomed Imaging. 2010:169–172. doi: 10.1109/ISBI.2010.5490386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Loerke J, Giesebrecht J, Spahn CMT. Multiparticle cryo-EM of ribosomes. Meth Enzym. 2010;483:161–177. doi: 10.1016/S0076-6879(10)83008-3. [DOI] [PubMed] [Google Scholar]
- 46.Shatsky M, Hall RJ, Nogales E, Malik J, Brenner SE. Automated multi-model reconstruction from single-particle electron microscopy data. J Struct Biol. 2010;170:98–108. doi: 10.1016/j.jsb.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Scheres SHW. A Bayesian view on cryo-EM structure determination. J Mol Biol. 2012;415:406–418. doi: 10.1016/j.jmb.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sigworth FJ. From cryo-EM, multiple protein structures in one shot. Nature Methods. 2007;4:20–21. doi: 10.1038/nmeth0107-20. [DOI] [PubMed] [Google Scholar]
- 49.Spahn CMT, Penczek PA. Exploring conformational modes of macromolecular assemblies by multiparticle cryo-EM. Cur Op Struct Biol. 2009;19:623–631. doi: 10.1016/j.sbi.2009.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mulder AM, Craig Yoshioka C, Beck AH, Bunner AE, Milligan RA, Potter CS, Carragher B, Williamson JR. Visualizing ribosome biogenesis: parallel assembly pathways for the 30S subunit. Science. 2010;330:673–677. doi: 10.1126/science.1193220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51••.Ratje AH, Loerke J, Mikolajka A, Brunner M, Hildebrand PW, Starosta AL, Donhofer A, Connell SR, Fucini P, Mielke T, Whitford PC, et al. Head swivel on the ribosome facilitates translocation by means of intrasubunit tRNA hybrid sites. Nature. 2010;468:713–716. doi: 10.1038/nature09547. A cryo-EM study of EF-G-bound ribosome complexes using classification of approximately 600,000 particles shows that translocation of tRNA on the 30S subunit is accompanied by the swivel movement of the 30S subunit head and is coupled, as expected, to a reverse rotation of the 30S-body. In the process, a newly discovered P/E tRNA configuration is formed as the tRNA maintains contact with the P site on the 30S subunit head while interacting with the E site on the 30S subunit platform. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gao N, Frank J. A library of RNA bridges. Nat Chem Biol. 2006;2:231–232. doi: 10.1038/nchembio0506-231. [DOI] [PubMed] [Google Scholar]
- 53.Yusupov MM, Yusupova GZh, Baucom A, Lieberman K, Earnest TN, Cate JHD, Noller HF. Crystal Structure of the Ribosome at 5.5 Å Resolution. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- 54.Tama F, Valle M, Frank J, Brooks CL., III Dynamic reorganization of the functionally active ribosome explored by normal mode analysis and cryo-electron microscopy. Proc Natl Acad Sci USA. 2003;100:9319–9323. doi: 10.1073/pnas.1632476100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cukras AR, Southworth DR, Brunelle JL, Culver GM, Green R. Ribosomal proteins S12 and S13 function as control elements for translocation of the mRNA:tRNA complex. Mol Cell. 2003;12:321–328. doi: 10.1016/s1097-2765(03)00275-2. [DOI] [PubMed] [Google Scholar]
- 56.Woese CR. Translation: in retrospect and prospect. RNA. 2001;7:1055–1067. doi: 10.1017/s1355838201010615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shasmal M, Chakraborty B, Sengupta J. Intrinsic molecular properties of the protein-protein bridge facilitate ratchet-like motion of the ribosome. Biochem Biophys Res Comm. 2010;399:192–197. doi: 10.1016/j.bbrc.2010.07.053. [DOI] [PubMed] [Google Scholar]
- 58.Munro JB, Altman RB, Tung C-S, Cate JHD, Sanbonmatsu KY, Blanchard SC. Spontaneous formation of the unlocked state of the ribosome is a multistep process. Proc Natl Acad Sci USA. 2010;107:709–714. doi: 10.1073/pnas.0908597107. [DOI] [PMC free article] [PubMed] [Google Scholar]