Abstract
With more than a hundred individual RNA and protein parts and a highly dynamic assembly and disassembly pathway, the spliceosome is arguably the most complicated macromolecular machine in the eukaryotic cell. This complexity has made kinetic and mechanistic analysis of splicing incredibly challenging. Yet recent technological advances are now providing tools for understanding this process in much greater detail. Ranging from genome-wide analyses of splicing and creation of an orthogonal spliceosome in vivo, to purification of active spliceosomes and observation of single molecules in vitro, such new experimental approaches are yielding significant insight into the inner workings of this remarkable machine. These experiments are rewriting the textbooks, with a new picture emerging of a dynamic, malleable machine heavily influenced by the identity of its pre-mRNA substrate.
The spliceosome snips and stitches RNAs
In eukaryotes, many genes are transcribed as precursors to messenger RNAs (pre-mRNAs) containing both introns and exons. To generate mature mRNAs, the introns must be snipped out and the exons stitched back together by the molecular tailor of the cell: the spliceosome. A fashion designer as well, the spliceosome can alter its snipping and stitching to produce many different mRNAs from a single bolt of pre-mRNA cloth. Such alternative splicing is particularly prevalent in multicellular organisms, where it has enabled evolution of highly complex proteomes without comparable increases in gene number. Deep sequencing has now revealed that >95% of human genes are alternatively spliced, often in a developmental, tissue-specific, or signal transduction-dependent manner [1]. The human genome contains >200,000 different introns, ranging in size from <100 to >700,000 nucleotides (nts), with median intron and exon lengths of 1800 and 123 nts, respectively. While typical human genes contain 7–9 introns, many hereditary disease-causing genes such as dystrophin and NF-1 have ten times this many [2]. To further compound the problem of piecing together the correct mRNA sequence, the 5′ and 3′ splice site (SS) consensus sequences contain nowhere near enough information to reliably define intron-exon boundaries [3], and the human splicing machinery can apparently utilize >2500 different 5′ SS signals [4].
Assigned the gargantuan task of recognizing and accurately processing such an overwhelming array of sequences, spliceosomes are arguably the most complicated macromolecular machines in the cell [5–7]. In budding yeast, spliceosomes comprise five small nuclear RNAs (the U1, U2, U4, U5, and U6 snRNAs) and ~100 different polypeptides [8], The human splicing machinery is even more complex. Along with the U1, U2, U4, U5, and U6 snRNAs, the major human spliceosome contains >300 different proteins [6]. Humans even possess a second splicing apparatus called the minor spliceosome containing the U11, U12, U4atac, U5, and U6atac snRNAs [9]. Proteomic analyses of purified spliceosomes have revealed remarkable evolutionary conservation, with 85% of yeast splicing proteins having a direct human counterpart. Thus, the yeast spliceosome likely represents the evolutionarily conserved ‘core’ of the splicing machinery [8]. This machinery has been greatly augmented in humans, likely to incorporate additional functionalities necessary for alternative splicing and exon definition (see below). In both systems, scores of polypeptides associate with one or more snRNAs to form stable ribonucleoprotein particles (snRNPs), while legions of others act as sequence-specific splicing regulators or transiently interacting helper proteins (called splicing factors) to both push the process along and exert quality control over the finished products. Platoons of other proteins serve as go-betweens to communicate with up- and downstream processes (e.g., transcription and translation) to ensure efficient flow of information from DNA to RNA to protein [10,11].
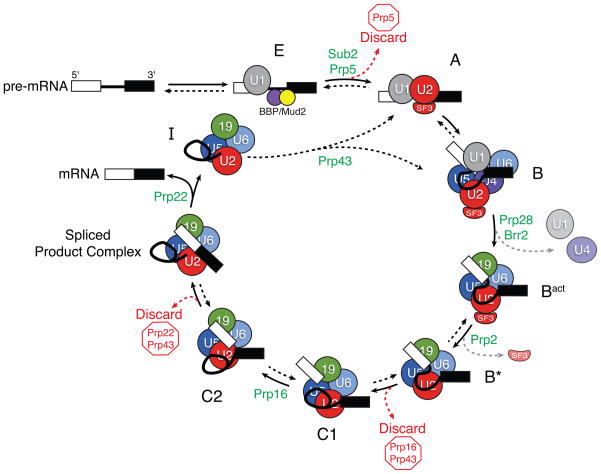
Mirroring its compositional complexity, the splicing machinery exhibits a remarkable degree of dynamic complexity. Over the past two and a half decades since the realization that splicing is carried out in multi-megadalton complexes, much work has gone into defining both the parts lists and the overall assembly and disassembly pathways of these complexes [5,6]. Most intensely studied have been the major human and the budding yeast spliceosomes. These spliceosomes are assembled from the U1 and U2 snRNPs, U4/U6.U5 tri-snRNP, and the Prp19 complex (a large protein-only subcomplex). Assembly of these major components proceeds step-wise on pre-mRNA consensus sequences defining the 5′ SS, branch point, and 3′ SS (Figure 1) [5]. Within the catalytically activated B* spliceosome, intron excision then occurs in two chemical steps (Figures 1 and 2). First, the phosphodiester bond at the 5′ exon/intron boundary (the 5′ SS) is cleaved by attack of the 2′ hydroxyl of the branch point adenosine to free the 5′ exon and form a lariat intron-3′ exon intermediate (C1 complex). Second, the 3′ hydroxyl of the liberated 5′ exon attacks the 3′ SS in C2 complex, resulting in exon ligation and freeing the lariat intron. After splicing, the subcomplexes are recycled and assembly begins anew on another intron, signifying that each spliceosome is a single turnover enzyme. Although both chemical steps of splicing are isoenergetic, numerous ATPases facilitate the structural transitions required for spliceosome assembly, activation, catalysis, and disassembly (Figure 1).
Figure 1.
A model for step-wise spliceosome assembly and catalysis. Specific spliceosomal complexes (E, A, B, and others) are identified according to the human nomenclature. Spliceosome assembly initiates by binding of the U1 snRNP to the 5′ SS and proteins [branch point bridging protein (BBP) and Mud2] to the branch site in E (early) complex. In an ATP-dependent reaction, U2 displaces BBP/Mud2 and binds to the branch site in A complex. B complex then forms by addition of the U4/U6.U5 tri-snRNP. Subsequent to assembly of B complex, catalytic activation requires several additional rearrangements. These include departure of U1 and U4 to form Bact complex (grey arrow); formation of catalytic structures between the pre-mRNA, U2, and U6 snRNAs; and destabilization of several U2 snRNP proteins (the SF3 complex, grey arrow) from the rest of the machinery to form B* complex [8,12,13]. C1 complex is formed after 5′ SS cleavage. For exon ligation, the spliceosome undergoes a conformational change into C2 complex. After the two chemical steps of splicing are complete, the spliceosome enters a disassembly and recycling pathway in which the spliced exons are released and the post-spliceosomal intron product complex (I) is disrupted. Multiple steps in the pathway are promoted by the presence of DExD/H-box ATPases (green). Some of these ATPases have also been implicated in fidelity checkpoints and control the use of discard pathways (red arrows and type) that prevent splicing. As discussed in the text, many of the steps in the pathway have now been shown to be reversible (double arrows), while others have not (single arrows).
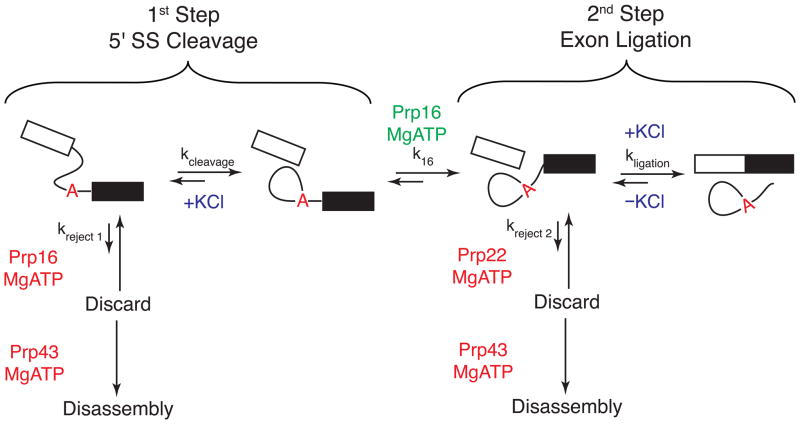
Figure 2.
The chemical steps of pre-mRNA splicing and kinetic proofreading. Splicing proceeds through two sequential transesterification reactions: 5′ SS cleavage and exon ligation. These reactions may be catalyzed by alternate conformations of the spliceosome according to the ‘two state’ model for spliceosomal catalysis, and ATP hydrolysis by Prp16 promotes interchange between the two states (k16). As in Figure 1, ATPase functions that promote splicing during these steps are shown in green and those that promote discard pathways are shown in red. The forward or reverse chemical reactions can also be promoted by the presence or absence of KCl (blue) [34]. On a substrate with a high probability of generating mRNA, the forward reaction pathways are favored (large arrows) while the reverse reactions and rejection pathways are disfavored (small arrows). On a poor splicing substrate (or in a malformed spliceosome), the rates of the forward reactions (kcleavage, k16, and kligation) may be slower than the reactions controlling entrance to the discard pathways (kreject 1 and kreject 2). In this case, Prp43 irreversibly disassembles the spliceosome. This represents a fidelity mechanism for splicing and a version of kinetic proofreading [25].
Most of our current understanding of splicing is derived from traditional in vivo and in vitro assays. In vivo studies have often tracked splicing of one or more reporter pre-mRNAs or coupled splicing to cell viability [14]. Conversely, in vitro assays often monitor rates at which stable spliceosome subcomplexes appear when analyzed by native gel electrophoresis or glycerol gradient centrifugation, and/or the time course of 5′ SS cleavage and exon ligation using radioactive ‘model’ pre-mRNA substrates [15]. In most cases, these assays have been carried out in a whole cell or nuclear extract (WCE or NE). Yet, while tremendous insight into the splicing machinery has been obtained by such methods, the difficulties of controlling reactions in vivo and the low abundance and heterogeneity of spliceosomes in extracts have proven significant obstacles for elucidating more detailed mechanisms.
In order to overcome these barriers, a number of laboratories have recently developed novel approaches for studying splicing. These methods include in vitro assembly and purification of active spliceosomes, microscopic visualization of single spliceosomes, and global analysis of pre-mRNA splicing in vivo. While each method is distinct, ranging from simultaneously quantifying splicing of hundreds of different pre-mRNAs in vivo to observing single pre-mRNA molecules in vitro, each provides a complementary and synergistic view that together are fundamentally changing the way we think about the splicing machinery. Focusing on recent studies of the yeast spliceosome, we highlight several new technologies and experimental approaches and discuss how each is significantly advancing our understanding of the highly dynamic and malleable molecular machine that is the spliceosome.
Purified spliceosomes and reaction reversibility
Many biochemical analyses require well-characterized enzymes and controlled conditions, making ‘purification’ one of the ‘Ten Commandments’ of enzymology [16]. With such a plethora of components to contend with, obeying this commandment was a seemingly insurmountable hurdle for spliceosome biochemists in the past. Recently, however, advances in affinity purification have made purifying active spliceosomes a reality, and several groups are now employing these methods to great effect.
The Lührmann group has succeeded in purifying fully-assembled and active spliceosomes from both human NE and yeast WCE [17,18]. These complexes were first assembled in extracts on pre-mRNAs containing several MS2 phage coat protein recognition RNA hairpins in the 5′ exon. This enabled subsequent spliceosome purification via amylose affinity chromatography with maltose-binding protein (MBP) fused to the MS2 protein. For the human spliceosome, the pre-mRNA substrate contained a 5′ exon and intron, but no 3′ SS or 3′ exon [17]. Spliceosomes formed on such pre-mRNAs carry out 5′ SS cleavage, but become stalled at the C1 or C2 stage due to absence of a 3′ SS (Figure 1). It was previously shown that such spliceosomes in unfractionated NE can perform bi-molecular exon ligation when supplied with an additional RNA containing a 3′ SS [19]. Now it has been shown that when purified, these intermediate-containing spliceosomes can carry out bi-molecular exon ligation without any additional protein factors, suggesting that they are predominantly in the C2 complex form [17] (Figure 1).
Active yeast spliceosomes were accumulated in WCE from cells carrying a temperature sensitive (Ts) mutation in the Prp2 spliceosomal ATPase [18]. In heat-inactivated prp2-1 extract, splicing stalls just prior to 5′ exon cleavage at the Bact stage [20,21] (Figure 1). Once purified, Bact was not itself competent for chemistry; 5′ SS cleavage required addition of wild type Prp2, recombinant Spp2 and Cwc25 proteins and ATP. Reconstitution of both 5′ SS cleavage and exon ligation additionally required four second step factors (Prp16, Prp18, Prp22, and Slu7). Thus, transitioning yeast Bact through both 5′ SS cleavage and exon ligation minimally requires ATP and seven transacting splicing factors. With this system established, it should now be possible to elucidate the exact functions of these seven factors in much greater detail.
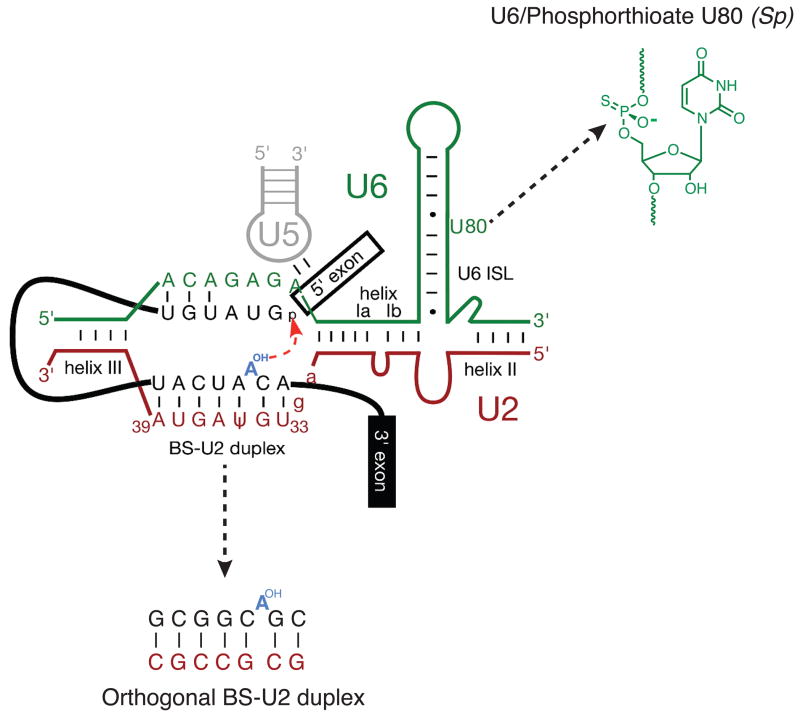
Assembly of spliceosomes in extracts allows for incorporation of site-specifically modified pre-mRNAs and snRNAs. For snRNAs, this is accomplished by first depleting the endogenous snRNA via targeted RNaseH cleavage, and then adding back an in vitro synthesized version containing the modification(s) of interest. The Staley lab recently used this approach to assemble spliceosomes with a synthetic U6 snRNA containing a single phosphorothioate linkage at the pro-Sp non-bridging oxygen of U80 (Figure 3). A phosphorothioate at this position had previously been shown to inhibit splicing just prior to 5′ SS cleavage [22], making it a likely candidate for Mg2+ coordination in the spliceosomal active site. Once assembled, the U6 phosphorothioate spliceosomes were isolated via a tandem affinity purification (TAP) tag [23] on spliceosomal protein Prp19 [24]. The addition of ‘soft’, thiophilic metal ions (Cd2+ and Mn2+) capable of coordinating the phosphorothioate triggered 5′ SS cleavage, indicating that the isolated species were indeed B* complexes poised for first step catalysis. Although Cd2+ and Mn2+ could rescue 5′ SS cleavage either in the presence or absence of ATP, Mg2+ only rescued splicing in the absence of ATP (albeit at a significantly reduced rate). While the slow reaction rate is consistent with Mg2+ being a poor phosphorothioate ligand, the ATP dependence suggests additional mechanisms at work. Koodathingal et al. hypothesized that in the presence of ATP, the reaction was being proofread by a spliceosomal ATPase that shunted slow spliceosomes into a discard pathway prior to 5′ SS cleavage. In the absence of ATP this discard pathway was unavailable, so the slow spliceosomes could cleave the 5′ SS at their leisure. This purified system thus exhibits ‘kinetic proofreading’ [25], in which the rate of a forward step in the spliceosome cycle is in competition with opening of a discard pathway by a spliceosomal ATPase (Figure 2).
Figure 3.
A model for the spliceosomal B* complex active site showing juxtaposition of several important groups during catalysis. In the work of Koodathingal et al., a site-specific phosphorthioate at position U80 in the U6 snRNA was used to stall spliceosomes prior to exon ligation [24]. Smith et al. created an orthogonal yeast spliceosome by mutating the branch site (BS)-U2 duplex [36]. This figure originated from reference [36] and is used with permission.
Using yeast genetics, it had previously been shown that mutations in the DExD/H-box ATPase Prp16 can promote splicing of suboptimal substrates in vivo, such as those containing branch point mutations [26,27]. This made Prp16 an attractive target for proofreading 5′ SS cleavage, in addition to its known role in promoting the transition between C1 and C2 complexes [28] (Figure 1). Using the purified U6/U80-phosphorothioate spliceosomes and recombinant Prp16, Koodathingal et al. were able to demonstrate both that Prp16 is directly responsible for this proofreading step and that the proofreading is reversible [24]. If spliceosomes can enter and exit the Prp16 discard pathway multiple times, why aren’t all spliceosomes in the discard pathway eventually pulled into productive splicing reactions? The key appears to be irreversible destruction of spliceosomes in the discard pathway by an additional DExD/H-box ATPase, Prp43 (Figures 1 and 2). Using a pulse-chase approach and glycerol gradient centrifugation, Staley and coworkers have now shown that discarded spliceosomes at the stages of 5′ SS cleavage or exon ligation are substrates for Prp43 [24,29]. Therefore, in addition to its roles in disassembling the I Complex (Figure 1) [30] and in ribosome biogenesis [31,32], Prp43 provides an irreversible step that prevents discarded spliceosomes from re-entering the splicing cycle.
In addition to reversible proofreading, we now know that the spliceosome can perform reverse splicing as well. While reverse splicing has long been known for self-splicing introns [33], experimental evidence for reversible spliceosome chemistry had proven elusive until recently. By V5-epitope tagging a mutant of Prp22 defective in catalyzing spliced exon release, Tseng and Cheng were able to isolate in vitro-assembled spliceosomes containing spliced exons and lariat intron product (spliced product complex, Figure 1) [34]. By altering the KCl concentration in the absence of ATP, these spliceosomes could be coaxed into reversing both exon ligation and 5′ SS cleavage to re-generate pre-mRNA. Surprisingly, the monovalent ion requirements for the two reverse reactions were different – whereas KCl inhibited exon ligation reversal, it promoted 5′ SS cleavage reversal. Forward exon ligation was also favored by KCl meaning that spliceosomes in which exon ligation was first reversed by removing KCl could subsequently re-ligate the exons upon KCl addition. These results are consistent with the spliceosome being able to toggle between different catalytic conformations that favor forward or reverse progress through one or the other chemical reaction (Figure 2) [34,35].
Probing the active site of an orthogonal spliceosome
The observation that spliceosomes can be toggled between two different catalytic states by simply varying the salt concentration suggests an extremely flexible active site. Query and co-workers have now developed a system wherein the extent of this active site flexibility can be probed in vivo using an orthogonal yeast spliceosome [36]. To construct their orthogonal spliceosome, the authors generated a second copy of U2 snRNA in yeast containing a highly mutated branch site binding region (e.g., 5′-UGΨAGUA-3′ → GCGCCGC) and a matching pre-mRNA reporter (5′-UACUAAC-3′ → GCGGCAGC) (Figure 3). This reporter is not recognized by endogenous U2 and can only be spliced in the presence of the mutant U2 snRNA. Consequently, splicing of the pre-mRNA reporter is orthogonal to the normal yeast splicing machinery, and the mutant U2 snRNA can be modified without affecting splicing of other cellular pre-mRNAs.
Using this system, Smith et al. were able to demonstrate that the spliceosome active site can accommodate (or adapt to) a remarkably large number of mutations in the branch site consensus [36]. In addition to finding that the exact sequence of the branch site/U2 snRNA duplex is unimportant for splicing, the authors discovered that the branch site nucleophile can be located at multiple positions within this duplex. Exactly which nucleotide serves as the branch point is a function of both nucleotide identity and nucleotide position relative to U2/U6 helix 1a (Figure 3).
These results are in agreement with a variety of other experiments supporting a highly flexible and dynamic spliceosome active site [37]. The Konarska and Query groups have together proposed a ‘two state’ model for the catalytic spliceosome in which different conformations promote either 5′ SS cleavage or exon ligation (Figure 2). Consistent with this model, certain mutations in the core spliceosomal protein Prp8 can either promote 5′ SS cleavage at the expense of exon ligation or vice versa [38,39]. These results suggest that Prp8 is a key player in modulating the equilibrium between first and second step conformations. Interconversion between these competing conformations likely requires a structural transition promoted by Prp16 [40,41]. The affinity purification methods described above will provide powerful tools for studying active site reorganization during splicing, as well as the functions of Prp8, Prp16 and other involved factors in much greater detail.
Dynamics of single spliceosomes
In the experiments described above, yeast genetics and in vitro splicing assays provided evidence for a conformationally flexible spliceosome with at least three reversible steps: 5′ SS cleavage, exon ligation, and discard by Prp16 during 5′ SS cleavage. None of these methods, however, is capable of providing quantitative kinetic or thermodynamic information about any discrete step in the splicing reaction. What was needed were time-resolved in vitro assays that could follow the dynamics of spliceosome component interactions with pre-mRNAs, spliceosome conformational changes, and/or splicing itself. One technique capable of addressing such questions is single molecule fluorescence microscopy. Single molecule methods can be used to follow complex reactions in real-time and, by definition, are well suited for studying low abundance enzymes [42,43]. However, given that single molecule experiments have traditionally employed highly purified components whereas in vitro spliceosome assembly must be carried out in WCE or NE, bringing single molecule approaches to bear on the splicing machinery was a truly formidable challenge.
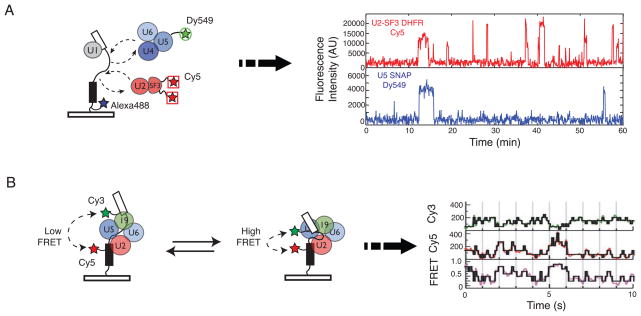
Two recent collaborations between spliceosome biochemists and single molecule biophysicists have now made real-time single molecule imaging of splicing reactions a reality. To follow spliceosome assembly in real-time, the Moore and Gelles groups implemented a single molecule total internal reflection fluorescence (TIRF) microscopy technique called co-localization of single molecules spectroscopy (CoSMoS) [44]. In this system, yeast genetic engineering was combined with chemical biology tools to fluorescently tag protein components of endogenous snRNPs in yeast WCE [45]. The comings and goings of the snRNPs on fluorescent, surface-tethered RP51A pre-mRNA molecules were subsequently recorded and analyzed. Use of spectroscopically- distinguishable fluorophores allowed multiple spliceosome components to be simultaneously monitored as they colocalized with the pre-mRNA substrate (Figure 4a). By quantitatively analyzing spliceosome assembly reactions on hundreds of individual pre-mRNA molecules, these experiments demonstrated that spliceosome assembly is highly ordered on this pre-mRNA and that pre-association of the snRNPs into some higher order structure is not required for splicing.
Figure 4.
Schematic for single molecule fluorescence analysis of splicing. (a) Hoskins et al. tethered fluorescent pre-mRNAs labeled with Alexa488 to a glass surface and then monitored how fluorescent spliceosome subcomplexes (e.g. U2-SF3 and U4/U6.U5 tri-snRNP) associated with the pre-mRNA. Fluctuations in fluorescence intensity indicated that the subcomplexes can associate multiple times with a given pre-mRNA. (b) By incorporating a FRET donor (Cy3) and acceptor (Cy5) pair into a surface-tethered pre-mRNA, Abelson et al. were able to show that pre-mRNA conformation reversibly fluctuated during splicing. These conformational changes are shown by changes in the FRET signal of single pre-mRNA molecules. Data in (a) and (b) are from references [45] and [49], respectively, and used with the authors’ permission.
In addition to reaction order, Hoskins et al. observed that all of the snRNPs and the Prp19 complex bound reversibly to the pre-mRNA [45]. In many cases, multiple (even dozens) of binding events were detected (Figure 4a). This implies that many interactions between spliceosomal subcomplexes and the pre-mRNA are non-productive and do not result in splicing. Whether or not these non-productive interactions stem from kinetic proofreading and Prp43-mediated disassembly remains to be seen; however, the results obtained are consistent with the presence of discard pathways (Figures 1 and 2).
Furthermore, by comparing the number of observed subcomplex pre-mRNA interactions with the extent of intron loss, Hoskins et al. also found that the probability of intron loss increases as the spliceosome assembles. These results contrast with the classical view of commitment during splicing in which the fate of the pre-mRNA is decided by initial (and irreversible) association of U1 with the 5′ SS [46]. Instead, the single molecule studies revealed that the probability of mRNA formation increases as the spliceosome passes successfully through each assembly stage and avoids discard pathways [45].
The above observation of low commitment to splicing upon U1 association highlights some of the intrinsic differences between single molecule and bulk measurements of spliceosome formation. In both methods, long-lived (>10 min) interactions between U1 and the pre-mRNA could be observed under conditions that favored formation of complexes previously described to be committed to splicing (e.g., no ATP) [45–47]. However, single molecule analysis revealed that these comprised the minority (<10%) of U1/pre-mRNA interactions and that most U1/pre-mRNA complexes (~75%) possessed lifetimes of <30 sec. Since bulk measurements of U1 association often rely on native gel analysis, it is likely these weaker complexes do not survive electrophoresis or run as ‘unspecific’ complexes [46,47]. Conversely, single molecule methods are limited only by camera detection efficiency, and so they can readily detect complexes with lifetimes as short as milliseconds. Although it is possible that U1 dissociation was accelerated by surface-tethering of the pre-mRNA, the work by Hoskins et al. agrees well with bulk measurements of U1 dynamics made using electrophoresis conditions designed to stabilize weaker U1/pre-mRNA interactions [48]. At a minimum, single molecule measurements have now indicated that long-lived interactions between the snRNPs and the pre-mRNA are not required for splicing. Further analysis of spliceosome assembly will certainly take advantage of the unique viewpoints offered by both single molecule and bulk solution assays.
In a complementary set of experiments, the Abelson/Guthrie and Walter groups used single molecule fluorescence resonance energy transfer (SM-FRET) to study pre-mRNA conformation during splicing [49]. By incorporating FRET acceptor and donor fluorophores into the exons of a splicing substrate derived from the Ubc4 gene [50], Abelson et al. were able to monitor changes in exon proximity during splicing. In this way, they uncovered an incredibly intricate series of conformational changes occurring during the reaction (Figure 4b). Under no conditions did this pre-mRNA appear to be locked into a specific conformation; instead, it toggled reversibly through numerous FRET states. Although no individual FRET state has yet been correlated with a specific stage in spliceosome assembly, such fluctuations could represent the presence or absence of a given spliceosome component and/or the spliceosome alternating between first or second step conformations. Now that single molecule analysis can be performed on reaction systems as complicated as WCE [51], many more insights will undoubtedly follow using the powerful tools of CoSMoS and/or SM-FRET to dissect the complex reaction pathways through which the splicing machinery must journey to generate spliced mRNAs.
Microarrays and the importance of pre-mRNA identity
Most of the experiments described above were carried out using one to three model pre-mRNAs that splice well both in vivo and in vitro: RP51A (containing a shortened form of the intron in ribosomal protein gene RPS17A), ACT1 (containing the full-length intron from the actin gene), and UBC4 (a very short full-length intron in a ubiquitin conjugating enzyme gene). However, given the intimate roles of the pre-mRNA in binding to proteins and snRNAs and providing the reactive nucleophiles for chemistry (Figure 3), an important question to ask is: which features of the splicing cycle are universal and which are specific to individual pre-mRNAs? Answering this question requires analysis of diverse transcripts, which has now been accomplished using splicing-dependent microarrays.
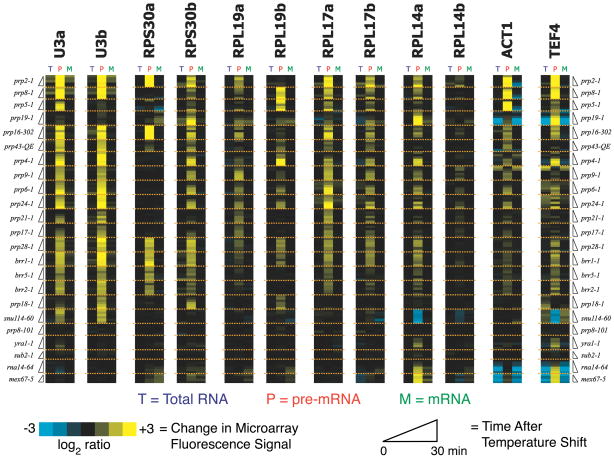
In the first analysis of this kind, the Ares group developed DNA oligonucleotide arrays that could differentiate between the unspliced and spliced RNAs synthesized from all ~250 yeast genes containing an intron [52]. Probing these arrays with cellular RNA isolated from various mutant yeast strains, this group demonstrated that specific subsets of pre-mRNAs are differentially affected by several nonessential splicing factors. Employing the same type of microarray, the Guthrie group subsequently analyzed the pan-intron effects of mutations in essential spliceosomal proteins [53]. Over the past few decades, a plethora of Ts or cold-sensitive (Cs) mutations in essential spliceosome proteins have been reported (e.g. the prp2-1 mutation described above). By analyzing the splicing responses of yeast pre-mRNAs when cells were shifted from permissive to non-permissive temperatures, Pleiss et al. were able to assess how loss of activity of a specific protein affected splicing of each pre-mRNA. They carried out their analysis on 18 different strains harboring mutations in spliceosome components predicted to affect almost every step in the spliceosome cycle (assembly, activation, catalysis, or disassembly). The results were quite striking: each pre-mRNA in yeast responded uniquely to mutations in core splicing factors (Figure 5) [53]. For example, whereas splicing of the well-characterized ACT1 transcript was strongly inhibited by the prp2-1 mutation, RPL19B intron splicing was unaffected.
Figure 5.
Microarray analysis of pre-mRNA splicing in yeast and the effects of mutations in core spliceosomal proteins. Each pre-mRNA in yeast (such as the subset of 12 shown here) can respond uniquely to mutations in core splicing factors. These responses can be identified by either increases (yellow bands) or decreases (blue bands) in pre-mRNA abundance relative to a wild type strain. This indicates that pre-mRNA identity can strongly influence multiple steps in splicing. Data are from reference [53] and used with the authors’ permission.
The implications of these findings are profound. Far from being a passive substrate upon which the spliceosome acts, each pre-mRNA can influence its own interaction with the splicing machinery to either promote or inhibit a given step in the pathway. Yeast can exploit this behavior to, for example, alter transcript-specific splicing responses during environmental stress [54,55]. In one intriguing mechanism, alternative splicing of the yeast APE2 gene appears to be governed by competing pre-mRNA structures that modulate 3′ SS selection in response to temperature [56]. Thus it appears that Saccharomyces cerevisiae has evolved mechanisms to modulate spliceosome activity as part of its biology, and like the Ts or Cs mutations, these mechanisms can be tailored to target certain transcripts. But even more important, each pre-mRNA:snRNP complex may need to be considered as its own unique spliceosome with different kinetic and/or thermodynamic properties. Thus, resolving which features of the splicing cycle are universal and which are specific to individual pre-mRNAs may not be at all straightforward. While it is likely that all spliceosomes share a common active site structure, the pathways by which those active sites are formed may be quite distinct.
Concluding remarks
Far from being the series of static complexes connected by one-way arrows as often appears in textbook pictures of the spliceosome cycle, we now know that the spliceosome is an extraordinarily dynamic and flexible machine. As described above, new technologies are providing ample evidence for reversible interactions and chemistry during splicing, as well as a highly flexible active site heavily influenced by the pre-mRNA substrate. As has been previously postulated [35,37,45], these results suggest that regulation of splicing can occur at any stage of the reaction. Indeed, it is possible that the only point at which the splicing decision becomes irreversible is in Prp22-catalyzed release of the mRNA product [57]. It is intriguing to speculate that some substrates may toggle repeatedly through the chemical steps of splicing and enter or exit discard pathways until either irreversible mRNA release by Prp22 or disassembly by Prp43 occurs. Consequently, the relative abundances and activities of Prp22 and Prp43 may be essential for controlling both fidelity and flux through the splicing pathway: high Prp22 activity relative to Prp43 may promote low fidelity but high turnover, whereas low Prp22 activity relative to Prp43 could increase fidelity at the expense of total mRNA output. Whether or not biology has learned to exploit these features remains to be seen.
The ability of yeast to regulate spliceosome assembly and the chemical steps of splicing at multiple stages also has significant implications for our understanding of alternative splicing in humans. In humans, spliceosome assembly initiates around exons (exon definition) and introns are subsequently identified by bringing together previously defined exons (SS pairing) [5]. The presence of reversible steps throughout the splicing pathway suggests that SS pairing may well be reversible. Spliceosome assembly could initiate around a particular set of exons only to have that spliceosome be discarded and reformed around an alternate set. Thus alternative splicing itself could be an extraordinarily dynamic process, with spliced exon release being the final arbiter in generating a particular isoform.
In humans, splice sites are defined on some pre-mRNAs during A Complex formation [58,59] (Figure 1). Yet it is not clear if this mechanism represents the rule or the exception. Some evidence suggests that more complicated mechanisms function on at least a subset of human pre-mRNAs [60,61]. While it is not yet formally known if kinetic proofreading and discard pathways are conserved beyond the yeast spliceosome, RNAi knockdown of spliceosomal DExD/H-box ATPases in Drosophila has shown that several (Sub2, Prp5, Brr2, and Prp22; Figure 1) can influence mRNA isoform generation [62]. This strongly supports the hypothesis that steps subsequent to SS pairing influence alternative splicing.
While recent years have seen much progress in our understanding of the spliceosome, many significant challenges remain. Foremost is the limited structural information available for either intact spliceosomes or their components [63]. More detailed structural information will be invaluable for elucidating the biochemistry of this remarkable machine, and preparation of samples for such structural elucidation will surely employ some of the affinity purification methods detailed in this review. Indeed, significant progress has recently been made in preparing and characterizing homogenous spliceosomes for structural characterization [64,65]. Also limiting is our kinetic understanding of steps not directly resulting in bond cleavage or formation. For example, kinetic proofreading and the use of discard pathways have been proposed for other steps in the splicing cycle besides that mediated by Prp16 [24,66–68]. However, no kinetic information for any of the spliceosomal ATPases in the context of an active spliceosome is yet known. Single molecule methods hold the promise of providing these missing parameters, as well as elucidating the discard and disassembly pathways. Recent advances in single molecule imaging in cells suggest that such measurements could even be made in vivo [69–72]. Finally, segregating the universal features of splicing away from those that are pre-mRNA specific remains to be fully explored. As demonstrated by the APE2 thermosensor [56] and microarray analysis [53], RNA sequence and secondary structure can profoundly influence the splicing fate of a pre-mRNA. Key to deciphering these mechanisms will be determination of the kinetic pathways for splicing of different transcripts in vitro and in vivo using the methods discussed in this review. Undoubtedly, many surprises yet await those of us who study this most complex and dynamic of macromolecular machines.
Acknowledgments
This work was supported by NIH awards R00-GM086471 (A.A.H.) and R01-GM053007 (M.J.M). M.J.M. is a Howard Hughes Medical Institute investigator.
Glossary
- Exon
regions of pre-mRNA that are ligated together by the spliceosome and often code for proteins
- Intron
the region of pre-mRNA excised by the spliceosome. Yeast pre-mRNAs usually contain a single intron of less than 1000 nts in length. Human pre-mRNAs can contain many introns ranging from a few dozen nts to >400,000 nts in size. Intronic RNA can serve as a source for other non-coding RNAs in the cell such as snoRNAs and micro RNAs
- Alternative splicing
the process by which the spliceosome can generate multiple mRNA isoforms from a single pre-mRNA. This is a fundamental mechanism for encoding genomic complexity in higher eukaryotes without an expansion in gene number
- snRNPs
small nuclear ribonucleoproteins. Complexes of small nuclear RNA (i.e. the U1, U2, U4, U5, and U6 snRNAs) and protein that form the core components of the spliceosome. The U1 and U2 snRNPs form separate particles, while the U4, U5, and U6 components join together in the U4/U6.U5 tri-snRNP
- Splice sites
the locations of 5′ SS cleavage and exon ligation at the exon/intron boundaries in a pre-mRNA. In yeast, a typical 5′ SS consensus sequence is n/GUAUGU, and the 3′ SS signal is YAG/n, where the intron sequences are noted in uppercase and the exon/intron boundary is noted by the diagonal. SS sequences in higher organisms can be much more degenerate
- Branch site
the region of the pre-mRNA intron containing the branch point. In yeast, this usually has the sequence UACUAAC
- Branch point
the nucleotide (usually an adenosine) in the branch site sequence that provides the nucleophile for 5′ SS cleavage and becomes the site of lariat formation. In yeast, the branch point is the 3′-most A (underlined) in the UACUAAC branch site consensus sequence
- DExD/H-box ATPases
RNA-binding enzymes that hydrolyze ATP to facilitate a number of biochemical processes. These enzymes can be divided into subfamilies based on conserved sequence motifs. The spliceosome contains 3 DEAD-box ATPases (Sub2, Prp5, and Prp28), 4 DEAH-box ATPases (Prp2, Prp16, Prp22, and Prp43), and one DEIH-box ATPase (Brr2). These ATPases are believed to facilitate structural transitions that occur during splicing, and some of these (Prp5, Prp16, Prp22, and Prp43) take part in discard pathways that enhance splicing fidelity
- Kinetic Proofreading
a model for increasing the fidelity of splicing in which spliceosomal ATPases control entry into a discard pathway based upon the relative kinetics of a given step during splicing (e.g. 5′ SS cleavage) and ATPase function (e.g. Prp16 activity)
- Two state model for splicing
a model for the catalytically activated spliceosome in which specific conformations exist in equilibrium that promote either 5′ SS cleavage or exon ligation. This equilibrium can be modulated by mutations in core spliceosome components, the pre-mRNA, or other factors
- CoSMoS
colocalization single molecule spectroscopy. A single molecule fluorescence microscopy technique in which biochemical reactions can be studied by watching multiple biomolecules, each labeled with a unique fluorophore, colocalize with a surface-tethered substrate
- smFRET
single molecule fluorescence resonance energy transfer. A single molecule fluorescence microscopy technique in which the proximity or environment of donor (e.g. Cy3) and acceptor (e.g. Cy5) fluorophores is monitored over time
- Splicing dependent microarray
a microarray method in which fluorescent probes are used to specifically detect pre-mRNA, mRNA, and total RNA obtained from a cell culture. This can be used to study changes in splicing that occur in vivo
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Nilsen TW, Graveley BR. Expansion of the eukaryotic proteome by alternative splicing. Nature. 2010;463:457–463. doi: 10.1038/nature08909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Scherer S. Guide to the Human Genome. 1. Cold Spring Harbor; 2010. [Google Scholar]
- 3.Lim LP, Burge CB. A computational analysis of sequence features involved in recognition of short introns. Proc Natl Acad Sci USA. 2001;98:11193–11198. doi: 10.1073/pnas.201407298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roca X, Krainer AR. Recognition of atypical 5′ splice sites by shifted base-pairing to U1 snRNA. Nat Struct Mol Biol. 2009;16:176–182. doi: 10.1038/nsmb.1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wahl MC, et al. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 6.Jurica MS, Moore MJ. Pre-mRNA splicing: awash in a sea of proteins. Mol Cell. 2003;12:5–14. doi: 10.1016/s1097-2765(03)00270-3. [DOI] [PubMed] [Google Scholar]
- 7.Nilsen TW. The spliceosome: the most complex macromolecular machine in the cell? Bioessays. 2003;25:1147–1149. doi: 10.1002/bies.10394. [DOI] [PubMed] [Google Scholar]
- 8.Fabrizio P, et al. The evolutionarily conserved core design of the catalytic activation step of the yeast spliceosome. Mol Cell. 2009;36:593–608. doi: 10.1016/j.molcel.2009.09.040. [DOI] [PubMed] [Google Scholar]
- 9.Steitz JA, et al. Where in the cell is the minor spliceosome? Proc Natl Acad Sci USA. 2008;105:8485–8486. doi: 10.1073/pnas.0804024105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moore MJ, Proudfoot NJ. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell. 2009;136:688–700. doi: 10.1016/j.cell.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 11.Alexander R, Beggs JD. Cross-talk in transcription, splicing and chromatin: who makes the first call? Biochem Soc Trans. 2010;38:1251–1256. doi: 10.1042/BST0381251. [DOI] [PubMed] [Google Scholar]
- 12.Lardelli RM, et al. Release of SF3 from the intron branchpoint activates the first step of pre-mRNA splicing. RNA. 2010;16:516–528. doi: 10.1261/rna.2030510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brow DA. Allosteric cascade of spliceosome activation. Annu Rev Genet. 2002;36:333–360. doi: 10.1146/annurev.genet.36.043002.091635. [DOI] [PubMed] [Google Scholar]
- 14.Lesser CF, Guthrie C. Mutational analysis of pre-mRNA splicing in Saccharomyces cerevisiae using a sensitive new reporter gene, CUP1. Genetics. 1993;133:851–863. doi: 10.1093/genetics/133.4.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stevens SW, Abelson J. Yeast pre-mRNA splicing: methods, mechanisms, and machinery. Meth Enzymol. 2002;351:200–220. doi: 10.1016/s0076-6879(02)51849-8. [DOI] [PubMed] [Google Scholar]
- 16.Kornberg A. Ten commandments of enzymology, amended. Trends Biochem Sci. 2003;28:515–517. doi: 10.1016/j.tibs.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 17.Bessonov S, et al. Isolation of an active step I spliceosome and composition of its RNP core. Nature. 2008;452:846–850. doi: 10.1038/nature06842. [DOI] [PubMed] [Google Scholar]
- 18.Warkocki Z, et al. Reconstitution of both steps of Saccharomyces cerevisiae splicing with purified spliceosomal components. Nat Struct Mol Biol. 2009;16:1237–1243. doi: 10.1038/nsmb.1729. [DOI] [PubMed] [Google Scholar]
- 19.Anderson K, Moore MJ. Bimolecular exon ligation by the human spliceosome. Science. 1997;276:1712–1716. doi: 10.1126/science.276.5319.1712. [DOI] [PubMed] [Google Scholar]
- 20.Lustig AJ, et al. The yeast RNA gene products are essential for mRNA splicing in vitro. Cell. 1986;47:953–963. doi: 10.1016/0092-8674(86)90810-x. [DOI] [PubMed] [Google Scholar]
- 21.Yean SL, Lin RJ. U4 small nuclear RNA dissociates from a yeast spliceosome and does not participate in the subsequent splicing reaction. Mol Cell Biol. 1991;11:5571–5577. doi: 10.1128/mcb.11.11.5571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yean SL, et al. Metal-ion coordination by U6 small nuclear RNA contributes to catalysis in the spliceosome. Nature. 2000;408:881–884. doi: 10.1038/35048617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Puig O, et al. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods. 2001;24:218–229. doi: 10.1006/meth.2001.1183. [DOI] [PubMed] [Google Scholar]
- 24.Koodathingal P, et al. The DEAH box ATPases Prp16 and Prp43 cooperate to proofread 5′ splice site cleavage during pre-mRNA splicing. Mol Cell. 2010;39:385–395. doi: 10.1016/j.molcel.2010.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hopfield JJ. Kinetic proofreading: a new mechanism for reducing errors in biosynthetic processes requiring high specificity. Proc Natl Acad Sci USA. 1974;71:4135–4139. doi: 10.1073/pnas.71.10.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Burgess SM, Guthrie C. A mechanism to enhance mRNA splicing fidelity: the RNA-dependent ATPase Prp16 governs usage of a discard pathway for aberrant lariat intermediates. Cell. 1993;73:1377–1391. doi: 10.1016/0092-8674(93)90363-u. [DOI] [PubMed] [Google Scholar]
- 27.Burgess S, et al. A putative ATP binding protein influences the fidelity of branchpoint recognition in yeast splicing. Cell. 1990;60:705–717. doi: 10.1016/0092-8674(90)90086-t. [DOI] [PubMed] [Google Scholar]
- 28.Schwer B, Guthrie C. A conformational rearrangement in the spliceosome is dependent on PRP16 and ATP hydrolysis. EMBO J. 1992;11:5033–5039. doi: 10.1002/j.1460-2075.1992.tb05610.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mayas RM, et al. Spliceosome discards intermediates via the DEAH box ATPase Prp43p. Proc Natl Acad Sci USA. 2010;107:10020–10025. doi: 10.1073/pnas.0906022107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arenas JE, Abelson JN. Prp43: An RNA helicase-like factor involved in spliceosome disassembly. Proc Natl Acad Sci USA. 1997;94:11798–11802. doi: 10.1073/pnas.94.22.11798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Combs DJ, et al. Prp43p is a DEAH-box spliceosome disassembly factor essential for ribosome biogenesis. Mol Cell Biol. 2006;26:523–534. doi: 10.1128/MCB.26.2.523-534.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bohnsack MT, et al. Prp43 bound at different sites on the pre-rRNA performs distinct functions in ribosome synthesis. Mol Cell. 2009;36:583–592. doi: 10.1016/j.molcel.2009.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Augustin S, et al. Reverse self-splicing of group II intron RNAs in vitro. Nature. 1990;343:383–386. doi: 10.1038/343383a0. [DOI] [PubMed] [Google Scholar]
- 34.Tseng C-K, Cheng S-C. Both Catalytic Steps of Nuclear Pre-mRNA Splicing Are Reversible. Science. 2008;320:1782–1784. doi: 10.1126/science.1158993. [DOI] [PubMed] [Google Scholar]
- 35.Smith DJ, Konarska MM. Mechanistic insights from reversible splicing catalysis. RNA. 2008 doi: 10.1261/rna.1289808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith DJ, et al. Insights into branch nucleophile positioning and activation from an orthogonal pre-mRNA splicing system in yeast. Mol Cell. 2009;34:333–343. doi: 10.1016/j.molcel.2009.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith DJ, et al. “Nought may endure but mutability”: spliceosome dynamics and the regulation of splicing. Mol Cell. 2008;30:657–666. doi: 10.1016/j.molcel.2008.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Query CC, Konarska MM. Suppression of multiple substrate mutations by spliceosomal prp8 alleles suggests functional correlations with ribosomal ambiguity mutants. Mol Cell. 2004;14:343–354. doi: 10.1016/s1097-2765(04)00217-5. [DOI] [PubMed] [Google Scholar]
- 39.Liu L, et al. Opposing classes of prp8 alleles modulate the transition between the catalytic steps of pre-mRNA splicing. Nat Struct Mol Biol. 2007;14:519–526. doi: 10.1038/nsmb1240. [DOI] [PubMed] [Google Scholar]
- 40.Konarska MM, et al. Repositioning of the reaction intermediate within the catalytic center of the spliceosome. Mol Cell. 2006;21:543–553. doi: 10.1016/j.molcel.2006.01.017. [DOI] [PubMed] [Google Scholar]
- 41.Smith DJ, et al. trans-splicing to spliceosomal U2 snRNA suggests disruption of branch site-U2 pairing during pre-mRNA splicing. Mol Cell. 2007;26:883–890. doi: 10.1016/j.molcel.2007.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Walter NG, et al. Do-it-yourself guide: how to use the modern single-molecule toolkit. Nat Meth. 2008;5:475–489. doi: 10.1038/nmeth.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gershenson A. Single molecule enzymology: watching the reaction. Curr Op Chem Biol. 2009;13:1–7. doi: 10.1016/j.cbpa.2009.06.011. [DOI] [PubMed] [Google Scholar]
- 44.Friedman LJ, et al. Viewing dynamic assembly of molecular complexes by multi-wavelength single-molecule fluorescence. Biophys J. 2006;91:1023–1031. doi: 10.1529/biophysj.106.084004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hoskins AA, et al. Ordered and dynamic assembly of single spliceosomes. Science. 2011;331:1289–1295. doi: 10.1126/science.1198830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Legrain P, et al. Early commitment of yeast pre-mRNA to the spliceosome pathway. Mol Cell Biol. 1988;8:3755–3760. doi: 10.1128/mcb.8.9.3755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Seraphin B, Rosbash M. Identification of functional U1 snRNA-pre-mRNA complexes committed to spliceosome assembly and splicing. Cell. 1989;59:349–358. doi: 10.1016/0092-8674(89)90296-1. [DOI] [PubMed] [Google Scholar]
- 48.Ruby SW. Dynamics of the U1 small nuclear ribonucleoprotein during yeast spliceosome assembly. J Biol Chem. 1997;272:17333–17341. doi: 10.1074/jbc.272.28.17333. [DOI] [PubMed] [Google Scholar]
- 49.Abelson J, et al. Conformational dynamics of single pre-mRNA molecules during in vitro splicing. Nat Struct Mol Biol. 2010;17:504–512. doi: 10.1038/nsmb.1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Abelson J, et al. Preparation of fluorescent pre-mRNA substrates for an smFRET study of pre-mRNA splicing in yeast. Meth Enzymol. 2010;472:31–40. doi: 10.1016/S0076-6879(10)72017-6. [DOI] [PubMed] [Google Scholar]
- 51.Crawford DJ, et al. Visualizing the splicing of single pre-mRNA molecules in whole cell extract. RNA. 2008;14:170–179. doi: 10.1261/rna.794808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Clark TA, et al. Genomewide analysis of mRNA processing in yeast using splicing-specific microarrays. Science. 2002;296:907–910. doi: 10.1126/science.1069415. [DOI] [PubMed] [Google Scholar]
- 53.Pleiss JA, et al. Transcript specificity in yeast pre-mRNA splicing revealed by mutations in core spliceosomal components. PLoS Biol. 2007;5:e90. doi: 10.1371/journal.pbio.0050090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pleiss JA, et al. Rapid, transcript-specific changes in splicing in response to environmental stress. Mol Cell. 2007;27:928–937. doi: 10.1016/j.molcel.2007.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bergkessel M, et al. Diverse environmental stresses elicit distinct responses at the level of pre-mRNA processing in yeast. RNA. 2011;17:1461–1478. doi: 10.1261/rna.2754011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Meyer M, et al. Deciphering 3′ss Selection in the Yeast Genome Reveals an RNA Thermosensor that Mediates Alternative Splicing. Mol Cell. 2011;43:1033–1039. doi: 10.1016/j.molcel.2011.07.030. [DOI] [PubMed] [Google Scholar]
- 57.Company M, et al. Requirement of the RNA helicase-like protein PRP22 for release of messenger RNA from spliceosomes. Nature. 1991;349:487–493. doi: 10.1038/349487a0. [DOI] [PubMed] [Google Scholar]
- 58.Lim SR, Hertel KJ. Commitment to splice site pairing coincides with A complex formation. Mol Cell. 2004;15:477–483. doi: 10.1016/j.molcel.2004.06.025. [DOI] [PubMed] [Google Scholar]
- 59.Kotlajich MV, et al. Spliceosome assembly pathways for different types of alternative splicing converge during commitment to splice site pairing in the A complex. Mol Cell Biol. 2009;29:1072–1082. doi: 10.1128/MCB.01071-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bonnal S, et al. RBM5/Luca-15/H37 regulates Fas alternative splice site pairing after exon definition. Mol Cell. 2008;32:81–95. doi: 10.1016/j.molcel.2008.08.008. [DOI] [PubMed] [Google Scholar]
- 61.Chen M, Manley JL. Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nat Rev Mol Cell Biol. 2009;10:741–754. doi: 10.1038/nrm2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Park JW, et al. Identification of alternative splicing regulators by RNA interference in Drosophila. Proc Natl Acad Sci USA. 2004;101:15974–15979. doi: 10.1073/pnas.0407004101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stark H, Lührmann R. Cryo-electron microscopy of spliceosomal components. Annual review of biophysics and biomolecular structure. 2006;35:435–457. doi: 10.1146/annurev.biophys.35.040405.101953. [DOI] [PubMed] [Google Scholar]
- 64.Ilagan J, et al. The role of exon sequences in C complex spliceosome structure. J Mol Biol. 2009;394:363–375. doi: 10.1016/j.jmb.2009.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wolf E, et al. Exon, intron and splice site locations in the spliceosomal B complex. EMBO J. 2009;28:2283–2292. doi: 10.1038/emboj.2009.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xu Y-Z, Query CC. Competition between the ATPase Prp5 and branch region-U2 snRNA pairing modulates the fidelity of spliceosome assembly. Mol Cell. 2007;28:838–849. doi: 10.1016/j.molcel.2007.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Staley JP, Guthrie C. An RNA switch at the 5′ splice site requires ATP and the DEAD box protein Prp28p. Mol Cell. 1999;3:55–64. doi: 10.1016/s1097-2765(00)80174-4. [DOI] [PubMed] [Google Scholar]
- 68.Mayas RM, et al. Exon ligation is proofread by the DExD/H-box ATPase Prp22p. Nat Struct Mol Biol. 2006;13:482–490. doi: 10.1038/nsmb1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schmidt U, et al. Real-time imaging of cotranscriptional splicing reveals a kinetic model that reduces noise: implications for alternative splicing regulation. The Journal of Cell Biology. 2011;193:819–829. doi: 10.1083/jcb.201009012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vargas DY, et al. Single-molecule imaging of transcriptionally coupled and uncoupled splicing. Cell. 2011;147:1054–1065. doi: 10.1016/j.cell.2011.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Larson DR, et al. Real-time observation of transcription initiation and elongation on an endogenous yeast gene. Science. 2011;332:475–478. doi: 10.1126/science.1202142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Huranová M, et al. The differential interaction of snRNPs with pre-mRNA reveals splicing kinetics in living cells. The Journal of Cell Biology. 2010;191:75–86. doi: 10.1083/jcb.201004030. [DOI] [PMC free article] [PubMed] [Google Scholar]