Abstract
Background
Familial dyskinesia with facial myokymia (FDFM) is an autosomal dominant disorder that is exacerbated by anxiety. In a five-generation family of German ancestry we previously mapped FDFM to chromosome 3p21-3q21. The 72.5 Mbp linkage region was too large for traditional positional mutation identification.
Objective
To identify the gene responsible for FDFM by exome resequencing of a single affected individual.
Design, Setting and Participants
We performed whole exome sequencing in one affected individual and used a series of bioinformatic filters, including functional significance and presence in dbSNP or 1000 Genomes project, to reduce the number of candidate variants. Co-segregation analysis was performed in 15 additional individuals in three generations.
Results
The exome contained 23428 single nucleotide variants, of which 9391 were missense, nonsense or splice site alterations. The critical region contained 323 variants, five of which were not present in one of the sequence-databases. Adenylate cyclase 5 (ADCY5) was the only gene in which the variant (c.2176G>A) was co-transmitted perfectly with disease status and was not present in 3510 control Caucasian exomes. This residue is highly conserved and the change is nonconservative and predicted to be damaging.
Conclusions
ADCY5 is highly expressed in striatum. Mice deficient in Adcy5 develop a movement disorder that is worsened by stress. We conclude that FDFM likely results from a missense mutation in ADCY5. This study demonstrates the power of a single exome sequence in combination with linkage information to identify causative genes for rare autosomal dominant Mendelian diseases.
Introduction
In 2001 we described a Caucasian family of German ancestry manifesting a disorder characterized by predominantly perioral and periorbital myokymia, and face, neck and upper limb dystonic/choreic movements (FDFM; OMIM 606703)1. Initially, the disorder had been described as familial essential (benign) chorea2. The onset of symptoms ranged from 2.5 to 19 years. Initially paroxysmal and precipitated or worsened by stress, the dyskinetic episodes become nearly constant by the end of the 3rd decade but in some individuals may diminish in frequency and severity in older ages. Autosomal dominant transmission of FDFM is supported by the presence of disease in all five generations, apparently complete penetrance in both males and females, and instances of male-to-male transmission.
By targeted genotyping, we excluded ten candidate genes chosen for their association with myokymia or chorea and two regions that contain single or clustered ion channel genes1. We mapped the disorder to a 71.73 cM region, corresponding to 72.5 Mb, on chromosome 3p21-3q213. Interrogation of such a large region, deemed by NCBI Entrez Gene, Build 37 to contain 253 annotated genes, became feasible by the recent development of “next generation” massively parallel sequencing technologies. We provide evidence that ADCY5 is a strong candidate gene for FDFM and demonstrate the power of exome sequencing to identify causative genes for autosomal dominant disorders.
Methods
Human Subjects
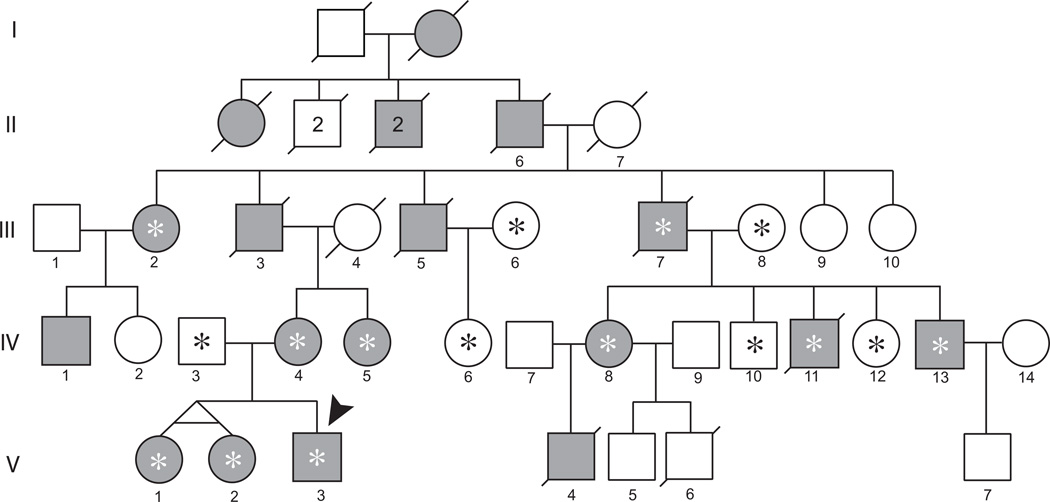
An updated pedigree is shown in Figure 1. Patients were evaluated and blood samples were collected from 16 individuals in three generations under a protocol approved by the Institutional Review Board of the University of Washington.
Figure 1.
Family with autosomal dominant familial dyskinesia with facial myokymia (FDFM). Shaded icons denote affected individuals; central asterisks denote individuals who provided DNA; an arrowhead marks the individual whose exome was sequenced.
Targeted Capture and Exome Sequencing
Genomic DNA was extracted from peripheral blood using standard procedures. Five µg of DNA from an affected member of the FDFM family (V-3 in Figure 1) was used to construct a shotgun sequencing library as described previously4. The shotgun library was hybridized to NimbleGen microarrays (Nimblegen_solution_V2refseq2010.HG19) for target enrichment, followed by washing, elution and additional amplification. We targeted all protein-coding regions as defined by RefSeq 36.3. Exome sequencing was performed on the Illumina GAIIx platform with paired-end 76 base reads. Sequence alignment and variant calling were performed against the reference human genome (NCBI 37/hg 19). For SNPs and indels, the calling and genotyping was done with the GATK Unified Genotyper. Annotations of variants were based on NCBI and UCSC databases using an in-house server (SeattleSeqAnnotation 131).
Candidates genes/variants were ranked on the basis of:
Coding region sequence effect. Missense, nonsense, coding indels and splice acceptor and donor sites were selected for further evaluation.
Variant frequency. Unique variants were selected based on non-overlap with variants present in dbSNP 131, the 1000 Genomes project, and 3510 control exomes of European ancestry from the UW Genome Sciences Genomic Resource Center (GSGRC) Exome Sequencing Projects (ESP). The UW Exome Variant Server catalogs variants identified through exome sequencing primarily of individuals with heart, lung or blood disorders.
Location within the linkage region. Variants in the 72.5Mb region on chromosome 3p21-3q21, bounded by D3S1582 and D3S3606 were further considered.
Conservation scores as estimated by UCSC PhastCons and Genomic Evolutionary Rate Profiling (GERP). PhastCons scores are based on sequence conservation in 17 vertebrate species5. The score from 0 to 1 denotes the probability that each nucleotide belongs to a conserved element. GERP scores range from −11.6 to 5.86 and are based on sequence conservation in 46 mammals6.
Potential effect of an amino acid substitution on protein structure and function as predicted by Sorting Intolerant From Tolerant7 and Polymorphism Phenotyping (PolyPhen-2)8,9.
Variant validation by Sanger sequencing
Capillary sequencing using customized primers was performed to confirm five unique coding variants in the linkage region and to investigate co-transmission with the FDFM phenotype in all family members. Primer sequences are shown in eTable 1. Genomic DNA was PCR amplified in a MJ Research DNA Engine Tetrad 2. Sequencing was performed on an ABI 3130XL sequencer using ABI BigDye Dye Terminator Cycle Sequencing Kits. Further analysis was performed with SeqScape 2.1 or DNASTAR Lasergene 8.1.
Evaluation for Copy Number Variation by Array Comparative Genomic Hybridization (aCGH)
To detect deletions and duplications, we utilized two array designs: (1) exon-focused 720×3 whole-exome NimbleGen microarray and (2) custom chromosome 3 tiling NimbleGen microarray with median probe spacing of 275 bp. All microarray hybridization experiments were performed as described previously10,11, using a single unaffected male (GM15724 from Coriell) as reference. CNV calls were made using a Hidden Markov Model-based algorithm12 and compared with CNVs from 5674 control individuals as previously reported13.
Results
Clinical history
Since description of the family and the disorder in 20011, two affected males have died (Individuals III-7 and IV-11). Reexamination of Individual IV-13 at age 50 years demonstrated some improvement of the neurologic manifestations since he was first evaluated. He still exhibited subtle choreic hand and arm movements and facial myokymia. At age 46 he was diagnosed with a severe dilated cardiomyopathy (left ventricular ejection fraction 10% at diagnosis and 15–20% with therapy). Cardiac catheterization detected non-occlusive atherosclerosis that was not felt to be sufficient to cause the global cardiac dysfunction. CT angiogram was negative for pulmonary emboli. Sleep apnea was diagnosed, but it could not be interpreted given the concurrent acute congestive heart failure. One year later, rest and regadenoson-stress sestamibi myocardial imagining showed normal perfusion except for a questionable inferior wall defect. He provided information that four of his neurologically affected relatives had also suffered from congestive heart failure (CHF). The age of onset of CHF and the age at and cause of death of his grandfather (II-6) were unknown. His father (III-7) had undergone coronary artery bypass grafting and died of CHF at age 83, but two uncles developed CHF at much earlier ages, one dying at age 63 (III-3) and the other in his 50s (III-5). Individual IV-11 died of cancer at 62. Records on these individuals were not available for our review.
Array CGH
To evaluate the possibility of a large copy number variation, aCGH was performed on DNA from individuals IV-8 and V-2. Two strategies for CNV detection were pursued. First, we utilized an exon-focused 720×3 whole-exome NimbleGen microarray to interrogate large CNVs (>150 kbp)10–12. Second, we targeted the entire chromosome 3 on a custom tiling NimbleGen microarray with median probe spacing of 275 bp. After filtering for variants identified in a large collection (~6,300) of population controls13, we assessed for rare, potentially pathogenic variants within and outside chromosome 3. No potentially pathogenic CNVs were detected individually or shared between the affected individuals. These results suggested that variants potentially below the resolution of our array or single nucleotide variants contribute to the observed phenotypes.
Multiply parallel sequencing
We generated 70 Mb of unpaired 76-bp sequencing reads from affected individual V-3. Of this total, 83.0% (58 Mb) were paired-end, 76 bp reads that passed the quality assessment and aligned to the human reference sequence, and 64.4% mapped to the targeted bases with a mean coverage of 68-fold. At this depth of coverage, 94.8% of the targeted bases were sufficiently covered to pass our thresholds for variant calling.
The exome of individual V-3 contained 23428 single nucleotide variants (SNPs), of which 9391 alter the coding sequence (Table 1). These coding SNPs consist of 9274 missense, 88 nonsense and 29 splice acceptor- or donor-site variants. In addition, 414 short insertions or deletions (indels) were detected. Based on the hypothesis that the mutation underlying this rare familial disease would be of sufficiently low prevalence in the general population that it would not be present in databases of variations, SNPs identified in the 1000 Genomes project or in dbSNP131 were removed. Application of this filter reduced the candidate gene pool to 715.
Table 1.
Variants detected in exome of individual V-3
| Whole exome | 72.5 Mb chr. 3 linkage region |
|
|---|---|---|
| Total single nucleotide variants | 23428 | 323 |
| Synonymous | 10500 | 130 |
| Coding-notMod31 | 240 | 0 |
| Intergenic | 837 | 13 |
| Intron | 1963 | 40 |
| Near gene (5´ and 3´) | 98 | 3 |
| 5´and 3´UTR | 399 | 4 |
| Nonsense | 88 | 2 |
| Splice site (5´ and 3´) | 29 | 1 |
| Missense | 9274 | 130 |
| Indels 2 | 414 | 9 |
Variant within an exon and translated, but number of coding bases is not a multiple of 3 and no attempt is made to rate as synonymous or not.
Short insertions or deletions; 79% of coding indels are 1–3 bp in length
Within the 72,504,463 bp region (UCSC, hg19) on chromosome 3 bounded by D3S1582 (54,696,005) and D3S3606 (127,200,468) there were 133 missense, nonsense, and splice site variants (Table 1). Of these variants, only five (SLC25A26, LMOD3, FILIP1L, WDR52, and ADCY5), all of which were heterozygous, were not annotated in either dbSNP131 or the 1000 Genome project dataset. In addition, there were nine indels in the linkage region, but all were present in dbSNP131. Details of the five novel missense variants, including conservation scores and predictions of effect on the protein, are shown in Table 2.
Table 2.
Novel missense variants in the FDFM linkage region detected in individual V-3
| Gene | Hg191 | Reference1 | Variant | Read Depth2 | Residue | PhastCons3 | GERP3 | PolyPhen-24 | SIFT4 |
|---|---|---|---|---|---|---|---|---|---|
| SLC25A2 | 66,287,056 | G | A | 61 | S41N | 0.717 | Not given | benign | benign |
| LMOD3 | 69,168,628 | C | T | 88 | G293D | 0.997 | 5.26 | damaging | benign |
| FILIP1L | 99,569,603 | T | C | 79 | Q306R | 0.999 | 4.22 | damaging | benign |
| WDR52 | 113,119,440 | C | T | 86 | V476M | 0.002 | −7.71 | benign | benign |
| ADCY5 | 123,038,601 | C | T | 118 | A726T | 1 | 4.68 | damaging | damaging |
Hg19 location (bp) and reference allele according to Human Genome build 37 (GRCh37/hg19); in all but SLC25A2, the reference allele is from the reverse strand.
Total number of reads for that base pair
Co-transmission Analysis
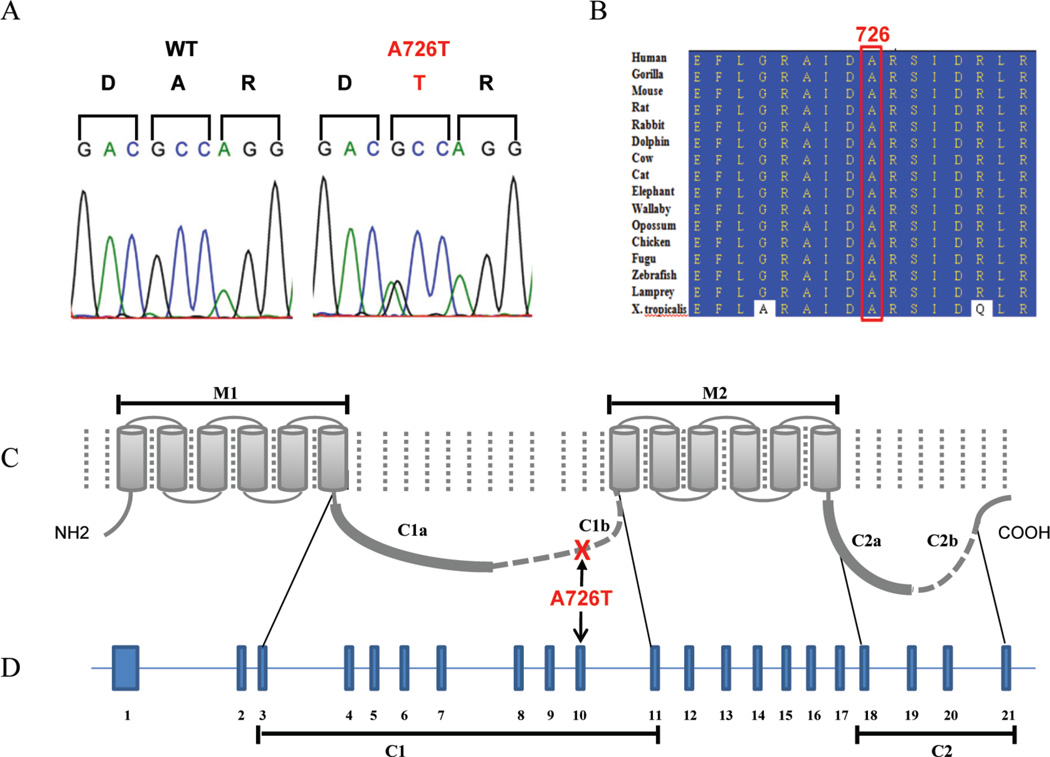
The five variants not present in either dbSNP or the 1000 Genomes project were sequenced in all available family members. Only the variants in ADCY5 and WDR52 associated completely with disease status (eTable 2). Because the variant in WDR52 was detected in 17/3493 Caucasian exomes in the UW GSGRC ESP and is neither conserved nor predicted to be damaging to the protein, it is deemed a rare polymorphism. The c.2176G>A transition (A726T) in adenylyl cyclase 5 (ADCY5) was predicted to alter protein function by both PolyPhen-2 and SIFT analysis and remains the sole candidate variant for FDFM (Figure 2A). Amino acid A726 is conserved in all species (Figure 2B), with GERP score 4.68 and PhastCons score 1.0.
Figure 2.
Missense mutation in ADCY5 identified in FDFM. A. Heterozygous c.2176G>A predicts p.A726T. B. Amino acid sequence alignment showing striking evolutionary conservation of AC5 at and around residue 726. C. Exonic organization of ADCY5 and relationship to primary structure of the protein. D. Secondary structure of AC5 showing domains.
Comment
The combination of linkage analysis and the exome sequence of a single affected individual revealed a strong candidate gene, ADCY5, for FDFM. Adenylyl cyclases (ACs) convert adenosine triphosphate (ATP) to cyclic adenosine-3´,5´-monophosphate (cAMP), the second messenger in a broad range of cellular activities. At least one AC isoform appears to participate in a large signaling complex with a β2 adrenergic receptor and a class C L-type Ca2+ channel14. There are six sequence dissimilar classes of ACs, but most bacterial and all eukaryotic ACs belong to Class III15. Nine of the 10 AC Class III isoforms are membrane bound and the 10th is soluble, structurally quite distinct, and testis specific15,16. The membrane-bound ACs are further divided into four subgroups on the basis of regulatory properties. Group III consists of AC5 and AC6, highly homologous forms that are activated by stimulatory guanosine triphosphate–binding proteins (G proteins; Gαss) and inhibited by inhibitory G proteins (Gαis) and calcium15,17–19.
The 21 exons of ADCY5 code for a 1261 amino acid protein that contains an intracellular N-terminus, two 6-unit membrane-spanning helices (M1 and M2) that flank the first of two intracellular cyclase homology domains (C1 and C2), and a short intracellular loop between the catalytic domains (Figure 2). AC activity is modulated through dopamine signaling20. Dopamine receptors are metabotropic G protein-coupled receptors. D1-dopamine receptors are stimulatory and D2-dopamine receptors are inhibitory. For activation, the catalytic domains of ACs must form a heterodimer that creates an ATP binding pocket21. Gαss activate the enzyme by promoting the interaction between C1 and C219. Gαis bind to C1 to impair this interaction. The AC5 variant we identified lies between C1 and M2. Although the Grantham score of 58 for this change places it in the moderately conservative class22,23, the substitution of polar hydrophilic threonine for nonpolar hydrophobic alanine might alter the structure of the binding pocket. It is possible that the change in conformation caused by the larger threonine residue for smaller alanine has an effect on the flexibility of the cytoplasic portion and thus alters enzyme activation or inactivation. Perhaps the mutation alters the enzyme’s affinity for inhibitory and/or stimulatory G proteins. Another possibility is that the mutant threonine residue could be an anomalous target for phosphorylation.
Expression data and animal models provide cogent support for the pathogenicity of the ADCY5 mutation for FDFM. AC5 is the major isoform in brain and heart and is especially predominant in the nucleus accumbens (NAc) and striatum, where it contributes more than 80% of AC activity24–26. The striatum and NAc are part of the mesolimbic dopaminergic system that is activated in response to stress27 Dopamine receptors are abundant in the striatum and NAc and these brain regions are major coordinators of movement. Mice deficient in AC5 develop a parkinsonian movement disorder that is worsened by stress26. Homozygous Adcy5 knockout mice manifest abnormal coordination by rotarod test, impaired locomotion in an open field test, decreased rearing actions in the home cage and bradykinesia on a pole test. Heterozygous Adcy5 +/− mice performed slightly worse than wildtype mice on these tests, but the impairment reached significance at the 5% level only for rearing frequency. The observation that D1- or D2-dopaminergic stimulation improved some but not all the motor deficits in the Adcy5 knockout mice suggests that other ACs present in striatum, including the highly homologous AC6, cannot fully replace the functions of AC526.
It is unclear what causes myokymia in human diseases. Abnormal firing of motor neurons or malfunction of peripheral nerves are possible explanations. The presence and activity of ADCY5 in motor neurons or the peripheral nervous system remains unknown.
Behavioral stress increases cAMP in the brain28 Mice deficient in AC5 displayed poor coping responses to restraint-induced stress, as evidenced by poor feeding and grooming, concomitant with over activation of the hippocampal-pituitary-adrenal (HPA) axis, as indicated by higher corticosterone levels29. This impaired stress tolerance was not affected by glucocorticoid receptor antagonists, but was abrogated by diazepam, a D1 dopamine receptor antagonist. The motor phenotype of the knockout mice is in some ways opposite that seen in FDFM where there is increased adventitious movement. Perhaps this reflects a gain of function effect of the mutation with respect to motor function. Similarly, exaggeration of the defect through stimulation of the cyclase pathway may account for the precipitating effect of stress in FDFM; alternatively, it may reflect a general phenomenon not specifically related to the ADCY5 mutation.
In the family we present, the multigenerational history of CHF, in some cases at relatively young ages and in one without clear etiology despite extensive workup, leads us to speculate that heart disease may be a component of FDFM. As with the neurologic phenotype, mouse models of cardiac disease suggest that the ADCY5 mutation in FDFM may have a gain of function effect. In the mouse models, disruption of Adcy5 is associated with longevity and protection against age-, stress- and catecholamine-related cardiomyopathy30–32, whereas overexpression results in cardiomyopathy33,34.
In summary, we present strong evidence that FDFM results from a missense mutation in ADCY5. By targeted linkage studies we had previously excluded 10 candidate genes/loci scattered in the genome and in our current study detected no pathogenic CNV. In the exome sequence, the FDFM linkage region contained only two novel coding variants that cosegregated with disease, and one of these was shown to be a rare polymorphism. The variant in ADCY5 affects a highly conserved residue, and the alteration is predicted to be damaging to protein function. The phenotypes of adcy5-null and overexpressing mice make this enzyme a credible candidate for FDFM. This study demonstrates the power of a single exome sequence in combination with linkage information to identify causative genes for rare autosomal dominant Mendelian diseases. Detection of mutations in additional families and functional studies of the variant may further support this conclusion.
Supplementary Material
Acknowledgments
Funding/Support: This research was supported by resources from the VISN-20 MIRECC VA Puget Sound Health Care System for T.D.B. and W.H.R., the American Recovery and Reinvestment Act (ARRA) funds through RC2 HG005608 from the National Human Genome Research Institute to D.A.N. and W.H.R., and R01NS069719 from the National Institute of Neurologic Diseases and Stroke to W.H.R. E.E.E. is an investigator of the Howard Hughes Medical Institute.
Role of the Sponsors: The sponsors had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Additional Contributions: We are grateful to the family for their participation in this study. John Wolff provided expert technical support. Drs. Guy Chan and Ning Zheng provided helpful information and advice about adenylyl cyclase biochemistry. We would like to thank and recognize the following ongoing studies that produced and provided exome variant calls for comparison: NHLBI Lung Cohort Sequencing Project (HL 1029230), NHLBI WHI Sequencing Project (HL 102924), and the Northwest Genomics Center (HL 102926). The University of Washington is a Huntington Disease (HDSA) Center of Excellence.
Footnotes
Authors’ Contributions: Study concept and design: Chen, Bird, Raskind. Acquisition of data: Chen, Matsushita, Robertson, Rieder, Girirajan, Antonacci, Lipe, Eichler, Nickerson, Bird, Raskind. Analysis and interpretation of data: Chen, Matsushita, Girirajan, Antonacci, Raskind. Drafting of the manuscript: Raskind. Critical revision of the manuscript for important intellectual content: Chen, Girirajan, Bird. Obtained funding: Eichler, Nickerson, Bird, Raskind. Administrative, technical and material support: Chen, Matsushita, Rieder, Robertson, Girirajan, Antonacci, Lipe. Study supervision: Bird and Raskind.
Financial Disclosure: T.D.B. and W.H.R. receive licensing fees from Athena Diagnostics.
On-Line Only Material: The eTables are available at http://www.archneurol.com.
Web Resources
- 1000 Genomes Project, http://www.1000genomes.org/page.php
- Database of Genomic Variants, http://projects.tcag.ca/variation/
- dbSNP homepage, http://www.ncbi.nlm.nih.gov/SNP/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/
- SeattleSeq Annotation, http://gvs.gs.washington.edu/SeattleSeqAnnotation/
- University of California Santa Cruz Human Genome Browser, http://genome.cse.ucsc.edu/cgi-bin/hgGateway
- PolyPhen-2 prediction of functional effects of human nsSNPs http://genetics.bwh.harvard.edu/pph2/
- NHLBI Exome Sequencing Project Exome Variant Server, http://evs.gs.washington.edu/EVS/
References
- 1.Fernandez M, Raskind W, Wolff J, et al. Familial dyskinesia and facial myokymia (FDFM): a novel movement disorder. Ann Neurol. 2001;49(4):486–492. [PubMed] [Google Scholar]
- 2.Bird TD, Hall JG. Additional information on familial essential (benign) chorea. Clinical genetics. 1978;14(5):271–272. doi: 10.1111/j.1399-0004.1978.tb02146.x. [DOI] [PubMed] [Google Scholar]
- 3.Raskind WH, Matsushita M, Peter B, et al. Familial dyskinesia and facial myokymia (FDFM): Follow-up of a large family and linkage to chromosome 3p21–3q21. Am J Med Genet B Neuropsychiatr Genet. 2009;150B(4):570–574. doi: 10.1002/ajmg.b.30879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ng SB, Turner EH, Robertson PD, et al. Hybrid Capture and Massively Parallel Sequencing of Twelve Human Exomes. Nature. 2009;461(7261):272–276. doi: 10.1038/nature08250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Siepel A, Bejerano G, Pedersen JS, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15(8):1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cooper GM, Stone EA, Asimenos G, Green ED, Batzoglou S, Sidow A. Distribution and intensity of constraint in mammalian genomic sequence. Genome Res. 2005;15(7):901–913. doi: 10.1101/gr.3577405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31(13):3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sunyaev S, Ramensky V, Koch I, Lathe W, 3rd, Kondrashov AS, Bork P. Prediction of deleterious human alleles. Hum Mol Genet. 2001;10(6):591–597. doi: 10.1093/hmg/10.6.591. [DOI] [PubMed] [Google Scholar]
- 9.Ramensky V, Bork P, Sunyaev S. Human non-synonymous SNPs: server and survey. Nucleic Acids Res. 2002;30(17):3894–3900. doi: 10.1093/nar/gkf493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Selzer RR, Richmond TA, Pofahl NJ, et al. Analysis of chromosome breakpoints in neuroblastoma at sub-kilobase resolution using fine-tiling oligonucleotide array CGH. Genes Chromosomes Cancer. 2005;44(3):305–319. doi: 10.1002/gcc.20243. [DOI] [PubMed] [Google Scholar]
- 11.Mefford HC, Muhle H, Ostertag P, et al. Genome-wide copy number variation in epilepsy: novel susceptibility loci in idiopathic generalized and focal epilepsies. PLoS Genet. 2010;6(5) doi: 10.1371/journal.pgen.1000962. e1000962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cooper GM, Zerr T, Kidd JM, Eichler EE, Nickerson DA. Systematic assessment of copy number variant detection via genome-wide SNP genotyping. Nat Genet. 2008;40(10):1199–1203. doi: 10.1038/ng.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Itsara A, Cooper GM, Baker C, et al. Population analysis of large copy number variants and hotspots of human genetic disease. Am J Hum Genet. 2009;84(2):148–161. doi: 10.1016/j.ajhg.2008.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davare MA, Avdonin V, Hall DD, et al. A beta2 adrenergic receptor signaling complex assembled with the Ca2+ channel Cav1.2. Science. 2001;293(5527):98–101. doi: 10.1126/science.293.5527.98. [DOI] [PubMed] [Google Scholar]
- 15.Linder JU. Class III adenylyl cyclases: molecular mechanisms of catalysis and regulation. Cell Mol Life Sci. 2006;63(15):1736–1751. doi: 10.1007/s00018-006-6072-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Buck J, Sinclair ML, Schapal L, Cann MJ, Levin LR. Cytosolic adenylyl cyclase defines a unique signaling molecule in mammals. Proc Natl Acad Sci U S A. 1999;96(1):79–84. doi: 10.1073/pnas.96.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Onda T, Hashimoto Y, Nagai M, et al. Type-specific regulation of adenylyl cyclase. Selective pharmacological stimulation and inhibition of adenylyl cyclase isoforms. J Biol Chem. 2001;276(51):47785–47793. doi: 10.1074/jbc.M107233200. [DOI] [PubMed] [Google Scholar]
- 18.Cooper DM. Molecular and cellular requirements for the regulation of adenylate cyclases by calcium. Biochem Soc Trans. 2003;31(Pt 5):912–915. doi: 10.1042/bst0310912. [DOI] [PubMed] [Google Scholar]
- 19.Beazely MA, Watts VJ. Regulatory properties of adenylate cyclases type 5 and 6: A progress report. Eur J Pharmacol. 2006;535(1–3):1–12. doi: 10.1016/j.ejphar.2006.01.054. [DOI] [PubMed] [Google Scholar]
- 20.Girault JA, Greengard P. The neurobiology of dopamine signaling. Arch Neurol. 2004;61(5):641–644. doi: 10.1001/archneur.61.5.641. [DOI] [PubMed] [Google Scholar]
- 21.Tesmer JJ, Sunahara RK, Gilman AG, Sprang SR. Crystal structure of the catalytic domains of adenylyl cyclase in a complex with Gsalpha.GTPgammaS. Science. 1997;278(5345):1907–1916. doi: 10.1126/science.278.5345.1907. [DOI] [PubMed] [Google Scholar]
- 22.Grantham R. Amino acid difference formula to help explain protein evolution. Science. 1974;185(4154):862–864. doi: 10.1126/science.185.4154.862. [DOI] [PubMed] [Google Scholar]
- 23.Li WH, Wu CI, Luo CC. Nonrandomness of point mutation as reflected in nucleotide substitutions in pseudogenes and its evolutionary implications. J Mol Evol. 1984;21(1):58–71. doi: 10.1007/BF02100628. [DOI] [PubMed] [Google Scholar]
- 24.Matsuoka I, Suzuki Y, Defer N, Nakanishi H, Hanoune J. Differential expression of type I, II, and V adenylyl cyclase gene in the postnatal developing rat brain. J Neurochem. 1997 Feb;68(2):498–506. doi: 10.1046/j.1471-4159.1997.68020498.x. [DOI] [PubMed] [Google Scholar]
- 25.Lee KW, Hong JH, Choi IY, et al. Impaired D2 dopamine receptor function in mice lacking type 5 adenylyl cyclase. J Neurosci. 2002;22(18):7931–7940. doi: 10.1523/JNEUROSCI.22-18-07931.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iwamoto T, Okumura S, Iwatsubo K, et al. Motor dysfunction in type 5 adenylyl cyclase-null mice. J Biol Chem. 2003;278(19):16936–16940. doi: 10.1074/jbc.C300075200. [DOI] [PubMed] [Google Scholar]
- 27.Finlay JM, Zigmond MJ. The effects of stress on central dopaminergic neurons: possible clinical implications. Neurochem Res. 1997;22(11):1387–1394. doi: 10.1023/a:1022075324164. [DOI] [PubMed] [Google Scholar]
- 28.Stone EA, John SM. Stress-induced increase of extracellular levels of cyclic AMP in rat cortex. Brain Res. 1992;597(1):144–147. doi: 10.1016/0006-8993(92)91516-h. [DOI] [PubMed] [Google Scholar]
- 29.Kim KS, Han PL. Mice lacking adenylyl cyclase-5 cope badly with repeated restraint stress. J Neurosci Res. 2009;87(13):2983–2993. doi: 10.1002/jnr.22119. [DOI] [PubMed] [Google Scholar]
- 30.Yan L, Vatner DE, O'Connor JP, et al. Type 5 adenylyl cyclase disruption increases longevity and protects against stress. Cell. 2007;130(2):247–258. doi: 10.1016/j.cell.2007.05.038. [DOI] [PubMed] [Google Scholar]
- 31.Okumura S, Takagi G, Kawabe J, et al. Disruption of type 5 adenylyl cyclase gene preserves cardiac function against pressure overload. Proc Natl Acad Sci U S A. 2003;100(17):9986–9990. doi: 10.1073/pnas.1733772100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Okumura S, Vatner DE, Kurotani R, et al. Disruption of type 5 adenylyl cyclase enhances desensitization of cyclic adenosine monophosphate signal and increases Akt signal with chronic catecholamine stress. Circulation. 2007;116(16):1776–1783. doi: 10.1161/CIRCULATIONAHA.107.698662. [DOI] [PubMed] [Google Scholar]
- 33.Hanoune J, Pouille Y, Tzavara E, et al. Adenylyl cyclases: structure, regulation and function in an enzyme superfamily. Molecular and cellular endocrinology. 1997;128(1–2):179–194. doi: 10.1016/s0303-7207(97)04013-6. [DOI] [PubMed] [Google Scholar]
- 34.Ho D, Yan L, Iwatsubo K, Vatner DE, Vatner SF. Modulation of beta-adrenergic receptor signaling in heart failure and longevity: targeting adenylyl cyclase type 5. Heart failure reviews. 2010;15(5):495–512. doi: 10.1007/s10741-010-9183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.