Abstract
Background and Objectives
A previously published study of antiretroviral pharmacokinetics in the female genital tract of HIV-infected women demonstrated differing degrees of female genital tract penetration among antiretrovirals. These blood plasma (BP) and cervicovaginal fluid (CVF) data were co-modelled for four antiretrovirals with varying CVF exposures.
Methods
Six paired BP and CVF samples were collected over 24 h, and antiretroviral concentrations determined using validated liquid chromatography (LC) with UV detection or LC-mass spectrometry analytical methods. For each antiretroviral, a BP model was fit using Bayesian estimation (ADAPT5), followed by addition of a CVF model. The final model was chosen based on graphical and statistical output, and then non-linear mixed-effects modelling using S-ADAPT was performed. Population mean parameters and their variability are reported. Model-predicated area under the concentration-time curve during the dosing interval (AUCτ) and exposure ratios of CVF AUCτ:BP AUCτ were calculated for each drug.
Results
The base model uses first-order absorption with a lag time, a two-compartment model, and a series of transit compartments that transfer the drug from BP to CVF. Protein-unbound drug transfers into CVF for efavirenz and atazanavir; total drug transfers for lamivudine and tenofovir. CVF follows a one-compartment model for efavirenz and atazanavir, and a two-compartment model for lamivudine and tenofovir. As expected, inter-individual variability was high. Model-predicted CVF AUCτ:BP AUCτ ratios are consistent with published results.
Conclusions
This is the first pharmacokinetic modelling of antiretroviral disposition in BP and CVF. These models will be further refined with tissue data, and used in clinical trials simulations to inform future studies of HIV pre-exposure prophylaxis in women.
Keywords: HIV-infections, pharmacokinetics, pharmacokinetic-modelling, population-pharmacokinetics, antiretrovirals, atazanavir, efavirenz, lamivudine, tenofovir
1. Introduction
With the availability of potent antiretroviral drugs, and no current HIV vaccine, the use of antiretrovirals as a prevention tool is an attractive option to reduce the global HIV epidemic. Recent data demonstrate that treating the index case with antiretrovirals reduces transmission by 96 % in heterosexual serodiscordant couples [1]. By inference, this is likely due to reducing HIV RNA in the genital tract by reducing HIV RNA in the blood to <400 copies/mL [2–4]. However, factors such as sexually transmitted infections and increased inflammation from non-infectious causes can result in HIV RNA discordance between blood and genital tract secretions [5–8]. It has also been suggested that the genital tract may be a sequestered site for HIV replication [9, 10]. Therefore, inadequate penetration of antiretrovirals into the genital tract has the potential for incomplete suppression of HIV RNA replication, the development of drug resistance, and an increase in the risk of sexual HIV transmission. In both the male and female genital tract, antiretroviral concentrations vary from a fraction of blood exposure to exposures several-fold higher than blood [11, 12]. The mechanism for this behaviour is unclear [12]. Pharmacokinetic modelling is a potential tool for understanding, and hence optimizing, drug behaviour in the genital tract.
This analysis extends previously published non-compartmental analysis and comparison of exposure in cervicovaginal fluid (CVF) relative to blood plasma (BP) for several antiretrovirals [13]. Agents were selected based on the number of women receiving the drug, and to represent varying penetration in CVF. Efavirenz represents a highly protein-bound (99.9 %) non-nucleoside reverse transcriptase inhibitor [14] with very low penetration into CVF. Among the protease inhibitors studied, atazanavir represents a moderately penetrative drug with high protein binding (86 %) [15]. Tenofovir and lamivudine, both nucleoside/nucleotide reverse transcriptase inhibitors, have low protein binding (<7 [16] and 40 % [17], respectively). Tenofovir shows similar exposures to BP in CVF, while lamivudine demonstrates higher exposures [13].
The aim of the current project is to develop a framework for future analysis of oral antiretroviral regimens to prevent heterosexual HIV transmission. Combining these models with concentration targets of efficacy will allow optimization of doses and dosing frequencies for antiretroviral prevention interventions.
2. Methods
2.1 Study Design and Conduct
A detailed description of study methods has been previously published [13]. Briefly, 27 HIV-infected pre-menopausal women without evidence of a sexually transmitted infection and initiating a new provider-selected antiretroviral regimen at the University of North Carolina (UNC) at Chapel Hill (Chapel Hill, NC, USA) were enrolled. Pharmacokinetic sampling was conducted around the first dose (FD) of the new regimen and approximately 30 days later, from November 2002 to May 2006. The multiple-dose (MD) visit was added after several subjects completed the protocol, and therefore not all subjects contribute data from both visits. Paired BP and directly aspirated CVF samples were taken pre-dose (time = 0), and 2, 4, 6, 12 and 24 h after observed dosing of the antiretroviral regimen. The study protocol was approved by the UNC Biomedical Institutional Review Board, and all subjects provided informed consent prior to study interventions. Drug concentrations were determined using high-performance liquid chromatography (HPLC) with UV detection or HPLC-tandem mass spectrometry methods available in the Center for AIDS Research Clinical Pharmacology and Analytical Chemistry Laboratory [18–22]. For the drugs of interest, the lower limit of quantitation (LLQ) in BP was 10 μg/L for efavirenz, lamivudine and tenofovir and 25 μg/L for atazanavir. In CVF, the LLQ was 5 μg/L for efavirenz, lamivudine and atazanavir, and 10 μg/L for tenofovir. Inter-day and intra-day percentage coefficient of variation (CV%) values of all assays were <15 %.
2.2 Pharmacokinetic Analysis
2.2.1 Structural Model Development
To develop the structural model for each drug, individual subject modelling was performed using the maximum a posteriori probability (MAP) Bayesian estimator [23] in ADAPT5 software (Biomedical Simulations Resource, Los Angeles, CA, USA) [24]. Prior means and variability around those means for BP were initially obtained from the literature for each drug [25–28]. Model development began with the first-dose BP data; once a satisfactory model [as assessed by Akaike’s Information Criterion (AIC) [29] and visual inspection of observed vs. fitted values] was found, Bayesian priors were refined and the CVF compartment was added to the model using empirical Bayesian priors. After each round of modelling, Bayesian priors were updated to reflect mean parameter and variance estimates from the data (iterative two-stage analysis). After first-dose BP and CVF were co-modelled, multiple dose data were added (BP and CVF added simultaneously). Since full 30-day dosing histories were not available, datasets were constructed using the limited available adherence and timing data with a variable number of doses between visits for each subject. For all drugs, multiple-dose data required a second set of model parameters to adequately describe the data (allowing for between-occasion variability).
Additive plus proportional residual error variance models, with the LLQ of the assay as the standard deviation intercept and twice the CV% of the assay as the initial estimate for the standard deviation slope, were used throughout; intercept values were fixed, and slope values were estimated. Through the in-built functionality in ADAPT5, the Beal M3 method for inclusion of data below the limit of quantitation (BLQ) was invoked [30]. A log-normal distribution was assumed for all estimated parameters.
2.2.2 Population Modelling Methods
After structural model development in ADAPT5, non-linear mixed effects modelling using the Monte-Carlo parametric expectation maximization (MCPEM, pmethod 4) algorithm of S-ADAPT with the S-ADAPT TRAN pre- and post-processing package [31, 32] was used to obtain population parameter estimates, inter-individual variability and estimates of parameter precision. Simplified datasets for each drug were constructed as follows to increase the speed of each modelling run. While still maintaining the timing of the samples around the dose of the drug, several days of dosing were administered after the first dose, and were uniform across subjects. Dosing allowed the drug to achieve steady-state conditions, based on five BP half-lives; this was 96 h of dosing for lamivudine and tenofovir, and 216 h of dosing for efavirenz and atazanavir. Following ADAPT5 results, parameters other than distributional clearances/rate constants were allowed to differ between visits, and were reduced to a single parameter when possible, based on similarities in mean values, post hoc individual estimates and inter-individual variability. Additionally, the between-occasion variability function in S-ADAPT was used, if appropriate. To account for two values for each parameter depending on the study period, the individual subject datasets were coded into groups, with group 1 values applied to the first dose, and group 2 values applied to all subsequent doses and the second pharmacokinetic visit. For initial model runs, 200 iterations were performed, with 400 iterations used for the final model.
In the residual error variance models, only the slope of the BP error variance model was estimated (SDSLBP); the intercepts were fixed at the LLQ and the CVF slope at twice the estimated CV% of the assay (0.3), as the model would not run with estimation of all error model parameters, and ADAPT5-estimated CVF error slopes were high for each drug (approximately 0.6). The Beal M3 method for BLQ data was invoked [30]. A log-normal distribution was assumed for all estimated parameters, and a full covariance matrix was used. Models were compared using the likelihood ratio test; decreases in objective function value were considered significant if they exceeded the critical value corresponding to a significance level of 0.05. Plots of the individual fitted concentrations compared with observed concentrations, weighted residuals, and individual subject observed versus predicted time-concentration profiles [33] were also examined to select the final model. Monte-Carlo simulations of 1,000 subjects using the final model parameter means and covariance matrix were performed in ADAPT5 for each drug to produce visual predictive checks (VPCs). VPC graphs were constructed using the 5th–95th percentiles and median values of the simulated data with SigmaPlot™ 11 (Systat Software Inc., Chicago, IL, USA).
2.2.3 Model-Predicted Cervicovaginal Fluid (CVF):Blood Plasma (BP) Area Under the Plasma Concentration-Time Curve (AUC) Ratios
To compare modelling results with the non-compartmental analysis-derived CVF exposure ratios for each drug, simulations in ADAPT5 were performed using subject post hoc estimates (generated using the mean of the Bayesian distribution) to generate an area under the concentration-time curve (AUC) during the dosing interval (AUCτ) in BP and CVF at both first and multiple doses by numerically integrating the relevant differential equations. As one model employed the protein-unbound fraction in blood (fu) to control drug transfer into the CVF, BP AUCτ values were multiplied by the estimated fu and compared to the CVF AUCτ estimate [14–17]. Correcting for protein binding in blood has been used in studies of drug penetration into cerebrospinal fluid (CSF) to assess measured CSF exposure compared to the theoretical amount of drug available (fu) to cross membranes [2, 9, 29]. To calculate median CVF:BP ratios, individual subject ratios were summarized.
3. Results
3.1 Subject Demographics
Subject demographics by drug studied are presented in Table 1. As typical antiretroviral regimens contain at least three drugs, each group of women receiving a given drug overlaps with women receiving other drugs. The population chosen for modelling represents 25 of the 27 women in the original non-compartmental analysis. Overall, the study population was predominantly African American (17/25 subjects), approaching middle-age (median age of 35 years) and antiretroviral-experienced (18/25 subjects). The median CD4+ cell count was 269 cells/mm3 at the first-dose visit. The median BP log HIV RNA concentration was 4.71 (51,209 copies/mL) at first dose, and ranged from the lower limit to the upper limit of the assay (<50 to >750,000 copies/mL). The median CVF log HIV RNA concentration was 4.2 (16,000 copies/mL), and ranged from the lower limit of the assay (<400 copies/mL) to 1,300,000 copies/mL. The median time between the first and second pharmacokinetic visits was 32.5 days (range: 20–154 days).
Table 1.
Subject demographics by drug
| Drug | EFV | ATV | 3TC | TFV |
|---|---|---|---|---|
| No. of subjects | 10 | 8 | 19 | 15 |
| Age, years | 33.5 (30–38) | 37 (33–42) | 36.0 (32.0–39.5) | 36.0 (32.0–43.0) |
| Weight, kg | 71.1 (67.7–74.1) | 69.0 (53.0–86.8) | 71.1 (63.5–94.7) | 68.9 (62.2–72.0) |
| Race, n | ||||
| African American/Black | 8 | 8 | 14 | 11 |
| Caucasian | 1 | 0 (0 %) | 4 | 3 |
| Other | 1 (Hispanic) | 0 (0 %) | 1 (Native American) | 1 (Hispanic) |
| ARV-naïve? [n (%)] | 5 (62.5 %) | 0 (0 %) | 6 (31.6 %) | 3 (20 %) |
| CD4+ cell count, first dose, cells/mm3 | 328 (212–446) | 191 (172–426) | 257 (183–422) | 244 (179–342) |
| Log BP HIV RNA, first dose | 4.30 (3.95–4.74) | 4.84 (4.63–5.04) | 4.71 (4.23–5.01) | 4.55 (3.78–5.04) |
| Log CVF HIV RNA, first dose | 2.60 (2.60–3.81) | 5.25 (3.90–5.99) | 4.42 (2.60–5.47) | 4.45 (2.60–5.47) |
| Background ARV regimen | 8/10 received 3TC; 1/10 received LPV/r; 1/10 received ATV/r | 5/10 received 3TC; 5/10 received TDF; 2/10 received 3TC/TDF; 1/10 received EFV | 8/19: PI-based regimen; 9/19: NNRTI-based regimen; 2/10: triple NRTI regimen | 9/15: PI-based regimen; 5/15: NNRTI-based regimen; 1/10: triple NRTI regimen |
| Dosage notes | All received 600 mg daily | 7/8 received 300 mg with ritonavir 100 mg; 1/8 received 400 mg daily | 10/19 received 300 mg daily; 9/19 received 150 mg twice daily | All received 300 mg of tenofovir disoproxil fumarate (136 mg of tenofovir) daily |
Values are expressed as median (IRQ) unless specified otherwise
3TC lamivudine, ARV antiretroviral, ATV atazanavir, ATV/r atazanavir/ritonavir, BP blood plasma, CVF cervicovaginal fluid, EFV efavirenz, IQR interquartile range, LPV/r lopinavir/ritonavir, NNRTI non-nucleoside reverse transcriptase inhibitor, NRTI nucleoside reverse transcriptase inhibitor, PI protease inhibitor, TDF tenofovir disoproxil fumarate, TFV tenofovir
3.2 Pharmacokinetic Modelling
3.2.1 Structural Models
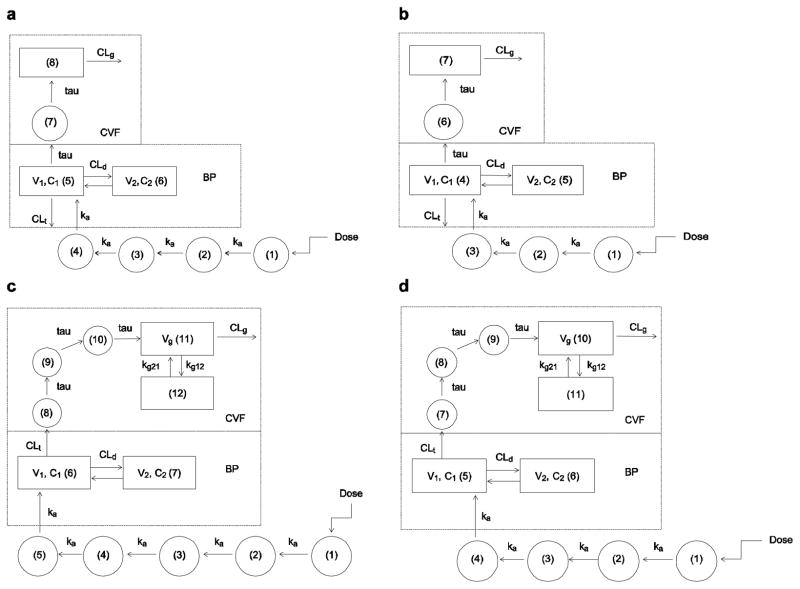
The structural models for each drug are depicted in Fig. 1. For efavirenz and atazanavir, a similar structural model was used, with atazanavir having one fewer absorptive compartments. A two-compartment model (central volume [V1], peripheral volume [V2]) with first-order elimination (total BP clearance [CLt]) and absorption via the first-order absorption rate constant ka and transit compartments between the site of drug administration and the central compartment was used to describe the BP data. Transfer of drug to the CVF occurred from the central compartment through a transit compartment via the rate constant tau; the amount of drug entering the CVF was the concentration in the BP central compartment multiplied by the estimated fu (0.01 for efavirenz, 0.14 for atazanavir). A one-compartment model with first-order elimination (CVF clearance [CLg]) was used to describe drug behaviour in the genital tract. The volume of the genital tract compartment was assumed to be 1 L to simplify the differential equations.
Figure 1.
Model schematics for efavirenz (a), atazanavir (b), lamivudine (c) and tenofovir (d). BP blood plasma, Cn compartment number n, CLd distributional clearance in blood plasma, CLg CVF clearance, CLt total BP clearance, CVF cervicovaginal fluid, ka first-order absorption rate constant, kgxy transfer rate constant from CVF compartment x to y, tau transit compartment transfer rate constant, V1 apparent volume of distribution of the central compartment, V2 apparent volume of distribution of the peripheral compartment, Vg apparent volume of distribution of the CVF compartment
The efavirenz/atazanavir model in BP was utilized for lamivudine and tenofovir. However, the efavirenz/atazanavir model in CVF did not provide satisfactory fits of the lamivudine/tenofovir data in CVF. Therefore, an alternate forcing function of drug transfer from BP to CVF was employed. The controlling factor of fu was removed, and clearance from the central compartment (CLt) moved drug into the CVF (Fig. 1c–d). In this model, volume of the CVF was not assumed to be 1 L, and was added as an estimated parameter (Vg) since a two-compartment model was used to describe the CVF disposition. Drug was cleared from Vg by CLg, and moved between the first and second CVF compartments via the first-order rate constants kg12 and kg21. For tenofovir, a dose of 136 mg was used, as this is the amount of tenofovir in the 300 mg dose of tenofovir disoproxil fumarate.
Model differential equations are available in the Online Resource.
3.2.2 Final Population Model Selection
Over multiple runs, we attempted to reduce the number of parameters from two parameter sets for the BP V1, BP V2, CLt, CLg, tau and ka. Table 2 describes the changes in minimum objective function value for these candidate models; for all drugs, a model with one set of parameters for both visits was the basis for comparison to models with separate parameters for each visit. Based on change in objective function value, a model using separate values for CLt, CLg, tau and ka was selected as the final model for efavirenz, lamivudine and tenofovir; a model using separate values for Clg and tau was selected for atazanavir. Final parameter estimates, inter-individual variability (CV%), and estimates of parameter precision (percentage standard error [SE%]) are presented in Table 3.
Table 2.
Collapsing of model parameters for each drug in S-ADAPT
| Model | Lamivudine | Atazanavir | Efavirenz | Tenofovir | ||||
|---|---|---|---|---|---|---|---|---|
| ΔOFV | Significant? | ΔOFV | Significant? | ΔOFV | Significant? | ΔOFV | Significant? | |
| Separate CLt, CLg, ka, tau | −368 | Yesa | −173 | Yes | −51.9 | Yesa | −258 | Yesa |
| Separate CLt, CLg, ka | −333 | Yes | −157 | Yes | −41.6 | Yes | NA | |
| Separate CLt, CLg, tau | −356 | Yes | NA | NA | −238 | Yes | ||
| Separate CLg, tau | −318 | Yes | −182 | Yesa | NA | NA | ||
| Separate ka, tau | −353 | Yes | −157 | Yes | −39.0 | Yes | NA | |
| Separate CLt, CLg | NA | NA | −22.5 | Yes | −211 | Yes | ||
| Separate CLg, tau; BOV on CLt, ka | −323 | Yes | −176 | Yes | NA | NA | ||
| Separate CLg, tau, CLt; BOV on ka | −358 | Yes | NA | NA | NA | |||
| Separate CLg | NA | −108 | Yes | NA | NA | |||
All comparisons are to a model with a single set of parameters for each drug
ΔOFV change in objective function value, BOV between-occasion variability, CLg cervicovaginal fluid clearance, CLt total blood plasma clearance, ka first-order absorption rate constant, NA not applicable, tau transit compartment transfer rate constant, – indicates not applicable
Final model selected
Table 3.
Final parameter estimates for each drug
| Parameter (units) | Efavirenz | Atazanavir | Lamivudine | Tenofovir |
|---|---|---|---|---|
| CLd (L/h) | 57.7 (71.2) [25.0] | 9.26 (111) [40.1] | 13.3 (75.1) [20.9] | 158 (61.3) [90.8] |
| CLg,FD (L/h) | 2.50 (73.2) [39.4] | 0.185 (198) [80.5] | 1.72 (113) [27.2] | 10.8 (17.2) [117] |
| CLg,MD (L/h) | 1.63 (171) [63.1] | 1.23 (208) [81.2] | 8.08 (112) [28.0] | 37.3 (96.7) [35.9] |
| CLt,FD (L/h) | 20.4 (50.4) [20.7] | 10.3 (114) [42.1] | 32.7 (43.6) [10.2] | 42.7 (147) [80.0] |
| CLt,MD (L/h) | 9.39 (80.7) [30.6] | NA | 28.5 (59.7) [14.8] | 71.3 (73.4) [57.9] |
| kg12 (h−1) | NA | NA | 1.69 (226) [54.5] | 0.525 (203) [63.6] |
| kg21 (h−1) | NA | NA | 0.209 (64.7) [22.2] | 0.323 (97.6) [60.7] |
| ka,FD (h−1) | 1.75 (52.7) [18.9] | 0.879 (79.5) [29.0] | 3.34 (42.6) [10.6] | 1.72 (61.1) [99.8] |
| ka,MD (h−1) | 0.945 (139) [54.0] | NA | 3.52 (85.1) [23.9] | 1.05 (69.8) [30.0] |
| tauFD (h−1) | 0.866 (208) [77.5] | 0.773 (145) [52.8] | 1.29 (74.0) [22.9] | 1.53 (106) [71.1] |
| tauMD (h−1) | 0.383 (156) [55.8] | 1.41 (216) [85.2] | 0.672 (179) [48.2] | 1.24 (103) [130] |
| V1 (L) | 45.5 (43.3) [16.1] | 92 (109) [39.4] | 85.7 (49.2) [11.7] | 187 (47.1) [44.7] |
| V2 (L) | 268 (30.8) [12.2] | 111 (105) [39.3] | 149 (77.2) [21.5] | 458 (85.3) [53.1] |
| Vg (L) | 2.55 | 10.4 | 2.55 (109) [30.4] | 4.75 (92.2) [47.2] |
| SDSLBP | 0.322 (NA) [14.0] | 0.418 (NA) [10.9] | 0.327 (NA) [7.78] | 0.343 (NA) [19.0] |
| SDSLCVF | 0.3 (NA) [NA] | 0.3 (NA) [NA] | 0.3 (NA) [NA] | 0.3 (NA) [NA] |
| SDINTBP | 10 (NA) [NA] | 25 (NA) [NA] | 5 (NA) [NA] | 10 (NA) [NA] |
| SDINTCVF | 0.1 (NA) [NA] | 5 (NA) [NA] | 5 (NA) [NA] | 5 (NA) [NA] |
All values are presented as mean (%CV) [SE%]
BP blood plasma, CLd distributional clearance in blood plasma, CV% coefficient of variation (inter-individual variability), CVF cervicovaginal fluid, FD first dose, ka first-order absorption rate constant, kgxy transfer rate constant from CVF compartment x to y, MD multiple dose, mean geometric mean for the log-normal probability density function, NA not applicable, SDINT standard deviation intercept term of the residual error model, SDSL standard deviation slope term of the residual error model, SE% estimates of parameter precision, presented as percentage standard error, tau transit compartment transfer rate constant, V1 volume of distribution of the central compartment, V2 volume of distribution of the peripheral compartment, Vg volume of distribution of the CVF compartment, – indicates not applicable
3.2.3 Individual Drug Model Fittings
3.2.3.1 Efavirenz
For efavirenz, six of ten women provided data at both visits, and all subjects received the standard dose of 600 mg by mouth daily in combination with other agents. In the efavirenz data set, 37 samples (19 % of data points) were BLQ. In the calculation of concentrations in CVF, the differing amounts of sample collected were accounted for with a multiplication factor, and because of this and the very low concentrations of efavirenz in CVF, an effective BLQ of 0.1 μg/L in CVF was used in the error model rather than the LLQ of the assay.
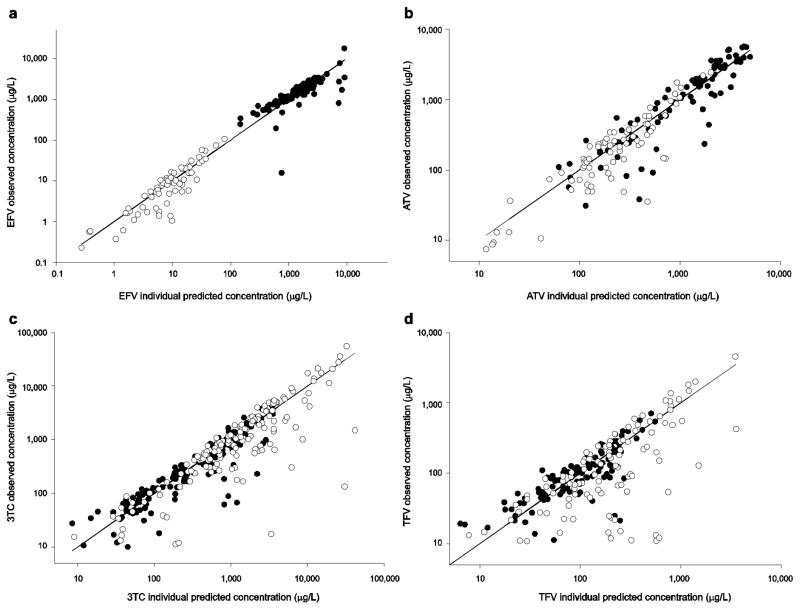
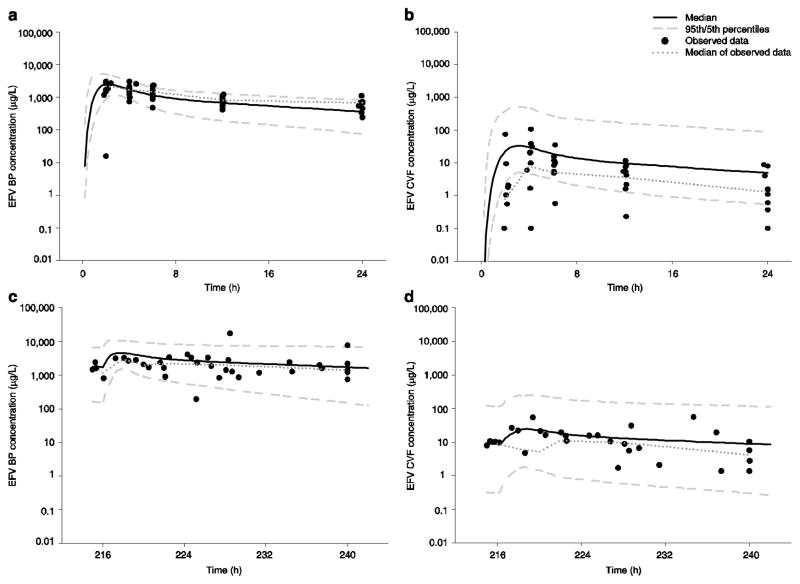
In Table 3, inter-individual variability was highest (>100 CV%) for parameters related to absorption (ka,MD) and drug transfer from BP to CVF (tauFD, tauMD), and these are also the least precise of the estimates. The individual predicted concentrations versus observed concentration plots for BP and CVF on a log scale are shown in Fig. 2a. For BP, the model over-predicted several low observed concentrations and under-predicted one high observed concentration. VPCs for BP and CVF are shown in Fig. 3 and are consistent with the individual fitted versus observed concentration plots. The confidence envelopes included >90 % of the observations, suggesting imprecision in the fitted parameters; the full covariance matrix was not able to be used in generating these plots.
Figure 2.
Individual predicted concentrations vs. observed data graphs for efavirenz (a), atazanavir (b), lamivudine (c) and tenofovir (d). The solid black line is the line of identity; blood plasma concentrations are depicted with filled circles, while the cervicovaginal fluid concentrations are depicted with open circles. 3TC lamivudine, ATV atazanavir, EFV efavirenz, TFV tenofovir
Figure 3.
Visual predictive checks for efavirenz at each sampling period in both blood plasma and cervicovaginal fluid using the 5th and 95th percentiles and median of 1,000 simulated subjects and observed data at both first dose and multiple dose: a blood plasma at first dose; b cervicovaginal fluid at first dose; c blood plasma at multiple dose; d cervicovaginal fluid at multiple dose. BP blood plasma, CVF cervicovaginal fluid, EFV efavirenz
3.2.3.2 Atazanavir
For this drug, all eight women provided data at both visits. Seven subjects received atazanavir 300 mg in combination with ritonavir 100 mg as a pharmacokinetic boosting agent; one subject received atazanavir 400 mg daily without ritonavir (unboosted). In the atazanavir dataset, 29 samples (15 % of data points) were BLQ.
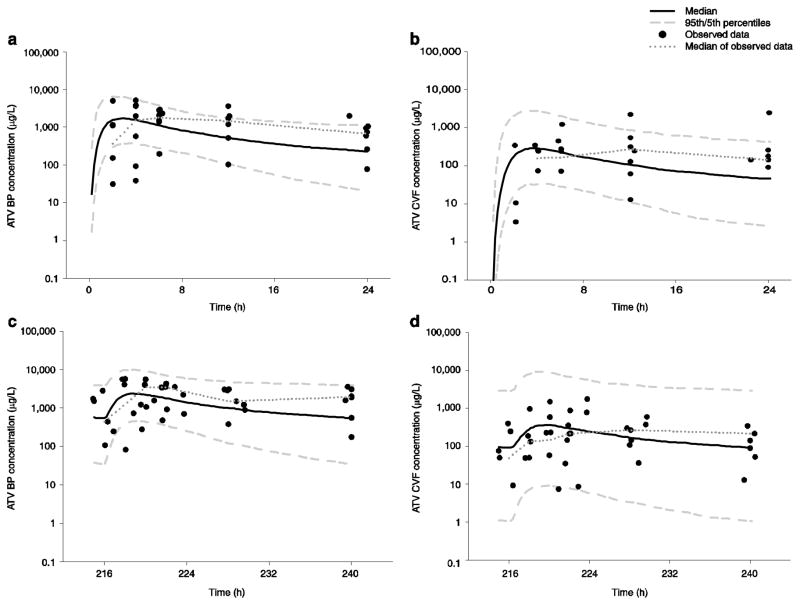
In Table 3, multiple parameters have variability estimates >100 %, suggesting some over-parameterization of the model. The individual fitted versus observed concentration plots for BP and CVF are shown in Fig. 2b, showing some over-prediction of the data, particularly in CVF. VPCs are shown in Fig. 4. Again, there was >90 % coverage within the 90 % confidence envelope; the full covariance matrix was not able to be used in generating these plots. No significant differences were noted when the subject receiving unboosted atazanavir was excluded.
Figure 4.
Visual predictive checks for atazanavir at each sampling period in both blood plasma and cervicovaginal fluid using the 5th and 95th percentiles and median of 1,000 simulated subjects and observed data at both first dose and multiple dose: a blood plasma at first dose; b cervicovaginal fluid at first dose; c blood plasma at multiple dose; d cervicovaginal fluid at multiple dose. ATV atazanavir, BP blood plasma, CVF cervicovaginal fluid
3.2.3.3 Lamivudine
Ten subjects received lamivudine 150 mg twice daily; five of those subjects provided data at both visits. The remaining nine subjects received lamivudine 300 mg once daily, and eight of those subjects provided data at both visits. CVF exposure ratios did not differ by regimen in the original analysis [13], and therefore the dosing regimens were combined for modelling. The complete lamivudine dataset contained 43 samples (12 % of data points) that were BLQ, 21 (48 %) of those from subjects receiving twice daily doses.
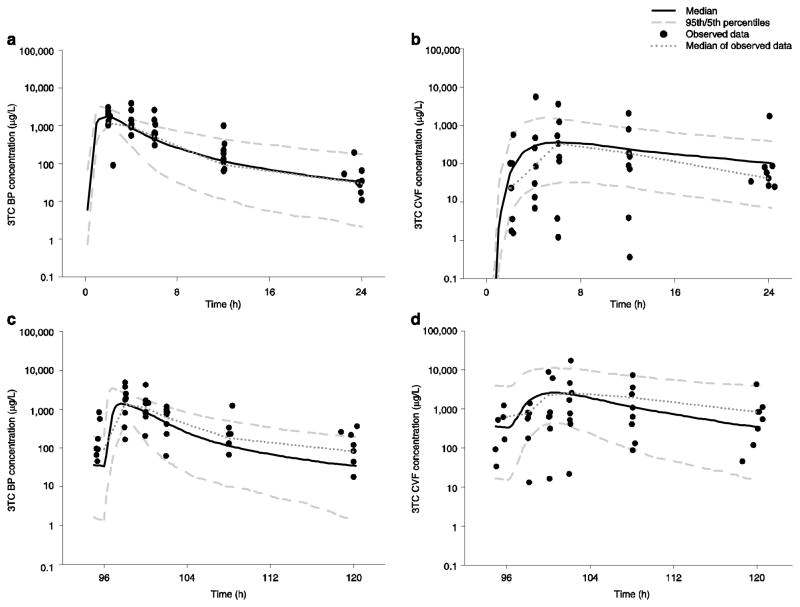
In Table 3, the parameters with the highest inter-individual variability were those of the CVF disposition, likely related to the highly variable nature of CVF concentrations and some model over-parameterization. The individual fitted versus observed concentration plots for BP and CVF are shown in Fig. 2c; the model predicts blood concentrations well, with over-prediction of several CVF observations. VPCs for subjects who received the drug once daily are shown in Fig. 5. Here, <90 % of the observed data were within the confidence envelope; parameters with >80 % correlation were included in the covariance matrix. Results were similar for those receiving the drug twice daily (data not shown).
Figure 5.
Visual predictive checks for lamivudine administered at 300 mg daily at each sampling period in both blood plasma and cervicovaginal fluid using the 5th and 95th percentiles and median of 1,000 simulated subjects and observed data at both first dose and multiple dose: a blood plasma at first dose; b cervicovaginal fluid at first dose; c blood plasma at multiple dose; d cervicovaginal fluid at multiple dose. 3TC lamivudine, BP blood plasma, CVF cervicovaginal fluid
3.2.3.4 Tenofovir
For this drug, ten of 15 women provided data at both visits, and all subjects received the standard dose of 300 mg of tenofovir disoproxil fumarate. The complete tenofovir dataset contained 81 samples (27 % of the data points) that were considered BLQ.
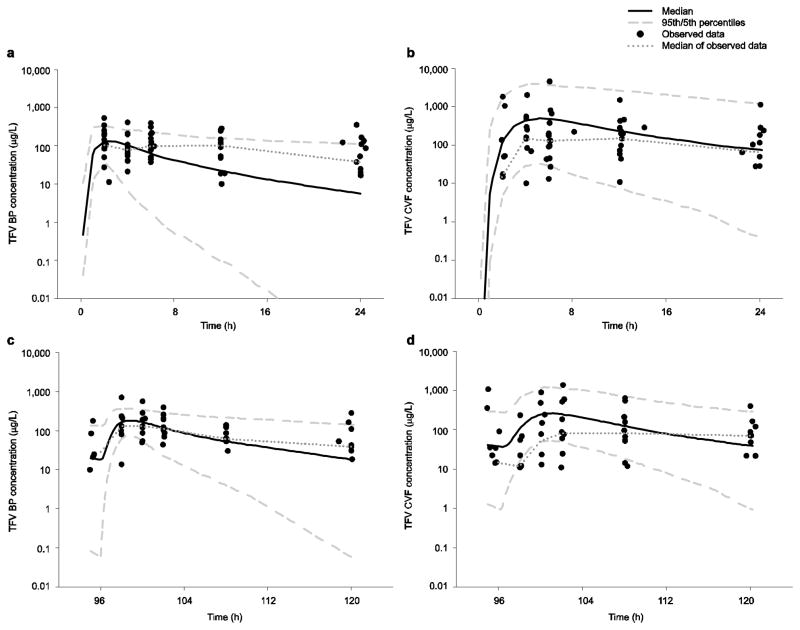
In Table 3, CLg and CLt increased by 27 and 29 L/h, respectively, between visits and CV% was >100 % for several parameters, and particularly high for kg12. Large increases in tenofovir clearance terms has been previously reported [34]. Most, but not all, subjects showed lower AIC values for the two-compartment CVF model, which increased the variability in kg12. The individual fitted versus observed concentration plots for BP and CVF are shown in Fig. 2d; for both BP and CVF, the model over-predicts lower BP and CVF observations. VPCs for BP and CVF are shown in Fig. 6 and are reflective of the model over-prediction, particularly in CVF after multiple doses.
Figure 6.
Visual predictive checks for tenofovir at each sampling period in both blood plasma and cervicovaginal fluid using the 5th and 95th percentiles and median of 1,000 simulated subjects and observed data at both first dose and multiple dose: a blood plasma at first dose; b cervicovaginal fluid at first dose; c blood plasma at multiple dose; d cervicovaginal fluid at multiple dose. BP blood plasma, CVF cervicovaginal fluid, TFV tenofovir
3.3 Model-Predicted CVF:BP AUC ratios
Model-predicted AUCτ values, ratios of AUCτ CVF:BP, and CVF:BP using unbound (free) BP AUCτ (fAUCτ) are presented in Table 4. For each drug, CVF:BP ratios were similar to previously published values. Ratios generated by using fAUCτ were different, as would be predicted by the range of fu. For efavirenz, the AUCτ CVF:fAUCτ BP ratios of 0.53 and 0.66 at first and multiple doses, respectively, indicate that approximately half of the unbound drug appears in the CVF. For atazanavir, the AUCτ CVF:fAUCτ BP ratios >1 indicate that unbound drug may concentrate in the genital tract. Less of an effect is seen for lamivudine and tenofovir, which have lower protein binding. Lamivudine demonstrated high penetration in CVF (ratios >4) and tenofovir demonstrated exposure in CVF similar to blood (ratios of approximately 1.6 for first and multiple doses, regardless of protein binding).
Table 4.
Median (25th, 75th percentiles) model-predicted values of the area under the concentration-time curve over a dosing interval for each drug in blood plasma and the estimated unbound drug area under the plasma concentration-time curve in blood plasma cervicovaginal fluid, at both first dose and multiple doses
| BP AUCτ (μg•h/L) | BP fAUCτ (μg•h/L) | CVF AUCτ (μg•h/L) | CVF AUCτ:BP AUCτ ratio | CVF AUCτ:BP fAUCτ ratio | |
|---|---|---|---|---|---|
| Efavirenz | |||||
| First dose | 24.5 (16.1, 30.3) | 0.245 (0.161, 0.303) | 0.0127 (0.0746, 0.193) | 0.00527 (0.00298, 0.0128) | 0.527 (0.298, 1.28) |
| Multiple dose | 46.7 (33.9, 106) | 0.470 (0.339, 1.06) | 0.262 (0.110, 0.579) | 0.00655 (0.0125, 0.000744) | 0.655 (0.0744, 1.25) |
| Atazanavir | |||||
| First dose | 25.3 (6.19, 43.9) | 3.55 (0.867, 6.15) | 5.66 (8.36, 2.12) | 0.299 (0.0846, 1.55) | 2.14 (0.604, 11.1) |
| Multiple dose | 54.6 (11.5, 68.1) | 7.64 (1.61, 9.54) | 5.53 (3.18, 11.4) | 0.173 (0.0719, 0.596) | 1.24 (0.514, 1.24) |
| Lamivudine 150 mg twice daily | |||||
| First dose | 3.86 (3.58, 4.20) | 2.31 (2.15, 2.52) | 10.0 (6.07, 48.9) | 2.77 (1.35, 1.62) | 4.62 (2.24, 27.0) |
| Multiple dose | 3.17 (2.71, 5.47) | 1.90 (1.62, 3.28) | 27.7 (14.5, 59.2) | 5.92 (4.29, 19.5) | 9.86 (7.15, 32.5) |
| Lamivudine 300 mg once daily | |||||
| First dose | 10.1 (9.49, 13.6) | 6.06 (5.70, 8.15) | 62.2 (10.4, 287) | 5.40 (0.976, 30.6) | 8.98 (1.63, 51.0) |
| Multiple dose | 12.9 (4.35, 17.9) | 7.73 (2.61, 10.7) | 36.1 (0.526, 99.2) | 2.81 (0.53, 9.97) | 4.67 (0.878, 16.6) |
| Tenofovir | |||||
| First dose | 2.56 (1.71, 4.68) | 2.38 (1.59, 4.35) | 4.18 (7.30, 18.1) | 1.57 (0.132, 13.7) | 1.68 (0.141, 14.7) |
| Multiple dose | 2.32 (1.21, 4.08) | 2.16 (1.13, 3.79) | 4.56 (1.78, 10.6) | 1.52 (0.722, 7.40) | 1.64 (0.776, 7.95) |
Ratios for CVF AUCτ:BP AUCτ and CVF AUCτ:BP fAUCτ are provided as median (25th, 75th percentiles) for each drug at each sampling visit
AUCτ area under the plasma concentration-time curve during the dosing interval, BP blood plasma, CVF cervicovaginal fluid, fAUCτ unbound drug area under the plasma concentration-time curve in BP
4. Discussion
Using data from a comprehensive investigation of antiretroviral pharmacokinetics in HIV-infected pre-menopausal women, these are the first pharmacokinetic models to describe the movement of drug from BP to CVF for efavirenz, atazanavir, lamivudine and tenofovir. Depending on the drug, either the unbound fraction in blood (efavirenz, atazanavir) or total drug clearance (lamivudine, tenofovir) was the forcing function for drug distribution from BP to CVF. This difference in model structure reflects the degree of protein binding in blood for these drugs, with highly bound drugs limited in penetration by the unbound fraction. We attempted to find a common model to accommodate the transfer from BP to CVF, but model predictions for efavirenz using the total-drug model resulted in unlikely parameter estimates for CVF parameters (i.e. Vg values >1,000 L), and V2 estimates increased by an order of magnitude. For atazanavir, AIC values in ADAPT5 were lower for most subjects using the unbound-fraction model. Lamivudine profiles were not well fitted with the unbound-fraction model, and for tenofovir, protein binding is so low (<7 %) that only the total-drug model was considered. Drug disposition in CVF also differed between efavirenz/atazanavir and tenofovir/lamivudine, with tenofovir/lamivudine best modelled using a two-compartment CVF model. This is consistent with behaviour recently observed in healthy volunteers receiving tenofovir, which suggests a long-lived cellular compartment with equilibrium between tenofovir and its diphosphate metabolite [35]. Equivalent data do not exist for lamivudine; however, emtricitabine, a fluorinated analogue of lamivudine, was co-administered with tenofovir and demonstrated similar behaviour [35].
In assessing the model-predicted AUCτ CVF:fAUCτ BP ratios for efavirenz and atazanavir and total CVF concentrations, 66 of 68 CVF samples with detectable efavirenz concentrations (97 %) were above the concentration producing 50 % inhibition of wild-type HIV virus (IC50) of 0.51 ng/mL [36] and 71 of 75 CVF samples with detectable atazanavir concentrations (95 %) were above the IC50 of 11 ng/mL [15]. Although a relationship between CVF penetration and protein binding was seen here, this does not universally hold true for other drugs in CVF [12]. Additionally, the protein binding of these drugs in the CVF is unknown, and as albumin and α1-acid glycoprotein concentrations are <1 % of plasma values [37], less drug may be bound in CVF, as has been demonstrated for maraviroc [38], potentially making more drug available for pharmacological action.
Pharmacokinetic modelling of antiretrovirals can be challenging, given multiple drugs used in combination, significant drug-drug interactions among antiretrovirals in a regimen as well as between antiretrovirals and drugs to treat co-morbid conditions, and the inherent high inter-individual variability in pharmacokinetics. This phenomenon has been well-documented for antiretrovirals in BP [39]. CVF pharmacokinetics appear to have even greater variability for the following reasons [11]. First, collection methods are more complex and less consistent from subject to subject for CVF. Second, drug-drug interactions among the antiretrovirals in a given subject’s regimen were not accounted for in the models. Third, relatively little is known about intra-individual variability in CVF; however, intra-individual variability of antiretrovirals in BP is known to be quite high, ranging from 25 % for nucleoside agents up to 76 % for indinavir, a protease inhibitor [39, 40]. This may partially explain the need for separate parameters to describe both study visits simultaneously, and the high variability in the estimated parameters. Finally, these drugs are also substrates of several drug transporters that may be relevant for controlling disposition into the genital tract [11].
Several limitations were present in the data. For each drug, several subjects in the data had difficult-to-fit concentration-time profiles, with no consistent pattern among subjects. Therefore, structural amendments to the models to improve fits were not possible. The large percentage of BLQ data also decreased the available data for model fitting. Previous reports in the literature suggest auto-induction by efavirenz during the first month of treatment [41]; however, this phenomenon was not observed here, and is a potential consequence of these limitations. Adding covariates to explain inter-individual variability is commonly employed in population pharmacokinetics, but was not possible here, given the homogeneity of the subject population. Each drug had limited representation: most participants were African American with a narrow range of ages. Although there was wide variability in bodyweight, bodyweight did not demonstrate a relationship with volume or clearance in exploratory data analysis (data not shown).
5. Conclusion
The presented pharmacokinetic models for efavirenz, atazanavir, lamivudine and tenofovir provided some insight into drug disposition into CVF. Using non-linear mixed-effects modelling, relatively sparse datasets were fitted to two basic models. High inter- and intra-individual variability were expected with antiretroviral drugs, particularly when measuring CVF concentrations, and pharmacokinetic parameter estimates show high inter-individual variability. Three of the four drugs required models using separate clearance, absorption and transfer terms for each dosing interval to best describe the data. These models provide the basis for future work in establishing relationships between drug concentrations in mucosal tissues and HIV replication. Moreover, this type of modelling will have increasing economic importance in optimizing antiretroviral therapy as treatment and prevention strategies move forward in limited resource environments.
Acknowledgments
The authors wish to thank the participating subjects, as well as the staff of the UNC Healthcare Infectious Disease Clinic for their assistance with recruitment. We also thank the nurses of the UNC CTRC for their assistance in conducting study visits. This work was presented in part at the 13th International Workshop on Clinical Pharmacology of HIV Therapy, 16–18 April 2012, Barcelona, Spain. Financial support for this work was provided by the UNC Center for AIDS Research (5P30AI050410-13 –J.B. Dumond, K.B. Patterson, A.D.M. Kashuba), the NC TraCS Institute (UL1RR025747 – J.B. Dumond) and the National Institute of Allergy and Infectious Diseases (K23AI093156 – J.B. Dumond; K23AI077355-KBP; K23AI54980 – A.D.M. Kashuba; U01AI095031 – A.D.M. Kashuba). These funding sources provided salary support and funds for research conduct, but did not have any input into study design, study analysis or reporting of study results.
Footnotes
Conflicts of Interest
A.D.M. Kashuba has received research funding and speaking honoraria from Bristol-Myers Squibb, Merck, Gilead and GlaxoSmithKline. K.B. Patterson has received research funding from GlaxoSmithKline. The other authors have no conflicts of interest that are directly relevant to the content of this study.
Author Contributions
J.B. Dumond: data collection, data analysis, including the original publication and current modelling applications, primary author of the original and current manuscript. M.R. Nicol: data analysis of atazanavir, critical review of the manuscript. R.N. Kendrick: data analysis of lamivudine, critical review of the manuscript. S.M. Garonzik: data analysis of the current modelling applications, critical review of the manuscript. K.B. Patterson: data collection, critical review of the original and current manuscript. M.S. Cohen: conceptual design of original study, critical review of the original and current manuscript. A. Forrest: data analysis of the current modelling applications, critical review of the manuscript. A.D.M. Kashuba: conceptual design of original study, data collection, data analysis of the original publication, funding of the original study, critical review of the original and current manuscript.
References
- 1.Cohen MS, Chen YQ, McCauley M, Gamble T, Hosseinipour MC, Kumarasamy N, et al. Prevention of HIV-1 infection with early antiretroviral therapy. N Engl J Med. 2011;365:493–505. doi: 10.1056/NEJMoa1105243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Quinn TC, Wawer MJ, Sewankambo N, Serwadda D, Li C, Wabwire-Mangen F, et al. viral load and heterosexual transmission of human immunodeficiency virus type 1. N Engl J Med. 2000;342:921–929. doi: 10.1056/NEJM200003303421303. [DOI] [PubMed] [Google Scholar]
- 3.Eron JJ, Jr, Smeaton LM, Fiscus SA, Gulick RM, Currier JS, Lennox JL, et al. The effects of protease inhibitor therapy on human immunodeficiency virus type 1 levels in semen (AIDS Clinical Trials Group Protocol 850) J Infect Dis. 2000;181:1622–1628. doi: 10.1086/315447. [DOI] [PubMed] [Google Scholar]
- 4.Cu-Uvin S, Caliendo AM, Reinert S, Chang A, Juliano-Remollino C, Flanigan TP, et al. Effect of highly active antiretroviral therapy on cervicovaginal HIV-1 RNA. AIDS. 2000;14:415–421. doi: 10.1097/00002030-200003100-00015. [DOI] [PubMed] [Google Scholar]
- 5.Launay O, Tod M, Tschöpe I, Si-Mohamed A, Bélarbi L, Charpentier C, et al. Residual HIV-1 RNA and HIV-1 DNA production in the genital tract reservoir of women treated with HAART: the prospective ANRS EP24 GYNODYN study. Antivir Ther. 2011;16:843–852. doi: 10.3851/IMP1856. [DOI] [PubMed] [Google Scholar]
- 6.Mitchell C, Hitti J, Paul K, Agnew K, Cohn SE, Luque AE, et al. Cervicovaginal shedding of HIV type 1 is related to genital tract inflammation independent of changes in vaginal microbiota. AIDS Res Hum Retroviruses. 2011;27:35–39. doi: 10.1089/aid.2010.0129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Henning TR, Kissinger P, Lacour N, Meyaski-Schluter M, Clark R, Amedee AM. Elevated cervical white blood cell infiltrate is associated with genital HIV detection in a longitudinal cohort of antiretroviral therapy-adherent women. J Infect Dis. 2010;202:1543–1552. doi: 10.1086/656720. [DOI] [PubMed] [Google Scholar]
- 8.Lorello G lPC, Pilon R, Zhang G, Karnauchow T, MacPherson P. Discordance in HIV-1 viral loads and antiretroviral drug concentrations comparing semen and blood plasma. HIV Med. 2009;10:548–554. doi: 10.1111/j.1468-1293.2009.00725.x. [DOI] [PubMed] [Google Scholar]
- 9.Kovacs A, Wasserman SS, Burns D, Wright DJ, Cohn J, Landay A, et al. Determinants of HIV-1 shedding in the genital tract of women. Lancet. 2001;358:1593. doi: 10.1016/S0140-6736(01)06653-3. [DOI] [PubMed] [Google Scholar]
- 10.Si-Mohamed A, Kazatchkine MD, Heard I, Goujon C, Prazuck T, Aymard G, et al. Selection of drug-resistant variants in the female genital tract of human immunodeficiency virus type 1-infected women receiving antiretroviral therapy. J Infect Dis. 2000;182:112–122. doi: 10.1086/315679. [DOI] [PubMed] [Google Scholar]
- 11.Else LJ, Taylor S, Back DJ, Khoo SH. Pharmacokinetics of antiretroviral drugs in anatomical sanctuary sites: the male and female genital tract. Antivir Ther. 2011;16:1149–1167. doi: 10.3851/IMP1919. [DOI] [PubMed] [Google Scholar]
- 12.Nicol MR, Kashuba ADM. Pharmacologic opportunities for HIV prevention. Clin Pharmacol Ther. 2010;88:598–609. doi: 10.1038/clpt.2010.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dumond JB, Yeh RF, Patterson KB, Corbett AH, Jung BH, Rezk NL, et al. Antiretroviral drug exposure in the female genital tract: implications for oral pre- and post-exposure prophylaxis. AIDS. 2007;21:1899–1907. doi: 10.1097/QAD.0b013e328270385a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bristol-Myers Squibb Company. Sustiva (efavirenz) full US prescribing information. Princeton: Bristol-Myers Squibb Company; 2009. [Google Scholar]
- 15.Bristol-Myers Squibb Company. Reyataz (atazanavir) full US prescribing information. Princeton: Bristol-Myers Squibb Company; 2009. [Google Scholar]
- 16.Gilead Sciences. Viread full US prescribing information. Foster City: Gilead Sciences; 2012. [Google Scholar]
- 17.ViiV Healthcase/GlaxoSmithKline. Epivir full US prescribing information. Research Triangle Park: ViiV Healthcase/GlaxoSmithKline; 2011. [Google Scholar]
- 18.Jung BH, Rezk NL, Bridges AS, Corbett AH, Kashuba ADM. Simultaneous determination of 17 antiretroviral drugs in human plasma for quantitative analysis with liquid chromatography–tandem mass spectrometry. Biomed Chromatogr. 2007;21:1095–1104. doi: 10.1002/bmc.865. [DOI] [PubMed] [Google Scholar]
- 19.Rezk NL, Crutchley RD, Kashuba ADM. Simultaneous quantification of emtricitabine and tenofovir in human plasma using high-performance liquid chromatography after solid phase extraction. J Chromatogr B Analyt Technol Biomed Life Sci. 2005;822:201–208. doi: 10.1016/j.jchromb.2005.06.019. [DOI] [PubMed] [Google Scholar]
- 20.Rezk NL, Tidwell RR, Kashuba ADM. Simultaneous determination of six HIV nucleoside analogue reverse transcriptase inhibitors and nevirapine by liquid chromatography with ultraviolet absorbance detection. J Chromatogr B Analyt Technol Biomed Life Sci. 2003;791:137–147. doi: 10.1016/s1570-0232(03)00224-1. [DOI] [PubMed] [Google Scholar]
- 21.Rezk NL, Tidwell RR, Kashuba ADM. High-performance liquid chromatography assay for the quantification of HIV protease inhibitors and non-nucleoside reverse transcriptase inhibitors in human plasma. J Chromatogr B Analyt Technol Biomed Life Sci. 2004;805:241–247. doi: 10.1016/j.jchromb.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 22.Rezk NLMS, Crutchley RD, Yeh RFP, Kashuba ADMP. Full validation of an analytical method for the HIV-protease inhibitor atazanavir in combination with 8 other antiretroviral agents and its applicability to therapeutic drug monitoring. Ther Drug Monit. 2006;28:517–525. doi: 10.1097/00007691-200608000-00006. [DOI] [PubMed] [Google Scholar]
- 23.D’Argenio DZ, Schumitzky A, Wang X. ADAPT 5 user’s guide: pharmacokinetic/pharmacodynamic systems analysis software. Los Angeles: Biomedical Simulations Resource; 2009. [Google Scholar]
- 24.D’Argenio DZ, Schumitzky A. A program package for simulation and parameter estimation in pharmacokinetic systems. Comput Programs Biomed. 1979;9:115–134. doi: 10.1016/0010-468x(79)90025-4. [DOI] [PubMed] [Google Scholar]
- 25.Beijnen JH, Huitema ADR, Kappelhoff BS, Meenhorst PL, Mulder JW, Prins JM, et al. Population pharmacokinetics of efavirenz in an unselected cohort of HIV-1-infected individuals. Clin Pharmacokinet. 2005;44:849–61. doi: 10.2165/00003088-200544080-00006. [DOI] [PubMed] [Google Scholar]
- 26.Colombo S, Buclin T, Cavassini M, Décosterd LA, Telenti A, Biollaz J, et al. Population pharmacokinetics of atazanavir in patients with human immunodeficiency virus infection. Antimicrob Agents Chemother. 2006;50:3801–3808. doi: 10.1128/AAC.00098-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jullien V, Tréluyer J-M, Rey E, Jaffray P, Krivine A, Moachon L, et al. Population pharmacokinetics of tenofovir in human immunodeficiency virus-infected patients taking highly active antiretroviral therapy. Antimicrob Agents Chemother. 2005;49:3361–3366. doi: 10.1128/AAC.49.8.3361-3366.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moore KHP, Yuen GJ, Hussey EK, Pakes GE, Eron JJ, Bartlett JA. Population pharmacokinetics of lamivudine in adult human immunodeficiency virus-infected patients enrolled in two phase III clinical trials. Antimicrob Agents Chemother. 1999;43:3025–3029. doi: 10.1128/aac.43.12.3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Akaike H. A new look at the statistical model identification. IEEE Trans Automat Control. 1974;19:716–723. [Google Scholar]
- 30.Beal SL. Ways to fit a PK model with some data below the quantification limit. J Pharmacokinet Pharmacodyn. 2001;28:481–504. doi: 10.1023/a:1012299115260. [DOI] [PubMed] [Google Scholar]
- 31.Bulitta J, Bingölbali A, Shin B, Landersdorfer C. Development of a new pre- and post-processing tool (SADAPT-TRAN) for nonlinear mixed-effects modeling in S-ADAPT. AAPS J. 2011;13:201–211. doi: 10.1208/s12248-011-9257-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bulitta J, Landersdorfer C. Performance and robustness of the Monte Carlo importance sampling algorithm using parallelized S-ADAPT for basic and complex mechanistic models. AAPS J. 2011;13:212–226. doi: 10.1208/s12248-011-9258-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Okusanya OO, Hamel J, Rubino CM. SADAPTPlot – an easy-to-use R package for S-ADAPT population analysis post-processing [poster]. American Conference on Pharmacometrics; 2011 Apr 4–7; San Diego. [Google Scholar]
- 34.Gagnieu M-C, Barkil ME, Livrozet J-M, Cotte L, Miailhes P, Boibieux A, et al. Population pharmacokinetics of tenofovir in AIDS patients. J Clin Pharmacol. 2008;48:1282–1288. doi: 10.1177/0091270008322908. [DOI] [PubMed] [Google Scholar]
- 35.Patterson KB, Prince HA, Kraft E, Jenkins AJ, Shaheen NJ, Rooney JF, et al. Penetration of tenofovir and emtricitabine in mucosal tissues: implications for prevention of HIV-1 transmission. Sci Transl Med. 2011;3:112re4. doi: 10.1126/scitranslmed.3003174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parkin NT, Hellmann NS, Whitcomb JM, Kiss L, Chappey C, Petropoulos CJ. natural variation of drug susceptibility in wild-type human immunodeficiency virus type 1. Antimicrob Agents Chemother. 2004;48:437–443. doi: 10.1128/AAC.48.2.437-443.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Salas Herrera IG, Pearson RM, Turner P. Quantitation of albumin and alpha-1-acid glycoprotein in human cervical mucus. Hum Exp Toxicol. 1991;10:137–139. doi: 10.1177/096032719101000209. [DOI] [PubMed] [Google Scholar]
- 38.Dumond JBP, Patterson KBMD, Pecha ALP, Werner REBS, Andrews EP, Damle BP, et al. Maraviroc concentrates in the cervicovaginal fluid and vaginal tissue of HIV-negative women. J Acquir Immune Defic Syndr. 2009;51:546–553. doi: 10.1097/QAI.0b013e3181ae69c5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bonnet B, Diquet B, Duval X, Goujard C, Katlama C, Legrand M, et al. High variability of indinavir and nelfinavir pharmacokinetics in HIV-infected patients with a sustained virological response on highly active antiretroviral therapy. Clin Pharmacokinet. 2005;44:1267–78. doi: 10.2165/00003088-200544120-00005. [DOI] [PubMed] [Google Scholar]
- 40.Nettles RE, Kieffer TL, Parsons T, Johnson J, Cofrancesco J, Jr, Gallant JE, et al. Marked intraindividual variability in antiretroviral concentrations may limit the utility of therapeutic drug monitoring. Clin Infect Dis. 2006;42:1189–1196. doi: 10.1086/501458. [DOI] [PubMed] [Google Scholar]
- 41.Zhu M, Kaul S, Nandy P, Grasela DM, Pfister M. Model-based approach to characterize efavirenz autoinduction and concurrent enzyme induction with carbamazepine. Antimicrob Agents Chemother. 2009;53:2346–2353. doi: 10.1128/AAC.01120-08. [DOI] [PMC free article] [PubMed] [Google Scholar]