Abstract
The movement of signal transduction enzymes in and out of multi-protein complexes coordinates the spatial and temporal resolution of cellular events. Anchoring and scaffolding proteins are key to this process because they sequester protein kinases and phosphatases with a subset of their preferred substrates. The protein kinase A-anchoring family of proteins (AKAPs), which target the cAMP-dependent protein kinase (PKA) and other enzymes to defined subcellular microenvironments, represent a well studied group of these signal-organizing molecules. In this report we demonstrate that the Rab27a GTPase effector protein MyRIP is a member of the AKAP family. The zebrafish homolog of MyRIP (Ze-AKAP2) was initially detected in a two-hybrid screen for AKAPs. A combination of biochemical, cell-based, and immunofluorescence approaches demonstrate that the mouse MyRIP ortholog targets the type II PKA holoenzyme via an atypical mechanism to a specific perinuclear region of insulin-secreting cells. Similar approaches show that MyRIP interacts with the Sec6 and Sec8 components of the exocyst complex, an evolutionarily conserved protein unit that controls protein trafficking and exocytosis. These data indicate that MyRIP functions as a scaffolding protein that links PKA to components of the exocytosis machinery.
When and where enzymes become active influences the cellular processes they control. As a result, sophisticated molecular mechanisms have evolved to manage the spatial and temporal synchronization of signal transduction pathways (1). Scaffolding and anchoring proteins target protein kinases and phosphatases to subcellular environments where they control the phosphorylation state of neighboring substrates (2–4). Protein phosphorylation events potentiate the relay of intracellular signals by propagating localized changes in cellular activity (5). Movement of enzymes in and out of these complexes contribute to the synchronization of signaling events (6). Prototypic examples of these signal-organizing molecules are the protein kinase A-anchoring proteins (AKAPs),2 which compartmentalize cAMP-dependent protein kinase (PKA) and other enzymes (6, 7).
The first AKAP was identified when it was demonstrated that type II PKA co-purified with microtubules and that the RII subunit bound to the microtubule-associated protein MAP2 (8). Many more of these RII-binding proteins have subsequently been detected by an overlay technique to screen nitrocellulose membranes of immobilized proteins using the purified RII subunit as a probe (9). This technique has been adapted to screen phage cDNA libraries and clone novel AKAPs (10–14). To date over 50 different AKAP forms have been identified that function to compartmentalize the type I and type II PKA holoenzymes (reviewed in Refs. 7 and 15). Two important principles have emerged from this work: AKAPs bind the R subunits through a common anchoring region (16–19), and each anchoring protein is targeted to a unique localization in a given cell type by an identifiable targeting motif (13, 20–24).
Another feature of AKAPs is their ability to interact with several signaling proteins (25, 26). By simultaneously tethering PKA with enzymes such as phosphatases, phosphodiesterases, small molecular weight GTPases, and other protein kinases, these multivalent anchoring proteins create focal points for the processing of intracellular messages (27–32). In this report we show that zebrafish and mammalian orthologs of the MyRIP (myosin Va-Rab27a-interacting protein) function as kinase A-anchoring proteins. Other studies reveal that MyRIP also interacts with the exocyst complex, an evolutionarily conserved octameric protein unit that is an integral element of the vesicle-docking machinery at sites of regulated and constitutive exocytosis (33–37).
EXPERIMENTAL PROCEDURES
Yeast Two-hybrid Assay
The 48-h zebrafish cDNA HybriZAP-2.1 XR library (kindly provided by Dr. David Ransom) was excised and amplified as detailed by Stratagene. A two-hybrid screen as described by Hollenberg et al. (38) was initiated using 1–45 Rze (a murine PKA RII ortholog) fused to the LexA-binding domain in pLexA vector as bait (pLexA Rze 1–45). The bait plasmid was introduced into the L40 yeast strain to screen a total of 2 × 106 transformants to ensure complete coverage of the library. Two-hybrid interactions were first detected by growth of the L40 strain on SD lacking histidine and supplemented with 3 mM 3-amino-1,2,4-triazole. All positive clones were further screened for lacZ expression by filter lift assay according to the manufacturer’s protocol (Stratagene). The LEU2-marked library plasmid was rescued, amplified in bacteria, digested with a variety of enzymes, and classed according to restriction pattern. Representatives from each class were retransformed into L40 along with the pLexA Rze 1–45 plasmid. In every case, retransformation was accompanied by complementation of the strain to His prototrophy. Library plasmids were then sequenced to identify the RII-interacting protein. In total, seven unique genes were represented.
Zebrafish Reverse Transcription-PCR and 5/3′-RACE
Total RNA was extracted from 24-, 48-, 72-, and 102-h zebrafish embryos using TRIzol (Invitrogen) and further purified using RNAeasy (Qiagen) according to the manufacturer’s protocols. Poly(A)+ mRNA was isolated from total RNA using Oligotex (Qiagen) and was used as a template for synthesis of single strand 5′-RACE and 3′-RACE cDNA libraries using the SMART RACE cDNA amplification kit (BD Biosciences, Clontech) according to the manufacturer’s protocols. 3′- and 5′-RACE PCR was carried out using the Advantage-GC 2 PCR kit (BD Biosciences, Clontech).
To clone the Rze, 3′-RACE was performed using nested primers 5′-GATCTCTGCACAACCCCTACTACTG-3′ and 5′-CACCATGAGTATTGAGATTCCAGTCGGG-3′, which were designed according to a murine PKA RII-like zebrafish EST sequence fw80b12.x1 (GenBank™ accession number BM317066). A 1.5-kb band was cloned into pENTER/SD/D (Invitrogen), and the resulting clone, Rze, was sequenced. The coding region is identical to a cDNA clone sequence (Gen-Bank™ accession number BC124725) that was submitted during the course of this study.
Zebrafish Ze-AKAP2 was cloned by 5′-RACE using nested primers 5′-GATTTCTAAGTCTCGCTCAATCTCTGATTGTC-3′ and 5′-CCTGTTTCTTCTCGACCTGCATCTCG-3′, which were designed according to the 1551-bp fragment obtained in the two-hybrid screen. PCR products were cloned into pCRII-TOPO using the TOPO TA cloning kit (Invitrogen) for sequencing, and the resulting clone, Ze-AKAP2, was sequenced (GenBank™ accession number EF694835).
Plasmids
Full-length MyRIP was PCR-amplified from a mouse cDNA library (BD Biosciences, Clontech) and cloned into pCRII-TOPO using primers 5′-CACCATGGGGGAAGCTGGACCTGTGGGG-3′ and 5′-TCAGTACATCACAGCTGACTCCAGATCAG-3′. For the mammalian expression constructs, full-length MyRIP was PCR-amplified and cloned directly into pcDNA3.1D/V5-His-TOPO (Invitrogen) using the same primers as above. The MyRIP fragments were PCR-amplified and cloned into pENTR/SD/D-TOPO for use as an entry clone in the Gateway system (Invitrogen). MyRIP fragments (1–153, 1–300, 153–856, and 490–856) were shuttled from pENTR/SD/D-TOPO into pcDNA3.1/nV5-DEST, an N-terminal V5-tagged destination vector for mammalian expression according to the manufacturer’s instructions. The remaining two MyRIP fragments (153–495 and 300–856) were PCR-amplified and cloned directly into pcDNA3.1D/V5-His-TOPO. pcDNA3.1D/V5-His-mouse MyRIP V197P, I206P, I236P, and I245P were generated by site-directed mutagenesis as per the manufacturer’s instructions (Stratagene) using primers 5′-GGCTGTGGCCCTACGACCGGCGAGGCCATCG-3′, 5′-GAAGAGGCCATCGAGGAGGCCCCTTCCAAAGCGAATC-3′, 5′-GAGCTAACTGAGGAGCCGGCCGGCACCATCCTG-3′, 5′-CCATCCTGCAGAGGATTCCAAGAAAGCAGAAGG-3′, and their complements, respectively. pcDNA3.1D/V5-His-Human MyRIP-FLAG Δ194–210, MyRIP Δ233–248 and MyRIP Δ194–210 Δ233–248 were also generated by site-directed mutagenesis using primers 5′-GTGATGGACACCTTGGCTGTGGACATAGGGGACAGCCTG-3′, 5′-CGGGACCACAAGGAGGAGCTAAAGAGCAAAAGTGAGCAGCAAG-3′, and their complements, respectively. pcDNA3.1/V5-His-AKAP-IS-GFP, pcDNA3.1/V5-His-Scrambled AKAP-IS-GFP, and pFOX.CMV/hGH (hGH sequence under control of the cytomegalovirus promoter) were generated in previous studies (39, 40). pET48 EK/LIC-HIS-MyRIP110–305, pGEX-4T1-Hs MyRIP 1–383, and pGEX-4T1-ZeAKAPs for expression in bacteria and pLexA Rze 1–45 for expression in yeast were generated by PCR amplification. Phogrin-EGFP was a gift from Dr. Guy A. Rutter (41), and pLexA Wave1 was a gift from Dr. Scott H. Soderling.
Cell Culture and Transfections
The clonal β-cell line INS-1(832/13) (42) was cultured (passages 48–60) in RPMI 1640 (Invitrogen) that contained 11 mM glucose and supplemented with 10% heat-inactivated fetal bovine serum, 10 mM Hepes, pH 7.4, 2 mM glutamine, 1 mM sodium pyruvate, 50 μM 2-mercaptoethanol, 100 units/ml penicillin, and 100 μg/ml streptomycin. The cells were grown in 10-cm tissue culture dishes at 37 °C in 5% CO2 and passaged every 5 days using 1 ml of 0.05% trypsin-EDTA. Transfections of INS-1 cells were performed on cells at 50–75% confluence using Lipofectamine 2000 (Invitrogen), according to the manufacturer’s instructions. The cells were harvested 24 h post-transfection.
HEK293 cells at 50–75% confluence were transfected using Lipofectamine Plus (Invitrogen), according to the manufacturer’s instructions. The cells were harvested 24 h post-transfection.
Antibodies
The following primary antibodies were used in this study: monoclonal anti-V5 and anti-V5 conjugated to horseradish peroxidase (Invitrogen), monoclonal anti-GFP (B2) and rabbit anti-PKA catalytic subunit (Santa Cruz Biotechnology), mouse anti-Sec6 (Stressgen), mouse anti-RIIα, PKA catalytic subunit, and anti-Sec8 (BD Biosciences). For indirect immunocytochemistry of INS-1 cells, the primary antibodies used were mouse anti-PKA RII (BD Biosciences), guinea pig anti-insulin (Chemicon), mouse anti-Sec8 and anti-Sec6 (Stressgen), secondary antibodies were Texas Red-conjugated donkey anti-rabbit IgG (Jackson ImmunoResearch), Alexa Fluor 488 goat anti-mouse IgG, Alexa Fluor 488 goat anti-mouse IgG, and Alexa Fluor 568 goat anti-guinea pig (Invitrogen and Molecular Probes).
Generation of MyRIP Antisera
Antibodies to MyRIP were generated in two rabbits using recombinant His-tagged mouse MyRIP (110–305) as the immunogen (Rockland), resulting in two antibody preparations, 7445 and 7839. For affinity purification, glutathione S-transferase-tagged human MyRIP (amino acids 1–383) was immobilized on resin (Affi-Gel 15) and incubated at 4 °C for 2 h with MyRIP antiserum (7445). The affinity resin was washed with 20 ml of PBS and 30 ml of PBS containing 0.5 M NaCl. Bound antibodies were eluted with 10 ml of 0.1 M glycine, pH 2.5; 1-ml fractions were collected; and 50 μl of Tris, pH 9.5, was added to each fraction.
Immunocytochemistry
INS-1(832/13) cells were cultured on glass coverslips coated with 0.2 mg/ml poly-L-lysine for 24 h. The cells were fixed in 4% paraformaldehyde and incubated 20 min at room temperature. The cells were washed once in PBS and incubated in staining solution (0.2% bovine serum albumin, 1% fish gelatin (Sigma), 0.1% Triton in PBS) overnight at 4 °C. Primary antibody (polyclonal MyRIP, 1:100; monoclonal PKA RII, 1:500; monoclonal Sec8, 1:100; monoclonal Sec6, 1:400; and polyclonal insulin, 1:500) in staining solution was then added for 2 h at room temperature, and the cells were washed three times with PBS for 10 min each. The cells were then incubated with the appropriate fluorescent secondary antibodies in the same manner. After three PBS washes, the cells were rinsed with water and mounted onto glass slides using ProLong Gold antifade reagent with DAPI (Molecular Probes).
AutoSpot Peptide Array and RII Overlay
Peptide arrays were synthesized on cellulose membranes with an Auto-Spot Robot ASP 222 (AbiMed) as described previously (44). RII overlays were performed using either [32P]Rze or murine [32P]RIIα as previously described (10).
Immunoprecipitations and PKA Activity Assay
The cell extracts were prepared by washing cells with cold PBS and followed by scraping cells in 1 ml of cold lysis buffer (25 mM Hepes, pH 7.5, 150 mM NaCl, 1 mM EDTA, 0.5% Triton X-100) plus protease inhibitors (1 tablet/10 ml complete mini, EDTA free protease mixture (Roche Applied Science)), and centrifuged at 16,000 × g for 15 min. 50 μl of prewashed protein G or A beads (50% slurry; Upstate) and 3 μg of antibody was added to the extract and incubated for 2 h at 4 °C. The beads were washed once with 1 ml of lysis buffer, three times with 1 ml of lysis buffer plus 1 M NaCl, and once with 1 ml lysis buffer. PKA kinase assays were performed as previously described (45). For immunoprecipitation of endogenous MyRIP, cell extracts from 10 tissue culture dishes (10-cm) were prepared as described above and pooled. Immunoprecipitations of MyRIP using the anti-MyRIP polyclonal antibody were carried out as described above.
hGH Secretion Assays
INS-1(832/13) cells in 10-cm dishes at 70% confluence were co-transfected with 60 nM MyRIP siRNA (Dharmacon Mouse MyRIP siGENOME Duplex) or 60 nM control siRNA (BLOCK IT Fluorescent oligonucleotide) and 10 μg of hGH-expressing vector (40) using Lipofectamine 2000. Twenty-four hours after transfection, the cells were plated in 24-well plates at 3.5 × 105 cells/well. After 48 h, the cells were washed once with 1 ml of Krebs-Ringer Hepes buffer (KRBH; 25 mM Hepes, pH 7.4, 5 mM NaHCO3, 1.2 mM MgSO4 (7H2O), 1.2 mM KH2PO4, 4.74 mM KCl, 125 mM NaCl, 1 mM CaCl2) supplemented with 2 mM glucose and preincubated for 1 h in 2 mM glucose KRBH at 37 °C and 5% CO2. The cells were then incubated 1 h in 1 ml of either 2 mM glucose KRBH, 20 mM glucose KRBH, 20 mM glucose KRBH supplemented with 1 μM forskolin, 2 mM glucose KRBH supplemented with 1 mM IBMX, 20 mM glucose KRBH supplemented with 1 mM IBMX, or 20 mM glucose KRBH supplemented with 1 μM forskolin and 1 mM IBMX. Culture supernatants were collected, centrifuged 10 min at 3000 rpm at 4 °C, and frozen at −80 °C until assayed for hGH. After collection of secretion samples, the cells were harvested in 2 M acetic acid containing 0.25% bovine serum albumin, subjected to three freeze/thaw cycles, and centrifuged at 20,000 × g for 20 min to remove cell debris. Analysis of hGH content in secretion samples and cell homogenates was performed using hGH enzyme-linked immunosorbent assay (Roche Applied Science). The data are presented as ratios of secreted hGH to total hGH content/well.
RESULTS
Characterization of Zebrafish AKAPs
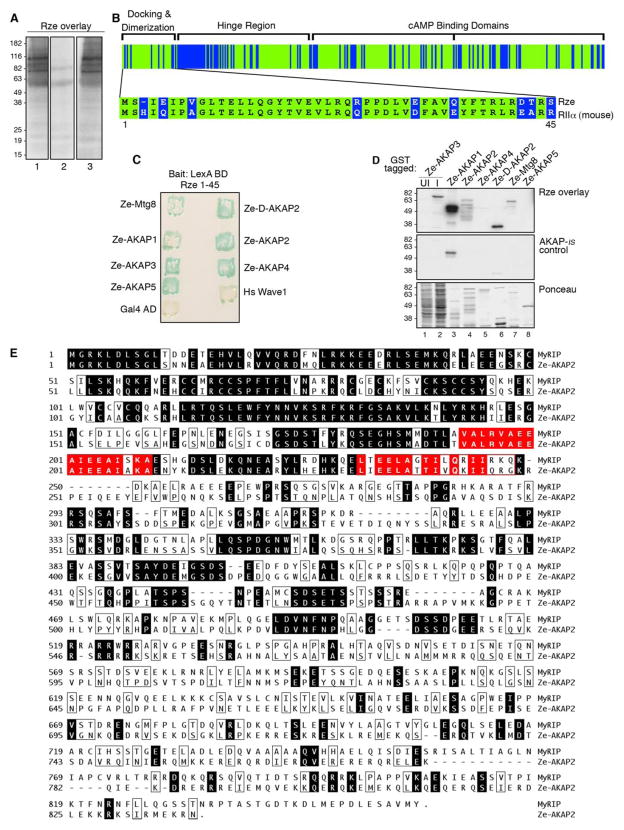
AKAPs are often classified by their ability to bind the regulatory subunits of PKA in a solid phase assay known as the RII overlay (9, 10). To determine whether AKAPs were expressed in developing zebrafish, two-day old embryo extracts were separated on SDS-PAGE gels, transferred to polyvinylidene difluoride membranes, and subjected to an overlay procedure using purified 32P-radiola-beled zebrafish regulatory subunit (Rze) as a probe. Several Rze-binding bands were detected that ranged in size from 60 to 200 kDa (Fig. 1A, lane 1). Mammalian AKAPs bind their R subunits through an amphipathic helix (16). These protein-protein interactions can be blocked with anchoring inhibitor peptides that compete for binding with the R subunit (39, 46). Preincubation with the anchoring inhibitor peptide, AKAP-IS, blocked Rze interaction with zebrafish AKAPs (Fig. 1A, lane 2). In contrast, preincubation with a control peptide (scrambled AKAP-IS) had no effect on the Rze/AKAP interactions (Fig. 1A, lane 3). Collectively, these results suggest that zebrafish express AKAPs and that the topology of the Rze/anchoring protein interface is similar to its mammalian counterparts. This latter view is also substantiated by evidence that zebrafish Rze is 71% identical to its mouse RII ortholog (Fig. 1B and Ref. 47).
FIGURE 1. Identification of Ze-AKAP2 as a putative AKAP.
A, zebrafish two-day embryo extracts were separated by SDS-PAGE and transferred to nitrocellulose. Binding of 32P-labeled zebrafish regulatory subunit (Rze) was assessed by overlay and was detected by autoradiography. Rze overlays were performed in the absence of inhibitor peptide (left), in the presence of 1 μM AKAP-IS anchoring inhibitor peptide (middle), or in the presence of 1 μM Scrambled AKAP-IS peptide (right). Molecular mass markers are indicated. B, a schematic representing the identity between Rze and murine RIIα (green). Functional domains of murine RIIα are labeled (inset). The sequence alignment of Rze 1– 44 and murine RIIa 1– 45 is indicated using the one-letter amino acid code. C, zebrafish Rze 1– 45/LexA binding domain was used as bait to screen a zebrafish two-day embryonic cDNA library using the yeast two-hybrid assay. The detection of positive colonies expressing Ze-AKAP2, other putative AKAPs, human Wave1 (positive control), and Gal4 AD (negative control) was assessed by β-galactosidase assay. The name of each AKAP is indicated. D, solid phase binding of Rze to glutathione S-transferase (GST) fragments of the putative AKAPs was assessed by RII overlay procedure in the absence of inhibitor peptide (top) or in the presence of 1 μM AKAP-IS (middle). Either induced (I; lane 2), uninduced bacterial extracts (UI; lane 1) or purified glutathione S-transferase fragments were used (lanes 3– 8). The names of the putative AKAPs are indicated above each lane. Ponceau staining of protein samples (bottom). E, amino acid sequence alignment of mouse MyRIP (top) with Ze-AKAP2 (bottom). The shaded regions depict sequence identity, and the boxed regions depict similarity. Putative PKA anchoring sites are indicated (red-shaded regions).
Biochemical and structural studies have defined the first 45 amino acids of RII as a docking and dimerization domain that forms the AKAP-binding site (18, 19, 48, 49). The corresponding region in Rze was used as bait to screen a zebrafish two-day-old embryonic cDNA library using the yeast two-hybrid assay (Fig. 1B, inset). Of the 1.9 × 106 transformants screened, 393 candidates activated both the lacZ and HIS reporters. In total, seven unique genes were represented (Fig. 1C). Two of these, Ze-Mtg8 and Ze-D-AKAP2, have mammalian homologs previously characterized as AKAPs (Fig. 1C and Refs. 50 and 51). The remaining five genes were named Ze-AKAP1–5 (zebrafish AKAP). Bacterially expressed protein fragments of each putative Ze-AKAP were screened for RII binding using the overlay assay. Five of the seven candidate proteins bound Rze (Fig. 1D, top panel). Control experiments confirmed that Rze binding was blocked upon preincubation with AKAP-IS (Fig. 1D, top and middle panels), whereas the addition of a scrambled peptide control did not affect the interaction with Rze (data not shown). Approximately equal amounts of each bacterially purified zebrafish protein were used in the overlay (Fig. 1D, bottom panel).
We chose to concentrate on a 517-amino acid fragment named Ze-AKAP2. Full-length Ze-AKAP2 was cloned from a zebrafish cDNA library by 5′-RACE. This cDNA encodes a protein of 856 amino acids that shares 35% identity with a human GTPase effector protein known as MyRIP (Ref. 52 and Fig. 1E). Most of the sequence homology lies in the N-terminal quarter of these proteins. The first 150 residues encompass a domain that binds to the GTPase Rab27a. This region is followed by two conserved sequences with similarity to PKA-anchoring sites on known AKAPs (Fig. 1E). A series of experiments confirmed that recombinant full-length Ze-AKAP2 functioned as an AKAP inside cells; however, when the Ze-AKAP2 gene expression was targeted for morpholino-mediated antisense knockdown, the modified zebrafish exhibited no obvious phenotype (data not shown). There are several possible explanations for this result, including the possibility that the presence of another gene compensates for the loss of Ze-AKAP2, or this anchoring protein may not play a role in early zebrafish development. Because we saw no obvious phenotype in zebrafish, we subsequently focused on the role of MyRIP as an anchoring protein in mammalian cells.
Characterization of Mouse MyRIP as an AKAP
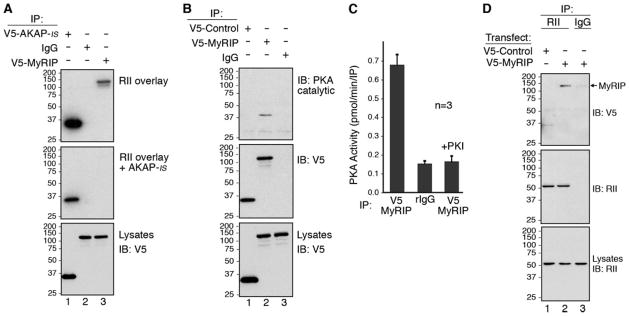
On the basis of protein sequence homology to Ze-AKAP2, we predicted that mouse MyRIP might be a PKA-anchoring protein (Fig. 1E). To test this hypothesis, a full-length mouse-coding region was cloned from a brain cDNA library by PCR, and a plasmid encoding V5-tagged MyRIP was transfected into HEK293 cells. Immune complexes were isolated using an antibody to the V5 epitope. Recombinant MyRIP interacted with PKA as assessed by RII overlay assay (Fig. 2A, top panel, lane 3). The addition of AKAP-IS blocked the RII/MyRIP interaction (Fig. 2A, middle panel, lane 3). Control immunoblots demonstrated that equal levels of each protein were expressed in the cell extracts (Fig. 2A, bottom panel). Other controls confirmed equal loading of proteins in each lane and that preincubation with a scrambled peptide control had no affect on the interaction with RII (data not shown). This result implies that MyRIP binds to RII in a manner similar to other known AKAPs.
FIGURE 2. Mammalian MyRIP is an AKAP.
A, HEK293 cells were transfected with plasmids to express V5-AKAP-IS (lane 1) or V5-MyRIP (lanes 2 and 3). Lysates were immunoprecipitated with an anti-V5 monoclonal antibody (lanes 1 and 3) or mouse IgG (lane 2). RII binding was assessed by overlay using 32P-labeled RIIα. RII overlays were performed in the absence of inhibitor peptide (top) or in the presence of 1 μM AKAP-IS anchoring inhibitor peptide (middle). Levels of V5-AKAP-IS (lane 1) or V5-MyRIP (lanes 2 and 3) in the lysates were determined by Western blot analysis using an anti-V5-horseradish peroxidase antibody (bottom). B, HEK293 cells were transfected with plasmids to express V5-Control (Scrambled AKAP-IS; lane 1) or V5-MyRIP (lanes 2 and 3). Lysates were immunoprecipitated with anti-V5 monoclonal antibody (lanes 1 and 2) or mouse IgG (lane 3) and analyzed by Western blot for the presence of PKA catalytic subunit (top) and V5-MyRIP (middle). Levels of V5-Control (lane 1) or V5-MyRIP (lanes 2 and 3) in the lysates were determined by Western blot analysis using an anti-V5-horseradish peroxidase antibody (bottom). C, HEK293 cells were transfected with plasmids to express V5-MyRIP. Lysates were immunoprecipitated with anti-V5 monoclonal antibody (columns 1 and 3) or mouse IgG (column 2). Co-purification of the PKA holoenzyme was measured as pmol/min/IP of 32P incorporation into the PKA substrate, kemptide using a filter paper binding assay (45). PKA activity was specifically blocked by the addition of 10 μM of the PKI (5–24) inhibitor peptide (column 3). n represents the amalgamated data from three independent experiments. D, HEK293 cells were transfected with plasmids to express V5-Control (Scrambled AKAP-IS; lane 1) or V5-MyRIP (lanes 2 and 3). Lysates were immunoprecipitated with a polyclonal antibody against RII (lanes 1 and 2) or rabbit IgG (lane 3) and analyzed by Western blot for the presence of V5-MyRIP (top) and RII (middle). Levels of PKA RII subunit in the lysates (lanes 1–3) were determined by Western blot analysis using an anti-RII monoclonal antibody (bottom). IB, immunoblot.
To further resolve the MyRIP/PKA interaction inside cells, a plasmid encoding V5-tagged MyRIP was transfected into HEK293 cells. V5 immune complexes were isolated, and co-precipitation of PKA was assessed by immunoblot using a polyclonal antibody against the catalytic subunit of the kinase (Fig. 2B, top panel). The catalytic subunit of PKA was detected in MyRIP immune complexes (Fig. 2B, top panel, lane 2) but not with a V5-tagged control protein or in the IgG control (Fig. 2B, top panel, lanes 1 and 3). Equal loading of proteins in each lane (Fig. 2B, middle panel, lanes 1 and 2) and equal levels of protein expression in the cell extracts (Fig. 2B, bottom panel) were confirmed by immunoblot using V5 antisera. PKA activity that co-precipitated with MyRIP was measured using the filter paper assay of Corbin and Reimann (45). PKA activity was enriched 4.3-fold ± 0.05 pmol/min/IP (n = 3) in MyRIP immune complexes when compared with IgG controls (Fig. 2C, first column), whereas kinase activity was blocked in the presence of the PKI (5–24) peptide, a specific inhibitor of the kinase (Ref. 53 and Fig. 2C, third column). In reciprocal studies, a polyclonal antibody was used to immunoprecipitate the RII subunit. Immunoblot analysis of RII immune complexes using a V5 antibody detected MyRIP associated with endogenous type II PKA (Fig. 2D, top panel, lane 2), but not with the V5-tagged control protein or in the IgG control (Fig. 2D, top panel, lanes 1 and 3). Equal loading of proteins in each lane (Fig. 2D, middle panel) and equal levels of protein expression in the cell extracts (Fig. 2D, bottom panel, lanes 1 and 2) were confirmed by immunoblot using RII antisera. Thus, we are able to conclude that recombinant murine MyRIP functions as a PKA-anchoring protein in cultured mammalian cells.
Mapping the RII-binding Sites in MyRIP
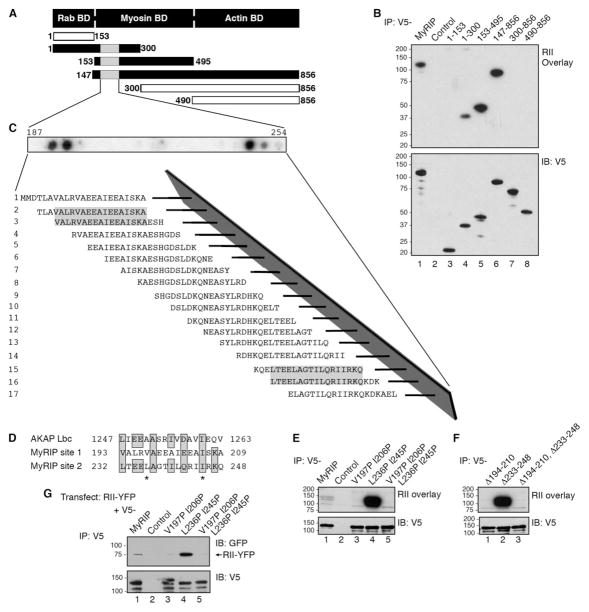
A defining characteristic of most AKAPs is a region of 14–18 residues that forms an amphipathic helix (16). The hydrophobic face of this helix binds with high affinity to the docking and dimerization domain of the R subunits (17–19). We surmised that this might also be true for MyRIP; thus six V5-tagged proteins encompassing distinct regions of MyRIP were generated (Fig. 3A). Residues 1–300, 153–495, and 147–856 of MyRIP bound RII as assessed by the overlay (Fig. 3B, top panel, lanes 4–6), suggesting that residues 153–300 were sufficient to interact with PKA. Control experiments confirmed that equal amounts of these MyRIP fragments were present in each experiment (Fig. 3B, bottom panel).
FIGURE 3. Mapping the RII-binding domain of MyRIP.
A, a schematic diagram that depicts the modular domains and fragments of MyRIP used to determine the location of the RII-binding domain. The first and last residue of each fragment is indicated. B, HEK293 cells were transfected with plasmids to express V5-MyRIP fragments. Lysates were immunoprecipitated with an anti-V5 monoclonal antibody (lanes 1 and 3– 8) or mouse IgG (lane 2). RII binding was assessed by overlay using 32P-labeled RIIα (top) and analyzed by Western blot for the presence of V5-MyRIP (bottom). C, AutoSpot peptide array was used for mapping of the RII-binding domains in MyRIP. A family of 20-mer peptides encompassing residues 187–254 were synthesized and immobilized to a membrane support. RII binding was assessed by 32P RII overlay assay, and binding was detected by autoradiography. The sequences of positive peptides are shaded. D, sequence comparison of the RII-binding domains from AKAP-Lbc with sites 1 and 2 on MyRIP. The first and last amino acid residue in each sequence is indicated. The amino acids replaced with prolines using site-directed mutagenesis are designated with stars. E, HEK293 cells were transfected with plasmids to express V5-MyRIP point mutants. Lysates were immunoprecipitated with an anti-V5 monoclonal antibody (lanes 1 and 3–5) or mouse IgG (lane 2), and RII binding was assessed by overlay using 32P-labeled RIIα (top) and analyzed by Western blot for the presence of V5-MyRIP (bottom). F, HEK293 cells were transfected with plasmids to express V5-MyRIP deletion mutants. Lysates were immunoprecipitated with an anti-V5 polyclonal antibody (lanes 1–3), and RII binding was assessed by overlay using 32P-labeled RIIα (top) and analyzed by Western blot for the presence of V5-MyRIP (bottom). G, HEK293 cells were co-transfected with plasmids to express YFP-RII and V5-MyRIP point mutants (lanes 1 and 3–5) or YFP-RII and pcDNA3.1-V5 vector (lane 2). Lysates were immunoprecipitated with a polyclonal V5 antibody and analyzed by Western blot for the presence of YFP-RII (top) and V5-MyRIP (bottom). IB, immunoblot.
Solid phase peptide array analysis was employed to more precisely map the RII-binding region of MyRIP. A family of 20-residue peptides (offset by three residues), spanning the MyRIP sequence from amino acids 187 to 254 was synthesized on a membrane support using an AutoSpot robot. RII binding was detected by the overlay assay (Fig. 3C). Interestingly, two RII-binding regions were detected that encompass amino acids 193–209 (site 1) and 232–248 (site 2) of MyRIP (Fig. 3C, shaded letters). Both regions are conserved in the zebrafish and mouse MyRIP forms (Fig. 1E, red highlighted regions) and are similar to corresponding regions on known AKAPs (Fig. 3D).
Previous studies have shown that introduction of amino acids with helix breaking side chains disrupt RII/AKAP interactions (14, 16, 39, 54). Therefore, proline residues were introduced within both putative RII-binding sites on MyRIP using a standard site-directed mutagenesis approach (Fig. 3D, letters marked with asterisks). The MyRIP V197P, I206P double mutant was unable to bind RII as assessed by the overlay assay (Fig. 3E, top panel, lane 3), suggesting that the site 1 helix contained critical determinants for PKA anchoring. In contrast, the corresponding mutations at positions 236 and 245 in site 2 increased the RII binding of MyRIP when compared with the wild-type protein (Fig. 3E, top panel, lanes 1 and 4). PKA anchoring was abolished in the MyRIP V197P, I206P, L236P, I245P quadruple mutant, where both putative PKA anchoring sites were perturbed (Fig. 3E, top panel, lane 5). Control experiments indicate that equal amounts of wild-type and mutant MyRIP proteins were present in each experiment (Fig. 3E, bottom panel).
Independent confirmation of these results was provided by analysis of MyRIP deletion mutants that lacked site 1, site 2, or both PKA-anchoring sites. Removal of residues 194–210 abolished RII binding to the MyRIP Δ194–210 mutant, whereas removal of site 2 enhanced RII binding to the MyRIP Δ233–248 mutant over controls (Fig. 3F, lanes 1 and 2). As expected, the MyRIP Δ194–210, Δ233–248 double mutant was unable to anchor PKA as assessed by the RII overlay (Fig. 3F, lane 3). Control experiments indicate that equal amounts of wild-type and mutant MyRIP proteins were present in each experiment (Fig. 3F, bottom panel).
Additional studies were conducted to further establish whether perturbation of site 2 on MyRIP produced a more efficient PKA-anchoring protein inside cells. Plasmids encoding V5-tagged MyRIP and YFP-tagged RII were co-transfected into HEK293 cells. V5 immune complexes were isolated, and co-purification of YFP-RII was assessed by immunoblot analysis using a monoclonal antibody to GFP (Fig. 3G). Yet again, more RII co-precipitated with the MyRIP L236P I245P mutant compared with the other mutants or the wild-type protein (Fig. 3G, top panel). Control experiments indicate that equal amounts of wild-type and mutant MyRIP proteins were present in all experiments (Fig. 3G, bottom panel). Collectively these studies demonstrate that the principal PKA-anchoring site on MyRIP is located between residues 194 and 210 and that residues 232–248 act in some way to negatively regulate RII binding.
Cellular Analysis of MyRIP
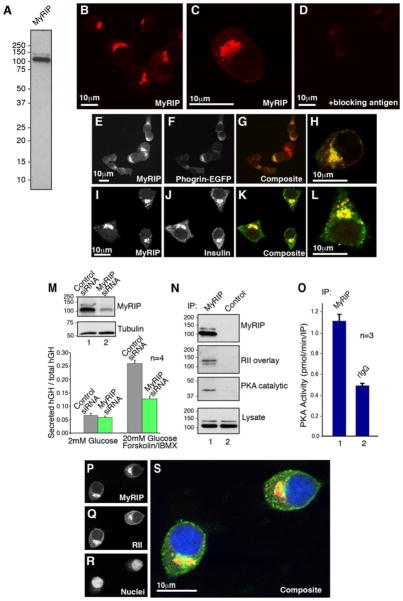
MyRIP has been previously implicated in the regulation of vesicle trafficking in a variety of cell types including insulin secretion from pancreatic β cells (55–57). To learn more about the cellular role of MyRIP, polyclonal antibodies were raised against murine MyRIP, and the affinity-purified antibody was used to screen a variety of tissues and cell lines. A protein doublet of 100 and ~125 kDa was specifically detected in each sample, including the pancreatic β-cell line INS-1(832/13) (Fig. 4A). Once the specificity of the antibody had been established, the cellular location of MyRIP was determined in INS-1 cells by immunohistochemistry (Fig. 4, B–D). MyRIP staining was asymmetrically distributed at the perinuclear regions of INS-1 cells (Fig. 4E) and partially overlapped with Phogrin-EGFP, an integral glycoprotein that is a marker for dense core secretory granules (Ref. 41 and Fig. 4F). Co-distribution of both signals is best observed in the composite images (Fig. 4, G and H). Moreover, a similar co-distribution was also seen between MyRIP and insulin (Fig. 4, I–L).
FIGURE 4. Cellular analysis of MyRIP in INS-1 cells.
A, INS-1(832/13) cell extracts were immunoblotted with affinity purified anti-MyRIP antibody. B–D, confocal immunofluorescence microscopy of INS-1 labeled with polyclonal antibodies against MyRIP (B and C) or against MyRIP in the presence of blocking peptide (D). E–H, confocal immunofluorescence microscopy of INS-1 cells transfected with a plasmid to express Phogrin-EGFP (F) and labeled with polyclonal antibodies against MyRIP (E). Composite images (G and H) indicate the cellular distribution of MyRIP (red) and Phogrin-GFP (green). I–L, confocal immunofluorescence microscopy of INS-1 labeled with polyclonal antibodies against MyRIP (I) and polyclonal antibodies against insulin (J). K and L, composite images indicates the cellular distribution of MyRIP (red) and Insulin (green). M, INS-1 cells were co-transfected with a plasmid that encodes hGH and either a MyRIP siRNA or a control siRNA. Silencing of MyRIP expression was determined by Western blot using an anti-MyRIP polyclonal antibody (inset, top). Control immunoblot showing equal expression of tubulin (inset, bottom). hGH is targeted to insulin-containing secretory granules and can be used to monitor exocytosis in a subpopulation of transfected cells (60, 85). After 72 h, the cells were treated with either 2 mM glucose or 20 mM glucose forskolin/IBMX, and exocytosis was measured by the release of hGH. n represents the amalgamated data from four independent experiments. N, INS-1 cell lysates were immunoprecipitated with anti-MyRIP polyclonal antibody and analyzed by Western blot for the presence MyRIP (top). Co-purification of MyRIP and the PKA holoenzyme was assessed by RII overlay using 32P-labeled RIIα (mid-upper) and by Western blot for the presence of PKA catalytic subunit (mid-lower). Levels of MyRIP in the lysates were determined by Western blot analysis using an anti-MyRIP polyclonal antibody (bottom). O, INS-1 lysates were immunoprecipitated with anti-MyRIP polyclonal antibody (column 1) or rabbit IgG (column 2). PKA activity (pmol/min/IP) was measured using a filter paper binding assay (45). n represents the amalgamated data from three independent experiments. P–S, confocal immunofluorescence microscopy of INS-1 cells labeled with polyclonal antibodies against (P) MyRIP, red, (Q) monoclonal antibody against RII, green, and (R) DAPI to label nuclei (blue). A composite image (S) indicates the cellular distribution of all three signals.
We have previously shown that AKAPs participate in the secretion of insulin from pancreatic β cells and related cell lines (22, 58, 59). Therefore, we wondered whether MyRIP could contribute to this process by anchoring PKA in INS-1 cells. RNA interference was used to suppress expression of the MyRIP gene in INS-1 cells (Fig. 4M, inset). Control immunoblots demonstrated that the expression of other AKAPs or PKA was not affected by the MyRIP siRNA (data not shown). Changes in insulin secretion were monitored by measuring the release of a marker protein hGH by enzyme-linked immunosorbent assay (56, 60). Suppression of MyRIP expression significantly reduced stimulation-dependent insulin released from these cells (Fig. 4M). In parallel studies, MyRIP immune complexes were isolated from INS-1 cells, and interaction with the PKA holoenzyme was confirmed by RII overlay and immunoblot using antibodies against the catalytic subunit (Fig. 4N, mid-lower panel). PKA activity was enriched 2.27-fold ±0.06 pmol/min/IP in MyRIP immune complexes when compared with IgG controls (Fig. 4O, n = 3). The co-precipitating PKA activity was selectively inhibited by the PKI 5–24 peptide (data not shown). Furthermore, indirect immunofluorescence detection of MyRIP and RII showed that the subcellular distribution of both proteins overlapped, and both signals were prominent at the perinuclear regions of INS-1 cells (Fig. 4, P–S). Taken together, these studies suggest that MyRIP functions as an AKAP and may place PKA in the vicinity of secretory granules to facilitate insulin release.
Interaction with Components of the Exocyst Complex
AKAPs are known to form anchored transduction complexes that cluster cAMP responsive enzymes with other signaling components. Likewise, a multimeric protein unit known as the exocyst complex participates in the targeting of secretory vesicles to sites of exocytosis (37). It has been shown that cAMP promotes insulin granule mobilization, and it has been suggested that phosphorylation of the exocytotic machinery by kinases such as PKA may enhance insulin granule mobilization (61). Therefore, it seemed logical to determine whether components of the exocyst complex interacted with MyRIP. Three lines of inquiry support this hypothesis.
First, the Sec8 component of the exocyst complex was detected by immunoblot in V5-MyRIP immune complexes isolated from INS-1 cells (Fig. 5A, top panel, lane 2). Sec8 was not detected in the IgG control (Fig. 5A, top panel, lane 1). Reciprocal experiments detected V5-MyRIP in Sec8 immune complexes isolated from INS-1 cells above the IgG control (Fig. 5B, top panel, lanes 1 and 2). Control immunoblots demonstrated that equal levels of each protein were expressed in the cell extracts (Fig. 5, A and B, bottom panels). Furthermore, indirect immunofluorescence detection of MyRIP and Sec8 showed that the subcellular distribution of both proteins overlapped and that both signals were prominent at the perinuclear regions of INS-1 cells (Fig. 5C).
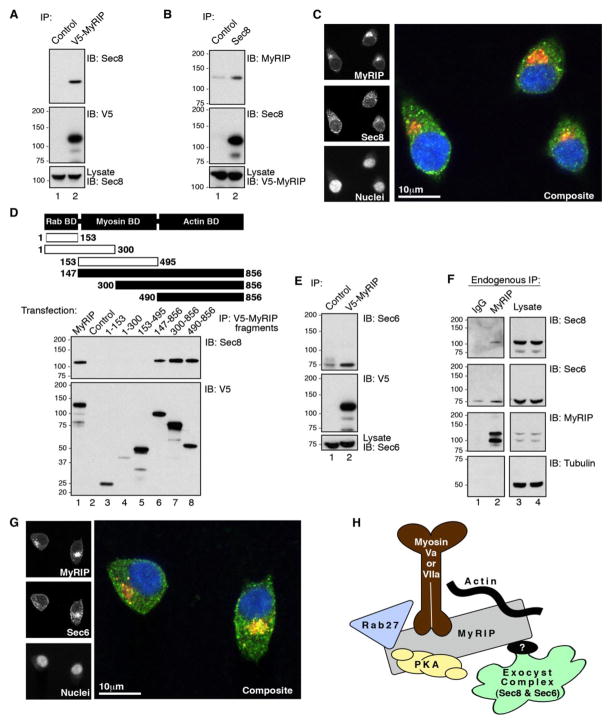
FIGURE 5. Interaction of MyRIP and exocyst components Sec6 and Sec8 in INS-1 cells.
A, INS-1 cells were transfected with plasmids to express V5-MyRIP. Lysates were immunoprecipitated with an anti-V5 monoclonal antibody (lane 2) or mouse IgG (lane 1) and analyzed by Western blot for the presence of Sec8 (top) and V5-MyRIP (middle). Levels of Sec8 in the lysates were determined by Western blot analysis using an anti-Sec8 monoclonal antibody (bottom). B, INS-1 cells were transfected with plasmids to express V5-MyRIP. Lysates were immunoprecipitated with an anti-Sec8 monoclonal antibody (lane 2) or mouse IgG (lane 1) and analyzed by Western blot for the presence of V5-MyRIP (top) and Sec8 (middle). Levels of V5-MyRIP in the lysates were determined by Western blot analysis using an anti-V5 monoclonal antibody (bottom). C, confocal immunofluorescence microscopy of INS-1 labeled with polyclonal antibodies against MyRIP (top left), monoclonal antibody against Sec8 (middle left), and DAPI (bottom left). A composite image indicates the cellular distribution of MyRIP (red), Sec8 (green), and DAPI (blue). D, a schematic diagram that depicts the modular domains and fragments of MyRIP used to determine the location of the Sec8 binding domain. The first and last residue of each fragment is indicated. INS-1 cells were transfected with plasmids to express V5-MyRIP fragments. Lysates were immunoprecipitated with an anti-V5 monoclonal antibody (lanes 1 and 3– 6) or mouse IgG (lane 2) and analyzed by Western blot for the presence of Sec8 (top) and V5-MyRIP (bottom). E, INS-1 cells were transfected with plasmids to express V5-MyRIP. Lysates were immunoprecipitated with an anti-V5 monoclonal antibody (lane 2) or mouse IgG (lane 1) and analyzed by Western blot for the presence of Sec6 (top) and MyRIP (middle). Levels of Sec6 in the lysates were determined by Western blot analysis using an anti-Sec6 monoclonal antibody (bottom). F, INS-1 lysates were immunoprecipitated with anti-MyRIP polyclonal antibody (lane 2) or rabbit IgG (lane 1). Immunoprecipitates and lysates were analyzed by Western blot for the presence endogenous Sec8 (top), endogenous Sec6 (mid-upper), endogenous MyRIP (mid-lower), and Tubulin (bottom). G, confocal immunofluorescence microscopy of INS-1 triple labeled with polyclonal antibodies against MyRIP (top left), monoclonal antibody against Sec6 (middle left), and DAPI (bottom left). A merged image indicates the cellular distribution of MyRIP (red), Sec6 (green), and DAPI (blue). H, a schematic diagram of the proposed topology of the MyRIP/Exocyst complex.
Second, mapping studies with a family of V5-tagged MyRIP fragments expressed in INS-1 cells demonstrated that residues 490–856 of the anchoring protein were sufficient to bind Sec8 (Fig. 5D, top panel, lanes 4–6). Control immunoblots demonstrated that equal levels of protein were expressed in each cell extract (Fig. 5D, bottom panel).
Third, a combination of cell-based and immunofluorescence experiments demonstrated that Sec6, another component of the exocyst complex, interacts with MyRIP. Western blot analysis demonstrated that endogenous Sec6 was detected in V5-MyRIP immune complexes isolated from INS-1 cells (Fig. 5E, top panel, lane 2) above IgG controls (Fig. 5E, top panel, lane 1). More importantly, related studies confirmed that endogenous Sec6 and Sec8 were both enriched in native MyRIP immune complexes isolated from INS-1 cells (Fig. 5F, mid-upper and mid-lower panels, lanes 1 and 2). Finally, indirect immunofluorescence detection of MyRIP and Sec6 showed that the subcellular distribution of both proteins overlapped, and both signals were prominent at the perinuclear regions of INS-1 cells (Fig. 3G). Together, these studies provide compelling evidence to support the notion that Sec8 and Sec6, two components of the exocyst complex, interact with the MyRIP scaffold.
DISCUSSION
In this report we demonstrate that MyRIP, a Rab GTPase effector protein previously implicated in the movement of secretory granules, also functions as an protein kinase A-anchoring protein. MyRIP (also known as Slac2c) belongs to a subfamily of three synaptotagmin-like proteins (Slps) that lack C2 domains (52, 62). Melanophilin/Slac2a, Slac2b, and MyRIP/Slac2c are modular proteins that each contain a conserved N-terminal domain that functions to recruit Rab proteins for a role in protein trafficking (52, 62–65). Additional features of MyRIP include a myosin Va/VII docking site and an actin-binding domain (52, 62). Our biochemical, cellular, and functional experiments may further distinguish MyRIP from other Slac2 family members by showing that it sequesters the PKA holoenzyme and interacts with components of the exocyst complex, a multimeric group of eight proteins that assist in the docking step of exocytosis (33, 37, 66). The regions of MyRIP that interact with PKA, Sec6, and Sec8 lie outside of the conserved N-terminal domain (Fig. 3A and Ref. 62); therefore, other MyRIP family members are not likely to bind PKA, Sec8, and Sec6, or if they do, it is not within the same sequence. Thus, MyRIP may position PKA and other signaling components such as Rab27a at sites of exocytosis to synchronize vesicle transport in response to second messenger and other cellular signals.
Initial evidence for the PKA anchoring function of MyRIP was provided when the zebrafish ortholog was identified in a two-hybrid screen for AKAPs using an N-terminal fragment of the Danio rerio regulatory subunit of PKA as bait. We and others have previously demonstrated that the first 45 residues of mammalian R subunits fold to form docking and dimerization domains that are sufficient for high affinity interaction with AKAPs (17–19, 48, 67). Analysis of the Rze-AKAP complex confirms that this protein-protein interaction is antagonized by anchoring inhibitor peptides such as AKAP-IS (39). This not only implies that zebrafish AKAPs interact with PKA in a manner that is reminiscent of their mammalian counterparts but also suggests that AKAP-IS and its derivatives could prove to be effective reagents that disrupt PKA anchoring in the zebrafish. Future studies are planned to use AKAP-IS as a reagent to establish the effect of disrupting PKA anchoring during zebrafish development.
A defining feature of most AKAPs is a single amphipathic helix with a hydrophobic face that forms an R subunit-binding surface. However, some anchoring proteins, such as AKAP220, AKAP350/450/cg-Nap, and BIG 2, have two or three RII subunit-binding surfaces that are distributed throughout the protein (68–71). Classically these regions are identified when amino acid residues with helix-breaking side chains, such as proline or glycine, are introduced into the hydrophobic face of the helix to abolish PKA anchoring (16, 22, 46, 72). Although MyRIP appears to be a conventional AKAP in certain respects, we have uncovered some atypical properties of this anchoring protein. Our mapping and peptide array analyses identified two adjacent PKA-anchoring domains located between residues 193 and 209 (site 1) and residues 232 and 248 (site 2) on MyRIP. Although both regions exhibit sequence similarity to a corresponding section of the prototypic AKAP Ht31 (now known as AKAP-Lbc; Ref. 73 and Fig. 3D), our combined data imply that only site 1 functions as a bona fide PKA-anchoring site. Interestingly, site 2 does not appear to be a conventional PKA-anchoring domain but instead may contribute to the functional regulation of site 1. This interpretation is based on evidence that mutation or removal of the MyRIP site 1 helix abolishes all PKA anchoring, yet R subunit binding is augmented when analogous changes are introduced within site 2 (Fig. 3, E and F). One explanation is that mutations in site 2 may induce intramolecular interactions within MyRIP that alter the context of site 1 to generate a more favorable RII-binding surface. Hence one could envisage a mechanism where protein binding or a post-translational modification of site 2 may induce partial unfolding of MyRIP to expose high affinity binding determinants within site 1. Interestingly, the intervening sequence between site 1 and site 2 on MyRIP is leucine-rich, and residues 193–222 are predicted to form a coiled-coil structure using the SMART data base, a protein secondary structure prediction algorithm (74). A similar leucine-rich structure has been detected within the PKA-anchoring domain of pericentrin, another anchoring protein that contains an extended and discontinuous RII-binding sequence (75). Thus, it is tempting to speculate that MyRIP and pericentrin form a subclass of AKAPs that utilize a nonconventional mechanism for PKA anchoring.
An intriguing finding of this report is that MyRIP serves as a scaffolding protein that recruits the exocyst complex, a molecular machine that has been implicated in the tethering of vesicles to sites of exocytosis (33, 37, 66). Recent data in pancreatic β cells suggest that the exocyst complex participates in the docking step of insulin vesicles and may facilitate their transfer from a reserve to a readily releasable pool at the plasma membrane (34). However, the exocyst complex is also detected in the perinuclear regions of other cell types where it is associated with either the Golgi or the microtubule-organizing centers (76–80). These latter reports are more compatible with our immunolocalization data in Fig. 4 (E–L), showing that the majority of MyRIP exhibits a punctate and perinuclear staining pattern that overlaps with markers for dense core secretory granules. It is worthy of note, however, that a small but significant fraction of MyRIP is uniformly distributed throughout the cytoplasm; thus some MyRIP may be complexed with the exocyst complex at the plasma membrane. Because MyRIP has already been shown to contribute to insulin secretion, our findings identify an additional step in this process that involves scaffolding to the exocyst complex. The PKA anchoring function of MyRIP may also be relevant to the control of insulin secretion because elevated intracellular cAMP promotes insulin granule mobilization and increases the size of the readily releasable pool of vesicles. Attempts to perform rescue experiments using siRNA-resistant forms of MyRIP (either that contain or lack the PKA-binding domain) have been inconclusive. The overexpression of MyRIP itself resulted in a reduction of exocytosis (data not shown). This is probably due to an excess of the scaffolding protein that segregates binding partners to promote a dominant-negative effect. In addition, we have previously shown that peptide-mediated disruption of PKA anchoring suppresses insulin secretion from pancreatic β cells and a variety of β cell-derived cell lines (22, 58, 81). Future studies will be directed toward determining whether MyRIP is the only AKAP that orchestrates insulin secretion or whether additional anchoring proteins contribute to the regulation of this multistep and complex cellular process.
Rudimentary mapping studies presented in Fig. 5D indicate that the exocyst component Sec8 binds to determinants within the C-terminal half of MyRIP. The same is likely to be true for Sec6, although it is not yet known whether the interaction between the exocyst complex and MyRIP is direct. The C-terminal region of MyRIP also encompasses an actin-binding sequence between residues 774 and 779 that is important for MyRIP interaction with the actin cytoskeleton (56). Furthermore, overexpression of the C-terminal half of MyRIP, which contains the actin-binding domain, reduces glucose-mediated insulin secretion in INS-1E cells (56, 57). This has led to the proposal that MyRIP facilitates secretory vesicle capture by tethering secretory vesicles to F-actin (62). However, in situ binding studies performed with mutants lacking the polyarginine motif suggest that this region is not required for association with the exocyst complex (data not shown). Thus, it would appear that Sec6 and/or Sec8 interact with other determinants located in the C-terminal region of MyRIP.
In conclusion, our data show that the Rab27a GTPase effector protein, MyRIP, functions as scaffolding protein that links protein kinase A to components of the exocyst complex. In recent years several reports have indicated that MyRIP also interacts with other proteins including Rab27a, F-actin, and the motor proteins myosin Va and myosin VIIa (52, 55–57, 82). A schematic diagram depicting all of the known proteins that interact with MyRIP is presented in Fig. 5H. As the number of MyRIP binding-partners increases, it seems likely that different protein combinations may modulate individual cellular processes. For example, a tripartite complex of Rab27a, MyRIP, and myosin VIIa tethers melanosomes to the actin cytoskeleton for a role in melanosome movement in retinal pigment epithelium cells (52, 83, 84). Now we propose another MyRIP complex that may include the PKA holoenzyme and the exocyst complex that may be responsible for the regulation of insulin granule mobilization or vesicle recruitment to the membrane. The only common functional element in these two examples is the anchoring protein itself, although Rab27a and myosin Va may also be in the complex. Hence cell type-specific expression of particular binding partners or the movement of components in and out of assembly points within an individual cell could conceivably drive formation of these customized molecular units.
Acknowledgments
We thank for Dr. David Ransom for the zebrafish cDNA library, Dr. Michael S. German for the pFOX.CMV/hGH construct, Dr. Guy A. Rutter for the Phogrin-EGFP construct, Dr. Scott H. Soderling for the pLexA Wave1 construct, and Dr. Wenbiao Chen for zebrafish embryos and the use of lab space and equipment. We thank Dr. Jennifer J. Carlisle Michel and colleagues in the Scott laboratory for critically evaluating the manuscript.
Footnotes
This work was supported by National Institutes of Health Grants 5 F32HL074647 (to A. S. G.) and GM48231 (to J. D. S.) and by a postdoctoral fellowship from the Canadian Institutes of Health Research (to S. A. H.).
The abbreviations used are: AKAP, protein kinase A-anchoring protein; PKA, cAMP-dependent protein kinase; RACE, rapid amplification of cDNA ends; PBS, phosphate-buffered saline; DAPI, 4′,6′-diamino-2-phenylindole; hGH, human growth hormone; siRNA, small interfering RNA; IBMX, isobutylmethylxanthine; YFP, yellow fluorescent protein; EGFP, enhanced green fluorescent protein; IP, immunoprecipitation.
References
- 1.Pawson T, Nash P. Science. 2003;300:445–452. doi: 10.1126/science.1083653. [DOI] [PubMed] [Google Scholar]
- 2.Inagaki N, Ito M, Nakano T, Inagaki M. Trends Biochem Sci. 1994;19:448–452. doi: 10.1016/0968-0004(94)90128-7. [DOI] [PubMed] [Google Scholar]
- 3.Printen JA, Brady MJ, Saltiel AR. Science. 1997;275:1475–1478. doi: 10.1126/science.275.5305.1475. [DOI] [PubMed] [Google Scholar]
- 4.Morrison DK. J Cell Sci. 2001;114:1609–1612. doi: 10.1242/jcs.114.9.1609. [DOI] [PubMed] [Google Scholar]
- 5.Hunter T. Cell. 2000;100:113–127. doi: 10.1016/s0092-8674(00)81688-8. [DOI] [PubMed] [Google Scholar]
- 6.Smith FD, Langeberg LK, Scott JD. Trends Biochem Sci. 2006;31:316–323. doi: 10.1016/j.tibs.2006.04.009. [DOI] [PubMed] [Google Scholar]
- 7.Wong W, Scott JD. Nat Rev Mol Cell Biol. 2004;5:959–971. doi: 10.1038/nrm1527. [DOI] [PubMed] [Google Scholar]
- 8.Theurkauf WE, Vallee RB. J Biol Chem. 1982;257:3284–3290. [PubMed] [Google Scholar]
- 9.Lohmann SM, DeCamili P, Enig I, Walter U. Proc Natl Acad Sci U S A. 1984;81:6723–6727. doi: 10.1073/pnas.81.21.6723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carr DW, Scott JD. Trends Biochem Sci. 1992;17:246–249. doi: 10.1016/0968-0004(92)90402-u. [DOI] [PubMed] [Google Scholar]
- 11.Bregman DB, Bhattacharyya N, Rubin CS. J Biol Chem. 1989;264:4648–4656. [PubMed] [Google Scholar]
- 12.Bregman DB, Hirsch AH, Rubin CS. J Biol Chem. 1991;266:7207–7213. [PubMed] [Google Scholar]
- 13.Coghlan VM, Langeberg LK, Fernandez A, Lamb NJC, Scott JD. J Biol Chem. 1994;269:7658–7665. [PubMed] [Google Scholar]
- 14.Lester LB, Coghlan VM, Nauert B, Scott JD. J Biol Chem. 1996;272:9460–9465. doi: 10.1074/jbc.271.16.9460. [DOI] [PubMed] [Google Scholar]
- 15.Tasken K, Aandahl EM. Physiol Rev. 2004;84:137–167. doi: 10.1152/physrev.00021.2003. [DOI] [PubMed] [Google Scholar]
- 16.Carr DW, Stofko-Hahn RE, Fraser IDC, Bishop SM, Acott TS, Brennan RG, Scott JD. J Biol Chem. 1991;266:14188–14192. [PubMed] [Google Scholar]
- 17.Newlon MG, Roy M, Morikis D, Carr DW, Westphal R, Scott JD, Jennings PA. EMBO J. 2001;20:1651–1662. doi: 10.1093/emboj/20.7.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gold MG, Lygren B, Dokurno P, Hoshi N, McConnachie G, Tasken K, Carlson CR, Scott JD, Barford D. Mol Cell. 2006;24:383–395. doi: 10.1016/j.molcel.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 19.Kinderman FS, Kim C, von Daake S, Ma Y, Pham BQ, Spraggon G, Xuong NH, Jennings PA, Taylor SS. Mol Cell. 2006;24:397–408. doi: 10.1016/j.molcel.2006.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Coghlan VM, Hausken ZE, Scott JD. Biochem Soc Trans. 1995;23:591–596. doi: 10.1042/bst0230592. [DOI] [PubMed] [Google Scholar]
- 21.Dell’Acqua ML, Faux MC, Thorburn J, Thorburn A, Scott JD. EMBO J. 1998;17:2246–2260. doi: 10.1093/emboj/17.8.2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fraser ID, Tavalin SJ, Lester LB, Langeberg LK, Westphal AM, Dean RA, Marrion NV, Scott JD. EMBO J. 1998;17:2261–2272. doi: 10.1093/emboj/17.8.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Westphal RS, Soderling SH, Alto NM, Langeberg LK, Scott JD. EMBO J. 2000;19:4589–4600. doi: 10.1093/emboj/19.17.4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alto NM, Soderling J, Scott JD. J Cell Biol. 2002;158:659–668. doi: 10.1083/jcb.200204081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klauck TM, Faux MC, Labudda K, Langeberg LK, Jaken S, Scott JD. Science. 1996;271:1589–1592. doi: 10.1126/science.271.5255.1589. [DOI] [PubMed] [Google Scholar]
- 26.Dodge K, Scott JD. FEBS Lett. 2000;476:58–61. doi: 10.1016/s0014-5793(00)01671-9. [DOI] [PubMed] [Google Scholar]
- 27.Schillace RV, Scott JD. Curr Biol. 1999;9:321–324. doi: 10.1016/s0960-9822(99)80141-9. [DOI] [PubMed] [Google Scholar]
- 28.Westphal RS, Tavalin SJ, Lin JW, Alto NM, Fraser ID, Langeberg LK, Sheng M, Scott JD. Science. 1999;285:93–96. doi: 10.1126/science.285.5424.93. [DOI] [PubMed] [Google Scholar]
- 29.Dodge KL, Khouangsathiene S, Kapiloff MS, Mouton R, Hill EV, Houslay MD, Langeberg LK, Scott JD. EMBO J. 2001;20:1921–1930. doi: 10.1093/emboj/20.8.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tasken KA, Collas P, Kemmner WA, Witczak O, Conti M, Tasken K. J Biol Chem. 2001;276:21999–22002. doi: 10.1074/jbc.C000911200. [DOI] [PubMed] [Google Scholar]
- 31.Carnegie GK, Smith FD, McConnachie G, Langeberg LK, Scott JD. Mol Cell. 2004;15:889–899. doi: 10.1016/j.molcel.2004.09.015. [DOI] [PubMed] [Google Scholar]
- 32.Dodge-Kafka KL, Soughayer J, Pare GC, Carlisle Michel JJ, Langeberg LK, Kapiloff MS, Scott JD. Nature. 2005;437:574–578. doi: 10.1038/nature03966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Novick P, Field C, Schekman R. Cell. 1980;21:205–215. doi: 10.1016/0092-8674(80)90128-2. [DOI] [PubMed] [Google Scholar]
- 34.Tsuboi T, Ravier MA, Xie H, Ewart MA, Gould GW, Baldwin SA, Rutter GA. J Biol Chem. 2005;280:25565–25570. doi: 10.1074/jbc.M501674200. [DOI] [PubMed] [Google Scholar]
- 35.Bajjalieh S. Nat Genet. 2004;36:216–217. doi: 10.1038/ng0304-216. [DOI] [PubMed] [Google Scholar]
- 36.Novick P, Zerial M. Curr Opin Cell Biol. 1997;9:496–504. doi: 10.1016/s0955-0674(97)80025-7. [DOI] [PubMed] [Google Scholar]
- 37.Munson M, Novick P. Nat Struct Mol Biol. 2006;13:577–581. doi: 10.1038/nsmb1097. [DOI] [PubMed] [Google Scholar]
- 38.Hollenberg SM, Sternglanz R, Cheng PF, Weintraub H. Mol Cell Biol. 1995;15:3813–3822. doi: 10.1128/mcb.15.7.3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alto NM, Soderling SH, Hoshi N, Langeberg LK, Fayos R, Jennings PA, Scott JD. Proc Natl Acad Sci U S A. 2003;100:4445–4450. doi: 10.1073/pnas.0330734100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goldfine ID, German MS, Tseng HC, Wang J, Bolaffi JL, Chen JW, Olson DC, Rothman SS. Nat Biotechnol. 1997;15:1378–1382. doi: 10.1038/nbt1297-1378. [DOI] [PubMed] [Google Scholar]
- 41.Pouli AE, Emmanouilidou E, Zhao C, Wasmeier C, Hutton JC, Rutter GA. Biochem J. 1998;333:193–199. doi: 10.1042/bj3330193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hohmeier HE, Mulder H, Chen G, Henkel-Rieger R, Prentki M, Newgard CB. Diabetes. 2000;49:424–430. doi: 10.2337/diabetes.49.3.424. [DOI] [PubMed] [Google Scholar]
- 43.Deleted in proof
- 44.Tegge WJ, Frank R. Methods Mol Biol. 1998;87:99–106. doi: 10.1385/0-89603-392-9:99. [DOI] [PubMed] [Google Scholar]
- 45.Corbin JD, Reimann EM. Methods Enzymol. 1974;38:287–294. doi: 10.1016/0076-6879(74)38044-5. [DOI] [PubMed] [Google Scholar]
- 46.Carr DW, Hausken ZE, Fraser ID, Stofko-Hahn RE, Scott JD. J Biol Chem. 1992;267:13376–13382. [PubMed] [Google Scholar]
- 47.Scott JD, Zoller MJ, Glaccum MB, Uhler MD, Helfman DM, McKnight GS, Krebs EG. Proc Natl Acad Sci U S A. 1987;84:5192–5196. doi: 10.1073/pnas.84.15.5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hausken ZE, Coghlan VM, Hasting CAS, Reimann EM, Scott JD. J Biol Chem. 1994;269:24245–24251. [PubMed] [Google Scholar]
- 49.Newlon MG, Roy M, Morikis D, Hausken ZE, Coghlan V, Scott JD, Jennings PA. Nat Struct Biol. 1999;6:222–227. doi: 10.1038/6663. [DOI] [PubMed] [Google Scholar]
- 50.Fukuyama T, Sueoka E, Sugio Y, Otsuka T, Niho Y, Akagi K, Kozu T. Oncogene. 2001;20:6225–6232. doi: 10.1038/sj.onc.1204794. [DOI] [PubMed] [Google Scholar]
- 51.Huang LJ, Durick K, Weiner JA, Chun J, Taylor SS. Proc Natl Acad Sci U S A. 1997;94:11184–11189. doi: 10.1073/pnas.94.21.11184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.El-Amraoui A, Schonn JS, Kussel-Andermann P, Blanchard S, Desnos C, Henry JP, Wolfrum U, Darchen F, Petit C. EMBO Rep. 2002;3:463–470. doi: 10.1093/embo-reports/kvf090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Scott JD, Fischer EH, Krebs EG. Proc Natl Acad Sci U S A. 1985;84:703–708. [Google Scholar]
- 54.Han JD, Baker NE, Rubin CS. J Biol Chem. 1997;272:26611–26619. doi: 10.1074/jbc.272.42.26611. [DOI] [PubMed] [Google Scholar]
- 55.Barral DC, Seabra MC. Pigment Cell Res. 2004;17:111–118. doi: 10.1111/j.1600-0749.2004.00138.x. [DOI] [PubMed] [Google Scholar]
- 56.Waselle L, Coppola T, Fukuda M, Iezzi M, El-Amraoui A, Petit C, Regazzi R. Mol Biol Cell. 2003;14:4103–4113. doi: 10.1091/mbc.E03-01-0022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ivarsson R, Jing X, Waselle L, Regazzi R, Renstrom E. Traffic. 2005;6:1027–1035. doi: 10.1111/j.1600-0854.2005.00342.x. [DOI] [PubMed] [Google Scholar]
- 58.Lester LB, Langeberg LK, Scott JD. Proc Natl Acad Sci U S A. 1997;94:14942–14947. doi: 10.1073/pnas.94.26.14942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lester LB, Faux MC, Nauert JB, Scott JD. Endocrinology. 2001;142:1218–1227. doi: 10.1210/endo.142.3.8023. [DOI] [PubMed] [Google Scholar]
- 60.Coppola T, Perret-Menoud V, Luthi S, Farnsworth CC, Glomset JA, Regazzi R. EMBO J. 1999;18:5885–5891. doi: 10.1093/emboj/18.21.5885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hisatomi M, Hidaka H, Niki I. Endocrinology. 1996;137:4644–4649. doi: 10.1210/endo.137.11.8895328. [DOI] [PubMed] [Google Scholar]
- 62.Fukuda M, Kuroda TS. J Biol Chem. 2002;277:43096–43103. doi: 10.1074/jbc.M203862200. [DOI] [PubMed] [Google Scholar]
- 63.Fukuda M, Saegusa C, Mikoshiba K. Biochem Biophys Res Commun. 2001;283:513–519. doi: 10.1006/bbrc.2001.4803. [DOI] [PubMed] [Google Scholar]
- 64.Kuroda TS, Fukuda M, Ariga H, Mikoshiba K. J Biol Chem. 2002;277:9212–9218. doi: 10.1074/jbc.M112414200. [DOI] [PubMed] [Google Scholar]
- 65.Matesic LE, Yip R, Reuss AE, Swing DA, O’Sullivan TN, Fletcher CF, Copeland NG, Jenkins NA. Proc Natl Acad Sci U S A. 2001;98:10238–10243. doi: 10.1073/pnas.181336698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lee MC, Miller EA, Goldberg J, Orci L, Schekman R. Annu Rev Cell Dev Biol. 2004;20:87–123. doi: 10.1146/annurev.cellbio.20.010403.105307. [DOI] [PubMed] [Google Scholar]
- 67.Newlon MG, Roy M, Hausken ZE, Scott JD, Jennings PA. J Biol Chem. 1997;272:23637–23644. doi: 10.1074/jbc.272.38.23637. [DOI] [PubMed] [Google Scholar]
- 68.Reinton N, Collas P, Haugen TB, Skalhegg BS, Hansson V, Jahnsen T, Tasken K. Dev Biol. 2000;223:194–204. doi: 10.1006/dbio.2000.9725. [DOI] [PubMed] [Google Scholar]
- 69.Witczak O, Skalhegg BS, Keryer G, Bornens M, Tasken K, Jahnsen T, Orstavik S. EMBO J. 1999;18:1858–1868. doi: 10.1093/emboj/18.7.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Takahashi M, Mukai H, Oishi K, Isagawa T, Ono Y. J Biol Chem. 2000;275:34592–34596. doi: 10.1074/jbc.M005285200. [DOI] [PubMed] [Google Scholar]
- 71.Li H, Adamik R, Pacheco-Rodriguez G, Moss J, Vaughan M. Proc Natl Acad Sci U S A. 2003;100:1627–1632. doi: 10.1073/pnas.0337678100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hoshi N, Langeberg LK, Scott JD. Nat Cell Biol. 2005;7:1066–1073. doi: 10.1038/ncb1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Diviani D, Soderling J, Scott JD. J Biol Chem. 2001;276:44247–44257. doi: 10.1074/jbc.M106629200. [DOI] [PubMed] [Google Scholar]
- 74.Letunic I, Copley RR, Pils B, Pinkert S, Schultz J, Bork P. Nucleic Acids Res. 2006;34:D257–D260. doi: 10.1093/nar/gkj079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Diviani D, Langeberg LK, Doxsey SJ, Scott JD. Curr Biol. 2000;10:417–420. doi: 10.1016/s0960-9822(00)00422-x. [DOI] [PubMed] [Google Scholar]
- 76.Vega IE, Hsu SC. J Neurosci. 2001;21:3839–3848. doi: 10.1523/JNEUROSCI.21-11-03839.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Vega IE, Hsu SC. Neuroreport. 2003;14:31–37. doi: 10.1097/00001756-200301200-00006. [DOI] [PubMed] [Google Scholar]
- 78.Barral Y, Mermall V, Mooseker MS, Snyder M. Mol Cell. 2000;5:841–851. doi: 10.1016/s1097-2765(00)80324-x. [DOI] [PubMed] [Google Scholar]
- 79.Shin DM, Zhao XS, Zeng W, Mozhayeva M, Muallem S. J Cell Biol. 2000;150:1101–1112. doi: 10.1083/jcb.150.5.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yeaman C, Grindstaff KK, Wright JR, Nelson WJ. J Cell Biol. 2001;155:593–604. doi: 10.1083/jcb.200107088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lester LB, Scott JD. Recent Prog Horm Res. 1997;52:409–430. [PubMed] [Google Scholar]
- 82.Desnos C, Schonn JS, Huet S, Tran VS, El-Amraoui A, Raposo G, Fanget I, Chapuis C, Menasche G, de Saint Basile G, Petit C, Cribier S, Henry JP, Darchen F. J Cell Biol. 2003;163:559–570. doi: 10.1083/jcb.200302157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lopes VS, Ramalho JS, Owen DM, Karl MO, Strauss O, Futter CE, Seabra MC. Traffic. 2007;8:486–499. doi: 10.1111/j.1600-0854.2007.00548.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kuroda TS, Fukuda M. J Biol Chem. 2005;280:28015–28022. doi: 10.1074/jbc.M501465200. [DOI] [PubMed] [Google Scholar]
- 85.Iezzi M, Escher G, Meda P, Charollais A, Baldini G, Darchen F, Wollheim CB, Regazzi R. Mol Endocrinol. 1999;13:202–212. doi: 10.1210/mend.13.2.0228. [DOI] [PubMed] [Google Scholar]