Abstract
Wnt/β-catenin is a neuroprotective pathway regulating cell fate commitment in the CNS and many vital functions of neurons and glia. Its dysregulation is linked to a number of neurodegenerative diseases. Wnt/β-catenin is also a repressor of HIV transcription in multiple cell types, including astrocytes, which are dysregulated in HIV-associated neurocognitive disorder. Given that HIV proteins can overcome host restriction factors and that perturbations of Wnt/β-catenin signaling can compromise astrocyte function, we evaluated the impact of HIV transactivator of transcription (Tat) on Wnt/β-catenin signaling in astrocytes. HIV clade B Tat, in primary progenitor-derived astrocytes and U87MG cells, inhibited Wnt/β-catenin signaling as demonstrated by its inhibition of active β-catenin, TOPflash reporter activity, and Axin-2 (a downstream target of Wnt/β-catenin signaling). Point mutations in either the core region (K41A) or the cysteine-rich region (C30G) of Tat abrogated its ability to inhibit β-catenin signaling. Clade C Tat, which lacks the dicysteine motif, did not alter β-catenin signaling, confirming that the dicysteine motif is critical for Tat inhibition of β-catenin signaling. Tat coprecipitated with TCF-4 (a transcription factor that partners with β-catenin), suggesting a physical interaction between these two proteins. Furthermore, knockdown of β-catenin or TCF-4 enhanced docking of Tat at the TAR region of the HIV long terminal repeat. These findings highlight a bidirectional interference between Tat and Wnt/β-catenin that negatively impacts their cognate target genes. The consequences of this interaction include alleviation of Wnt/β-catenin-mediated suppression of HIV and possible astrocyte dysregulation contributing to HIV neuropathogenesis.
Introduction
HIV enters the brain during acute infection as infected leukocytes cross the blood–brain barrier and seed the CNS with virus (Fox, 2008; Valcour et al., 2012). Even with effective combined antiretroviral therapy, it is estimated that ≥50% of HIV-infected individuals experience some degree of HIV-associated neurocognitive disorder (HAND) (Heaton et al., 2010). Defining cellular and molecular mechanisms driving HIV-mediated neuropathogenesis are vital in understanding the basis for this comorbidity and devising novel strategies targeting HIV in the CNS.
Astrocytes are infected by HIV and likely represent a significant CNS viral reservoir. HIV DNA is detected in astrocytes at a frequency that depends on their proximity to perivascular macrophages and severity of HAND (Churchill et al., 2009), although productive replication is restricted by multiple mechanisms (Li et al., 2002; Ong et al., 2005; Zhang et al., 2005). We identified the Wnt/β-catenin pathway as a potent repressor of HIV replication in astrocytes, specifically through the action of downstream effectors TCF-4 and β-catenin (Carroll-Anzinger et al., 2007; Henderson et al., 2012; Narasipura et al., 2012), and reported that inflammatory mediators such as IFN-γ that downregulate Wnt/β-catenin signaling promote HIV productive replication in astrocytes (Li et al., 2011).
The HIV transactivator of transcription (Tat) is vital for efficient transcription. Without Tat, HIV replication is repressed due to repressive chromatin architecture as well as a defect in transcription elongation. Tat induces chromatin remodeling at the HIV promoter and recruits a positive elongation complex (pTEFb) that phosphorylates RNA polymerase II, allowing for efficient transcription. Tat can be detected in the serum of HIV-infected individuals in nanogram ranges (Westendorp et al., 1995), despite the instability and relatively short half-life of Tat in culture (Passiatore et al., 2009). Local concentrations of Tat may be considerably higher, particularly in compartments such as the CNS where there is evidence for chronic, low-level HIV replication that drives inflammation and production of neurotoxic viral proteins (Pomerantz, 2003; González-Scarano and Martín-García, 2005).
HIV has evolved multiple mechanisms to evade host restriction factors to enhance viral release (Douglas et al., 2009), evade CD8+ T-cell responses (Blagoveshchenskaya et al., 2002), prevent undesirable mutations (Sheehy et al., 2002), or inhibit interferon responses (Yan et al., 2010). We determined whether HIV employs a similar strategy to counteract inhibition by β-catenin/TCF-4. We focused on Tat because (1) Tat enhances activity of GSK3β, which would likely disrupt β-catenin signaling (Maggirwar et al., 1999; Sui et al., 2006); (2) Tat is secreted and is internalized by noninfected cells (Helland et al., 1991; Marcuzzi et al., 1992; Ensoli et al., 1993; Debaisieux et al., 2012); and (3) Tat has a well established role in promoting HIV neuropathogenesis by enhancing oxidative stress in target cells and inducing gliosis in astrocytes, contributing to neurodegeneration.
Materials and Methods
Cell culture.
U87MG and U251MG astroglioma cell lines were obtained from the National Institutes of Health AIDS Research and Reference Reagent Program and the American Type Culture Collection, respectively. They were propagated in DMEM (Invitrogen) supplemented with 10% heat-inactivated fetal bovine serum (Sigma-Aldrich) and 1% penicillin–streptomycin (Invitrogen) in 5% CO2 humidified atmosphere at 37°C. Progenitor-derived astrocytes (PDAs) were generated from neural progenitor cells as previously described (Lamba et al., 2009). Briefly, progenitor cells (provided by Dr. Eugene Major, National Institute of Neurological Disorders and Stroke–National Institutes of Health, Bethesda, MD) were seeded on poly-d-lysine-coated T-75 tissue culture flasks at 2 × 106 cells/flask and maintained in progenitor medium consisting of Neurobasal media (Invitrogen) supplemented with 0.5% bovine albumin (Sigma-Aldrich), NSF (neurosurvival factor) (Lonza), N2 components (Invitrogen), 25 ng/ml bFGF (fibroblast growth factor), 20 ng/ml EGF (epidermal growth factor) (R&D Systems), 50 μg/ml gentamicin (Lonza), and 2 mm l-glutamine (Invitrogen). To induce differentiation, progenitor medium was replaced with PDA medium containing DMEM supplemented with 10% heat-inactivated fetal bovine serum, 2 mm l-glutamine, and 50 μg/ml gentamicin. Cultures were >90% positive for glial fibrillary acidic protein (GFAP) after 30 d of differentiation. Both adherent primary cells and cell lines were removed by treatment with 1 mm EDTA for 5 min with gentle scraping or pipetting multiple times.
Flow cytometry.
Intracellular staining of cells to detect β-catenin by flow cytometry was performed as previously described (Li et al., 2011), using an antibody that specifically detects ABC (active β-catenin) that is not phosphorylated at Ser37 and Thr41 (US Biological). Data were acquired in an LSR II flow cytometer (BD Biosciences) and analyzed with FlowJo software (Tree Star).
Plasmid transfections.
Transfections were performed with indicated plasmids using TransIT-LT1 reagent as per manufacturer's instruction (Mirus Bio LLC). Cells were ∼60–70% confluent at the time of transfections. The TOPflash construct (Millipore) to detect Wnt/β-catenin signaling contains multiple TCF/LEF binding sites tied to a luciferase reporter. Renilla construct is an internal control for transfection efficiency and contains cDNA encoding Renilla luciferase under the control of a CMV promoter for constitutive expression. pCDNA3.1 vector (Invitrogen) was included throughout to equalize DNA added per transfection condition. Tat expression constructs Tat101 (Addgene plasmid 14654), TatK41A (Addgene plasmid 14665), and TatC30G (Addgene plasmid 14656) were provided courtesy of Dr. Matija Peterlin (University of California, San Francisco, San Francisco, CA). The following reagents were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases–National Institutes of Health: pSV2tat72 (Tat72) from Dr. Alan Frankel (University of California, San Francisco, San Francisco, CA); and pCTatBL43.CS (wild-type clade C) from Dr. Udaykumar Ranga (Jawaharlal Nehru Centre for Advanced Scientific Research, Jakkur, Bangalore, India). The TatΔ49–59 has been previously described (Li et al., 2008) and was a kind gift from Dr. Avindra Nath (National Institute of Neurological Disorders and Stroke–National Institutes of Health, Bethesda, MD).
Dual luciferase reporter assay.
Twenty-four hours after transfection, culture medium was removed, cells were gently washed with PBS once, and 100 μl of passive lysis buffer was added and incubated at 37°C for 10–12 min. Cells were lysed by pipetting up and down several times and spun at 5000 rpm for 4 min to remove debris, and 10–20 μl was used to assay for luciferase activity using dual luciferase reporter assay (Promega) in a single injector luminometer. Total protein concentration was estimated using Pierce BCA protein assay kit (Thermo Fisher Scientific). Relative light units were normalized to micrograms/milliliter protein or to cotransfected Renilla luciferase, as indicated. Graphs were plotted from data obtained as a mean of three independent experiments with SD as error bars.
Protein immunoprecipitation and Western blot.
For standard Western blot (WB), PDAs were treated with recombinant Tat (rTat) (10 ng/ml) for 12 h, then lysed using standard RIPA buffer and prepared for Western blot using antibody directed against active (hypophosphorylated) β-catenin (Sigma-Aldrich). For immunoprecipitation, PDA cultures were first cross-linked with 1.0% formaldehyde (Sigma-Aldrich) to preserve protein–protein interactions, and then lysed with RIPA buffer. Lysates were then precleared by incubation with magnetic A/G beads (Thermo Fisher Scientific) and a nontargeting mouse IgG (Cell Signaling) for 1 h. Lysates were then incubated with magnetic A/G beads and with either IgG control or antibody against hemagglutinin (HA) tag (Abcam) overnight at 4°C with rotation. Beads were washed extensively with TBS supplemented with 2 m urea, followed by elution with low pH buffer, pH 2.9 (GE Healthcare). Additionally, to minimize nonspecific interference from detection of antibody heavy chain in Western blotting, HA tag antibodies and their cognate control antibodies were cross-linked to magnetic A/G beads by incubation with 200 mm triethanolamine (GE Healthcare) containing 50 mm DMP (dimethyl pimelimidate dihydrochloride) (Sigma-Aldrich) before immunoprecipitation. For Western blotting, lysates or immunoprecipitated samples were separated on a 10% SDS-PAGE gel, transferred to a nitrocellulose membrane, blocked with Superblock (Thermo Fisher Scientific) containing 0.1% Tween 20 (T20) for 1 h, incubated with the indicated antibody overnight at 4°C (total β-catenin; Sigma-Aldrich; 1:5000; TCF-4; Cell Signaling; 1:1000) in Superblock–0.1% T20. Membranes were washed extensively with Tris-buffered saline with Tween 20 (TBST) and incubated with secondary antibody conjugated to horseradish peroxidase (HRP) (1:50,000 in Superblock–0.1% Tween 20) for 45 min at room temperature. Membranes were again washed extensively in TBST and developed with SuperSignal West Femto maximum sensitivity substrate according to manufacturer's instructions (Thermo Fisher Scientific). WB for GFAP was performed using mouse monoclonal anti-GFAP antibody (Cell Signaling) and secondary HRP-linked anti-mouse antibody (Thermo Fisher Scientific). EAAT2/GLT-1 WB used a rabbit anti-human EAAT2 antibody from Cell Signaling. Glutamine synthetase WB used rabbit anti-glutamine synthetase antibody (Thermo Fisher Scientific) and secondary HRP-linked anti-rabbit antibody (Cell Signaling).
Glutamate uptake assay.
Glutamate uptake in PDAs was measured using a glutamate assay kit purchased from BioVision. Briefly, PDAs were treated with the GLT-1 inhibitor dl-TBOA (Tocris Bioscience) at 100 μm or with vehicle (DMSO). At 1 h after treatment, PDAs were spiked with glutamate at a final concentration of 1.2 mm. At 10 min after glutamate addition, 10 ml of supernatant was used to measure the amount of glutamate using a colorimetric and spectrophotometry at λ = 450 nm method.
Quantitative real-time PCR and RT-PCR.
RNA was isolated from PDAs using the RNeasy Mini kit (QIAGEN). Subsequently, cDNA was synthesized using Quantitect reverse transcription kit (QIAGEN). Real-time RT-PCR was performed using a Quantitect SYBR Green PCR kit (QIAGEN) in a 7500 Real-Time PCR System (Applied Biosystems) using 7500 software, version 2.0.1. Melting-curve analysis was performed to ensure the amplification of a single product. Primers used were as follows: Axin2-F, 5′-ACAACAGCATTGTCTCCAAGCAGC, and Axin2-R, 5′-GCGCCTGGTCAAACATGATGGAAT; and GAPDH-F, 5′-CTTCAACGACCACTTTGT, and GAPDH-R, 5′-TGGTCCAGGGGTCTTACT. For chromatin immunoprecipitation, primers used were TAR1-F, 5′-AGCTTTCTACAAGGGACTTTCCGC, and TAR1-R, 5′-ATTGAGGCTTAAGCAGTGGGTTCC. Fold change was calculated by relative quantification using the comparative CT method with GAPDH (RT-PCR) or nontargeting IgG (ChIP) as control. Relative quantification (RQ) = 2−ΔΔCt, where ΔCt = CtTarget − CtControl, and ΔΔCt = ΔCttreated − ΔCtuntreated.
Chromatin immunoprecipitation assay.
Chromatin immunoprecipitation (ChIP) assays were performed from U87MG astrocytoma cells using the Magna ChIP A/G kit (Millipore). Cells were seeded to 60–70% confluency, treated as described in the figure legends, and then processed for ChIP beginning with cross-linking proteins to DNA by 1.0% formaldehyde. Chromatin was sonicated six times for 10 s each, generating DNA fragments of ∼500–1000 bp. The sonicated supernatants containing the DNA were diluted with ChIP dilution buffer (0.01% SDS, 1% Triton X-100, 1.2 mm EDTA, 16.7 mm Tris-HCl, pH 8.1, and 167 mm NaCl) to a total volume of 500 μl plus 2.5 μl of protease inhibitor mixture, and precleared by rotating for 1 h at 4°C with magnetic protein A/G beads and isotype control antibody (Cell Signaling). The lysate was transferred to a fresh tube. Supernatant (5 μl) was reserved for input, and then 5 μg of control IgG or anti-HA tag were added to the reaction mixture with magnetic protein A/G beads. After extensive washes with low salt, high salt, LiCl, and TE buffers, the immune complexes were treated with ChIP elution buffer (NaCl, SDS, Tris hydrochloride, and proteinase K) at 62°C for 2 h, followed by 10 min at 95°C to reverse cross-linking. Afterward, DNA was precipitated with ethanol solution and isolated by spin column purification. Purified DNA was then amplified by real-time PCR using primer pairs that flank the transcription initiation site of the HIV-1 long terminal repeat (LTR), as described above. Data were analyzed according to the comparative CT method and were normalized to IgG control and reported as fold change in binding relative to control siRNA.
Statistical analysis.
Descriptive statistics and graphical analyses were used as appropriate. Group means were compared by ANOVA and post hoc tests, when the data were distributed normally. When the data were not normally distributed, nonparametric analysis was performed. All tests assumed a two-sided significance level of 0.05.
Results
HIV-1 Tat diminishes β-catenin signaling
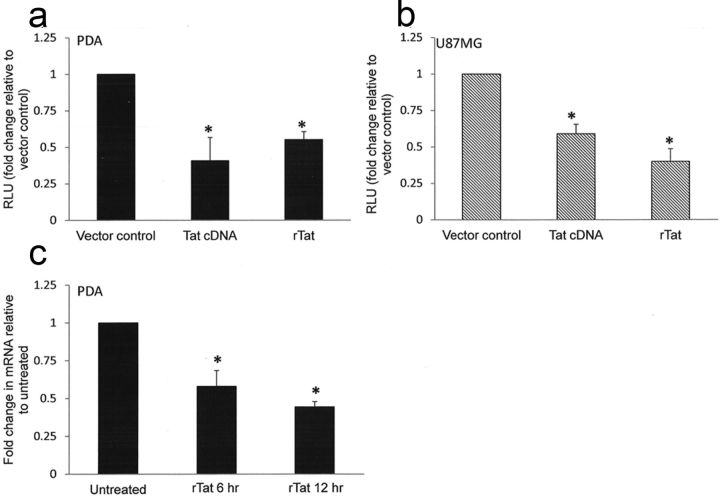
We previously demonstrated that astrocytes exhibit robust β-catenin signaling, which mediates inhibition of HIV transcription (Carroll-Anzinger et al., 2007; Li et al., 2011; Henderson et al., 2012; Narasipura et al., 2012). To determine whether HIV has evolved a microbial adaptation mechanism to counteract the effects of β-catenin/TCF-4 on its replication, we evaluated the impact of HIV Tat on β-catenin signaling. We monitored β-catenin-dependent signaling using TOPflash construct which contains multiple TCF/LEF binding sites linked to minimal thymidine kinase (TK) promoter and luciferase gene. We performed these experiments in PDAs as well as astrocytic cell lines (U87MG, U251MG) to show that results are consistent between cell types and are not limited to fetal astrocytes. PDAs exhibit prototypical characteristics of astrocytes including expression of GFAP, glutamate transporter 1/EAAT2, glutamate synthase, and are capable of glutamate uptake in a standard glutamate uptake assay (Fig. 1a–d). Transfection with a Tat expression plasmid encoding full-length (101 aa) clade B Tat reduced the activity of the β-catenin reporter TOPflash by 59% in PDA (Fig. 2a) and 41% in the astrocytoma cell line U87MG (Fig. 2b). Similarly, treatment with rTat protein (86 aa form) reduced TOPflash activity by 45% in PDA (Fig. 2a) and 60% in U87MG (Fig. 2b). To confirm that Tat reduces canonical Wnt signaling, PDAs were treated with rTat for 1–48 h, and then RNA was isolated to detect mRNA levels of Wnt target gene Axin-2. As shown in Figure 2c, rTat significantly decreased axin2 mRNA at 6 and 12 h after treatment in PDAs. These results indicate that both endogenously expressed and exogenously added HIV Tat can downregulate β-catenin-dependent signaling in primary progenitor-derived astrocytes and in an astrocytic cell line.
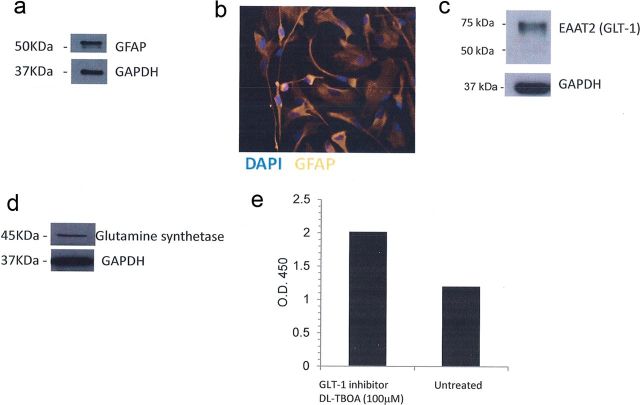
Figure 1.
Prototypical characteristics of astrocytes exhibited by human PDAs. Expression of GFAP was evaluated by Western blot (a) and immunofluorescence (b), EAAT2/GLT-1 (c), and glutamate synthetase (d) was evaluated in PDAs by Western blot. GAPDH was used as a loading control. In e, glutamate uptake in PDAs was measured using a glutamate assay from BioVision (Milpitas, CA) with or without the addition of a GLT-1 inhibitor, dl-TBOA, at 100 μm.
Figure 2.
Tat inhibits β-catenin-dependent signaling in astrocytes. Primary PDAs (a) or U87MG (b) were transfected with the Wnt/β-catenin reporter TOPflash with either vector control (pcDNA3.1) or a plasmid encoding full-length Tat (Tat cDNA) overnight. Some cells received vector control followed by treatment with rTat overnight. Renilla luciferase was included in all conditions as an internal control for transfection efficiency. Cells were then lysed and dual luciferase assay performed. Results are normalized to Renilla luciferase and are reported as fold change in relative light units (RLU) compared with vector control. All TOPflash experiments were also performed with the negative control FOPflash, which contains mutated TCF/LEF binding sites. FOPflash luciferase readings did not exceed pcDNA background levels. c, PDAs were treated with rTat (10 ng/ml) or were left untreated for 6 or 12 h, followed by RNA extraction, conversion to cDNA and quantitative RT-PCR to detect axin2 levels. Results are reported as fold change in mRNA expression compared with untreated control. Data represent at least three experiments. The asterisks denote p < 0.05 relative to vector control as measured by two-tailed Student's t test. Error bars indicate SD.
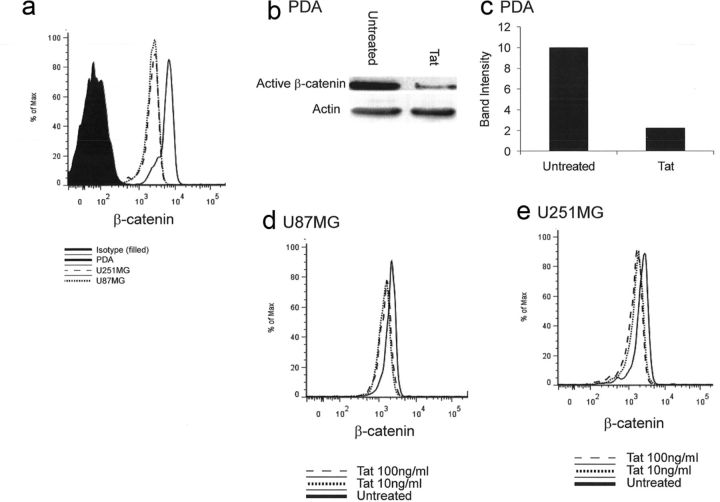
Astrocytes exhibit robust expression of the central mediator of this pathway, β-catenin (Fig. 3a). To determine the effect of Tat on β-catenin, we monitored active β-catenin level by flow cytometry in response to Tat treatment. Treatment with recombinant Tat (clade B, 86 aa form) at 10 ng/ml was sufficient to diminish active β-catenin by 20% in two astrocytoma cell lines, U87MG (Fig. 3d) and U251MG (Fig. 3e). Due to the difficulty of preparing primary astrocytes for flow cytometry, PDA β-catenin levels were evaluated by Western blot. β-Catenin levels were significantly reduced in Tat-treated PDA (Fig. 3b,c).
Figure 3.
Tat diminishes active β-catenin in astrocytes. a, U87MG, U251MG, or primary PDAs were cultured for 24 h, and then subjected to flow cytometry to detect active β-catenin. b, PDAs were treated with rTat (10 ng/ml) for 12 h, and then lysed and prepared for Western blot using antibodies directed against active β-catenin or actin. c, Densitometry of Western blot detected in b normalized to actin loading control. d, e, U87MG (d) or U251MG (e) were left untreated or treated with rTat at 10 or 100 ng/ml for 12 h. Cells were then subjected to flow cytometry to detect active β-catenin levels. Representative examples of at least three experiments are shown.
Tat effects on β-catenin signaling are independent of Tat exon 2 and its basic domain
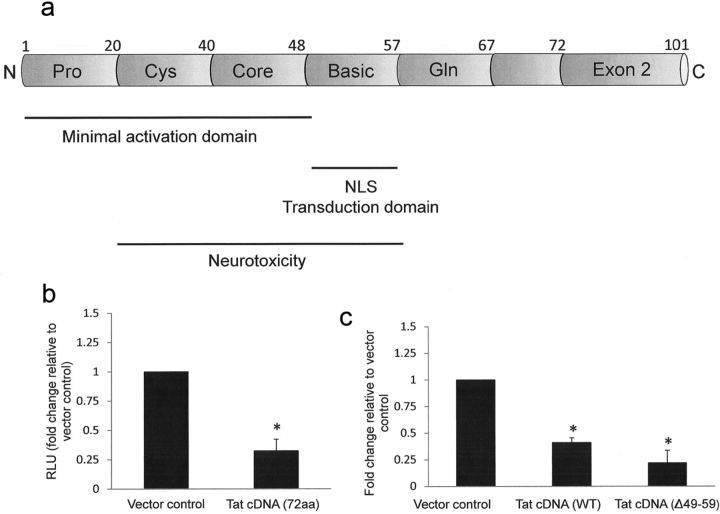
Tat exists in multiple forms, including a full-length (101 aa) form translated from two exons, a 72 aa splice variant that is encoded by the first exon, and an 86 aa form produced by certain laboratory passaged HIV strains. Tat length can vary depending on the HIV isolate (Li et al., 2009). Additionally, intracellular Tat is cleaved at the C terminus by calpains (Passiatore et al., 2009). The functional domains of Tat are summarized in Figure 4a. Within exon 1 (1–48 aa) is the minimal HIV transactivation domain, which promotes Tat binding to the transactivation-responsive (TAR) region to accelerate the rate of HIV transcription. Translocation of Tat into the nucleus is mediated by the nuclear localization signal (NLS) located within the basic domain (amino acids 49–59). The second exon of Tat (amino acids 73–101) contributes to optimal transactivation (Li et al., 2009) and may be involved in internalization (Ma and Nath, 1997), but its other functions are largely undefined. To identify domain(s) of Tat that are involved in downregulation of β-catenin signaling, we transfected PDA with TOPflash reporter with or without an expression plasmid encoding the 72 aa form of Tat. Tat (72 aa) reduced β-catenin dependent signaling by ∼67% in PDA (Fig. 4b), indicating that residues 73–101 are not involved in the ability of Tat to downregulate β-catenin signaling. We next sought to determine the impact of Tat localization on its ability to downregulate β-catenin signaling. Evidence from several groups indicates that the basic region of Tat (amino acids 48–59) is vital for nuclear import, and that mutation or deletion of arginine residues within this domain results in cytoplasmic localization of Tat (Hauber et al., 1989; Orsini and Debouck, 1996; Meredith et al., 2009). Transfection of PDA with a Tat construct deleted in the basic domain (Tat cDNA Δ48–56) reduced TOPflash activity by ∼78% (Fig. 4c), indicating that the basic domain is not required for Tat-mediated inhibition of β-catenin signaling.
Figure 4.
Tat effects on β-catenin are independent of exon 2 and the basic region. a, Structure of HIV-1 Tat showing functional domains. Pro, Proline-rich; Cys, cysteine-rich; Gln, glycine-rich; NLS, nuclear localization sequence. b, PDAs were transfected with TOPflash reporter with either vector control (pcDNA3.1) or a plasmid encoding the 72 aa form of Tat. Twelve hours later, cells were lysed, and dual luciferase assay was performed as in Figure 2. Results are reported as relative light units (RLU) normalized to Renilla luciferase. c, PDAs were cotransfected with TOPflash reporter and the indicated control or Tat construct (full-length 101 aa or Δ49–59). Twenty-four hours later, cells were lysed, and dual luciferase activity was measured. Results in b are reported as relative light units (RLU) normalized to micrograms protein. Data represent at least three experiments. The asterisks denote p < 0.05 relative to vector control as measured by two-tailed Student's t test. Error bars indicate SD.
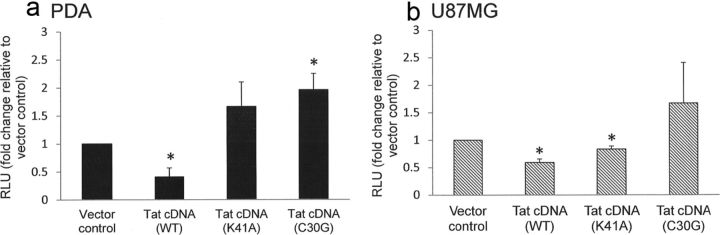
The ability of Tat to downregulate β-catenin signaling is dependent on its intact core and cysteine-rich domains
We next sought to evaluate whether the core region (amino acids 38–48) or cysteine-rich region (amino acids 22–37) of Tat are involved in inhibition of β-catenin signaling. Introducing a point mutation into the core domain (K41A) or a mutation at Cys30 to glycine (C30G) abrogated the ability of Tat to downregulate β-catenin signaling in PDAs (Fig. 5a). The Tat C30G construct enhanced β-catenin activity by approximately twofold in PDAs (Fig. 5a). While Tat C30G also abrogated Tat inhibition of β-catenin signaling in U87MG, it did not increase TOPflash activity above baseline (Fig. 5b). The K41A Tat construct reduced TOPflash activity in U87MG by 16.6% (Fig. 5b), which, while statistically significant, is unlikely to be biologically relevant. These findings suggest that there are differences in domain requirements for Tat-mediated downregulation of β-catenin between cell lines and primary cells. Nonetheless, these data demonstrate that the intact core and cysteine-rich domains of Tat are important for its ability to inhibit β-catenin signaling in primary progenitor derived astrocytes.
Figure 5.
Downregulation of β-catenin signaling by Tat requires intact core and cysteine-rich domains. PDAs (a) or U87MG (b) were cotransfected with TOPflash reporter and the indicated control or Tat construct. Twelve hours later, cells were lysed and dual luciferase assay performed as in Figure 2. Results are reported as relative light units (RLU) normalized to Renilla luciferase. Data represent at least three experiments. The asterisks denote p < 0.05 relative to vector control as measured by two-tailed Student's t test. Error bars indicate SD.
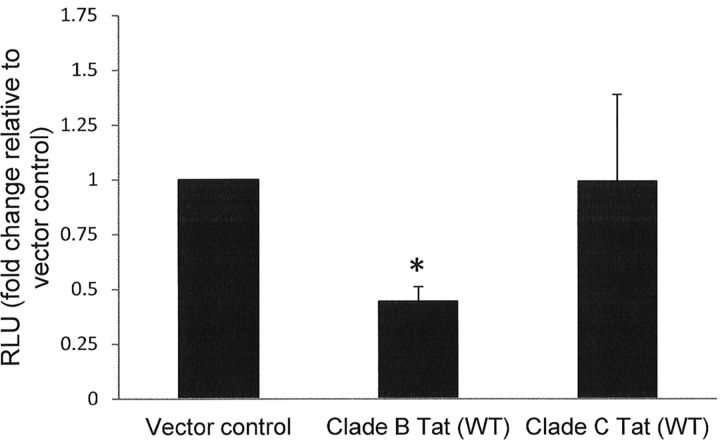
Tat from HIV clade C does not downregulate β-catenin signaling
Clade B Tat contains a dicysteine motif (C30C31) at positions 30–31 that is absent in clade C (C30S31) but is highly conserved in non-C clades. We observed that a point mutation at position 30 abolished the ability of Tat to inhibit β-catenin signaling (Fig. 5). We next evaluated whether clade C Tat, which contains a cysteine residue at position 30 only, inhibits β-catenin signaling. Transfection of PDA with a Tat expression plasmid derived from clade C did not reduce β-catenin signaling (Fig. 6), suggesting that the dicysteine motif is important for inhibition of β-catenin signaling.
Figure 6.
Clade C Tat does not inhibit TOPflash activity in astrocytes. PDAs were cotransfected with TOPflash reporter and either vector control or the indicated Tat expression plasmid. Twenty-four hours later, cells were lysed, and dual luciferase assay was performed as in Figure 2. Results are reported as relative light units (RLU) normalized to micrograms protein. Data represent at least three experiments. The asterisks denote p < 0.05 relative to vector control as measured by two-tailed Student's t test. Error bars indicate SD.
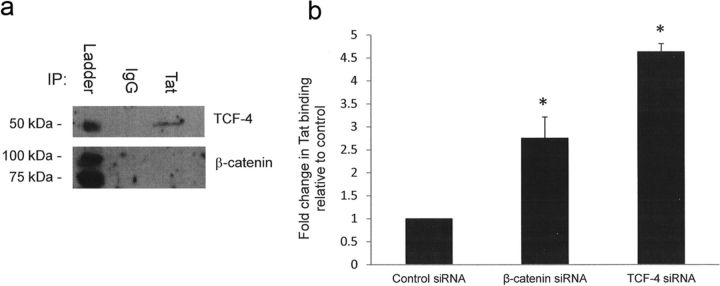
Knockdown of TCF-4 and β-catenin increase docking of Tat at the TAR region of the HIV LTR
We previously showed that knockdown of TCF-4 or β-catenin increases the docking and processivity of RNA polymerase II (Pol II) on the HIV LTR in astrocytes (Narasipura et al., 2012). Tat-mediated transactivation of the LTR is required for efficient transcription by Pol II. Therefore, we evaluated the consequences of Tat/β-catenin/TCF-4 interaction on HIV transcription. We first established whether Tat/TCF-4/β-catenin directly associate with each other in vivo. As shown in Figure 7a, Tat coprecipitated with TCF-4 but not β-catenin. To determine the impact of TCF-4/β-catenin on Tat binding to the TAR element, U87MG were knocked down for β-catenin or TCF-4 and transfected with LTR and Tat constructs, followed by chromatin immunoprecipitation to detect Tat. Knockdown of β-catenin or TCF-4 increased binding of Tat at the TAR region by 2.3- and 3.5-fold, respectively (Fig. 7b). Collectively, these results indicate that TCF-4 associates with Tat and that TCF-4 and β-catenin inhibit the ability of Tat to transactivate the HIV LTR.
Figure 7.
Knockdown of β-catenin or TCF-4 enhances docking of Tat on TAR region of the HIV LTR. a, PDAs were transfected with an HA-Tat construct. At 48 h after transfection, cells were cross-linked with 1.0% formaldehyde and lysed. β-Catenin or TCF-4 was immunoprecipitated from the PDA lysate, followed by Western blot using the indicated antibodies. b, U87MG were transfected with the indicated siRNA. At 48 h after knockdown, cells were transfected with LTR-luciferase construct as well as a construct that encodes an HA-tagged clade B Tat for an additional 24 h (72 h total knockdown). Cells were then cross-linked with formaldehyde, and chromatin immunoprecipitation was performed using IgG control or anti-HA antibody. Results are normalized to IgG control antibody and are reported as fold change in Tat-HA binding relative to control siRNA-treated samples. Results represent at least three experiments. The asterisks denote p < 0.05 relative to control as measured by two-tailed Student's t test. Error bars indicate SD.
Discussion
We provide evidence here to show that HIV Tat inhibits Wnt/β-catenin signaling in astrocytes. Robust expression of Wnt/β-catenin is an important regulator of the extent of HIV productive replication in astrocytes (Carroll-Anzinger et al., 2007; Li et al., 2011; Henderson et al., 2012; Narasipura et al., 2012). Inflammatory mediators such as IFN-γ enhance HIV productive replication in astrocytes by antagonizing β-catenin signaling through a stat-3-dependent mechanism (Li et al., 2011). Wnt/β-catenin is thus a host restriction factor for HIV in astrocytes. HIV has many mechanisms to overcome host restriction factors, such as the action of Vif on APOBEC3G (Sheehy et al., 2002) or Vpu on tetherin (Douglas et al., 2009). Tat inhibition of β-catenin signaling represents a mechanism for viral evasion of the suppressive effect of β-catenin signaling on HIV transcription. β-Catenin/TCF-4 inhibit both basal and Tat-mediated transactivation of the HIV LTR (Henderson et al., 2012; Narasipura et al., 2012). We recently identified several TCF-4 binding sites on the HIV promoter (Henderson et al., 2012). We further demonstrated that β-catenin signaling inhibits HIV basal transcription by inducing the formation of a repressive complex composed of TCF-4, β-catenin, and the nuclear matrix binding protein SMAR1 on the LTR at −143 nt from the +1 transcription initiation site. The complex pulls the HIV DNA into the nuclear matrix and render it inaccessible to transcription machinery (Henderson et al., 2012). Although β-catenin/TCF-4 signaling also inhibits Tat-mediated transactivation of the HIV LTR, the mechanism does not involve the −143 site (Henderson et al., 2012). Rather, we show here that Tat binding to TCF-4 sequesters Tat away from the TAR element, a required association for accelerated rate of transcription. Likewise, Tat binding to TCF-4 sequesters TCF-4 away from its cognate target genes such as Axin-2. We were unable to demonstrate an association between Tat and β-catenin, which may indicate that Tat itself does not directly bind to β-catenin, but does not rule out that β-catenin through its association with TCF-4 may be part of this Tat binding complex.
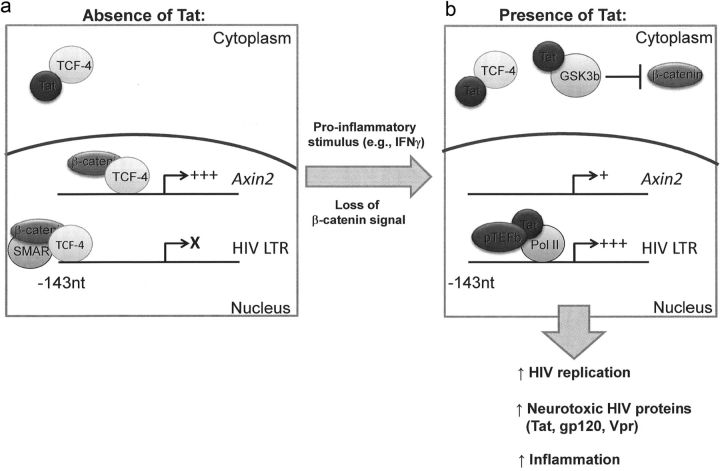
Based on the data presented here and other recently published (Li et al., 2011; Henderson et al., 2012; Narasipura et al., 2012), we suggest a model of bidirectional inhibition in which β-catenin and TCF-4 repress basal and Tat transactivation of the HIV LTR through distinct mechanisms and are in turn antagonized by HIV Tat (Fig. 8). Under basal LTR activity (without significant Tat level), the TCF-4/β-catenin/SMAR1 complex is associated with the HIV LTR and transcription is low or silent. Low levels of Tat may be produced but are primarily retained in the cytoplasm by association with TCF-4. When β-catenin signaling is disrupted, for example by proinflammatory mediators (IFN-γ) or any other signal that downregulates the β-catenin pathway, this complex is disrupted and LTR activity increases. If this spike in promoter activity is sufficient to allow for Tat concentrations to reach a threshold level, Tat will (1) allow for efficient viral replication and spread of HIV virions in the CNS; and (2) antagonize β-catenin signaling through mutual binding/inhibition with TCF-4 and enhanced degradation of β-catenin to maintain a permissive state for HIV replication. Therefore, Tat-mediated effects on the β-catenin pathway would not only increase HIV replication in infected cells, but could potentially cause dysregulation of uninfected cells and enhance susceptibility of bystanders to HIV infection as well.
Figure 8.
Model of Tat/β-catenin interaction in astrocytes. β-Catenin and TCF-4 repress basal and Tat transactivation of the HIV LTR through distinct mechanisms and are in turn antagonized by HIV Tat. a, Under basal LTR activity (without significant Tat level), the TCF-4/β-catenin/SMAR1 complex represses LTR activity and transcription is low or silent. Low levels of Tat may be produced but are primarily retained in the cytoplasm by association with TCF-4. b, When β-catenin signaling is disrupted by proinflammatory mediators (IFN-γ) or any other signal that downregulates the β-catenin pathway, this complex is disrupted and LTR activity increases. Once the level of Tat reaches a certain threshold, Tat will (1) transactivate the HIV LTR by inducing chromatin remodeling and recruiting positive transcription elongation factor b (pTEFb), allowing for efficient viral replication; and (2) antagonize β-catenin signaling through mutual binding/inhibition with TCF-4 and enhanced degradation of β-catenin to maintain a permissive state for HIV replication. The broader biological consequences of productive infection in astrocytes include raising the CNS viral load and increasing production of neurotoxic HIV proteins (Tat, gp120, Vpr), potentially leading to neuronal injury and neuroinflammation.
In support of our proposed model, the ability of Tat to downregulate β-catenin signaling is independent of its nuclear localization signal. TatΔ49–59, which lacks a nuclear localization sequence in the basic domain (Truant and Cullen, 1999) and is sequestered in the cytoplasm of transfected cells (Li et al., 2008), also reduced TOPflash activity in primary astrocytes even more effectively than wild-type Tat. These findings indicate that the nuclear localization of Tat is not required for its effect on β-catenin signaling and point to a mechanism of mutual inhibition in which both Tat and TCF-4 are sequestered in the cytoplasm away from their transcriptional cofactors leading to reduced expression of β-catenin (Axin-2) and HIV targets (Fig. 7). A mutation in the core domain (K41A) abrogated the ability of Tat to downregulate β-catenin signaling. Furthermore, mutation of Cys30 to glycine abrogated the ability of Tat to diminish TOPflash activity. This finding prompted us to examine the relevance of the cysteine-rich region of Tat (amino acids 22–37) in Tat-mediated inhibition of β-catenin signaling. This region is strongly associated with neurotoxicity (Nath et al., 1996; Mishra et al., 2008). Specifically, a dicysteine motif (C30C31) that is highly conserved in almost all HIV clades is involved in dysregulation and/or apoptosis of neurons (Mishra et al., 2008). An intact dicysteine motif is required for Tat-induced activation of the NMDA receptor that can lead to excitotoxicity of neurons (Li et al., 2008). Interestingly, the dicysteine motif is absent in clade C Tat, which contains a serine substitution at position 31 (C30S31). Correspondingly, clade C Tat has severely attenuated neurotoxicity in vitro (Mishra et al., 2008), although there is not yet strong evidence that points to decreased incidence of HIV-associated dementia or other neurocognitive disorders in regions where clade C predominates (Constantino et al., 2011; Joska et al., 2011). Clade C Tat did not reduce TOPflash activity in astrocytes, which indicates that the dicysteine motif is involved in inhibiting β-catenin signaling.
There are several possible alternative mechanisms of Tat interface with components of the β-catenin pathway leading to signaling inhibition. Tat binds to low-density lipoprotein (LRP) (Liu et al., 2000; Eugenin et al., 2007), a coreceptor for the Wnt/β-catenin pathway, and leads to its internalization, which will then sequester it away from Wnt ligands that initiate β-catenin signaling. Tat also enhances the activity of GSK3β, albeit in neurons (Maggirwar et al., 1999; Sui et al., 2006), which leads to the phosphorylation of β-catenin, which tags it for proteosomal degradation. Indeed, given that levels of active β-catenin are reduced at 12 h after Tat treatment, it is likely that increased degradation of β-catenin due to phosphorylation by GSK3β is at least partly responsible for reduction in signaling.
Collectively, these findings highlight the β-catenin pathway as an important restriction factor for HIV replication in astrocytes and identify a mechanism of viral adaptation (Tat) to diminish β-catenin signaling in these cells. Investigating the interplay between HIV and the β-catenin pathway in the presence or absence of Tat will lead to greater understanding of the mechanisms that regulate HIV in astrocytes and the biologic consequences of perturbed β-catenin signaling that may contribute to the role of astrocytes in HIV-associated neuropathogenesis.
Footnotes
This work was supported by National Institutes of Health Grants R01 NS060632 (L.A.-H.) and F31 NS071999 (L.J.H.), and the Chicago Developmental Center for AIDS Research (Grant P30 AI 082151) supported by National Institute of Allergy and Infectious Diseases, National Cancer Institute, National Institute of Mental Health, National Institute on Drug Abuse, National Institute of Child Health and Human Development, National Heart, Lung, and Blood Institute, and National Center for Complementary and Alternative Medicine.
References
- Blagoveshchenskaya AD, Thomas L, Feliciangeli SF, Hung CH, Thomas G. HIV-1 Nef downregulates MHC-I by a PACS-1- and PI3K-regulated ARF6 endocytic pathway. Cell. 2002;111:853–866. doi: 10.1016/s0092-8674(02)01162-5. [DOI] [PubMed] [Google Scholar]
- Carroll-Anzinger D, Kumar A, Adarichev V, Kashanchi F, Al-Harthi L. Human immunodeficiency virus-restricted replication in astrocytes and the ability of gamma interferon to modulate this restriction are regulated by a downstream effector of the Wnt signaling pathway. J Virol. 2007;81:5864–5871. doi: 10.1128/JVI.02234-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill MJ, Wesselingh SL, Cowley D, Pardo CA, McArthur JC, Brew BJ, Gorry PR. Extensive astrocyte infection is prominent in human immunodeficiency virus-associated dementia. Ann Neurol. 2009;66:253–258. doi: 10.1002/ana.21697. [DOI] [PubMed] [Google Scholar]
- Constantino AA, Huang Y, Zhang H, Wood C, Zheng JC. HIV-1 clade B and C isolates exhibit differential replication: relevance to macrophage-mediated neurotoxicity. Neurotox Res. 2011;20:277–288. doi: 10.1007/s12640-011-9241-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debaisieux S, Rayne F, Yezid H, Beaumelle B. The ins and outs of HIV-1 Tat. Traffic. 2012;13:355–363. doi: 10.1111/j.1600-0854.2011.01286.x. [DOI] [PubMed] [Google Scholar]
- Douglas JL, Viswanathan K, McCarroll MN, Gustin JK, Früh K, Moses AV. Vpu directs the degradation of the human immunodeficiency virus restriction factor BST-2/Tetherin via a βTrCP-dependent mechanism. J Virol. 2009;83:7931–7947. doi: 10.1128/JVI.00242-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ensoli B, Buonaguro L, Barillari G, Fiorelli V, Gendelman R, Morgan RA, Wingfield P, Gallo RC. Release, uptake, and effects of extracellular human immunodeficiency virus type 1 Tat protein on cell growth and viral transactivation. J Virol. 1993;67:277–287. doi: 10.1128/jvi.67.1.277-287.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eugenin EA, King JE, Nath A, Calderon TM, Zukin RS, Bennett MV, Berman JW. HIV-tat induces formation of an LRP-PSD-95- NMDAR-nNOS complex that promotes apoptosis in neurons and astrocytes. Proc Natl Acad Sci U S A. 2007;104:3438–3443. doi: 10.1073/pnas.0611699104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox HS. Virus-host interaction in the simian immunodeficiency virus-infected brain. J Neurovirol. 2008;14:286–291. doi: 10.1080/13550280802132824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Scarano F, Martín-García J. The neuropathogenesis of AIDS. Nat Rev Immunol. 2005;5:69–81. doi: 10.1038/nri1527. [DOI] [PubMed] [Google Scholar]
- Hauber J, Malim MH, Cullen BR. Mutational analysis of the conserved basic domain of human immunodeficiency virus tat protein. J Virol. 1989;63:1181–1187. doi: 10.1128/jvi.63.3.1181-1187.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaton RK, Clifford DB, Franklin DR, Jr, Woods SP, Ake C, Vaida F, Ellis RJ, Letendre SL, Marcotte TD, Atkinson JH, Rivera-Mindt M, Vigil OR, Taylor MJ, Collier AC, Marra CM, Gelman BB, McArthur JC, Morgello S, Simpson DM, McCutchan JA, et al. HIV-associated neurocognitive disorders persist in the era of potent antiretroviral therapy: CHARTER Study. Neurology. 2010;75:2087–2096. doi: 10.1212/WNL.0b013e318200d727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helland DE, Welles JL, Caputo A, Haseltine WA. Transcellular transactivation by the human immunodeficiency virus type 1 tat protein. J Virol. 1991;65:4547–4549. doi: 10.1128/jvi.65.8.4547-4549.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson LJ, Narasipura SD, Adarichev V, Kashanchi F, Al-Harthi L. Identification of novel T cell factor 4 (TCF-4) binding sites on the HIV long terminal repeat which associate with TCF-4, β-catenin, and SMAR1 to repress HIV transcription. J Virol. 2012;86:9495–9503. doi: 10.1128/JVI.00486-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joska JA, Westgarth-Taylor J, Myer L, Hoare J, Thomas KG, Combrinck M, Paul RH, Stein DJ, Flisher AJ. Characterization of HIV-associated neurocognitive disorders among individuals starting antiretroviral therapy in South Africa. AIDS Behav. 2011;15:1197–1203. doi: 10.1007/s10461-010-9744-6. [DOI] [PubMed] [Google Scholar]
- Lamba S, Ravichandran V, Major EO. Glial cell type-specific subcellular localization of 14-3-3 zeta: an implication for JCV tropism. Glia. 2009;57:971–977. doi: 10.1002/glia.20821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Liu Y, Park IW, He JJ. Expression of exogenous Sam68, the 68-kilodalton SRC-associated protein in mitosis, is able to alleviate impaired Rev function in astrocytes. J Virol. 2002;76:4526–4535. doi: 10.1128/JVI.76.9.4526-4535.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Huang Y, Reid R, Steiner J, Malpica-Llanos T, Darden TA, Shankar SK, Mahadevan A, Satishchandra P, Nath A. NMDA receptor activation by HIV-Tat protein is clade dependent. J Neurosci. 2008;28:12190–12198. doi: 10.1523/JNEUROSCI.3019-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Li G, Steiner J, Nath A. Role of Tat protein in HIV neuropathogenesis. Neurotox Res. 2009;16:205–220. doi: 10.1007/s12640-009-9047-8. [DOI] [PubMed] [Google Scholar]
- Li W, Henderson LJ, Major EO, Al-Harthi L. IFN-gamma mediates enhancement of HIV replication in astrocytes by inducing an antagonist of the beta-catenin pathway (DKK1) in a STAT 3-dependent manner. J Immunol. 2011;186:6771–6778. doi: 10.4049/jimmunol.1100099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Jones M, Hingtgen CM, Bu G, Laribee N, Tanzi RE, Moir RD, Nath A, He JJ. Uptake of HIV-1 tat protein mediated by low-density lipoprotein receptor-related protein disrupts the neuronal metabolic balance of the receptor ligands. Nat Med. 2000;6:1380–1387. doi: 10.1038/82199. [DOI] [PubMed] [Google Scholar]
- Ma M, Nath A. Molecular determinants for cellular uptake of Tat protein of human immunodeficiency virus type 1 in brain cells. J Virol. 1997;71:2495–2499. doi: 10.1128/jvi.71.3.2495-2499.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maggirwar SB, Tong N, Ramirez S, Gelbard HA, Dewhurst S. HIV-1 Tat-mediated activation of glycogen synthase kinase-3beta contributes to Tat-mediated neurotoxicity. J Neurochem. 1999;73:578–586. doi: 10.1046/j.1471-4159.1999.0730578.x. [DOI] [PubMed] [Google Scholar]
- Marcuzzi A, Weinberger J, Weinberger OK. Transcellular activation of the human immunodeficiency virus type 1 long terminal repeat in cocultured lymphocytes. J Virol. 1992;66:4228–4232. doi: 10.1128/jvi.66.7.4228-4232.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meredith LW, Sivakumaran H, Major L, Suhrbier A, Harrich D. Potent inhibition of HIV-1 replication by a Tat mutant. PLoS One. 2009;4:e7769. doi: 10.1371/journal.pone.0007769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra M, Vetrivel S, Siddappa NB, Ranga U, Seth P. Clade-specific differences in neurotoxicity of human immunodeficiency virus-1 B and C Tat of human neurons: significance of dicysteine C30C31 motif. Ann Neurol. 2008;63:366–376. doi: 10.1002/ana.21292. [DOI] [PubMed] [Google Scholar]
- Narasipura SD, Henderson LJ, Fu SW, Chen L, Kashanchi F, Al-Harthi L. Role of β-catenin and TCF/LEF family members in transcriptional activity of HIV in astrocytes. J Virol. 2012;86:1911–1921. doi: 10.1128/JVI.06266-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nath A, Psooy K, Martin C, Knudsen B, Magnuson DS, Haughey N, Geiger JD. Identification of a human immunodeficiency virus type 1 Tat epitope that is neuroexcitatory and neurotoxic. J Virol. 1996;70:1475–1480. doi: 10.1128/jvi.70.3.1475-1480.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong CL, Thorpe JC, Gorry PR, Bannwarth S, Jaworowski A, Howard JL, Chung S, Campbell S, Christensen HS, Clerzius G, Mouland AJ, Gatignol A, Purcell DF. Low TRBP levels support an innate human immunodeficiency virus type 1 resistance in astrocytes by enhancing the PKR antiviral response. J Virol. 2005;79:12763–12772. doi: 10.1128/JVI.79.20.12763-12772.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orsini MJ, Debouck CM. Inhibition of human immunodeficiency virus type 1 and type 2 Tat function by transdominant Tat protein localized to both the nucleus and cytoplasm. J Virol. 1996;70:8055–8063. doi: 10.1128/jvi.70.11.8055-8063.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passiatore G, Rom S, Eletto D, Peruzzi F. HIV-1 Tat C-terminus is cleaved by calpain 1: implication for Tat-mediated neurotoxicity. Biochim Biophys Acta. 2009;1793:378–387. doi: 10.1016/j.bbamcr.2008.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomerantz RJ. Reservoirs, sanctuaries, and residual disease: the hiding spots of HIV-1. HIV Clin Trials. 2003;4:137–143. doi: 10.1310/80jh-148k-nadq-u927. [DOI] [PubMed] [Google Scholar]
- Sheehy AM, Gaddis NC, Choi JD, Malim MH. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature. 2002;418:646–650. doi: 10.1038/nature00939. [DOI] [PubMed] [Google Scholar]
- Sui Z, Sniderhan LF, Fan S, Kazmierczak K, Reisinger E, Kovács AD, Potash MJ, Dewhurst S, Gelbard HA, Maggirwar SB. Human immunodeficiency virus-encoded Tat activates glycogen synthase kinase-3beta to antagonize nuclear factor-kappaB survival pathway in neurons. Eur J Neurosci. 2006;23:2623–2634. doi: 10.1111/j.1460-9568.2006.04813.x. [DOI] [PubMed] [Google Scholar]
- Truant R, Cullen BR. The arginine-rich domains present in human immunodeficiency virus type 1 Tat and Rev function as direct importin beta-dependent nuclear localization signals. Mol Cell Biol. 1999;19:1210–1217. doi: 10.1128/mcb.19.2.1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valcour V, Chalermchai T, Sailasuta N, Marovich M, Lerdlum S, Suttichom D, Suwanwela NC, Jagodzinski L, Michael N, Spudich S, van Griensven F, de Souza M, Kim J, Ananworanich J, RV254/SEARCH 010 Study Group Central nervous system viral invasion and inflammation during acute HIV infection. J Infect Dis. 2012;206:275–282. doi: 10.1093/infdis/jis326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westendorp MO, Frank R, Ochsenbauer C, Stricker K, Dhein J, Walczak H, Debatin KM, Krammer PH. Sensitization of T cells to CD95-mediated apoptosis by HIV-1 Tat and gp120. Nature. 1995;375:497–500. doi: 10.1038/375497a0. [DOI] [PubMed] [Google Scholar]
- Yan N, Regalado-Magdos AD, Stiggelbout B, Lee-Kirsch MA, Lieberman J. The cytosolic exonuclease TREX1 inhibits the innate immune response to human immunodeficiency virus type 1. Nat Immunol. 2010;11:1005–1013. doi: 10.1038/ni.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Liu Y, Henao J, Rugeles MT, Li J, Chen T, He JJ. Requirement of an additional Sam68 domain for inhibition of human immunodeficiency virus type 1 replication by Sam68 dominant negative mutants lacking the nuclear localization signal. Gene. 2005;363:67–76. doi: 10.1016/j.gene.2005.06.043. [DOI] [PubMed] [Google Scholar]