Abstract
Nematodes are ubiquitous organisms that have a significant global impact on ecosystems, economies, agriculture, and human health. The applied importance of nematodes and the experimental tractability of many species have promoted their use as models in various research areas, including developmental biology, evolutionary biology, ecology, and animal-bacterium interactions. Nematodes are particularly well suited for investigating host associations with bacteria because all nematodes have interacted with bacteria during their evolutionary history and engage in a diversity of association types. Interactions between nematodes and bacteria can be positive (mutualistic) or negative (pathogenic/parasitic) and may be transient or stably maintained (symbiotic). Furthermore, since many mechanistic aspects of nematode-bacterium interactions are conserved their study can provide broader insights into other types of associations, including those relevant to human diseases. Recently, genome-scale studies have been applied to diverse nematode-bacterial interactions, and have helped reveal mechanisms of communication and exchange between the associated partners. In addition to providing specific information about the system under investigation, these studies also have helped inform our understanding of genome evolution, mutualism, and innate immunity. In this review we will discuss the importance and diversity of nematodes, 'omics' studies in nematode-bacterial systems, and the wider implications of the findings.
Keywords: Steinernema, Heterorhabditis, Laxus, Brugia, Caenorhabditis elegans, Xenorhabdus, Photorhabdus, Wolbachia
Introduction
Nematodes are among the most abundant and diverse organisms on the planet, comprising as many as 1 million species in 12 clades and numerically accounting for as much as 80% of all animals (Lambshead and Boucher, 2003; Holterman et al., 2006). They have been found in all trophic levels within a wide range of environments, including 1 kilometer beneath the Earth's surface (Ettema, 1998; De Ley, 2006; Borgonie et al., 2011). As such, they have a global impact on ecosystems, economies, and human health. Many nematodes are viewed as targets for eradication because of their devastating effects on agriculture and health (Perry and Randolph, 1999; Bird and Kaloshian, 2003; Chitwood, 2003). In particular, parasitic nematodes known as helminths cause a wide range of diseases in humans and animals, and it is estimated that greater than 10 percent of the world’s population is at risk for helminthic infection every year (Crompton, 1999). Two severe forms of helminth-caused disease, lymphatic filariasis (elephantiasis) and onchocerciasis (river blindness), are due to infection by filarial nematodes (Taylor et al., 2010). An estimated 150 million people suffer from these two diseases, with another billion at risk (Molyneux et al., 2003; Taylor et al., 2010). The devastating impact of parasitic nematodes on human productivity and health has spurred efforts to develop treatments and preventions by elucidating parasite biology using new technologies (Kumar et al., 2007; Mitreva et al., 2007; Taylor et al., 2011).
Despite their sinister reputation, parasitic nematodes also can have many beneficial impacts on human interests and health. For example entomopathogenic nematodes (EPNs), such as steinernematids and heterorhabditids, are commercially used as biological control agents for crop pests (Grewal et al., 2005). Also, human-parasitic nematodes are being tested for therapeutic use in many autoimmune diseases (Summers et al., 2005; Schneider and Ayres, 2008; Liu et al., 2009; Kuijk and van Die, 2010; Correale and Farez, 2011).
The simplicity, tractability, and conserved genes of many nematode species have supported their use as models for diverse biological processes, including human diseases, aging, immunity, development, ecology, evolution, and host-bacterial interactions (Aboobaker and Blaxter, 2000; Couillault and Ewbank, 2002; Goodrich-Blair, 2007; Mitreva et al., 2009; Markaki and Tavernarakis, 2010; Neher, 2010; Xu and Kim, 2011). This last phenomenon - the intimate associations between two of the most speciose organisms on the planet - will be the focus of the remainder of this review.
Nematode-bacterium associations can be beneficial (mutualistic) or harmful (pathogenic/parasitic) and can range from facultative, temporary interactions to stably maintained, long-term symbioses. Bacteria can be a potential food source for nematodes (Poinar and Hansen, 1986). Bacterivory only occurs in select nematode species and can be non-specific (such as in Caenorhabditis elegans (Freyth et al., 2010) or specialized. In specialized interactions, the nematodes preferentially depend on select genera or species of bacteria, and these bacteria may be purposefully introduced or raised by the nematode (Ott et al., 1991; Goodrich-Blair, 2007).
As well as being a food source, bacteria can be pathogens of nematodes. Many of these are the same or similar to pathogens of humans, which has spurred the use of C. elegans as a model host of human infectious diseases (Couillault and Ewbank, 2002; Waterfield et al., 2008; Irazoqui et al., 2010; Pukkila-Worley and Ausubel, 2012). In addition to trophic and pathogenic interactions, bacteria can serve as mutualists by aiding nematodes in development, defense, reproduction and nutrient acquisition (Poinar and Hansen, 1986; Zhou et al., 2002; Goodrich-Blair and Clarke, 2007; Musat et al., 2007; Slatko et al., 2010; Hansen et al., 2011; Foster et al., in press).
In recent years, 'omic' studies, high-throughput analyses of whole cell, organism and population-wide data sets, have begun to reveal the mechanistic underpinnings of many nematode-bacteria interactions. Genome sequencing has opened the door for transcriptomics examining nematode and bacterium transcriptional profiles as well as for proteomics to identify and quantify proteins in complex mixtures (Malmstrom et al., 2011). While these types of 'omics' studies have been applied to only a few of myriad nematode-bacterium associations, the findings have been integral to the understanding of other aspects of nematode biology and are paving the way for comparative analyses with non-nematode symbioses.
Model systems of nematode-bacterium symbiosis
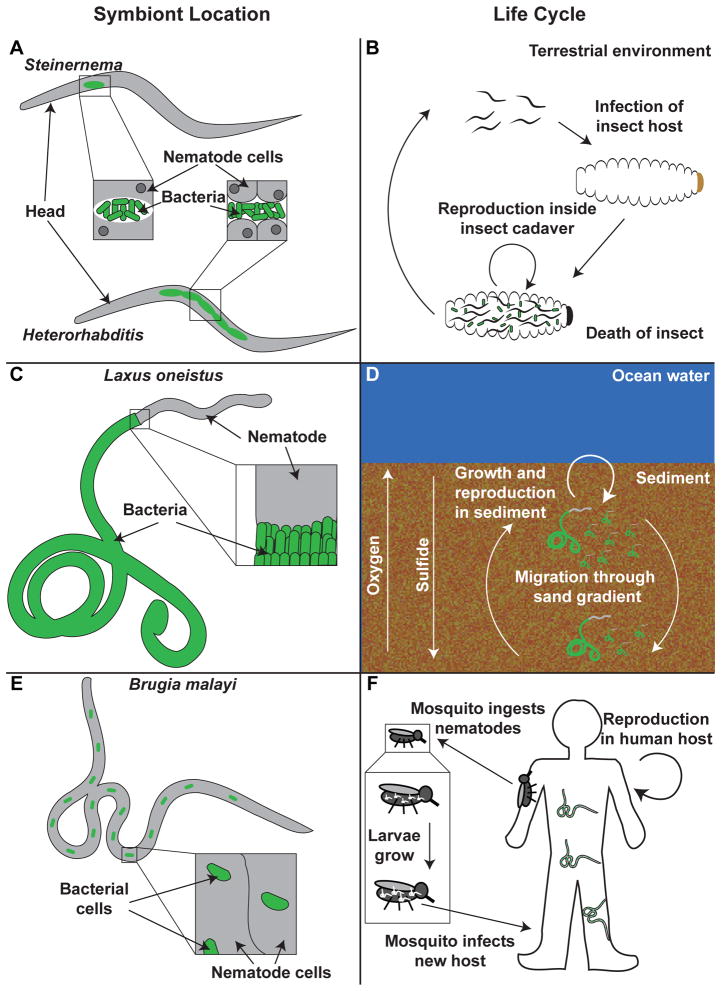
Nematodes and their bacterial associates exist in marine, freshwater, soil, and plant or animal host environments. The most exhaustively studied of the nematodes, C. elegans, is a terrestrial nematode whose relationships with bacteria are predatory (Brenner, 1974), defensive (Tan and Shapira, 2011), and possibly commensal (Portal-Celhay and Blaser, 2011). The long experimental history of C. elegans has made it an unparalleled model of numerous biological processes (Blaxter, 2011; Xu and Kim, 2011), including bacterial pathogenesis and host immunity (Irazoqui et al., 2010; Tan and Shapira, 2011; Pukkila-Worley and Ausubel, 2012). This body of work also has facilitated the advancement of nematode-bacterium associations in which the nematode and bacteria engage in specific, persistent, mutualistic relationships. We emphasize three such associations here: terrestrial entomopathogenic nematodes associated with Xenorhabdus and Photorhabdus bacteria, Laxus oneistus marine nematodes with thiotrophic surface-colonizing bacteria, and parasitic filarial nematodes colonized by intracellular Wolbachia symbionts (Fig. 1) (Table 1).
Figure 1.
Schematic Overview of Model Nematode-Bacterium Symbioses: Symbiont Location and Life Cycle>
A: Xenorhabdus and Photorhabdus (green) are located within infective juveniles (environmental stage) of Steinernema and Heterorhabditis nematodes respectively. The bacteria are located in the lumen between intestinal epithelial cells (grey with dark grey nuclei) (insets).
B: The infective juveniles of Sterinernema and Heterorhabditis parasitize insect hosts. The nematodes and bacteria kill the insect and reproduce within the insect cadaver. The nematodes then re-associate with their bacterial symbiont and migrate away from the cadaver into the environment to seek new hosts.
C: The ecotsymbiont (green) of Laxus oneistus (grey) is located on the outside of all nematode life stages. The bacteria are arranged in a perpendicular fashion to the exterior of the nematode (inset).
D. L. oneistus nematodes grow and reproduce in the sediment of the sea floor. Their thiotrophic ectosymbiont profits from nematode migrations in oxygen and sulfide gradients (see text for more details).
E. The Wolbachia symbiont (green) of Brugia malayi nematodes (grey) are localized to the hypodermal cells of the lateral chords in all nematode life stages and the reproductive tissues of females. These bacteria are intracellular (inset).
F. B. malayi is transmitted to a human host through a mosquito vector. The nematodes undergo reproduction within the human host and produce a larval stage that can be taken up by new mosquitoes. Larval stages grow within the mosquito and can then infect new human hosts when the mosquito takes a blood meal.
Table 1.
’Omics’ studies applied to nematode-bacterium symbioses. Only those nematode-bacterium associations discussed in this review for which there are available 'omics' data are listed.
| Symbiosis1 | Nematode ‘omics’ | References | Bacterium ‘omics’ | References |
|---|---|---|---|---|
| Parasites of invertebrates | ||||
| Steinernema (Clade 10) - Xenorhabdus | Genomes and transcriptomes of S. carpocapsae, S. scapterisci, S. monticolum, S. feltiae, and S. glaseri | Dillman et al., in preparation) | Genomes of X. nematophila and X. bovienii | (Latreille et al., 2007) (Chaston et al., 2011) |
| Heterorhabditis (Clade 9) - Photorhabdus | Genome and transcriptome of H. bacteriophora | (Ciche, 2007) (Harris et al., 2010) (Bai et al., in preparation) (Bai et al., 2009) (Hao et al., 2012) |
Genomes of P. luminescens and P. asymbiotica Proteomes of P. luminescens TT01 variants Transcriptome of P. luminescens TT01 variants |
(Duchaud et al., 2003) (Gaudriault et al., 2006) (Gaudriault et al., 2008) (Ogier et al., 2010) (Wilkinson et al., 2009) (Derzelle et al., 2004) (Turlin et al., 2006) (Lanois et al., 2011) |
| Parasites of vertebrates | ||||
| Brugia malayi (Clade 8) - Wolbachia | Genome sequence Transcriptomes Proteomes |
(Bennuru et al., 2011) (Bennuru et al., 2009) (Choi et al., 2011) (Ghedin et al., 2009) (Ghedin et al., 2007) |
Genome Proteome |
(Foster et al., 2005) (Bennuru et al., 2009) (Bennuru et al., 2011) |
| Free living nematodes | ||||
| Laxus oneistus (Stilbonematinae, Clade 4) | Transcriptomes | (Bulgheresi, 2012a) (Bulgheresi, 2012b) |
Draft genome of L. oneistus ectosymbiont | Available upon request at http://rast.nmpdr.org/rast.cgi |
| Plant-parasitic nematodes | ||||
| Meloidogyne incognita (Clade 12) | Genome | (Abad et al., 2008) | NA | NA |
| Meloidogyne hapla (Clade 12) | Genome | (Opperman et al., 2008) | NA | NA |
Clades refer to those defined by (Holterman et al., 2006).
Entomopathogenic nematodes (EPNs) and bacteria
At least two genera of nematodes, Steinernema and Heterorhabditis, have evolved symbiotic associations with gammaproteobacteria, Xenorhabdus and Photorhabdus respectively, that allow them to kill insects and utilize the cadavers as food sources (Dillman et al., 2012). A specialized infective stage of EPNs carries a population of the symbiont within the intestine, and releases them upon invasion of an insect host. There, the bacteria contribute to killing the insect, help degrade the insect cadaver for nutrients, and protect the cadaver from opportunists. Once the insect resources are consumed, the EPN progeny nematodes develop into the colonized infective stage and emerge to hunt for a new insect host (Fig. 1) (Herbert and Goodrich-Blair, 2007; Clarke, 2008). There are three species recognized within the Photorhabdus genus: P. temperata, P. luminescens, and P. asymbiotica. The last was isolated originally from human wounds, but recently was discovered to colonize, like the other species, a heterorhabditid nematode host, of which there are 18 recognized species (Nguyen, 2010; Stock and Goodrich-Blair, 2012). In contrast, there are 22 species of Xenorhabdus (Tailliez et al., 2010; Tailliez et al., 2011) that colonize one or more of the >70 known species of Steinernema nematodes (Nguyen, 2010; Stock and Goodrich-Blair, 2012). In both types of associations, the bacteria and nematodes can be cultivated independently or together, and molecular genetic techniques are available for the bacteria and, in some cases, for the nematodes (Ciche and Sternberg, 2007; Goodrich-Blair, 2007; Clarke, 2008). This technical tractability has enabled the use of EPNs and bacteria as models of mutualism, virulence, evolution, behavior, ecology, and drug discovery (Clarke, 2008; Ram et al., 2008; Adhikari et al., 2009; Bode, 2009; Richards and Goodrich-Blair, 2009; Eleftherianos et al., 2010; Hallem et al., 2011; Bashey et al., 2012). Furthermore, since these nematode-bacterium complexes are pathogenic toward a wide but varying range of insects, an additional goal in studying EPNs is improving their use in biological control of insect pests (Stock, 2004). In particular, investigators have focused on identifying nematode traits associated with host range and successful parasitism to help improve the field efficacy of EPNs, and on identifying products of the entomopathogenic bacteria with insecticidal properties, efforts facilitated by sequencing of both bacterial and nematode genomes (Duchaud et al., 2003; Ciche, 2007; Wilkinson et al., 2009; Chaston et al., 2011; Bai et al., in preparation; Dillman et al., in preparation).
Laxus oneistus symbiosis
Stilbonematids occur in marine sand and establish ectosymbioses with thiotrophic gammaproteobacteria (Ott et al., 2004b, a; Bulgheresi, 2011). Based on 18S rRNA-gene based phylogeny, stilbonematids form a monophyletic, distinct group of closely related genera within the Desmodoridae (Kampfer et al., 1998; Bayer et al., 2009). Stilbonematids are hypothesized to trophically depend on their ectosymbionts and these in turn are assumed to profit from nematode migrations through the sulfide gradient in the marine sediment (Fig. 1) (Ott et al., 1991). Stilbonematid sexual reproductive biology and development are poorly known. Two distinctive morphological characters unifying all stilbonematids are a poorly muscularized pharynx and the presence of unique epidermal organs called glandular sense organs (GSOs) (Bauer-Nebelsick et al., 1995). GSOs appear to play a key role in the ectosymbiosis as they express a Ca2+-dependent lectin (C-type lectin) that mediates ectosymbiont aggregation and host attachment (Bulgheresi et al., 2006; Bulgheresi et al., 2011). Each GSO is composed of at least two gland cells and a sensory neuron (Bauer-Nebelsick et al., 1995). Secretory products from the gland cells accumulate into a canal that crosses the epidermis and cuticle and terminating in a hollow bristle (seta). Therefore, with the cuticle being like a sieve, a continuum exists between each GSO and the nematode surface.
L. oneistus ectosymbiont cells are rod shaped and aligned perpendicularly to the nematode surface, forming an epithelium-like monolayer (Fig. 1). Notably, the cuticle thins at the bacterial coat onset (Urbancik et al., 1996). The bacteria belong to the marine oligochaete and nematode thiotrophic symbiont (MONTS) cluster, which comprises 16S rRNA-gene sequences retrieved from gammaproteobacterial sulfur-oxidizers associated to these invertebrates, as well as sequences of environmental origin (Heindl et al., 2011). The closest cultivable relatives of MONTS members are free-living sulfur purple bacteria (Chromatiaceae). Beside their 16S rRNA-gene based phylogenetic placement, uptake of 14C bicarbonate (Schiemer et al., 1990) and the presence of RuBisCo enzymatic activity indicate Laxus ectosymbiont autotrophy (Polz et al., 1992). Enzymatic activity of ATP sulphurylase and sulphite oxidase, the presence of elemental sulfur in symbiotic but not in aposymbiotic L. oneistus (Polz et al., 1992) and the cloning of the symbiont's aprA gene, encoding the alpha subunit of adenosine-5-phosphosulfate (APS) reductase (Bayer et al., 2009) indicate sulfur-oxidation capability. Moreover, metabolic studies suggest denitrification capability (Hentschel et al., 1999). The available genome draft (Table 1) confirms nitrate respiration and suggests additional nitrite respiration and ammonia assimilation capabilities.
A distinguishing quality of stilbonematids is their ability to form monospecific ectosymbioses. The fact that host and ectosymbiont can be easily separated from one another makes stilbonematids an excellent system for dissecting the molecular base of symbiosis-specificity. Indeed, both host-secreted and microbe- associated molecular patterns (MAMPs) identified through 'omics' can be expressed in vitro and directly tested on these nematode-bacteria consortia. In addition, L. oneistus represents an example of how the study of nematode-bacterial associations can have direct impacts in solving societal problems; the C-type lectin mentioned above is structurally and functionally similar to a human HIV-1 receptor, and could also block viral infection of human immune cells (Nabatov et al., 2008).
Filaria nematode-Wolbachia symbiosis
Wolbachia are alphaproteobacteria belonging to the order Rickettsiales and closely related to Anaplasma, Ehrlichia and Rickettsia. Wolbachia are perhaps the most abundant of all intracellular bacteria, being found in filarial nematodes and arthropods, with around 70% of insects species colonized (Hilgenboecker et al., 2008; Werren et al., 2008). It remains unresolved if the Wolbachia bacteria present in different hosts or different invertebrate phyla represent distinct bacterial species or strains (Pfarr et al., 2007). These maternally inherited, intracellular bacteria are generally considered reproductive parasites of arthropods due to the various reproductive manipulations they induce (cytoplasmic incompatibility, parthenogenesis induction, feminization, male killing) that serve to promote the reproductive success of infected females and the spread of Wolbachia through populations (Werren et al., 2008). However, there is recent evidence that Wolbachia may confer fitness advantages to arthropods in certain situations. For example, Wolbachia increases resistance to viral pathogens in both fruit flies and mosquitoes and may be involved in nutritional provisioning in times of metabolic stress (Schneider and Chambers, 2008; Teixeira et al., 2008; Brownlie et al., 2009; Moreira et al., 2009; Osborne et al., 2009; Bian et al., 2010; Glaser and Meola, 2010). The Wolbachia found in most filarial nematode species are believed to be obligate mutualists and have shared a long stable co-existence with their worm hosts (Foster et al., in press). Clearance of Wolbachia with antibiotics has dire consequences for the nematode host with disrupted development, blockage of embryogenesis and eventual death of the worm (Taylor et al., 2005; Foster et al., in press). Consequently, Wolbachia represent a major new drug target for control of filarial diseases and doxycycline has been used in several clinical trials in Africa and Asia (Taylor et al., 2010).
Within filarial nematodes the Wolbachia are found in the hypodermal cells of the lateral chords of both sexes and in the ovaries, oocytes and developing intrauterine embryonic stages of females. Wolbachia are present in all developmental stages of the worm but undergo extensive multiplication within a week of the nematode transitioning from its insect vector to the mammalian host (Fig. 1). Wolbachia titer increases further as the larvae develop to adulthood and as the oocytes and embryonic stages become infected (Fenn and Blaxter, 2004; McGarry et al., 2004). These observations suggest a molecular crosstalk that serves to regulate Wolbachia titer. Complete genome sequences of both Brugia malayi (causes lymphatic filariasis) and its Wolbachia endosymbiont are available (Foster et al., 2005; Ghedin et al., 2007) and have facilitated subsequent microarray, transcriptomic and proteomic studies (Table 1) that are beginning to tease apart aspects of the filarial nematode- Wolbachia symbiosis.
'Omic' insights into nematode-bacterial mutualism
Experimental systems of nematode-bacterium mutualism provide an opportunity to test existing symbiosis theory (Douglas, 2008) including how benefits and costs within each association vary depending on environment and partner, how specific partners are transmitted between generations, how the development of cheating is prevented or maintained at acceptable levels, how tolerance or avoidance of host immunity is achieved, and how symbiosis impacts the evolution of an organism. Through numerous approaches, including 'omics', these questions are beginning to be answered in several nematode-bacterium symbioses.
Nutrient provisioning between filarial nematodes and Wolbachia symbionts
Filarial nematodes depend on their Wolbachia symbiont for normal development, embryogenesis and viability raising the hypothesis that the bacteria may provide essential nutrients to their nematode host (Taylor et al., 2005; Foster et al., in press). In turn, Wolbachia bacteria are unable to be cultured outside host cells, indicating it too receives some nutritive benefit from its host. The genome sequences of B. malayi (Ghedin et al., 2007) and its Wolbachia endosymbiont (Foster et al., 2005) together with transcriptomic approaches (see below) have revealed several candidate examples of metabolic provisioning between the bacterium and its nematode host. Wolbachia is very limited in its production of amino acids but encodes several proteases and importers, which presumably enable the bacterium to grow on host-derived amino acids. Surprisingly, B. malayi lacks genes required for de novo synthesis of purines and pyrimidines but maintains salvage pathways; conversely Wolbachia has retained de novo synthesis but lacks nucleotide salvage pathways. Similarly, B. malayi is deficient in genes required for biosynthesis of heme, riboflavin and FAD while Wolbachia, despite having a streamlined genome typical of intracellular bacteria, has retained these biosynthetic capabilities.
Experimental studies based on these genomic insights suggest the Wolbachia heme pathway may indeed be critical for the B. malayi host (Wu et al., 2009). Furthermore, a microarray study that compared gene expression in tetracycline-treated Litomosoides sigmodontis (a closely related filarial worm) to untreated worms that retained their Wolbachia showed higher expression in treated worms of a nematode heme-binding globin as well as several heme- and riboflavin-containing respiratory chain components encoded by the mitochondrion (Strubing et al., 2010). These transcriptional changes were not observed in a filarial nematode that naturally lacks Wolbachia suggesting that the responses observed in L. sigmodontis were a true consequence of Wolbachia clearance. These results highlight the power of genomics to focus experimentation on key specific testable hypotheses.
L. sigmodontis expression of genes involved in translation, transcription, protein folding/sorting, structure, motility, metabolism, signaling and immunomodulation were also affected by Wolbachia clearance (Strubing et al., 2010). Broadly similar changes were observed in a comparable microarray experiment conducted in B. malayi (Ghedin et al., 2009). In this study, genes in certain classes (e.g., signaling) showed a bimodal pattern of regulation: they were up-regulated soon after antibiotic treatment started, then quickly down-regulated, before becoming up-regulated again after the end of treatment (Ghedin et al., 2009). Since antibiotics affect embryogenesis in advance of worm viability, the authors postulated that early changes in gene transcript levels reflect disruption of the embryo program while later transcriptional changes are the result of reduction of the Wolbachia load in the hypodermis (Ghedin et al., 2009). Although the cDNA preparation selected against Wolbachia transcripts, some were detectable. As might be expected, Wolbachia probes that hybridized almost exclusively showed down-regulation following antibiotic treatment. However, three Wolbachia genes (hypothetical, short-chain alcohol dehydrogenase and stress-induced morphogen) were up-regulated following treatment (Strubing et al., 2010) although the significance of this observation is not understood.
The relative costs and benefits of bacterial association can be influenced by the developmental stage of the organisms, and therefore key insights can be gained by monitoring through transcriptomics aspects of mutualism throughout the life cycles of the associates. Expressed sequence tag (EST) sequencing from 25 cDNA libraries made from different life-cycle stages of B. malayi has produced over 25,000 sequences that cluster to nearly 10,000 genes. Similar datasets are available for other filarial nematode species (Elsworth et al., 2011; Blaxter, 2012). In addition, a recent comprehensive RNASeq transcriptomic profiling of seven different life-cycle stages of B. malayi (Choi et al., 2011) will be invaluable for tracking the temporal transcription of nematode genes predicted to be involved in the symbiotic relationship with Wolbachia. Stage specific proteomic studies on B. malayi (Bennuru et al., 2009) and its excreted/secreted proteins (Bennuru et al., 2011) have confirmed production of about two-thirds of the predicted proteome and validated about half of the genes annotated as hypothetical. Of note, Wolbachia proteins were also found amongst the excretory/secretory products, suggesting integration of nematode and bacterial physiology. A recent genome-wide computational prediction of protein-protein interactors in six species of parasitic nematodes, including B. malayi as well as the free-living C. elegans was undertaken to highlight interactors as candidate drug targets (Taylor et al., 2011). This study did not include the Wolbachia proteome with the Brugia dataset but prediction of the Wolbachia-Brugia interactome is highly warranted given their likely physiological integration. Based on the hypothesis that outer membrane proteins such as Wolbachia surface protein (WSP) might interact with nematode proteins, WSP was used to bind B. malayi protein extracts, for panning a Brugia cDNA library and for ELISA and pull-down assays (Melnikow et al., 2011). One Brugia protein annotated as hypothetical was identified by all approaches and provides the first example of a Brugia-Wolbachia interacting protein pair. Thus, the combination of transcriptomic and proteomic data from the host nematode and it’s symbiont allows detailed investigation of the presence and abundance of nematode and Wolbachia gene products throughout the life-cycle and will lead to enhanced understanding of the host-bacterial interactome and the symbiosis in general.
Specificity in the EPN-bacteria symbiosis
Photorhabdus and Xenorhabdus bacteria are closely related to each other phylogenetically, and both infect a similar range of insect hosts, but each associates with an EPN from a different clade (Table 1). Both bacteria make similar symbiotic contributions to the fitness of their nematode hosts: helping establish infection in insects, defending the insect host from predators and competitors, and promoting normal nematode development (Goodrich-Blair and Clarke, 2007). However, comparative analyses of the four sequenced bacterial genomes (P. luminescens, P. asymbiotica, X. nematophila, and X. bovienii) (Duchaud et al., 2003; Wilkinson et al., 2009) revealed these similar fitness traits are the product of convergent evolution (Chaston et al., 2011). For example, each symbiont limits the growth of competitor microbes, but does so through the production of different types of antimicrobial compounds (Chaston et al., 2011). In contrast, the genes involved in entomopathogenicity, such as those encoding insecticidal toxins, appear to be conserved among the four bacterial species. Based on the apparent convergent evolution of genes involved in nematode-association and conservation of those involved in insect virulence, this study also predicted which bacterial genes may be involved in either of these symbiotic behaviors (Chaston et al., 2011). The analysis was based on the idea that genes present in both Xenorhabdus and Photorhabdus but absent in non-insect pathogens may be enriched for those that encode activities necessary for killing and digesting insects. Similarly, genes that are unique to either Xenorhabdus or Photorhabdus should be enriched for those that are necessary for interactions with the nematode host. The study found 243 X. nematophila genes common to Xenorhabdus and Photorhabdus but absent in non-insect pathogens, including many with predicted roles in pathogenesis, and 290 genes specific to Xenorhabdus. Perhaps not surprisingly, genes of unknown function predominate in the latter “nematode host interaction” category, suggesting that bacterial genes involved in nematode interactions remain to be functionally characterized (Chaston et al., 2011). Further application of proteomic and “panning” approaches, such as those described above for Wolbachia-filaria interactions would be useful for exploring this set of potential host-interaction genes.
In addition to comparative genome approaches, genome sequencing of EPN symbionts facilitated genetic screens that lent insights into the biology involved in host-microbe interactions. As with all mutualistic symbiotic associations a key component of the EPN-bacterium symbiosis is transmission of the bacterial symbiont to the next generation. In EPNs this occurs by bacterial colonization of the intestines of progeny infective juveniles and carriage to the next insect host. Bacterial colonization of the infective juvenile stage can be highly selective, such that in some EPN-bacterium associations only one species of bacterium is capable of colonizing a particular species of nematode (Goodrich-Blair, 2007; Clarke, 2008). Transposon mutagenesis screens in both X. nematophila and P. luminescens have revealed novel genes involved in this specificity (Heungens et al., 2002; Easom et al., 2010; Somvanshi et al., 2010). In one study, nine X. nematophila genes essential for normal colonization of the infective stage of S. carpocapsae nematodes were identified. Three of these genes, nilA, B, and C, are encoded together on a 3.5-kb locus (Heungens et al., 2002). Further study revealed that this locus is not present in other Xenorhabdus bacterial symbionts and is sufficient to confer colonization of S. carpocapsae on naturally non-colonizing bacteria, establishing for the first time a genetic element conferring host range expansion in an animal-bacterial association (Cowles and Goodrich-Blair, 2008). nilB is similar to genes found in animal associated microbes, including mucosal pathogens (Heungens et al., 2002; Bhasin et al., 2012), supporting the idea that common molecules or mechanisms maintain many host-bacterial interactions regardless of whether the outcome of the interaction is mutualistic or pathogenic (McFall-Ngai et al., 2010). The function of NilB, a surface exposed outer membrane protein (Bhasin et al., 2012) remains unclear, but analysis of the EPN symbiont genome sequences has provided some clues. Relaxed search parameters revealed that each of the four sequenced genomes of EPN symbionts, including X. nematophila itself, encodes a NilB-like protein in a conserved genomic context. Adjacent genes are predicted to encode TonB-like transporters and TonB-dependent receptors, involved in metabolite transport across the membrane. This finding leads to the hypothesis that NilB and NilB-like proteins may be involved in transport of a class of molecules that varies among different nematode hosts, allowing their function to dictate host range specificity (Bhasin et al., 2012). Alternatively, the NilB-like orthologs may play a role in other aspects of the EPN symbiont biology, such as insect virulence.
Consistent with the latter hypothesis, screens for P. luminescens mutants defective in colonizing their nematode host H. bacteriophora did not reveal the NilB-like ortholog, nor any of the other colonization genes identified in X. nematophila (Heungens et al., 2002; Easom et al., 2010; Somvanshi et al., 2010). This finding further supports the convergent abilities of Xenorhabdus and Photorhabdus to mutualistically associate with their respective nematodes (Chaston et al., 2011). Putative P. luminescens nematode colonization genes revealed by mutant screens include those involved in lipopolysaccharide metabolism, fimbriae biosynthesis, and regulation (Easom et al., 2010; Somvanshi et al., 2010). Subsequent microarray work established that the colonization gene hdfR encodes a transcription factor that regulates more than 100 genes, including many involved in metabolic processes. Nematodes co-cultivated with the hdfR mutant display a developmental lag, suggesting that hdfR is required for normal nematode development (Easom and Clarke, 2011). As the roles of bacteria in EPN development are elucidated, it will be particularly interesting to compare these findings to those in the filarial nematode-Wolbachia associations to determine if common themes are revealed.
Another avenue toward elucidating the molecular dynamics of nematode-bacterium mutualism is identification of genes that are expressed specifically during association. Such an approach has been applied to P. luminescens and P. temperata. Using selective capture of transcribed sequences (SCOTS), 106 P. temperata transcripts were identified to have altered levels when cells were grown in liquid culture versus colonizing the nematode host (An and Grewal, 2010). The authors identified genes involved in cell surface structure, regulation, stress response, nucleic acid modification, transport, and metabolism, and found that half of the transcriptional changes overlap with that of the bacterial starvation response (An and Grewal, 2010). This overlap as well as the metabolic shifts that occur in sugar metabolism and amino acid biosynthesis indicate that it is likely that the nematode is a nutrient poor environment. The authors hypothesized that this could be a mechanism by which the nematode controls the bacterial population (An and Grewal, 2010), which again echoes the potential of filarial nematodes to control their Wolbachia symbiont titer (Fenn and Blaxter, 2004; McGarry et al., 2004).
Comparative-omics to elucidate the molecular dialogue between host and symbiont
Nematodes likely interact with their symbiont partners through immune pathways. For example, nematode immunity may be down-regulated by the symbiont that may in turn produce antimicrobials to protect the immuno-depressed host from pathogens. Alternatively, the symbiont may induce, but be resistant to, nematode immunity. Also, the nematode may immunologically tolerate the symbiont (Schneider and Ayres, 2008). In each of these scenarios the nematode resistance, response or tolerance to microbes and the relevant immune pathways must be identified to fully unravel the molecular dialogue between host and symbiont.
The C. elegans immune system
Our knowledge of nematode innate immune defense derives primarily from C. elegans and its interactions with pathogens (Alper et al., 2007; Schulenburg et al., 2008; Irazoqui et al., 2010; Ewbank and Zugasti, 2011; Tan and Shapira, 2011). C. elegans does not have circulating immune cells. Therefore, if behavioral avoidance cannot spare it from deleterious microorganisms (Pradel et al., 2007), it relies on epithelial immunity to respond to pathogens. Three principal pathways activate distinct but overlapping sets of immune effectors: the p38 mitogen-activated protein kinase (MAPK) pathway, the insulin/IGF-1 signaling (IIS) pathway, and a transforming growth factor-beta (TGF-beta) pathway. Despite their undisputed role in mammalian immunity, C. elegans epithelial immunity does not rely on Toll-like receptors (Pujol et al., 2001). Moreover, many genes encoding Toll-NF-κB pathway components are absent from all the available nematode genomes (Irazoqui et al., 2010). C. elegans can discern between non-pathogenic and pathogenic, but also between different classes of microbes. The specificity of this customized immune response may arise at the recognition level or at the effector level. It may also be achieved through differential immune regulation (e.g. different microbes cause a different degree of activation of one or more signaling pathway(s) or a different integration among pathways) (Schulenburg et al., 2008).
In C. elegans p38 MAPK-mediated epidermal immunity, the binding of an unknown ligand to an unknown receptor results in successive activation of heterotrimeric G protein, protein kinase(s) C, and the p38 MAPK module. Activation of the module results in the expression of antimicrobial peptide-encoding genes such as nlp-29. Additionally, neuronally secreted DBL-1 may also ignite epidermal immunity, though the identity of the DBL-1 secreting neurons is unknown. In this case, DBL-1 receptor-regulated Smad proteins would activate (an) unknown transcription factor(s), which, in turn, would switch on transcription of antimicrobials such as caenacins in the epidermal cell.
C. elegans intestinal immunity differs from epithelial immunity; in the latter the p38 MAPK pathway (Kim et al., 2002) is integrated with the neuronally activated TGF-beta-pathway whereas, in the former, it is integrated with the insulin/IGF-1 signaling (IIS) pathway (Garsin et al., 2003). The IIS pathway is also neuronally activated and it is a conserved regulator of metabolism, stress resistance, and immune homeostasis (Becker et al., 2010; Peng, 2010). Activation of the insulin/IGF-1 receptor DAF-2 by insulin-like ligands triggers a phosphorylation cascade involving lipid and serine/threonine kinases. These phosphorylation events lead to the cytoplasmic retention of the transcription factor homologue DAF-16. If DAF-2 is not activated, or if its function is reduced, DAF-16 is translocated into the nucleus and this triggers the expression of antimicrobial genes, such as those encoding lysozymes and saposin-like proteins. DAF-16 was long hypothesized to be the only transcription factor capable of conferring pathogen resistance and it is probably the most crucial stress protective transcription factor (Tan and Shapira, 2011). In the recently described C. elegans model for persistent intestinal colonization, daf-2 mutants exhibited reduced colonization by E. coli, while daf-16 mutants showed increased colonization, but neither mutation appeared to influence the competitive advantage of Salmonella relative to E. coli for colonization (Portal-Celhay and Blaser, 2011), indicating these factors generally influence colonization, but do not necessarily contribute to specificity.
Since no nematode has been as extensively tested as C. elegans, it is unclear how different nematodes respond to microbial challenge. A comparison of current genome or transcriptome nematode databases reveals many regulatory components of the epithelial pathway described above and other immunity pathways seem to be conserved across nematodes (Table 2, Appendix 1, and Supplemental Table 1, http://www.biolbull.org/content/supplemental). Indeed both DAF-2 and DAF-16 appear to have orthologs in every nematode species examined, highlighting their critical roles in nematode biology. The increasing availability and decreasing costs of ‘omic’ techniques promises that nematode immunity will slowly but surely be revealed, answering such questions as how nematodes respond to the physical presence of bacterial cells on their cuticle, how they recognize one type of bacterium from another (and therefore select for beneficial associates while defending against pathogens), and how they control symbiont populations.
Table 2.
Orthology analysis of selected proteins known to play a significant role in the immunity of C. elegans. The protein names for C. elegans are given in the leftmost column with protein descriptions given in the second column from the left. The number of proteins found in orthology clusters with the proteins in the leftmost column are labeled under each species examined. The orthology analysis was run using the species listed across the top and also included B. xylophilus and P. pacificus as non-parasitic nematodes as well as Nasonia vitripennis, the parasitoid wasp, as an arthropod outgroup (see Appendix 1 for details). All protein data was taken from whole genome releases except for L. oneistus, for which protein data from transcriptomics was used. All individual orthology results and the protein identifiers for the clustered orthologs can be found in Supplemental Table 1 (http://www.biolbull.org/content/supplemental). Those nematodes with limited or no free-living stages are indicated with grey stars (
 ) above their names.
) above their names.
| C. elegans protein | Protein description | C. elegans | L. oneistus | S. carpocapsae |
 B. malayi
B. malayi
|
 A. suum
A. suum
|
 T. spiralis
T. spiralis
|
||
|---|---|---|---|---|---|---|---|---|---|
| TGF-beta | DBL-1 | TGF-beta ligand | 1 | 1 | 1 | 1 | 1 | 0 | |
| SMA-6 | Type I TGF-beta receptor | 1 | 1 | 1 | 2 | 1 | 1 | ||
| SMA-2 & SMA-3 | Smad protein | 3 | 2 | 3 | 3 | 3 | 2 | ||
| SMA-4 | Smad protein | 1 | 1 | 1 | 3 | 2 | 1 | ||
| Insulin/IGF-1 | GOA-1 | G protein alfa subunit | 1 | 1 | 1 | 0 | 1 | 1 | |
| DGK-1 | Diacylglycerol kinase beta | 1 | 2 | 1 | 4 | 1 | 1 | ||
| INS-7 | Insulin/IGF-1-like peptide | 8 | 0 | 0 | 0 | 0 | 0 | ||
| Epithelial cell | DAF-2 | Insulin/IGF-1 receptor | 1 | 1 | 3 | 4 | 2 | 2 | |
| AGE-1 | Phosphotidylinositol 3-kinase | 1 | 3 | 1 | 1 | 1 | 0 | ||
| AKT-1 | Rac Ser/Thr protein kinase | 1 | 3 | 0 | 0 | 0 | 0 | ||
| AKT-2 | Rac Ser/Thr protein kinase | 1 | 1 | 1 | 1 | 1 | |||
| SGK-1 | Serum/glucocorticoid regulated kinase 1 | 1 | 2 | 1 | 0 | 1 | 0 | ||
| DAF-16 | FOXO family transcription factor | 1 | 2 | 2 | 2 | 1 | 1 | ||
| p38 MAPK pathway | Epidermal immunity | RACK-1 | G protein beta subunit | 1 | 1 | 2 | 1 | 1 | 1 |
| PLC-3 | Phospholipase C gamma | 1 | 1 | 2 | 1 | 1 | 1 | ||
| PKC-3 | Protein kinase C iota type | 1 | 1 | 1 | 2 | 1 | 1 | ||
| GPA-12 | G protein alpha subunit | 1 | 0 | 1 | 1 | 1 | 1 | ||
| NIPI-3 | Tribbles homolog 1 (TRB-1) | 1 | 1 | 1 | 1 | 1 | 0 | ||
| Intestinal immunity | EGL-30† | G protein G(q) alpha subunit | 1 | 2 | 1 | 2 | 2 | 1 | |
| EGL-8† | Phospholipase C beta homolog | 1 | 0 | 2 | 2 | 1 | 2 | ||
| DKF-2 | Ser/Thr protein kinase D | 1 | 2 | 1 | 2 | 1 | 1 | ||
| RAB-1 | Ras-related GTPase Rab-1A | 1 | 5 | 1 | 1 | 1 | 2 | ||
| NSY-1 | ASK1 MAPKKK | 1 | 2* | 2 | 2 | 1 | 1 | ||
| SEK-1 | MKK3, MKK6, MAPKK | 1 | 4 | 1 | 1 | 1 | 1 | ||
| PMK-1 | p38 MAPK | 1 | 3 | 1 | 1 | 1 | 1 | ||
| Other immune effectors | SPP-10 | Saposin-like protein | 1 | 3 | 2 | 1 | 1 | 0 | |
| LYS-8 | Lysozyme | 5 | 1 | 2 | 1 | 0 | 0 | ||
| LYS-4,5,6, & 10 | Lysozyme | 4 | 5 | 2 | 0 | 1 | 0 | ||
| CLEC-48 & 50 | C type domain-containing proteins (CTLD) | 3 | 23 | 2 | 1 | 1 | 0 | ||
| CLEC-178 | CTLD | 1 | 3 | 0 | 0 | 1 | 0 | ||
| CLEC-56 | CTLD | 5 | 1 | 2 | 0 | 0 | 0 | ||
| CLEC-3,10, & 11 | CTLD | 43 | 3 | 0 | 0 | 0 | 0 | ||
| CLEC-150 | CTLD | 1 | 1 | 0 | 0 | 0 | 0 | ||
| FIP-1-like | FIDR protein | 1 | 1 | 0 | 0 | 0 | 0 |
EGL-30 and EGL-8 are known to be involved in both the Insulin/IGF-1 pathway as well as intestinal immunity in C. elegans.
No NSY ortholog was identified in L. oneistus; L. oneistus proteins identified in this cluster are orthologs of human TFG-beta activated kinase MAPKKK7.
L. oneistus immunity pathways putatively involved in symbiosis
Transcriptomics have revealed potential immune pathways functioning in the L. oneistus-bacterium symbiosis. A manual search of adult L. oneistus transcriptomic data (Bulgheresi, 2012a) for immunity genes based on the C. elegans annotation (Harris et al., 2010) indicates that L. oneistus expresses the p38 MAPK module (Table 2). Putative p38 MAPK module activators expressed in L. oneistus include heterotrimeric G protein component beta RACK-1 and protein kinase C PLC-3, as well as a Tribbles homolog 1 (C. elegans NIPI-3). The presence of DBL-1 transcripts in the L. oneistus transcriptome may indicate neuronally secreted DBL-1 triggers the epidermal TGF-beta pathway, the basic components of which are also expressed by L. oneistus. At present, it is not known whether or not the p38 MAPK and the TGF-beta pathways may be triggered in L. oneistus epidermal cells by bacteria contacting the worm's surface. Although epidermal cells underlying an intact cuticle may be insensitive to microbes attached to it, there is a continuum between each GSO lumen and the nematode surface (see background on L. oneistus above). Moreover, the gland and neuronal cells making up each GSO are in direct contact with one another. Therefore, it is very tempting to speculate that the GSO gland cells may mount an immune response instead of – or in addition to – the epidermal cells, and that the GSO neuronal cells may locally modulate their response.
It has long been hypothesized that adult stilbonematids feed on their symbionts, and while this has not yet been observed (Ott et al., 1991) it remains possible that at some developmental stages the ectosymbiont is present, undigested, in the L. oneistus gut. This is even likelier in light of the fact that in contrast to C. elegans, stilbonematids do not possess a grinder that can efficiently crush ingested bacteria (Hoschitz et al., 2001). How might adult L. oneistus intestinal cells react to and limit bacterial proliferation? They express a DFK-2 ortholog, and this kinase could activate the p38 MAPK cascade. Additionally, a neuronally activated ISS pathway might play a role in mediating microbial recognition in the gut (Table 2).
L. oneistus appears to constitutively express signaling pathway components necessary to react to the presence of its ectosymbiont. In particular, transcripts encoding all the members of the TGF-β pathway, which is central in C. elegans epidermal immunity, are present. Secondly, more conservation seems to exist among signaling pathways working in C. elegans and L. oneistus than among the downstream effectors that they regulate (Table 2). These are, notoriously, poorly conserved and it is therefore likely that investigating diverse systems will provide greater insights into host responses to and selectivity for bacteria and will enable the discovery of novel antimicrobials.
Contrasting immunity in free-living and host-associated nematodes
While many nematodes, like L. oneistus and the EPNs, are either free-living or have free-living stages, there are also nematodes such as B. malayi, Ascaris suum, and Trichinella spiralis that complete most of their life cycles within animal hosts and have less exposure to bacterial diversity. For example, Wolbachia are intracellular, mostly restricted to B. malayi reproductive tissue and hypodermal chords, and likely to have existed in a long-term evolutionarily stable relationship with their nematode host. A recent report documented low numbers of Wolbachia in the excretory – secretory canal of B. malayi raising a potential mechanism for release of Wolbachia to the nematode surface or surrounding tissue (Landmann et al., 2010). Wolbachia have also been observed in the intestinal wall of a related filarial nematode (Ferri et al., 2011). Any effects the bacteria in these locations might have on epidermal or intestinal immunity are unknown. There is extensive transcriptomic data for seven different life cycle stages of B. malayi (Choi et al., 2011), which reveals that all the predicted immunity related genes indicated in Table 2 are transcribed with the exception of the MAP kinase kinase, MEK-2.
The lifestyle features of nematodes such as B. malayi, A. suum, and T. spiralis might be expected to result in a very reduced spectrum of nematode immune defense mechanisms. However, the repertoire of immune regulators seems to be broadly conserved across the phylum (Table 2). When looking more specifically at the abundance of immune effectors such as lysozymes, defensin-like ABF proteins, thaumatins, and C-type lectin domain-containing proteins (CTLDs) (Table 3), there seems to be more of a pattern. Nematodes that are either free-living or have free-living stages seem to possess a greater abundance and diversity of both general and adaptively specific immune regulators than those with limited or no free-living stage (Tables 2 and 3). The orthology analysis seems to suggest that the evolution of immune effectors has been sculpted to the lifestyle of each nematode, with those lineages encountering a potentially broader array of microbes having experienced expansions in these protein families. Notably, T. spiralis, an intracellular mammalian parasite with no free-living stage, shows a high level of conservation of immune regulators, but a contraction of immune effector protein families (Tables 2 and 3). B. malayi and A. suum, though closely related phylogenetically, differ in the presence and abundance of immune effector orthologs. A. suum, which lives in the intestine and likely experiences more bacterial interactions, is armed with more immune effectors than B. malayi, which resides in the lymphatic system. It is is possible that this difference in the presence and abundance of immune effectors could result from incomplete sequencing, significant sequence divergence of orthologs such that they are no longer detectable by sequence similarity, the evolution of different immune effectors not orthologous to the C. elegans ones, or the expansion of these gene families in C. elegans. Nevertheless, is tempting to speculate that the diversity of effectors present in the genome positively correlates with the nematode's exposure to microbes and the consequent need for immunity.
Table 3.
A broad protein orthology analysis of all known C. elegans proteins in the listed immune effector categories. For example, there are 265 C. elegans proteins labeled CLEC (1–266 with no protein assigned as CLEC-200), but not all of these have been functionally shown to play a role in immunity. This table shows the total number of clusters generated by an orthology analysis including the species listed across the top as well as the parasitoid wasp, N. vitripennis, as an arthropod outgroup. Those nematodes with limited or no free-living stages are indicated with grey stars (
 ) above their names. See Appendix 1 for analysis methods. All the individual protein names from individual species, identified as orthologs can be found in Supplemental Table 2 (http://www.biolbull.org/content/supplemental).
) above their names. See Appendix 1 for analysis methods. All the individual protein names from individual species, identified as orthologs can be found in Supplemental Table 2 (http://www.biolbull.org/content/supplemental).
| Immune effector | # of clusters | C. elegans | P. pacificus | B. xylophilus | S. carpocapsae |
 B. malayi
B. malayi
|
 A. suum
A. suum
|
 T. spiralis
T. spiralis
|
|---|---|---|---|---|---|---|---|---|
| Lysozymes | 3 | 15 | 14 | 14 | 8 | 2 | 2 | 0 |
| Antimicrobial caenacins | 0 | 11 | 0 | 0 | 0 | 0 | 0 | 0 |
| Caenopores or saposin-like | 9 | 23 | 6 | 2 | 7 | 1 | 4 | 1 |
| Neuropeptide-like proteins (NLPs) | 18 | 47 | 9 | 10 | 15 | 7 | 11 | 1 |
| Thaumatins (THNs) | 1 | 8 | 1 | 1 | 1 | 0 | 0 | 0 |
| Defensin-like ABF proteins | 2 | 6 | 3 | 0 | 0 | 0 | 5 | 0 |
| C type lectin domain-containing proteins (CTLD) | 34 | 265 | 66 | 4 | 15 | 3 | 19 | 0 |
Orthology analysis across several genomes suggests that some immune effectors are lineage specific. For example there is no evidence for orthologs of any of the 11 antimicrobial caenacins of C. elegans (Table 3). Similarly, orthologs of C. elegans genes encoding potentially antimicrobial neuropeptide-like proteins nlp-29, -31 or -33 or other candidate antimicrobial nlp genes encoding a YGGYG motif (nlp-24 through -33) (Gravato-Nobre and Hodgkin, 2005; McVeigh et al., 2008) were not identified (Supplemental Table 2, http://www.biolbull.org/content/supplemental). Interestingly, genes encoding antimicrobial proteins also appear absent in the necromenic nematode Pristionchus pacificus and the migratory endo-plant-parasitic nematode Bursaphelechus xylophilus despite their both having free-living stages (Table 3). In fact, in a survey of 33 nematode EST datasets, orthologs of the 3 C. elegans nlp genes encoding antimicrobials were not found. Sequences with YGGYG motifs were identified, albeit sporadically and predominantly only in representatives of nematode clades 9–12 (Gravato-Nobre and Hodgkin, 2005; McVeigh et al., 2008). Although the bulk of diversity within Nematoda remains to be explored, B. malayi, A. suum, L. oneistus and T. spiralis belong to clades 8, 8, 4, and 2 respectively, indicating that although preliminary, analyses including these species span a considerable segment of the phylum (Holterman et al., 2006). Therefore, the absence of known antimicrobial-encoding nlp genes in B. malayi, A. suum, L. oneistus and T. spiralis suggests that they are an immune adaptation that is unique to C. elegans.
Although our orthology analysis described above relies on knowledge of the C. elegans immune system, it does suggest that 'omics'-acquired data can provide provocative hypotheses including how transient bacterial exposure, symbiosis, and environmental adaptation affect the evolution of nematode immune effectors and other immune pathways.
Exploring parasitism, pathogenesis, and competition through 'omics'
To date almost 700,000 nematode ESTs have been generated, representing about 230,000 genes from 62 nematode species (Elsworth et al., 2011). Sequencing of ESTs from diverse nematodes offers a powerful approach towards uncovering candidate drug targets, lineage-specific parasitic traits as well as conserved features of parasitism. For example, transcriptomics have been used to identify S. carpocapsae and H. bacteriophora nematode genes that may be involved in parasitism. In one study, subtractive hybridization was used to enrich for ESTs expressed by a virulent wild isolate of H. bacteriophora relative to a less virulent wild isolate. This approach revealed 87 ESTs differentially regulated between the strains that may contribute to pathogenesis, almost half of which lacked similarity to sequences in the public database (Hao et al., 2012). In the S. carpocapsae study, investigators sequenced ESTs from the infective stage exposed to insect hemolymph. Of the 1592 unique transcripts, 37% lacked similarity to database sequences (Hao et al., 2010). In both the H. bacteriophora and S. carpocasae studies, among those that do have significant similarity to database sequences are those predicted to be involved in signaling (e.g. G protein), metabolism (e.g. fatty acid catabolism), stress response (e.g. heat shock and oxidative stress-response proteins), and host-parasite interactions (e.g. protease inhibitors, chitinases and lectins). To identify proteins specific to parasitism, Bai et al. (2009) sequenced a library of 31,485 expressed sequence tags (EST) of the EPN H. bacteriophora TT01 (Bai et al., 2009), and classified these ESTs based on their presence in parasitic nematodes and absence in free-living nematodes. This approach yielded 554 genes as candidates for being involved in the parasitic life style of the heterorhabditid nematodes. Again, the majority of these (412) have no matches to known proteins in the public sequence database.
In another study, transcriptome comparison of inbred, laboratory-cultured lines with deteriorated parasitism traits relative to those of parental lines was used to identify potential parasitism genes in H. bacteriophora using microarrays against 15,220 EST probes (Bilgrami et al., 2006; Adhikari et al., 2009). Genes that showed differential expression in the two nematode lines were enriched in metabolism, signal transduction, virulence, and longevity, with the ratio of primary to secondary metabolism being lower in the inbred strain. One of the genes present in higher levels in the inbred line relative to the parent line was nitric oxide synthase interacting protein, predicted to be a negative regulator of NO production (Adhikari et al., 2009). Since NO may be involved in nematode virulence (e.g., it is present in filarial nematode excretory products that inhibit immune cell proliferation (Pfarr et al., 2001)) down regulation of NO might be one contributor to decreased virulence in insects of the inbred line relative to the parent line. Similarly, a microarray study comparing mosquito vectored third stage larvae of the filarial nematode B. malayi to those maintained in culture found numerous differentially expressed genes (Li et al., 2009). Transcripts from mosquito-derived nematodes were enriched for those encoding stress resistance and immune modulation (such as cysteine proteinase inhibitors, which were also identified in H. bacteriophora ESTs), while genes differentially expressed by cultured nematodes were enriched for cell growth and molting (Li et al., 2009). In a recurring theme, of the B. malayi mosquito-derived nematode-specific transcripts, 28% were of unknown function and may represent novel virulence determinants (Li et al., 2009).
The studies described above highlight that while ’omics’ can focus the attention of researchers toward likely genes of interest, comprehensive understanding of molecular and cellular processes can only come from in depth genetic and biochemical analyses. Since many candidate parasitism genes lack significant homologs in the database are therefore absent from the genetically tractable model organism C. elegans, investigations into their function must necessarily be conducted in nematode parasites. Therefore, it is critical to continue developing tools such as transformation and RNA interference that are necessary to investigate gene function in a broader array of nematode genera. Furthermore, there is a need for in-depth comparative analyses of transcriptome datasets from diverse nematode systems to facilitate the identification of conserved and diverged mechanisms by which parasitic nematodes overcome their hosts' immune defenses.
The bacterial symbiont partners can also contribute to parasitism. For example, EPNs rely on their bacterial symbionts to help kill the insect host and to support reproduction in the cadaver. These bacterial symbionts can themselves be bona fide insect pathogens, capable of killing insects within several hours after injection into the insect blood cavity (Eleftherianos et al., 2006; Richards and Goodrich-Blair, 2009). Comparative transcriptomics have been applied to identify P. luminescens TT01 genes potentially involved in insect pathogenesis. Genes differentially regulated between a virulent strain (TT01α) and an attenuated phenotypic variant included those encoding toxins, secreted enzymes, and proteins involved in oxidative stress (Lanois et al., 2011). An et al. 2009 used “Selective Capture of Transcribed Sequences” to identify X. koppenhoeferi and P. temperata genes expressed more highly during infection of insects compared to laboratory growth, in an effort to identify virulence factors commonly and distinctly used by these bacteria. Both bacteria displayed in vivo up-regulation of genes involved in stress response genes, toxin production (tcaC), hemolysins, fatty acid biosynthesis (reminiscent of the H. bacteriophora ESTs identified in more virulent strains described above), and metal transport. These authors further analyzed their data using a pathway-building program (PathwayStudio, Ariadne, Rockville, MD) to reveal patterns and pathways involved in virulence of the two EPN bacteria. Continued mapping of both nematode and bacterial metabolic pathways induced during infection has the potential to reveal metabolic integration in the symbiosis.
One of the mutualistic services provided by the bacteria to their nematode partners is protection of the cadaver from scavengers and opportunistic organisms that may compete for nutrients. The Xenorhabdus and Photorhabdus genera therefore offer tremendous potential as a source of anti-insecticidal, antimicrobial, and other bioactive molecules, and genomics has opened numerous doors to the discovery of novel metabolites. To date, the genomes of four EPN bacterial symbionts have been sequenced and analyzed from a comparative perspective (Duchaud et al., 2003; Latreille et al., 2007; Wilkinson et al., 2009; Ogier et al., 2010; Chaston et al. 2011). These sequences revealed numerous loci predicted to encode secondary metabolites with potential pharmaceutical and agricultural uses (Bode, 2009). There are at least 23 biosynthetic gene clusters in P. luminescens TT01 (Duchaud et al., 2003; Bode, 2009), primarily non-ribosomal peptide synthetases (NRPS). Similarly P. asymbiotica encodes a rich diversity of NRPS or polyketide synthetase loci (Wilkinson et al., 2009). This potential for secondary metabolite production was revealed through genome sequencing and belies the few compounds that were known based on experimental approaches. Also, while some molecules had been biochemically characterized, the genes encoding them had not been identified, precluding detailed analysis of their synthesis and efforts to engineer high output production. Genome sequences have provided an invaluable resource for identification of genes responsible for secondary metabolite production. For example, the genes responsible for synthesis of xenematide, a molecule with anti-microbial activity, were bioinformatically predicted (Crawford et al., 2011).
Genomic analyses reveal bacterial contributions to nematode genome evolution
Lateral gene transfer between nematodes and bacteria
Nematode-bacterium symbioses have contributed to our understanding of genome evolution, including genome plasticity and microbial-eukaryotic lateral gene transfer (LGT). LGT between eukaryotes and prokaryotes has been documented through transcriptome and genome analysis of plant parasitic nematodes (Scholl and Bird, 2011) with genes encoding glucanases and pectate lyases that are absent in other animals but are similar to those of rhizosphere bacteria. These genes are fully integrated into the genomes, with introns and mRNA processing typical of eukaryotes. They are prevalent among plant-parasitic nematodes such as Meloidogyne incognita and M. hapla, suggesting ancient acquisition (Scholl et al., 2003; Scholl and Bird, 2011). Investigations on the genomes of, Pristionchus pacificus (Dieterich et al., 2008) and Bursaphelenchus xylophilus (Kikuchi et al., 2011), provide additional compelling evidence that LGT of microbial genes is a component of nematode evolution (Mayer et al., 2011).
Bacterial symbionts that are closely associated with the germ-line of their hosts are most likely to contribute to LGT events, and therefore it might not be surprising to find evidence of LGT in nematodes symbiotically associated with such bacteria. Indeed, fragments of Wolbachia DNA appear to be present in non-coding regions of the filarial nematodes Onchocerca volvulus, O. ochengi (Fenn et al., 2006), B. malayi and Dirofilaria immitis (Dunning Hotopp et al., 2007). Further evidence comes from 454 pyrosequencing that identified Wolbachia genes in two naturally Wolbachia-free filarial nematodes: Acanthocheilonema viteae and Onchocerca flexuosa (McNulty et al., 2010). Based on the hypothesis that the ancestor of extant filarids in the Onchocercinae and Dirofilariinae was in a symbiosis with Wolbachia (Casiraghi et al., 2004), the authors posited that the presence of Wolbachia DNA in these uncolonized symbionts is evidence of former infection and ancient LGT. That these genes might play some functional role in the nematode is supported by evidence that some of the Wolbachia sequences are expressed in specific tissues (McNulty et al., 2010). The presence of Wolbachia DNA in the genomes of many filarial nematodes raises intriguing possibilities about the role of symbiont DNA in shaping nematode evolution. In addition to Wolbachia-derived fragments, filarial nematodes also contain a functional ferrochelatase gene (last step in heme biosynthesis) that includes introns and a mitochondrial targeting signal but appears to be the result of horizontal transfer from a Rhizobiales bacterium (Slatko et al., 2010).
Since cross-kingdom LGT horizontal gene transfer from bacteria to nematodes has been revealed in worms from several different clades, this method of gene acquisition may be commonplace among nematodes, or at least in chromadorean nematodes, and may represent an additional route by which nematodes may gain essential functions - a symbiosis of sorts.
Insights into bacterial genome evolution revealed by comparative genomics
Aspects of genome evolution have been explored through comparative analysis of non-core regions of the genomes of the EPN symbionts Photorhabdus spp. and Xenorhabdus spp. Flexible genome regions, or regions of genome plasticity (RGP) are defined as DNA sequences that are absent from one or more genomes being analyzed. In Photorhabdus and Xenorhabdus comparisons, as much as 60% of the genomic content falls into this class (Ogier et al., 2010). Analysis of these regions revealed that regions of genomic plasticity are made up of modules that can be shuffled by recombination and are proposed to be the actual units of genome plasticity. Indeed, the authors show that a P. luminescens TTO1-derived strain that had been associated with lab-reared nematodes had several deletions within RGP compared to the reference strain. These deletions encompassed modules, rather than entire RGP, and appeared to result from a single block deletion event.
Another observation of this study was that P. asymbiotica, the species isolated from human wounds, has a higher proportion, relative to the other Photorhabdus and Xenorhabdus genomes, of RGP that do not have canonical markers of mobile genetic elements (e.g. they lack transposases, insertion elements, or genes encoding DNA modification enzymes). The authors suggest that understanding the functions of genes encoded on these regions might give insights into the evolution of P. asymbiotica as a human pathogen (Ogier et al., 2010).
Conclusions
The biology of nematode-bacterial symbiotic associations is far-reaching and fundamental. Although in its infancy, the broad knowledge gained by “omics” studies in diverse biological disciplines including symbiosis, evolution, immunology, infectious disease and secondary metabolism, is already remarkable. These studies have revealed several key interactions that are common within nematode – bacterial interactions, such as bacterial contribution to nematode development and genome content. However, they also highlight that while the themes are common, the molecular mechanisms underlying them are likely specific to the system, as in the immune pathways involved in direct communication. While large-scale data-generating ‘omics’-style experiments have been critical for identifying important themes and mechanisms, they are only a starting place, producing many provocative hypotheses that remain to be functionally tested. As such, it is important that the necessary tools for subsequent mechanistic explorations (e.g. transformation and RNAi) continue to be developed in diverse nematode systems. As more data sets from previously unexplored clades of the phylum are produced, continued conversation between systems will be critical to further our understanding of conserved and unique patterns in the evolving relationships between nematodes and the bacteria they encounter.
Supplementary Material
Acknowledgments
Discussions relevant to this paper were facilitated by the NEMASYM (Nematode-Bacterium Symbioses) Research Coordination Network (NSF- IOS 0840932 to SPS), which was also used to support publication costs. ARD was supported by a United States Public Health Service Training Grant (T32GM07616). K.E.M. was supported by National Institutes of Health (NIH) National Research Service Award T32 AI55397. JMF and BES are supported by New England Biolabs, Inc. PWS is an Investigator with the Howard Hughes Medical Institute. HGB was supported by NSF (IOS-0920631 and IOS- 0950873). S.B. is very grateful to Mark Blaxter and Stephen Bridgett for sequencing, assembling and making publically available the Laxus oneistus transcriptome. S.B. is supported by the Austrian Science Fund (FWF) grant P224701.
Footnotes
The official version is available at http://www.biolbull.org/content/223/1/85.full.
Literature Cited
- Abad P, Gouzy J, Aury JM, Castagnone-Sereno P, Danchin EG, Deleury E, Perfus-Barbeoch L, Anthouard V, Artiguenave F, Blok VC, Caillaud MC, Coutinho PM, Dasilva C, De Luca F, Deau F, Esquibet M, Flutre T, Goldstone JV, Hamamouch N, Hewezi T, Jaillon O, Jubin C, Leonetti P, Magliano M, Maier TR, Markov GV, McVeigh P, Pesole G, Poulain J, Robinson-Rechavi M, Sallet E, Segurens B, Steinbach D, Tytgat T, Ugarte E, van Ghelder C, Veronico P, Baum TJ, Blaxter M, Bleve-Zacheo T, Davis EL, Ewbank JJ, Favery B, Grenier E, Henrissat B, Jones JT, Laudet V, Maule AG, Quesneville H, Rosso MN, Schiex T, Smant G, Weissenbach J, Wincker P. Genome sequence of the metazoan plant-parasitic nematode Meloidogyne incognita. Nat Biotechnol. 2008;26:909–915. doi: 10.1038/nbt.1482. [DOI] [PubMed] [Google Scholar]
- Aboobaker AA, Blaxter ML. Medical significance of Caenorhabditis elegans. Ann Med. 2000;32:23–30. doi: 10.3109/07853890008995906. [DOI] [PubMed] [Google Scholar]
- Adhikari BN, Lin CY, Bai X, Ciche TA, Grewal PS, Dillman AR, Chaston JM, Shapiro-Ilan DI, Bilgrami AL, Gaugler R, Sternberg PW, Adams BJ. Transcriptional profiling of trait deterioration in the insect pathogenic nematode Heterorhabditis bacteriophora. BMC Genomics. 2009;10:609. doi: 10.1186/1471-2164-10-609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alper S, McBride SJ, Lackford B, Freedman JH, Schwartz DA. Specificity and complexity of the Caenorhabditis elegans innate immune response. Mol Cell Biol. 2007;27:5544–5553. doi: 10.1128/MCB.02070-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An R, Grewal PS. Molecular mechanisms of persistence of mutualistic bacteria Photorhabdus in the entomopathogenic nematode host. PLoS One. 2010;5:e13154. doi: 10.1371/journal.pone.0013154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai X, Adams BJ, Ciche TA, Clifton S, Gaugler R, Hogenhout SA, Spieth J, Sternberg PW, Wilson RK, Grewal PS. Transcriptomic analysis of the entomopathogenic nematode Heterorhabditis bacteriophora TTO1. BMC Genomics. 2009;10:205. doi: 10.1186/1471-2164-10-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashey F, Young SK, Hawlena H, Lively CM. Spiteful interactions between sympatric natural isolates of Xenorhabdus bovienii benefit kin and reduce virulence. J Evol Biol. 2012;25:431–437. doi: 10.1111/j.1420-9101.2011.02441.x. [DOI] [PubMed] [Google Scholar]
- Bauer-Nebelsick M, Blumer M, Urbancik W, Ott JA. The glandular sensory organ of Desmodoridae (Nematoda)- ultrastructure and phylogenetic implications. Invert Biol. 1995;114:211–219. [Google Scholar]
- Bayer C, Heindl NR, Rinke C, Lucker S, Ott JA, Bulgheresi S. Molecular characterization of the symbionts associated with marine nematodes of the genus Robbea. Environ Microbiol Rep. 2009;1:136–144. doi: 10.1111/j.1758-2229.2009.00019.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker T, Loch G, Beyer M, Zinke I, Aschenbrenner AC, Carrera P, Inhester T, Schultze JL, Hoch M. FOXO-dependent regulation of innate immune homeostasis. Nature. 2010;463:369–373. doi: 10.1038/nature08698. [DOI] [PubMed] [Google Scholar]
- Bennuru S, Meng Z, Ribeiro JM, Semnani RT, Ghedin E, Chan K, Lucas DA, Veenstra TD, Nutman TB. Stage-specific proteomic expression patterns of the human filarial parasite Brugia malayi and its endosymbiont Wolbachia. Proc Natl Acad Sci USA. 2011;108:9649–9654. doi: 10.1073/pnas.1011481108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennuru S, Semnani R, Meng Z, Ribeiro JM, Veenstra TD, Nutman TB. Brugia malayi excreted/secreted proteins at the host/parasite interface: stage- and gender-specific proteomic profiling. PLoS Negl Trop Dis. 2009;3:e410. doi: 10.1371/journal.pntd.0000410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhasin A, Chaston JM, Goodrich-Blair H. Mutational analyses reveal overall topology and functional regions of NilB, a bacterial outer membrane protein required for host-association in a model animal-bacterial mutualism. J Bacteriol. 2012;194:1763–1776. doi: 10.1128/JB.06711-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian G, Xu Y, Lu P, Xie Y, Xi Z. The endosymbiotic bacterium Wolbachia induces resistance to dengue virus in Aedes aegypti. PLoS Pathog. 2010;6:e1000833. doi: 10.1371/journal.ppat.1000833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilgrami AL, Gaugler R, Shapiro-Ilan D, Adams BJ. Source of trait deterioration in entomopathogenic nematodes Heterorhabditis bacteriophora and Steinernema carpocapsae during in vivo culture. Nematol. 2006;8:397–409. [Google Scholar]
- Bird DM, Kaloshian I. Are roots special? Nematodes have their say. Physiol Mol Plant Pathol. 2003;62:115–123. [Google Scholar]
- Blaxter M. Nematodes: the worm and its relatives. PLoS Biol. 2011;9:e1001050. doi: 10.1371/journal.pbio.1001050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaxter M. [Accessed January 7, 2012.];Nembase4: Nematode Transcriptome Analyses. 2012 http://www.nematodes.org/nembase4/overview.shtml.
- Bode HB. Entomopathogenic bacteria as a source of secondary metabolites. Curr Opin Chem Biol. 2009;13:224–230. doi: 10.1016/j.cbpa.2009.02.037. [DOI] [PubMed] [Google Scholar]
- Borgonie G, Garcia-Moyano A, Litthauer D, Bert W, Bester A, van Heerden E, Moller C, Erasmus M, Onstott TC. Nematoda from the terrestrial deep subsurface of South Africa. Nature. 2011;474:79–82. doi: 10.1038/nature09974. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownlie JC, Cass BN, Riegler M, Witsenburg JJ, Iturbe-Ormaetxe I, McGraw EA, O'Neill SL. Evidence for metabolic provisioning by a common invertebrate endosymbiont, Wolbachia pipientis, during periods of nutritional stress. PLoS Pathog. 2009;5:e1000368. doi: 10.1371/journal.ppat.1000368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulgheresi S. Calling the roll on Laxus oneistus immune defense molecules. Symbiosis. 2011;55:127–135. doi: 10.1007/s13199-012-0157-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulgheresi S. [Accessed October 27, 2011.];Adult Laxus oneistus 454 transcriptome. 2012a http://genepool.bio.ed.ac.uk/GP_Partigene/2008075_SilviaBulgheresi/
- Bulgheresi S. [Accessed October 27, 2011.];Adult Laxus oneistus ESTs. 2012b http://www.nematodes.org/NeglectedGenomes/NEMATODA/Laxus_oneistus/index.html.
- Bulgheresi S, Gruber-Vodicka HR, Heindl NR, Dirks U, Kostadinova M, Breiteneder H, Ott JA. Sequence variability of the pattern recognition receptor Mermaid mediates specificity of marine nematode symbioses. ISME J. 2011;5:986–998. doi: 10.1038/ismej.2010.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulgheresi S, Schabussova I, Chen T, Mullin NP, Maizels RM, Ott JA. A new C-type lectin similar to the human immunoreceptor DC-SIGN mediates symbiont acquisition by a marine nematode. Appl Environ Microbiol. 2006;72:2950–2956. doi: 10.1128/AEM.72.4.2950-2956.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casiraghi M, Bain O, Guerrero R, Martin C, Pocacqua V, Gardner SL, Franceschi A, Bandi C. Mapping the presence of Wolbachia pipientis on the phylogeny of filarial nematodes: evidence for symbiont loss during evolution. Int J Parasitol. 2004;34:191–203. doi: 10.1016/j.ijpara.2003.10.004. [DOI] [PubMed] [Google Scholar]
- Chaston JM, Suen G, Tucker SL, Andersen AW, Bhasin A, Bode E, Bode HB, Brachmann AO, Cowles CE, Cowles KN, Darby C, de Léon L, Drace K, Du Z, Givaudan A, Herbert Tran EE, Jewell KA, Knack JJ, Krasomil-Osterfeld KC, Kukor R, Lanois A, Latreille P, Leimgruber NK, Lipke CM, Liu R, Lu X, Martens EC, Marri PR, Médigue C, Menard ML, Miller NM, Morales-Soto N, Norton S, Ogier J-C, Orchard SS, Park D, Park Y, Qurollo BA, Sugar DR, Richards GR, Rouy Z, Slominski B, Slominski K, Snyder H, Tjaden BC, van der Hoeven R, Welch RD, Wheeler C, Xiang B, Barbazuk B, Gaudriault S, Goodner B, Slater SC, Forst S, Goldman BS, Goodrich-Blair H. The entomopathogenic bacterial endosymbionts Xenorhabdus and Photorhabdus: Convergent lifestyles from divergent genomes. PLoS One. 2011;6:e27909. doi: 10.1371/journal.pone.0027909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitwood DJ. Research on plant-parasitic nematode biology conducted by the United States Department of Agriculture-Agricultural Research Service. Pest Manag Sci. 2003;59:748–753. doi: 10.1002/ps.684. [DOI] [PubMed] [Google Scholar]
- Choi YJ, Ghedin E, Berriman M, McQuillan J, Holroyd N, Mayhew GF, Christensen BM, Michalski ML. A deep sequencing approach to comparatively analyze the transcriptome of lifecycle stages of the filarial worm, Brugia malayi. PLoS Negl Trop Dis. 2011;5:e1409. doi: 10.1371/journal.pntd.0001409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciche T. The biology and genome of Heterorhabditis bacteriophora. WormBook. 2007:1–9. doi: 10.1895/wormbook.1.135.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciche TA, Sternberg PW. Postembryonic RNAi in Heterorhabditis bacteriophora: a nematode insect parasite and host for insect pathogenic symbionts. BMC Dev Biol. 2007;7:101. doi: 10.1186/1471-213X-7-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke DJ. Photorhabdus: a model for the analysis of pathogenicity and mutualism. Cell Microbiol. 2008;10:2159–2167. doi: 10.1111/j.1462-5822.2008.01209.x. [DOI] [PubMed] [Google Scholar]
- Correale J, Farez MF. The impact of parasite infections on the course of multiple sclerosis. J Neuroimmunol. 2011;233:6–11. doi: 10.1016/j.jneuroim.2011.01.002. [DOI] [PubMed] [Google Scholar]
- Couillault C, Ewbank JJ. Diverse bacteria are pathogens of Caenorhabditis elegans. Infect Immun. 2002;70:4705–4707. doi: 10.1128/IAI.70.8.4705-4707.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowles CE, Goodrich-Blair H. The Xenorhabdus nematophila nilABC genes confer the ability of Xenorhabdus spp. to colonize Steinernema carpocapsae nematodes. J Bacteriol. 2008;190:4121–4128. doi: 10.1128/JB.00123-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford JM, Portmann C, Kontnik R, Walsh CT, Clardy J. NRPS substrate promiscuity diversifies the Xenematides. Org Lett. 2011;13:5144–5147. doi: 10.1021/ol2020237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crompton DW. How much human helminthiasis is there in the world? J Parasitol. 1999;85:397–403. [PubMed] [Google Scholar]
- De Ley P. A quick tour of nematode diversity and the backbone of nematode phylogeny. WormBook. 2006:1–8. doi: 10.1895/wormbook.1.41.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derzelle S, Ngo S, Turlin E, Duchaud E, Namane A, Kunst F, Danchin A, Bertin P, Charles JF. AstR-AstS, a new two-component signal transduction system, mediates swarming, adaptation to stationary phase and phenotypic variation in Photorhabdus luminescens. Microbiology. 2004;150:897–910. doi: 10.1099/mic.0.26563-0. [DOI] [PubMed] [Google Scholar]
- Dieterich C, Clifton SW, Schuster LN, Chinwalla A, Delehaunty K, Dinkelacker I, Fulton L, Fulton R, Godfrey J, Minx P, Mitreva M, Roeseler W, Tian H, Witte H, Yang SP, Wilson RK, Sommer RJ. The Pristionchus pacificus genome provides a unique perspective on nematode lifestyle and parasitism. Nat Genet. 2008;40:1193–1198. doi: 10.1038/ng.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillman AR, Chaston JM, Adams BJ, Ciche TA, Goodrich-Blair H, Stock SP, Sternberg PW. An entomopathogenic nematode by any other name. PLoS Pathog. 2012;8:e1002527. doi: 10.1371/journal.ppat.1002527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas AE. Conflict, cheats and the persistence of symbioses. New Phytol. 2008;177:849–858. doi: 10.1111/j.1469-8137.2007.02326.x. [DOI] [PubMed] [Google Scholar]
- Duchaud E, Rusniok C, Frangeul L, Buchrieser C, Givaudan A, Taourit S, Bocs S, Boursaux-Eude C, Chandler M, Charles JF, Dassa E, Derose R, Derzelle S, Freyssinet G, Gaudriault S, Medigue C, Lanois A, Powell K, Siguier P, Vincent R, Wingate V, Zouine M, Glaser P, Boemare N, Danchin A, Kunst F. The genome sequence of the entomopathogenic bacterium Photorhabdus luminescens. Nat Biotechnol. 2003;21:1307–1313. doi: 10.1038/nbt886. [DOI] [PubMed] [Google Scholar]
- Dunning Hotopp JC, Clark ME, Oliveira DC, Foster JM, Fischer P, Munoz Torres MC, Giebel JD, Kumar N, Ishmael N, Wang S, Ingram J, Nene RV, Shepard J, Tomkins J, Richards S, Spiro DJ, Ghedin E, Slatko BE, Tettelin H, Werren JH. Widespread lateral gene transfer from intracellular bacteria to multicellular eukaryotes. Science. 2007;317:1753–1756. doi: 10.1126/science.1142490. [DOI] [PubMed] [Google Scholar]
- Easom CA, Clarke DJ. Environ Microbiol. 2011. HdfR is a regulator in Photorhabdus luminescens that modulates metabolism and symbiosis with the nematode Heterorhabditis. [DOI] [PubMed] [Google Scholar]
- Easom CA, Joyce SA, Clarke DJ. Identification of genes involved in the mutualistic colonization of the nematode Heterorhabditis bacteriophora by the bacterium Photorhabdus luminescens. BMC Microbiol. 2010;10:45. doi: 10.1186/1471-2180-10-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eleftherianos I, ffrench-Constant RH, Clarke DJ, Dowling AJ, Reynolds SE. Dissecting the immune response to the entomopathogen Photorhabdus. Trends Microbiol. 2010;18:552–560. doi: 10.1016/j.tim.2010.09.006. [DOI] [PubMed] [Google Scholar]
- Eleftherianos I, Marokhazi J, Millichap PJ, Hodgkinson AJ, Sriboonlert A, ffrench-Constant RH, Reynolds SE. Prior infection of Manduca sexta with non-pathogenic Escherichia coli elicits immunity to pathogenic Photorhabdus luminescens: roles of immune-related proteins shown by RNA interference. Insect Biochem Mol Biol. 2006;36:517–525. doi: 10.1016/j.ibmb.2006.04.001. [DOI] [PubMed] [Google Scholar]
- Elsworth B, Wasmuth J, Blaxter M. NEMBASE4: the nematode transcriptome resource. Int J Parasitol. 2011;41:881–894. doi: 10.1016/j.ijpara.2011.03.009. [DOI] [PubMed] [Google Scholar]
- Ettema CH. Soil nematode diversity: species coexistence and ecosystem function. J Nematol. 1998;30:159–169. [PMC free article] [PubMed] [Google Scholar]
- Ewbank JJ, Zugasti O. C. elegans: model host and tool for antimicrobial drug discovery. Dis Models Mech. 2011;4:300–304. doi: 10.1242/dmm.006684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenn K, Blaxter M. Quantification of Wolbachia bacteria in Brugia malayi through the nematode lifecycle. Mol Biochem Parasitol. 2004;137:361–364. doi: 10.1016/j.molbiopara.2004.06.012. [DOI] [PubMed] [Google Scholar]
- Fenn K, Conlon C, Jones M, Quail MA, Holroyd NE, Parkhill J, Blaxter M. Phylogenetic relationships of the Wolbachia of nematodes and arthropods. PLoS Pathog. 2006;2:e94. doi: 10.1371/journal.ppat.0020094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferri E, Bain O, Barbuto M, Martin C, Lo N, Uni S, Landmann F, Baccei SG, Guerrero R, de Souza Lima S, Bandi C, Wanji S, Diagne M, Casiraghi M. New insights into the evolution of Wolbachia infections in filarial nematodes inferred from a large range of screened species. PLoS One. 2011;6:e20843. doi: 10.1371/journal.pone.0020843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster J, Ganatra M, Kamal I, Ware J, Makarova K, Ivanova N, Bhattacharyya A, Kapatral V, Kumar S, Posfai J, Vincze T, Ingram J, Moran L, Lapidus A, Omelchenko M, Kyrpides N, Ghedin E, Wang S, Goltsman E, Joukov V, Ostrovskaya O, Tsukerman K, Mazur M, Comb D, Koonin E, Slatko B. The Wolbachia genome of Brugia malayi: endosymbiont evolution within a human pathogenic nematode. PLoS Biol. 2005;3:e121. doi: 10.1371/journal.pbio.0030121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster JM, Hoerauf A, Slatko BE, Taylor MJ. The molecular biology, immunology and chemotherapy of Wolbachia bacterial endosymbionts of filarial nematodes. in Parasitic Nematodes. In: Kennedy MW, Harnett W, editors. Molecular biology, Biochemistry and Immunology. CABI; Wallingford, UK: in press. [Google Scholar]
- Freyth K, Janowitz T, Nunes F, Voss M, Heinick A, Bertaux J, Scheu S, Paul RJ. Reproductive fitness and dietary choice behavior of the genetic model organism Caenorhabditis elegans under semi-natural conditions. Mol Cells. 2010;30:347–353. doi: 10.1007/s10059-010-0125-9. [DOI] [PubMed] [Google Scholar]
- Garsin DA, Villanueva JM, Begun J, Kim DH, Sifri CD, Calderwood SB, Ruvkun G, Ausubel FM. Long-lived C. elegans daf-2 mutants are resistant to bacterial pathogens. Science. 2003;300:1921. doi: 10.1126/science.1080147. [DOI] [PubMed] [Google Scholar]
- Gaudriault S, Duchaud E, Lanois A, Canoy AS, Bourot S, Derose R, Kunst F, Boemare N, Givaudan A. Whole-genome comparison between Photorhabdus strains to identify genomic regions involved in the specificity of nematode interaction. J Bacteriol. 2006;188:809–814. doi: 10.1128/JB.188.2.809-814.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudriault S, Pages S, Lanois A, Laroui C, Teyssier C, Jumas-Bilak E, Givaudan A. Plastic architecture of bacterial genome revealed by comparative genomics of Photorhabdus variants. Genome Biol. 2008;9:R117. doi: 10.1186/gb-2008-9-7-r117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghedin E, Hailemariam T, DePasse JV, Zhang X, Oksov Y, Unnasch TR, Lustigman S. Brugia malayi gene expression in response to the targeting of the Wolbachia endosymbiont by tetracycline treatment. PLoS Negl Trop Dis. 2009;3:e525. doi: 10.1371/journal.pntd.0000525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghedin E, Wang S, Spiro D, Caler E, Zhao Q, Crabtree J, Allen JE, Delcher AL, Guiliano DB, Miranda-Saavedra D, Angiuoli SV, Creasy T, Amedeo P, Haas B, El-Sayed NM, Wortman JR, Feldblyum T, Tallon L, Schatz M, Shumway M, Koo H, Salzberg SL, Schobel S, Pertea M, Pop M, White O, Barton GJ, Carlow CK, Crawford MJ, Daub J, Dimmic MW, Estes CF, Foster JM, Ganatra M, Gregory WF, Johnson NM, Jin J, Komuniecki R, Korf I, Kumar S, Laney S, Li BW, Li W, Lindblom TH, Lustigman S, Ma D, Maina CV, Martin DM, McCarter JP, McReynolds L, Mitreva M, Nutman TB, Parkinson J, Peregrin-Alvarez JM, Poole C, Ren Q, Saunders L, Sluder AE, Smith K, Stanke M, Unnasch TR, Ware J, Wei AD, Weil G, Williams DJ, Zhang Y, Williams SA, Fraser-Liggett C, Slatko B, Blaxter ML, Scott AL. Draft genome of the filarial nematode parasite Brugia malayi. Science. 2007;317:1756–1760. doi: 10.1126/science.1145406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaser RL, Meola MA. The native Wolbachia endosymbionts of Drosophila melanogaster and Culex quinquefasciatus increase host resistance to West Nile virus infection. PLoS One. 2010;5:e11977. doi: 10.1371/journal.pone.0011977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich-Blair H. They've got a ticket to ride: Xenorhabdus nematophila-Steinernema carpocapsae symbiosis. Curr Opin Microbiol. 2007;10:225–230. doi: 10.1016/j.mib.2007.05.006. [DOI] [PubMed] [Google Scholar]
- Goodrich-Blair H, Clarke DJ. Mutualism and pathogenesis in Xenorhabdus and Photorhabdus: two roads to the same destination. Mol Microbiol. 2007;64:260–268. doi: 10.1111/j.1365-2958.2007.05671.x. [DOI] [PubMed] [Google Scholar]
- Gravato-Nobre MJ, Hodgkin J. Caenorhabditis elegans as a model for innate immunity to pathogens. Cell Microbiol. 2005;7:741–751. doi: 10.1111/j.1462-5822.2005.00523.x. [DOI] [PubMed] [Google Scholar]
- Grewal PS, Ehlers RU, Shapiro-Ilan DI, editors. Nematodes as Biocontrol Agents. CABI Publishing; Wallingford, UK: 2005. [Google Scholar]
- Hallem EA, Dillman AR, Hong AV, Zhang Y, Yano JM, DeMarco SF, Sternberg PW. A sensory code for host seeking in parasitic nematodes. Curr Biol. 2011;21:377–383. doi: 10.1016/j.cub.2011.01.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen RD, Trees AJ, Bah GS, Hetzel U, Martin C, Bain O, Tanya VN, Makepeace BL. A worm's best friend: recruitment of neutrophils by Wolbachia confounds eosinophil degranulation against the filarial nematode Onchocerca ochengi. Proc Biol Sci. 2011;278:2293–2302. doi: 10.1098/rspb.2010.2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao YJ, Montiel R, Abubucker S, Mitreva M, Simoes N. Transcripts analysis of the entomopathogenic nematode Steinernema carpocapsae induced in vitro with insect haemolymph. Mol Biochem Parasitol. 2010;169:79–86. doi: 10.1016/j.molbiopara.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao YJ, Montiel R, Lucena MA, Costa M, Simoes N. Genetic diversity and comparative analysis of gene expression between Heterorhabditis bacteriophora Az29 and Az36 isolates: uncovering candidate genes involved in insect pathogenicity. Exp Parasitol. 2012;130:116–125. doi: 10.1016/j.exppara.2011.12.001. [DOI] [PubMed] [Google Scholar]
- Harris TW, Antoshechkin I, Bieri T, Blasiar D, Chan J, Chen WJ, De La Cruz N, Davis P, Duesbury M, Fang R, Fernandes J, Han M, Kishore R, Lee R, Muller HM, Nakamura C, Ozersky P, Petcherski A, Rangarajan A, Rogers A, Schindelman G, Schwarz EM, Tuli MA, Van Auken K, Wang D, Wang X, Williams G, Yook K, Durbin R, Stein LD, Spieth J, Sternberg PW. WormBase: a comprehensive resource for nematode research. Nucleic Acids Res. 2010;38:D463–467. doi: 10.1093/nar/gkp952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heindl NR, Gruber-Vodicka HR, Bayer C, Luecker S, Ott JA, Bulgheresi S. First detection of thiotrophic symbiont phylotypes in the pelagic marine environment. FEMS Microbiol Ecol. 2011 doi: 10.1111/j.1574-6941.2011.01096.x. [DOI] [PubMed] [Google Scholar]
- Hentschel U, Berger EC, Bright M, Felbeck H, Ott JA. Metabolism of nitrogen and sulfur in ectosymbiotic bacteria of marine nematodes (Nematoda, Stilbonematinae) MEPS. 1999;183:149–158. [Google Scholar]
- Herbert EE, Goodrich-Blair H. Friend and foe: the two faces of Xenorhabdus nematophila. Nat Rev Microbiol. 2007;5:634–646. doi: 10.1038/nrmicro1706. [DOI] [PubMed] [Google Scholar]
- Heungens K, Cowles CE, Goodrich-Blair H. Identification of Xenorhabdus nematophila genes required for mutualistic colonization of Steinernema carpocapsae nematodes. Mol Microbiol. 2002;45:1337–1353. doi: 10.1046/j.1365-2958.2002.03100.x. [DOI] [PubMed] [Google Scholar]
- Hilgenboecker K, Hammerstein P, Schlattmann P, Telschow A, Werren JH. How many species are infected with Wolbachia?--A statistical analysis of current data. FEMS Microbiol Lett. 2008;281:215–220. doi: 10.1111/j.1574-6968.2008.01110.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holterman M, van der Wurff A, van den Elsen S, van Megen H, Bongers T, Holovachov O, Bakker J, Helder J. Phylum-wide analysis of SSU rDNA reveals deep phylogenetic relationships among nematodes and accelerated evolution toward crown clades. Mol Biol Evol. 2006;23:1792–1800. doi: 10.1093/molbev/msl044. [DOI] [PubMed] [Google Scholar]
- Hoschitz M, Bright M, Ott JA. Ultrastructure and reconstruction of the pharynx of Leptonemella juliae (Nematoda, Adenophorea) Zoomorphol. 2001;121:95–107. [Google Scholar]
- Irazoqui JE, Urbach JM, Ausubel FM. Evolution of host innate defence: insights from Caenorhabditis elegans and primitive invertebrates. Nat Rev Immunol. 2010;10:47–58. doi: 10.1038/nri2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampfer S, Sturmbauer C, Ott CJ. Phylogenetic analysis of rDNA sequences from adenophorean nematodes and implications for the Adenophorea-Secernetea controversy. Invert Biol. 1998;117:29–36. [Google Scholar]
- Kikuchi T, Cotton JA, Dalzell JJ, Hasegawa K, Kanzaki N, McVeigh P, Takanashi T, Tsai IJ, Assefa SA, Cock PJ, Otto TD, Hunt M, Reid AJ, Sanchez-Flores A, Tsuchihara K, Yokoi T, Larsson MC, Miwa J, Maule AG, Sahashi N, Jones JT, Berriman M. Genomic insights into the origin of parasitism in the emerging plant pathogen Bursaphelenchus xylophilus. PLoS Pathog. 2011;7:e1002219. doi: 10.1371/journal.ppat.1002219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH, Feinbaum R, Alloing G, Emerson FE, Garsin DA, Inoue H, Tanaka-Hino M, Hisamoto N, Matsumoto K, Tan MW, Ausubel FM. A conserved p38 MAP kinase pathway in Caenorhabditis elegans innate immunity. Science. 2002;297:623–626. doi: 10.1126/science.1073759. [DOI] [PubMed] [Google Scholar]
- Kuijk LM, van Die I. Worms to the rescue: can worm glycans protect from autoimmune diseases? IUBMB Life. 2010;62:303–312. doi: 10.1002/iub.304. [DOI] [PubMed] [Google Scholar]
- Kumar S, Chaudhary K, Foster JM, Novelli JF, Zhang Y, Wang S, Spiro D, Ghedin E, Carlow CK. Mining predicted essential genes of Brugia malayi for nematode drug targets. PLoS One. 2007;2:e1189. doi: 10.1371/journal.pone.0001189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambshead PJD, Boucher G. Marine nematode deep-sea biodiversity - hyperdiverse or hype? J Biogeogr. 2003;30:475–485. [Google Scholar]
- Landmann F, Foster JM, Slatko B, Sullivan W. Asymmetric Wolbachia segregation during early Brugia malayi embryogenesis determines its distribution in adult host tissues. PLoS Negl Trop Dis. 2010;4:e758. doi: 10.1371/journal.pntd.0000758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanois A, Pages S, Bourot S, Canoy AS, Givaudan A, Gaudriault S. Transcriptional analysis of a Photorhabdus sp. variant reveals transcriptional control of phenotypic variation and multifactorial pathogenicity in insects. Appl Environ Microbiol. 2011;77:1009–1020. doi: 10.1128/AEM.01696-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latreille P, Norton S, Goldman BS, Henkhaus J, Miller N, Barbazuk B, Bode HB, Darby C, Du Z, Forst S, Gaudriault S, Goodner B, Goodrich-Blair H, Slater S. Optical mapping as a routine tool in bacterial genome sequencing. BMC Genomics. 2007;8:321. doi: 10.1186/1471-2164-8-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li BW, Rush AC, Mitreva M, Yin Y, Spiro D, Ghedin E, Weil GJ. Transcriptomes and pathways associated with infectivity, survival and immunogenicity in Brugia malayi L3. BMC Genomics. 2009;10:267. doi: 10.1186/1471-2164-10-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Sundar K, Mishra PK, Mousavi G, Liu Z, Gaydo A, Alem F, Lagunoff D, Bleich D, Gause WC. Helminth infection can reduce insulitis and type 1 diabetes through CD25- and IL-10-independent mechanisms. Infect Immun. 2009;77:5347–5358. doi: 10.1128/IAI.01170-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malmstrom L, Malmstrom J, Aebersold R. Quantitative proteomics of microbes: Principles and applications to virulence. Proteomics. 2011;11:2947–2956. doi: 10.1002/pmic.201100088. [DOI] [PubMed] [Google Scholar]
- Markaki M, Tavernarakis N. Modeling human diseases in Caenorhabditis elegans. Biotechnol J. 2010;5:1261–1276. doi: 10.1002/biot.201000183. [DOI] [PubMed] [Google Scholar]
- Mayer WE, Schuster LN, Bartelmes G, Dieterich C, Sommer RJ. Horizontal gene transfer of microbial cellulases into nematode genomes is associated with functional assimilation and gene turnover. BMC Evol Biol. 2011;11:13. doi: 10.1186/1471-2148-11-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFall-Ngai M, Nyholm SV, Castillo MG. The role of the immune system in the initiation and persistence of the Euprymna scolopes--Vibrio fischeri symbiosis. Semin Immunol. 2010;22:48–53. doi: 10.1016/j.smim.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGarry HF, Egerton GL, Taylor MJ. Population dynamics of Wolbachia bacterial endosymbionts in Brugia malayi. Mol Biochem Parasitol. 2004;135:57–67. doi: 10.1016/j.molbiopara.2004.01.006. [DOI] [PubMed] [Google Scholar]
- McNulty SN, Foster JM, Mitreva M, Dunning Hotopp JC, Martin J, Fischer K, Wu B, Davis PJ, Kumar S, Brattig NW, Slatko BE, Weil GJ, Fischer PU. Endosymbiont DNA in endobacteria-free filarial nematodes indicates ancient horizontal genetic transfer. PLoS One. 2010;5:e11029. doi: 10.1371/journal.pone.0011029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVeigh P, Alexander-Bowman S, Veal E, Mousley A, Marks NJ, Maule AG. Neuropeptide-like protein diversity in phylum Nematoda. Int J Parasitol. 2008;38:1493–1503. doi: 10.1016/j.ijpara.2008.05.006. [DOI] [PubMed] [Google Scholar]
- Melnikow E, Xu S, Liu J, Li L, Oksov Y, Ghedin E, Unnasch TR, Lustigman S. Interaction of a Wolbachia WSP-like protein with a nuclear-encoded protein of Brugia malayi. Int J Parasitol. 2011;41:1053–1061. doi: 10.1016/j.ijpara.2011.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitreva M, Smant G, Helder J. Role of horizontal gene transfer in the evolution of plant parasitism among nematodes. Methods Mol Biol. 2009;532:517–535. doi: 10.1007/978-1-60327-853-9_30. [DOI] [PubMed] [Google Scholar]
- Mitreva M, Zarlenga DS, McCarter JP, Jasmer DP. Parasitic nematodes - from genomes to control. Vet Parasitol. 2007;148:31–42. doi: 10.1016/j.vetpar.2007.05.008. [DOI] [PubMed] [Google Scholar]
- Molyneux DH, Bradley M, Hoerauf A, Kyelem D, Taylor MJ. Mass drug treatment for lymphatic filariasis and onchocerciasis. Trends Parasitol. 2003;19:516–522. doi: 10.1016/j.pt.2003.09.004. [DOI] [PubMed] [Google Scholar]
- Moreira LA, Iturbe-Ormaetxe I, Jeffery JA, Lu G, Pyke AT, Hedges LM, Rocha BC, Hall-Mendelin S, Day A, Riegler M, Hugo LE, Johnson KN, Kay BH, McGraw EA, van den Hurk AF, Ryan PA, O'Neill SL. A Wolbachia symbiont in Aedes aegypti limits infection with dengue, Chikungunya, and Plasmodium. Cell. 2009;139:1268–1278. doi: 10.1016/j.cell.2009.11.042. [DOI] [PubMed] [Google Scholar]
- Musat N, Giere O, Gieseke A, Thiermann F, Amann R, Dubilier N. Molecular and morphological characterization of the association between bacterial endosymbionts and the marine nematode Astomonema sp. from the Bahamas. Environ Microbiol. 2007;9:1345–1353. doi: 10.1111/j.1462-2920.2006.01232.x. [DOI] [PubMed] [Google Scholar]
- Nabatov AA, de Jong MA, de Witte L, Bulgheresi S, Geijtenbeek TB. C-type lectin Mermaid inhibits dendritic cell mediated HIV-1 transmission to CD4+ T cells. Virol. 2008;378:323–328. doi: 10.1016/j.virol.2008.05.025. [DOI] [PubMed] [Google Scholar]
- Neher DA. Ecology of plant and free-living nematodes in natural and agricultural soil. Annu Rev Phytopathol. 2010;48:371–394. doi: 10.1146/annurev-phyto-073009-114439. [DOI] [PubMed] [Google Scholar]
- Nguyen KB. [Accessed January 16, 2012.];Steinernema, Neosteinernema species, names and authorities. 2010 http://entnem.ifas.ufl.edu/nguyen/morph/namespp.HTM.
- Ogier JC, Calteau A, Forst S, Goodrich-Blair H, Roche D, Rouy Z, Suen G, Zumbihl R, Givaudan A, Tailliez P, Medigue C, Gaudriault S. Units of plasticity in bacterial genomes: new insight from the comparative genomics of two bacteria interacting with invertebrates, Photorhabdus and Xenorhabdus. BMC Genomics. 2010;11:568. doi: 10.1186/1471-2164-11-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opperman CH, Bird DM, Williamson VM, Rokhsar DS, Burke M, Cohn J, Cromer J, Diener S, Gajan J, Graham S, Houfek TD, Liu Q, Mitros T, Schaff J, Schaffer R, Scholl E, Sosinski BR, Thomas VP, Windham E. Sequence and genetic map of Meloidogyne hapla: A compact nematode genome for plant parasitism. Proc Natl Acad Sci USA. 2008;105:14802–14807. doi: 10.1073/pnas.0805946105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne SE, Leong YS, O'Neill SL, Johnson KN. Variation in antiviral protection mediated by different Wolbachia strains in Drosophila simulans. PLoS Pathog. 2009;5:e1000656. doi: 10.1371/journal.ppat.1000656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott JA, Bright M, Bulgheresi S. Marine microbial thiotrophic ectosymbioses. Oceanogr Mar Biol Ann Rev. 2004a;42:95–118. [Google Scholar]
- Ott JA, Bright M, Bulgheresi S. Symbiosis between marine nematodes and sulfur-oxidizing chemoautotrophic bacteria. Symbiosis. 2004b;36:103–126. [Google Scholar]
- Ott JA, Novak R, Schiemer F, Hentschel U, Nebelsick M, Polz M. Tackling the sulfide gradient: A novel strategy involving marine nematodes and chemoautotrophic ectosymbionts. Marine Ecol. 1991;12:261–279. [Google Scholar]
- Peng SL. Forkhead transcription factors in chronic inflammation. Int J Biochem Cell Biol. 2010;42:482–485. doi: 10.1016/j.biocel.2009.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry BD, Randolph TF. Improving the assessment of the economic impact of parasitic diseases and of their control in production animals. Vet Parasitol. 1999;84:145–168. doi: 10.1016/s0304-4017(99)00040-0. [DOI] [PubMed] [Google Scholar]
- Pfarr K, Foster J, Slatko B, Hoerauf A, Eisen JA. On the taxonomic status of the intracellular bacterium Wolbachia pipientis: should this species name include the intracellular bacteria of filarial nematodes? Int J Syst Evol Microbiol. 2007;57:1677–1678. doi: 10.1099/ijs.0.65248-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfarr KM, Qazi S, Fuhrman JA. Nitric oxide synthase in filariae: demonstration of nitric oxide production by embryos in Brugia malayi and Acanthocheilonema viteae. Exp Parasitol. 2001;97:205–214. doi: 10.1006/expr.2001.4613. [DOI] [PubMed] [Google Scholar]
- Poinar GO, Jr, Hansen EL. Associations between nematodes and bacteria. Helminthol Abst B. 1986;55:61–79. [Google Scholar]
- Polz MF, Felbeck H, Novak R, Nebelsick M, Ott JA. Chemoautotrophic, sulfur-oxidizing symbiotic bacteria on marine nematodes: Morphological and biochemical characterization. Microb Ecol. 1992;24:313–329. doi: 10.1007/BF00167789. [DOI] [PubMed] [Google Scholar]
- Portal-Celhay C, Blaser MJ. Competition and resilience between founder and introduced bacteria in the Caenorhabditis elegans gut. Infect Immun. 2011 doi: 10.1128/IAI.05522-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradel E, Zhang Y, Pujol N, Matsuyama T, Bargmann CI, Ewbank JJ. Detection and avoidance of a natural product from the pathogenic bacterium Serratia marcescens by Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2007;104:2295–2300. doi: 10.1073/pnas.0610281104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pujol N, Link EM, Liu LX, Kurz CL, Alloing G, Tan MW, Ray KP, Solari R, Johnson CD, Ewbank JJ. A reverse genetic analysis of components of the Toll signaling pathway in Caenorhabditis elegans. Curr Biol. 2001;11:809–821. doi: 10.1016/s0960-9822(01)00241-x. [DOI] [PubMed] [Google Scholar]
- Pukkila-Worley R, Ausubel FM. Immune defense mechanisms in the Caenorhabditis elegans intestinal epithelium. Curr Op Immunol. 2012 doi: 10.1016/j.coi.2011.10.004. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ram K, Preisser EL, Gruner DS, Strong DR. Metapopulation dynamics override local limits on long-term parasite persistence. Ecol. 2008;89:3290–3297. doi: 10.1890/08-0228.1. [DOI] [PubMed] [Google Scholar]
- Richards GR, Goodrich-Blair H. Masters of conquest and pillage: Xenorhabdus nematophila global regulators control transitions from virulence to nutrient acquisition. Cell Microbiol. 2009;11:1025–1033. doi: 10.1111/j.1462-5822.2009.01322.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiemer F, Novak R, Ott JA. Metabolic studies on thiobiotic free-living nematodes and their symbiotic microorganisms. Marine Biol. 1990;106:129–137. [Google Scholar]
- Schneider DS, Ayres JS. Two ways to survive infection: what resistance and tolerance can teach us about treating infectious diseases. Nat Rev Immunol. 2008;8:889–895. doi: 10.1038/nri2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider DS, Chambers MC. Rogue insect immunity. Science. 2008;322:1199–1200. doi: 10.1126/science.1167450. [DOI] [PubMed] [Google Scholar]
- Scholl EH, Bird DM. Computational and phylogenetic validation of nematode horizontal gene transfer. BMC Biol. 2011;9:9. doi: 10.1186/1741-7007-9-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholl EH, Thorne JL, McCarter JP, Bird DM. Horizontally transferred genes in plant-parasitic nematodes: a high-throughput genomic approach. Genome Biol. 2003;4:R39. doi: 10.1186/gb-2003-4-6-r39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulenburg H, Hoeppner MP, Weiner J, 3rd, Bornberg-Bauer E. Specificity of the innate immune system and diversity of C-type lectin domain (CTLD) proteins in the nematode Caenorhabditis elegans. Immunobiol. 2008;213:237–250. doi: 10.1016/j.imbio.2007.12.004. [DOI] [PubMed] [Google Scholar]
- Slatko BE, Taylor MJ, Foster JM. The Wolbachia endosymbiont as an anti-filarial nematode target. Symbiosis. 2010;51:55–65. doi: 10.1007/s13199-010-0067-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somvanshi VS, Kaufmann-Daszczuk B, Kim KS, Mallon S, Ciche TA. Photorhabdus phase variants express a novel fimbrial locus, mad, essential for symbiosis. Mol Microbiol. 2010;77:1021–1038. doi: 10.1111/j.1365-2958.2010.07270.x. [DOI] [PubMed] [Google Scholar]
- Stock SP. Insect parasitic nematodes: from lab curiosities into model organisms. J Invertebr Pathol. 2004;89:57–66. doi: 10.1016/j.jip.2005.02.011. [DOI] [PubMed] [Google Scholar]
- Stock SP, Goodrich-Blair H. Nematode parasites, pathogens and associates of insects and invertebrates of economic importance. In: Lacey LA, editor. Manual of Techniques in Invertebrate Pathology. Elsevier Press; 2012. pp. 375–425. [Google Scholar]
- Strubing U, Lucius R, Hoerauf A, Pfarr KM. Mitochondrial genes for heme-dependent respiratory chain complexes are up-regulated after depletion of Wolbachia from filarial nematodes. Int J Parasitol. 2010;40:1193–1202. doi: 10.1016/j.ijpara.2010.03.004. [DOI] [PubMed] [Google Scholar]
- Summers RW, Elliott DE, Urban JF, Jr, Thompson RA, Weinstock JV. Trichuris suis therapy for active ulcerative colitis: a randomized controlled trial. Gastroenterol. 2005;128:825–832. doi: 10.1053/j.gastro.2005.01.005. [DOI] [PubMed] [Google Scholar]
- Tailliez P, Laroui C, Ginibre N, Paule A, Pages S, Boemare N. Phylogeny of Photorhabdus and Xenorhabdus based on universally conserved protein-coding sequences and implications for the taxonomy of these two genera. Proposal of new taxa: X. vietnamensis sp. nov., P. luminescens subsp. caribbeanensis subsp. nov., P. luminescens subsp. hainanensis subsp. nov., P. temperata subsp. khanii subsp. nov., P. temperata subsp. tasmaniensis subsp. nov., and the reclassification of P. luminescens subsp. thracensis as P. temperata subsp. thracensis comb. nov. Int J Syst Evol Microbiol. 2010;60:1921–1937. doi: 10.1099/ijs.0.014308-0. [DOI] [PubMed] [Google Scholar]
- Tailliez P, Pages S, Edgington S, Tymo LM, Buddie AG. Description of Xenorhabdus magdalenensis sp. nov., the symbiotic bacterium associated with Steinernema australe. Int J Syst Evol Microbiol. 2011 doi: 10.1099/ijs.0.034322-0. [DOI] [PubMed] [Google Scholar]
- Tan MW, Shapira M. Genetic and molecular analysis of nematode-microbe interactions. Cell Microbiol. 2011;13:497–507. doi: 10.1111/j.1462-5822.2011.01570.x. [DOI] [PubMed] [Google Scholar]
- Taylor CM, Fischer K, Abubucker S, Wang Z, Martin J, Jiang D, Magliano M, Rosso MN, Li BW, Fischer PU, Mitreva M. Targeting protein-protein interactions for parasite control. PLoS One. 2011;6:e18381. doi: 10.1371/journal.pone.0018381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor MJ, Bandi C, Hoerauf A. Wolbachia bacterial endosymbionts of filarial nematodes. Adv Parasitol. 2005;60:245–284. doi: 10.1016/S0065-308X(05)60004-8. [DOI] [PubMed] [Google Scholar]
- Taylor MJ, Hoerauf A, Bockarie M. Lymphatic filariasis and onchocerciasis. Lancet. 2010;376:1175–1185. doi: 10.1016/S0140-6736(10)60586-7. [DOI] [PubMed] [Google Scholar]
- Teixeira L, Ferreira A, Ashburner M. The bacterial symbiont Wolbachia induces resistance to RNA viral infections in Drosophila melanogaster. PLoS Biol. 2008;6:e2. doi: 10.1371/journal.pbio.1000002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turlin E, Pascal G, Rousselle JC, Lenormand P, Ngo S, Danchin A, Derzelle S. Proteome analysis of the phenotypic variation process in Photorhabdus luminescens. Proteomics. 2006;6:2705–2725. doi: 10.1002/pmic.200500646. [DOI] [PubMed] [Google Scholar]
- Urbancik W, Bauer-Nebelsick M, Ott JA. The ultrastructure of the cuticle of Stilbonematinae (Nematoda, Desmodoridae). I. The somatic cuticle. Zoomorphol. 1996;116:51–64. [Google Scholar]
- Waterfield NR, Sanchez-Contreras M, Eleftherianos I, Dowling A, Yang G, Wilkinson P, Parkhill J, Thomson N, Reynolds SE, Bode HB, Dorus S, Ffrench-Constant RH. Rapid Virulence Annotation (RVA): identification of virulence factors using a bacterial genome library and multiple invertebrate hosts. Proc Natl Acad Sci USA. 2008;105:15967–15972. doi: 10.1073/pnas.0711114105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werren JH, Baldo L, Clark ME. Wolbachia: master manipulators of invertebrate biology. Nat Rev Microbiol. 2008;6:741–751. doi: 10.1038/nrmicro1969. [DOI] [PubMed] [Google Scholar]
- Wilkinson P, Waterfield NR, Crossman L, Corton C, Sanchez-Contreras M, Vlisidou I, Barron A, Bignell A, Clark L, Ormond D, Mayho M, Bason N, Smith F, Simmonds M, Churcher C, Harris D, Thompson NR, Quail M, Parkhill J, Ffrench-Constant RH. Comparative genomics of the emerging human pathogen Photorhabdus asymbiotica with the insect pathogen Photorhabdus luminescens. BMC Genomics. 2009;10:302. doi: 10.1186/1471-2164-10-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B, Novelli J, Foster J, Vaisvila R, Conway L, Ingram J, Ganatra M, Rao AU, Hamza I, Slatko B. The heme biosynthetic pathway of the obligate Wolbachia endosymbiont of Brugia malayi as a potential anti-filarial drug target. PLoS Negl Trop Dis. 2009;3:e475. doi: 10.1371/journal.pntd.0000475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Kim SK. The early bird catches the worm: new technologies for the Caenorhabditis elegans toolkit. Nat Rev Genet. 2011;12:793–801. doi: 10.1038/nrg3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Kaya HK, Heungens K, Goodrich-Blair H. Response of ants to a deterrent factor(s) produced by the symbiotic bacteria of entomopathogenic nematodes. Appl Environ Microbiol. 2002;68:6202–6209. doi: 10.1128/AEM.68.12.6202-6209.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.