Abstract
Introduction:
Bacterial nonspecific acid phosphohydrolases (NSAPs) or phosphatases are group of enzymes secreted as soluble periplasmic proteins or retained as membrane bound lipoproteins that are usually able to dephosphorylate a broad array of structurally unrelated organic phosphoesters (nucleotides, sugar phosphates, phytic acid etc.) to acquire inorganic phosphate (Pi) and organic byproducts. They exhibit optimal catalytic activity at acidic to neutral pH values. On the basis of amino acid sequence relatedness, phosphatase are grouped into different molecular families namely Class A, Class B and Class C acid phosphatase respectively.
Results and discussion:
In this article out of thirty three sequences, twenty six belonging to each of the three classes of bacterial acid phosphatase and seven belonging to archaeal phosphoesterases were analyzed using various tools of bioinformatics. Phylogenetic analysis, dot plot comparisons and motif analysis were done to identify a number of similarities and differences between three classes of bacterial acid phosphatases and archaeal phosphoesterases. In this research we have attempted to decipher evolutionary relationship between three classes of bacterial acid phosphatase and archaeal phosphoesterases using bioinformatics approach.
Key words: Non-specific acid phosphatase, archaeal phosphoesterase, NSAP, evolution.
1. INTRODUCTION
Phosphatases constitute a diverse group of enzymes that hydrolyze phosphoesters in various kinds of substrates under different conditions (1). Based on the criteria such as specificity and optimum pH, phosphatase can be classified into several families, one of which is the family of bacterial non-specific acid phosphohydrolases or phosphatases (NSAPs). In general, acid phosphatases hydrolyze phosphate esters via the two-step mechanism shown in Scheme 1 (1).
NSAPs are found to be widely distributed among many Gram-positive, Gram-negative bacterial species and eukaryotes. NSAPs are considered physiologically important because they either help the cell to utilize the organic phosphoesters that cannot cross the cytoplasmic membrane thus providing the cell with essential nutrients (2) or have functional importance in gene expression (3). Some of these enzymes can be secreted outside the cell, where they are either released in soluble form or retained as membranebound proteins where they can dephosphorylate a broad array of structurally unrelated phosphoester substrates and exhibit optimal catalytic activity at acidic to neutral pH values (Rossolini et al., 1998) (4). In addition to their role in P acquisition, phosphohydrolases regulate cellular metabolism, are involved in signal transduction and may also be critical to bacterial pathogenicity (5, 6, 7).
NSAPs are usually grouped into three types (Classes A, B and C) on the basis of amino acid sequence relatedness (8, 9, 10, 11). The structural criterion has led to the definition of various molecular families of phosphohyrolases for which the signature sequence motifs have been identified (12). NSAPs have been detected in several microbial taxa, and the enzymes of different classes can be produced by the same bacterial species (4). Class A NSAPs are secreted monomeric to oligomeric proteins containing a polypeptide component of approximately 25-27 kDa (8, 13, 14, 15, 16). This group of enzymes has recently been demonstrated to share some conserved sequence motifs with other bacterial and eukaryotic phosphatases, suggesting that the conserved residues could be essential for catalytic activity and possibly part of the active site of these enzymes (17). The Zymomonas mobilis PhoC-Zm enzyme represents the major Pi-irrepressible acid phosphohydrolases and was the first sequenced class A enzyme (13).
Class B NSAPs are secreted homotetrameric metallo-proteins containing a 25-kDa polypeptide component (10, 11, 18) that are completely unrelated to class A enzymes at the sequence level. The Salmonella enterica AphA-Se enzyme was the first class B NSAP purified and characterized in detail (18, 19). Class C NSAPs are secreted bacterial lipoproteins that contain a polypeptide component with a molecular mass of approximately 30kDa and share conserved sequence motifs (11, 20). At the sequence level Class C appear to be related, although distantly, to Class B NSAPs and also to some plant acid phosphophydrolases (4). This represents the first example of family bacterial NSAPs that has a relatively close eukaryotic counter-part. The first identified Class C enzyme was OlpA enzyme of Chryseobacterium meningosepticum (21). All the three class contain conserved signature sequence motifs (22).
These structural similarities together with dephosphorylating activity exhibited by bacterial, eukaryotic and archaeal phosphatases support the definition of this phosphatase motif and the inclusion of all of these enzymes into a molecular superfamily of phosphohydrolases which is indicated as “DDDD” due to the couple of invariant aspartate residues present in each domain. (4). The invariant residues could be essential for the phosphohydrolase catalytic activity of these enzymes and part of active site (22). Thus, the similar catalytic mechanism of all the bacterial acid phosphatase, archaeal phosphatase and archaeal membrane bound phosphoesterases belonging to phosphohydrolase family would seem to suggest us that they evolved from a common ancestor. In this article, we analyzed the sequences of the three classes of bacterial acid phosphatase and archaeal phosphoesterase to determine their evolution using bioinformatics tools.
2. METHODS
The sequences for bacterial acid phosphatases and archaeal phosphoesterases were obtained from the NCBI website (http://www.ncbi.nlm.nih.gov/). From the available several acid phosphatase sequences, only the bacterial and archaeal acid phosphatases were selected whereas the viral and eukaryotic sequences were excluded. A total of 26 sequences belonging to bacterial acid phosphatase and 7 sequences belonging to archaeal phosphoesterases were meticulously chosen based on the size of sequences, substrate specificity, structural criterion, functional differences, biophysical features, distribution of NSAP’s/ organism origin, signature sequence motifs, conserved structural motifs. We obtained 10 sequences belonging to bacterial Class A acid phosphatases, 7 sequences belonging to bacterial Class B acid phosphatases, 9 sequences belonging to bacterial Class C acid phosphatases, and 7 sequences belonging to archaeal phosphoesterases. Structural and phylogenetic relationships have existed among various bacterial NSAP’s and some other bacterial and eukaryotic phosphohydrolases. Here for the first time we tried to analyze the evolution of bacterial acid phosphatases and archaeal phosphoesterases using bioinformatics tools.
Similarity motifs were identified in all the sequences using MEME Motif discovery tool. All the settings were set to default, except for the maximum number of Motifs which was increased from three to thirteen (23). The phylogenetic tree and motif analysis were then used to construct dot plots. The position of a specific amino acid motif in the selected protein sequence was found by dot plots.
In order to compare the similarity and differences in the sequences of each class of bacterial acid phosphatases and archaeal phosphoesterases the dot matcher program was used to construct dot plots. The similarity in the protein sequences can be easily accessed from dot plots simply by seeing the diagonal fragment in between the X and Y axis of a graph, which is constructed by using data matrix, distance matrix and chi squared analysis (24). Here similar sequences show a diagonal line whereas in dissimilar sequences this line is absent or highly fragmented. We first constructed the dot plots by using sequences belonging to same class bacterial acid phosphatase and archaeal phosphoesterases and then used each sequences from different class using different combinations of different class of bacterial and archaeal phosphoesterases each time. The parameters of the program were mostly set at default except for the window size of 10 and threshold of 23 (25).
The selected sequences were obtained in FASTA format and then aligned by using Clustal X (Thompson et al., 1997) (26). Neighbour joining method was used to construct the phylogenetic tree from the sequences which were aligned using PHYLIP (27). The phylogenetic tree was then bootstrapped in order to see how well the sequences related to each other. Finally, treeview was used to see their position in each clade and the study of bacterial acid phosphatase and archaeal phosphoesterases were related by evolution (28).
3. RESULTS
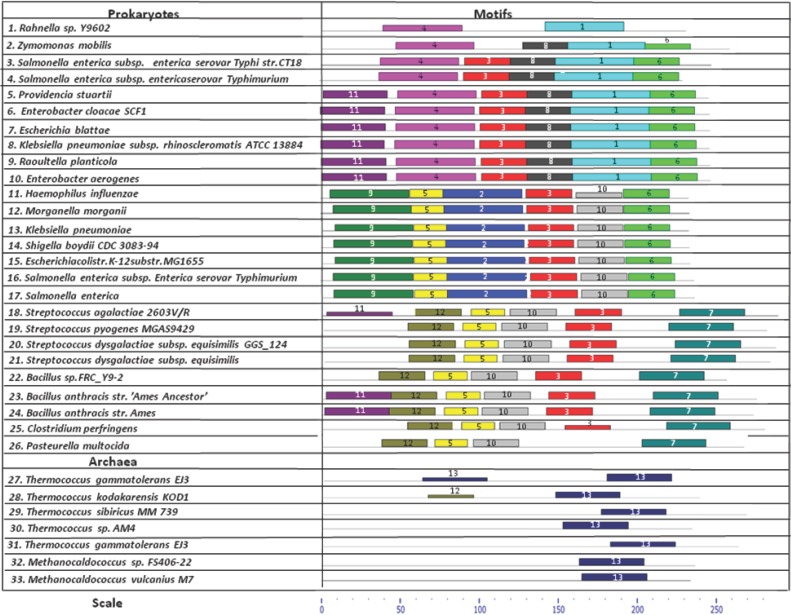
Analysis of motifs of bacterial acid phosphatases and archaeal phosphoesterases reveals distinct features in all three classes of bacterial acid phosphatases and archaeal phosphoesterases. As seen in Figure 1a, Class A acid phosphatase consisting of sequences 1 (Rahnella sp. Y9602) through 10 (Enterobacter aerogenes) share a common Motif 1 and Motif 4. Class B acid phosphatase consisting of sequences 11 (Haemophilus influenza) through 17 (Salmonella enterica) share a common Motif 2 and Motif 9. Class C acid phosphatase consisting of sequences 18 (Streptococcus agalactiae 2603V/R) through 26 (Pasteurella multocida) share common Motif 7 and Motif 12. Sequences 27 (Thermococcus gammatolerans EJ3) through 33 (Methanocaldococcus vulcanius M7) belonging Archaeal phosphoesterases have only one common Motif 13. Motif 5 and Motif 10 is common to Class B and Class C acid phosphatase. Motif 6 is common to Class A acid phosphatase from sequences 2 through 10 and Class B acid phosphatase from sequences 11 through 17. Motif 3 is common to sequences 3 through 10 of Class A acid phosphatase, sequences 11 through 17 of Class B acid phosphatase and sequences 18 through 25 of Class C acid phosphatase. (Figure 1b).
Figure 1a.
The motifs present in all 33 sequences of the three classes of bacterial acid phosphatase and archaeal phosphoesterases are shown in above table. to the left are the names of all 26 bacterial species whose acid phosphatases and 7 archaeal species whose phosphoesterases sequences were taken for analysis. sequences 1 through 10 represent bacterial Class A acid phosphatases, sequences 11 through 17 represent bacterial Class B acid phosphatases, sequences 18 through 26 represent bacterial Class C acid phosphatases and sequences 27 through 33 represent archaeal phosphoesterases. thirteen different motifs are shown in 33 sequences. some of the sequences share some conserved sequence motifs whereas some of the motifs are uniquely present in each class of bacterial acid phosphatase and archaeal phosphoesterases. the height of the motif “block” is proportional to -log(p-value), truncated at the height for a motif with a p-value of 1e-10.
Figure 1b.
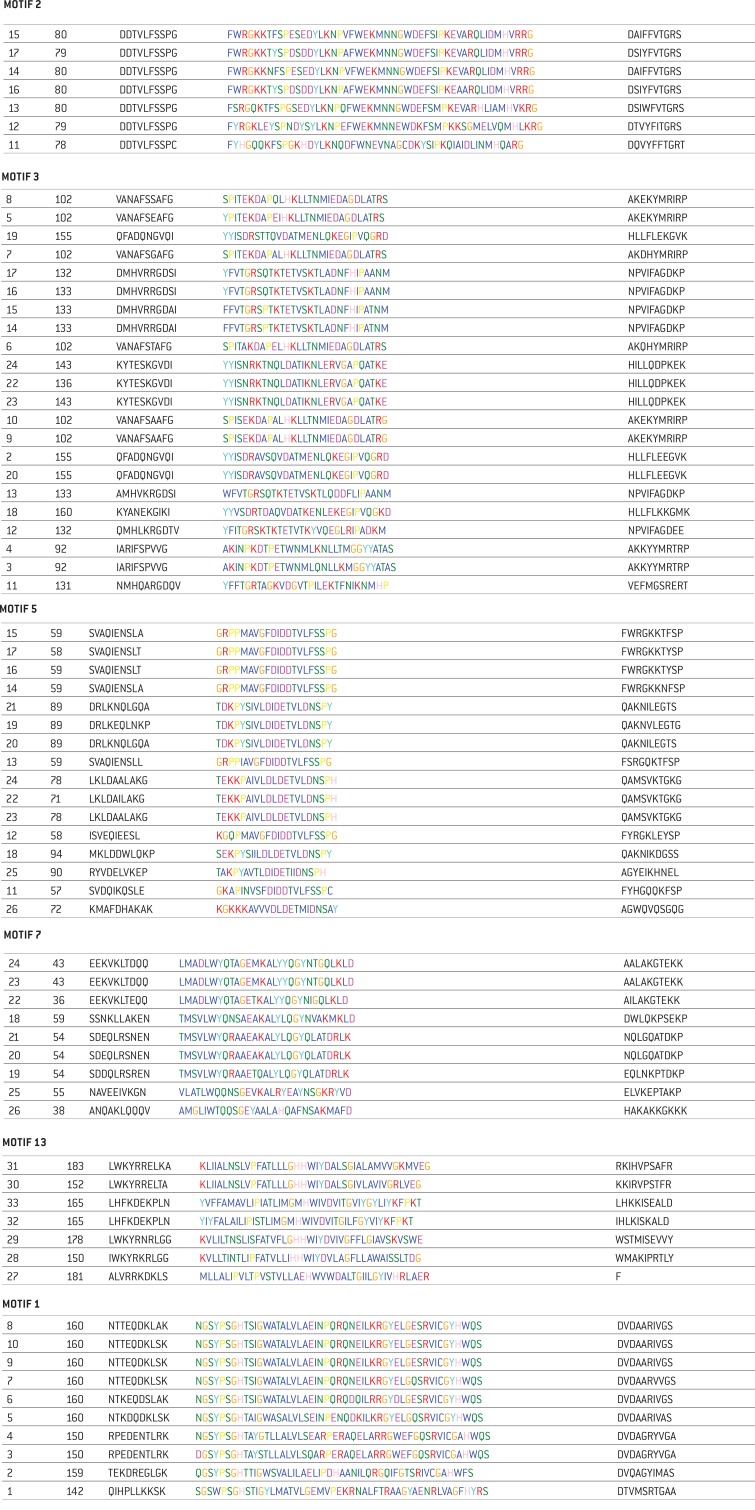
The sequences of motifs 1, 2, 3, 5, 7, 13 are shown in the figure above. The numbers to the left of each protein sequence corresponds with the number and name of the organism in table of Fig. 1a. The second number represents the sequence length of the enzyme acid phosphatase and phosphoesterase and third column shows the sequence with the motifs. For example in Motif 2, the first bacterium is number 15, Escherichia coli str. K-12 substr. MG1655 has a length of 80 amino acids which is shown in the sequence with its motif.
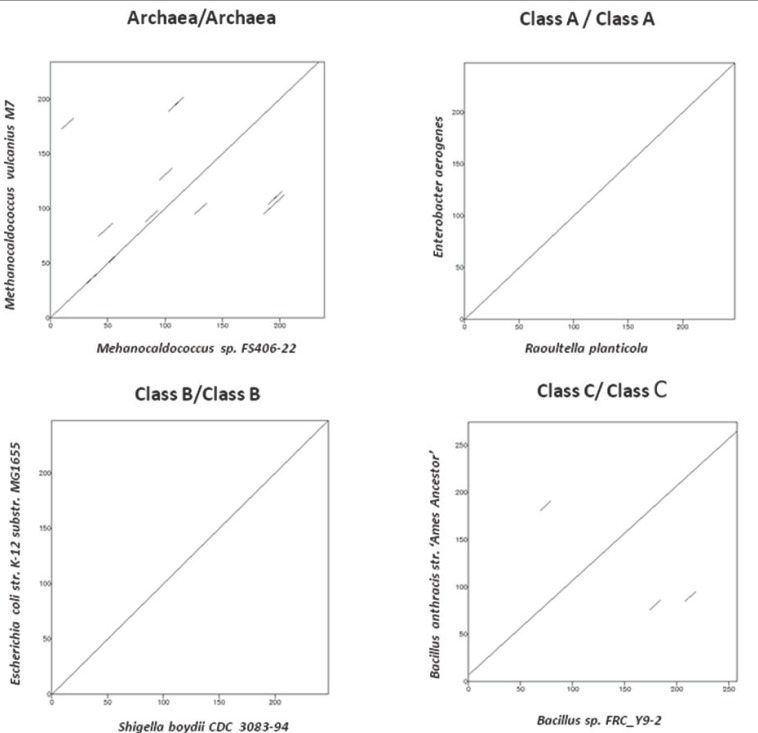
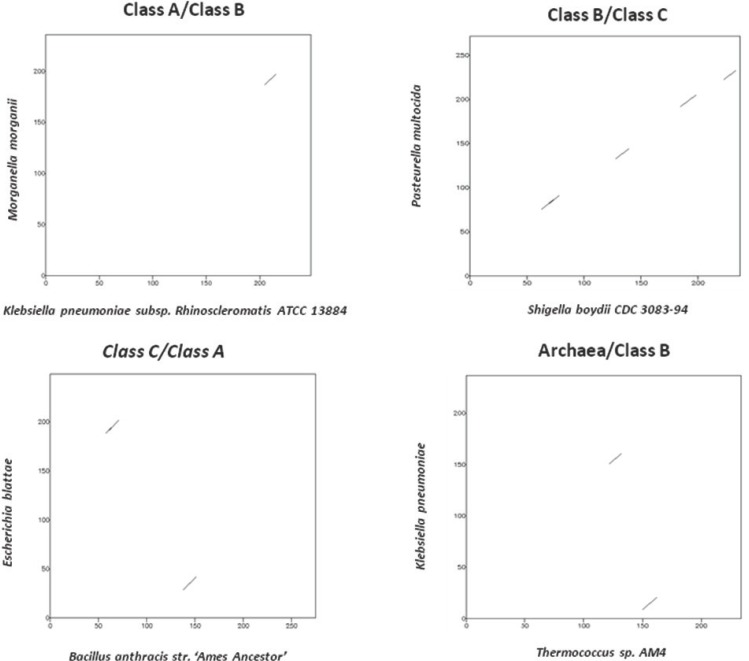
The degree of similarity between the protein sequences of the three classes of bacterial acid phosphatase and archaeal phosphoesterase can be seen by comparing the dot plots in Figure 2a. The dot plots constructed with sequences belonging to same class shows linear graph whereas the dot plots constructed between two different bacterial acid phosphatase classes and archaeal phosphoesterase class show a high degree of dissimilarity or fragmentation as seen in Figure 2b. These findings correlate with the motifs discovered which are shared only by that particular class.
Figure 2a.
The dot plot comparison of sequences belonging to same classes resulted in collinear diagonal fragment. the dot plots shown are within the same class of bacterial acid phosphatase ClassA/ClassA, ClassB/ClassB, ClassC/ClassC and archaeal phoshoesterases, Archaea/Archaea.
Figure 2b.
The dot plot comparison of sequences belonging to different classes such as ClassA/ClassB, ClassB/ClassC, ClassC/ClassA, Archaea/ClassB resulted in a plot with non- collinear fragments.
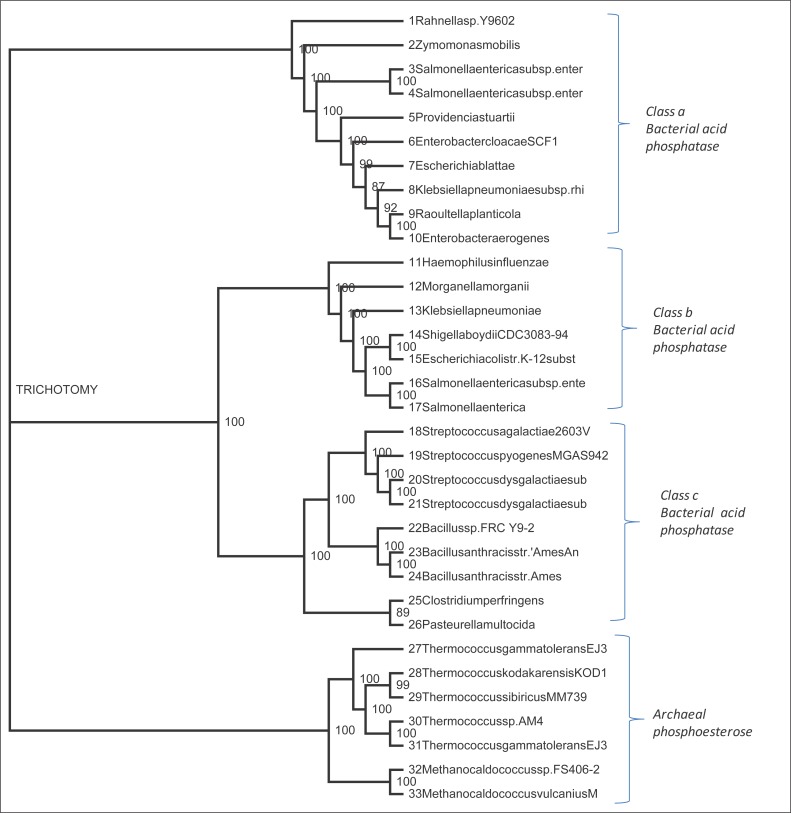
As seen in Figure 3, the phylogenetic analysis of three classes of bacterial acid phosphatase and archaeal phosphoesterase resulted in the formation of a tree with three distinct clades for ClassA, ClassB-ClassC and archaeal phosphoesterase. As we can see from the tree that the bootstrap values of three classes of bacterial acid phosphatase and archaeal phosphoesterase is mostly above ninety, we can infer that bacterial acid phosphatase and archaeal phosphoesterase share a common ancestor.
Figure 3.
A Phylogenetic tree constructed by using amino acid sequences belonging to all the tree classes of bacterial acid phosphatase and archaeal phosphoesterase. The name of the bacteria with the class it belongs to makes the first and second clade and archaea that makes the third clade is shown on the right hand side of the cladogram. The scores seen on the tree show the degree of sequence similarity between the sequences of each class. The names of bacteria and archaea along with the accession numbers in parentheses are as follows: The numbers 1 to10 represent Class A bacterial acid phosphatase. Numbers 11 to 17 represent Class B acid phosphatase, numbers 18 to 26 represent Class C acid phosphatase and number 27-33 represent Archaeal phosphoesterase. 1. Rahnella sp. Y9602 (ADW72173.1), 2. Zymomonas mobilis (AAA27700.1), 3. Salmonella enterica subsp. enterica serovar Typhi str. CT18 (NP _458615.1), 4. Salmonella enterica subsp. enterica serovar Typhimurium (CAA41760.1), 5. Providencia stuartii (CAA46032.1), 6. Enterobacter cloacae SCF1(YP _003942142.1), 7. Escherichia blattae (BAA84942.1) , 8. Klebsiella pneumoniae subsp. rhinoscleromatis ATCC 13884 (ZP_06016217.1), 9. Raoultella planticola (BAB18918.1), 10. Enterobacter aerogenes (ABW37174.1), 11. Haemophilus influenza (CAA68889.1), 12. Morganella morganii (CAA55131.1), 13. Klebsiella pneumoniae (AAL59317.1), 14. Shigella boydii CDC 3083-94 (ACD06580.1), 15. Escherichia coli str. K-12 substr. MG1655 (CAA60534.1), 16. Salmonella enterica subsp. enterica serovar Typhimurium (AAW22868.1), 17. Salmonella enterica (CAB40974.1), 18. Streptococcus agalactiae 2603V/R (NP _688757.1), 19. Streptococcus pyogenes MGAS9429 (YP _597336.1), 20. Streptococcus dysgalactiae subsp. equisimilis GGS_124 (YP _002997631.1), 21. Streptococcus dysgalactiae subsp. equisimilis (CAA73175.1), 22. Bacillus sp. FR C_Y9-2(ABO69628.1), 23. Bacillus anthracis str. ‘Ames Ancestor’ (YP _021394.1), 24. Bacillus anthracis str. Ames (NP _846955.1), 25. Clostridium perfringens (ACB11490.1), 26. Pasteurella multocida (ACM68930.1), 27. Thermococcus gammatolerans EJ3 (YP _002960201.1), 28. Thermococcus kodakarensis KOD 1 (BAD84554.1), 29. Thermococcus sibiricus MM 739 (YP _002993470.1), 30. Thermococcus sp. AM4 (ZP_04880406.1), 31. Thermococcus gammatolerans EJ3 (YP _002959503.1), 32. Methanocaldococcus sp. FS 406-22 (YP _003458929.1), 33. Methanocaldococcus vulcanius M7 (YP _003247262.1).
4. DISCUSSION
Multiple sequence alignment was performed on 26 bacterial acid phosphatase and 7 archaeal phosphoesterase giving sufficient evidence to conclude that they have a common evolutionary origin. The phylogenetic tree shows three distinct clades; one clade belonging to Class A bacterial acid phosphate, second clade belonging to Class B and Class C being closely related (4); third clade belonging to archaeal phosphoesterases providing a definitive evidence of common ancestral origin of bacterial acid phosphatase and archaeal phosphoesterase with divergent evolution. Our analysis found sequence 4 (Salmonella enterica subsp. enterica serovar Typhimurium) is a Class A acid phosphatase and also Class B acid phosphatase as seen in sequence 16. This shows that enzymes of different classes is produced by same bacterial species (8, 10, 14, 29).
Membrane bound type 2 phosphatidic acid phosphatase (PAP2) shares sequence motifs with a soluble vanadium-dependent chloroperoxidase of known structure. These regions are also conserved in other soluble and membrane bound proteins including bacterial acid phosphatases, mammalian glucose-6-phosphatases and the Drosophila developmental protein Wunen. This shows that similar arrangement of the catalytic residues specifies the active site within the soluble and membrane spanning domains (30). Vanadate and vanadate derivatives have been employed to interrogate a range of enzymes that interact with phosphorylated substrates (31). Acid phosphatase enzymes have evolved to accommodate vanadate as a redox cofactor (32, 33).
In this article, archaeal phosphatase belonging to vanadium-dependent haloperoxidase superfamily (Thermococcus gammatolerans EJ3) representing sequence 27 has be included as well as membrane-bound phosphoesterase, belonging to PAP2 superfamily (Thermococcus gammatolerans EJ3) representing sequence 31 is also studied. Our analysis found conserved sequence motif 13 in the soluble and membrane bound phosphatase enzyme. Also, sequence 32 (Methanocaldococcus sp. FS406-22) phosphoesterase PAphosphatase related protein and sequence 33 (Methanocaldococcus vulcanius M7) a phosphoesterase PA-phosphatase related protein sharing conserved sequence motif 13 with all of the other archaeal membrane bound phosphoesterases and soluble phosphatase. This analysis suggests that membrane-associated PAP2 like proteins share conserved structural elements and similar catalytic mechanism with related soluble globular enzymes (haloperoxidases, bacterial acid phosphatases, ATP diphosphohydrolase). Assuming divergent evolution, this implies that catalytic activity was somehow retained during relocation of an ancestral enzyme from the membrane to a water-soluble environment, or vice versa (30).
The analyses of the different motifs show that bacterial Class A has conserved sequence motifs 1 and 4. Bacterial Class B has conserved sequence motifs 2 and 9; bacterial Class C has unique motifs 7 and 12; archaeal phosphoesterase has unique Motif 13. Class A, B, and C share common Motif 3 and Class A and B share common Motif 5 and Class B and C share a common Motif 5 and 10. Archaeal phosphoesterase do not share any common Motifs with any of the bacterial Class A, B and C acid phosphatases except sequence 28 (Thermococcus kodakarensis KOD1) share Motif 12 with Class C bacterial acid phosphatase. It suggests that it must have evolved and adapted separately but seem to have a common ancestral origin. The high bootstrap scores also reveal the common ancestral origin of bacterial acid phosphatase and archaeal phosphoesterases.
The dot-plots which were used as a comparative tool between the two sequences showed a high degree of similarity within members of the same class. However, dot plots between members of different classes resulted in multiple fragments without solid collinear lines suggesting no similarity between sequences of different classes. These results are consistent with the motif analysis where each class of bacterial acid phosphatase and archaeal phosphoesterases has unique conserved motifs and share only one motif between all the three classes of acid phosphatase. Thus by comparing the dot plots, motif analysis, protein sequences, phylogenetic analysis we could conclude that the three classes of bacterial acid phosphatase and archaeal phosphoesterases have evolved separately but have a common ancestry.
In this research we considered only the bacterial and archaeal sequences. Further research may be done by comparing all other soluble globular and membrane associated proteins to get a more comprehensive picture of the evolution of phosphatases. This knowledge will be useful to correlate bacterial, eukaryotic and archaeal world. This study may also offer insight into the study of immobilization of toxic uranium U(VI) mediated by the intrinsic phosphatase activity of naturally occurring bacteria isolated from contaminated subsurface soils (Martinez et.al., 2007) (34). It also may be promising towards bioremediation of contaminated soils, ground water and waste streams using bacteria and archaea.
Conflict of interest
None declared.
REFERENCES
- 1.Vincent JB, Crowder MW, Averill BA. Hydrolysis of phosphate monoesters: a biological problem with multiple chemical solutions. Trends Biochem Sci. 1992;17:105–110. doi: 10.1016/0968-0004(92)90246-6. [DOI] [PubMed] [Google Scholar]
- 2.Beacham IR. Periplasmic enzymes in gram-negative bacteria. Int J Biochem. 1979;10:877–883. doi: 10.1016/0020-711x(79)90117-4. [DOI] [PubMed] [Google Scholar]
- 3.Carmany DO, Hollingsworth K, Mc-Cleary WR. Genetic and biochemical studies of phosphatase activity of PhoR. J Bacteriol. 2003;185:1112–1115. doi: 10.1128/JB.185.3.1112-1115.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rossolini GM, Schippa S, Riccio ML, Berlutti F, Macaskie LE, Thaller MC. Bacterial nonspecific acid phosphohydrolases: physiology, evolution and use as tools in microbial biotechnology. Cell Mol Life Sci. 1998;54:833–850. doi: 10.1007/s000180050212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reilly TJ, Baron GS, Nano FE, Kuhlenschmidt MS. Characterization and sequencing of a respiratory burst-inhibiting acid phosphatase from Francisella tularensis. J Biol Chem. 1996;271:10973–10983. doi: 10.1074/jbc.271.18.10973. [DOI] [PubMed] [Google Scholar]
- 6.Wanner BL. Phosphorus assimilation and control of the phosphate regulon. In: Neidhardt FC, Curtiss R III, Ingraham JL, Lin ECC, Low KB, Magasanik B, et al., editors. Escherichia Coli and Salmonella, Cellular and Molecular Biology. 2nd. Vol. 1. Washington, DC, USA: American Society for Microbiology Press; 1996. pp. 1357–1381. [Google Scholar]
- 7.Berlutti F, Casalino M, Zagaglia C, Fradiani PA, Visca P, Nicoletti M. Expression of the virulence plasmid-carried apyrase gene (apy) of enteroinvasive Escherichia coli and Shigella flexneri is under the control of H-NS and the VirF and VirB regulatory cascade. Infect Immun. 1998;66:4957–4964. doi: 10.1128/iai.66.10.4957-4964.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thaller MC, Berlutti F, Schippa S, Lombardi G, Rossolini GM. Characterization and sequence of PhoC, the principal phosphate-irrepressible acid phosphatase of Morganella morganii. Microbiology. 1994;140:1341–1350. doi: 10.1099/00221287-140-6-1341. [DOI] [PubMed] [Google Scholar]
- 9.Thaller MC, Berlutti F, Schippa S, Iori P, Passariello C, Rossolini GM. Heterogeneous patterns of acid phosphatases containing low molecular mass polypeptides in members of the family Enterobacteriaceae. In1 J Syst Bacteriol. 1995a;45:255–261. [Google Scholar]
- 10.Thaller MC, Lombardi G, Berlutti F, Schippa S, Rossolini GM. Cloning and characterization of the NapA acid phosphatase/phosphotransferase of Morganella morganii: identification of a new family of bacterial acid-phosphatase-encoding genes. Microbiology. 1995b;141:147–154. doi: 10.1099/00221287-141-1-147. [DOI] [PubMed] [Google Scholar]
- 11.Thaller MC, Schippa S, Bonci A, Cresti S, Rossolini GM. Identification of the gene (aphA) encoding the class B acid phosphatase/phosphotransferase of Escherichia coli MG1655 and characterization of its product. FEMS Microbiol Lett. 1997a;146:191–198. doi: 10.1111/j.1574-6968.1997.tb10192.x. [DOI] [PubMed] [Google Scholar]
- 12.Bairoch A, Bucher P, Hofmann K. The PROSITE database, its status in 1995. Nucleic Acids Res. 1995;24:189–196. doi: 10.1093/nar/24.1.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pond JL, Eddy CK, Mackenzie KF, Conway T, Borecky DJ, lngram LO. Cloning, sequencing, and characterization of the principal acid phosphatase the phoC+ product, from Zymomonus mobilis. J Bacteriol. 1989;171:767–774. doi: 10.1128/jb.171.2.767-774.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kasahara M, Nakata A, Shinagawa H. Molecular analysis of the Salmonella typhimurum phoN gene, which encodes nonspecific acid phosphatase. J Bacteriol. 1991;173:6770–6775. doi: 10.1128/jb.173.21.6760-6765.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bhargava T, Datta S, Ramachandran V, Ramakrishnan R, Roy RK, Sankaran K, Subrahmanyam YVBK. Virulent Shigella codes for a soluble apyrase: Identification, characterization and cloning of the gene. Curr Sci. 1995;68:293–300. [Google Scholar]
- 16.Uchiya K, Tohsuji M, Nikai T, Sugihara H, Sasakawa C. Identification and characterization of phoN-Sf, a gene on the large plasmid of Shigella flexneri 2a encoding a nonspecific phosphatase. J Bacteriol. 1996;178:4548–4554. doi: 10.1128/jb.178.15.4548-4554.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stukey J, Carman GM. Identification of a novel phosphatase sequence motif. Protein Sci. 1997;6:469–472. doi: 10.1002/pro.5560060226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Uerkvitz W. Periplasmic nonspecific phosphatase II from Salmonella typhimurium LT2. J Biol Chem. 1988;263:15823–15830. [PubMed] [Google Scholar]
- 19.Uerkvitz W, Beck CF. Periplasmic phosphatases in Salmonella typhimurium LT2. A biochemical, physiological and partial genetic analysis of three nucleoside monophosphate dephosphorylating enzymes. J Biol Chem. 1981;256:382–389. [PubMed] [Google Scholar]
- 20.Green BA, Farley JE, Quinn-Dey T, Deich RA, Zlotnick GW. The e (P4) outer membrane protein of Haemophilus influenzae: biologic activity of anti-e serum and cloning and sequencing of the structural gene. Infect Immun. 1991;59:3191–3198. doi: 10.1128/iai.59.9.3191-3198.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bloch KC, Nadarajah R, Jacobs R. Chryseobacterium meningosepticum: an emerging pathogen among immunocompromised adults. Medicine. 1997;76:30–41. doi: 10.1097/00005792-199701000-00003. [DOI] [PubMed] [Google Scholar]
- 22.Thaller MC, Schippa S, Rossolini GM. Conserved sequence motifs among bacterial, eukaryotic and archaeal phosphatases that define a new phosphohydrolase superfamily. Protein Sci. 1998;7:1647–1652. doi: 10.1002/pro.5560070722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bailey TL, Williams N, Misleh C, Li WW. MEME: discovering and analyzing DNA and protein sequence motifs. Nucl Acids Res. 2006;34:W369–373. doi: 10.1093/nar/gkl198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Landes C, Henaut A, Risler J. Dot-Plot comparison by multivariate analysis (DOCMA): A tool for classifying protein sequences. Bioinformatics. 1998;9:191–196. doi: 10.1093/bioinformatics/9.2.191. [DOI] [PubMed] [Google Scholar]
- 25.Rice P, Longden I. Emboss: the European Molecular Open Software Suite. Trends in Genet. 16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 26.Thompson J, Gibson T, Plewniak F, Jeanmougin F, Higgins D. The Clustal X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucl Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Felsenstein J. PHYLIP Phylogeny Inference Package. Cladistics. 1989;5:164–166. [Google Scholar]
- 28.Page R. TREEVIEW: An application to display phylogenetic trees on personal computers. Computer Applications in the Biosciences. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- 29.Groisman EA, Saier MH, Ochman H. Horizontal transfer of a phosphatase gene as evidence for mosaic structure of the Salmonella genome. EMBO. 1992;11:1309–1316. doi: 10.1002/j.1460-2075.1992.tb05175.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Neuwald AF. An unexpected structural relationship between integral membrane phosphatase and soluble haloperoxidases. Protein Sci. 1997;6:1764–1767. doi: 10.1002/pro.5560060817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Crans DC, Smee JJ, Gaidamauskas E, Yang L. The chemistry and biochemistry of vanadium and the biological activities exerted by vanadium compounds. Chem Rev. 2004;104:849–902. doi: 10.1021/cr020607t. [DOI] [PubMed] [Google Scholar]
- 32.Littlechild J, Garcia-Rodriguez E, Dalby A, Isupov M. Structural and functional comparisons between vanadium haloperoxidase and acid phosphatase enzymes. J Mol Recognit. 2002;15:291–296. doi: 10.1002/jmr.590. [DOI] [PubMed] [Google Scholar]
- 33.Wever R, Hemrika W. In: Handbook of Metalloproteins. Messerschmidt A, Huber R, Wieghardt K, Poulos T, editors. Chichester, United Kingdom: John Wiley & Sons Ltd; 2001. pp. 1417–1428. [Google Scholar]
- 34.Martinez RJ, Beazely MJ, Taillefert M, Arakaki AK, Skoinick J, Sobecky PA. Aerobic uranium(VI) bioprecipitation by metal-resistant bacteria isolated from radionuclide- and metal contaminated subsurface soils. Environ Microbiol. 2007;12:3122–3133. doi: 10.1111/j.1462-2920.2007.01422.x. [DOI] [PubMed] [Google Scholar]