Abstract
Background
The rise of high mountain chains is widely seen as one of the factors driving rapid diversification of land plants and the formation of biodiversity hotspots. Supporting evidence was reported for the impact of the rapid rise of the Andean mountains but this hypothesis has so far been less explored for the impact of the “roof of the world”. The formation of the Himalaya, and especially the rise of the Qinghai–Tibetan Plateau in the recent 20 million years, altered the monsoon regimes that dominate the current climates of South East Asia. Here, we infer the hypothesis that the rise of Himalaya had a strong impact on the plant diversity in the biodiversity hotspot of the Southwest Chinese Mountains.
Results
Our analyses of the diversification pattern of the derived fern genus Lepisorus recovered evidence for changes in plant diversity that correlated with the strengthening of South East Asian monsoon. Southwest China or Southwest China and Japan was recovered as the putative area of origin of Lepisorus and enhancing monsoon regime were found to shape the early diversification of the genus as well as subsequent radiations during the late Miocene and Pliocene.
Conclusions
We report new evidence for a coincidence of plant diversification and changes of the climate caused by the uplift of the Himalaya. These results are discussed in the context of the impact of incomplete taxon sampling, uncertainty of divergence time estimates, and limitations of current methods used to assess diversification rates.
Keywords: Diversification pattern, East Asian monsoon, Himalaya, LASER, Lepisorus
Background
The origin of the uneven distribution of plant diversity, in particular with biodiversity hotspots [1], has been increasingly explored by employing phylogenetic methods to reconstruct the history of selected lineages. The majority of these studies addressed the origin of the Andean (e.g., [2-4]), Great Cape region (e.g., [5,6]) and Madagascan (e.g., [7,8]) biodiversity hotspots. These studies not only assembled evidence for the evolution of biodiversity of these particular hotspots but also laid out the foundation for a generalized theory on the history of biodiversity hotspots by classifying the attribution of diversification events into mature and rapid radiations [9]. Besides some shared features, the unique geographic and historical aspects, e.g. isolation by distance, relative constant climate, and major orogenic events, resulted in unique patterns of biodiversity assembly in these hotspots.
Current macro-evolutionary studies have given remarkably little attention to the origin of the biodiversity hotspots in Southeast (SE) Asia such as the southwest Chinese mountain region [1]. However, the origin of the later biodiversity hotspot is widely attributed to the rise of the Himalaya and the subsequent formation of the Qinghai-Tibetan Plateau (QTP), which is frequently referred as “the roof of the world” (e.g., [10-16]). In this respect, this hotspot is comparable to the Andean biodiversity hotspot attributed to the rise of the Andes (e.g., [2-4]). The rise of the Himalaya was onset by the collision of the Indian plate with the Asian plate around 50-55 million years ago (Ma) and is seen widely as one of the largest orogenic events in the history of Earth with dramatic implications on the climates of Central and East Asia. Despite some controversy about the exact timing of the uplift of the Himalaya [17-22], geological estimates date the start of this process unequivocally within the late Tertiary/mid-Miocene (ca. 15-10 Ma) [23] , and episodes of uplift probably continued throughout the late Pliocene (ca. 3 Ma) and well into the Quaternary [24-26]. Major impacts of the latest uplift of the Himalaya on the evolution of life in Southwest China has been suggested because the steep mountain ranges not only caused the formation of desert-like habitats in central Asia, but also changed drastically the East Asian monsoon [21,22,27-29]. The exact climate history of these events is still subject of controversy [18,21], but existing evidence suggests an initiation of the East Asian monsoon in the late Oligocene followed by several periods of strengthening in the Miocene (e.g., ~15 Ma & ~ 8 Ma) and a putative abrupt strengthening in the Pliocene / Pleistocene periods (~3 Ma) [30,31]. Various scenarios have been discussed [17,18,20-22,28] including the hypothesis of an abrupt weakening of the East Asian summer monsoon around 7.5 Ma [32]. Despite the ambiguity concerning the exact pattern, consensus exists concerning an onset of the current monsoon regimes in the Miocene and its fluctuation during the late Miocene, Pliocene and Pleistocene. The resulting changes in the amount and seasonal distribution of precipitation have likely supported a high rate of diversity turnovers including radiations of various plant lineages that compose the unique biodiversity of this region today.
Ferns are well suited to address the impact of climatic change modulating not only the maximum amount of precipitation but also its annual distribution. Ferns contribute a substantial component of the extant plant diversity in this region of the world. Recent studies rejected long-held assumptions of antiquity of fern diversity and established the post-Cretaceous origin of the majority of ferns [33-35]. Fern diversity is positively correlated with precipitation and most of the extant diversity is found in tropical mountain ranges [36]. In this context, the high fern species diversity in the SE Asia is highly remarkable and may reflect the combination of favorable climatic conditions, landscape structures and geographical history of this region [37]. A further advantage of ferns is their ability of dispersal and reproduction independent from biological interactions, which have a strong influence on the lineage history of most angiosperms [38]. Here, we focus on the derived fern genus Lepisorus that includes mainly epiphytic or saxicolous plants. The genus shows an accumulation of its species diversity in this region and some species display an adaptation to seasonality of the precipitation by having a leafless dormancy phase [39,40].
Hypotheses concerning the diversification rate changes are now regularly studied using DNA-based phylogenies despite limitations of the analytical approaches (e.g., [41-45]). Taxonomic sampling appears to be one of the major sources of errors, especially when the sampling covers <80% and/ or is non-random [42]. Thus, we sampled here more than 90% of the currently recognized species diversity of Lepisorus adopting the up-to-date classification [46-48], avoided a bias by sampling mainly deeper nodes and used simulations to assess the impact of incomplete taxon sampling. To our advantage, the taxonomy and phylogeny of the genus has been addressed in several recent studies and we consider here the most recent treatments [46-51]. These studies provided us with a rather comprehensive understanding of the species diversity and phylogenetic structure of this genus, although some uncertainty still exists concerning the number of species in this lineage that originated by primary speciation (speciation via cladogenese). Relatively little evidence for secondary speciation (speciation via polyploidy) was reported for this genus [52]. Rates of secondary speciation cannot be estimated using the established phylogenetic methods of diversification rate analyses because these analyses incorporate only the rate of primary speciation, meaning speciation by lineages splits via cladogenese events. These issues need to be considered in the interpretation of the results on the diversification of plants in which polyploidization is frequent [53]. We also consider the impact of phylogenetic and related uncertainties. Thus, we used several methods to study the rate of diversification [54-56] and employed Bayesian Dispersal-Vicariance analyses [57,58] to reconstruct ancestral ranges of lineages.
Results
Divergence time estimates
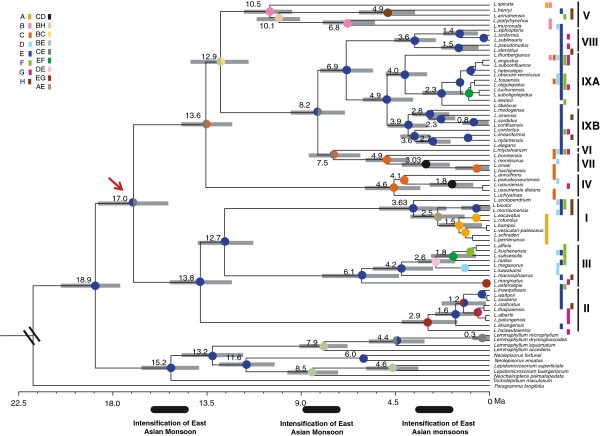
In total, nine clades (I-IX) were resolved in the genus (Figure 1), and for consistency they were marked in the same order as in our previous report [39]. The initial diversification of the genus Lepisorus dated back to around 17 Ma followed by several main cladogenese events in the period between 8 to 3 Ma (Figure 1). Lepisorus clades I and II showed crown-group divergences around 4 to 3 Ma that follow periods of phylogenetic stasis. The crown node age of clade V (= the former genus Belvisia) dated back to 10.5 Ma (Figure 1). Four clades (I to IV) had a crown group age of around or less than 6 Ma, although the stem group age dated back to more than 12 Ma.
Figure 1.
Combined chronogram and biogeographic analysis of the paleotropical genus Lepisorus. The tree is obtained from the BEAST analysis, with branches proportional to absolute age (in millions of years) calculated from median branch length of 10,000 Bayesian trees. The tree was calibrated at the most basal node (Paragramma sister to the remaining lepisoroid ferns) with 19.6 Ma (based on Schuettpelz and Pryer, 2009; Schneider et al., 2010) as a log-normal distributed calibration, which is indicated with the sign “\\”. The well-resolved nine clades are indicated as I – IX (Wang et al. 2010a). Node charts show the relative probabilities of alternative ancestral distribution obtained by statistical dispersal-vicariance analysis (S-DIVA) optimization over the 10,000 Bayesian trees. Present ranges for each species are given after the species name and detailed information is given in Additional file 2: Table S1. Areas used in the biogeographic analysis: A, Africa; B, Tropical Asia; C, Japan; D, Taiwan; E, Southwest China; F, Hainan; G, most northern part of China; H, most southern part of China. The color gradients shown in the top left corner correspond to the regions. The red arrow represents the “maximum shift point”. The black shaded boxes under the time scale correspond to the flux phases of East Asian monsoon.
Ancestral area reconstruction
Southwest (SW) China, or a combined area including SW China and Japan (both 50% probability), were recovered as the putative areas of origin of Lepisorus (Figure 1). Subsequent range expansions had been discovered to be coinciding with the crown group diversification, e.g. clade V colonizing tropical East Asia, clade IV and VII inhabiting Taiwan and Japan. Noticeably, the species occurring in Taiwan were probably migrated from Japan (Figure 1), while the ancestors of these island clades were likely colonizers from the China mainland. The ancestral distribution ranges of clades I, III, VIII, IX were inferred to be correlated with the SW China region. The members of clade II occurred up to ca. 5,000 m. Clade I showed evidence of a rapid radiation in the Afromadagascan region starting around 1.5 Ma and the ancestors of the radiated species were originately migrated from Southwest China (Figure 1).
Patterns of diversification
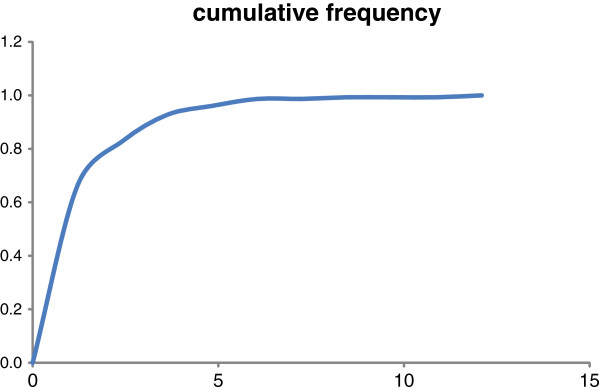
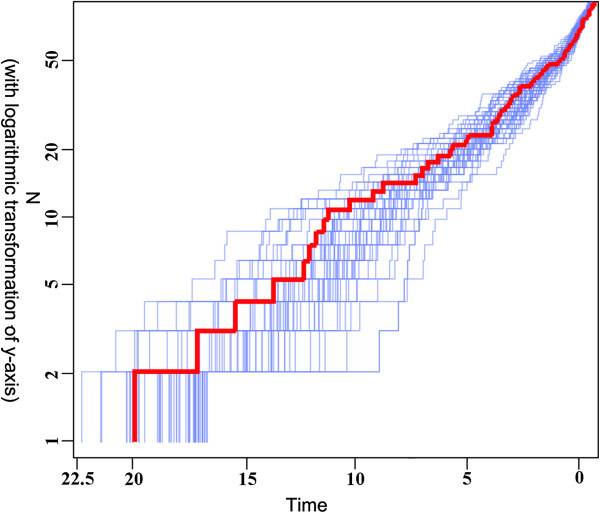
Exploring the divergence rates using the Constant-Rate test (CR) resulted in a positive γ-value of 2.638 with p = 0.996. This value did not reject constant rate but suggested instead an accumulation of branching events closer to the tips of the phylogeny. A Monte Carlo CR test (MCCR) indicated that the γ-value is unlikely caused by incomplete taxon sampling (Additional file 1: Figure S1). This result was inconsistent with the result recovered using the Venditti’s method [56]; the observed curve of cumulative density distribution of branch length corresponded to variable-rates cumulative distribution function (Figure 2). Lineage-through-time plots (LTTP) and multiple-lineage-through-time plots indicated an initial rapid radiation followed by a reduction of assembling diversity, which was subsequently replaced by an increase of the diversification rate towards the current time (Figure 3). The same trend was recovered in statistical analyses using Rabosky’s methods as implemented in LASER. The non-constant diversification rate was supported because the model assuming two rates fit better than the model assuming a single rate (ΔAIC2 rates vs. 1 rate = 9.161). The LASER result suggested an increase of diversification rate in Lepisorus after its establishment as indicated by the “best shift point” with a maximum log-likelihood value of -140.4435 (Figure 1). The maximum shift-point was correlated with the first-phase increase of the diversification rate recovered in LTTP (Figure 3), while the increase of the diversification rate in the last 4 million years was not correlated to a single shift point, but it instead may be created by the accumulation of lineages in clades I, II, and IX.
Figure 2.
Cumulative density of branch lengths for the phylogeny taken from a posterior sample of Lepisorus. The results generated by the method of Venditti et al. (2010) correspond to variable-rates cumulative distribution function.
Figure 3.
Multiple lineage-through-time plots (MLTTP) for the genus Lepisorus based on 50 randomly sampled trees from the tree pool output from the Bayesian dating analysis. The bold curve indicates the lineage-through-time plots (LTTP) for the consensus chronogram. The curve suggests two phases of shift in the diversification rate although the Constant Rate was not rejected with positive γ-value as 2.638 and p = 0.996 in LASER and APE analyses.
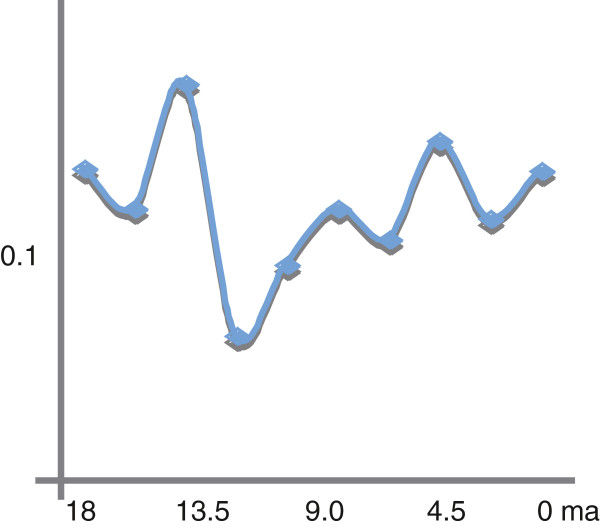
The hypothesis of two phases of increased diversification rate was also recovered by plotting the difference of species-birth (appearance of new species) per time intervals of 0.9 Ma. The origin of Lepisorus was followed by a period of increased diversification rate at about 13.5 Ma. The second phase of species-birth events was determined to date back to about 4 Ma (Figure 4).
Figure 4.
Estimates fluctuation of diversification rates through time by considering the newly appearing species (Δn) against equal time intervals. The calculation considers the number of newly appearing species versus the number of existing lineages at the beginning of the interval.
Discussion
Dealing with uncertainty
Considering the results of this study, we need to take into account different kinds of uncertainty that may prevent to recover the diversification pattern of Lepisorus. Firstly, our dating of the lepisoroid ferns was calibrated with estimates obtained in previous analyses and thus the error of these studies holds the potential to create misleading results [33-35]. However, we lack any alternative as a result of the absence of lepisoroids in the fossil record. This is consistent with the generally poor fossil record of Polypodiaceae [34,59]. To alleviate the limitations of this approach we implemented the lognormal calibration model [60].
A further uncertainty is created by the fact that the presented estimates of divergence rates are based exclusively on extant taxa. Several authors have pointed out the limitations of divergence rate estimates applying extant taxa alone, especially in the context of estimating the extinction rate (e.g., [41-43]). Therefore, we considered our results with caution and employed several methods to explore the evidence, although Rabosky’s method provides likely the most robust test. In general, the recovered pattern points toward two periods of increased diversification rate which appears to coincide with time periods in which the climate in SW China may have considerably changed (Figures 1, 4). The rather long branches of clades (e.g. clades I to IV) showing an increased rate around 6-3 Ma may be interpreted as an indicator for replacement of older species by newly established species. Studies incorporating exclusively extant taxa are ill equipped to differentiate between the lack of speciation events along long branches or the replacement as the result of species turn-over. Our argument is based on the assumption of a relative constant speciation rate along all branches and thus long branches were not expected. Interestingly, only clade V (= the former genus Belvisia) shows an unambiguous signal for constant accumulation of species through time. At the same time, this clade is the only one with a mainly tropical distribution range.
Finally, the uncertainty of the taxonomy of Lepisorus is a major challenge. Different opinions exist about the number of species. We follow here the most recent revisions, which tend to reduce the number of recognized species [46,47]. The reduced number of accepted species is caused by the careful re-evaluation of the information provided by morphological characters and considering exhaustive number of specimens [40,46,47]. Some of the ignored species may be the result of secondary speciation via polyploidization. Allopolyploid speciation has been documented for Lepisorus thunbergianus[52] and our recent results suggest speciation via polyploidy as a mode of evolution in the Lepisorus clathratus complex [61]. Current methods of diversification rate estimates can deal exclusively with primary speciation events (cladogeneses) and the contribution of speciation events via reticulation cannot be assessed. Thus, we need to keep in mind that our study considers only speciation events that are based on cladogenese events despite the fact that many lineages of plants assemble their species diversity partly via polyploidy [53].
We also need to consider the sensitivity of the different methods employed to explore the diversification pattern. The constant-rate test of Pybus & Harvey [54] did not provide significant evidence to reject a constant diversification rate, although the positive γ-value may indicate a clustering of the branching events towards the recent. In contrast, the method of Venditti et al. [56] suggests a slow-down of the diversification rate. Both methods are rather simplistic and limit undoubtedly their explanatory power [62]. Parameter-rich approaches [55] are likely more powerful to reconstruct complex patterns of diversification rate changes such as multiple deviations from a constant rate. As a payback, these methods may be especially problematic in the context of changes affected by the extinction rate [45]. Remarkably, LASER analysis detected the early diversification of Lepisorus (Figure 1, red arrow) as the main shift point of the diversification rate, but it did not detect nodes corresponding to the crown groups of clades I II, IV and IX as possible shift-points. In our case, the hypothesis of multiple-changes of the diversification rates appears to be most consistent with the results recovered by different methods to infer divergence rates.
Evidence for the impact of changing monsoon climates on fern diversity
Our results suggest two periods of increased diversification rate exist in the genus Lepisorus. They likely coincide with time periods suggested to be crucial phases of changes in the evolution of the East Asian monsoon (Figure 1). The initial radiation of Lepisorus is accordant with the proposed phase of strengthening of the East Asian monsoon at around 15 Ma, whereas the second period of diversification rate increase dates back to around 6-5 Ma, which lags behind an intensification of the East Asian monsoons [27] (Figure 1). This change of the climate is correlated with the rise of the southern part of the QTP [17,18,20,27,29]. All the clades showing the increase of diversification rates share the SW Chinese Mountains as their likely area of origin and centre of biodiversity only with the putative exception of clade II. The L. clathratus clade, clade II, is of particular interest because these ferns tend to grow in very high altitudes with up to 5,000 m [40]. Furthermore, clades I and II show a unique adaptation to strong seasonal climates in form of summer-green versus the ever-green leaves [39]. In these clades, the ferns undergo a dormant phase in the winter after they have shed their leaves in the late autumn.
The assembled evidences may fit best with the hypothesis of species turn-over events in which taxa with different climatic preferences were replaced with the taxa that are more adapted to the new climatic conditions. The clades with SW China as the origin centre share long branches that may reflect the replacement of previously established taxa with taxa adapted to the new climatic regime. The most outstanding exception is clade V, which corresponds to the former genus Belvisia. These taxa occur mainly in tropical SE Asia and thus the impact of the monsoon regimes is expected to be limited on their biodiversity. Thus, the more constant accumulation of species diversity shown by ‘Belvisia’ may reflect the more conservative nature of the climates in tropical SE Asia.
Conclusions
The presented results outline the direction of the research aiming to explore the impact of the uplift of the Qinghai-Tibetan Plateau on the diversification of plant lineages occurring in SE Asia. Our results are consistent with reports on other lineages of plants such as the eudicot genera Caragana[16] and Rheum[15], the conifer genus Juniperus[63] or animals such as spiny frogs [10]. Most importantly, our results show evidence that the impact is not limited to plants occurring on the plateau and Asian interior areas affected by aridification. Instead, the uplift may have had a strong impact on the plant life in whole SE Asia with the exception of areas strongly influenced by maritime climates such as Japan and Taiwan. These areas may be the refuge of taxa adapted to less seasonal monsoon climates. These new findings suggest the need to study more extensively the impact of the rise of the Himalaya on the assembly and maintenance of biodiversity in SE Asia [64]. Previous studies were often focused only on genera that colonized the newly founded mountain ranges (e.g., [15,63,65]), whereas the impact on the plant diversity in the lower altitudes of SE Asia were widely ignored.
Methods
Datasets, taxon sampling, phylogenetic analyses
We assembled sequences of four chloroplast genome regions, rbcL, rbcL-atpB intergenic spacer sequences (IGS), rps4 plus rps4-trnS IGS, and trnL-F region including the trnL intron and trnL-trnF IGS using sequences generated in previous studies [39,40,51]. A few additional sequences of expanded sampled species (L. nudus Ching, L. nylamensis Ching et S. K. Wu, L. perrierianus Ching, L. schraderi (Mett.) Faden, L. rotundus Ching and L. vesiculari-peleaceus (Hieron.) Pic. Serm.) were obtained using the protocols published in these studies. In total, the dataset included 65 out of the ca. 70 currently known species of Lepisorus plus 11 species of other lepisoroid fern genera (See Additional file 2). The taxonomy follows the most recent treatments [39,40,46-51,66]. The sampling was designed to cover more than 90% of the species diversity of Lepisorus and represent its geographic range without any obvious bias. All alignment were generated manually in Maclade 4.08 [67] and ambiguous regions excluded from further analyses.
Initially, the dataset was explored by carefully designed phylogenetic analyses using PAUP 4.0 [68] for maximum parsimony, PHYML 3.0 [69] for maximum likelihood and MrBayes 3.1.2 [70] for Bayesian Inference of phylogeny as described in Wang et al. [39]. The model of sequence evolution was determined using jModelTest [71] and a likelihood ratio test (LHR) was carried out to test for the presence of a molecular clock. No evidence for topological heterogeneity was discovered and all subsequent analyses were carried out with a combined dataset.
Divergence time estimates
Bayesian approaches with uncorrelated relaxed clock model were employed to estimate the divergence times of the lepisoroid ferns with focus on the genus Lepisorus. The LHR test results supported the use of the relaxed clock model. All divergence time estimates were carried out using BEAST 1.6.2 ( http://beast/bioed.ac.uk) [72]. The model selected with jModelTest was implemented, but parameters were estimated simultaneously with the BEAST analyses. Several BEAST analyses were carried out and their results were summarized utilizing TRACER 1.5 ( http://beast.bio.ed.ac.uk) and TREEANNOTATOR, part of the BEAST package. Markov chains were run for 10,000,000 generations with every 10,000 generations sampled and at least a 10% burn-in phase. The final analyses were performed with a relaxed molecular clock [73], birth-death model, and calibration of the split of Paragramma and the remaining lepisoroid ferns [39] with a lognormal distribution model with a shift of 19.6 ma. This node age was obtained from divergence time estimate carried out with a comprehensive sampling throughout the tree of ferns [35]. Unfortunately, this study did not provide confidence intervals for nodes age estimates. The obtained age estimates were consistent with divergence time estimates of Polypodiaceae and the limited fossil record of the Polypodiaceae [33,34,59] (see also discussion). As a further confirmation, we calculated the nucleotide substitution rates for several clades and compared them with previously reported substitution rates of the chloroplast genome [74,75]. The rates were within the expected range. Shown chronograms were calculated using median clade credibility tree plus 95% confidence intervals.
Ancestral area reconstruction
Distribution data for each species were obtained from available floristic or taxonomic treatments [39,40,46-51,66] plus information obtained from selected herbaria (BM, PE). The distribution range of Lepisorus was divided into eight regions of endemism (A-H) following general guidelines to define those areas. Bayesian Divergence Vicariance analyses (Bayes-DIVA) [58] were carried out using S-DIVA [76]. We used all trees in the plateau phase of the BEAST analysis (see above). The maximum area number at each node was set as two in the final analyses but alternative analyses with three or more maximal areas were carried out for comparison. We chose Bayes-DIVA considering its ability to infer ancestral biogeopraphic ranges under consideration of phylogenetic uncertainty [57,58]. The analyses were carried out with and without incorporation of the outgroup taxa. The waste majority of the outgroup taxa occurs exclusively in southwest China. Thus, the results of the analyses with and without outgroup taxa were consistent.
Diversification rates
To investigate patterns of diversification through time, we used five approaches. First, we explored evidence for non-constant diversification rate in Lepisorus using the Constant Rate (CR) birth death model test [54] as implemented in several programs including Gammastatistics V.10 ( http://www.oekologie.biologie.uni-mainz.de/people/evi/home/html) and in R applications ( http://www.R-project.org): APE [77], GEIGER [78] and LASER [55]. A MCCR test using simulations of phylogenies was employed to account for the impact of missing species [54,79]. Second, we estimated the cumulative density distribution of branch length [56]. Length of the branches were detached from the calibrated consensus tree and divided into ten length classes. Frequency number of each class was calculated and plotted as cumulative frequency against branch length classes. Third, the diversification rates were visualized by generating multiple lineage-through-time plots (MLTTP) for 50 randomly selected trees from the tree pool output from BEAST using the R packages APE and GEIGER, plus lineage-through-time plots (LTTP) for the consensus chronogram with a mean node age. Fourth, we used Rabosky’s LASER software [55] in R to determine whether the observed pattern of diversification through time fits best to a simple model of a constant diversification rate or to complex models with variable diversification rates. The best fitting rate model, maximum shift points and the threshold of shift points were estimated according to the maximum log-likelihood values using Akaike Information Criterion (AIC) weights and Delta-AIC scores [55]. Fifth, we visualized changes of the diversification rate by plotting the difference of number of newly appearing species (Δn) against the setting time intervals (Δt) as 0.9 Ma. The value of Δn was then transferred into changes of diversification rate per time interval by considering the number of species at the beginning of the interval. The limitations of the outgroup sampling were considered by exploring the impact of different sampling densities. However, the sister lineage of Lepisorus is considerable less species-rich [51]. All the results were compared to the suggested periods of changes in the monsoon regimes and major geological events [17-20,27].
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
ZXC, HS and XQP designed the study. WL and ZXC collected materials. WL finished molecular experiments. WL and HS analyzed data. WL, XQP, ZXC, and HS wrote the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Figure S1. Histogram of a null distribution of gamma statistic on incompletely sampled trees using simulation. The red arrow demonstrates the retrieved gamma value, which is outside of the 95% confidence interval, and thus indicates that the incomplete sampling does not bias the gamma statistic.
Table S1. Information regarding taxon names, collecting localities, voucher numbers, Genbank accession numbers and distributional ranges for taxa included in the analyses. Distributional ranges are coded as follows: A, Africa; B, Tropical Asia; C, Japan; D, Taiwan; E, Southwest China; F, Hainan; G, most northern part of China; H, most southern part of China.
Contributor Information
Li Wang, Email: liwang@ibcas.ac.cn.
Harald Schneider, Email: h.schneider@nhm.ac.uk.
Xian-Chun Zhang, Email: zhangxc@ibcas.ac.cn.
Qiao-Ping Xiang, Email: qpxiang@ibcas.ac.cn.
Acknowledgements
This project was financially supported by the National Natural Science Foundation of China Grant (NSFC No. 31070196, No. 30870167 and No. 31070182) and the senior visiting professorship grant by the Chinese Academy of Sciences to H. Schneider.
References
- Myers N, Mittermeier RA, Mittermeier CG, da Fonesca GAB, Kent J. Biodiversity hotspots for conservation priorities. Nature. 2000;403(6772):853–858. doi: 10.1038/35002501. [DOI] [PubMed] [Google Scholar]
- Antonelli A, Nylander JAA, Persson C, Sanmartínc I. Tracing the impact of the Andean uplift on neotropical plant evolution. Proc Natl Acad Sci USA. 2009;106(24):9749–9754. doi: 10.1073/pnas.0811421106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias M, Joron M, Willmott K, Silva-Brandão KL, Kaiser V, Arias CF, Piñerez LMG, Uribe S, Brower AVZ, Freitas AVL. Out of the Andes: patterns of diversification in clearwing butterflies. Mol Ecol. 2009;18(3):1716–1729. doi: 10.1111/j.1365-294X.2009.04149.x. [DOI] [PubMed] [Google Scholar]
- Hoorn C, Wesselingh FP, ter Steege H, Bermudez MA, Mora A, Sevink J, Sanmartin I, Sanchez-Meseguer A, Anderson CL, Figueiredo JP, Jaramillo C, Riff D, Negri FR, Hooghiemstra H, Lundberg J, Stadler T, Saerkinen T, Antonelli A. Amazonia through time: Andean uplift, climate change, landscape evolution, and biodiversity. Science. 2010;330(6006):927–931. doi: 10.1126/science.1194585. [DOI] [PubMed] [Google Scholar]
- Linder HP. The radiation of the cape flora, Southern Africa. Biol Rev (Camb) 2003;78(4):597–638. doi: 10.1017/s1464793103006171. [DOI] [PubMed] [Google Scholar]
- Richardson JE, Weitz FM, Fay MF, Cronk QCB, Linder HP, Reeves G, Chase MW. Rapid and recent origin of species richness in the cape flora of South Africa. Nature. 2001;412(6843):181–183. doi: 10.1038/35084067. [DOI] [PubMed] [Google Scholar]
- Janssen T, Bystriakova N, Rakotondrainibe F, Coomes D, Labat JN, Schneider H. Neoendemism in Madagascan scaly tree ferns results from recent, coincident diversification bursts. Evolution. 2008;62(8):1876–1889. doi: 10.1111/j.1558-5646.2008.00408.x. [DOI] [PubMed] [Google Scholar]
- Vences M, Wollenberg KC, Vieites DR, Lees DC. Madagascar as a model region of species diversification. Trends Ecol Evol. 2009;24(8):456–465. doi: 10.1016/j.tree.2009.03.011. [DOI] [PubMed] [Google Scholar]
- Linder HP. Plant species radiation: where, when, why? Phil Trans R Soc B. 2008;363(1506):3097–3105. doi: 10.1098/rstb.2008.0075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Che J, Zhou WW, Hu JS, Yan F, Papenfuss TJ, Wake DB, Zhang YP. Spiny frogs (Paini) illuminate the history of the Himalayan region and Southeast Asia. Proc Natl Acad Sci USA. 2010;107(31):13765–13770. doi: 10.1073/pnas.1008415107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ST, Xing YW, Su T, Zhou ZK, Dilcher DL, Soltis DE. Phylogeopraphic analysis reveals significant spatial genetic structure if Incarvellia sinensis as a product of mountain building. BMC Plant Biol. 2012;12:58. doi: 10.1186/1471-2229-12-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo XG, He SP, Zhang YG. Phylogeny and biogeography of Chinese sisorid catfishes re-examined using mitochondrial cytochrome b and 16S rRNA gene sequences. Mol Phylogenet Evol. 2005;35(2):344–362. doi: 10.1016/j.ympev.2004.12.015. [DOI] [PubMed] [Google Scholar]
- Liu JQ, Wang YJ, Wang AL, Hideaki O, Abbott RJ. Radiation and diversification within the Ligularia-Cremanthodium-Parasenecio complex (Asteraceae) trigerred by uplift of the Qinghai-Tibetan Plateau. Mol Phylogenet Evol. 2006;38(1):31–49. doi: 10.1016/j.ympev.2005.09.010. [DOI] [PubMed] [Google Scholar]
- Qu Y, Lei F. Comparative phylogeography of two endemic birds of the Tibetan plateau, the white-rumped snow finch (Onychostruthus taczanowskii) and the Hume’s ground tit (Pseudopodoces humilis) Mol Phylogenet Evol. 2009;51(2):312–326. doi: 10.1016/j.ympev.2009.01.013. [DOI] [PubMed] [Google Scholar]
- Sun YS, Wang A, Wan DS, Wang Q, Liu JQ. Rapid radiation of Rheum (Polygonaceae) and parallel evolution of morphological traits. Mol Phylogenet Evol. 2012;63(1):150–158. doi: 10.1016/j.ympev.2012.01.002. [DOI] [PubMed] [Google Scholar]
- Zhang ML, Fritsch PW. Evolutionary response of Caranga (Fabaceae) to Quinghai-Tibetan plateau uplift and Asian interior aridification. Plant Syst Evol. 2010;288(3–4):191–199. [Google Scholar]
- Harris N. The elevation history of the Tibetan plateau and its implications for the Asian monsoon. Palaeogeogr Palaeoclimatol Palaeoecol. 2006;241(1):4–15. doi: 10.1016/j.palaeo.2006.07.009. [DOI] [Google Scholar]
- Passey BH, Ayliffe LK, Kaakinen A, Zhang ZQ, Eronen JT, Zhu YM, Zhou LP, Cerling TE, Fortelius M. Stengthened East Asian summer monsoons during a period of high-latitute warmth? isotopic evidence from Mio-Pliocene fossil mammals and soild carbonates from Northern China. Earth Planet Sci Lett. 2009;277(3–4):443–452. [Google Scholar]
- Spicer RA, Harris NBW, Widdowson M, Herman AB, Guo S, Valdes PJ, Wolfe JA, Kelley SP. Constant elevation of southern Tibet over the past 15 million years. Nature. 2003;421(6923):622–624. doi: 10.1038/nature01356. [DOI] [PubMed] [Google Scholar]
- Sun XJ, Wang PX. How old is the Asian monsoon system?-palaeobotanical records from China. Paleogeogr Paleoclimatol Paleoecol. 2005;222(3–4):181–222. [Google Scholar]
- Yao YF, Bruch AA, Mosbrugger V, Li CS. Quantitative reconstruction of Miocene climate patterns and evolution in Southern China based on plant fossils. Palaeogeogr Palaeoclimatol Palaeoecol. 2011;304(3–4):291–307. [Google Scholar]
- Zheng HB, McPowell CA, Red DK, Wang JL, Wang PX. Late Miocene and mid-Pliocene enhancement of the East Asian monsoon as viewed from the land and sea. Global Planet Change. 2004;41(3–4):147–155. [Google Scholar]
- Royden LH, Burchfiel BC, van der Hilst RD. The geological evolution of the Tibetan Plateau. Science. 2008;321(5892):1054–1058. doi: 10.1126/science.1155371. [DOI] [PubMed] [Google Scholar]
- Li JJ, Wen SX, Zhang QS, Wang FB, Zheng BX, Li BY. A discussion on the period, amplitude and type of the uplift of the Qinghai-Xizang Plateau. Sci Sin. 1979;22:1314–1328. [Google Scholar]
- Qiu YX, Fu CX, Comes HP. Plant molecular phylogeography in China and adjacent regions: tracing the genetic imprints of Quaternary climate and environmental change in the world’s most diverse temperate flora. Mol Phylogenet Evol. 2011;59(1):225–244. doi: 10.1016/j.ympev.2011.01.012. [DOI] [PubMed] [Google Scholar]
- Zhou SZ, Wang XL, Wang J, Xu LB. A preliminary study on timing of the oldest Pleistocene glaciation in Qinghai–Tibetan Plateau. Quatern Intern. 2006;154-155:44–51. [Google Scholar]
- An Z, Kutzbach JE, Prell WL, Porter SC. Evolution of Asian monsoons and phased uplift of the Himalayan-Tibetan Plateau since Late Miocene times. Nature. 2001;411(6833):62–66. doi: 10.1038/35075035. [DOI] [PubMed] [Google Scholar]
- Li FJ, Rousseau DD, Wu NQ, Hao QZ, Pei YP. Late Neogene evolution of the East Asian monsoon revealed by terrestrial mollusk record in Western Chinese Loess Plateau: from winter to summer dominated sub-regime. Earth Planet Sci Lett. 2008;274(3–4):439–447. [Google Scholar]
- Liu X, Yin ZY. Sensitivity of East Asian monsoon climate to the uplift of the Tibetan Plateau. Palaeogeogr Palaeoclimatol Palaeoecol. 2002;183(3–4):223–225. [Google Scholar]
- Jacques FMB, Go SX, Xing YW, Huang YJ, Liu YS, Ferguson DK, Zhou ZK. Qunatitive reconstruction of the late Miocene monsoon climates of southwest China: a case study of the lincang flora from Yunnan province. Palaeogeogr Palaeoclimatol Palaeoecol. 2011;304(3–4):318–327. [Google Scholar]
- Wan S, Li AC, Clift PD, Stuut JBW. Development of the East Asian monsoon: mineralogical and sedimentological records in the northern South China Sea since 20 Ma. Palaeogeogr Palaeoclimatol Palaeoecol. 2007;254(3–4):561–582. [Google Scholar]
- Steinke S, Groeneveld J, Johnstone H, Rendle-Bühringa R. East Asia summer monsoon weakening after 75 Ma: evidence from combined planktonic foraminifera Mg/Ca and δ18O (ODP site 1146; northernS China Sea) Paleogeogr Paleoclimatol Paleoecol. 2010;289(1–4):33–43. [Google Scholar]
- Schneider H, Schuettpelz E, Pryer KM, Cranfill R, Magallón S, Lupia R. Ferns diversified in the shadow of angiosperms. Nature. 2004;428(6982):553–557. doi: 10.1038/nature02361. [DOI] [PubMed] [Google Scholar]
- Schneider H, Kreier HP, Janssen T, Otto E, Muth H, Heinrichs J. In: Evolution in action. Glaubrecht M, editor. Berlin: Springer; 2010. Key innovations versus key opportunities: identifying causes of rapid radiations in derived ferns; pp. 61–76. [Google Scholar]
- Schuettpelz E, Pryer KM. Evidence for a Cenozoic radiation of ferns in an angiosperm-dominated canopy. Proc Natl Acad Sci USA. 2009;106(27):11200–11205. doi: 10.1073/pnas.0811136106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreft H, Jetz W, Mutke J, Barthlott W. Contrasting environmental and regional effects on global pteridophyte and seed plant diversity. Ecography. 2010;33(2):408–419. doi: 10.1111/j.1600-0587.2010.06434.x. [DOI] [Google Scholar]
- Chen SB, Jiang GM, Ouyang ZY, Xu WH, Xiao Y. Relative importance of water, energy, and heterogeneity in determining regional pteridophyte and see plant richness in China. J Syst Evol. 2011;49(2):95–107. doi: 10.1111/j.1759-6831.2011.00120.x. [DOI] [Google Scholar]
- Schnitzler J, Barraclough TG, Boatwright JS, Goldblatt P, Manning JC, Powell MP, Rebelo T, Savolainen V. Causes of plant diversification in the cape biodiversity hotspot of South Africa. Syst Biol. 2011;60(3):343–357. doi: 10.1093/sysbio/syr006. [DOI] [PubMed] [Google Scholar]
- Wang L, Qi XP, Xiang QP, Heinrichs J, Schneider H, Zhang XC. Phylogeny of the paleotropical fern genus Lepisorus (Polypodiaceae, Polypodiopsida) inferred from four chloroplast genome regions. Mol Phylogenet Evol. 2010;54(1):211–225. doi: 10.1016/j.ympev.2009.08.032. [DOI] [PubMed] [Google Scholar]
- Wang L, Wu ZQ, Bystriakova N, Ansell SW, Xiang QP, Heinrichs J, Schneider H, Zhang XC. Phylogeography of the Sino-Himalayan fern Lepisorus clathratus on “the roof of the world”. PLoS One. 2011;6:e25896. doi: 10.1371/journal.pone.0025896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brock CD, Harmon LJ, Alfaro ME. Testing for temporal variation in diversification rates when sampling is incomplete and nonrandom. Syst Biol. 2011;60(4):416–419. doi: 10.1093/sysbio/syr007. [DOI] [PubMed] [Google Scholar]
- Cusimano N, Renner SS. Slowdowns in diversification rates from real phylogenies may not be real. Syst Biol. 2010;59(4):458–464. doi: 10.1093/sysbio/syq032. [DOI] [PubMed] [Google Scholar]
- Liow LH, Quental TB, Marshall CR. When can decreasing diversification rates be detected with molecular phylogenies and the fossil record. Syst Biol. 2010;59(6):646–659. doi: 10.1093/sysbio/syq052. [DOI] [PubMed] [Google Scholar]
- Quental TB, Marshall CR. Diversity dynamics: molecular phylogenies need the fossil record. Trends Ecol Evol. 2010;25(8):434–441. doi: 10.1016/j.tree.2010.05.002. [DOI] [PubMed] [Google Scholar]
- Rabosky DL. Extinction rates should not be estimated from molecular phylogenies. Evolution. 2010;64(6):1816–1824. doi: 10.1111/j.1558-5646.2009.00926.x. [DOI] [PubMed] [Google Scholar]
- Qi XP, Zhang XC. Taxonomic revision of Lepisorus (J Sm) Ching sect Lepisorus (Polypodiaceae) from China. Acta Phytotax Sinica. 2009;47(6):581–598. doi: 10.1111/j.1759-6831.2009.00056.x. [DOI] [Google Scholar]
- Qi XP, Zhang XC, Wei R. Taxonomic revision of Lepisorus sect Sclerophyllon (Polypodiaceae) from China. Acta Bot Yunn Supp. 2010;XVII:55–64. [Google Scholar]
- Zink MJ. Systematics of the fern genus Lepisorus (J Smith) Ching (Polypodiaceae, Lepisoreae) Zurich: PhD Dissertation University Zurich; 1993. [Google Scholar]
- Hovenkamp P. An account of the fern genus Belvisia Mirbel (Polypodiaceae) Blumea. 1993;37:511–527. [Google Scholar]
- Liu QR, Ming GH, Ge Y, Zhang XC. A taxonomic revision of Lepisorus sect Hymenophyton (Polypodiaceae) from China. J Syst Evol. 2008;46(6):906–915. [Google Scholar]
- Wang L, Wu ZQ, Xiang QP, Heinrichs J, Schneider H, Zhang XC. A molecular phylogeny and a revised classification of tribe Lepisoreae (Polypodiaceae) based on an analysis of four plastid DNA regions. Bot J Linn Soc. 2010;162(1):28–38. doi: 10.1111/j.1095-8339.2009.01018.x. [DOI] [Google Scholar]
- Shinohara W, Ushio Y, Seo A, Nakato N, Kono M, Udoh H, Tobe H, Murakami N. Evidence for hybrid orign and segmental allopolyploidy in eutetraploid and aneutetraploid Lepisorus thunbergianus (Polypodiaceae) Syst Bot. 2010;35(1):20–29. doi: 10.1600/036364410790862498. [DOI] [Google Scholar]
- Wood TE, Takebayashi N, Barker MS, Mayrose I, Greenspoon PB, Rieseberg LH. The frequency of polyploid speciation in vascular plants. Proc Natl Acad Sci USA. 2009;106(33):13869–13875. doi: 10.1073/pnas.0811575106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pybus OG, Harvey PH. Testing macro-evolutionary models using incomplete molecular phylogenies. Proc R Soc B. 2000;267(1459):2267–2272. doi: 10.1098/rspb.2000.1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabosky DK. LASER: a maximum likelihood toolkit for detecting temporal shifts in diversification rates from molecular phylogenies. Evol Bioinform. 2006;2:257–260. [PMC free article] [PubMed] [Google Scholar]
- Venditti C, Meade A, Pagel M. Phylogenies reveal new interpretation of speciation and the red queen. Nature. 2010;463(7279):349–352. doi: 10.1038/nature08630. [DOI] [PubMed] [Google Scholar]
- Buerki S, Forrest F, Alvarez N, Nylander JAA, Arrigo N, Sanmartin I. An evaluation of new parsimony-based versus parametric inference methods in biogeography: a case study using the globally distributed plant family Sapindaceae. J Biogeogr. 2011;38(3):531–550. doi: 10.1111/j.1365-2699.2010.02432.x. [DOI] [Google Scholar]
- Nylander JAA, Olson U, Alstrom P, Sanmartin I. Accounting for phylogenetic uncertainty in biogeography: a Bayesian approach to dispersal-vicariance analysis of the trushes (Aves: Turdus ) Syst Biol. 2008;57(2):257–268. doi: 10.1080/10635150802044003. [DOI] [PubMed] [Google Scholar]
- Van Uffelen GA. Fossil polypodiaceae and their spores. Blumea. 1991;36:253–272. [Google Scholar]
- Ho SYW. Calibrating molecular estimates of substitution rages and divergence times in birds. J Avian Biol. 2007;38(4):409–414. [Google Scholar]
- Wang L, Schneider H, Wu ZQ, He LJ, Zhang XC, Xiang QP. Indehiscent sporangia enable the accumulation of local fern diversity at the Qinghai-Tibetan Plateau. BMC Evol Biol. 2012;12:158. doi: 10.1186/1471-2148-12-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertheim JO, Sanderson MJ. Estimating diversification rates; how useful are divergence times? Evolution. 2011;65(2):309–320. doi: 10.1111/j.1558-5646.2010.01159.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao KS, Hao G, Li JQ, Adams RP, Milne RI. Diversification and biogeography of Juniperus (Cupressaceae): variable diversification rates and multiple intercontinental dispersals. New Phytol. 2010;188(2):254–272. doi: 10.1111/j.1469-8137.2010.03351.x. [DOI] [PubMed] [Google Scholar]
- Lopez-Pujol J, Zhang FM, Sun HQ, Ying TS, He S. Mountains of southern china as “plant museums” and “plant cradles”. Mount Res Devel. 2011;31(3):261–269. doi: 10.1659/MRD-JOURNAL-D-11-00058.1. [DOI] [Google Scholar]
- Wang YL, Susanna A, von Rabb-Straube E, Milne R, Liu JQ. Island-like radiation of Saussurea (Asteraceae: Cardueae) triggered by uplifts of the Qinghai-Tibetan Plateau. Biol J Linn So. 2009;97(6):863–903. [Google Scholar]
- Hovenkamp P. Lepisorus in Malesia. Blumea. 1998. pp. 109–115.
- Maddison DR, Maddison WP. MacClade 408 sinauer associates, Sunderland, mass. 2005.
- Swofford DL. PAUP* phylogeneitc analysis using parsimony (*and other methods) version 4. Sunderland, Mass: Sinauer Associates; 2002. [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hirdijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59(3):307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F. MrBayes: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17(8):754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Posada D. jModelTest: phylogenetic model averaging. Mol Biol Evol. 2008;25(7):1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Ho SYW, Philips MJ, Rambaut A. Relaxed phylogenetics and dating with confidence. PLoS Biol. 2006;4:699. doi: 10.1371/journal.pbio.0040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willyard A, Syring J, Gernandt DS, Liston A, Cronn R. Fossil calibration of molecular divergence infers a moderate mutation rate and recent radiations of Pinus. Mol Biol Evol. 2007;24(1):90–101. doi: 10.1093/molbev/msl131. [DOI] [PubMed] [Google Scholar]
- Wolfe KH, Li WH, Sharp PM. Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. Proc Natl Acad Sci USA. 1987;84(24):9054–9058. doi: 10.1073/pnas.84.24.9054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Harris AJ, He XJ. S-DIA (statistical dispersal-vicariance analysis): a tool for inferring biogeographic histories. Mol Phylogenet Evol. 2010;56(3):848–850. doi: 10.1016/j.ympev.2010.04.011. [DOI] [PubMed] [Google Scholar]
- Paradis E, Claude J, Strimmer K. APE: analyses of phylogenetics and evolution in R language. Bioinfor. 2004;20(2):289–290. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- Harmon LJ, Weit J, Brock C, Glor RE, Challenger W. GEIGER: investigating evolutionary radiations. Bioinfor. 2008;24(1):129–131. doi: 10.1093/bioinformatics/btm538. [DOI] [PubMed] [Google Scholar]
- Rabosky DK, Lovette IJ. Density-dependent diversification in North American wood warblers. Proc R Soc B. 2008;275(1649):2363–2371. doi: 10.1098/rspb.2008.0630. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Histogram of a null distribution of gamma statistic on incompletely sampled trees using simulation. The red arrow demonstrates the retrieved gamma value, which is outside of the 95% confidence interval, and thus indicates that the incomplete sampling does not bias the gamma statistic.
Table S1. Information regarding taxon names, collecting localities, voucher numbers, Genbank accession numbers and distributional ranges for taxa included in the analyses. Distributional ranges are coded as follows: A, Africa; B, Tropical Asia; C, Japan; D, Taiwan; E, Southwest China; F, Hainan; G, most northern part of China; H, most southern part of China.