Abstract
Background
Adiponectin is reported to be related to the development of chronic obstructive pulmonary disease (COPD). Genetic variants in the gene encoding adiponectin (ADIPOQ) have been reported to be associated with adiponectin level in several genome–wide linkage and association studies. However, relatively little is known about the effects of ADIPOQ gene variants on COPD susceptibility. We determined the frequencies of single-nucleotide polymorphisms (SNPs) in ADIPOQ in a Chinese Han population and their possible association with COPD susceptibility.
Methods
We conducted a case–control study of 279 COPD patients and 367 age- and gender-distribution-matched control subjects. Seven tagging SNPs in ADIPOQ, including rs710445, rs16861205, rs822396, rs7627128, rs1501299, rs3821799 and rs1063537 were genotyped by SNaPshot. Association analysis of genotypes/alleles and haplotypes constructed from these loci with COPD was conducted under different genetic models.
Results
The alleles or genotypes of rs1501299 distributed significantly differently in COPD patients and controls (allele: P = 0.002, OR = 1.43 and 95%CI = 1.14–1.79; genotype: P = 0.008). The allele A at rs1501299 was potentially associated with an increased risk of COPD in all dominant model analysis (P = 0.009; OR: 1.54; 95%CI: 1.11–2.13), recessive model analyses (P = 0.015; OR: 1.75; 95% CI: 1.11–2.75) and additive model analyses (P = 0.003; OR: 2.11; 95% CI: 1.29–3.47). In haplotype analysis, we observed haplotypes AAAAACT and GGACCTC had protective effects, while haplotypes AGAACTC, AGGCCTC, GGAACTC, GGACACT and GGGCCTC were significantly associated with the increased risk of COPD.
Conclusions
We conducted the first investigation of the association between the SNPs in ADIPOQ and COPD risk. Our current findings suggest that ADIPOQ may be a potential risk gene for COPD. Further studies in larger groups are warranted to confirm our results.
Introduction
Chronic obstructive pulmonary disease (COPD) is a major global disease that has been predicted to be the third leading cause of mortality worldwide by the year 2020 [1], and it is estimated to affect nearly 8.2% of the Chinese adult population [2]. Cigarette smoking is the major environmental risk factor for COPD; however, only approximately 15% of smokers develop clinically relevant airflow obstruction [3] . The variation in the susceptibility to cigarette smoke, in combination with the familial inheritance pattern of COPD, suggests that there may be a genetic component to the development of COPD [4]. The associations between COPD and polymorphisms in genes with potential importance in COPD pathogenesis have been investigated [5]; however, only α1-antitrypsin has been unequivocally identified as relevant to the development of COPD. Recently, polymorphisms in the CHRNA3-CHRNA5-IREB2, HHIP, and FAM13A loci have been found to be associated with COPD by genome-wide association studies (GWAS) [6]–[8].
There is increasing evidence of systemic inflammation in patients with COPD. Several disease biomarkers have been found to be helpful in assessing systemic and local inflammation, including interleukin (IL) 6, IL-8 and C-reactive protein (CRP) [9], [10]. Adiponectin is a secretary 30 kD protein synthesized by adipocytes in healthy subjects. Its role in inflammation is controversial, as its plasma concentration decreases in diseases such as metabolic syndrome and type II diabetes [11], but increases in some inflammatory diseases such as rheumatoid arthritis and systemic lupus erythematosus [12], [13]. Animal studies, for instance, suggest that reduced expression of adiponectin is associated with development of emphysema and is associated with the pathogenesis of cachexia and osteoporosis [14]. However, human studies indicate that circulating adiponectin levels are raised in COPD and associated with poor health outcomes including increased risk of exacerbations [15]–[17]. Thus, the relationship of adiponectin to COPD outcomes remains uncertain. Genetic factor accounts for about 40–70% of the variation in adiponectin levels [18]. Genetic variants in the gene encoding adiponectin (ADIPOQ) have been reported to be associated with adiponectin levels in several genome-wide linkage and association studies [18]–[20]. However, inconsistent findings on the association of genetic variants of ADIPOQ with adiponectin levels have been reported [18]–[22], which could be due to a difference in ethnic populations, single nucleotide polymorphism (SNP) selection, and study power.
With these considerations in mind, we hypothesized that polymorphisms in the ADIPOQ gene might modulate susceptibility to COPD. To test this hypothesis, we investigated the association of common genetic variants in the ADIPOQ gene with the risk of COPD in a Chinese Han population.
Materials and Methods
Ethics Statement
The use of human tissue and the protocol in this study were strictly conformed to the principles expressed in the Declaration of Helsinki and were approved by the Ethical Committee of the West China Hospital, Sichuan University, and written informed consent was obtained from all subjects before their participation in the study. The investigator explained the nature, purpose and risks of the study and provided the subject with a copy of the information sheet.
Subjects
As described previously [23], 279 patients with COPD and 367 age-matched non-COPD control subjects were recruited for this study. The subjects in both groups were unrelated ethnic Han Chinese individuals recruited from Chengdu city or surrounding regions in the Sichuan province of western China. All subjects underwent physical examinations including chest x-ray, anthropometric measurements including body mass index (BMI), assessment of lung function, and blood sampling. The recruitment and the clinical analyses were conducted at the department of respiratory medicine in West China Hospital of Sichuan University; clinical analyses were performed according to the Global Initiative for Chronic Obstructive Lung Disease (GOLD) criteria [24]. COPD patients were enrolled when they suffered from cough, sputum production and dyspnea at least upon exertion and showed chronic irreversible airflow limitation defined by an FEV1 (forced expiratory volume in 1s) to FVC (forced vital capacity) ratio <70%, and FEV1 predicted <80% after the inhalation of a β2-agonist. Patients were excluded from this study if they had other significant respiratory diseases, such as bronchial asthma, bronchiectasis, lung cancer, or pulmonary tuberculosis based on their chest x-ray test.
The age-matched non-COPD control subjects were volunteers who came to the West China Hospital of Sichuan University for physical examination only. The inclusion criteria for controls were as follows: (1) FEV1/FVC ratio >70%, FEV1% and FVC% predicted >80% and (2) without pulmonary disease. Individuals were excluded if they had a history of chronic lung disease, atopy, an acute pulmonary infection in the 4 weeks before assessment for this study, or a family history of COPD.
Biochemical Measurements
Blood samples were collected at baseline from patients and controls after an overnight fast. Plasma separated from cells by centrifugation at 500 g for 10 min at room temperature was used for lipid, glucose and adiponectin analyses. The plasma levels of total cholesterol, triglycerides and glucose were determined with an enzymatic kit (Boehringer Mannheim) and calibrated with a plasma calibrator. Circulating total adiponectin level was measured by the enzyme-linked immunosorbent assay method (Quantikine, R&D Systems, Minneapolis, MN, USA).
SNP Selection and Genotyping
Genotype data of the Chinese population for the ADIPOQ region were obtained from the HapMap website (http://www.hapmap.org/), and tag SNPs were selected using the Tagger software implemented in the Haploview software [25], with an r2 threshold of 0.8 and minor allele frequencies (MAF) of 0.1. There were seven tagging SNPs (rs710445, rs16861205, rs822396, rs7627128, rs1501299, rs3821799 and rs1063537), which captured all the fifteen SNPs from 3-kb region upstream to 1-kb downstream of the gene (position 188,040,157–188,059,946 bp, GenBank accession number NM_004797.3, NCBI build 36).
Genomic DNA was extracted from peripheral blood leukocytes using a commercial extraction kit (Bioteke Corporation, Beijing, China) according to the manufacturer's instructions. SNPs were genotyped using the ABI SNaPshot method (Applied Biosystems, CA, USA). The genomic regions of interest were amplified by primers shown in Table 1. The PCR products were then purified by incubating with shrimp alkaline phosphatase (SAP) and exo-nuclease I (Exo I). The purified PCR products were used as the templates for SNaPshot reaction using the specific SNaPshot primers (Table 1). 3 µl of pooled PCR products, 1 µl of pooled SNaPshot primers and 1 µl of deionized water were incubated in a GeneAmp 9600 thermal cycler by 25 cycles at 96°C for 10 s, 50°C for 5 s, and 60°C for 30 s, and finally 60°C for 30 s. Then, 1 U of SAP was added to SNaPshot product and incubated at 37°C for an hour to deactivate the enzyme. The SNaPshot reaction products were mixed with Hi-Di formamide and GeneScan-120 LIZ internal size standard (Applied Biosystems), and analyzed on an ABI 3130 Genetic Analyzer (Applied Biosystems, CA, USA). The data were analyzed by the software of GeneMapper 4.0. Genotype analysis was performed in a blinded manner so that the staff was unaware of the cases or control status. For quality control, a 10% masked random sample of cases and controls was tested repetitively by different investigators and all the results were completely concordant.
Table 1. Seven SNPs in the ADIPOQ gene in the study.
| SNPs | Position (build 37.3) | Alleles | Primers (5′-3′) | |
| PCR | SNaPshot | |||
| rs710445 | 186,561,518 | G/A | ctgaggcaggagaattgtctg | taatgagataaaatgagaaaagcctggcat |
| acaaccaggatgcctcagaa | ||||
| rs16861205 | 186,561,634 | G/A | cctggcatatagtggcaact | cctgctcgccccagtgagtgctgtttct |
| ggatccatgtcctctaacac | ||||
| rs822396 | 186,566,877 | A/G | tcatggaacccaggctgatc | ttcccttagggtaggagaaagagatctttattttt |
| ggaaacaggaggagagaatc | ||||
| rs7627128 | 186,568,799 | C/A | cagagacattcttggagttg | agatgagttggatgtgccacgtgaagagggtagtatagaa |
| gacaaacctactcctctgtg | ||||
| rs1501299 | 186,571,123 | A/C | cagcaaagccaaagtcttgg | gctttctccctgtgtctaggccttagttaataatgaatg |
| tggtgagaagggtgagaaag | ||||
| rs3821799 | 186,571,486 | T/C | cca gtggcattcaaccacat | tgtgcaaggctctgttggtggttac |
| ccttgaagccttcattcttc | ||||
| rs1063537 | 86,574,075 | C/T | gtctccttgagtaccaacag | ttctctcaggagacaataactccagtgatgtt |
| atcgcttgaacctgggagg | ||||
Statistical Analyses
The demographic and clinical data between the COPD patients and the control subjects were compared using the χ2 test and Student’s t-test. A two-sided significance level of α <0.05 was used for all significant tests. Statistical analyses were performed in SPSS version 17.0 and Microsoft Excel. A multiple logistic regression analysis using BMI and glucose as covariates was done to correct the significant p-value of adiponectin.
The Hardy-Weinberg equilibrium (HWE) test using two-sided χ2 analysis was done for each SNP among cases and controls. Differences in the distribution of genotypes or alleles under different genetic models (including dominant, recessive and additive models) between the COPD patients and the controls were estimated by using the χ2 test. Odds ratios (OR) and 95% CIs were calculated by unconditional logistic regression analyses [26], [27]. Correction for multiple testing was performed by the SNP spectral decomposition method (SNPSpD) [28]. Under this method, the effective number of independent marker loci (MeffLi) was 7, and the experimental-wide significance threshold to keep type 1 error rate at 5% was 0.0098.
Pairwise linkage disequilibrium (LD) estimation and haplotype reconstruction were performed using SHEsis (http://analysis.bio-x.cn) [29]. For haplotype analysis, only haplotypes with a frequency >3% in at least one group were tested. We also used Haploview 4.2 [25] to estimate LD.
Results
General Characteristics of the Subjects
The baseline characteristics, biochemical features and the results of the pulmonary function tests for the 279 patients with COPD and 367 control subjects were presented in Table 2. All patients had FEV1 values <80% of predicted and thus were diagnosed with moderate-to-severe COPD according to the Global Initiative for Chronic Obstructive Lung Disease [24] (classification of severity: mild = FEV1≥80% of predicted; moderate = FEV1≥50% to <80% of predicted; severe = FEV1≥30% to <50% of predicted; and very severe = FEV1<30% of predicted). The COPD cases and control subjects did not significantly differ in sex, age or smoking history. The FEV1, FEV1/predicted and FEV1/FVC were significantly lower in the COPD patients compared with the controls (P<0.01). Compared to control subjects, the COPD patients showed statistically higher glucose concentrations (p<0.01), however, they were still within the normal range. In addition, the COPD patients showed a significantly lower BMI (22.03±2.27 kg/m2 vs. 23.91±2.48 kg/m2, p<0.01) and a 1.4-fold higher adiponectin levels (8.54±0.66 µg/ml vs 6.12±0.57 µg/ml; p<0.01). The adiponectin levels remained significantly higher in COPD patients after further adjusting for BMI and glucose levels (p<0.01).
Table 2. Characteristics of COPD patients and control subjects.
| Variable | Controls (n = 367) | Cases (n = 279) | P |
| Age, years | 65±8 | 63±9 | NS |
| Sex (Men/Women) | 323/44 | 239/40 | NS |
| BMI(kg/m2) | 23.91±2.48 | 22.03±2.27 | <0.01 |
| Total cholesterol(mmol/l) | 4.87±0.22 | 4.96±0.47 | NS |
| Triglycerides(mmol/l) | 1.24±0.57 | 1.19±0.48 | NS |
| Glucose(mmol/l) | 5.13±0.12 | 5.86±0.16 | <0.01 |
| Adiponectin(µg/ml) | 6.12±0.57 | 8.54±0.66 | <0.01 |
| Smoking history | |||
| 0–20 pack years | 88 | 75 | NS |
| ≥20 pack years | 279 | 204 | NS |
| FEV1 | 1.87±0.60 | 0.97±0.32 | <0.01 |
| FEV1 of predicted, % | 93.7±3.4 | 46.0±0.4 | <0.01 |
| FEV1/FVC, % | 78.0±4.6 | 49.2±8.3 | <0.01 |
Data are presented as means ± SD.
NS, no significant difference (P>0.05).
BMI, body mass index.
FEV1, forced expiratory volume in 1 second; FVC, forced vital capacity.
Pack years = (number of cigarettes smoked per day × number of years smoked)/20.
Distribution of the SNPs in ADIPOQ between COPD Patients and Controls
Seven SNPs in ADIPOQ, including rs710445, rs16861205, rs822396, rs7627128, rs1501299, rs3821799 and rs1063537, were screened in all 279 patients with COPD and 367 controls using the SNaPshot method. The genotype and allele frequencies of each SNP in both COPD patients and controls were presented in Table 3. All of the tested SNPs didn’t significantly deviate from that expected for a Hardy-Weinberg equilibrium (HWE) in the COPD patients and controls (Table 3, all P values were higher than 0.05), illustrating that our subjects presented the source population well.
Table 3. Distributions of the ADIPOQ SNPs in COPD patients and controls.
| SNP | Group | Genotype (freq.%) | P | HWEa P | Allele (freq.%) | P | OR [95%CI]b | |||
| GG | AG | AA | G | A | ||||||
| rs710445 | COPD | 75(26.9) | 147(52.7) | 57(20.4) | 0.953 | 0.331 | 297(53.2) | 261(46.8) | 0.910 | 1.01 |
| Control | 102(27.8) | 189(51.5) | 76(20.7) | 0.501 | 393(53.5) | 341(46.5) | [0.81–1.26] | |||
| rs16861205 | GG | AG | AA | G | A | |||||
| COPD | 155(55.6) | 112(40.1) | 12(4.3) | 0.165 | 0.137 | 422(75.6) | 136(24.8) | 0.132 | 1.22 | |
| Control | 230(62.6) | 121(33.0) | 16(4.4) | 0.968 | 581(79.2) | 153(20.8) | [0.94–1.59] | |||
| rs822396 | AA | AG | GG | A | G | |||||
| COPD | 169(60.6) | 101(36.2) | 9 (3.2) | 0.103 | 0.188 | 439(78.7) | 119(21.3) | 0.133 | 1.24 | |
| Control | 249(67.8) | 104(28.3) | 14 (3.8) | 0.451 | 602(82.0) | 132(18.0) | [0.94–1.63] | |||
| rs7627128 | CC | AC | AA | C | A | |||||
| COPD | 119(42.7) | 133(47.7) | 27 (9.7) | 0.998 | 0.244 | 371(66.5) | 187(33.5) | 0.960 | 1.01 | |
| Control | 157(42.8) | 175(47.7) | 35 (9.5) | 0.167 | 489(66.6) | 245(33.4) | [0.80–1.27] | |||
| rs1501299 | CC | AC | AA | C | A | |||||
| COPD | 92(33.0) | 139(49.8) | 48(17.2) | 0.008 | 0.715 | 323(57.9) | 235(42.1) | 0.002 | 1.43 | |
| Control | 158(43.1) | 170(46.3) | 39(10.6) | 0.499 | 486(66.2) | 248(33.8) | [1.14–1.79] | |||
| rs3821799 | TT | TC | CC | T | C | |||||
| COPD | 110(39.4) | 128(45.9) | 41(14.7) | 0.993 | 0.705 | 348(62.4) | 210(37.6) | 0.970 | 0.99 | |
| Control | 145(39.5) | 167(45.5) | 55(15.0) | 0.544 | 457(62.3) | 277(37.7) | [0.79–1.25] | |||
| rs1063537 | CC | CT | TT | C | T | |||||
| COPD | 131(47.0) | 126(45.2) | 22 (7.8) | 0.823 | 0.271 | 388(69.5) | 170(30.5) | 0.648 | 1.06 | |
| Control | 181(49.3) | 157(42.8) | 29 (7.9) | 0.531 | 519(70.7) | 215(29.3) | [0.83–1.35] | |||
a. HWE: Hardy-Weinberg equilibrium. b. OR: odds ratio; CI: confidence interval.
We compared the differences in frequency distributions of genotypes or alleles of every SNP between COPD patients and controls by χ2 test. As shown in Table 3, significant differences in allele or genotype frequencies were observed between COPD patients and controls at rs1501299 (allele: P = 0.002, OR = 1.43 and 95%CI = 1.14–1.79; genotype: P = 0.008).
Association of Genotypes with COPD under Different Genetic Models
For each SNP, if one allele frequency is relatively lower compared to another one, it is recognized as the minor allele (Table 3). We assumed that the minor allele of each SNP was a risk allele compared to the wild type allele. We compared the genotype frequencies of every polymorphism between groups under the dominant, recessive and additive genetic models, respectively. As shown in Table 4, the rs1501299 was observed to be associated with COPD risk by all dominant model analysis (P = 0.009; OR: 1.54; 95%CI: 1.11–2.13), recessive model analyses (P = 0.015; OR: 1.75; 95% CI: 1.11–2.75) and additive model analyses (P = 0.003; OR: 2.11; 95% CI: 1.29–3.47).
Table 4. Association between ADIPOQ SNPs and the risk of COPD under different genetic models.
| SNP | Genetic model | P | OR [95%CI] | |
| rs710445 | Dominant | (AG+AA) vs GG | 0.797 | 1.05 [0.74–1.49] |
| Recessive | AA vs.(GG+AG) | 0.931 | 0.98 [0.67–1.45] | |
| Additive | AG vs. GG | 0.765 | 1.06 [0.73–1.53] | |
| AA vs. GG | 0.932 | 1.02 [0.65–1.61] | ||
| rs16861205 | Dominant | (AG+AA) vs GG | 0.068 | 1.34 [0.98–1.84] |
| Recessive | AA vs.(GG+AG) | 0.971 | 0.99 [0.46–2.12] | |
| Additive | AG vs. GG | 0.058 | 1.37 [0.99–1.91] | |
| AA vs. GG | 0.787 | 1.11 [0.51–2.42] | ||
| rs822396 | Dominant | (AG+GG) vs. AA | 0.055 | 1.37 [0.99–1.90] |
| Recessive | GG vs. (AG+AA) | 0.689 | 0.84 [0.36–1.97] | |
| Additive | AG vs. AA | 0.036 | 1.43 [1.02–2.00] | |
| GG vs. AA | 0.902 | 0.95 [0.40–2.24] | ||
| rs7627128 | Dominant | (AC+AA) vs. CC | 0.974 | 1.01 [0.73–1.38] |
| Recessive | AA vs. (CC+AC) | 0.952 | 1.02 [0.60–1.72] | |
| Additive | AC vs. CC | 0.987 | 1.00 [0.72–1.39] | |
| AA vs. CC | 0.95 | 1.02 [0.58–1.77] | ||
| rs1501299 | Dominant | (AC+AA) vs. CC | 0.009 | 1.54 [1.11–2.13] |
| Recessive | AA vs. (CC+AC) | 0.015 | 1.75 [1.11–2.75] | |
| Additive | AC vs. CC | 0.051 | 1.40 [0.99–1.98] | |
| AA vs. CC | 0.003 | 2.11 [1.29–3.47] | ||
| rs3821799 | Dominant | (TC+CC) vs TT | 0.983 | 1.00 [0.73–1.38] |
| Recessive | CC vs. (TT+TC) | 0.918 | 0.98 [0.63–1.52] | |
| Additive | TC vs. TT | 0.952 | 1.01 [0.72–1.42] | |
| CC vs. TT | 0.942 | 0.98 [0.61–1.58] | ||
| rs1063537 | Dominant | (CT+TT) vs. CC | 0.551 | 1.01 [0.81–1.50] |
| Recessive | TT vs. (CC+CT) | 0.994 | 1.00 [0.56–1.78] | |
| Additive | CT vs. CC | 0.533 | 1.11 [0.80–1.53] | |
| TT vs. CC | 0.877 | 1.05 [0.58–1.91] | ||
OR: odds ratio; CI: confidence interval.
rs1501299 is Associated with Plasma Adiponectin Levels
We investigated the relationship between rs1501299 and plasma adiponectin levels in both COPD patients and control subjects (Figure 1). Compared to subjects with homozygote CC at rs1501299, the subjects with homozygote AA had significantly higher adiponectin levels after adjusting for BMI and glucose levels (6.82±0.52 µg/ml vs. 5.85±0.57 µg/ml in controls, 9.13±0.49 µg/ml vs. 8.27±0.53 µg/ml in COPD patients, P <0.05 in both groups).
Figure 1. The allele effect of rs1501299 on plasma adiponectin levels.
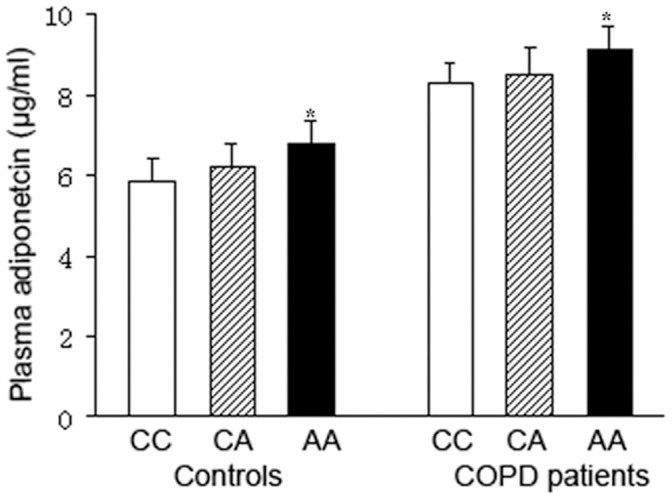
Values were expressed as means ± SD. P <0.05 vs. subjects with CC genotype.
Linkage Disequilibrium (LD) between SNPs and Haplotype Analysis
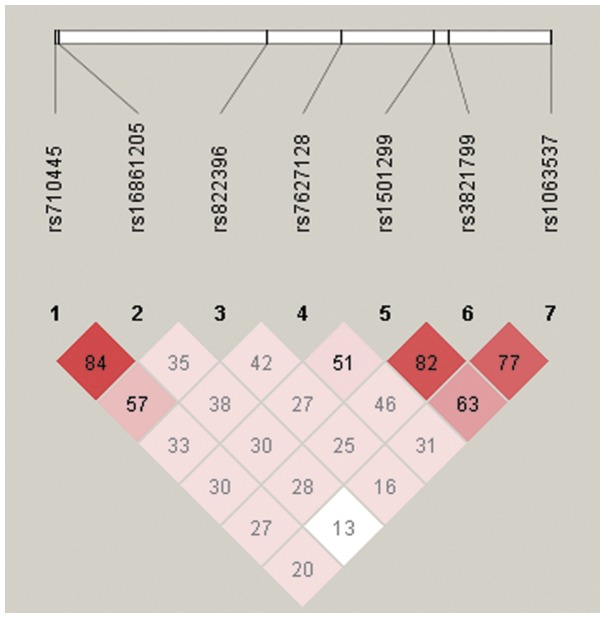
The extent of linkage disequilibrium in pairwise combinations of alleles in different SNP was estimated by means of maximum likelihood from the genotype frequency in the COPD and control groups. Pairwise LD between the seven SNPs was shown in Table 5 and Figure 2. Based on LD determinations, two blocks with moderate LD were detected: block 1 is composed of rs710445 and rs16861205, block 2 is composed of rs1501299, rs3821799 and rs1065537.
Table 5. Pairwise linkage disequilibrium analysis of SNPs of the ADIPOQ gene.
| D' | rs16861205 | rs822396 | rs7627128 | rs1501299 | rs3821799 | rs1063537 |
| rs710445 | 0.846 | 0.573 | 0.337 | 0.308 | 0.277 | 0.203 |
| rs16861205 | – | 0.358 | 0.380 | 0.302 | 0.284 | 0.130 |
| rs822396 | – | – | 0.424 | 0.273 | 0.250 | 0.160 |
| rs7627128 | – | – | – | 0.511 | 0.467 | 0.318 |
| rs1501299 | – | – | – | – | 0.820 | 0.639 |
| rs3821799 | – | – | – | – | – | 0.775 |
D': linkage disequilibrium coefficient.
Figure 2. Linkage disequilibrium (LD) plots for ADIPOQ.
The LD plots were generated by Haploview 4.2. Polymorphisms are identified by their dbSNP rs numbers, and their relative positions are marked by vertical lines within the white horizontal bar. The numbers within squares indicate the D’ value, expressed as a percentile.
We estimated the frequencies of haplotypes constructed from phased multi-locus genotypes in ADIPOQ. The haplotypes with a frequency higher than 3% in at least one group were involved in the haplotype analysis (Table 6). The global result for block 1 (rs710445 and rs16861205) was: χ2 = 22.99 while df = 3, P = 4.16×10−5. The global result for block 2 (rs1501299, rs3821799 and rs1065537) was: χ2 = 79.33 while df = 5, P = 7.66×10−15. The overall frequency distribution of haplotype composed of all seven SNPs was significantly different between cases and controls (total global χ2 = 159.35 while df = 10, P = 4.40×10−29).
Table 6. Frequencies of pairwise haplotype constructed by SNPs in ADIPOQ.
| Block | Haplotypea,b | freq (case) | freq(control) | χ2 | Fisher's P | OR [95%CI]c,d |
| 1 | AA | 0.204 | 0.213 | 0.16 | 0.686 | 0.95 [0.72–1.24] |
| AG | 0.264 | 0.252 | 0.26 | 0.613 | 1.07 [0.83–1.37] | |
| GA | 0.040 | 0.004 | 22.14 | 2.61×10−6 | 11.54 [3.20–41.57] | |
| GG | 0.492 | 0.532 | 1.98 | 0.160 | 0.85 [0.69–1.06] | |
| Global | 22.99 | 4.16×10−5 | ||||
| 2 | ACC | 0.068 | 0.128 | 12.40 | 4.33×10−4 | 0.50 [0.34–0.74] |
| ACT | 0.279 | 0.193 | 13.19 | 2.85×10−4 | 1.62 [1.25–2.10] | |
| ATC | 0.064 | 0.017 | 18.87 | 1.43×10−5 | 3.85 [2.01–7.37] | |
| CCT | 0.014 | 0.032 | 4.48 | 0.054 | 0.42 [0.19–1.06] | |
| CTC | 0.552 | 0.537 | 0.21 | 0.645 | 1.05 [0.84–1.32] | |
| CTT | 0.002 | 0.068 | 36.16 | 1.97×10−9 | 0.02 [0.01–0.12] | |
| Global | 79.33 | 7.66×10−15 | ||||
| Total | AAAAACT | 0.014 | 0.032 | 6.47 | 0.011 | 0.36 [0.16–0.82] |
| AAGAACT | 0.040 | 0.038 | 0.12 | 0.725 | 0.90 [0.51–1.60] | |
| AGAAACT | 0.025 | 0.037 | 2.91 | 0.088 | 0.56 [0.29–1.10] | |
| AGAACTC | 0.037 | 0.012 | 6.76 | 0.009 | 2.80 [1.25–6.27] | |
| AGACCTC | 0.078 | 0.064 | 0.06 | 0.802 | 1.06 [0.68–1.64] | |
| AGGCCTC | 0.051 | 0.005 | 22.74 | 1.91×10−6 | 8.73 [3.02–25.20] | |
| GGAAACT | 0.048 | 0.023 | 3.76 | 0.053 | 1.84 [0.99–3.43] | |
| GGAACTC | 0.050 | 0.010 | 15.98 | 6.48×10−5 | 4.76 [2.06–11.00] | |
| GGACACT | 0.092 | 0.008 | 46.57 | 9.89×10−12 | 12.03 [4.96–29.21] | |
| GGACCTC | 0.212 | 0.359 | 72.32 | 4.00×10−15 | 0.29 [0.22–0.39] | |
| GGGCCTC | 0.034 | 0.000 | 21.95 | 2.87×10−6 | – | |
| Global | 159.35 | 4.40×10−29 |
a. The order of SNPs from left to right is: rs710445 and rs16861205 for block 1; rs1501299, rs3821799 and rs1063537 for block 2; rs710445, rs16861205, rs822396, rs7627128, rs1501299, rs3821799 and rs1063537 for total.
b. Only haplotypes with a frequency >3% in at least one group were listed.
c. OR: odds ratio; CI: confidence interval.
d. The OR could not be calculated for the haplotype GGGCCTC, because of the zero value in the population.
The results of the association between the ADIPOQ haplotype and the risk of COPD were listed in Table 6. Haplotype GA in block 1 was found to be associated with an increased risk of COPD (OR = 11.54, 95%CI = 3.20–41.57, P = 2.61×10−6). In block 2, two haplotypes were observed to be associated with the risk of COPD (ACT: OR = 1.62, 95%CI = 1.20–2.10, P = 2.85×10−4; ATC: OR = 3.85, 95%CI = 2.01–7.37, P = 1.43×10−5), while haplotypes ACC and CTT were protective from COPD (ACC: OR = 0.50, 95%CI = 0.34–0.74, P = 4.33×10−4; CTT: OR = 0.02, 95%CI = 0.01–0.12, P = 1.97×10−9). Global haplotype association analyses showed that five haplotypes, including AGAACTC, AGGCCTC, GGAACTC, GGACACT and GGGCCTC, were significantly associated with the risk of COPD (all OR>1.00, and P<0.01). In addition, two protective haplotypes AAAAACT (OR = 0.36, 95%CI = 0.16–0.82, P = 0.011) and GGACCTC (OR = 0.29, 95%CI = 0.22–0.39, P = 4.00×10−15), which were associated with a decreased risk of COPD.
Discussion
Adiponectin, an adipose tissue-derived cytokine, has important roles in insulin sensitization, cardioprotection, and anti-inammatory processes. Plasma adiponectin level is negatively correlated with body mass index (BMI), glucose, insulin and triglyceride levels and is positively associated with high-density lipoprotein cholesterol (HDL-C) concentration and insulin-stimulated glucose disposal [30]. Although its action on the respiratory system is not fully known, expression of adiponectin and its functional receptors on airway epithelium have been reported [31]. There have been several clinical studies reporting on the relationship between circulating adiponectin and COPD, and elevation of plasma adiponectin level was found in patients with stable and acute exacerbation of COPD [15]–[17], [32]. The most bioactive form of adiponectin is a 400-kDa high molecular weight (HMW) complex [33], which was also found to be increased dramatically in COPD patients in a recent report [34].
In this case-control study in a Han Chinese population, we evaluated the possible association of ADIPOQ polymorphisms with susceptibility to COPD. To the best of our knowledge, this study was the first investigation of the association between the SNPs in ADIPOQ and COPD risk. Our current findings suggested that rs1501299 associated with the risk of COPD. In comparison with allele G at rs1501299, the allele A could increase the risk of COPD under all dominant, recessive and additive genetic models. The polymorphism rs1501299, also known as C276A, in ADIPOQ has been found to be in association with adiponectin levels in diverse population [35]–[37], and its A allele is associated with higher adiponectin. One report showed that serum adiponectin levels were higher in COPD patients than in control subjects and were associated with weight loss and systemic inflammation as assessed by circulating tumor necrosis factor-α (TNF-α) levels [15]. A recently study showed that serum adiponectin was associated with all-cause mortality in COPD patients [38]. However, this association is conflicted in animal models. In one study, hypoadiponectinemia in ADIPOQ gene-knockout mice was associated with emphysema-like changes in the lungs and extrapulmonary features including systemic inflammation, obesity, and osteoporosis [14], while another study reported the opposite effect in which mice deficient in adiponectin were protected from emphysema when they were exposed to cigarette smoke [39]. In our current study, the association of rs1501299 in ADIPOQ and COPD might be modulated by its effects on the adiponection levels. However, rs1501299 is in intron 2 of the ADIPOQ gene and does not have a known function. It is probably a marker of some other variant affecting adiponectin expression. We assessed the extent of linkage disequilibrium (LD) between rs1501299 and other SNPs in exons, 3'UTR and promoter regions of the ADIPOQ gene in Han Chinese in Beijing (CHB) using HapMap project data (Figure S1), which indicated that rs1501299 was in a LD block encompassing the whole 3'UTR of the ADIPOQ gene, but whether and how far such block extends beyond the gene boundaries remain to be determined [40].
In addition to the genotype analysis, our study also adopted a haplotype based approach. Haplotype analysis, in which several SNPs within the same gene are evaluated simultaneously, can provide more information than a single SNP and thus elevates the statistical power of the analysis [41]. Using this approach, we provided strong support that ADIPOQ variations contributed to the susceptibility to COPD. LD analysis showed that some SNPs in ADIPOQ gene were in moderate LD, and some haplotypes with low frequency were found to affect the risk of COPD dramatically. Haplotypes AAAAACT and GGACCTC were associated with a reductive risk of COPD, while haplotypes AGAACTC, AGGCCTC, GGAACTC, GGACACT and GGGCCTC increased the risk of developing COPD, indicating the complexity of ADIPOQ gene in the development of COPD. This might be attributable to the complex genetic determinants of plasma adiponectin levels. In addition to the SNP in ADIPOQ gene, other gene loci such as 14q13 have been reported to affect plasma adiponectin value and play a much bigger role [40]. Recently, Genome wide association study (GWAS) for genetic markers in determining plasma adiponectin value in Korean population reported that genetic variants in CDH13 on chromosome 16, but not genetic variants in the ADIPOQ gene, influence adiponectin levels [42]. However, another GWAS using plasma adiponectin as a quantitative trait demonstrated the ADIPOQ gene as the only major gene for plasma adiponectin in Caucasian population [20].
We are aware that the significant results in this study could prove to be false positives because of the relatively small sample size. 279 COPD patients and 367 control subjects were not relatively large among COPD association studies published to date, further studies using larger populations are needed. But even with a larger sample, the functional and biological impacts of the described polymorphisms would require further study. Possible gene-gene and gene-environment interactions also pose a challenge for genetic analysis of COPD association studies. We selected SNPs with MAF higher than 10% in the Han Chinese population (CHB) using HapMap project data, this is not suited for situations where genetic architecture is such that multiple rare disease-causing variants contribute significantly to disease risk. Recent studies demonstrate that identification of rare variants may lead to critically important insights about disease etiology through implication of new genes and/or pathways [43], [44]. The rare variants in the ADIPOQ gene should be investigated to clarify their susceptibility to the development of COPD.
In conclusion, our comprehensive analysis of SNPs in the ADIPOQ gene suggests that ADIPOQ genotypes and haplotypes are associated with COPD risk. These findings indicate that the genetic variants of ADIPOQ gene play a complex role in the development of COPD, and that interactions of loci in ADIPOQ gene may be more important than a single locus. Our findings in this study provided new evidence for the association between SNPs and haplotypes of ADIPOQ gene and the risk of COPD.
Supporting Information
Relative position of SNPs and LD map for ADIPOQ in the Han Chinese population (CHB) using HapMap project data. This figure shows that strong LD was observed between rs1501299 and the SNPs in the 3′UTR of ADIPOQ.
(TIF)
Acknowledgments
We are grateful to all the participants in this study.
Funding Statement
This work was financially supported by the Natural Science Foundation of China (no. 31071108,30973161), the Program for New Century Excellent Talents in University (NCET-10-0600), the Scientific Research Foundation for Returning Overseas Chinese Scholars, and the Chinese Ministry of Education (2011-508-4-3). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Murray CJ, Lopez AD (1996) Evidence-based health policy–lessons from the Global Burden of Disease Study. Science 274: 740–743. [DOI] [PubMed] [Google Scholar]
- 2. Zhong N, Wang C, Yao W, Chen P, Kang J, et al. (2007) Prevalence of chronic obstructive pulmonary disease in China: a large, population-based survey. Am J Respir Crit Care Med 176: 753–760. [DOI] [PubMed] [Google Scholar]
- 3. Davis RM, Novotny TE (1989) The epidemiology of cigarette smoking and its impact on chronic obstructive pulmonary disease. Am Rev Respir Dis 140: S82–84. [DOI] [PubMed] [Google Scholar]
- 4. Khoury MJ, Beaty TH, Tockman MS, Self SG, Cohen BH (1985) Familial aggregation in chronic obstructive pulmonary disease: use of the loglinear model to analyze intermediate environmental and genetic risk factors. Genet Epidemiol 2: 155–166. [DOI] [PubMed] [Google Scholar]
- 5. Castaldi PJ, Cho MH, Cohn M, Langerman F, Moran S, et al. (2010) The COPD genetic association compendium: a comprehensive online database of COPD genetic associations. Hum Mol Genet 19: 526–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cho MH, Boutaoui N, Klanderman BJ, Sylvia JS, Ziniti JP, et al. (2010) Variants in FAM13A are associated with chronic obstructive pulmonary disease. Nat Genet 43: 200–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pillai SG, Ge D, Zhu G, Kong X, Shianna KV, et al. (2009) A genome-wide association study in chronic obstructive pulmonary disease (COPD): identification of two major susceptibility loci. PLoS Genet 5: e1000421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wilk JB, Chen TH, Gottlieb DJ, Walter RE, Nagle MW, et al. (2009) A genome-wide association study of pulmonary function measures in the Framingham Heart Study. PLoS Genet 5: e1000429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kolsum U, Roy K, Starkey C, Borrill Z, Truman N, et al. (2009) The repeatability of interleukin-6, tumor necrosis factor-alpha, and C-reactive protein in COPD patients over one year. Int J Chron Obstruct Pulmon Dis 4: 149–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yamamoto C, Yoneda T, Yoshikawa M, Fu A, Tokuyama T, et al. (1997) Airway inflammation in COPD assessed by sputum levels of interleukin-8. Chest 112: 505–510. [DOI] [PubMed] [Google Scholar]
- 11. Kadowaki T, Yamauchi T, Kubota N, Hara K, Ueki K, et al. (2006) Adiponectin and adiponectin receptors in insulin resistance, diabetes, and the metabolic syndrome. J Clin Invest 116: 1784–1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Otero M, Lago R, Gomez R, Lago F, Dieguez C, et al. (2006) Changes in plasma levels of fat-derived hormones adiponectin, leptin, resistin and visfatin in patients with rheumatoid arthritis. Ann Rheum Dis 65: 1198–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rovin BH, Song H, Hebert LA, Nadasdy T, Nadasdy G, et al. (2005) Plasma, urine, and renal expression of adiponectin in human systemic lupus erythematosus. Kidney Int 68: 1825–1833. [DOI] [PubMed] [Google Scholar]
- 14. Nakanishi K, Takeda Y, Tetsumoto S, Iwasaki T, Tsujino K, et al. (2011) Involvement of endothelial apoptosis underlying chronic obstructive pulmonary disease-like phenotype in adiponectin-null mice: implications for therapy. Am J Respir Crit Care Med 183: 1164–1175. [DOI] [PubMed] [Google Scholar]
- 15. Tomoda K, Yoshikawa M, Itoh T, Tamaki S, Fukuoka A, et al. (2007) Elevated circulating plasma adiponectin in underweight patients with COPD. Chest 32: 135–140. [DOI] [PubMed] [Google Scholar]
- 16. Kirdar S, Serter M, Ceylan E, Sener AG, Kavak T, et al. (2009) Adiponectin as a biomarker of systemic inflammatory response in smoker patients with stable and exacerbation phases of chronic obstructive pulmonary disease. Scand J Clin Lab Invest 69: 219–224. [DOI] [PubMed] [Google Scholar]
- 17. Chan KH, Yeung SC, Yao TJ, Ip MS, Cheung AH, et al. (2010) Elevated plasma adiponectin levels in patients with chronic obstructive pulmonary disease. Int J Tuberc Lung Dis 14: 1193–1200. [PubMed] [Google Scholar]
- 18. Guo X, Saad MF, Langefeld CD, Williams AH, Cui J, et al. (2006) Genome-wide linkage of plasma adiponectin reveals a major locus on chromosome 3q distinct from the adiponectin structural gene: the IRAS family study. Diabetes 55: 1723–1730. [DOI] [PubMed] [Google Scholar]
- 19. Heid IM, Wagner SA, Gohlke H, Iglseder B, Mueller JC, et al. (2006) Genetic architecture of the APM1 gene and its influence on adiponectin plasma levels and parameters of the metabolic syndrome in 1,727 healthy Caucasians. Diabetes 55: 375–384. [DOI] [PubMed] [Google Scholar]
- 20. Heid IM, Henneman P, Hicks A, Coassin S, Winkler T, et al. (2010) Clear detection of ADIPOQ locus as the major gene for plasma adiponectin: results of genome-wide association analyses including 4659 European individuals. Atherosclerosis 208: 412–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hivert MF, Manning AK, McAteer JB, Florez JC, Dupuis J, et al. (2008) Common variants in the adiponectin gene (ADIPOQ) associated with plasma adiponectin levels, type 2 diabetes, and diabetes-related quantitative traits: the Framingham Offspring Study. Diabetes 57: 3353–3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chuang LM, Chiu YF, Sheu WH, Hung YJ, Ho LT, et al. (2004) Biethnic comparisons of autosomal genomic scan for loci linked to plasma adiponectin in populations of Chinese and Japanese origin. J Clin Endocrinol Metab 89: 5772–5778. [DOI] [PubMed] [Google Scholar]
- 23. Xu SC, Kuang JY, Liu J, Ma CL, Feng YL, et al. (2012) Association between fibroblast growth factor 7 and the risk of chronic obstructive pulmonary disease. Acta Pharmacol Sin 33: 998–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.GOLD Executive Committee (2011) Global Strategy for the Diagnosis, Management and Prevention of chronic obstructive pulmonary disease, Global Initiative for Chronic Obstructive Lung Disease (GOLD). Available: http://www.goldcopd.org. Accessed 2012 Oct 31.
- 25. Barrett JC, Fry B, Maller J, Daly MJ (2005) Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21: 263–265. [DOI] [PubMed] [Google Scholar]
- 26. Bland JM, Altman DG (2000) Statistics notes. The odds ratio. BMJ 320: 1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Altman DG, Bland JM (2011) How to obtain the confidence interval from a P value. BMJ 343: d2090. [DOI] [PubMed] [Google Scholar]
- 28. Nyholt DR (2004) A simple correction for multiple testing for single-nucleotide polymorphisms in linkage disequilibrium with each other. Am J Hum Genet 74: 765–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shi YY, He L (2005) SHEsis, a powerful software platform for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci. Cell Res 15: 97–98. [DOI] [PubMed] [Google Scholar]
- 30. Diez JJ, Iglesias P (2010) The role of the novel adipocyte-derived protein adiponectin in human disease: an update. Mini Rev Med Chem 10: 856–869. [DOI] [PubMed] [Google Scholar]
- 31. Miller M, Cho JY, Pham A, Ramsdell J, Broide DH (2009) Adiponectin and functional adiponectin receptor 1 are expressed by airway epithelial cells in chronic obstructive pulmonary disease. J Immunol 182: 684–691. [DOI] [PubMed] [Google Scholar]
- 32. Krommidas G, Kostikas K, Papatheodorou G, Koutsokera A, Gourgoulianis KI, et al. (2010) Plasma leptin and adiponectin in COPD exacerbations: associations with inflammatory biomarkers. Respir Med 104: 40–46. [DOI] [PubMed] [Google Scholar]
- 33. Brochu-Gaudreau K, Rehfeldt C, Blouin R, Bordignon V, Murphy BD, et al. (2010) Adiponectin action from head to toe. Endocrine 37: 11–32. [DOI] [PubMed] [Google Scholar]
- 34. Daniele A, De Rosa A, Nigro E, Scudiero O, Capasso M, et al. (2012) Adiponectin oligomerization state and adiponectin receptors airway expression in chronic obstructive pulmonary disease. Int J Biochem Cell Biol 44: 563–569. [DOI] [PubMed] [Google Scholar]
- 35. Woo JG, Dolan LM, Deka R, Kaushal RD, Shen Y, et al. (2006) Interactions between noncontiguous haplotypes in the adiponectin gene ACDC are associated with plasma adiponectin. Diabetes 55: 523–529. [DOI] [PubMed] [Google Scholar]
- 36. Vasseur F, Helbecque N, Dina C, Lobbens S, Delannoy V, et al. (2002) Single-nucleotide polymorphism haplotypes in the both proximal promoter and exon 3 of the APM1 gene modulate adipocyte-secreted adiponectin hormone levels and contribute to the genetic risk for type 2 diabetes in French Caucasians. Hum Mol Genet 11: 2607–2614. [DOI] [PubMed] [Google Scholar]
- 37. Qi L, Li T, Rimm E, Zhang C, Rifai N, et al. (2005) The +276 polymorphism of the APM1 gene, plasma adiponectin concentration, and cardiovascular risk in diabetic men. Diabetes 54: 1607–1610. [DOI] [PubMed] [Google Scholar]
- 38. Yoon H, Li Y, Man SF, Tashkin D, Wise RA, et al. (2012) The complex relationship of serum adiponectin to COPD outcomes. Chest 142: 1311–2173. [DOI] [PubMed] [Google Scholar]
- 39. Miller M, Pham A, Cho JY, Rosenthal P, Broide DH (2010) Adiponectin-deficient mice are protected against tobacco-induced inflammation and increased emphysema. Am J Physiol Lung Cell Mol Physiol 299: L834–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Menzaghi C, Ercolino T, Salvemini L, Coco A, Kim SH, et al. (2004) Multigenic control of serum adiponectin levels: evidence for a role of the APM1 gene and a locus on 14q13. Physiol Genomics 19: 170–174. [DOI] [PubMed] [Google Scholar]
- 41. Morris RW, Kaplan NL (2002) On the advantage of haplotype analysis in the presence of multiple disease susceptibility alleles. Genet Epidemiol 23: 221–233. [DOI] [PubMed] [Google Scholar]
- 42. Jee SH, Sull JW, Lee JE, Shin C, Park J, et al. (2010) Adiponectin concentrations: a genome-wide association study. Am J Hum Genet 87: 545–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cole JW, Stine OC, Liu X, Pratap A, Cheng Y, et al. (2012) Rare variants in ischemic stroke: an exome pilot study. PLoS One 7: e35591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Nelson MR, Wegmann D, Ehm MG, Kessner D, St Jean P, et al. (2012) An Abundance of Rare Functional Variants in 202 Drug Target Genes Sequenced in 14,002 People. Science 337: 100–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Relative position of SNPs and LD map for ADIPOQ in the Han Chinese population (CHB) using HapMap project data. This figure shows that strong LD was observed between rs1501299 and the SNPs in the 3′UTR of ADIPOQ.
(TIF)