Abstract
Since Panayiotis Gennadius first identified the whitefly, Aleyrodes tabaci in 1889, there have been numerous revisions of the taxonomy of what has since become one of the world's most damaging insect pests. Most of the taxonomic revisions have been based on synonymising different species under the name Bemisia tabaci. It is now considered that there is sufficient biological, behavioural and molecular genetic data to support its being a cryptic species complex composed of at least 34 morphologically indistinguishable species. The first step in revising the taxonomy of this complex involves matching the A. tabaci collected in 1889 to one of the members of the species complex using molecular genetic data. To do this we extracted and then amplified a 496 bp fragment from the 3′ end of the mitochondrial DNA cytochrome oxidase one (mtCOI) gene belonging to a single whitefly taken from Gennadius' original 1889 collection. The sequence identity of this 123 year-old specimen enabled unambiguous assignment to a single haplotype known from 13 Mediterranean locations across Greece and Tunisia. This enabled us to unambiguously assign the Gennadius A. tabaci to the member of the B. tabaci cryptic species complex known as Mediterranean or as it is commonly, but erroneously referred to, as the ‘Q-biotype’. Mediterranean is therefore the real B. tabaci. This study demonstrates the importance of matching museum syntypes with known species to assist in the delimitation of cryptic species based on the organism's biology and molecular genetic data. This study is the first step towards the reclassification of B. tabaci which is central to an improved understanding how best to manage this globally important agricultural and horticultural insect pest complex.
Introduction
In 1889, Panayiotis Gennadius, an Inspector of Agriculture, was sent to Agrinio, Greece to identify the small fly-like pest that was devastating tobacco crops there. After consulting with Filippo Silvestri and Adolfo Targioni Tozzetti, two of the leading experts on whiteflies at that time, it was determined that the pest was an undescribed species of whitefly and subsequently named, Aleyrodes tabaci Gennadius, later transferred to the genus Bemisia [1], [2]. For 104 years, the whitefly was considered to be a single, morphologically invariable cosmopolitan species, having an enormous host range and the ability to transmit a wide variety of plant viruses [3], [4]. From the late 1880s to the early 1980s, outbreaks were sporadic and relatively small, but this changed in the mid-1980s with widespread outbreaks occurring across the south western USA [5]. This was odd because this pest was well known across the region as a minor pest yet here it was destroying crops. Researchers concluded that while there were no morphological differences, there were sufficient molecular and biological differences to indicate that the outbreak pest was a different species [6]. This touched off a debate that has continued until today [7]–[10].
Species names are a tool for communicating our knowledge of species diversity and should make communication easier and clearer [11]. Nomina dubia, names of doubtful or unknown application as defined by the International Code of Zoological Nomenclature (ICZN, 1999), are problematic and hinder the communication of knowledge. These names arise when species are described based on a small number of specimens that are either entirely destroyed, uninformative or lost, and have original descriptions that are largely incomplete and do not provide characters needed for identification [12]. To avoid nomina dubia, the integrative taxonomy approach utilizes multiple lines of evidence to assist species delimitation and has been promoted as a solution [13]. The integrative taxonomic approach undertaken by the B. tabaci community has provided several lines of evidence indicating that a complete revision of the species complex is necessary. This evidence is summarized in [4] and draws upon a range of molecular, biological and behavioral data, all of which supports the conclusion that B. tabaci is a cryptic species complex. However, before initiating a taxonomic revision, extreme care needs to be taken so as to avoid further inaccurate species identifications [14], [15] and in the case of B. tabaci, this involves reconsidering the identity of the various species that have been synonymised under the name B. tabaci [16].
The debate over B. tabaci taxonomy began in January 1993 when Science [6] published a study that provided evidence suggesting that the whitefly causing considerable damage to food and fiber crops in the south western USA, while morphologically indistinguishable from B. tabaci (Gennadius), displayed considerable molecular and biological differences which together provided evidence that it was a distinct species and it was subsequently named B. argentifolii [6], [17]. In an attempt to draw a conclusion as to whether or not this was valid, Barinaga [7] posed the following question, “Is devastating whitefly invader really a new species?”, but by the article's end the search for a consensus remained elusive. A 1995 review of the subject again came to no conclusion [3].
Whether B. tabaci is a complex species or a species complex may seem a somewhat esoteric argument, but this is a major pest of some of the most important food and fiber crops. Ten percent of all known plant pathogenic viruses are transmitted by B. tabaci [18], [19]. The key staple crops cassava and sweetpotato are severely damaged by the viruses transmitted by this vector compounding the problem facing the achievement of food self-sufficiency across Africa, Asia and Central and South America [20]. Major cash crops in developing countries such as cotton, chilli and tomato are also devastated adding considerably to the financial burden borne by poor smallholder farmers in Africa, Asia and Central and South Americas as they borrow money to plant crops that are then subsequently wiped out [21], [22]. The answer as to whether we are dealing with a single vector species or complex of related, but different species will have a significant bearing on the applicability and transferability of management practices between regions where the pest occurs. These practices usually depend on insect biology, behavior, natural enemy interactions and responses to agricultural chemicals; what works for one may be ineffective for another [23].
The taxonomic identity of B. tabaci has a complicated history which has contributed to the confusion surrounding its identity. Whitefly taxonomy relies primarily on morphological characters of the fourth instar [24]. However, the morphology of the fourth instar in the case of B. tabaci was shown to be extremely plastic [24]. This meant that many of the morphological features used for identification of B. tabaci and 19 other Bemisia species that have been synonymized with it, have turned out to be highly plastic and a response to the host plant. This has no doubt contributed to the serial redescriptions and may well have been exacerbated by geographic isolation between the different taxonomists of the time, resulting in a lack of information exchange and the consequential assignment of multiple names to a globally distributed insect. As a result, between 1900 and 1978, 19 species of whiteflies were synonymised with B. tabaci [2], [16], [23], [25].
Since 1993, the body of knowledge surrounding this pest has grown to a point that in 2010 there was sufficient molecular data based on mitochondrial cytochrome oxidase I (mtCOI) to enable a compelling argument to be raised in support of the species complex argument [26]. This was later supported by a further study that applied a wider range of species delimitation metrics [11]. Using these metrics, it was proposed that rather than one species, B. tabaci was composed of at least 28 different morphologically indistinguishable species, all separated by at least 3.5% divergence in mtCOI [4]. To date 34 species have been delimited using these metrics [4], [27], [28]. In addition, delimitation is supported not only by available molecular evidence, but also all available mating compatibility studies. These have shown that crosses between individuals identified as different species using the delimitation metrics are reproductively isolated to the point that in most cases copulation does not occur and where it does, the resulting progeny are either sterile or reproductively inferior to their parents [29]–[45]. Given the debate is settling in favor of a species complex, thought now needs to be given to how best to revise the taxonomy of the complex. A key part of this revision will be to link named species that were synonymised under B. tabaci to the species identified using the delimitation metrics [14], [15]. The first step is to start at the beginning and determine the identity of the whitefly collected from tobacco in Agrinio, Greece, in 1889 [1]. This study shows how we succeeded in extracting DNA from an intact whitefly that was part of the 1889 collection and then amplifying the mtCOI and unambiguously matching it to a known member of the B. tabaci complex, and thus determining its true identity.
Results
We succeeded in amplifying five mtCOI fragments that assembled as a 469 bp of continuous sequence (GenBank JX268596) after primer site removal (Fig. 1). When aligned against the 34 consensus sequences representing the delimited species within the complex, the sequence was a 100% match with the consensus sequence for the Mediterranean species (Species names e.g. Mediterranean, Middle East-Asia Minor 1 are currently being used until a formal process of renaming occurs and are those referred to in [26]). This species is commonly referred to as the Q biotype, although the biotype appellation is now largely discredited. The consensus sequence for Mediterranean was assembled from 87 different haplotypes all of which cluster within Mediterranean. Apart from Mediterranean, three other species in the complex occur in the Mediterranean region, Middle East-Asia Minor 1, Italy, and Sub-Saharan Africa 2 and differ by 8.0%, 15.7% and 22.0%, respectively [26]. Mediterranean belongs to a cluster containing two close relatives in addition to Middle East-Asia Minor 1; Middle East-Asia Minor 2 and Indian Ocean which differ by 5.7% and 9.8%, respectively (Fig. 2) [26]. The magnitude of these differences and the tight spread of within species variation (<3.5% divergence) enable unambiguously assignment of the Gennadius sequence to Mediterranean.
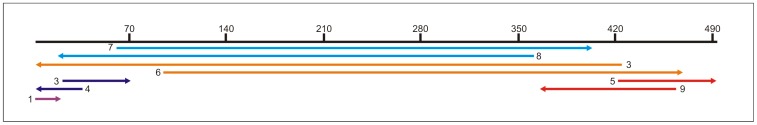
Figure 1. Schematic diagram detailing sequencing of the Gennadius 1889 Bemisia tabaci mtCOI amplicons using primers listed in Table 1.
The final 496 bp contig was assembled from five amplicons (primer pairs 1+2; 3+4; 5+9; 3+6; 7+8).
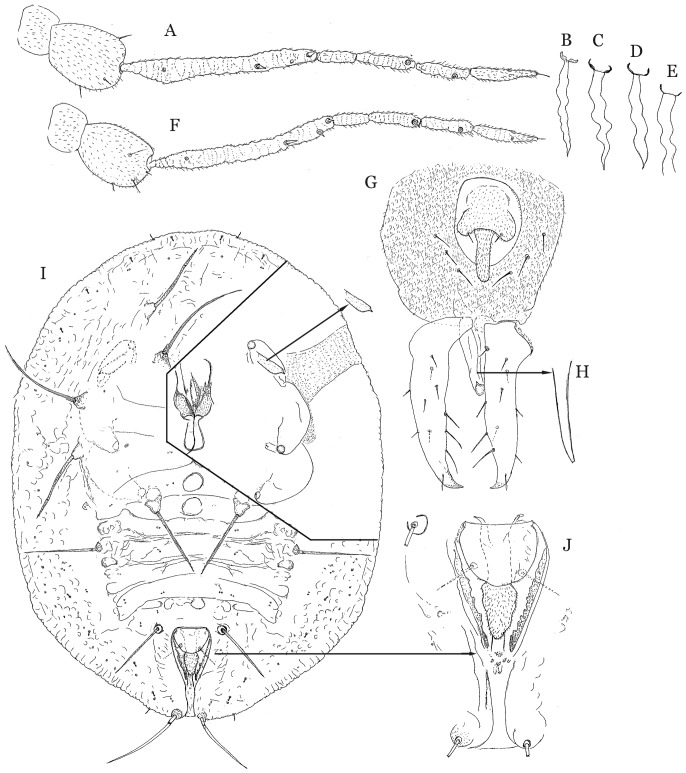
Figure 2. Illustrations of a puparium (I, J), variations in the cement gland of three adult females (C–E) and lateral view of aedeagus of an adult male (H) from Bemisia tabaci specimens collected by Gennadius from tobacco in Agrinio, Greece in 1889.
In addition, illustrations of the antenna (A) and cement gland (B) of a female and the antenna (F) and posterior apex of an adult male (G) of specimens collected in China and determined to be Mediterranean based on DNA analysis.
The sequence was then aligned against each of the haplotypes used to construct the Mediterranean consensus sequence. There was a 100% match with one haplotype only. When compared against those sequences in GenBank using a BLAST search, 13 GenBank records were recovered, HM807578 from Tunisia and DQ365857, DQ365858, DQ365860, DQ365862, DQ365863, DQ365865–DQ365871 all of which come from Greece [46]. One of these, DQ365865, came from a collection at Agrinio [47], 123 years after it was originally collected by Gennadius.
Illustrations are presented of a puparium (Fig. 2 I, J), variations in the cement gland of adult 3 adult females (Fig. 2 C, D, E), and lateral view of aedeagus of an adult male (H) from Bemisia tabaci specimens collected by Gennadius. In addition, illustrations were made of the antenna (Fig. 2 A) and cement gland (Fig. 2 B) of a female and the antenna (Fig. 2 F) and posterior apex of an adult male (Fig. 2 G) of specimens collected in China and determined to be Mediterranean based on DNA analysis. All of the puparia collected by Gennadius that were examined have 6 pairs of long, dorsal setae arising from tubercles, in addition to the long pair of A8 setae located lateral to the anterior margin of the vasiform orifice and a long pair of caudal setae (Fig. 2 I, J). However, the length of the dorsal setae has been shown to be extremely variable often due to the characteristics of the leaf on which the puparium is found. We hope to find morphological characters in the adult female and/or male, such as the shape and size of the cement gland, aedeagus and claspers, body setation and/or antennal characters that will be useful in distinguishing Bemisia tabaci and its look-alikes.
Discussion
The differences in mtCOI between the different putative species make the assignment of the B. tabaci that Gennadius collected in 1889, unambiguous. We can therefore state without doubt that Mediterranean is the real B. tabaci. This species has now begun its own global journey of invasion spreading from its Mediterranean home range to at least 10 different countries [4], [48].
There have been several studies that have succeeded in amplifying mtCOI from museum specimens [49]. Only one has used members from the Hemiptera and these were samples that were less than four years old [50]. However, mtCOI has been amplified from beetles from museum collections dating back to 1820. In that study, of the 20 specimens assayed, five were as old as or older than our specimen and all yielded fragments [51]. So, our amplification of mtCOI from a member of the Hemiptera represents the oldest record.
To answer the original question posed by Barinaga [7] “Is the devastating whitefly invader really a new species?” the answer is yes, it is a new species. Further, Perring and co-workers [6] were right, they did have two different species. However, their assignment of the name B. tabaci to the indigenous American species (referred to as type A and later as the A biotype and now as New World) was incorrect as we now know that that this one belongs to the New World cluster of species [17], [26], [28]. But, in a way the ledger has now been squared. The true B. tabaci was not the one that invaded the USA in the early 1990s, that was Middle East – Asia Minor 1 (MEAM1, formerly known as the B biotype or B. argentifolii), but rather the one first detected in the USA in December 2004 [48] more than 10 years after MEAM1 was detected and initiated the debate.
Materials and Methods
Sample Preparation
Specimens from Gennadius' syntype series are held as slide-mounted material in the Smithsonian National Museum of Natural History along with a collection of intact nymphs and adults on tobacco leaves, also collected from Agrinio, Greece in 1889 by Gennadius. The samples were provided free of charge by Gregory A. Evans (co-author) of the U.S. Department of Agriculture in Beltsville, MD.
We took the following precautions to assure that the sample being tested had no chance of contamination. The laboratory and the building housing the laboratory are stand alone and have never been involved with B. tabaci research. Furthermore, B. tabaci does not occur in the Australian Capital Territory where the CSIRO research facility undertaking the research is located. A single dried adult from the Gennadius 1889 collection was placed into 100% ethanol for 2 days. This adult had the unexpanded wings typical of a newly emerged adult and is most likely to have emerged from a nymph on the leaf. It was then transferred into a sterile 1.5 mL Eppendorf tube to air-dry for 10 minutes at room temperature. 50 µL of Invitrogen's UltraPure water was added to the insect and crushed using a pre-sealed sterile p200 tip and immediately used for PCR amplification.
Primer design, DNA amplification and sequencing
PCR primers (Table 1) were designed using the mtCOI sequences derived from [26]. Primers were designed using the Oligo Primer Analysis Software v7.17 (Molecular Biology Insights, Inc., Cascade, CO 80809 USA) with the aim to minimise primer dimmers and hair-pin loops formation, reduced false priming sites, have an individual primer Tm of between 60°C to 72°C and to generate amplicons that ranged from 103 bp to 608 bp depending on primer pair combinations. All PCR reactions were carried out in a 25 µL reaction volume, and consisted of 2.5 µL of gDNA template, 1× ThermoPol Reaction Buffer (NEB), 0.625 u of DNA polymerase (NEB), 1.25 mM of dNTP's (Qiagen), and 0.4 mM of each forward and reverse primers [52]. A negative control that substituted the gDNA template with equal volume of UltraPure water was included in all PCR reaction preparations. PCR thermo-cycle profile consisted of an initial denaturing step at 95°C for 5 minutes, followed by 37cycles of template denaturing-primer annealing-template extension step of 95°C (30 seconds) - 54°C (30 seconds) – 72°C (45 seconds), and an additional template extension cycle at 72°C for 5 minutes. PCR amplicons were refrigerated at 10°C until the electrophoresis step, and at −20°C post electrophoresis. The presence of PCR amplicons were ascertained by running 6 µL of PCR reaction volume mixed with 1 µL of 6× Loading Dye (NEB) on 1.5% 1×TAE agarose gel at 90 V for 75 min. All agarose gels were pre-stained with GelRed (Biotium) following supplier's recommendation and viewed over a UV-transilluminator.
Table 1. Primer names, sequences and combinations used to obtain 496 bp of the mtCOI sequence from the Gennadius 1889 Bemisia tabaci specimen.
| Name | Primer sequence (5′→3′) | Primer Pairs | Amplicon (bp) | |
| 1 | G1889-30F26 | ATAATTTATGCTATANTRACTATYGG | 1+2 | 103 |
| 2 | G1889-110R23 | AAGTGAARTAAGCTCGAGTATCT | ||
| 3 | G1889-85F25 | AYATATTTACWGTTGGRATAGATGT | 3+4 | 103 |
| 4 | G1889-163R25 | GCAAGCCAACTAAAAATTTTAATTC | ||
| 5 | G1889-466F24 | TGTTYATTGGAGTAAAYTTAACTT | 5+9 | 172 |
| 6 | G1889-577R24 | TACTYAAAATCCTRCCCGCARAAG | 3+6 | 516 |
| 7 | G1889-127F25 | TCACTTCAGCTACTATGATTATTGC | 7+8 | 388 |
| 8 | G1889-493R22 | AAACCAAGAAAATGCTGAGGAA | ||
| 9 | G1889-606R32 | TCTAAAACRATAAATAAAAAATAAATAACAGA |
Sanger sequencing of PCR amplicons was carried out by direct 1∶10 dilution with Ultra-Pure water, and using 2.5 µL of the diluted PCR amplicon in a final 10 µL sequencing reaction volume that consisted of 0.16 mM of appropriate PCR primer, 1 µL of BigDye reaction mix, and 0.75× BigDye sequencing Buffer (Applied Biosystems). Sequencing profile was 1 cycle at 96°C for 2 minutes, followed by 30 cycles of 96°C for 10 seconds, 50°C for 5 seconds, and 60°C for 4 minutes, and incubation at 4°C at the end of the PCR cycle. Post sequencing reaction clean-up was by adding 80 µL 75% room temperature isopropanol to each sample and spin at 18,400 rcf for 10 minutes, followed by washing with 250 µL of 75% room temperature ethanol and spin at 18,400 rcf for 5 minutes. Ethanol was discarded and samples dried on 55°C heating block for 10 minutes. Sequencing of all samples was by the ANU's ACRF Biomolecular Resource Facility. We used the Staden sequence assembly and analysis package to check for sequencing errors and construct our contig [53].
Sequence identification
We used BioEdit to aligned against each of the consensus sequences that represent the 34 different putative species within the B. tabaci complex that have so far been identified [26]–[28]. The consensus sequences were designed from those haplotypes that were assigned to a particular putative species using Kimura 2 Parameter, Rosenberg's reciprocal monophyly (P(AB)), Rodrigo's (P(randomly distinct)), genealogical sorting index, and general mixed Yule- coalescent [11], [26]. The sequence was also compared against those in GenBank using BLAST.
Acknowledgments
We thank David Yeates and John La Salle for helpful comments to improve earlier versions of the manuscript.
Funding Statement
No funds external to CSIRO were used for this research. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Gennadius P (1889) Disease of the tobacco plantations in the Trikonia. The aleurodid of tobacco. Ellenike Georgia 5: 1–3. [Google Scholar]
- 2. Takahashi R (1936) Some Aleyrodidae, Aphididae, Coccidae (Homoptera) and Thysanoptera from Micronesia. Tenthredo 1: 109–120. [Google Scholar]
- 3. Brown JK, Frohlich DR, Rosell RC (1995) The sweetpotato or silverleaf whiteflies: biotypes of Bemisia tabaci or a species complex? Annu Rev Entomol 40: 511–534. [Google Scholar]
- 4. De Barro PJ, Liu SS, Boykin LM, Dinsdale AB (2011) Bemisia tabaci: A Statement of Species Status. Annu Rev Entomol 56: 1–19. [DOI] [PubMed] [Google Scholar]
- 5. Perring TM, Cooper A, Kazmer DJ, Shields C, Shields J (1991) New strain of sweetpotato whitefly invades California vegetables. Calif Agric 45: 10–12. [Google Scholar]
- 6. Perring TM, Cooper AD, Rodriguez RJ, Farrar CA, Bellows TS Jr (1993a) Identification of a whitefly species by genomic and behavioral studies. Science 259: 74–77. [DOI] [PubMed] [Google Scholar]
- 7. Barinaga M (1993) Is devastating whitefly invader really a new species? Science 259: 30. [DOI] [PubMed] [Google Scholar]
- 8. Bartlett AC, Gawel NJ (1993) Determining whitefly species. Science 261: 1333–1334. [DOI] [PubMed] [Google Scholar]
- 9. Campbell BC, Duffus JE, Baumann P (1993) Determining whitefly species. Science 261: 1333–1335. [DOI] [PubMed] [Google Scholar]
- 10. Perring TM, Farrar CA, Cooper AD, Bellows TS Jr (1993b) Determining whitefly species. Science 261: 1334–1335. [DOI] [PubMed] [Google Scholar]
- 11. Boykin LM, Armstrong KF, Kubatko L, De Barro P (2012) Species delimitation and global biosecurity. Evol Bioinformatics 8: 1–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dayrat B (2005) Towards integrative taxonomy. Biol J Linn Soc 85: 407–415. [Google Scholar]
- 13. Schlick-Steiner BC, Steiner FM, Seifert B, Stauffer C, Christian E, et al. (2010) Integrative taxonomy: a multisource approach to exploring biodiversity. Annu Rev Entomol 55: 421–438. [DOI] [PubMed] [Google Scholar]
- 14. Bortolus A (2008) Error cascades in the biological sciences: the unwanted consequences of using bad taxonomy in ecology. Ambio 37: 114–118. [DOI] [PubMed] [Google Scholar]
- 15. Lis JA, Lis B (2011) Is accurate taxon identification important for molecular studies? Several cases of faux pas in pentatomid bugs (Hemiptera: Heteroptera: Pentatomoidea). Zootaxa 2932: 47–50. [Google Scholar]
- 16.Mound LA, Halsey SM (1978) Whitefly of the world. British Museum (Natural History), London; and John Wiley, Chichester.
- 17. Bellows TS Jr, Perring TM, Gill RJ, Headrick DH (1994) Description of a species of Bemisia (Homoptera: Aleyrodidae). Ann Entomol Soc Am 87: 195–206. [Google Scholar]
- 18.Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA (2005) Virus taxonomy: VIIIth Report of the International Committee on Taxonomy of Viruses. San Diego: Elsevier Academic Press. 1162 p.
- 19. Fauquet CM, Briddon RW, Brown JK, Moriones E, Stanley J, et al. (2008) Geminivirus strain demarcation and nomenclature. Arch Virol 153: 783–821. [DOI] [PubMed] [Google Scholar]
- 20. Thresh JM, Otim-Nape GW, Legg JP, Fargette D (1997) African cassava mosaic virus disease: The magnitude of the problem. Afr J Root Tuber Crops 2: 13–18. [Google Scholar]
- 21. Oliviera MRV, Henneberry TJ, Anderson P (2001) History, current status, and collaborative research projects for Bemisia tabaci . Crop Prot 20: 709–723. [Google Scholar]
- 22. De Barro PJ, Hidayat SH, Frohlich D, Subandiyah S, Ueda S (2008) A virus and its vector, pepper yellow leaf curl virus and Bemisia tabaci, two new invaders of Indonesia. Biol Invasions 10: 411–433. [Google Scholar]
- 23. Perring TM (2001) The Bemisia tabaci species complex. Crop Prot 20: 725–737. [Google Scholar]
- 24. Mound LA (1963) Host-correlated variation in Bemisia tabaci (Gennadius) (Homoptera: Aleyrodidae). Proc R Entomol Soc Lond A 38: 171–180. [Google Scholar]
- 25. Russell LM (1958) Synonyms of Bemisia tabaci (Gennadius) (Homoptera, Aleyrodidae). Bull Brooklyn Entomol Soc 52: 122–123. [Google Scholar]
- 26. Dinsdale A, Cook L, Riginos C, Buckley YM, De Barro P (2010) Refined global analysis of Bemisia tabaci (Hemiptera: Sternorrhyncha: Aleyrodoidea: Aleyrodidae) mitochondrial cytochrome oxidase 1 to identify species level genetic boundaries. Ann Entomol Soc Am 103: 196–208. [Google Scholar]
- 27. Hu J, De Barro P, Zhao H, Wang J, Nardi F, Liu SS (2011) An extensive field survey combined with a phylogenetic analysis reveals rapid and widespread invasion of two alien whiteflies in China. PLoS One 6: 1–9 e16161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Alemandri V, De Barro P, Bejerman N, Argüello Caro EB, Dumón AD, et al. (2012) Species Within the Bemisia tabaci (Hemiptera: Aleyrodidae) complex in soybean and bean crops in Argentina. J Econ Entomol 105: 48–53. [DOI] [PubMed] [Google Scholar]
- 29. Costa HS, Brown JK, Sivasupramaniam S, Bird J (1993) Regional distribution, insecticide resistance, and reciprocal crosses between the A-biotype and B-biotype of Bemisia tabaci . Insect Sci Appl 14: 255–266. [Google Scholar]
- 30. Bedford ID, Briddon RW, Brown JK, Rosell RC, Markham PG (1994) Geminivirus transmission and biological characterization of Bemisia tabaci (Gennadius) biotypes from different geographic regions. Ann Appl Biol 125: 311–325. [Google Scholar]
- 31. Byrne FJ, Cahill M, Denholm I, Devonshire AL (1995) Biochemical identification of interbreeding between B-type and non B-type strains of tobacco whitefly Bemisia tabaci . Biochem Genet 33: 13–23. [DOI] [PubMed] [Google Scholar]
- 32. De Barro PJ, Hart PJ (2000) Mating interactions between two biotypes of the whitefly, Bemisia tabaci (Hemiptera: Aleyrodidae) in Australia. Bull Entomol Res 90: 103–112. [DOI] [PubMed] [Google Scholar]
- 33. Maruthi MN, Colvin J, Seal S (2001) Mating compatibility, life-history traits, and RAPD-PCR variation in Bemisia tabaci associated with the cassava mosaic disease pandemic in East Africa. Entomol Exp Appl 99: 13–23. [Google Scholar]
- 34. Maruthi MN, Colvin J, Thwaites RM, Banks GK, Gibson G, et al. (2004) Reproductive incompatibility and cytochrome oxidase I gene sequence variability among host-adapted and geographically separate Bemisia tabaci populations (Hemiptera: Aleyrodidae). Syst Ent 29: 560–568. [Google Scholar]
- 35. Mallet J (2005) Hybridization as an invasion of the genome. Trends Ecol Evol 20: 229–237. [DOI] [PubMed] [Google Scholar]
- 36. Omondi BA, Sseruwagi P, Obeng-Ofori D, Danquah EY, Kyerematen RA (2005) Mating interactions between okra and cassava biotypes of Bemisia tabaci (Homoptera: Aleyrodidae) on eggplant. Int J Trop Insect Sci 25: 159–167. [Google Scholar]
- 37. Liu SS, De Barro PJ, Xu J, Luan JB, Zang LS, et al. (2007) Asymmetric mating interactions drive widespread invasion and displacement in a whitefly. Science 318: 1769–1772. [DOI] [PubMed] [Google Scholar]
- 38. Zang LS, Liu SS (2007) A comparative study on mating behavior between the B biotype and a non-B biotype of Bemisia tabaci (Hemiptera: Aleyrodidae) from Zhejiang, China. J Insect Behav 20: 157–171. [Google Scholar]
- 39. Dalmon A, Halkett F, Granier M, Delatte H, Peterschmitt M (2008) Genetic structure of the invasive pest Bemisia tabaci: evidence of limited but persistent genetic differentiation in glasshouse populations. Heredity 100: 316–325. [DOI] [PubMed] [Google Scholar]
- 40. Elbaz M, Lahav N, Morin S (2010) Evidence for pre-zygotic reproductive barrier between the B and Q biotypes of Bemisia tabaci (Hemiptera: Aleyrodidae). Bull Entomol Res 100: 581–590. [DOI] [PubMed] [Google Scholar]
- 41. Xu J, Liu SS, De Barro P (2010) Reproductive incompatibility among genetic groups of Bemisia tabaci supports the proposition that the whitefly is a cryptic species complex. Bull Entomol Res 100: 359–366. [DOI] [PubMed] [Google Scholar]
- 42. Sun DB, Xu J, Luan JB, Liu SS (2011b) Reproductive incompatibility between the B and Q biotypes of the whitefly Bemisia tabaci: genetic and behavioural evidence. Bull Entomol Res 101: 211–220. [DOI] [PubMed] [Google Scholar]
- 43. Wang P, Sun DB, Qiu BL, Liu SS (2011) The presence of six cryptic species of the whitefly Bemisia tabaci complex in China as revealed by crossing experiments. Insect Sci 18: 67–77. [Google Scholar]
- 44. Luan JB, Liu SS (2012) Differences in mating behavior lead to asymmetric mating interactions and consequential changes in sex ratio between an invasive and an indigenous whitefly. Integrative Zool 7: 1–15. [DOI] [PubMed] [Google Scholar]
- 45. Liu SS, Colvin J, De Barro PJ (2012) Species concepts as applied to the whitefly Bemisia tabaci systematics: how many species are there?” Species concepts as applied to the whitefly Bemisia tabaci systematics: how many species are there? J Integrative Agric 11: 176–186. [Google Scholar]
- 46. Tsagkarakou A, Mouton L, Kristoffersen JB, Dokianakis E, Grispou M, et al. (2012) Population genetic structure and secondary endosymbionts of Q Bemisia tabaci (Hemiptera: Aleyrodidae) from Greece. Bull Entomol Res 102: 353–365. [DOI] [PubMed] [Google Scholar]
- 47. Tsagkarakou A, Tsigenopoulos CS, Gorman K, Lagnel J, Bedford ID (2007) Biotype status and genetic polymorphism of the whitefly Bemisia tabaci (Hemiptera: Aleyrodidae) in Greece: mitochondrial DNA and microsatellites. Bull Entomol Res 97: 29–40. [DOI] [PubMed] [Google Scholar]
- 48. Dalton R (2006) Whitefly infestations: the Christmas invasion. Nature 443: 898–900. [DOI] [PubMed] [Google Scholar]
- 49. Jinbo U, Kato T, Iti M (2011) Current progress in DNA barcoding and future implications for entomology. Entomol Sci 14: 107–124. [Google Scholar]
- 50. Castalanelli MA, Severtson DL, Brumley CJ, Szito A, Foottit RG, et al. (2010) J Asia Pacific Entomol 13: 243–248. [Google Scholar]
- 51. Thomsen PF, Elias S, Gilbert MTP, Haile J, Munch K, et al. (2009) Non-Destructive Sampling of Ancient Insect DNA. PLoS ONE 4 4: e5048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Tay WT, Behere GT, Heckel DG, Lee SF, Batterham P (2008) Exon-primed intron-crossing (EPIC) PCR markers of Helicoverpa armigera (Lepidoptera: Noctuidae). Bull Entomol Res 98: 509–518. [DOI] [PubMed] [Google Scholar]
- 53. Staden R, Beal KF, Bonfield JK (2000) The Staden package, 1998. Methods Mol Biol 132: 115–130. [DOI] [PubMed] [Google Scholar]