Abstract
While the mammalian heart has low, but functionally significant, levels of telomerase expression, the cellular population responsible remains incompletely characterized. This study aimed to identify the cell types responsible for cardiac telomerase activity in neonatal, adult, and cryoinjured adult hearts using transgenic mice expressing green fluorescent protein (GFP), driven by the promoter for murine telomerase reverse transcriptase (mTert), which is a necessary and rate-limiting component of telomerase. A rare population of mTert-GFP-expressing cells was identified that possessed all detectable cardiac telomerase RNA and telomerase activity. It was heterogeneous and included cells coexpressing markers of cardiomyocytic, endothelial, and mesenchymal lineages, putative cardiac stem cell markers, and, interestingly, cardiomyocytes with a differentiated phenotype. Quantification using both flow cytometry and immunofluorescence identified a significant decline in mTert-GFP cells in adult animals compared to neonates (∼9- and ∼20-fold, respectively). Cardiac injury resulted in a ∼6.45-fold expansion of this population (P<0.005) compared with sham-operated controls. This study identifies the cells responsible for cardiac telomerase activity, demonstrates a significant diminution with age but a marked response to injury, and, given the relationship between telomerase activity and stem cell populations, suggests that they represent a potential target for further investigation of cardiac regenerative potential.—Richardson, G. D., Breault, D., Horrocks, G., Cormack, S., Hole, N., Owens, W. A. Telomerase expression in the mammalian heart.
Keywords: mTert, transgenic, cardiac, stem cell, regeneration
Telomerase is a ribonucleoprotein complex typically expressed in cellular populations capable of extended replication, such as germ cells, tumor cells, and stem cells. It is best known for its ability to maintain the telomeric ends of chromosomes, which are ordinarily shortened with cell division, leading to cellular senescence and death. Telomerase reverse transcriptase (TERT) is the catalytic subunit of the telomerase complex, is transcriptionally regulated (distinguishing it from the other components of the complex) (1), and is the rate-limiting component of telomerase activity (ref.2; reviewed in ref. 3). Increasing evidence suggests that TERT has functions in stem cell proliferation and differentiation (4), including transcriptional regulation of mitogenic and angiogenic genes (5, 6). TERT also binds mitochondrial DNA and has functions in cell respiration, apoptosis, and protection against oxidative stress (7).
Cardiac telomerase activity is detectable at the earliest stages of life and is down-regulated in adult rat myocardium (8). Evidence points to an important role for telomerase activity in the development and function of the normal heart; TERC−/− mice that lack telomerase function develop cardiac abnormalities, including dilated cardiomyopathy and reduced angiogenic potential (reviewed in ref. 9). In contrast, forced telomerase expression leads to prolonged cardiomyocyte cycling and hypertrophy (10). Telomere dysfunction in the cardiac stem cell population is suggested to be a contributing factor to hearts suffering from aging and end-stage failure in mice and dogs (11–13). Telomerase activity has been used as a defining feature of stem cell populations and demonstrated in some descriptions of putative cardiac stem cells (14).
Demonstration of TERT expression and the source of cardiac telomerase activity is challenging. The telomeric repeat amplification protocol (TRAP) assay, while specific, is limited in sensitivity, which can be exacerbated by the effects of tissue digestion and cell sorting. This is important in antigenically defined cardiac stem cell populations, where cell numbers are typically low. While immunohistochemistry is an appealing alternative, there is controversy regarding the specificity of antibodies (15).
To enable the identification and isolation of TERT mRNA-expressing cells in vivo, we previously generated a transgenic mouse, in which the murine TERT (mTert) promoter drives green fluorescent protein (GFP) expression. In this model, mTert-GFP expression marks telomerase activity within embryonic, hematopoietic, and intestinal stem cells, germ cells (16–18), and induced pluripotent stem (iPS) cells (19). We hypothesized that it would enable the in vivo identification of the population responsible for the telomerase activity previously described in cardiac tissue, with the potential to demonstrate whether these cells have a role in a cardiac regenerative response to injury.
We identified an infrequent population of nonhematopoietic cells in the heart containing all of the detectable telomerase activity. This population was found in the highest quantity in the neonatal heart and declined on maturation to adulthood. Immunofluorescent studies demonstrated that mTert is expressed in a number of different cell lineages, including those expressing recognized stem cell characteristics. Although mTert-expressing cells are extremely rare in the adult heart (∼0.012%), there was a localized increase in the number of mTert-GFP cells in the response to an acute myocardial injury.
This study provides a comprehensive description of the cell types responsible for telomerase activity in the heart in vivo. Further examination of this population will provide a better understanding of the function of both telomerase activity and TERT signaling in the heart.
MATERIALS AND METHODS
Ethics statement
Ethical approval of animal work carried out in this project has been authorized by the Newcastle University Ethics Committee and was covered by Project License PPL/60-3876, approved by the UK Home Office.
Transgenic animals
Generation and initial phenotypic characterization of the mTert-GFP mice have been published previously (17–19).
Cardiac cell harvesting
Hearts were removed from adult and neonatal animals killed by approved methods, finely minced, and incubated at 37°C for 30 min in MEM containing collagenase II (0.1%; Worthington Biochemical, Lakewood, NJ, USA) and DNase (0.6 mg/ml) to disaggregate into a single-cell suspension. Red blood cells were depleted with red blood cell lysis buffer.
Flow cytometry and mTert-GFP quantification
Native GFP expression was detectable by flow cytometry without the need for antibody labeling. For all flow cytometry experiments, the mouse lineage antibody cocktail (BD Pharmingen, Oxford, UK) specific to CD3e chain, CD11b, CD45R/B220, and erythroid cells (Ly-76/TER-119), Ly-6G, and Ly-6C, was used to exclude contamination by hematopoietic lineage. To exclude nonviable cells, 7-aminoactinomycin D (7-AAD; BD Pharmingen) was added prior to flow cytometery. Flow cytometry was performed using a FACSAria (BD Biosystems, Oxford, UK). To characterize expression of putative stem cell antigens, cells were further labeled with fluorochrome-conjugated antibodies specific to Sca-1/Ly6A/E (clone D7-phycoerythrin). Expression analysis was performed on a BD FACSCalibur (BD Biosystems), running FlowJo software (Tree Star, Ashland, OR, USA). Hearts from mTert-GFP transgenic animals were studied by flow cytometry at postnatal day 3 (n=4) and 3 mo (n=26) and 12 mo of age (n=6).
Reverse transcriptase–polymerase chain reaction (RT-PCR)
Equal numbers of hematopoietic lineage negative (Lin−), GFP-expresssing (GFP+), and Lin−, GFP absent (GFP−) viable, hematopoietic lineage-depleted cells were separated from transgenic mTert-GFP neonatal hearts using flow cytometry. mRNA was isolated, cDNA was transcribed, and PCR was performed using Invitrogen PCR Supermix with the following primers: TERT-F, 5′-GGATTGCCACTGGCTCCG-3′, and TERT-R, 5′-TGCCTGACCTCCTCTTGTGAC-3′ (20); and GAPDH-F, 5′-TGTGCAGTGCCAGCCTCGTC-3′, and GAPDH-R, 5′-TGACCAGGCGCCCAATACGG-3′.
TRAP
Flow cytometry was used to isolate viable, hematopoietic lineage-depleted cells from hearts and viable cells from the bone marrow (n=5) of mTert-GFP transgenic animals, and these were subjected to TRAP using the TRAPeze RT telomerase detection kit (Millipore, Bedford, MA, USA), as per manufacturer's instructions. This assay quantifies telomerase activity by measuring real-time fluorescence emission using real-time quantitative RT-PCR (qRT-PCR). Cells (1×104) for each population were lysed in 20 μl of 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS) lysis buffer. Telomerase activity, in terms of the addition of telomeric repeats (GGTTAG) onto the 3′ end of a substrate oligonucleotide (TS) during an initial reaction, was then calculated by real-time qRT-PCR. Relative values were calculated by comparing the average Ct values from each sample against a standard curve generated using a TSR8 control template. Telomerase-expressing cells (provided with the kit) and GFP+ cells isolated from bone marrow, previously shown to be telomerase active (17) served as a positive control, and heat-inactivated cell extracts served as a negative control. Assays were performed on an Applied Biosystems 7500 Fast real-time PCR System (Applied Biosystems, Foster City, CA, USA). The baseline was set to a no-TSR8 control.
Immunofluorescence
Hearts were removed from freshly killed animals, fixed in 4% paraformaldehyde in PBS at 4°C overnight, and cyroprotected with 30% sucrose in PBS. The samples were embedded in TissueTek optimal cutting temperature (OCT) compound (Agar Scientific, Essex, UK), snap-frozen in liquid nitrogen, and stored at −80°C. Paraformaldehyde-fixed cryosections (10 μm) were labeled with the primary antibodies, and secondary Alexa Fluor-conjugated antibodies (Molecular Probes; Invitrogen, Carlsbad, CA, USA) were used for detection. Primary antibody: anti-α-smooth muscle actin (α-SMA; clone ab5694), anti-CD45 (clone ab19592), anti-C-kit (clone ab5506), anti-GATA4 (phosphor S105; clone ab5245), anti-GFP (clone ab6662), anti-islet 1 (clone ab20670), anti-Sca-1/Ly6A/E (E13 161-7; clone ab51317), anti-cardiac troponin I (CTnI; clone ab58544), and anti-vimentin (clone ab24525) (Abcam, Cambridge, UK); anti-CD31 (PECAM-1; clone Mec13.3; BD Pharmingen); anti-Nkx2.5 (clone H-114; Santa Cruz Biotechnology; Autogen Bioclear, Wiltshire, UK); anti-phospho-histone H3 (Ser10; Upstate, Millipore, Billerica, MA, USA); anti-Ki67 (NCL-Ki67p; Novocastra, Newcastle-on-Tyne, UK). Nuclei were labeled with 0.2 μg/ml 4′,6-diamidino-2-phenylindole (DAPI). For analysis, an Axioimager M1 fluorescence microscope (Carl Zeiss, Oberkochen, Germany) running OpenLab software (Improvision, Coventry, UK) or the Leica SP5 laser-scanning confocal system (Leica Microsystems, Wetzlar, Germany) was used. To rule out staining artifacts creating false colocalization events, all primary and Alexa Fluor-conjugated secondary antibodies used were also analyzed individually. The fluorescence signals for each fluorochrome-conjugated antibody used was only observed in the appropriate fluorescence channel. Colocalization of multiple signals was verified by creating Z-stack images using the Leica SP5 laser-scanning confocal system; cells were only considered coexpressing if the fluorescent signals localized to a single DAPI-labeled nucleus throughout the Z stack. Specificity of all secondary antibodies and the absence of autofluorescence were tested by omitting primary controls. These controls gave undetectable background signal. For all immunofluorescence studies, GFP signal was amplified using an anti-GFP antibody. The specificity of this antibody was evaluated on wild-type littermate controls. Wild-type cardiac sections displayed no reactivity to this GFP antibody.
Adult mTert-GFP cell counts were performed in a manner similar to the cellular counting method described by Walsh et al. (21). Myocardial tissue was cryosectioned laterally in 10-μm sections. Sections were separated by intervals of 50 μm, and all mTert-GFP-expressing cells were counted in each section, creating a representative cross section that included all regions of the heart. The percentage of GFP-expressing cells was obtained by dividing the total number of GFP cells per section by the total number of cells in each section; cells were defined by the nucleus label DAPI. Quantification of total nuclei was obtained by multiplying cell density by section area. Both the density of nuclei and area of the section were quantified using digital image analysis (ImageJ; U.S. National Institutes of Health; http://rsbweb.nih.gov/ij/). Approximately 1.8 × 106 nuclei/heart with a total of 5.4 × 106 nuclei analyzed for each antibody were investigated. The results are presented as means ± se. The significance of the differences was determined with the use of Student's t test. While this method does not account for binucleation, as described by Walsh et al. (21), it has the advantage of enabling more extensive coexpression analyses, while still demonstrating relative proportions of cell types and changes in them, if not absolute numbers in individual hearts.
To assess expression of antigenic markers by mTert-GFP-expressing cardiac cells, hematopoietic cells were excluded from the analysis. Hearts from mTert-GFP animals were labeled with antibodies to GFP and CD45. No cells coexpressing mTert-GFP and CD45 were detected in extravascular tissues, and only a small number of mTert-GFP- and CD45-coexpressing cells were observed within the lumen of the larger blood vessels and the chambers of atria and ventricles. The latter cells had typical mononucleocyte morphology, suggestive of a hematopoietic lineage, and were excluded from further analysis on this basis (Supplemental Fig. S6).
Induced myocardial cryoinjury
Myocardial cryoinjury was performed as described by Leferovich et al. (22). This model of injury, as opposed to coronary ligation, was chosen aiming to generate a regenerative response in a reproducible manner with low mortality, rather than replicate a myocardial infarction per se, with the associated ischemic and pathophysiological phenomena. At 14 d after the injury, the hearts were removed and analyzed by immunofluorescence as described above. A total of 9 mTert-GFP animals underwent cryoinjury; controls either underwent a sham procedure (abdomen opened but no cryoinjury) or did not undergo any surgical procedure. Quantification of the mTert-GFP population in the injured and sham-injured animal hearts was determined by immunofluorescence, as described for adult hearts above.
Statistics
Data are presented as means ± se. Groups were compared using Student's t test. When comparing multiple groups, data were analyzed by analysis of variance (ANOVA). A value of P < 0.05 was considered statistically significant.
RESULTS
mTert mRNA expression and telomerase activity are restricted to the GFP-expressing population
Previous work has demonstrated that mTert-GFP expression correlates closely with the native mTert mRNA transcripts and telomerase activity within all tissue systems investigated (17–19). We sought to confirm that GFP transgene expression maps to mTert mRNA expression and telomerase activity in Lin− cardiac cells.
Relative to a housekeeping gene, Lin−, GFP+ cells exhibited the highest levels of mTert transcript, whereas no mTert mRNA expression was observed in the Lin−, GFP− population (Fig. 1A). mTert mRNA expression was also analyzed in digested but unsorted mTert-GFP transgenic hearts. In unsorted heart cells, expression was low but detectable. TRAP demonstrated that telomerase activity was restricted to the GFP+ population of Lin− cardiac cells; telomerase activity in the Lin−, GFP+ cardiac population was at a level comparable to that of GFP+ bone marrow cells, previously identified as containing all bone marrow telomerase and stem cell activity (17). Telomerase activity was undetectable in the GFP− cardiac population (Fig. 1B).
Figure 1.
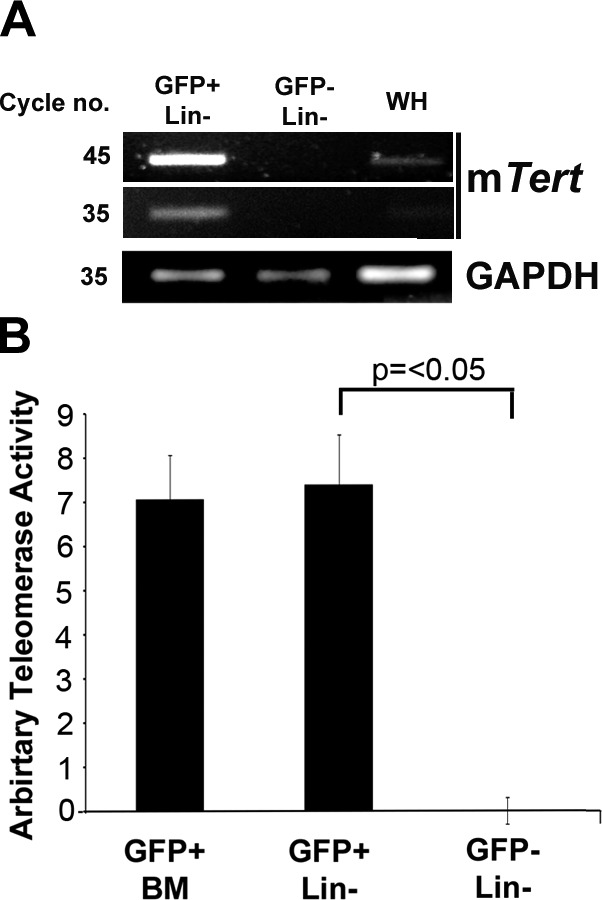
GFP expression maps to TERT transcripts and telomerase activity in the heart. A) Comparison of mTert mRNA expression in Lin−, gated GFP+, and GFP− cells compared to GAPDH. Amplified product was assessed at 35 and 45 cycles to clarify comparative expression. All products were of correct size. WH, whole unsorted heart. B) Comparison of telomerase activity in viable Lin−, GFP+, and GFP− cells via real-time qPCR-based TRAP assay. Bars represent means ± se of ≥3 independent experiments.
mTert-GFP is expressed in cell populations in the heart that change in proportion over 12 mo
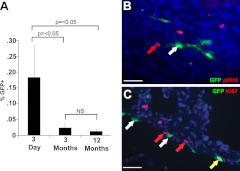
We investigated changes in the number of mTert-expressing cells in the hearts from mTert-GFP transgenic animals by flow cytometry at 3 d, 3 mo, and 12 mo of age (Fig. 2A). The Lin− population accounted for ∼98% of total cells at all ages. At 3 d of age, 0.18 ± 0.10% of the cardiac Lin− cells expressed mTert-GFP; this population decreased at 3 mo to 0.02 ± 0.01% (P<0.05), and the number of mTert-GFP-expressing cells continued to decline to 0.01 ± 0.01% at 12 mo (not statistically significant).
Figure 2.
A) Number of mTert-expressing cells is dynamic during development and aging. FACS quantification of mTert-expressing cells within the viable Lin− populations in 3-d, 3-mo, and 12-mo GFP transgenic hearts. Bars represent means ± se of ≥4 independent experiments. B, C) mTert-GFP expression is not restricted to actively cycling cells. Green, mTert-GFP; red, pHH3 (B) or Ki67 (C); blue, DAPI (nuclei). White arrows indicate GFP only; red arrows indicate Ki67 or pHH3 only; yellow arrows indicate coexpression. Scale bars = 25 μm.
Quantification by immunofluorescence identified that 0.24 ± 0.08% of cells (as defined in Materials and Methods) in neonatal hearts expressed mTert-GFP. In adult (>3 mo) mTert-GFP transgenic hearts, this method demonstrated GFP expression in 0.012 ± 0.003% of total cardiac cells.
Telomerase expression is associated with cellular proliferation (23); however, in the intestinal crypt, mTert expression identifies a population of slow-cycling stem cells that generally lack expression of the proliferation marker Ki67 (18). Therefore, we determined expression of Ki67 and phospho-histone H3 (pHH3) in mTert-expressing cells. In the neonatal heart, 0.75 ± 0.25 and 1.4 ± 0.35% of total cardiac cells expressed pHH3 or Ki67, respectively. Although no mTert-GFP cells were observed coexpressing pHH3 (Fig. 2B and Supplemental Fig. S1), 5.71 ± 1.63% coexpressed Ki67 (Fig. 2C and Supplemental Fig. S1). This may represent the more restricted cell cycle expression of pHH3 than Ki67 (24, 25). The Ki67 coexpression levels are consistent with those in the intestine and suggest that, as in the intestine, cardiac mTert-expressing cells are a largely quiescent population.
mTert-GFP-expressing cells in the heart express a range of antigenic markers
Ablation of telomerase activity influences the biology of a number of cardiac lineages, reducing angiogenic potential and resulting in abnormalities in cardiomyocyte size, number, and proliferative potential, and enhancing myocyte death (26). Therefore, we localized and quantified mTert-GFP expression in relation to lineage-specific antigens.
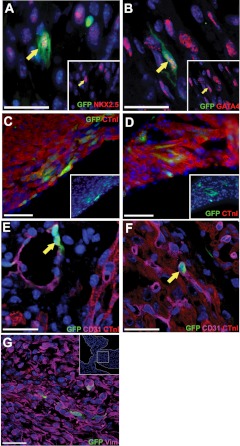
mTert-GFP+ cells coexpressing the cardiac transcription factors Nkx2.5 or GATA4 were identified within the myocardium (Fig. 3A, B). mTert-GFP+, GATA4+, or Nkx2.5+ cells had morphologies ranging from cells lacking sarcomeric structures to those of more mature cardiomyocytes. Association with a cardiomyocytic lineage was confirmed with CTnI coexpression. mTert-GFP+ CTnI+ cells were observed in the compact layer of each ventricle (Fig. 3C) and atrial wall (Fig. 3D). As mTert-GFP+ cells were observed in the cardiac vasculature, hearts were stained for CD31, a marker of endothelial cells. mTert-GFP+ CD31+ cells were identified in two distinct locations: within the endothelial layer of the lumen of the coronary vasculature (Fig. 3E) and within the myocardium of the ventricular walls (Fig. 3F). The latter were round or oval-shaped, relatively small, and otherwise featureless, with a large nuclear to cytoplasmic ratio. A proportion of these cells had a histological distribution, suggesting a relationship with the smaller (CD31-expressing) vessels in the myocardium. However, individual mTert-GFP+ CD31+ cells were also observed, which displayed no such association.
Figure 3.
In the neonatal heart, mTert-GFP cells are a heterogeneous population and express genes associated with cardiomyocyte endothelial and fibroblastic lineages. A–D) mTert-GFP+ NKx2.5+ (A) and mTert-GFP+ GATA4+ cells (B) within myocardium, mTert-GFP+ CTnI+ cells in the ventricle (C), and mTert-GFP+ CTnI+ cells in the atria (D). Insets: lower-magnification images. E) Vascular structures expressing CD31 were identified containing mTert-GFP-expressing cells. F) mTert-GFP+ CD31+-coexpressing cells in the myocardium. G) mTert-GFP+ vimentin (Vim)+ cells in the atrioventricular valve. Inset: lower-magnification image and the region of AV shown in the main figure. Green, mTert-GFP; red, GATA4, Nkx2.5, or CTnI; magenta, CD31 or vimentin; blue, DAPI (nuclei). Yellow arrows indicate coexpressing cells. Scale bars = 25 μm.
With respect to the mesenchymal or fibroblast lineages, mTert-GFP+ vimentin+ cells lacking expression of CD31 were identified. mTert-GFP+ cells expressing fibroblast-specific protein 1 were also identified (Supplemental Fig. S2). These cells had a fibroblastic morphology and were found in the atrial septum and fibrous skeleton of the heart, related to the atrioventricular valves (Fig. 3G).
Immunofluorescent analysis of adult hearts revealed a similar profile in terms of the expression of markers of the investigated lineages (Supplemental Fig. S3), although with a reduced frequency.
Regardless of the antibody combinations used, mTert-GFP+ cells were identified that failed to coexpress any of the investigated antigens. These cells were distributed throughout the myocardium (Supplemental Fig. S4A) and epicardium (Supplemental Fig. S4B).
These data demonstrate that the mTert-GFP cell population is heterogeneous and represents a very rare subpopulation of cardiomyocytic, endothelial, and fibroblast lineages, including cells with a mature phenotype.
Cryoinjury to the adult myocardium is associated with an increase in mTert-GFP-expressing cell numbers
We investigated mTert-GFP expression in myocardial injury, using a cryoinjury model to generate a reproducible injury with low mortality and enabling examination of mTert-GFP+ cells in a localized area surrounded by relatively healthy myocardium.
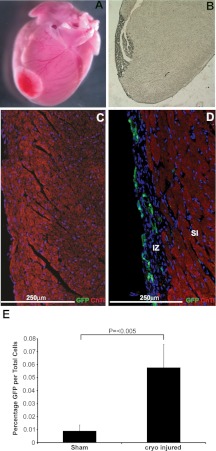
At 14 d following cryoinjury, a distinct “injury zone” was apparent where CTnI-expressing cardiomyocytes had been destroyed with relatively normal tissue adjacent (Fig. 4A–D). This allowed identification of the area of greatest cellular damage and comparisons between the injury zone and the surrounding area.
Figure 4.
Cryoinjured hearts contain a localized injury with an increase in mTert-GFP-expressing cells. A) Heart at 14 d after cryoinjury showing a localized injury with normal surrounding tissue. B) Unstained mouse heart demonstrating full depth of the tissue damage. Injured tissue penetrated the myocardium through to the ventricular cavity. C) Sham-injured animals showed a consistent expression of CTnI throughout the myocardium, CTnI only being absent from the outermost epicardial cell layer. D) Hearts at 14 d after cryoinjury had a localized injury zone (IZ) lacking CTnI expression with adjacent subinjury zone (SI). Green, mTert-GFP; red, CTnI; blue, DAPI (nuclei). Scale bars = 250 μm. E) Percentage of mTert-GFP cells in sham-injured and cyroinjured hearts at 14 d after injury. Values are means ± se of ≥3 independent experiments.
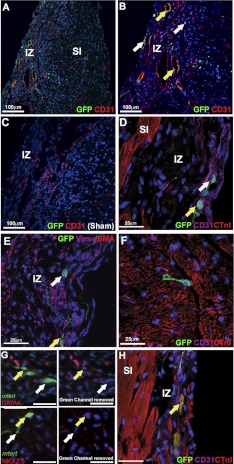
Comparison of the total percentage of mTert-GFP+ cells in the cyroinjured hearts identified a significant 6.45-fold (P<0.005) increase compared to control adult hearts (Figs. 4 and 5 and Supplemental Fig. S5). Quantification of the mTert-GFP-expressing cells within the injury zone demonstrated that 4.52 ± 1.09% of cells expressed GFP (Fig. 4E), an ∼500-fold increase in mTert-GFP-expressing cells when compared to the same region in controls (P<0.005). No mTert-GFP+ cells were found to costain with the hematopoietic marker CD45 (data not shown). mTert-GFP+ CD31+ cells were identified both within vascular-like structures in the injury zone (Fig. 5A–C) and as less structurally organized cells throughout the injury zone (Fig. 5D). mTert-GFP+ Vim+ cells were also found in the injury zone (Fig. 5E). As in the uninjured hearts, cells lacking the expression of all investigated antigens were identified in the injury zone and in the periphery of the injury (Fig. 5F).
Figure 5.
In the adult heart, all mTert-GFP cell phenotypes described in neonates were found at 14 d after cryoinjury. A) mTert-GFP cells in the injury zone (IZ) and subinjury (SI) zone. B) In the IZ, CD31 expression shows the presence of structures resembling blood vessels, which contain mTert-GFP+ CD31+ cells (yellow arrows). Single mTert-GFP-expressing CD31− cells were also observed in the epicardium and myocardium of the IZ, as well as the SI zone (white arrows). C) Cryoinjured wild-type littermate controls expressed a similar CD31 profile to mTert-GFP animals, but no GFP reactivity was observed. D) mTert-GFP+ CTnI− and mTert-GFP+ CD31− cells were observed in the SI myocardium. E, F) Higher-magnification images of the IZ showing mTert-GFP+ Vim+ (E) and mTert-GFP+ CD31 (F) cells. G) A population of mTert-GFP cells within the IZ express cardiac transcription factors GATA4 and Nkx2.5. H) Cells with mature cardiomyocyte morphologies expressing mTert-GFP and CTnI are found within the IZ (yellow arrows). A–D) Green, mTert-GFP; red, CD31 or CTnI; blue, DAPI (nuclei). E–F) Green, mTert-GFP; red, CTnI, α-SMA, GATA4, or Nkx2.5; magenta, CD31 or vimentin; blue, DAPI (nuclei). White arrows indicate mTert-GFP only; yellow arrows indicate mTert-GFP cells coexpressing investigated markers. Scale bars = 25 μm or as indicated.
Regarding cardiac transcription factors, mTert-GFP+ GATA4+ and/or Nkx2.5+ cells were identified in relation to the area of injury (Fig. 5G). Very infrequently, cells that expressed CnTI in a sarcomeric pattern with a low level of GFP expression were seen at the periphery of the injury and within the injury zone (Fig. 5H and Supplemental Fig. S5).
Relationship between mTert-GFP expression and markers of putative cardiac stem cells
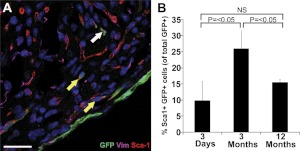
Within the heart, cells expressing Sca-1, c-kit, and Isl1 have been associated with cardiac stem cell activity. In neonatal mTert-GFP transgenic hearts, cells coexpressing mTert-GFP and Sca-1 were observed (Fig. 6A) and quantified by fluorescence-activated cell sorting (FACS) analysis. Sca-1-expressing and -nonexpressing populations were identified within the Lin− mTert-GFP-expressing population. In 3-d-old neonates, 9.9 ± 5.6% of the mTert-GFP-expressing population coexpressed Sca-1, significantly increasing to 26.0 ± 6.6% (P<0.05) at 3 mo of age, and subsequently significantly decreasing to 15.5 ± 0.9% (P<0.05) at 12 mo of age (Fig. 6B and Supplemental Fig. S6). Although cells expressing c-kit or Isl1 were observed in neonatal mTert-GFP hearts, none expressing mTert-GFP were found (Supplemental Fig. S7B, C).
Figure 6.
Sca-1 is expressed in a subpopulation of neonatal cardiac mTert-GFP-expressing cells. A) mTert-GFP+ Sca-1+ cells in the myocardium. Green, mTert-GFP; red, Sca-1; magenta, vimentin; blue, DAPI (nuclei). White arrow indicates mTert-GFP only, yellow arrows indicate mTert-GFP and Sca-1 coexpression. Scale bars = 25 μm. B) Quantification of Sca-1-coexpressing cells within the viable Lin− mTert+ population by flow cytometry in mTert-GFP transgenic hearts at 3 d, 3 mo, and 12 mo. NS, not significant. Data represent means ± sd of ≥4 independent experiments.
DISCUSSION
The use of this mTert-GFP reporter mouse enables the demonstration of telomerase expression at a single-cell level, overcoming the problems associated with conventional immunohistochemistry. We demonstrate that in the heart, mTert-GFP is expressed by a heterogeneous population containing all detectable telomerase activity and TERT mRNA and includes cells of endothelial, fibroblast, and cardiomyocytic lineages, as well as populations with characteristics of putative cardiac stem cells. mTert-expressing cells were rare, in the order of several hundred cells per heart, and highest in neonates, indicating that the reported decrease in telomerase activity after birth (27) is at least in part due to this reduction in mTert cell number. The demonstration of a marked, localized increase in number of telomerase-expressing cells in response to injury in the adult heart suggests a role for these cells in regeneration.
It has been suggested that cardiac telomerase activity is restricted to cardiomyocytes. Cardiomyocyte-enriched cultures, prepared by enzymatic digestion of rat ventricle, were shown to possess telomerase activity by PCR-TRAP assay, and an immunohistological approach using a canine model suggested that TERT is expressed in a surprisingly high proportion of cardiomyocytes but undetectable in cardiac endothelial cells or fibroblasts (28). Earlier studies using the TRAP assay failed to demonstrate telomerase activity in smooth muscle and fibroblast cell lines (29). The presence of telomerase activity in endothelial cells continues to be debated (see ref. 30 for review).
Our data demonstrating mTert expression in endothelial cells is consistent with other reports that identified low levels of telomerase expression in fresh endothelial cultures (31, 32), confirming that they express telomerase in vivo. The increase in CD31-coexpressing cells following injury is consistent with an angiogenic response, supported by studies demonstrating increased endothelial Tert mRNA and telomerase activity in response to FGF-2-induced proliferation (32). Studies are planned to characterize the role of telomerase-expressing endothelial cells in angiogenesis.
With respect to fibroblasts, there is evidence that quiescent fibroblasts possess low levels of telomerase activity that increases in response to stimuli, for example, with bleomycin-induced lung injury (33) and reprogramming (19). In this study, cells with a fibroblastic phenotype were observed to express mTert; although infrequent in the normal heart, this population was found in association with the fibrous skeleton, the apparent fibroblastic response to injury further suggested a mesenchymal lineage. Consistent with the reported loss of telomerase expression on myofibroblast differentiation, myofibroblasts, identified by α-SMA, did not demonstrate mTert-GFP expression (34).
Although this study contradicts the suggestion that cardiac endothelial cells and fibroblasts do not express telomerase, it is unlikely to significantly underestimate total numbers of telomerase-expressing cells. Independent methods of quantification demonstrate absolute numbers with similar magnitudes, even when the inevitable cell loss associated with digestion and sorting is taken into consideration. The lack of telomerase activity and RNA in the GFP− population, and previous validation of this model, suggests strongly that there is unlikely to be a significant population of telomerase-competent cells not expressing the transgene. Given the low number of cells expressing fibroblast or endothelial markers, particularly in adult hearts, failure to identify them by conventional immunohistochemistry or TRAP assays is unsurprising; the difference in numbers of telomerase-expressing cardiomyocytes may reflect issues of antibody specificity, species differences, and/or the injury models used.
The localized expansion of populations of mTert-GFP-expressing cells in the adult heart, together with the fidelity of this model in the identification of stem cell populations (17, 18), suggests a subpopulation of mTert cells representing a stem or progenitor population. This is reinforced by mTert-GFP expression found in association with cardiac transcription factors NKX2.5 and GATA4, factors required for cardiomyocyte development and previously described as a hallmark of native cardiac stem cells. Descriptions of native cardiac stem cells, particularly those using Sca-1 as a marker, have resulted in interest in telomerase expression; Oh et al. (14) reported that all of the detectable cardiac telomerase activity was restricted to Sca-1-expressing cells in myocyte-depleted adult murine cell suspensions. We also identified Sca-1+ mTert-GFP cells, although the majority of mTert-GFP+ cells were Sca-1−. A different transgenic model driving reporter gene expression from an mTert promoter demonstrated a relationship between Sca-1 and mTert expression (35), albeit in clonally derived cell lines. Sca-1 is not an exclusive marker of cardiac stem cells and is expressed on microvascular endothelial cells (14), and we acknowledge that a proportion of the Sca1, mTert-GFP-coexpressing population will represent this. These data highlight the potential use of this mouse in the study of telomerase biology in the context of cardiac stem cell populations, although lineage-tracing models will provide a more definitive understanding of the contribution they make to various components of the heart. The finding of mTert-GFP expression in putative cardiac stem cell populations supports other studies and is compatible with the assertion that telomerase expression is a feature of stem cells, germ cells, and tumors.
Although this model allows for the identification of telomerase-expressing cells with high specificity and sensitivity, it cannot determine the role of telomerase in them, nor can the regenerative and replicative competency of the mTert-GFP populations be determined. Although all the detectable telomerase activity was restricted to the mTert-GFP-expressing population, it is possible that a proportion of this population does not possess telomerase catalytic activity, and, in fact, expresses it in its signaling role. Regardless of the mechanism, telomerase expression has been shown to maintain myocyte growth, survival, and turnover (28, 36).
The expression of mTert in a population of mature cardiomyocytes questions previous assertions that mTert is restricted to germ, stem, or tumor cells and suggests a number of hypotheses. It may represent a population of mature cardiomyocytes capable of replication (37–39), where mTert acts to maintain telomere length in its classic role. This is somewhat supported by the higher frequency of these cells in neonates, in which such a proliferative potential has been demonstrated (21, 40, 41). It could, however, be the result of GFP perdurance, identifying cells that have recently differentiated from more immature cells, perhaps stem cells. This would still support the utility of mTert-GFP expression in identifying a population responsible, at least in part, for cardiomyocyte development and regeneration. Finally, the expression of mTert in differentiated cardiomyocytes, where cell division is undoubtedly very infrequent, may represent its signaling role rather than telomere maintenance. As such activity is typically related to pathways of cell survival, this too would be consistent with the increase in expression following injury (36, 42). Lineage tracing studies are planned to examine these hypotheses.
Reduced telomerase activity and reduced telomere length have been associated with the aging male heart (11) and heart disease (8, 43). The effect of telomerase loss in the heart demonstrates a role for the low-level telomerase activity previously identified in this organ. As this model demonstrates that telomerase activity is restricted to the mTert-GFP cell population, it suggests a role for these cells, or a proportion of them, in the maintenance of normal cardiac function. We suggest that this activity arises from subpopulations of a range of cell types, associated with all components of the heart, which, although representing only a small proportion, clearly have a fundamental role. Further studies are planned to dissect their functions in this and related models, particularly in response to injury.
Supplementary Material
Acknowledgments
This work was supported by the British Heart Foundation, project grant PG/06/110/21521, U.S. National Institutes of Health grant R01 DK084056 (D.T.B), and the Harvard Stem Cell Institute (D.T.B).
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
- α-SMA
- α-smooth muscle actin
- CTnI
- cardiac troponin I
- DAPI
- 4,6-diamidino-2-phenylindole dihydrochloride
- FACS
- fluorescence-activated cell sorting
- GFP
- green fluorescent protein
- Lin−
- hematopoietic lineage negative
- mTert
- murine telomerase reverse transcriptase
- pHH3
- phospho histone H3
- qRT-PCR
- quantitative reverse transcriptase–polymerase chain reaction
- TERT
- telomerase reverse transcriptase
- TRAP
- telomeric repeat amplification protocol
REFERENCES
- 1. Blackburn E. H. (2005) Telomeres and telomerase: their mechanisms of action and the effects of altering their functions. FEBS Lett. 579, 859–862 [DOI] [PubMed] [Google Scholar]
- 2. Meyerson M., Counter C. M., Eaton E. N., Ellisen L. W., Steiner P., Caddle S. D., Ziaugra L., Beijersbergen R. L., Davidoff M. J., Liu Q., Bacchetti S., Haber D. A., Weinberg R. A. (1997) hEST2, the putative human telomerase catalytic subunit gene, is up-regulated in tumor cells and during immortalization. Cell 90, 785–795 [DOI] [PubMed] [Google Scholar]
- 3. Madonna R., De Caterina R., Willerson J. T., Geng Y. J. Biologic function and clinical potential of telomerase and associated proteins in cardiovascular tissue repair and regeneration. Eur. Heart J. 32, 1190–1196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Stewart S. A., Hahn W. C., O'Connor B. F., Banner E. N., Lundberg A. S., Modha P., Mizuno H., Brooks M. W., Fleming M., Zimonjic D. B., Popescu N. C., Weinberg R. A. (2002) Telomerase contributes to tumorigenesis by a telomere length-independent mechanism. Proc. Natl. Acad. Sci. U. S. A. 99, 12606–12611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Smith L. L., Coller H. A., Roberts J. M. (2003) Telomerase modulates expression of growth-controlling genes and enhances cell proliferation. Nat. Cell Biol. 5, 474–479 [DOI] [PubMed] [Google Scholar]
- 6. Zaccagnini G., Gaetano C., Della Pietra L., Nanni S., Grasselli A., Mangoni A., Benvenuto R., Fabrizi M., Truffa S., Germani A., Moretti F., Pontecorvi A., Sacchi A., Bacchetti S., Capogrossi M. C., Farsetti A. (2005) Telomerase mediates vascular endothelial growth factor-dependent responsiveness in a rat model of hind limb ischemia. J. Biol. Chem. 280, 14790–14798 [DOI] [PubMed] [Google Scholar]
- 7. Ahmed S., Passos J. F., Birket M. J., Beckmann T., Brings S., Peters H., Birch-Machin M. A., von Zglinicki T., Saretzki G. (2008) Telomerase does not counteract telomere shortening but protects mitochondrial function under oxidative stress. J. Cell Sci. 121, 1046–1053 [DOI] [PubMed] [Google Scholar]
- 8. Borges A., Liew C. C. (1997) Telomerase activity during cardiac development. J. Mol. Cell. Cardiol. 29, 2717–2724 [DOI] [PubMed] [Google Scholar]
- 9. Wong L. S., Oeseburg H., de Boer R. A., van Gilst W. H., van Veldhuisen D. J., van der Harst P. (2009) Telomere biology in cardiovascular disease: the TERC-/- mouse as a model for heart failure and ageing. Cardiovasc. Res. 81, 244–252 [DOI] [PubMed] [Google Scholar]
- 10. Oh H., Taffet G. E., Youker K. A., Entman M. L., Overbeek P. A., Michael L. H., Schneider M. D. (2001) Telomerase reverse transcriptase promotes cardiac muscle cell proliferation, hypertrophy, and survival. Proc. Natl. Acad. Sci. U. S. A. 98, 10308–10313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Leri A., Quaini F., Kajstura J., Anversa P. (2001) Myocyte death and myocyte regeneration in the failing human heart. Ital. Heart. J. 2(Suppl. 3), 12S–14S [PubMed] [Google Scholar]
- 12. Sussman M. A., Anversa P. (2004) Myocardial aging and senescence: where have the stem cells gone? Annu. Rev. Physiol. 66, 29–48 [DOI] [PubMed] [Google Scholar]
- 13. Torella D., Rota M., Nurzynska D., Musso E., Monsen A., Shiraishi I., Zias E., Walsh K., Rosenzweig A., Sussman M. A., Urbanek K., Nadal-Ginard B., Kajstura J., Anversa P., Leri A. (2004) Cardiac stem cell and myocyte aging, heart failure, and insulin-like growth factor-1 overexpression. Circ. Res. 94, 514–524 [DOI] [PubMed] [Google Scholar]
- 14. Oh H., Bradfute S. B., Gallardo T. D., Nakamura T., Gaussin V., Mishina Y., Pocius J., Michael L. H., Behringer R. R., Garry D. J., Entman M. L., Schneider M. D. (2003) Cardiac progenitor cells from adult myocardium: homing, differentiation, and fusion after infarction. Proc. Natl. Acad. Sci. U. S. A. 100, 12313–12318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wu Y. L., Dudognon C., Nguyen E., Hillion J., Pendino F., Tarkanyi I., Aradi J., Lanotte M., Tong J. H., Chen G. Q., Segal-Bendirdjian E. (2006) Immunodetection of human telomerase reverse-transcriptase (hTERT) re-appraised: nucleolin and telomerase cross paths. J. Cell Sci. 119, 2797–2806 [DOI] [PubMed] [Google Scholar]
- 16. Armstrong L., Lako M., Lincoln J., Cairns P. M., Hole N. (2000) mTert expression correlates with telomerase activity during the differentiation of murine embryonic stem cells. Mech. Dev. 97, 109–116 [DOI] [PubMed] [Google Scholar]
- 17. Breault D. T., Min I. M., Carlone D. L., Farilla L. G., Ambruzs D. M., Henderson D. E., Algra S., Montgomery R. K., Wagers A. J., Hole N. (2008) Generation of mTert-GFP mice as a model to identify and study tissue progenitor cells. Proc. Natl. Acad. Sci. U. S. A. 105, 10420–10425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Montgomery R. K., Carlone D. L., Richmond C. A., Farilla L., Kranendonk M. E., Henderson D. E., Baffour-Awuah N. Y., Ambruzs D. M., Fogli L. K., Algra S., Breault D. T. Mouse telomerase reverse transcriptase (mTert) expression marks slowly cycling intestinal stem cells. Proc. Natl. Acad. Sci. U. S. A. 108,, 179–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Stadtfeld M., Maherali N., Breault D. T., Hochedlinger K. (2008) Defining molecular cornerstones during fibroblast to iPS cell reprogramming in mouse. Cell Stem Cell 2, 230–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gonzalez-Suarez E., Geserick C., Flores J. M., Blasco M. A. (2005) Antagonistic effects of telomerase on cancer and aging in K5-mTert transgenic mice. Oncogene 24, 2256–2270 [DOI] [PubMed] [Google Scholar]
- 21. Walsh S., Ponten A., Fleischmann B. K., Jovinge S. Cardiomyocyte cell cycle control and growth estimation in vivo—an analysis based on cardiomyocyte nuclei. Cardiovasc. Res. 86, 365–373 [DOI] [PubMed] [Google Scholar]
- 22. Leferovich J. M., Bedelbaeva K., Samulewicz S., Zhang X. M., Zwas D., Lankford E. B., Heber-Katz E. (2001) Heart regeneration in adult MRL mice. Proc. Natl. Acad. Sci. U. S. A. 98, 9830–9835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Greider C. W. (1998) Telomerase activity, cell proliferation, and Cancer Proc. Natl. Acad. Sci. U. S. A. 95, 90–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Endl E., Gerdes J. (2000) The Ki-67 protein: fascinating forms and an unknown function. Exp. Cell Res. 257, 231–237 [DOI] [PubMed] [Google Scholar]
- 25. Juan G., Traganos F., James W. M., Ray J. M., Roberge M., Sauve D. M., Anderson H., Darzynkiewicz Z. (1998) Histone H3 phosphorylation and expression of cyclins A and B1 measured in individual cells during their progression through G2 and mitosis. Cytometry 32, 71–77 [DOI] [PubMed] [Google Scholar]
- 26. Blasco M. A. (2002) Mouse models to study the role of telomeres in cancer, aging and DNA repair. Eur. J. Cancer 38, 2222–2228 [DOI] [PubMed] [Google Scholar]
- 27. Leri A., Malhotra A., Liew C. C., Kajstura J., Anversa P. (2000) Telomerase activity in rat cardiac myocytes is age and gender dependent. J. Mol. Cell. Cardiol. 32, 385–390 [DOI] [PubMed] [Google Scholar]
- 28. Leri A., Barlucchi L., Limana F., Deptala A., Darzynkiewicz Z., Hintze T. H., Kajstura J., Nadal-Ginard B., Anversa P. (2001) Telomerase expression and activity are coupled with myocyte proliferation and preservation of telomeric length in the failing heart. Proc. Natl. Acad. Sci. U. S. A. 98, 8626–8631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhu X., Kumar R., Mandal M., Sharma N., Sharma H. W., Dhingra U., Sokoloski J. A., Hsiao R., Narayanan R. (1996) Cell cycle-dependent modulation of telomerase activity in tumor cells. Proc. Natl. Acad. Sci. U. S. A. 93, 6091–6095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Erusalimsky J. D., Skene C. (2009) Mechanisms of endothelial senescence. Exp. Physiol. 94, 299–304 [DOI] [PubMed] [Google Scholar]
- 31. Kurz D. J., Erusalimsky J. D. (2003) Role of telomerase in human endothelial cell proliferation. Arterioscler. Thromb. Vasc. Biol. 23, e54. [DOI] [PubMed] [Google Scholar]
- 32. Kurz D. J., Hong Y., Trivier E., Huang H. L., Decary S., Zang G. H., Luscher T. F., Erusalimsky J. D. (2003) Fibroblast growth factor-2, but not vascular endothelial growth factor, upregulates telomerase activity in human endothelial cells. Arterioscler. Thromb. Vasc. Biol. 23, 748–754 [DOI] [PubMed] [Google Scholar]
- 33. Liu T., Nozaki Y., Phan S. H. (2002) Regulation of telomerase activity in rat lung fibroblasts. Am. J. Respir. Cell Mol. Biol. 26, 534–540 [DOI] [PubMed] [Google Scholar]
- 34. Liu T., Hu B., Chung M. J., Ullenbruch M., Jin H., Phan S. H. (2006) Telomerase regulation of myofibroblast differentiation. Am. J. Respir. Cell Mol. Biol. 34, 625–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tateishi K., Ashihara E., Takehara N., Nomura T., Honsho S., Nakagami T., Morikawa S., Takahashi T., Ueyama T., Matsubara H., Oh H. (2007) Clonally amplified cardiac stem cells are regulated by Sca-1 signaling for efficient cardiovascular regeneration. J. Cell Sci. 120, 1791–1800 [DOI] [PubMed] [Google Scholar]
- 36. Leri A., Franco S., Zacheo A., Barlucchi L., Chimenti S., Limana F., Nadal-Ginard B., Kajstura J., Anversa P., Blasco M. A. (2003) Ablation of telomerase and telomere loss leads to cardiac dilatation and heart failure associated with p53 upregulation. EMBO J. 22, 131–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Marino T. A., Haldar S., Williamson E. C., Beaverson K., Walter R. A., Marino D. R., Beatty C., Lipson K. E. (1991) Proliferating cell nuclear antigen in developing and adult rat cardiac muscle cells. Circ. Res. 69, 1353–1360 [DOI] [PubMed] [Google Scholar]
- 38. Kajstura J., Zhang X., Reiss K., Szoke E., Li P., Lagrasta C., Cheng W., Darzynkiewicz Z., Olivetti G., Anversa P. (1994) Myocyte cellular hyperplasia and myocyte cellular hypertrophy contribute to chronic ventricular remodeling in coronary artery narrowing-induced cardiomyopathy in rats. Circ. Res. 74, 383–400 [DOI] [PubMed] [Google Scholar]
- 39. Anversa P., Palackal T., Sonnenblick E. H., Olivetti G., Meggs L. G., Capasso J. M. (1990) Myocyte cell loss and myocyte cellular hyperplasia in the hypertrophied aging rat heart. Circ. Res. 67, 871–885 [DOI] [PubMed] [Google Scholar]
- 40. Beltrami A. P., Barlucchi L., Torella D., Baker M., Limana F., Chimenti S., Kasahara H., Rota M., Musso E., Urbanek K., Leri A., Kajstura J., Nadal-Ginard B., Anversa P. (2003) Adult cardiac stem cells are multipotent and support myocardial regeneration. Cell 114, 763–776 [DOI] [PubMed] [Google Scholar]
- 41. Porrello E. R., Mahmoud A. I., Simpson E., Hill J. A., Richardson J. A., Olson E. N., Sadek H. A. Transient regenerative potential of the neonatal mouse heart. Science 331, 1078–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Oh H., Wang S. C., Prahash A., Sano M., Moravec C. S., Taffet G. E., Michael L. H., Youker K. A., Entman M. L., Schneider M. D. (2003) Telomere attrition and Chk2 activation in human heart failure. Proc. Natl. Acad. Sci. U. S. A. 100, 5378–5383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yamaguchi Y., Nozawa K., Savoysky E., Hayakawa N., Nimura Y., Yoshida S. (1998) Change in telomerase activity of rat organs during growth and aging. Exp. Cell Res. 242, 120–127 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.