Abstract
Phospholipase Cβ1 (PLCβ1) is a G-protein-regulated enzyme whose activity results in proliferative and mitogenic changes in the cell. We have previously found that in solution PLCβ1 binds to the RNA processing protein translin-associated factor X (TRAX) with nanomolar affinity and that this binding competes with G proteins. Here, we show that endogenous PLCβ1 and TRAX interact in SK-N-SH cells and also in HEK293 cells induced to overexpress PLCβ1. In HEK293 cells, TRAX overexpression ablates Ca2+ signals generated by G protein-PLCβ1 activation. TRAX plays a key role in down-regulation of proteins by small, interfering RNA, and PLCβ1 overexpression completely reverses the 2- to 4-fold down-regulation of GAPDH by siRNA in HEK293 and HeLa cells as seen by an ∼4-fold recovery in both the transcript and protein levels. Also, down-regulation of endogenous PLCβ1 in HEK293 and HeLa cells allows for an ∼20% increase in siRNA(GAPDH) silencing. While PLCβ1 overexpression results in a 50% reversal of cell death caused by siRNA(LDH), it does not affect cell survival or silencing of other genes (e.g., cyclophilin, Hsp90, translin). PLCβ1 overexpression in HEK293 and HeLa cells causes a 30% reduction in the total amount of small RNAs. LDH and GAPDH are part of a complex that promotes H2B synthesis that allows cells to progress through the S phase. We find that PLCβ1 reverses the cell death and completely rescues H2B levels caused by siRNA knockdown of LDH or GAPDH. Taken together, our study shows a novel role of PLCβ1 in gene regulation through TRAX association.—Philip, F., Guo, Y., Aisiku, O., Scarlata, S. Phospholipase Cβ1 is linked to RNA interference of specific genes through translin-associated factor X.
Keywords: G protein signaling, RNA transcription, histone H2B
The Gαq family of G proteins, which are activated through many hormones and neurotransmitters to transmembrane receptors, generates cell signals primarily through the activation of phospholipase Cβ (PLCβ) enzymes (for reviews see refs. 1, 2). These enzymes catalyze the hydrolysis of the signaling lipid phosphatidylinositol 4,5-bisphosphate [PI(4,5)P2] to produce the second messengers inositol 1,4,5-trisphosphate and diacylglycerol, which stimulate the release of Ca2+ from intracellular stores and activate protein kinase C, respectively, and lead to mitogenic and proliferative changes in the cell. The four known PLCβ subtypes differ in their sensitivity to G proteins and their tissue distribution. PLCβ1 is highly expressed in neuronal tissue and is the most sensitive to Gαq. PLCβ1 resides predominantly on the plasma membrane, where it is associated with Gαq (3). In addition, PLCβ1 can be found in the cytosol and the nucleus. The nuclear localization of PLCβ1 depends on the state of cellular differentiation, cell cycle, and environmental conditions (see refs. 4, 5), which may be triggered by the exposure of PLCβ1's nuclear localization signal (NLS) and/or phosphorylation (6). The regulation and function of PLCβ1 in these alternate cellular compartments are unknown.
We have found recently that the protein translin-associated factor X (TRAX) is a strong binding partner of PLCβ1 (7). Interaction between the two proteins occurs through the helical C-terminal domain of PLCβ1, which is also the binding region of Gαq, and excess TRAX can displace Gαq from PLCβ1, resulting in deactivation of the enzyme. TRAX forms strong complexes with translin (8). Translin is a single-stranded DNA- and RNA-binding protein (9) with proposed functions in chromosomal translocations in lymphoid cells, and mRNA transport and storage in brain and testis (10). TRAX binds translin in the cytosol, which allows the complex to partition into the nucleus through TRAX's NLS (11). Recently, it was found that TRAX and translin form an octamer that activates RNA-induced silencing complexes (RISCs; refs. 12–14). In humans, siRNA down-regulation occurs through a pathway in which the duplex siRNA is guided onto the protein Argonaute-2 (Ago2) of RISC (see ref. 15). C3PO, a complex consisting of 2 TRAX:6 translin molecules, promotes RISC activity by facilitating siRNA unwinding and removal of the cleaved passenger strand. In humans, siRNA activity can be reconstituted by the TRAX-translin octamer and Ago2 (12), and unlike Drosphilia, Dicer-2 (Dcr2) is not required (13).
While PLCβ1 and TRAX have different cellular functions, knockout mice of these proteins show similar neurological symptoms (e.g., refs. 16–18). In addition, knocking out TRAX has been reported to down-regulate gene expression of several G protein regulated genes, including two Gαq-coupled receptors (17, 19). We have shown here that PLCβ1 associates to TRAX in cells, affecting its RISC activity. This effect is seen by the reversal of siRNA down-regulation by overexpressed PLCβ1 of genes to two metabolic enzymes, lactate dehydrogenase (LDH) and glyceraldehyde 3-phosphate dehydrogenase (GAPDH), that promote the transcription of histone H2B (20, 21) allowing cells to progress through the S phase. Our studies suggest a novel link between Gαq, and transcriptional regulation and cell cycle progression through PLCβ-TRAX interactions.
MATERIALS AND METHODS
Materials
Human embryonic kidney (HEK)293-β1 and HEK293-TAP-PLCβ1 cells and enhanced yellow fluorescent protein (eYFP)-PLCβ1 were generous gifts from Dr. Loren Runnels (University of Medicine and Dentistry of New Jersey, Piscataway, NJ, USA). These cells were prepared by introducing PLCβ1 tagged with a 38-aa strepavidin-binding peptide on the N terminus (Sigma, St. Louis, MO, USA) using the Flp-In system (Invitrogen, Carlsbad, CA, USA). It has been shown that the fluorescent tagged PLCβ1 shows wild-type behavior in transfected cells. Mouse monoclonal and rabbit polyclonal PLCβ1 antibodies and mouse monoclonal and goat polyclonal primary antibodies for TRAX were obtained from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA, USA). An additional mouse monoclonal antibody for TRAX was from BD Biosciences (San Jose, CA, USA). Primary antibodies for Dcr2 (rabbit) and Ago-2 (mouse) were from Abcam, Inc. (Cambridge, MA, USA).
Cell culture
HEK293-β1 cells were cultured in Dulbecco's modified Eagle's medium (DMEM) with 10% fetal bovine serum at 37°C with 5% CO2.
eYFP-PLCβ1, Trax–enhanced cyan fluorescent protein (eCFP), or Gαq-RC-eCFP were transfected into HEK293 cells with FuGene-HD (Promega, Madison, WI, USA) using the manufacturer's protocol or by electroporation using a protocol adapted from Maniatis and colleagues (22) as described previously (3). Proteins were viewed by immunofluorescence using the methodology described in ref. 23.
Western blotting
The cell lysates were subjected to gel electophoresis, followed by the transfer of the samples to a nitrocellulose membrane. The membrane was then incubated for 1 h with 1% BSA to block the nonspecific binding. The primary antibody, diluted according to the manufacturer's instructions in Tris-buffered saline with 0.5% Tween (TBST), was added to the membrane and incubated for 1 h. This was followed by 3 washes of 15 min each in TBST. After the addition of horseradish peroxidase-conjugated secondary antibody for 1 h, the membrane was washed 3 times for 15 min each in TBST. The membrane was either exposed to a photographic film or subjected to imaging after a chemiluminescence reaction (Pierce, Inc., Rockford, IL, USA).
Pulldown studies
HEK293-β1 cells, treated with tetracycline for 24 h to express PLCβ1 with a streptavidin peptide tag, were washed twice with ice-cold PBS and lysed with 150 mM NaCl, 20 mM HEPES (pH 7.4), 2 mM MgCl2, 5 mM β-mercaptoethanol, 1 mM PMSF, 10 μg/ml leupeptin, and 10 μg/ml aprotinin by a homogenizer. The lysate was added to streptavidin beads and rotated for 1 h at 4°C. The beads were washed with the lysis buffer and eluted with buffer containing 50 mM Tris-HCl (pH 8.0), 150 mM KCl, 2 mM EDTA, 5 mM β-mercaptoethanol, 100 μg/ml BSA, and 2 mM biotin.
Coimmunoprecipitation
To confirm the results on TRAX pulldown by PLCβ1 through the streptavidin-based system, we performed a coimmunoprecipitation experiment using PLCβ1 antibodies on SK-N-SH cells. The cells were lysed by passing the cells 10 times through a 25-gauge needle, followed by 20 strokes of homogenization in lysis buffer containing 50 mM NaCl, 20 mM HEPES, 2 mM MgCl2, 5 mM β-mercaptoethanol, and protease inhibitors. The cell lysates were incubated overnight at 4°C with PLCβ1 antibodies that were prebound to proteinA-Sepharose beads. After washing the beads 3 times in lysis buffer, the proteins were eluted from the beads in electrophoresis sample buffer at 95°C for 5 min.
Separation of nuclear and cytosolic cell fractions
HEK293 cells were harvested by centrifugation at 600 g for 5 min. Cells were resuspended in 5 vol of PBS at 4°C and centrifuged 5 min at 600 g. The washed pellet was resuspended in 10 vol of a hypotonic buffer solution (Active Motif Inc., Carlsbad, CA, USA) and placed on ice for 10 min. Cells were checked for swelling under a phase-contrast microscope. Once 90% had swollen, they were pipetted into a prechilled Dounce-type homogenizer and subjected to 10–12 upward strokes. This process was continued until <10% whole cells remained. The suspension was centrifuged at 1000 g for 3 min to pellet the nuclei, and the pellet was resuspended in 10 vol of the hypotonic buffer solution. This latter step was repeated twice. Verification of the nuclear and cytosolic extraction was additionally performed using a commercial kit from Thermo Scientific (Waltham, MA, USA) and following the manufacturer's protocol.
siRNA studies
siRNA for GAPDH, hsp90, cyclophilin A, translin, PLCβ1, and negative control were purchased from Dharmacon Inc. (Lafayette, CO, USA) The purchased siRNAs are from the company's On-Target Plus products, consisting of a mixture of four different RNA sequences that target different regions of the specific mRNA with a low probability of off-target interactions. Proteins were down-regulated following the manufacturer's protocol. PLCβ1expression in HEK293-β1 was induced by 1 μg/ml tetracycline for 40–50% confluence 24 h before knockdown. Untreated cells were used as control. Live cells were counted at 12, 24, 48, and 72 h postknockdown and were lysed after harvesting. Electrophoresis was done on both detached cells and attached cell lysate to check for protein expression.
Ca+2 measurements
Tetracycline-treated HEK293-β1 cells that were transfected with eCFP-Gαq and/or TRAX-eCFP (to assess transfection efficiency) and harvested after 48 h. Cellular calcium levels were determined using Fura-2-AM (Invitrogen) using an ISS spectrofluorometer (ISS Inc., Champaign, IL, USA) as described previously (24).
RT- PCR
RNA was extracted by the RNeasy mini system (Qiagen, Valencia, CA, USA) followed by cDNA synthesis with the use of QuantiTect Reverse Transcriptase (Qiagen). The primers were from Applied Biosciences (Foster City, CA, USA). Quantitative real-time PCR was performed on the cDNA samples using TaqMan method (Applied Biosciences) on an MJ Research Opticon II system (MJ Research. St. Bruno, QC, Canada). The GAPDH expression levels in each sample were normalized to β-actin or ubiquitin levels. The relative expressions of GAPDH mRNA levels were calculated using the comparative CT method (2−ΔΔCt; see ref. 25).
RNA gel electrophoresis
Electrophoretic mobility of RNA samples of same concentration was monitored on a 2% agarose gel. Ethidium bromide fluorescence intensities of bands lower than 600 bp were quantified for both groups using ImageJ (U.S. National Institutes of Health, Bethesda, MD, USA) and compared using a paired t test.
RESULTS
PLCβ1 and TRAX are associated in HEK293 cells
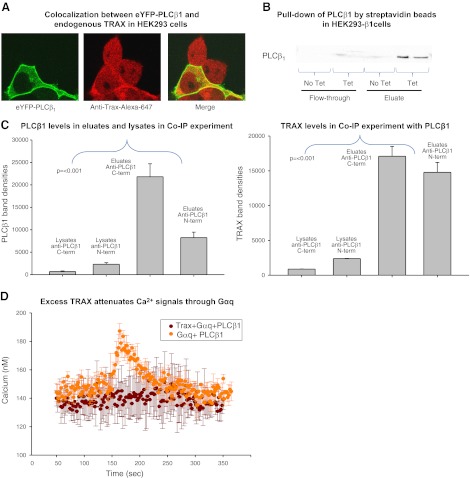
We have found previously that purified PLCβ1 and TRAX strongly associate in solution, and endogenous as well as overexpressed PLCβ1 and TRAX are colocalized in the cytosol and nucleus of C6 glial cells and Neuro2A cells (7). Here, we used wild-type HEK293 (wtHEK29) cells or ones containing a PLCβ1 gene whose expression is under regulation of a tetracycline promoter (HEK293-β1; see Materials and Methods). Figure 1A shows the colocalization of eYFP-PLCβ1 in transfected wtHEK293 cells and endogenous TRAX, visualized using a polyclonal antibody. In accord with previous studies (3), PLCβ1 localizes primarily to the plasma membrane but still has a significant cytosolic population. Alternately, TRAX is found in the cytosol and nucleus but not on the plasma membrane. The absence of TRAX on the plasma membrane is thought to be due to a lack of intrinsic membrane affinity, as well as competition for PLCβ1 binding sites by Gαq (7). Cellular association of PLCβ1 and TRAX was verified by probing for TRAX after immobilizing PLCβ1, which has a streptavidin peptide tag that can bind to streptavidin beads, expressed in tetracycline-treated HEK293-β1 cells (Fig. 1B). To support the idea that endogenous PLCβ1 and TRAX associate in cells, we carried out coimmunoprecipitation studies in SK-N-SH cells that express easily detectable levels of these proteins. The results in Fig. 1C suggest association between the proteins. In another series of studies, we used Förster resonance energy transfer to verify the association of fluorescent-tagged PLCβ1 and TRAX expressed in HEK293 cells; these results are included in Supplemental Data.
Figure 1.
Evidence that PLCβ1 and TRAX associate in HEK293 cells. A) HEK293 cells expressing eYFP-PLCβ1 (left panel), and immunostained with anti-TRAX using an Alexa647 secondary antibody (middle panel). The figure is representative of 12 or more images and 4 independent studies. No bleed-through between the YFP and Alexa647 channels was detected. Degree of colocalization (yellow) of the two proteins is found in the cytosol and nucleus (right panel). B) Pulldown assay in which endogenous TRAX and PLCβ1, tagged on the N terminus with a strepavidin-binding peptide that was induced to express in HEK293-β1 cells, were copurified using beads coated with streptavidin (see Materials and Methods) and eluted in buffer containing biotin. C) Band intensity of PLCβ1 and TRAX pulled down by PLCβ1 through coimmunoprecipitation (Co-IP) in SKNSH cells. Two different PLCβ1 antibodies (Santa Cruz Biotechnology), one with an epitope at the C terminus and another at the N terminus, were used to pull down PLCβ1. Levels of PLCβ1 in the eluates were significantly higher than those in the lysates, indicating that PLC was being pulled down by the antibodies, with the C-terminal antibody having a higher affinity for PLCβ1. Higher levels of TRAX in the eluates indicate that TRAX was being concentrated by PLCβ1. We observed similar results from HEK cells (data not shown). D) Change in cellular Ca2+ with stimulation by 10 μM carbachol; cells overexpressing PLCβ1 and Gαq-eCFP (n=3) are shown in orange. Calcium levels in cells expressing Trax-eCFP, PLCβ1, and Gαq-eCFP (n=3) did not increase on stimulation and are shown in brown.
In vitro studies showed that TRAX competes with Gαq for PLCβ1 binding and attenuates its activation (7). To determine whether this competition occurs in cells, we transfected tetracycline-treated HEK293-β1 cells with TRAX and cotransfected with Gαq to provide enough G protein to activate overexpressed PLCβ1. We found that TRAX eliminates Ca2+ signals produced by carbachol stimulation (Fig. 1D) supporting the idea that TRAX binds to PLCβ1 intracellularly to reverse Gαq activation.
Gαq stimulation promotes movement of a population of TRAX to the nucleus
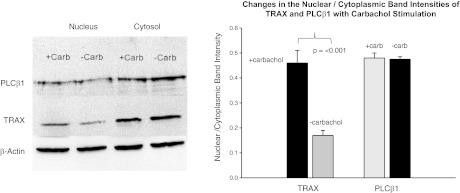
In addition to its role in RNA silencing in the cytosol, TRAX has proposed roles in functions in the nucleus (10). Since Gαq and TRAX bind to the same region of PLCβ1, and since the affinity between PLCβ1 and Gαq increases ∼20-fold with activation, activating Gαq should disrupt TRAX-PLCβ1 complexes, which can result in changes in TRAX localization. This idea was tested by treating HEK293-β1 cells with tetracycline to induce PLCβ1 expression, stimulating with carbachol for 5 min or 30 min (both gave identical results), and following localization by cell fractionation and Western blotting. While we could not detect movement of PLCβ1, we found that a population of TRAX moved to the nucleus (Fig. 2). This result correlates well with the observation that carbachol stimulation results in a 46% drop in TRAX-PLCβ1 colocalization that may accompany movement of TRAX with stimulation (Table 1), and also immunofluorescence studies support changes in TRAX properties with Gαq activation (Supplemental Data). These studies suggest that TRAX localization is responsive to G protein activation.
Figure 2.
Movement of TRAX to the nucleus with cell stimulation. Left panel: Western blot results of nuclear and cytosolic fractions of TRAX and PLCβ1 from HEK293 cells untreated or treated with 1μM carbachol and incubated for 5 min before lysing and fractionating. Treatment for 30 min gave identical results. Image is a composite of 4 independent experiments and was carried out by digitizing the band density and normalizing each band to actin loading controls. Right panel: comparison of the nuclear/cytosolic intensity of cells (see Materials and Methods) stained with Alexa488 anti-TRAX monoclonal (Santa Cruz Biotechnology) for 46–58 cells before and 5 min after carbachol stimulation.
Table 1.
Summary of colocalization studies
| Colocalization | Treatment |
||||
|---|---|---|---|---|---|
| GAPDH siRNA | Hsp90 siRNA | Carbachol stimulation | Adenosine stimulation | Control | |
| PLC-Trax | 1.65 ± 0.13 | 1.91 ± 0.18 | 0.46 ± 0.12 | 1 ± 0.19 | |
| PLC-Ago2 | 1.64 ± 0.2 | 1.06 ± 0.13 | 0.95 ± 0.05 | 0.95 ± 0.05 | 1 ± 0.1 |
| PLC-Dcr | 0.8 ± 0.07 | 1.04 ± 0.12 | 1.01 ± 0.06 | 1 ± 0.05 | |
Data are averages of multiple images with each image consisting of 1–4 cells. Number of images analyzed for each group varies from 9 to 20. Data represent either multiple group comparison or compilation of pairwise independent experiments, all normalized to control group. For comparisons between multiple groups, 1-way ANOVA was used; for pairwise comparisons, t test was used.
TRAX has been found to bind to the long cytosolic tail of adenosine 2A receptors. These receptors are coupled to Gαi/o subunits. We tested whether activation of these receptors would similarly result in changes in TRAX localization. Stimulation of HEK293-β1 untreated or treated with tetracycline with adenosine did not affect the localization of TRAX (Supplemental Data). In addition, TRAX does not appear to affect the cellular localization of PLCβ1 in the basal and stimulated states of tetracycline-treated HEK293-β1 cells, suggesting that TRAX interaction does not significantly alter the ratio of nuclear to cytoplasmic PLCβ1 (see Fig. 4 and Supplemental Data).
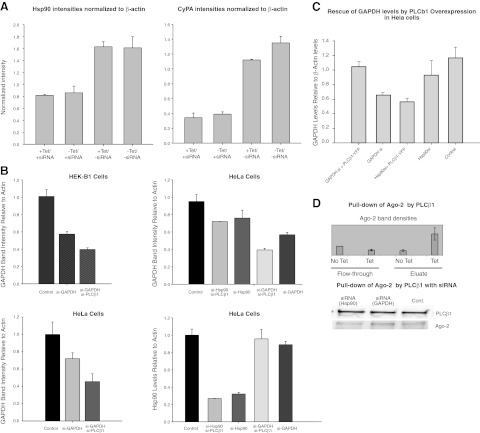
Figure 4.
PLCβ1 expression does not affect siRNA down-regulation of several proteins. A) Left panels: identical study as described in Fig. 3, except using siRNAs to cyclophilin and Hsp90; n = 4. B) Reduction in PLCβ1 levels promotes siRNA(GAPDH) activity in uninduced HEK-β1 and HeLa cells. Western blot band intensities of GAPDH and Hsp90 of cells with various siRNA treatment; n = 2. C) Control study showing that PLCβ1 overexpression reverses siRNA(GAPDH) in HeLa cells. D) Top panel: level of Ago-2 pulled down by PLCβ1 in tetracycline vs. untreated cells; n = 2. Bottom panel: Western blot bands corresponding to PLCβ1 that were pulled down by streptavidin beads from HEK293 cells induced for PLCβ1 expression, and treated with either siRNA(GAPDH), siRNA(Hsp90) or no siRNA. Error bars = sd.
PLCβ1 overexpression reverses the activity of specific siRNAs
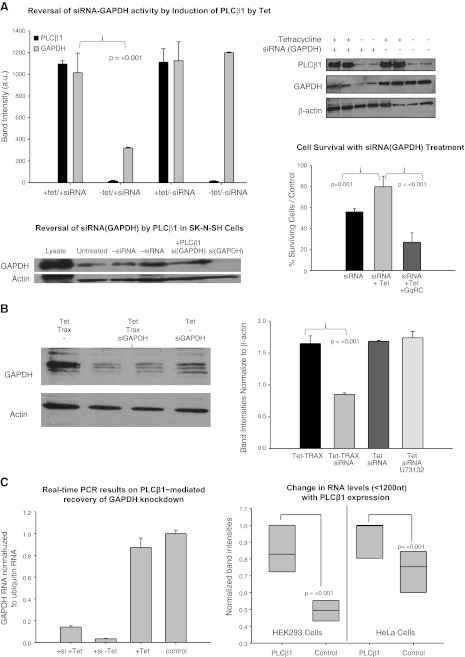
TRAX plays a key role in RNA silencing, and we tested whether overproduction of PLCβ1 would affect its regulation of RISC and the efficacy of RNA interference. To this end, we down-regulated the gene for the housekeeping enzyme, GAPDH, and determined the effect of PLCβ1 overproduction in HEK293-β1 cells. We found that siRNA(GAPDH) treatment resulted in an increase in cell death. However, when we treated the cells with tetracycline to induce PLCβ1 expression, survival of the siRNA(GAPDH)-treated cells largely recovered (Fig. 3A, bottom right panel). A similar result of PLCβ1-mediated rescue of GAPDH knockdown was also seen in Hela cells (Fig. 3C). Coexpression of a constitutively active form of Gαq, Gαq(RC) (26), which would bind PLCβ1 and compete with TRAX association, reversed this recovery (Fig. 3A, bottom right panel). Western blot analysis of the different cell groups showed that the protein level of GAPDH in the siRNA-treated cells was restored when PLCβ1 was overexpressed (Fig. 3A). This result was unchanged when cells were additionally treated with a phospholipase C inhibitor (10 μM U73122), suggesting that the reversal of siRNA(GAPDH) by PLCβ1 is independent of its phospholipase activity (Fig. 3B). Quantitative RT-PCR studies show the GAPDH transcript levels are reduced with siRNA treatment but show the same recovery of siRNA down-regulation by overexpression of PLCβ1 (Fig. 3C, left panel). A global effect of PLCβ1 on lower-molecular-weight RNAs is seen when PLCβ1 is overexpressed in HEK29 cells by tetracycline treatment or in HeLa cells by transient transfection (Fig. 3C, right panel), supporting a relationship between this enzyme and RNA transcription or processing.
Figure 3.
Increased expression of PLCβ1 abolishes siRNA (GAPDH) activity. A) Top panels: HEK293-TAP-PLCβ1 cells were treated with siRNA(GAPDH) and/or tetracycline (tet) to induce PLCβ1 expression. Cells treated only with siRNA(GAPDH) had diminished protein expression, which was reversed in cells also treated with tetracycline. All bands were normalized according to the actin loading control, n = 3. Error bars = sd. Bottom left panel: Representative Western blot showing the rescue of GAPDH levels in SKNSH cells with PLCβ1-eYFP overexpression (n=4). Bottom right panel: degree of cell survival of siRNA(GAPDH)-treated cells as normalized by untreated controls, showing the rescue of cell death by PLCβ1 overexpression and the reversal of this effect by cotransfection with the constitutively active Gαq(RC). B) A similar study, except cells were transiently transfected with eCFP-TRAX with tetracycline treatment to restore siRNA(GAPDH) activity, as seen by a reduction in GAPDH expression. Inclusion of 10 μM of the PLC inhibitor U73122 did not affect the reversal of siRNA(GAPDH) by PLCβ1 overexpression. Bands were normalized using the actin loading control, n = 3. Error bars = sd. C) Left panel: relative levels of GAPDH mRNA, as determined by RT-PCR (see Materials and Methods) for showing a recovery of GAPDH mRNA with PLCβ1 overexpression in siRNA treated cells where the levels were normalized to ubiquitin mRNA (n=3, P=0.016). Right panel: change in the levels of small RNAs (i.e., <1600 nt) with PLCβ1 overexpression either by tetracycline treatment of HEK293-β1 cells or transient transfection of HeLa cells (36 h). Bands were quantified by Image J and normalized to the highest value. Each set was compared using a paired t test; n = 6.
If overproduction of PLCβ1 reverses siRNA(GAPDH) activity through its interaction with TRAX, then overproduction of TRAX should reverse this effect. We tested this idea by overexpressing TRAX in HEK293-β1 cells and determining the amount of GAPDH expressed in tetracycline-treated cells in the presence and absence of siRNA(GAPDH). We found that TRAX overproduction rescued siRNA(GAPDH) down-regulation (Fig. 3B) in the presence of elevated PLCβ1. The results in Fig. 3A, B show a connection between PLCβ1/TRAX and siRNA(GAPDH) activity.
To test whether PLCβ1 overproduction could reverse siRNA knockdown of genes encoding other proteins, we repeated the above study using siRNA of two other ubiquitous proteins, Hsp90 and cyclophilin A. Unlike GAPDH, down-regulation of these proteins by 50–80% did not affect growth or survival of HEK293-β1 cells. Also unlike GAPDH, induction of PLCβ1 did not reverse the down-regulation of these proteins (Fig. 4A). We tested whether PLCβ1 could rescue the down-regulation of translin (data not shown). We found that overexpression of PLCβ1 did not change the translin levels, suggesting that PLCβ1 selectively rescues GAPDH levels.
Down-regulation of PLCβ1 promotes siRNA(GAPDH) activity
To support the idea that PLCβ1 overproduction reverses siRNA activity of siRNA(GAPDH), we down-regulated PLCβ1 in uninduced HEK293-β1 cells and in HeLa cells and measured the potency of siRNA(GAPDH). We found that when PLCβ1 is down-regulated, siRNA down-regulation of GAPDH, is more robust (Fig. 4B). It is notable that treatment with siRNA(Hsp90) reduces the protein level of GAPDH, although si(RNA)PLCβ1 does not increase this effect. PLCβ1 does not interfere with RNA silencing of Hsp90 (Fig. 4B). As a control, we show that overexpression of PLCβ1 reverses siRNA(GAPDH) in this cell line (Fig. 4C).
Our results suggest that PLCβ1 affects TRAX-associated RISC activity toward some genes but not others. Since siRNA activity is mediated by the protein Ago2, we tested whether PLCβ1 associates with Ago2 using a pulldown assay in HEK293-β1 cells. We found that the association between these proteins in nontransfected cells and in cells transfected with different siRNAs differs (Fig. 4D). These data support the idea that PLCβ1 is associated with RISC proteins but that its expression only affects siRNA regulation of specific genes.
PLCβ1 and TRAX interact with RISC components differently on GAPDH vs. Hsp90 knockdown
It is possible that the effect of PLCβ1 on GAPDH down-regulation vs. Hsp90, cyclophilin A, and translin reflects differences in RISC assembly. To this end, we compared the colocalization between PLCβ1, TRAX, and two other RISC components, Ago2 and Dcr 2, in the presence of siRNA(GAPDH) and siRNA(Hsp90). The results are shown in Table 1. Treatment with Hsp90 siRNA showed an increase in colocalization between PLCβ1 and TRAX, but PLCβ1/Ago2 and PLCβ1/Dcr2 colocalization remained constant. Alternately, treatment with siRNA(GAPDH) affected the colocalization of all pairs. Interestingly, the colocalization between PLCβ1 and Ago2 increased even though less Ago2 is seen in pulldown studies (see Fig. 4), suggesting a reorganization of the complex with siRNA(GAPDH). These results strongly suggest that differences in RISC composition may underlie differences in the ability of PLCβ1 to reverse siRNA activity.
PLCβ1 can affect histone H2B synthesis through GAPDH/LDH regulation
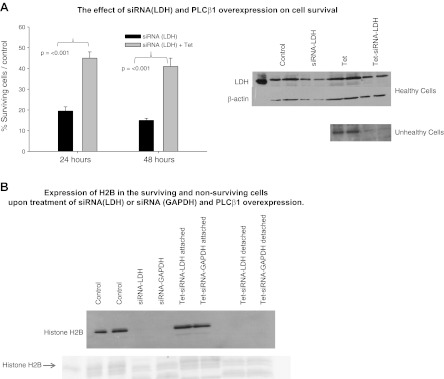
Aside from its classic metabolic function, GAPDH also resides in a complex with another metabolic enzyme, LDH, that promotes expression of histone H2B (20). We reasoned that PLCβ1 may specifically target the siRNA activity of these functionally related enzymes. To this end, we transfected HEK293-β1 cells with siRNA(LDH [type A]) and found that a large portion of the siRNA-treated cells died as compared to control siRNA-treated cells (Fig. 5A). However, induction of PLCβ1 expression increased the percentage of surviving cells, suggesting that PLCβ1 reversed the deleterious effects of siRNA(LDH) treatment. Using Western blot analysis and immunofluorescence (Supplemental Data), we found that the surviving cells had normal LDH levels and that LDH could not be detected in the nonsurviving cells (Fig. 5A).
Figure 5.
PLCβ1 increases cell survival in siRNA(LDH)-treated cells. A) Left panel: HEK293-TAP-PLCβ1 cells were treated with siRNA(LDH) and/or tetracycline (tet) to induce PLCβ1 expression and were counted 24 and 48 h after treatment; n = 4. Error bars = sd. Right panel: Western blot analysis showing the presence of LDH in cells that survived siRNA (LDH) treatment and the absence of LDH in nonsurviving cells. B) Linkage of siRNA(LDH) or siRNA (GAPDH) to H2B expression. Western blot and Coomassie-stained gel showing that down-regulation of LDH and GAPDH eliminates H2B expression in the detached cells as compared to attached (i.e., healthy) cells that express H2B; n=3. Protein loading was assessed by Ponceau staining (see Supplemental Fig. S6).
Transcription of histone H2B is required for DNA synthesis, and so we determined the changes in its with siRNA(LDH) or siRNA(GAPDH) treatment of HEK293-β1 cells. We note that 85% or more of the cells were easily detached from the dish, as compared to control cells or cells treated with siRNA to cyclophilin or Hsp90. As shown in Fig. 5B, down-regulating LDH or GAPDH is associated with the absence of H2B. Treating the cells with tetracycline increased the fraction of cells strongly attached to the dish to almost 50% as compared to control. These attached cells were found to express normal levels of H2B. In contrast, no H2B could be detected in the detached cells. These results support the idea that overexpression of PLCβ1 prevents cell death that occurs by down-regulation of H2B resulting from siRNA knockdown of LDH and GAPDH.
DISCUSSION
In this study, we have shown that PLCβ1 is not only the main effector of Gαq-associated receptors in mediating Ca2+ signals but can also affect the siRNA activity of genes for two metabolic enzymes, GAPDH and LDH, through its interaction with TRAX. Interestingly, overproduction of PLCβ1 has been shown to rescue NIH3T3 cells from death from oxidative stress (27), supporting a connection between PLCβ1levels and metabolism. The role of PLCβ1 in RNAi activity is surprising since the only clue of a nuclear function of this enzyme comes from its localization in nuclear speckles (refs, 4, 28; for review see ref. 29), and studies implicating it in cell cycle regulation through cyclin D1 (30). Although the connection between TRAX and PLCβ1 in cells is surprising, we note that studies of TRAX-knockout mice connect expression of this protein to two Gαq-related receptors (17, 19). It is also notable and that both TRAX- and PLCβ1-knockout mice have similar neurological disorders (e.g., refs. 16–18).
TRAX has been shown to regulate the cellular localization of its established binding partner translin through its NLS (8, 11). We find that excess TRAX quenches the PLCβ1/Gαq-induced Ca2+ on cell stimulation, suggesting that TRAX does not change the cellular localization of PLCβ1 but does compete with Gαq for PLCβ1 association. In addition, Gαq activation of PLCβ1 allows a population of TRAX to move to the nucleus to participate in ssDNA/RNA processing, presumably due to a loss of PLCβ1-TRAX interactions. The observation that a Gαq not a Gαi/o agonist affects TRAX localization suggests that TRAX movement is specific to the Gαq/PLCβ pathway.
TRAX and translin have similar helical conformations, as seen in the human and Drosophila structures (12, 13), and the C terminus of PLCβ1 (31), where the two proteins interact (7), crystallizes as an intertwined helical dimer. In cells, expression of TRAX and translin are closely coupled, and both are required for cell proliferation (8, 11). Whether cell proliferation is connected to the RISC activity of TRAX/ translin is not clear. The suggestion that PLCβ1 may be proliferative under some circumstances comes from the observation that its deletion is directly linked to acute myeoloid leukemia (32).
Our results show that the level of PLCβ1 affects the cellular amount of small RNAs and that it can reverse the siRNA activity of two cellular proteins involved in metabolism. We have found that PLCβ1 reduces the nuclease activity of C3PO and TRAX by an amount that depends on RNA sequence and structure (unpublished results). This reduction could stem from removing TRAX from C3PO, displacement of TRAX or translin from the octamer, and/or changing the composition of RISC. We gained insight into this question by monitoring changes in the colocalization between different components of RISC. In human cells, RISC activity can be reconstituted by Ago-2 and C3PO, while in Drosophila cells, Dcr2 is required (12, 13). We find a significant increase in PLCβ1/Ago2 colocalization with siRNA(GAPDH) treatment but not with siRNA(Hsp90) treatment, which is consistent with the participation of PLCβ1 in GAPDH regulation. These results lead to the intriguing suggestion that different RISCs form when different genes are being regulated.
Both GAPDH and LDH have been shown to be part of octamer-binding factor 1 coactivator in S phase (OCA-S), which binds to the H2B promoter in S phase in when histone expression occurs (20, 21). The inclusion of GAPDH and LDH in H2B transcriptional regulation connects histone expression to the metabolic state of the cell, and H2B expression is linked to NAD+/NADH levels. We speculate that high levels of cytosolic PLCβ1 prevent down-regulation of LDH and GAPDH through TRAX, thereby allowing for histone synthesis and cell cycle progression. One of the many possible reasons for the selective nature of PLCβ1 for LDH and GAPDH rescue could be its participation in RISC activity of proteins involved in metabolic pathways vs. stress (Hsp90) and inflammatory (cyclophilin A) pathways of the cell, although more studies are required to test this idea. The link between PLCβ1 and LDH and GAPDH regulation correlates well with the observation that PLCβ1 rescues cells from death due to oxidative stress (27).
Our findings, summarized in Fig. 6, show that PLCβ1 and G protein signaling are linked to gene regulation. This link generates many interesting questions about the details of this regulation such as the role of translin in PLCβ1-TRAX association, the effect of PLCβ1 on RNA processing by RISC proteins, the type and number of genes that might be regulated by TRAX or by PLCβ1-TRAX through RNA interference, and the regulation of different genes by different RISCs. We are currently testing the role of translin in PLCβ1-mediated interference of RISC activity. It is notable that RISC assembly may occur on endosomes (33), and it is possible that RISC composition might vary in different cellular compartments or with the activation state of the cell. The potential link between PLCβ1 and the metabolic enzymes LDH and GAPDH is also intriguing. While much more needs to be learned, these observations show that transcriptional processes may be closely linked to G-protein signals.
Figure 6.
Summary of the results. In the basal state, PLCβ1 binds to TRAX, but on activation the complex dissociates, allowing PLCβ1 to catalyze PI(4,5)P2 hydrolysis and downstream signaling events. PLCβ1 is also involved in TRAX-dependent RISC-mediated down-regulation of the metabolic proteins GAPDH and LDH. Down-regulation of these latter proteins eliminates H2B synthesis that ultimately leads to cell death. Alternately, PLCβ1 does not affect RNA interference of Hsp90, cyclophilin, or translin.
Supplementary Material
Acknowledgments
The authors are grateful to Loren Runnels (University of Medicine and Dentistry of New Jersey, Piscataway, NJ, USA) for the HEK293-TAP-PLCβ1 cells, to Catherine Berlot (Geisinger Research Center, Danville, PA, USA) for the Gαq(RC) construct, to Izolda Mileva (Stony Brook University) for her assistance in the RT-PCR studies, and to Urszula Golebiewska and Mario Rebecchi (Stony Brook University) for critically reading this paper.
This work was supported by U.S. National Institutes of Health grant GM53132.
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
- Ago2
- Argonaute-2
- Dcr2
- Dicer 2
- eCFP
- enhanced cyan fluorescent protein
- eYFP
- enhanced yellow fluorescent protein
- GAPDH
- glyceraldehyde 3-phosphate dehydrogenase
- HEK
- human embryonic kidney
- LDH
- lactate dehydrogenase
- NLS
- nuclear localization signal
- PI(4,5)P2
- phosphatidylinositol 4,5-bisphosphate
- PLCβ
- phospholipase Cβ
- RISC
- RNA-induced silencing complex
- TRAX
- translin-associated factor X
REFERENCES
- 1. Suh P., Park J., Manzoli L., Cocco L., Peak J., Katan M., Fukami K., Kataoka T., Yun S., Ryu S. (2008) Multiple roles of phosphoinositide-specific phospholipase C isozymes. BMB Reports 41, 415–434 [DOI] [PubMed] [Google Scholar]
- 2. Rebecchi M. J., Pentyala S. N. (2000) Structure, function, and control of phosphoinositide-specific phospholipase C. Physiol. Rev. 80, 1291–1335 [DOI] [PubMed] [Google Scholar]
- 3. Dowal L., Provitera P., Scarlata S. (2006) Stable association between G alpha(q) and phospholipase C beta 1 in living cells. J. Biol. Chem. 281, 23999–24014 [DOI] [PubMed] [Google Scholar]
- 4. Cocco L., Faenza I., Fiume R. M., Gilmour S. R., Manzoli F. A. (2006) Phosphoinositide-specific phospholipase C (PI-PLC) β1 and nuclear lipid-dependent signaling. Biochim. Biophys. Acta 1761, 509–521 [DOI] [PubMed] [Google Scholar]
- 5. Cocco L., Martelli A. M., Gilmour R. S., Rhee S. G., Manzoli F. A. (2001) Nuclear phospholipase C and signaling. Biochim. Biophys. Acta 1530, 1–14 [DOI] [PubMed] [Google Scholar]
- 6. Xu A., Suh P. G., Marmy-Conus N., Pearson R. B., Seok O. Y., Cocco L., Gilmour R. (2001) Phosphorylation of nuclear phospholipase Cb1 by extracellular signal-regulated kinase mediates the mitogenic action of insulin-like growth factor 1. Mol. Cell. Biol. 21, 2981–2990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Aisiku O. R., Runnels L. W., Scarlata S. (2010) Identification of a novel binding partner of phospholipase Cβ1: translin-associated factor X. PLoS ONE 5, e15001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yang S., Cho Y. S., Chennathukuzhi V. M., Underkoffler L. A., Loomes K., Hecht N. B. (2004) Translin-associated factor X is post-transcriptionally regulated by its partner protein TB-RBP, and both are essential for normal cell proliferation. J. Biol. Chem. 279, 12605–12614 [DOI] [PubMed] [Google Scholar]
- 9. Wang J., Boja E. S., Oubrahim H., Chock P. B. (2004) Testis brain ribonucleic acid-binding protein/translin possesses both single-stranded and double-stranded ribonuclease activities. Biochemistry 43, 13424–13431 [DOI] [PubMed] [Google Scholar]
- 10. Jaendling A., McFarlane R. J. (2010) Biological roles of translin and translin-associated factor-X: RNA metabolism comes to the fore. Biochem. J. 429, 225–234 [DOI] [PubMed] [Google Scholar]
- 11. Cho Y. S., Chennathukuzhi V. M., Handel M. A., Eppig J., Hecht N. B. (2004) The relative levels of translin-associated factor X (TRAX) and testis brain RNA-binding protein determine their nucleocytoplasmic distribution in male germ cells. J. Biol. Chem. 279, 31514–31523 [DOI] [PubMed] [Google Scholar]
- 12. Ye X., Huang N., Liu Y., Paroo Z., Huerta C., Li P., Chen S., Liu Q., Zhang H. (2011) Structure of C3PO and mechanism of human RISC activation. Nat. Struct. Mol. Biol. 18, 650–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tian Y., Simanshu D. K., Ascano M., Diaz-Avalos R., Park A. Y., Juranek S. A., Rice W. J., Yin Q., Robinson C. V., Tuschl T., Patel D. J. (2011) Multimeric assembly and biochemical characterization of the Trax–translin endonuclease complex. Nat. Struct. Mol. Biol. 18, 658–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Liu Y., Ye X., Jiang F., Liang C., Chen D., Peng J., Kinch L. N., Grishin N. V., Liu Q. (2009) C3PO, an endoribonuclease that promotes RNAi by facilitating RISC activation. Science 325, 750–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kawamata T., Tomari Y. (2010) Making RISC. Trends Biochem. Sci. 35, 368–376 [DOI] [PubMed] [Google Scholar]
- 16. McOmish C. E., Burrows E. L., Howard M., Hannan A. J. (2008) PLC-β1 knockout mice as a model of disrupted cortical development and plasticity: behavioral endophenotypes and dysregulation of RGS4 gene expression. Hippocampus 18, 824–834 [DOI] [PubMed] [Google Scholar]
- 17. Chennathukuzhi V., Stein J. M., Abel T., Donlon S., Yang S., Miller J. P., Allman D. M., Simmons R. A., Hecht N. B. (2003) Mice deficient for testis-brain RNA-binding protein exhibit a coordinate loss of TRAX, reduced fertility, altered gene expression in the brain, and behavioral changes. Mol. Cell. Biol. 23, 6419–6434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Stein J. M., Bergman W., Fang Y., Davison L., Brensinger C., Robinson M. B., Hecht N. B., Abel T. (2006) Behavioral and neurochemical alterations in mice lacking the RNA-binding protein translin. J. Neurosci. 26, 2184–2196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Li Z., Wu Y., Baraban J. M. (2008) The Translin/Trax RNA binding complex: clues to function in the nervous system. Biochim. Biophys. Acta 1779, 479–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zheng L., Roeder R. G., Luo Y. (2003) S Phase activation of the histone H2B promoter by OCA-S, a coactivator complex that contains GAPDH as a key component. Cell 114, 255–266 [DOI] [PubMed] [Google Scholar]
- 21. Dai R.-P., Yu F.-X., Goh S.-R., Chng H.-W., Tan Y.-L., Fu J.-L., Zheng L., Luo Y. (2008) Histone 2B (H2B) expression is confined to a proper NAD+/NADH redox status. J. Biol. Chem. 283, 26894–26901 [DOI] [PubMed] [Google Scholar]
- 22. Sambrook J., Fritsch E. F., Maniatis T. (1989) DNA transfection by electroporation. In Molecular Cloning: A Laboratory Manual (Irwin N., ed) Chap. 16.54, Cold Spring Harbor Press, Plainview, New York, USA [Google Scholar]
- 23. Guo Y., Golebiewska U., Scarlata S. (2011) Modulation of Ca2+ activity in cardiomyocytes through caveolae-gαq interactions. Biophys. J. 100, 1599–1607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Guo Y., Rebecchi M., Scarlata S. (2005) Phospholipase Cbeta2 binds to and inhibits phospholipase Cdelta1. J. Biol. Chem. 280, 1438–1447 [DOI] [PubMed] [Google Scholar]
- 25. Livak K. J., Schmittgen T. D. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 26. Hughes T. E., Zhang H., Logothetis D., Berlot C. H. (2001) Visualization of a functional Gaq-green fluorescent protein fusion in living cells. J. Biol. Chem. 276, 4227–4235 [DOI] [PubMed] [Google Scholar]
- 27. Lee Y. H., Kim S.-Y., Kim J.-R., Yoh K.-T., Baek S.-H., Kim M. J., Ryu S. H., Suh P.-G., Kim J.-H. (2000) Overexpression of phospholipase Cβ-1 protects NIH3T3 cells from oxidative stress-induced cell death. Life Sci. 67, 827–837 [DOI] [PubMed] [Google Scholar]
- 28. Cocco L., Martelli A. M., Vitale M., Falconi M., Barnabei O., Stewart Gilmour R., Manzoli F. A. (2002) Inositides in the nucleus: regulation of nuclear PI-PLCbeta1. Adv. Enzyme Regul. 42, 181–193 [DOI] [PubMed] [Google Scholar]
- 29. Lamond A. I., Spector D. L. (2003) Nuclear speckles: a model for nuclear organelles. Nat. Rev. Mol. Cell Biol. 4, 605–612 [DOI] [PubMed] [Google Scholar]
- 30. Faenza I., Matteucci A., Bavelloni A., Marmiroli S., Martelli A. M., Gilmour R. S., Suh P. G., Manzoli L., Cocco L. (2002) Nuclear PLCbeta(1) acts as a negative regulator of p45/NF-E2 expression levels in friend erythroleukemia cells. Biochim. Biophys. Acta 1589, 305–310 [DOI] [PubMed] [Google Scholar]
- 31. Singer A., Waldo G., Harden T. K., Sondek J. (2002) A unique fold of phospholipase C-beta mediates dimerization and interaction with GalphaQ. Nat. Struc. Biol. 9, 32–36 [DOI] [PubMed] [Google Scholar]
- 32. Follo M. Y., Finelli C., Clissa C., Mongiorgi S., Bosi C., Martinelli G., Baccarani M., Manzoli L., Martelli A. M., Cocco L. (2009) Phosphoinositide-phospholipase C β1 mono-allelic deletion is associated with myelodysplastic syndromes evolution into acute myeloid leukemia. J. Clin. Oncol. 27, 782–790 [DOI] [PubMed] [Google Scholar]
- 33. Siomi H., Siomi M. C. (2009) RISC hitches onto endosome trafficking. Nat. Cell Biol. 11, 1049–1051 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.