Abstract
The idea that aging follows a predetermined sequence of events, a program, has been discredited by most contemporary authors. Instead, aging is largely thought to occur due to the accumulation of various forms of molecular damage. Recent work employing functional genomics now suggests that, indeed, certain facets of mammalian aging may follow predetermined patterns encoded in the genome as part of developmental processes. It appears that genetic programs coordinating some aspects of growth and development persist into adulthood and may become detrimental. This link between development and aging may occur due to regulated processes, including through the action of microRNAs and epigenetic mechanisms. Taken together with other results, in particular from worms, these findings provide evidence that some aging changes are not primarily a result of a build-up of stochastic damage but are rather a product of regulated processes. These processes are interpreted as forms of antagonistic pleiotropy, the product of a “shortsighted watchmaker,” and thus do not assume aging evolved for a purpose. Overall, it appears that the genome does, indeed, contain specific instructions that drive aging in animals, a radical shift in our perception of the aging process.—de Magalhães, J. P. Programmatic features of aging originating in development: aging mechanisms beyond molecular damage?
Keywords: epigenetics, functional genomics
The idea that aging follows a somewhat predetermined sequence of events, a program, while debated for decades (1–7), has been discredited by most contemporary authors and evolutionary aging theorists, in particular. At present, the overwhelming view among experts is that aging derives from multiple forms of molecular damage that steadily accumulate with age and lead to the functional and physiological decline characterizing aging (8–13). Although single-gene manipulations in model organisms can delay aging (14), the prevailing hypothesis is that they do so by improving maintenance and repair functions that slow down damage accumulation (10, 12). Likewise, the idea that aging originates in development, also called the developmental theory of aging, has been largely discarded (8, 15). Even though in invertebrates, and in worms, in particular, links between development and aging have been reported (16–18), their relevance to mammals has not been established.
Recent results from mice, monkeys, and humans show a strong overlap between gene expression and DNA methylation changes during aging and changes occurring during development (19–21). Taken together, these findings suggest that, indeed, some aspects of mammalian aging may follow predetermined patterns encoded in the genome as part of developmental processes and provide the strongest evidence to date that aging in mammals is at least partly programmed, suggesting a new layer of mechanisms of aging. Here, I review these recent findings and put forward a model of how programmatic features originating in development may contribute to aging. Because, unlike other authors (1), my model does not assume aging to have evolved for a purpose (i.e., I do not defend that aging is adaptive), it can be reconciled with evolutionary theories of aging and, in particular, with antagonistic pleiotropy.
TRANSCRIPTIONAL AND EPIGENETIC CHANGES DURING MAMMALIAN AGING ORIGINATING IN DEVELOPMENT
Changes in gene expression during mammalian aging have been traditionally seen as the result of damage accumulation and related transcriptional responses (reviewed in ref. 13). Several recent studies, however, suggest that such changes may be, in part, the result of genetically regulated processes or predetermined mechanisms. Somel et al. (19) measured mRNA, microRNA (miRNA), and protein expression levels in the prefrontal cortex of humans and rhesus macaques over their lifetime. They found that the majority of genes and miRNAs differentially expressed during aging are also differentially expressed during development. Some aging changes represent reversals, while others represent extensions of developmental patterns. The authors then link gene expression changes to specific miRNAs and transcription factors, showing the action of regulatory interactions throughout the life span, and, thus, suggesting a link between developmental regulation and aging (19).
Working with mice, Lui et al. (21) also employed gene expression profiling to compare changes during aging with those occurring during development in the kidney, liver, and lung. The same researchers had earlier found evidence for a genetic program coordinating growth deceleration (22) and focused their analysis on the period of juvenile growth deceleration. Like Somel et al. (19), Lui et al. (21) found that many age-dependent gene expression changes, including those in cell-cycle-related genes, originate in development and in particular during the period of juvenile growth deceleration. These authors argue that the genetic program that coordinates growth deceleration during development persists into adulthood and may contribute to age-related physiological and functional decline (21).
At the epigenetic level, evidence is also emerging of regulated developmental processes having detrimental roles during aging. Working in mouse muscle, brain and liver, Takasugi (20) found a correlation between DNA methylation changes in the juvenile-to-adult period and those in the adult-to-aged period. In other words, DNA methylation changes after adulthood were often a reflection of same direction changes during development (20). Because DNA methylation changes are tissue-specific, which Takasugi also observed, this emphasizes the role of regulatory mechanisms rather than random damage accumulation. Takasugi's interpretation is that DNA methylation changes in adults appear to be “an extension of more prominent changes during growth, rather than a process of deterioration, which starts after adulthood” (20).
PROGRAMMATIC FEATURES OF AGING
In invertebrates, it has been known for decades that progression through normal development can trigger or at least accelerate aging, and preventing developmental transitions can often extend life span (17, 18). This is clear in C. elegans in which developmental arrest via the alternative dauer pathway greatly extends life span. Moreover, the rapid decline and death following reproduction of the Pacific salmon have also been interpreted by some authors as a case of programmed aging (1, 4), even if it can be seen as an exception to the rule (23). The recent results in mice, monkeys, and humans are remarkable because they suggest the same may be true for mammals as well, even if to a smaller extent. As such, these findings represent a shift in paradigm with far-reaching implications on the theoretical nature of the aging process and for tackling aging and age-related diseases.
Although the recent results do not prove that aging is caused by sequential changes in gene expression or DNA methylation, they provide strong evidence that gene expression and epigenetic changes during aging are not primarily a result of stochastic damage but rather are a product of regulated processes. Some of the gene expression and epigenetic changes originating in development may well turn out to be detrimental during aging or reflect developmentally triggered changes, for instance, in cell proliferation or differentiation, that alter gene expression patterns. Either way, it seems that not only in invertebrates but also in mammals, aging has a programmatic component, which originates in developmental mechanisms. The gene expression patterns and the underlying regulatory factors identified link developmental processes to aging in a nonrandom way; the complex patterns observed (e.g., the reversal in gene expression changes at specific time points in the life span observed by Somel et al.; ref. 19) cannot be explained by the null hypothesis of a lifelong accumulation of damage. This is akin to results from worms showing that a large fraction of age-related gene expression changes are regulated by three specific transcription factors (16), which follow from earlier observations in invertebrates suggesting that changes in gene expression with age are not merely the outcome of accumulated damage but can entail developmentally timed transcriptional regulation in adults (24). The fact that genes involved in the progression of the developmental program are also involved in age-related gene expression supports the case for developmental genes and processes also being important causal factors in aging.
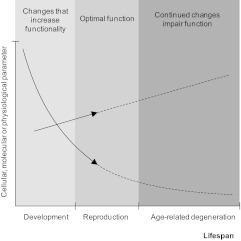
Results at other biological levels also suggest a link between aging and development. Network analyses hint at a relationship in mammals between the genetics of aging and development (25). It has long been known that neuroendocrine changes during aging reflect to some degree changes during development (5). Similarly, presbyopia (or far-sightedness) is thought to derive from the continual growth of eye lenses (26, 27), which can be seen as an example (albeit admittedly unique) of physiological changes determined by the genetic program that continue into adulthood and eventually become detrimental. Developmental pathways may promote changes in adult tissues that predispose to cancer (28), and it was previously hypothesized that processes that decrease brain plasticity early in life may continue later in life and contribute to cognitive aging (29). Functional genomics has also revealed an up-regulation with age of genes related to blood coagulation, which may equally be seen as an extension from early developmental processes (30). Lastly, RNA editing levels have been reported to decline with age in a gene-specific fashion in the human brain (31). Since RNA editing is tightly regulated during development, and in some cases, it is believed to represent regulated processes (32), it is possible that RNA editing alterations late in life in the human brain represent an extension of developmental processes that are under genetic regulation. Therefore, the emerging picture is one of multiple processes whose trajectories throughout the life span are set during development and become detrimental later in life (Fig. 1). In addition to molecular changes, developmental processes could result in cell- or tissue-level deregulation that does not entail molecular damage; in fact, many of the aging gene expression changes observed may reflect developmentally triggered changes in cell populations, in tissue microenvironment, or in systemic factors.
Figure 1.
Developmental programs affecting aging. Hypothetical developmental processes that result in a given molecular, cellular, or physiological change with biological time contribute to increase or optimize function during the reproductive period. The hypothesis advocated in this work is that these processes, whose trajectories are set during development, continue after maturity and contribute to dysfunction later in life.
The involvement of miRNAs suggested by Somel et al. (19) is another fascinating angle. In worms, miRNAs have been previously shown to regulate developmental timing and life span, in line with the idea of an intrinsic clock mechanism (33). The work of Somel et al. (19) extends to mammals the possibility of a developmental clock modulating aging and suggests a role for miRNAs in this process. Given the mounting research on miRNAs, including in human longevity (34), it is tantalizing to speculate that many miRNAs regulate some aspects of aging in specific or across multiple tissues.
Another emerging area of potential relevance to programmed aging is epigenetics. Studies in humans have shown that early development can cause epigenetic marks that persist in adulthood (35), and epigenetic changes during aging are starting to be linked to development. Working in human dermal fibroblasts, Koch et al. (36) found evidence that aging entails regulated developmental mechanisms at the level of DNA methylation. This follows from the same researchers' previous work on mesenchymal stromal cells showing age-related methylation changes in developmental genes (37). Methylation patterns observed during aging in differentiation and developmental genes, such as homeobox genes, were interpreted as evidence that developmental processes regulated by specific epigenetic modifications contribute to aging (37). Human aging-related methylation changes have also been shown to occur preferentially in developmental gene promoters (38). As such, epigenetic regulation of gene expression is a candidate master regulatory mechanism of gene expression changes with age. The recent results showing similar trajectories of epigenetic changes during development and aging support this idea (20). In the future, it will be interesting to study how broader epigenetic alterations early in life are maintained or modified during aging in more tissues and organisms.
EVOLUTIONARY INTERPRETATIONS AND IMPLICATIONS: THE “SHORTSIGHTED WATCHMAKER”
Selection may establish a given trajectory for developmental processes (e.g., epigenetic changes, cellular differentiation, gene expression changes), yet these processes may continue beyond maturity and become detrimental later in life (Fig. 1). Evolutionarily, these would be cases of antagonistic pleiotropy (39), a well-established concept in the evolutionary theory of aging. Although the idea that aging has programmatic features rooted in the genome is a new paradigm for research on metazoan aging, it does not conflict with the evolutionary theory of aging; these deleterious features can be seen as specific examples of age-dependent pleiotropic decline (4). The patterns observed by these recent studies are consistent with the following model: they were optimized for reproduction and then, because of lack of selection, they extend past the age at which they are optimally useful, finally becoming detrimental late in life. In other words, there is no evidence that these patterns were selected for their detrimental effects during aging (i.e., group selection) but are rather the product of a shortsighted watchmaker. The genetic programs continue to be executed later in life because there is insufficient selection to change or stop them. Although some programs may involve active processes that contribute to dysfunction or disease (e.g., presbyopia), others may cause a gradual decrease in some biological parameter, like cessation of growth or terminal differentiation of a particular cell type. Nevertheless, it is the residual effects of these developmental programs that become detrimental.
One of the great mysteries of the biology of aging is the diversity of life spans and aging rates between different species, including similar species, like mammals (26, 40). The great differences in rates of aging, even when animals are kept in captivity, suggest some level of genetic regulation (41), although the underlying causes remain unknown. If the timing of aging is to some degree set by developmental processes, then this would help explain the great variability of lifespans observed in nature. Across species, a very strong correlation exists between the time it takes an organism to reach maturity and how long that organism can expect to live afterward (42, 43). Therefore, the reason a mouse normally lives only 2 and up to 4 yr after reaching maturity while a human may live over 100 may be in part because the developmental processes that have a negative effect later in life progress slower in humans than in mice. Within mice and rats, growth and maturation have also been, respectively, negatively and positively associated with longevity (44, 45).
If an organism's development is seen as building a house, then developmental processes (which could have a physiological, cellular, or molecular basis) are all the construction workers who perform tasks like laying the bricks, plastering the walls, and carpeting the floors. One would expect these workers to follow a blueprint that would tell them when to stop. But imagine that the blueprint only tells some of the workers when to start and the pace at which to perform their tasks. Provided these are well timed, the workers can construct a functional house with everything in place at a given time, but what happens next? The bricklayer may continue laying bricks after the house is finished, which at first is only a minor inconvenience until living space is substantially diminished, and even the structural integrity of the house is threatened. Likewise, ever-increasing layers of carpets will eventually prevent doors from opening, and ultimately, nobody will be able to get in or out of the house. The programmatic features of aging that originate in development are like instructions to construction workers that tell them the pace at which they must work to build a house that reaches its functional peak, much like reproduction, but do not tell them when to stop and thus eventually their essential tasks early in the house's construction become detrimental and ultimately destroy the house. When looking across species, the pace at which the house is built is inversely proportional to its functionality, and crucially, it will be directly proportional to how quickly it will decay and crumble.
A critical paradigm of aging research is caloric restriction (CR), which was interestingly initially discovered by Clive McCay and colleagues as a test of the hypothesis that stunted growth extends life span (45, 46). Given that CR delays growth and development, it has been hypothesized that CR works, at least in part, by delaying the progression of the genetic program (4, 21). The fact that CR can extend life span even if started well after maturity is irrelevant to the shortsighted watchmaker hypothesis. Going back to the house construction analogy, CR slows down the construction workers and hence the house's functional decline, independent of whether they have finished the house, though of course an earlier slowing down will lead to a greater extension in functionality.
EXPERIMENTAL PERSPECTIVES FROM MICE TO HUMANS
Curran and Ruvkun (47) performed a screen for life-extension effects among genes essential for worm development by inactivating the genes in adult animals. They found multiple genes that extend life span when inactivated postdevelopmentally, and even observed stronger life-extension effects than those observed in traditional screens (47). Such results clearly support the thesis of this work, though a similar approach in mammals is currently not feasible. Because programmatic features of aging are both subtle and intrinsically linked to growth and development in mammals, testing the hypothesis put forward herein via manipulations of genes or pathways involved is extremely complicated, for any gross manipulation (e.g., gene knockout) is likely to cause problems prior to adulthood. The use of conditional knockouts in mice may allow researchers to overcome this problem and test the role of genes identified thus far in high-throughput experiments in a more specific fashion. Unfortunately, the cost of life span experiments in mice impedes the sort of large-scale approaches that are possible in invertebrates, and thus, a better understanding of these programmatic features of aging is necessary to better prioritize candidate genes for further experiments. Nonetheless, to my knowledge, the proof of concept that adult inactivation of developmental genes in mammals extends life span has not been demonstrated, and this would be a crucial experiment even if shown for a single gene.
The recent explosion in functional genomics and systems biology offers opportunities to begin to model the genetic programs that determine who we are and how we develop. It is my view that the systematic analysis of the genome coupled with the integration of these models with aging changes at different biological levels could help us begin to unravel these mechanisms and develop more precise genetic interventions (e.g., using the rich tools available for mouse genetics) to test the role of developmental genes in mammalian aging. I think the timing of developmental processes that have an impact on aging is set by complex gene networks, and to understand these, we will need to understand the intrinsic, complex nature of human biology. Unraveling the intracellular developmental programs also merits further attention (48).
Extreme life extension (nearly 10-fold) has been observed in worms with a prolonged developmental time (49). Although slow development is often associated with pathological processes in mammals, a few intriguing paradigms may be suitable for further study. Chief among these is the case of a teenage patient who has the physical appearance of an infant and whose study may provide clues about the genetic determinants of the pace of development (6, 50). Some mouse strains may also be suitable for further experiments, such as the POSCH-2 strain that exhibits delayed reproduction (51), and to my knowledge its aging process has not been studied.
If human aging is not merely the outcome of a build-up of stochastic damage, and some developmental programs contribute to aging, then this may increase our chances to identify therapeutic targets both for longevity enhancement and disease prevention. Assuming that there are specific instructions in the genome that drive aging, then this opens new avenues for interfering with such instructions in adulthood to combat age-related diseases. It is tempting to speculate that, for example, specific transcriptional regulators may be identified that can be suitable drug targets to retard or halt some forms of age-related decline.
CONCLUSIONS
It is well established that some modulators of aging, like endocrine mechanisms and TOR, have pleiotropic effects (4, 5, 7). The recent functional genomics results, however, suggest a regulatory level in aging-related changes that alters our interpretation of aging. Even if they were not selected for such a purpose, it appears that the genome does, indeed, contain instructions that drive aging in animals. This clearly contradicts the prevailing paradigm and views of many experts (8).
The model put forth herein does not explain all facets of aging. Some forms of damage, such as damage to the DNA (52), are still likely major causal factors in mammalian aging, and many age-related changes, like clots in arteries and misfolded proteins in the brain, are likely due to the accumulation of junk. What the new results in mice, monkeys, and humans reveal is an additional and underappreciated layer of aging mechanisms, consisting of developmental mechanisms gone awry at later ages, and highly suggestive signs of these mechanisms at work have now been detected by experimental genomic approaches. Therefore, I do not think existing evidence supports the idea that aging is programmed but rather that aging has programmatic features. These are cases of antagonistic pleiotropy, of a shortsighted watchmaker, not of a purpose.
In summary, it appears that aging is due, in part, to genetic programs that evolved to regulate development but progress into adulthood and gradually cause dysfunction. Understanding these programmatic contributions to aging and following the leads of this new paradigm in the study of aging and senescence might keep scientists busy for decades to come—even given the current and growing power of modern genomics (53). But we need not look so far into the future since we face many immediate questions: What are the specific transcriptional modules that regulate these programmatic features of aging? Why is there a reversal of some age-related gene expression changes and progression of others? How do these transcriptional changes influence, and how are they influenced by, cellular and physiological changes? How do they interact with environmental factors like diet? Can we identify miRNAs, transcription factors, and epigenetic alterations high in the hierarchy of these alterations, which we can then test experimentally? Crucially, if we can unravel the regulatory elements of these programmatic features, can we manipulate them by diet, drugs, or other therapy?
Acknowledgments
The author thanks Pete Estep and George Martin for comments on previous drafts.
The author is also grateful for support from the UK Biotechnology and Biological Sciences Resource Council, the Wellcome Trust, the Ellison Medical Foundation, and a Marie Curie International Reintegration grant within EC-FP7 for supporting the work in his laboratory.
Footnotes
- CR
- caloric restriction
- miRNA
- microRNA
REFERENCES
- 1. Longo V. D., Mitteldorf J., Skulachev V. P. (2005) Programmed and altruistic ageing. Nat. Rev. Genet. 6, 866–872 [DOI] [PubMed] [Google Scholar]
- 2. Bredesen D. E. (2004) The non-existent aging program: how does it work? Aging Cell 3, 255–259 [DOI] [PubMed] [Google Scholar]
- 3. Comfort A. (1964) Ageing: The Biology of Senescence, Routledge & Kegan Paul, London [Google Scholar]
- 4. De Magalhaes J. P., Church G. M. (2005) Genomes optimize reproduction: aging as a consequence of the developmental program. Physiology 20, 252–259 [DOI] [PubMed] [Google Scholar]
- 5. Finch C. E. (1976) The regulation of physiological changes during mammalian aging. Q. Rev. Biol. 51, 49–83 [DOI] [PubMed] [Google Scholar]
- 6. Walker R. F. (2011) Developmental theory of aging revisited: focus on causal and mechanistic links between development and senescence. Rejuv. Res. 14, 429–436 [DOI] [PubMed] [Google Scholar]
- 7. Blagosklonny M. V. (2006) Aging and immortality: quasi-programmed senescence and its pharmacologic inhibition. Cell Cycle 5, 2087–2102 [DOI] [PubMed] [Google Scholar]
- 8. Hayflick L. (2007) Entropy explains aging, genetic determinism explains longevity, and undefined terminology explains misunderstanding both. PLoS Genet. 3, e220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Holliday R. (2006) Aging is no longer an unsolved problem in biology. Ann. N. Y. Acad. Sci. 1067, 1–9 [DOI] [PubMed] [Google Scholar]
- 10. Kirkwood T. B. (2008) Understanding ageing from an evolutionary perspective. J. Intern. Med. 263, 117–127 [DOI] [PubMed] [Google Scholar]
- 11. Partridge L. (2010) The new biology of ageing. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365, 147–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rattan S. I. (2006) Theories of biological aging: genes, proteins, and free radicals. Free. Radic. Res. 40, 1230–1238 [DOI] [PubMed] [Google Scholar]
- 13. Kirkwood T. B., Melov S. (2011) On the programmed/non-programmed nature of ageing within the life history. Curr. Biol. 21, R701–707 [DOI] [PubMed] [Google Scholar]
- 14. Kenyon C. J. (2010) The genetics of ageing. Nature 464, 504–512 [DOI] [PubMed] [Google Scholar]
- 15. Miller R. A. (1999) Kleemeier award lecture: are there genes for aging? J. Gerontol. A Biol. Sci. Med. Sci. 54, B297–B307 [DOI] [PubMed] [Google Scholar]
- 16. Budovskaya Y. V., Wu K., Southworth L. K., Jiang M., Tedesco P., Johnson T. E., Kim S. K. (2008) An elt-3/elt-5/elt-6 GATA transcription circuit guides aging in C. elegans. Cell 134, 291–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Klass M., Hirsh D. (1976) Non-ageing developmental variant of Caenorhabditis elegans. Nature 260, 523–525 [DOI] [PubMed] [Google Scholar]
- 18. Miller S. E., Hadfield M. G. (1990) Developmental arrest during larval life and life-span extension in a marine mollusc. Science 248, 356–358 [DOI] [PubMed] [Google Scholar]
- 19. Somel M., Guo S., Fu N., Yan Z., Hu H. Y., Xu Y., Yuan Y., Ning Z., Hu Y., Menzel C., Hu H., Lachmann M., Zeng R., Chen W., Khaitovich P. (2010) MicroRNA, mRNA, and protein expression link development and aging in human and macaque brain. Genome Res. 20, 1207–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Takasugi M. (2011) Progressive age-dependent DNA methylation changes start before adulthood in mouse tissues. Mech. Ageing Dev. 132, 65–71 [DOI] [PubMed] [Google Scholar]
- 21. Lui J. C., Chen W., Barnes K. M., Baron J. (2010) Changes in gene expression associated with aging commonly originate during juvenile growth. Mech. Ageing Dev. 131, 641–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lui J. C., Forcinito P., Chang M., Chen W., Barnes K. M., Baron J. (2010) Coordinated postnatal down-regulation of multiple growth-promoting genes: evidence for a genetic program limiting organ growth. FASEB J. 24, 3083–3092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Austad S. N. (2004) Is aging programed? Aging Cell 3, 249–251 [DOI] [PubMed] [Google Scholar]
- 24. McCarroll S. A., Murphy C. T., Zou S., Pletcher S. D., Chin C. S., Jan Y. N., Kenyon C., Bargmann C. I., Li H. (2004) Comparing genomic expression patterns across species identifies shared transcriptional profile in aging. Nat. Genet. 36, 197–204 [DOI] [PubMed] [Google Scholar]
- 25. De Magalhaes J. P., Toussaint O. (2004) GenAge: a genomic and proteomic network map of human ageing. FEBS Lett. 571, 243–247 [DOI] [PubMed] [Google Scholar]
- 26. Finch C. E. (1990) Longevity, Senescence, and the Genome, The University of Chicago Press, Chicago and London [Google Scholar]
- 27. Hayflick L. (1994) How and Why We Age, Ballantine Books, New York [Google Scholar]
- 28. Vijg J., Campisi J. (2008) Puzzles, promises and a cure for ageing. Nature 454, 1065–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. De Magalhaes J. P., Sandberg A. (2005) Cognitive aging as an extension of brain development: a model linking learning, brain plasticity, and neurodegeneration. Mech. Ageing Dev. 126, 1026–1033 [DOI] [PubMed] [Google Scholar]
- 30. De Magalhaes J. P., Curado J., Church G. M. (2009) Meta-analysis of age-related gene expression profiles identifies common signatures of aging. Bioinformatics 25, 875–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nicholas A., de Magalhaes J. P., Kraytsberg Y., Richfield E. K., Levanon E. Y., Khrapko K. (2010) Age-related gene-specific changes of A-to-I mRNA editing in the human brain. Mech. Ageing Dev. 131, 445–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Higuchi K., Kitagawa K., Kogishi K., Takeda T. (1992) Developmental and age-related changes in apolipoprotein B mRNA editing in mice. J. Lipid Res. 33, 1753–1764 [PubMed] [Google Scholar]
- 33. Boehm M., Slack F. (2005) A developmental timing microRNA and its target regulate life span in C. elegans. Science 310, 1954–1957 [DOI] [PubMed] [Google Scholar]
- 34. Gombar S., Jung H. J., Dong F., Calder B., Atzmon G., Barzilai N., Tian X. L., Pothof J., Hoeijmakers J. H., Campisi J., Vijg J., Suh Y. (2012) Comprehensive microRNA profiling in B-cells of human centenarians by massively parallel sequencing. BMC Genomics 13, 353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Heijmans B. T., Tobi E. W., Stein A. D., Putter H., Blauw G. J., Susser E. S., Slagboom P. E., Lumey L. H. (2008) Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proc. Natl. Acad. Sci. U. S. A. 105, 17046–17049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Koch C. M., Suschek C. V., Lin Q., Bork S., Goergens M., Joussen S., Pallua N., Ho A. D., Zenke M., Wagner W. (2011) Specific age-associated DNA methylation changes in human dermal fibroblasts. PloS One 6, e16679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bork S., Pfister S., Witt H., Horn P., Korn B., Ho A. D., Wagner W. (2010) DNA methylation pattern changes upon long-term culture and aging of human mesenchymal stromal cells. Aging Cell 9, 54–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rakyan V. K., Down T. A., Maslau S., Andrew T., Yang T. P., Beyan H., Whittaker P., McCann O. T., Finer S., Valdes A. M., Leslie R. D., Deloukas P., Spector T. D. (2010) Human aging-associated DNA hypermethylation occurs preferentially at bivalent chromatin domains. Genome Res. 20, 434–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Williams G. C. (1957) Pleiotropy, natural selection, and the evolution of senescence. Evolution 11, 398–411 [Google Scholar]
- 40. Austad S. N. (2009) Comparative biology of aging. J. Gerontol. A Biol. Sci. Med. Sci. 64, 199–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. De Magalhaes J. P. (2003) Is mammalian aging genetically controlled? Biogerontology 4, 119–120 [DOI] [PubMed] [Google Scholar]
- 42. de Magalhaes J. P., Costa J., Church G. M. (2007) An analysis of the relationship between metabolism, developmental schedules, and longevity using phylogenetic independent contrasts. J. Gerontol. A Biol. Sci. Med. Sci. 62, 149–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ricklefs R. E. (2010) Life-history connections to rates of aging in terrestrial vertebrates. Proc. Natl. Acad. Sci. U. S. A. 107, 10314–10319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yuan R., Meng Q., Nautiyal J., Flurkey K., Tsaih S. W., Krier R., Parker M. G., Harrison D. E., Paigen B. (2012) Genetic coregulation of age of female sexual maturation and lifespan through circulating IGF1 among inbred mouse strains. Proc. Natl. Acad. Sci. U. S. A. 109, 8224–8229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rollo C. D. (2002) Growth negatively impacts the life span of mammals. Evol. Dev. 4, 55–61 [DOI] [PubMed] [Google Scholar]
- 46. McCay C. M. (1933) Is longevity compatible with optimum growth? Science 77, 410–411 [DOI] [PubMed] [Google Scholar]
- 47. Curran S. P., Ruvkun G. (2007) Lifespan regulation by evolutionarily conserved genes essential for viability. PLoS Genet. 3, e56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Raff M. (2006) The mystery of intracellular developmental programmes and timers. Biochem. Soc. Trans. 34, 663–670 [DOI] [PubMed] [Google Scholar]
- 49. Ayyadevara S., Alla R., Thaden J. J., Shmookler Reis R. J. (2008) Remarkable longevity and stress resistance of nematode PI3K-null mutants. Aging Cell 7, 13–22 [DOI] [PubMed] [Google Scholar]
- 50. Walker R. F., Pakula L. C., Sutcliffe M. J., Kruk P. A., Graakjaer J., Shay J. W. (2009) A case study of “disorganized development” and its possible relevance to genetic determinants of aging. Mech. Ageing Dev. 130, 350–356 [DOI] [PubMed] [Google Scholar]
- 51. Biddle F. G., Eden S. A., Rossler J. S., Eales B. A. (1997) Sex and death in the mouse: genetically delayed reproduction and senescence. Genome 40, 229–235 [DOI] [PubMed] [Google Scholar]
- 52. Freitas A. A., de Magalhaes J. P. (2011) A review and appraisal of the DNA damage theory of ageing. Mutation Res. 728, 12–22 [DOI] [PubMed] [Google Scholar]
- 53. De Magalhaes J. P., Finch C. E., Janssens G. (2010) Next-generation sequencing in aging research: emerging applications, problems, pitfalls and possible solutions. Ageing Res. Rev. 9, 315–323 [DOI] [PMC free article] [PubMed] [Google Scholar]